Abstract
Prolonged skeletal unloading through bedrest results in bone loss similar to that observed in elderly osteoporotic patients, but with an accelerated timeframe. This rapid effect on weight-bearing bones is also observed in astronauts who can lose up to 2% of their bone mass per month spent in Space. Despite the important implications for Spaceflight travelers and bedridden patients, the exact mechanisms involved in disuse osteoporosis have not been elucidated. Parathyroid hormone-related protein (PTHrP) regulates many physiological processes including skeletal development, and has been proposed as a mechanosensor. To investigate the role of PTHrP in microgravity-induced bone loss, trabecular and calvarial osteoblasts (TOs and COs) from Pthrp +/+ and -/- mice were subjected to actual Spaceflight for 6 days (Foton M3 satellite). Pthrp +/+, +/- and -/- osteoblasts were also exposed to simulated microgravity for periods varying from 6 days to 6 weeks. While COs displayed little change in viability in 0g, viability of all TOs rapidly decreased in inverse proportion to PTHrP expression levels. Furthermore, Pthrp+/+ TOs displayed a sharp viability decline after 2 weeks at 0g. Microarray analysis of Pthrp+/+ TOs after 6 days in simulated 0g revealed expression changes in genes encoding prolactins, apoptosis/survival molecules, bone metabolism and extra-cellular matrix composition proteins, chemokines, insulin-like growth factor family members and Wnt-related signalling molecules. 88% of 0g-induced expression changes in Pthrp+/+ cells overlapped those caused by Pthrp ablation in normal gravity, and pulsatile treatment with PTHrP1-36 not only reversed a large proportion of 0g-induced effects in Pthrp+/+ TOs but maintained viability over 6-week exposure to microgravity. Our results confirm PTHrP efficacy as an anabolic agent to prevent microgravity-induced cell death in TOs.
Introduction
Bone loss due to osteoporosis (OP) is the most common cause of fractures among the elderly. Age-related OP elicits fractures with minimal trauma in up to 40% of women and 20% of men over the age of fifty and is a major health and fiscal burden [1, 2]. OP can also develop in younger individuals as a consequence of premature menopause or skeletal unloading. In the absence of gravity load on weight-bearing bones, the equilibrium between osteoblastic bone formation and osteoclastic bone resorption is uncoupled and the balance is shifted towards resorption, resulting in bone loss. Skeletal disuse can occur after prolonged bedrest on Earth but is also encountered in the weightless conditions of Space [3]. During orbital flight, astronauts are exposed to gravity forces below the physiological detection threshold (≤ 10-5g). Even a short-duration exposure to microgravity results in profound metabolic changes, and the lack of mechanical loading causes rapid demineralization in bones of the lower body leading to development of a disuse osteoporotic-like phenotype [4]. Microgravity can cause a remarkable loss of up to 2% of load-bearing bone mineral density (BMD) per month spent in Space, compared to an average 1% per year observed on Earth (starting from approximately 24–30 years of age when peak bone mass is attained) [5]. Microgravity-induced bone loss shows no sign of stabilization while at 0g and displays a highly-variable speed of skeletal recovery upon return to Earth [6–10]. The causes of this rapidly-progressing syndrome have not been elucidated, and only partially-effective countermeasures have been developed [11].
Skeletal loss has severe implications for long-time inhabitants of the International Space Station and will be a hindrance to spatial exploration since up to one half of bone mass could be lost during a three-year trip to Mars, resulting in mission-compromising fractures as well as complications from renal stones caused by skeleton-released calcium [5, 12–14]. The mechanosensors which control bone loss during unloading are not known, and the striking difference in microgravity-induced mineral loss between bone derived from endochondral formation (eg. trabecular bone) and bone derived from membraneous formation (calvarial bone) has also not been fully analysed. Elucidation of the causes of skeletal loss at 0g is further complicated by the fact that the two main cellular components regulating bone turnover, the osteoblasts and osteoclasts, are differently affected by microgravity. Osteoblasts exposed to 0g undergo decreases in cellular integrity with modified microtubule structure, focal adhesions and increasingly fragmented nuclei, while osteoclasts present higher numbers of discrete resorption pits and increased cellular activity [15]. Spaceflight countermeasures to bone loss include exercise and resistive training, artificial musculoskeletal loading, electrical stimulation and vibration treatment, as well as dietary additions of vitamin D, K, calcium and various drugs [16]. While physical activity in Space helps to counter muscle atrophy, it does not prevent bone loss and none of these measures fully compensate for skeletal degradation [10, 13, 14]. In long-duration bedrest studies which mimic microgravity’s physiological effects, treatment of male subjects with the anti-resorptive bisphosphonate drugs alendronate or pamidronate (osteoclast inhibitors) prevented some, but not all, bone loss [17, 18]. Furthermore, skeletal bone formed during bisphosphonate administration does not display the mechanical integrity of normal bone [6, 19] and high doses of anti-resorptive drugs may inhibit formation in unloaded bone [20]. Consequently, an alternative to be investigated is the use of anabolic substances such as parathyroid hormone (PTH) and parathyroid hormone-related hormone (PTHrP) which promote bone formation rather than inhibit resorption [21, 22].
PTHrP is a secreted factor expressed in almost all normal fetal and adult tissues. It acts as an autocrine, paracrine or intracrine factor in a wide range of developmental and physiological processes, including skeletal development and growth [23, 24]. The 13 N-terminal amino acids of PTHrP are highly-homologous to those of PTH, a characteristic that allows PTHrP to activate their common type 1 PTH/PTHrP receptor. PTH and PTHrP differ in the remainder of their sequence, a fact which accounts at least in part for the distinct properties of the two molecules [23, 24]. The major role played by PTHrP in bone development was made evident through Pthrp gene deletion which caused profound abnormalities in the development of the cartilaginous growth plate [25, 26], a phenotype not seen in mice with PTH deletion [27]. Heterozygous mice with only one null Pthrp allele (Pthrp +/-) developed premature OP due to decreased bone formation linked to enhanced apoptosis of osteogenic cells [28]. Knock-out mice with a selective Pthrp deletion targeting only the osteoblasts further recapitulated this severe osteoporotic phenotype and importantly, exogenous application of PTH or PTHrP prevented apoptosis and associated bone loss [29]. PTH1-34 (teriparatide, Forteo™) is currently the only approved anabolic drug for treatment of osteoporosis but may cause bone resorption, hypercalcemia, nausea, muscle cramps and other adverse side-effects. In contrast, a study conducted on post-menopausal women showed that the 1–36 N-terminal peptide of PTHrP did not activate bone resorption (based on bone marker responses, although changes in BMD were not measured) and acted as a pure anabolic agent while causing none of the adverse effects observed with teriparatide [30]. Although intermittent treatment with PTHrP causes an anabolic response, continuous treatment causes paradoxical downregulation of osteogenic genes resulting in skeletal catabolism [31, 32].
Decreased expression levels of PTHrP have been reported in rat long bones and cultured osteoblasts following exposure to microgravity [33], and loss of PTHrP may underlie at least part of the pathophysiology of microgravity-induced OP. We consequently tested the hypothesis that PTHrP could counter actual or simulated microgravity-induced deleterious effects in osteoblasts, and we investigated a timeline of viability for osteoblasts in weightlessness which would predispose astronauts to impaired bone formation. We also examined the efficacy of exogenous PTHrP1-36 peptide administration in preventing osteoblast viability loss in microgravity conditions. For these studies, we used trabecular and calvarial osteoblasts isolated one day before birth from bones of Pthrp +/+, +/-, or -/- mice. The cells in adherent conditions were subjected to Spaceflight aboard the Foton M3 satellite or to simulated microgravity in a Synthecon rotary cell culture system (RCCS) for 6-day experiments, with or without intermittent PTHrP 1–36 treatment. We observed that PTHrP expression confers resistance to microgravity-induced cell death, that a very large proportion of the genes affected by microgravity is common to those affected by Pthrp gene deletion, and that intermittent treatment with the PTHrP1-36 peptide reverses the 0g effects on TO viability.
Materials and Methods
Animal Ethics Statement
All experiments were carried out in compliance with regulations of the McGill University Institutional Animal Care Committee (Animal permit: 5210) which specifically approved this study.
The mice were housed in conventional (non-barrier) animal facilities at 21°C with light-dark cycles of 12 hours, and were fed Bacon Softies™ soft pellets (Bio Serv) and water from drinking bottles ad libitum.
Animals
Mice with various levels of Pthrp gene ablation (wild-type Pthrp +/+, heterozygous Pthrp +/-, and homozygous Pthrp -/-) in a C57BL/6 background [26] were used to obtain osteoblasts. Because homozygous animals (Pthrp -/-) exhibit perinatal lethality due to skeletal and pulmonary problems [26], osteoblasts were collected from fetal mice of all types one day before birth. Briefly, Pthrp +/- males were bred with Pthrp +/- females, pregnant females were sacrificed by cervical dislocation one day before partum and the fetal mice collected and sacrificed by decapitation. The animals were immediately surface-sterilized in 70% ethanol, tail samples were preserved for genotyping, and femoral and cranium bones extracted immediately to obtain trabecular and calvarial osteoblast cells respectively.
Preparation of osteoblasts and cell culture
The trabecular and calvarial cells were obtained by a method modified after [34]. Briefly, calvarial (cranium, derived from membraneous bone formation) and trabecular (femur, derived from endochondral bone formation) bones were dissected from the animals and predigested in 6-well culture plates for 15 min at 37°C with gentle rotation in digestion medium (TCDA) made immediately before use: trypsin 0.05%, type I collagenase (2 mg/ml, Gibco), DNAse I (0.3 mg/ml, Sigma), penicillin streptomycin (100 U/ml). Bones were transferred to new 6-well plates with fresh TCDA, chopped into small slivers, particles were transferred to a third plate with TCDA and incubated at 37°C 15 min with rotation. Finally, the bone fragments were transferred to a fourth plate containing MAF medium (α-MEM with 15% FBS, penicillin/streptomycin 100 U/ml, fungizone 0.25 ug/ml) and incubated without rotation at 37°C in a humidified cell incubator (5% CO2). Only the cells emerging from the bone fragments at this step were kept for experimental purposes. The MAF medium was changed every two days and cells were transferred to 6-well plates when confluent. Aliquots were incubated with AGD differenciation medium (α-MEM 15% FBS, penicillin/streptomycin 100U/ml, fungizone 0.25 ug/ml, ascorbic acid 50 ug/ml, glycerophosphate 10 mM and dexamethasone 0,1 uM final) [35] or MAF as control to test for differenciation markers. The osteoblast primary cultures were filtered through sterile nylon cell strainers (100 microns, BD Biosciences), passaged twice, and 500,000 cells (minimum) were frozen per vial in 90% FBS, 10% DMSO using cryojars (ethanol/ dry ice) for 4 hours at -80°C. The cell vials were transferred to liquid nitrogen storage after 2 days. All culture reagents were from Wisent.
Genotyping
Tail segments were incubated in lysis buffer (100 mM Tris-HCl 8.0, 5 mM EDTA 8.0, 0.2% SDS, 200 mM NaCl, 0.2 mg/ml proteinase K) at 55°C for 16 hours. Genomic DNA was extracted with phenol/CHCl3, ethanol-precipitated and resuspended in TE buffer. Aliquots were digested with PvuII, separated by agarose electrophoresis, and blots hybridized with a PTHrP-specific probe [26].
Culture for Space mission bioreactors
The available surface on culture slides in the Foton M3 mission biomodules was extremely limited, and allowed testing of the Pthrp +/+ and -/- genotypes only. Cultured primary osteoblasts of both genotypes from calvarial and trabecular origin were exposed to AGD differenciation medium for 4–5 days then trypsinised, counted and cells were attached at very high density (25,000 cells/cm2, or 6,500 cells/compartment in 4-compartment slides) to the growth surfaces of sterile CC2 glass slides (Nunc). These slides were handled in a completely sterile fashion and placed inside 10cm culture dishes in a normal culture incubator (5% CO2) for a 4-day pre-attachment period in AGD medium. The CC2 slides superstructures were removed under sterile conditions, the glass slides inserted into the sterile mission biomodules and connected to the feeding syringes and waste exhaust lines of the automated experimental trays (Fig 1A, CALM Technologies, Kingston, Canada). Two identical trays were sealed and powered by battery to maintain 37°C temperature and provide medium changes every 48 hours. One tray was kept at 1g at the European Space Research and Technology Centre (ESTEC, Noordwijk, The Netherlands) while the twin tray (for 0g) was transported over a period of 4 days to Baikonour and installed aboard the Roscosmos (Russian Federal Space Agency) Foton M3 satellite which was launched into orbital flight. (http://esamultimedia.esa.int/docs/foton/FOTON-M3_brochure.pdf).
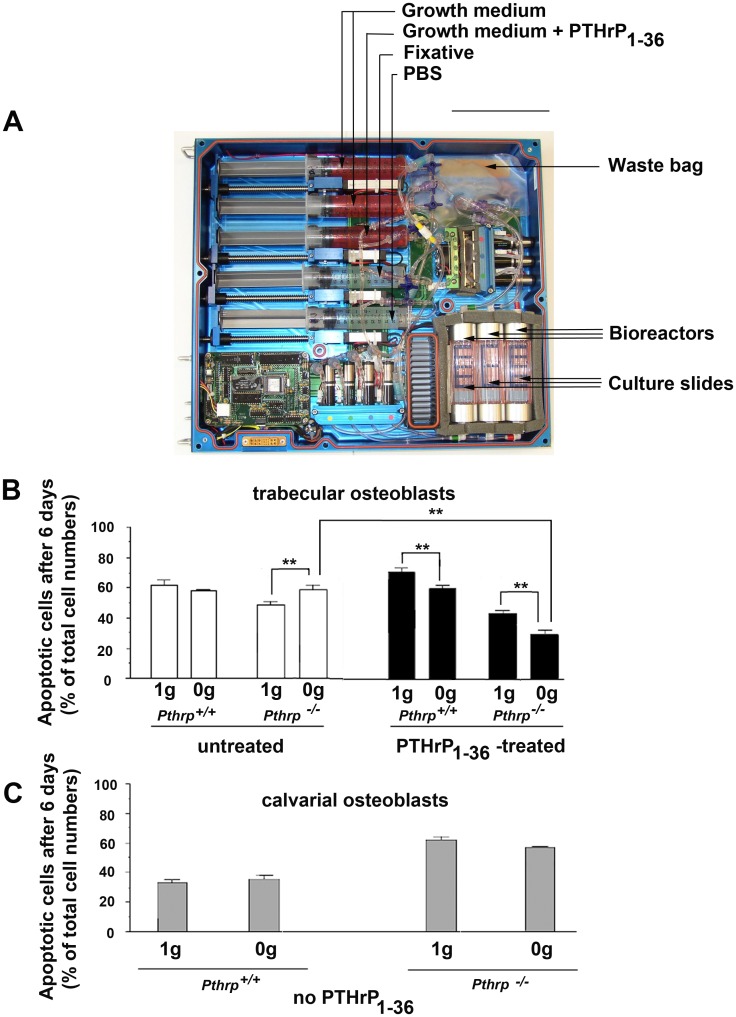
Fig 1. Endogenous PTHrP protects trabecular osteoblasts from Space microgravity-induced apoptosis. (6-day experiments).
(A) Automated flight-ready tray for conducting cell culture experiments aboard Foton M3 satellite. Osteoblasts were attached onto CC2 culture slides inside three bioreactors (lower right) in a sealed pathway. (B) Proportion of apoptotic cells in trabecular osteoblasts in bioreactors after 6 days in Space microgravity (0g) or in normal Earth gravity (1g). ** p < 0.01. White bars: untreated; black bars: 2-h daily treatment with PTHrP1-36 10-8M. (C) Proportion of apoptotic cells in calvarial osteoblasts in bioreactors after 6 days in Space microgravity (0g) or in normal Earth gravity (1g). No PTHrP1-36 treatment was conducted on calvarial cells. Scale bar A: 15 cm.
Experimental flight conditions
Experimental conditions were automatically started one day after reaching orbital flight. Cells were fed fresh AGD growth medium every day, or received a daily 2-hour treatment with 10-8M PTHrP peptide amino acids 1–36, (Bachem, Germany) in AGD, followed by a phosphate-buffered saline (PBS) rinse, then fresh AGD medium. Intermittent PTHrP1-36 treatment was used as it provides anabolic effects on bone whereas continuous treatment does not [36]. After 6 days of experimentation, the cells were fixed with 1% glutaraldehyde in situ on the slides within the 3 bioreactors, then stored in PBS at 15°C. After retrieval from the satellite, slides were stored in PBS at 4°C until assays. Experimental conditions were identical for both 0g and 1g trays (described in S1 Table).
Apoptosis assay
Mission slides were analysed for apoptosis events using the single-stranded DNA (ssDNA) detection technique [37]. The glass slides with glutaraldehyde-fixed cells were heated for 20 minutes at 75°C in neat formamide, rinsed with H2O, blocked with 3% non-fat dry milk for 1h at room temperature, reacted with a monoclonal antibody to ssDNA (1:10) coupled with horseradish peroxidase (Chemicon), then colored with freshly-prepared diaminobenzidine (DAB) and counter-stained with hematoxylin. Fixed cells positive for DAB (i.e.ssDNA) were apoptotic at the time of in-flight fixation, cells stained with hematoxylin were viable at the same moment. 11 optical fields were counted on each slide and the proportion of apoptotic cells to non-apoptotic cells at the time of fixation was determined.
Simulated microgravity in rotary cell culture system
The flight data was completed, confirmed and expanded using the Synthecon Revolving Cell Culture System (RCCS) Earth-based microgravity simulator consisting of a horizontally-rotating axis connected to a sterile 10-ml disposable growth chamber called High Aspect Ratio Vessel (HARV) with a CO2-permeable membrane (Fig 2A, Synthecon, Houston, TX). The equipment was used inside a standard CO2 incubator while connected to an external speed regulator and power supply. Because free, non-adherent cells would rapidly undergo anoikis-induced apoptosis [38], osteoblasts grown in AGD medium were trypsinised, counted, and aliquots of 106 cells were pre-attached to sterile MicroHex microcarriers (Nunc) using 1 ml of 50 cm2/ml MicroHex per 106 cells, and incubated 4 days in AGD medium at 30°C in culture plates coated with sterile 4% agarose (Gibco) to prevent attachment [39]. The medium was changed every second day. Primary osteoblasts attach to the surface of the microcarriers and develop in a manner similar to osteoblasts plated on standard culture plasticware except for the fact that the cells create three-dimensional connections with more than one plastic surface, resulting in microcarrier-cell aggregates (Fig 2B and 2C). Within 24 hours of attachment at 1g, the cell/microcarrier structures assembled into clumps visible to the naked eye which mimicked the more natural cellular environment found in a living organism. After 5 days, MicroHex-attached osteoblasts (5 x 106) were transferred to 10-ml sterile Synthecon HARV chambers containing AGD medium which were placed inside a standard cell incubator (37°C, 5% CO2). The units were rotated at optimized speed (20 rpm) to maintain the constant free fall state characteristic of orbital flight and which is the cause of weightlessness. The rotation simulated 0g while avoiding shear forces due to excessive rotation and collision with vessel walls. Two chambers were rotated simultaneously on the same rotation axis, one containing the untreated control receiving daily medium change, the other receiving a daily 2-h PTHrP1-36 10-8M treatment followed by fresh medium. Cells on microcarriers but growing in AGD medium in agarose-coated plasticware in a standard cell culture incubator were used as 1g controls and received the same regimen of treatments or medium changes. Triplicate experiments were conducted with conditions identical to those aboard the satellite. At the end of the 6th day, cells were collected, trypsinized to release them from the microcarriers, and aliquots analysed for viability by Trypan Blue analysis. Treatment and media change conditions for 6-week experiments were identical to those lasting 6 days.
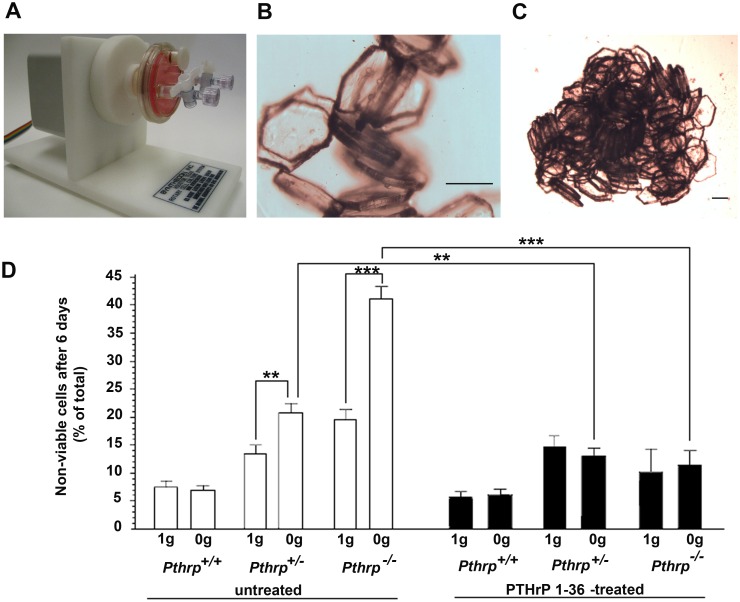
Fig 2. Allelic effect of endogenous PTHrP levels in simulated microgravity, and compensation by exogenous PTHrP 1–36 (6-day experiments).
(A) Microgravity simulation apparatus (RCCS) with one rotating culture unit (HARV). (B) MicroHex carriers with attached trabecular osteoblasts in culture. (C) Cell-induced MicroHex aggregate in culture (occurs for both trabecular and calvarial osteoblasts). (D) Trabecular osteoblast viability after 6 days on MicroHex carriers in normal gravity (1g) or in simulated microgravity (0g) (triplicate 6-day experiments). White bars: untreated; black bars: 2-h daily treatment with PTHrP1-36 10-8M. ** p < 0.01, *** p < 0.001. Scale bars: B, C: 200 μm.
Trypan Blue viability test
During long-term experiments, sampling from the HARV rotating chamber was conducted at weekly intervals. Aliquots of cell-microcarriers aggregate were quickly collected in sterile fashion from the momentarily-immobilised HARV, and transferred to sterile microfuge tubes. The aggregates were washed with growth medium then trypsinized in tubes kept horizontally for 20 minutes at 37°C. After cell detachment, the tubes were put vertically in racks to sediment the microcarriers, and the supernatant collected. Microcarriers were rinsed twice with growth medium to collect residual cells, the trypsinised combined volumes were centrifuged, the cells resuspended in growth medium and stained with Trypan Blue for viability [40].
RNA extraction and microarrays
Collected cells from duplicate experiments were lysed on MicroHex microcarriers using Trizol (InVitrogen, Carlsbad, CA) according to manufacturer’s intructions, frozen at -80°C and sent for RNA extraction and microarray analysis to the Centre d’Innovation de Génome Québec, (Montréal, QC). Illumina Mouse Ref-8 v2.0 slides (25,697 probes) were hybridized with trabecular osteoblast samples (Pthrp +/+ and Pthrp -/-) from experiments in 1g or in simulated microgravity for 6 days, conducted with or without intermittent (2-h daily) PTHrP 1–36 treatment (8 different conditions in total). Data pre-processing and analysis of expressions levels results were conducted using the FlexArray software version 1.5 from Génome Québec [41] http://genomequebec.mcgill.ca/FlexArray. Normalization was performed with the Lumi package and the data were analyzed using EB Wright and Simon statistical analysis. We retained only gene lists with probe values of p ≤ 0.05 for analysis. Cluster analysis was performed with the Database for Annotation, Visualization and Integrated Discovery online functional annotation tool [42] and by manual searches. Venn diagrams were made using FunRich analysis tool [43]. Microarray data was deposited in the NCBI GEO database and is available under accession number GSE 78980.
Analysis software, statistics
Statistical analyses of viability experiments were conducted using GraphPad Prism 4.0 for Windows (GraphPad Software, San Diego, CA), using one-way ANOVA with Bonferroni’s multiple comparison post-test. Results were considered significant at p ≤ 0.05.
Results
Endogenous PTHrP and exogenous PTHrP1-36 protect trabecular osteoblasts from Space microgravity-induced apoptosis
In order to investigate the role of PTHrP in bone cells exposed to space microgravity, trabecular and calvarial osteoblasts obtained from bones of Pthrp -/- fetal mice and Pthrp +/+ littermates [26] were attached to CC2 culture slides and inserted into sterile bioreactors connected online to the automated experimental trays (Fig 1A). The 6-day experiment involved a daily 2-hour treatment with 10-8 M PTHrP 1–36 peptide, or a control with growth medium change only for both the 0g and 1g trays (S1 Table). After return to Earth, an analysis of apoptotic cells was conducted by the ssDNA method [37]. A comparison of Pthrp +/+ TOs in the 1g tray with equivalent cells aboard the Foton3 satellite indicates that a 6-day exposure to microgravity causes no significant induction of apoptosis. For Pthrp -/- TOs, however, a 20.4 ± 5.0% increase in apoptotic cells was observed at 0g over the 6-day period compared to the equivalent Pthrp -/- cells which had remained on Earth (Fig 1B white bars). Intermittent treatment with 10-8M PTHrP 1–36 peptide reduced the number of apoptotic cells in 0g compared to 1g by 11.6 ± 4.1% and 50.2% ± 5.4% for +/+ and -/- TOs respectively (Fig 1B black bars) (p < 0.01). In contrast, over the 6-day period, osteoblasts of calvarial origin (COs) showed little difference in apoptosis due to microgravity exposure although Pthrp -/- COs displayed elevated apotosis proportions compared to +/+ COs regardless of gravity conditions (Fig 1C). COs were not treated with PTHrP due to the limited reaction volumes available within the experimental trays. These data implicate PTHrP in TO survival in short-term microgravity exposure, and show that in the absence of endogenous PTHrP, intermittent treatment with exogenous PTHrP 1–36 reduces the number of cells undergoing apoptosis.
Allelic effect of endogenous PTHrP levels in simulated microgravity, and compensation by exogenous PTHrP 1–36
In order to reproduce and expand Space flight results on the role of endogenous and exogenous PTHrP in trabecular octeoblasts, we used the Revolving Cell Culture System Earth-based microgravity simulator from Synthecon (Fig 2A). TOs from Pthrp +/+, Pthrp +/- and Pthrp -/- fetal animals attached to sterile MicroHex microcarriers (Fig 2B) spontaneously transformed into three-dimensional structures within 24 h (Fig 2C) and these aggregates remained intact throughout the experiment. The low negative impact of our design is evident in the excellent survival of various types of osteoblasts in the Synthecon chambers for extended periods of time (see next section).
Cell-microcarrier aggregates were collected after 6 days, trypsinised and analysed by Trypan blue for viability. After 6 days, Pthrp+/+ TOs presented a low (7.5±1.1%) percentage of non-viable cells in both 0g and 1g experimental set-ups. Heterozygous Pthrp +/- TOs displayed a higher percentage of non-viable cells (22% ± 2.1 at 0g compared to 13.5±1.9% at 1g) p < 0.01, and Pthrp -/- TOs were greatly affected by weightlessness (non-viable cells: 43±3.1% at 0g compared to 20 ± 2.5% at 1g) (Fig 2D white bars) p < 0.001. Intermittent treatment with PTHrP 1–36 made little difference at 1g or 0g for Pthrp+/+ TOs but reduced the percentage of non-viable cells from 22.1 to 12.7 ± 4% at 0g in the heterozygous TOs. In Pthrp -/- cells, PTHrP 1–36 intermittent treatment decreased non-viable cells from 20±3.2% to 11±2.5% at 1g, but there was a striking reduction (43±4.2% to 11±5.0%) in numbers of non-viable cells at 0g (Fig 2D black bars) p < 0.001. These data confirm the results obtained in Space: there is little to no effect of microgravity on Pthrp+/+ cell viability in 0g over 6 days, but incremental loss of viability as Pthrp levels of expression decrease. Intermittent PTHrP1-36 maintains or increases viability, especially in Pthrp-/- TOs. The simulated microgravity experiments demonstrate again the greater vulnerability of Pthrp-ablated trabecular osteoblasts to microgravity and point to a definite allelic effect for Pthrp ablation.
Long-term exposure to simulated microgravity is detrimental to wild-type trabecular osteoblasts but compensated for by exogenous PTHrP1-36 treatment
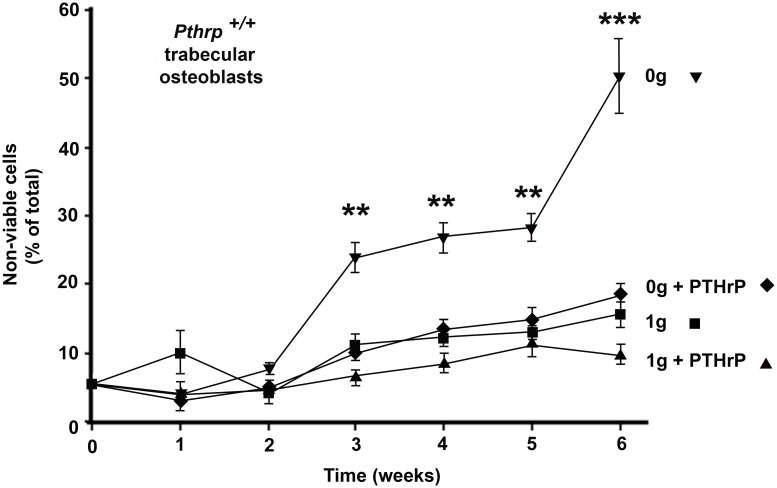
In order to investigate the effect of longer microgravity exposure on wild-type TOs, 6-week experiments were conducted with wild-type Pthrp+/+ TO cells attached to microcarriers and exposed to RCCS-simulated microgravity (0g) or to normal gravity (1g). Incubation conditions, media changes and intermittent daily PTHrP 1–36 treatments (10-8 M) were identical to those described above. Samples of osteoblast aggregates on microcarriers were drawn from 1g and 0g experimental set at weekly intervals, trypsinized, and cell viability assessed by Trypan blue staining. The three-dimensional cell aggregates on micro-carriers provide in vitro growth conditions that are closer to those found in vivo compared to single layer cultures [44] and allow excellent survival of wild-type trabecular osteoblasts in normal gravity over the course of 6 weeks; wild-type Pthrp+/+ TOs attached to MicroHex carriers easily survived 6 weeks in culture plates in normal 1g gravity, with only a 10–15% overall loss in viability (Fig 3). In contrast, Pthrp+/+ TOs on MicroHex carriers exposed to simulated microgravity (0g) started to lose viability after 2 weeks and presented a striking 50% non-viable cells after 6 weeks. Importantly, a 2-h daily treatment with PTHrP1-36 over the 6-week period reduced this loss of viability to 15%, similar to that of cells left at 1g. As expected from results observed in Fig 2, Pthrp-/- TOs did not survive past week 2 (not shown). These results suggest that microgravity-induced molecular events leading to cell death occur early in wild-type TOs and that intermittent treatment with PTHrP 1–36 peptide maintains viability in these cells over the course of 6 weeks.
Fig 3. Long-term exposure to simulated microgravity is detrimental to wild-type trabecular osteoblasts but compensated for by exogenous intermittent PTHrP1-36 treatment.
Pthrp+/+ trabecular osteoblasts at 1g (■) or 0g (▼). Pthrp+/+ trabecular osteoblasts treated with PTHrP1-36 10-8M.at 1g (▲) or 0g (♦). (triplicate 6-week experiments). Viability estimated by Trypan blue. ** p < 0.01, *** p < 0.001.
Long-term exposure to simulated microgravity has little negative effect on calvarial osteoblasts
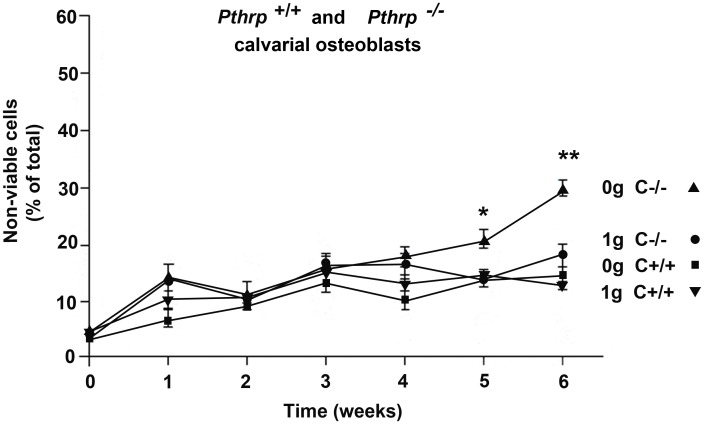
Long-term experiments (6-weeks) were conducted in the RCCS equipment with calvarial osteoblasts from Pthrp +/+ and -/- mice. CO cells attached to MicroHex carriers produced three-dimensional structures similar to those observed in trabecular experiments. Experimental 0g and 1g conditions were identical to above except that no exogenous PTHrP 1–36 treatment was conducted. Fig 4 shows that COs from wild type Pthrp +/+ mice maintain comparable viability in 0g and 1g for at least 6 weeks (10 to 12% non-viable cells). As suggested by results from Fig 1C, Pthrp -/- COs are more sensitive to 0g but the percentage of non-viable cells after 6 weeks at 0g is only 13% above that of Pthrp -/- calvarial cells at 1g. These results indicate that wild-type COs are little affected by a 6-week exposure to microgravity, but that PTHrP nevertheless plays some role in resistance to weightlessness in cranium-derived osteoblasts since Pthrp -/- calvarial cells slowly lose viability in 0g.
Fig 4. Long-term exposure to simulated microgravity has little negative effect on calvarial osteoblasts.
Phtrp+/+ calvarial osteoblasts at 1g (▼) or 0g (■). Pthrp-/- calvarial osteoblasts at 1g (●) or 0g (▲). (triplicate 6-week experiments). No exogenous PTHrP1-36 treatment was applied in this experiment. Viability estimated by Trypan blue. * p < 0.05, ** p < 0.01.
Microarray analysis conducted on trabecular osteoblasts: effect of 6-day exposure to microgravity, effect of Pthrp ablation, and reversal by intermittent PTHrP 1–36 treatment
Microgravity-induced adverse effects appear early on in TOs exposed to weightless conditions. Since these early events are crucial to all subsequent physiological changes, expression microarrays after 6 days at 0g were analysed in order to identify the genes involved. Microarray analysis (Illumina Mouse Ref8) was conducted on Pthrp+/+ wild type TOs exposed to 6 days of simulated microgravity to check for early gene expression changes that lead to microgravity-induced alterations detectable in the following weeks. 0g-induced changes in Pthrp+/+ TOs were also compared with those resulting from Pthrp gene ablation. Reversal was examined upon intermittent treatment with the PTHrP1-36 peptide (10-8M) of Pthrp-ablated and 0g-exposed TO cells.
In Pthrp+/+ trabecular osteoblasts, a 6-day period in simulated microgravity significantly upregulated 59 genes (fold change > 2.0, p < 0.05, complete gene list in S2A Table), and downregulated 129 genes (fold change < 0.5, p < 0.05, complete gene list in S2B Table) compared to Pthrp+/+ TOs which remained at 1g (total number of genes significantly affected by 6 days at 0g = 188). Pthrp gene ablation (i.e. comparison of Pthrp+/+ and Pthrp-/- TOs at 1g) significantly upregulated 270 genes (fold change > 2.0, p < 0.05, complete genelist in S3A Table) and downregulated 493 genes (fold change < 0.5, p < 0.05, complete gene list in S3B Table)(total number of genes significantly affected by Pthrp ablation = 763).
With this information, a parallel was established between the effects of microgravity exposure on wild-type Pthrp+/+ TOs, and the changes in gene expression occurring after Pthrp ablation at 1g. 52 upregulated and 115 downregulated genes were found to be common between 0g exposure and Pthrp ablation effects (Fig 5A and 5B, intersects, complete list of common genes in Tables 1 and 2). In all, 88% of the genes modified by microgravity were similarly affected by Pthrp ablation. Clustering analysis suggests the common genes are mainly related to the following metabolic categories: prolactins (up: Plf2, Mrpplf3), genes involved in bone growth, mineralization and bone morphogenic protein (BMP) metabolism (up: Aqp5; Grem1, Inhba, Nbl1, Cryab, Ank, CD44, Scx, Mustn1, Tnfrsf11b (osteoprotegerin), Bdnf, Stmn2; down: Ptn (pleiotrophin), Hp, Scara5, Itm2a, Ptn, Dlk1, Ramp2, Dab, Eno1, CD14, Hemp1, Lgmn, Txnip, Ptx3), apoptosis/survival (down: Clu, Rbm3), extracellular matrix components (up: Timp3, Prelp, Ctgf, Col7a1; down: Lum, Dcn, H19, Nid2, Col3a1, Dpt, Ctsc), Wnt signaling (up: Gpc1, Nkd2; down: Sfrp 2 and 1), IGF signaling (down: Igfbp5 and 3, Igf2), heat-shock proteins (up: Hspb1 and 8), chemokines (down: Cxcl12 and 4, Ccr5), cell cycle control (up: Ccng1, down: Cdkn1c), as well as a variety of histone genes. Importantly, none of the significantly-affected genes presented a contradictory direction change in expression level between Pthrp ablation or microgravity treatment.
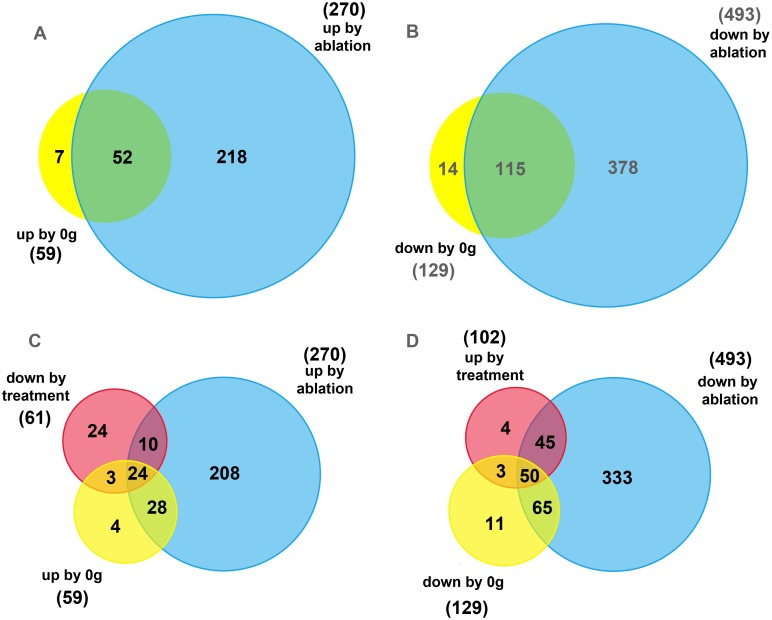
Fig 5. Venn diagrams illustrating the number of trabecular osteoblast genes significantly up- or downregulated by microgravity exposure (6 days) or by Pthrp ablation, and reversed by PTHrP1-36 treatment.
(A) 52 genes are upregulated by both 0g (yellow) and Pthrp ablation (blue). Fold change > 2.0, p < 0.05. (B) 115 genes are downregulated by both 0g (yellow) and Pthrp ablation (blue). Fold change < 0.5, p < 0.05. (C) 24 genes are upregulated by both 0g (yellow) and Pthrp ablation (blue), and are also downregulated (central intersect) by PTHrP1-36 treatment (red). Fold change < 0.65, p < 0.05. (D) 50 genes are downregulated by both 0g (yellow) and Pthrp ablation (blue), and upregulated (central intersect) by PTHrP1-36 treatment (red). Fold change >1.5, p < 0.05.
Table 1. Genes (52) upregulated by both microgravity and Pthrp ablation.
| Target ID | Definition (all probes: Mus musculus) | RefSeq ID | Fold change 0g | Fold change ablation | C |
|---|---|---|---|---|---|
| PLF2 | prolactin family 2, subfamily c, member 3 (Prl2c3), mRNA. | NM_011118.1 | 12.81098 | 20.67081 | P |
| MRPPLF3 | prolactin family 2, subfamily c, member 4 (Prl2c4), mRNA. | NM_011954.2 | 11.76853 | 19.04649 | P |
| AQP5 | aquaporin 5 (Aqp5), mRNA.PREDICTED: similar to aquaporin 5 (LOC100046616), mRNA. | NM_009701.4XM_001476512.1 | 7.7909763.627969 | 25.586929.547247 | A |
| GREM1 | gremlin1 (Grem1) mRNA. | NM_011824.1 | 4.151464 | 5.17375 | B |
| GPC1 | glypican 1 (Gpc1), mRNA. | NM_016696.3 | 4.13393 | 4.008327 | W |
| ALDH3A1 | aldehyde dehydrogenase family 3, subfamily A1 (Aldh3a1), mRNA. | NM_007436.1 | 3.821048 | 2.733907 | |
| ASS1 | argininosuccinate synthetase 1 (Ass1), mRNA. | NM_007494.3 | 3.663361 | 2.781254 | |
| NKD2 | naked cuticle 2 homolog (Drosophila) (Nkd2), mRNA. | NM_028186.3 | 3.63063 | 9.964373 | W |
| NQO1 | NAD(P)H dehydrogenase, quinone 1 (Nqo1), mRNA. | NM_008706.4 | 3.439677 | 9.410905 | |
| INHBA | inhibin beta-A (Inhba), mRNA. | NM_008380.1 | 3.374437 | 6.03861 | B |
| HIST1H4H | histone cluster 1, H4h (Hist1h4h), mRNA. | NM_153173.2 | 3.122563 | 6.525515 | H |
| HSPB1 | heat shock protein 1 (Hspb1), mRNA. | NM_013560.1 | 2.937195 | 3.686913 | HS |
| CRIP2 | cysteine rich protein 2 (Crip2), mRNA. | NM_024223.1 | 2.87678 | 4.260978 | |
| TIMP3 | tissue inhibitor of metalloproteinase 3 (Timp3), mRNA | NM_011595.2NM_011595.2 | 2.8337662.168426 | 4.7180244.573879 | E |
| ESD | esterase D/formylglutathione hydrolase (Esd), mRNA. | NM_016903.2 | 2.617979 | 4.782361 | |
| PRELP | proline arginine-rich end leucine-rich repeat (Prelp), mRNA. | NM_054077.3 | 2.563206 | 2.682429 | E |
| 1110032E23RIK | RIKEN cDNA 1110032E23 gene (1110032E23Rik), mRNA | NM_133187.2 | 2.532211 | 5.501027 | |
| GDF15 | growth differentiation factor 15 (Gdf15), mRNA. | NM_011819.1 | 2.506674 | 2.841855 | T |
| AHRR | aryl-hydrocarbon receptor repressor (Ahrr), mRNA. | NM_009644.2 | 2.366026 | 2.51242 | |
| NBL1 | neuroblastoma, suppression of tumorigenicity 1 (Nbl1), mRNA. | NM_008675.1 | 2.362421 | 2.214473 | B |
| CRYAB | crystallin, alpha B (Cryab), mRNA. | NM_009964.1 | 2.332638 | 5.135748 | B |
| FABP3 | fatty acid binding protein 3, muscle and heart (Fabp3), mRNA. | NM_010174.1 | 2.32906 | 7.70136 | |
| MDM2 | transformed mouse 3T3 cell double minute 2 (Mdm2), mRNA. | NM_010786.2NM_010786.3 | 2.318342.246531 | 2.8090142.71183 | |
| KCTD10 | potassium channel tetramerisation domain containing 10 (Kctd10), mRNA. | NM_026145.3 | 2.279135 | 2.72172 | IC |
| SGK | serum/glucocorticoid regulated kinase 1 (Sgk1), mRNA. | NM_011361.1 | 2.240079 | 2.362139 | |
| ANK | progressive ankylosis (Ank), mRNA. | NM_020332.3 | 2.228785 | 9.115022 | B |
| EPHX1 | epoxide hydrolase 1, microsomal (Ephx1), mRNA. | NM_010145.2 | 2.215451 | 2.907278 | |
| WHRN | whirlin (Whrn), transcript variant 3, mRNA. | NM_001008792.1 | 2.202689 | 2.327795 | |
| CD44 | CD44 antigen (Cd44), transcript variant 2, mRNA. | NM_001039150.1 | 2.195092 | 4.927895 | B |
| CTGF | connective tissue growth factor (Ctgf), mRNA. | NM_010217.1 | 2.193812 | 3.29075 | E |
| SCX | scleraxis (Scx), mRNA. | NM_198885.2 | 2.185468 | 4.251521 | B |
| MUSTN1 | musculoskeletal, embryonic nuclear protein 1 (Mustn1), mRNA. | NM_181390.1NM_181390.2 | 2.1853642.109783 | 3.9570883.099647 | B |
| HIST1H2BJ | histone cluster 1, H2bj (Hist1h2bj), mRNA. | NM_178198.1 | 2.173656 | 2.755197 | H |
| EEF1A2 | eukaryotic translation elongation factor 1 alpha 2 (Eef1a2), mRNA. | NM_007906.2 | 2.153269 | 2.293173 | A |
| TNFRSF11B | tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) (Tnfrsf11b), mRNA. | NM_008764.3 | 2.129809 | 9.058214 | B |
| HOXC6 | homeo box C6 (Hoxc6), mRNA. | NM_010465.2 | 2.107019 | 2.215212 | |
| 2300002D11RIK | RIKEN cDNA 2300002D11 gene (2300002D11Rik), mRNA. | NM_001081156.1 | 2.097322 | 4.2774 | |
| BDNF | brain derived neurotrophic factor (Bdnf), transcript variant 3, mRNA. | NM_001048141.1 | 2.091327 | 7.461118 | B |
| HSPB8 | heat shock protein 8 (Hspb8), mRNA. | NM_030704.1 | 2.090601 | 2.42948 | H |
| PDGFA | platelet-derived growth factor, alpha chain (Pdgfa) mRNA. | 2.084386 | 2.631637 | PD | |
| CCNG1 | cyclin G1 (Ccng1), mRNA. | NM_009831.2 | 2.083259 | 3.109058 | CC |
| HIST1H2BM | histone cluster 1, H2bm (Hist1h2bm), mRNA. | NM_178200.1 | 2.072647 | 2.824912 | H |
| HIST1H2BH | histone cluster 1, H2bh (Hist1h2bh), mRNA. | NM_178197.1 | 2.069317 | 2.693571 | H |
| HIST1H2BF | histone cluster 1, H2bf (Hist1h2bf), mRNA. | NM_178195.1 | 2.066524 | 2.55937 | H |
| COL7A1 | collagen, type VII, alpha 1 (Col7a1), mRNA. | NM_007738.3 | 2.044317 | 3.129226 | E |
| A630005A06RIK | TBC1 domain family, member 2 (Tbc1d2), mRNA. | NM_198664.3 | 2.044002 | 3.518042 | |
| GSTP1 | glutathione S-transferase, pi 1 (Gstp1), mRNA. | NM_013541.1 | 2.039079 | 2.672256 | |
| TNFRSF12A | tumor necrosis factor receptor superfamily, member 12a (Tnfrsf12a), mRNA. | NM_013749.1 | 2.026671 | 3.449432 | |
| SNRPN | SNRPN upstream reading frame (Snurf), mRNA. | NM_033174.2 | 2.014161 | 3.736806 | |
| HIST1H2BK | histone cluster 1, H2bk (Hist1h2bk), mRNA. | NM_175665.1 | 2.012974 | 2.669902 | H |
| STMN2 | stathmin-like 2 (Stmn2), mRNA. | NM_025285.2 | 2.007183 | 3.763786 | B |
| DOS | downstream of Stk11 (Dos), mRNA. | NM_015761.2 | 2.00261 | 3.474366 |
Genes (52) upregulated by both microgravity and Pthrp ablation. All fold changes > 2. All p values < 0.05. Multiple entries for a gene indicate multiple probes. C: Clustering: A: apoptosis/survival, B: bone metabolism and bone morphogenic proteins, CC: cell cycle, E: extracellular matrix, H: histone, HS: heat shock, IC: ion channel, P: prolactins, PD: Platelet-derived growth factor, T: tumor growth factor, W: Wnt.
Table 2. Genes (115) downregulated by both microgravity and Pthrp ablation.
| Target ID | Definition (all probes: Mus musculus) | RefSeq ID | Fold change 0g | Fold change ablation | C |
|---|---|---|---|---|---|
| SFRP2 | secreted frizzled-related protein 2 (Sfrp2), mRNA. | NM_009144.1 | 0.1316429 | 0.096523 | W |
| PTN | pleiotrophin (Ptn), mRNA. | NM_008973.2 | 0.1557338 | 0.047141 | B |
| IGFBP5 | insulin-like growth factor binding protein 5 (Igfbp5), mRNA. | NM_010518.2 | 0.1603545 | 0.2130594 | I |
| 1500015O10RIK | RIKEN cDNA 1500015O10 gene (1500015O10Rik), mRNA. | NM_024283.2 | 0.1804316 | 0.0975587 | |
| IGF2 | insulin-like growth factor 2 (Igf2), mRNA. | NM_010514.2 | 0.2085582 | 0.026108 | I |
| CLU | PREDICTED: similar to clusterin (LOC100046120), mRNA. | XM_001475611.1 | 0.213665 | 0.47421 | A |
| CXCL12 | chemokine (C-X-C motif) ligand 12 (Cxcl12), transcript variant 3, mRNA. | NM_013655.2NM_021704.2NM_001012477.1 | 0.21502010.22348560.236011 | 0.28323790.40775260.3778157 | CH |
| HP | haptoglobin (Hp), mRNA. | NM_017370.1NM_017370.1 | 0.22221020.4034571 | 0.18333460.4167181 | B |
| C3 | complement 3 (C3) mRNA. | NM_009778.1 | 0.2325967 | 0.1254136 | |
| SCARA5 | scavenger receptor class A, member 5 (putative) (Scara5), mRNA. | NM_028903.1 | 0.2351416 | 0.1405516 | B |
| LUM | lumican (Lum), mRNA. | NM_008524.1 | 0.235224 | 0.0573447 | E |
| MRC1 | mannose receptor, C type 1 (Mrc1), mRNA. | NM_008625.1 | 0.253038 | 0.0815048 | |
| 4930583H14RIK | RIKEN cDNA 4930583H14 gene (4930583H14Rik), mRNA. | NM_026358.2 | 0.2575598 | 0.1684547 | |
| 2310061N23RIK | interferon, alpha-inducible protein 27 (Ifi27), mRNA. | NM_029803.1 | 0.2600992 | 0.0590283 | |
| DCN | decorin (Dcn), mRNA. | NM_007833.4NM_007833.4 | 0.28897940.3837174 | 0.06403410.0548521 | E |
| H19 | H19 fetal liver mRNA (H19), non-coding RNA. | NR_001592.1 | 0.2906019 | 0.0360464 | E |
| PPP1R3C | protein phosphatase 1, regulatory (inhibitor) subunit 3C (Ppp1r3c), mRNA. | NM_016854.2 | 0.2954025 | 0.1777717 | |
| RBM3 | PREDICTED: similar to RNA binding motif protein 3 (LOC100043257), mRNA. | XM_001480197.1 | 0.2991486 | 0.2378549 | A |
| APOD | PREDICTED: similar to apolipoprotein D (LOC100047583), mRNA. | XM_001479138.1 | 0.2996465 | 0.1742532 | A |
| ITM2A | integral membrane protein 2A (Itm2a), mRNA. | NM_008409.2 | 0.2997355 | 0.138948 | B |
| D930038M13RIK | ABI gene family, member 3 (NESH) binding protein (Abi3bp), transcript variant 1, mRNA. | NM_178790.3 | 0.3036303 | 0.2955018 | B |
| PFKL | phosphofructokinase, liver, B-type (Pfkl), mRNA. | NM_008826.3 | 0.3054363 | 0.2514627 | |
| OLFML1 | olfactomedin-like 1 (Olfml1), mRNA. | NM_172907.2 | 0.3123395 | 0.3050197 | |
| HIST1H2AD | histone cluster 1, H2ad (Hist1h2ad), mRNA. | NM_178188.3 | 0.3149356 | 0.0061305 | |
| DLK1 | delta-like 1 homolog (Drosophila) (Dlk1), mRNA. | NM_010052.4 | 0.3273854 | 0.1513731 | B |
| HIST1H2AK | histone cluster 1, H2ak (Hist1h2ak), mRNA. | NM_178183.1 | 0.3372404 | 0.0596174 | H |
| 2310056P07RIK | family with sequence similarity 162, member A (Fam162a), mRNA. | NM_027342.1 | 0.3391683 | 0.2567712 | |
| HIST1H2AF | histone cluster 1, H2af (Hist1h2af), mRNA. | NM_175661.1 | 0.3396376 | 0.067387 | H |
| SLC1A3 | solute carrier family 1 (glial high affinity glutamate transporter), member 3 (Slc1a3), mRNA. | NM_148938.2 | 0.3399492 | 0.1181837 | |
| PSCDBP | cytohesin 1 interacting protein (Cytip), mRNA. | NM_139200.4 | 0.3403133 | 0.1328767 | |
| NID2 | nidogen 2 (Nid2), mRNA. | NM_008695.2 | 0.3477556 | 0.3321977 | E |
| TGFBI | transforming growth factor, beta induced (Tgfbi), mRNA. | NM_009369.1 | 0.3514812 | 0.0792747 | T |
| HIST1H2AH | histone cluster 1, H2ah (Hist1h2ah), mRNA. | NM_175659.1 | 0.352093 | 0.0777217 | |
| FMO1 | flavin containing monooxygenase 1 (Fmo1), mRNA. | NM_010231.2 | 0.3568025 | 0.3366457 | |
| EGLN3 | EGL nine homolog 3 (C. elegans) (Egln3), mRNA. | NM_028133.1 | 0.3569504 | 0.3106277 | |
| DPEP2 | dipeptidase 2 (Dpep2), mRNA. | NM_176913.3 | 0.35774720.3965119 | 0.14003250.18665 | |
| COL3A1 | collagen, type III, alpha 1 (Col3a1), mRNA. | NM_009930.1 | 0.3599768 | 0.2207009 | E |
| 1200009O22RIK | RIKEN cDNA 1200009O22 gene (1200009O22Rik), mRNA. | NM_025817.3 | 0.3616481 | 0.3116025 | |
| HIST1H2AO | histone cluster 1, H2ao (Hist1h2ao), mRNA. | NM_178185.1 | 0.3618695 | 0.057145 | H |
| AGTR1A | angiotensin II receptor, type 1a (Agtr1a), mRNA. | NM_177322.2 | 0.3640873 | 0.3477361 | |
| LDB2 | LIM domain-binding protein 2 (Ldb2) mRNA. | NM_010698.2 | 0.3694307 | 0.2963222 | |
| DPT | dermatopontin (Dpt), mRNA. | NM_019759.2 | 0.369822 | 0.2973928 | E |
| 6330406I15RIK | RIKEN cDNA 6330406I15 gene (6330406I15Rik), mRNA. | NM_027519.1NM_027519.3 | 0.37182430.3991528 | 0.19828060.2041744 | |
| RASSF4 | Ras association (RalGDS/AF-6) domain family member 4 (Rassf4), mRNA. | NM_178045.3NM_178045.3 | 0.3723380.3845508 | 0.36712360.1214973 | |
| PDGFRA | platelet derived growth factor receptor, alpha polypeptide (Pdgfra), transcript variant 1, mRNA. | NM_011058.2 | 0.3755132 | 0.394926 | PD |
| HIST1H2AN | histone cluster 1, H2an (Hist1h2an), mRNA. | NM_178184.1 | 0.3776532 | 0.0945669 | H |
| ZCCH5 | Zinc finger, CCHC domain-containing 5 (Zcchc5), mRNA | NM_199468.2 | 0.3839341 | 0.4813192 | |
| IGFBP3 | insulin-like growth factor binding protein 3 (Igfbp3), mRNA. | NM_008343.2 | 0.3878084 | 0.3767934 | I |
| AOC3 | amine oxidase, copper containing 3 (Aoc3), mRNA. | NM_009675.1 | 0.390921 | 0.102008 | |
| KNG1 | kininogen 1 (Kng1), mRNA. | NM_023125.2 | 0.3914529 | 0.2682237 | |
| SERPING1 | serine (or cysteine) peptidase inhibitor, clade G, member 1 (Serping1), mRNA. | NM_009776.1 | 0.3939361 | 0.2493484 | |
| SFRP1 | secreted frizzled-related protein 1 (Sfrp1), mRNA. | NM_013834.1 | 0.3966504 | 0.1332278 | W |
| EMR1 | EGF-like module containing, mucin-like, hormone receptor-like sequence 1 (Emr1), mRNA. | NM_010130.3 | 0.4002873 | 0.0452258 | |
| SIRPB1 | signal-regulatory protein beta 1 (Sirpb1), transcript variant 3, mRNA. | NM_001002898.1 | 0.4006563 | 0.250706 | |
| CAR9 | carbonic anhydrase 9 (Car9), mRNA. | NM_139305.1 | 0.401926 | 0.3807879 | |
| SRPX | sushi-repeat-containing protein (Srpx), mRNA. | NM_016911.4NM_016911.4 | 0.40437110.4169736 | 0.12827860.1438187 | |
| LBP | lipopolysaccharide binding protein (Lbp), mRNA. | NM_008489.2 | 0.4046987 | 0.3596355 | |
| 2310006J04RIK | Ankyrin repeat domain 37 (Ankrd 37), mRNA | NM_001039562.1 | 0.4075139 | 0.4461992 | |
| CCR5 | chemokine (C-C motif) receptor 5 (Ccr5), mRNA. | NM_009917.2NM_009917.2NM_009917.4 | 0.40916990.44612370.4461237 | 0.31747080.32485210.3880605 | CH |
| RAMP2 | receptor (calcitonin) activity modifying protein 2 (Ramp2), mRNA. | NM_019444.2 | 0.4105468 | 0.2878545 | B |
| LSP1 | PREDICTED: predicted gene, ENSMUSG00000043795 (ENSMUSG00000043795), mRNA. | XM_001480835.1 | 0.4114556 | 0.1243922 | |
| CDKN1C | cyclin-dependent kinase inhibitor 1C (P57) (Cdkn1c), mRNA. | NM_009876.3 | 0.4116614 | 0.347614 | CC |
| CORO1A | coronin, actin binding protein 1A (Coro1a), mRNA. | NM_009898.2NM_009898.2NM_009898.2 | 0.41277540.44359450.4970813 | 0.08707340.05964910.1696803 | |
| C1QA | complement component 1, q subcomponent, alpha polypeptide (C1qa), mRNA. | NM_007572.2 | 0.4128893 | 0.1927809 | |
| SLC2A1 | solute carrier family 2 (facilitated glucose transporter), member 1 (Slc2a1), mRNA. | NM_011400.2 | 0.4167042 | 0.2991707 | |
| GPR23 | G protein-coupled receptor 23 (Gpr23), mRNA. | NM_175271.2 | 0.4254305 | 0.4447173 | |
| PDK1 | pyruvate dehydrogenase kinase, isoenzyme 1 (Pdk1), nuclear gene encoding mitochondrial protein, mRNA. | NM_172665.3 | 0.4255101 | 0.3667602 | |
| CTSC | cathepsin C (Ctsc), mRNA. | NM_009982.2 | 0.4265272 | 0.0758005 | E |
| OASL2 | 2'-5' oligoadenylate synthetase-like 2 (Oasl2), mRNA. | NM_011854.1 | 0.4272569 | 0.1159377 | |
| C1QB | complement component 1, q subcomponent, beta polypeptide (C1qb), mRNA. | NM_009777.2 | 0.4278048 | 0.0245339 | |
| DAB2 | disabled homolog 2 (Drosophila) (Dab2), transcript variant 2, mRNA. | NM_023118.1NM_001008702.1 | 0.42782040.4450885 | 0.17925610.1341114 | B |
| RSPO3 | R-spondin 3 homolog (Xenopus laevis) (Rspo3), mRNA. | NM_028351.2 | 0.4293909 | 0.2896171 | W |
| MFAP2 | microfibrillar-associated protein 2 (Mfap2), mRNA. | NM_008546.2NM_008546.2NM_008546.2 | 0.42967080.47591550.4945922 | 0.18086140.25899670.2757687 | B |
| GMFG | glia maturation factor, gamma (Gmfg), transcript variant 1, mRNA. | NM_022024.2NM_022024.2 | 0.43023620.4870403 | 0.203090.2355645 | |
| SELENBP1 | PREDICTED: hypothetical protein LOC100044204 (LOC100044204), mRNA. | XM_001471696.1 | 0.433163 | 0.2613255 | |
| MEST | mesoderm specific transcript (Mest), mRNA. | NM_008590.1NM_008590.1 | 0.43492520.4556044 | 0.31959320.3918639 | |
| NDRG1 | N-myc downstream regulated gene 1 (Ndrg1), mRNA. | NM_008681.2 | 0.4352534 | 0.3870171 | |
| MS4A6D | membrane-spanning 4-domains, subfamily A, member 6D (Ms4a6d), mRNA. | NM_026835.2 | 0.4373044 | 0.0358791 | |
| ALOX5AP | arachidonate 5-lipoxygenase activating protein (Alox5ap), mRNA. | NM_009663.1 | 0.4387523 | 0.0317102 | |
| HIST1H2AG | histone cluster 1, H2ag (Hist1h2ag), mRNA. | NM_178186.2 | 0.4395184 | 0.2645484 | H |
| 1300002F13RIK | ERBB receptor feedback inhibitor 1 (Errfi1), mRNA. | NM_133753.1 | 0.4395548 | 0.2132049 | |
| EVI2A | ecotropic viral integration site 2a (Evi2a), transcript variant 2, mRNA. | NM_010161.3 | 0.4402088 | 0.1280774 | |
| CXCL4 | chemokine (C-X-C motif) ligand 4 (Cxcl4), mRNA. | NM_019932.2 | 0.4419611 | 0.180652 | CH |
| 1190002H23RIK | RIKEN cDNA 1190002H23 gene (1190002H23Rik), mRNA. | NM_025427.2 | 0.4430251 | 0.3481645 | |
| ENO1 | enolase 1, alpha non-neuron (Eno1), mRNA. | NM_023119.1 | 0.4461543 | 0.4872021 | B |
| LYZ | lysozyme 1 (Lyz1), mRNA. | NM_013590.3 | 0.4473266 | 0.3851333 | |
| CD52 | CD52 antigen (Cd52), mRNA. | NM_013706.1 | 0.44963 | 0.0749331 | |
| NDRL | N-myc downstream gene 1 (Ndrl), mRNA. | 0.4504366 | 0.4056498 | ||
| CYP7B1 | cytochrome P450, family 7, subfamily b, polypeptide 1 (Cyp7b1), mRNA. | NM_007825.3 | 0.450876 | 0.1997939 | |
| PIRA4 | paired-Ig-like receptor A4 (Pira4), mRNA. | NM_011091.1 | 0.4527747 | 0.0263051 | |
| RARRES2 | retinoic acid receptor responder (tazarotene induced) 2 (Rarres2), mRNA. | NM_027852.1 | 0.4537163 | 0.3671236 | |
| CD14 | CD14 antigen (Cd14), mRNA. | NM_009841.3 | 0.4582975 | 0.1117494 | B |
| ALDH3B1 | aldehyde dehydrogenase 3 family, member B1 (Aldh3b1), mRNA. | NM_026316.2 | 0.4627661 | 0.3188771 | |
| KCNAB2 | potassium voltage-gated channel, shaker-related subfamily, beta member 2 (Kcnab2), mRNA. | NM_010598.2 | 0.4659567 | 0.1179869 | |
| C1QG | complement component 1, q subcomponent, C chain (C1qc), mRNA. | NM_007574.2 | 0.468035 | 0.3163972 | |
| GALNT9 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9 (Galnt9), mRNA. | NM_198306.1 | 0.4683551 | 0.3906375 | |
| ADD3 | adducin 3 (gamma) (Add3), mRNA. | NM_013758.2 | 0.4687887 | 0.306828 | |
| ZFP36L1 | zinc finger protein 36, C3H type-like 1 (Zfp36l1), mRNA. | NM_007564.4 | 0.4703194 | 0.2280471 | |
| HEMP1 | NCK associated protein 1 like (Nckap1l), mRNA. | NM_153505.4 | 0.4709411 | 0.0826084 | B |
| 2810046M22RIK | NM_026621.1 | 0.4778584 | 0.4976117 | ||
| LGMN | legumain (Lgmn), mRNA. | NM_011175.2 | 0.4770529 | 0.2286424 | B |
| TXNIP | thioredoxin interacting protein (Txnip), transcript variant 1, mRNA. | NM_001009935.2 | 0.4827267 | 0.3327381 | B |
| EHD4 | EH-domain containing 4 (Ehd4), mRNA. | NM_133838.4 | 0.483588 | 0.3327381 | E |
| EMR1 | EGF-like module containing, mucin-like, hormone receptor-like sequence 1 (Emr1), mRNA. | NM_010130.1 | 0.4839607 | 0.1299728 | |
| ARHGAP20 | Rho GTPase activating protein 20 (Arhgap20), mRNA. | NM_175535.3 | 0.4859805 | 0.4102931 | |
| PTX3 | pentaxin related gene (Ptx3) mRNA | NM_008987.2 | 0.4867727 | 0.1136763 | B |
| NCF4 | neutrophil cytosolic factor 4 (Ncf4), mRNA. | NM_008677.1 | 0.4881307 | 0.1080301 | |
| PFC | complement factor properdin (Cfp), mRNA. | NM_008823.3 | 0.488737 | 0.0720321 | |
| SMARCA1 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 (Smarca1), mRNA. | NM_053123.3 | 0.4911543 | 0.3176144 | |
| FCER1G | Fc receptor IgE high affinity 1 gamma polypeptide (Fcerg) mRNA. | NM_010185.2 | 0.4915371 | 0.0163625 | |
| CENPA | centromere protein A (Cenpa), mRNA. | NM_007681.2 | 0.4943203 | 0.0808812 | |
| 1200002N14RIK | RIKEN cDNA 1200002N14 gene (1200002N14Rik), mRNA. | NM_027878.2 | 0.4949315 | 0.207954 | |
| TK1 | thymidine kinase 1 (Tk1), mRNA. | NM_009387.1 | 0.4950169 | 0.1742371 | |
| ALDOA | aldolase A, fructose-bisphosphate (Aldoa), mRNA. | NM_007438.3 | 0.4959253 | 0.4941576 | |
| SLPI | secretory leukocyte peptidase inhibitor (Slpi), mRNA. | NM_011414.2 | 0.4988716 | 0.2429238 |
Genes (115) downregulated by both microgravity and Pthrp ablation. All fold changes < 0.5. All p values < 0.05. Multiple entries for a gene indicate multiple probes. C: Clustering: A: apoptosis/survival, B: bone metabolism and bone morphogenic proteins, CC: cell cycle, CH: chemokines, E: extracellular matrix, H: histone, I: Insulin-like growth factor, PD: Platelet-derived growth factor, T: tumor growth factor, W: Wnt.
Because the PTHrP1-36 anabolic agent was observed to compensate for weightlessness and reverse the effects of microgravity exposure in TOs (Figs 1, 2 and 3), microarray analysis was conducted on Pthrp+/+ TOs at 0g treated intermittently with 10-8M PTHrP1-36 for 6 days. The expression levels of 102 genes were upregulated and 61 downregulated (total 163, fold change >1.5 or <0.65, p value< 0.05, complete list in S4A and S4B Table). Although the treatment peptide corresponds to only 36 amino acids from the N-terminus of PTHrP and not the whole sequence, we expect reversal of some effects induced by microgravity and/or Pthrp ablation. Accordingly, 27 of the 0g-upregulated genes (45.7%), and 34 of the ablation-upregulated genes (12.5%) were downregulated by PTHrP1-36 treatment. Of those, 24 genes were common to all three conditions (Fig 5C, listed in Table 3). 53 of the 0g-downregulated (41%) and 95 of the ablation-downregulated genes (19.2%) were upregulated by PTHrP1-36 treatment. Of those, 50 genes were common to all three conditions (Fig 5D, listed in Table 4). Clustering analysis revealed the same functional gene categories as above. None of the significantly-affected genes presented a contradictory direction change in expression level between PTHrP1-36 treatment and Pthrp ablation or microgravity treatment (S1 Fig).
Table 3. Genes (24) upregulated by both Pthrp ablation and microgravity and downregulated by PTHrP1-36 treatment.
| Target ID | Definition (all probes: Mus musculus) | RefSeqID | Fold change PTHrP 1–36 treatment | C |
|---|---|---|---|---|
| AQP5 | aquaporin 5 (Aqp5), mRNA.PREDICTED: similar to aquaporin 5 (LOC100046616), mRNA. | NM_009701.4XM_001476512.1 | 0.37050890.4987479 | A |
| TNFRSF11B | tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) (Tnfrsf11b), mRNA. | NM_008764.3 | 0.4687011 | B |
| HSPB1 | heat shock protein 1 (Hspb1), mRNA. | NM_013560.1 | 0.4871401 | HS |
| MRPPLF3 | prolactin family 2, subfamily c, member 4 (Prl2c4), mRNA. | NM_011954.2 | 0.5052176 | P |
| NKD2 | naked cuticle 2 homolog (Drosophila) (Nkd2), mRNA. | NM_028186.3 | 0.5150308 | W |
| PLF2 | prolactin family 2, subfamily c, member 3 (Prl2c3), mRNA. | NM_011118.1 | 0.5191368 | P |
| BDNF | brain derived neurotrophic factor (Bdnf), transcript variant 3, mRNA. | NM_001048141.1 | 0.5334656 | B |
| TIMP3 | tissue inhibitor of metalloproteinase 3 (Timp3), mRNA. | NM_011595.2NM_011595.2 | 0.53376130.6267908 | E |
| CRYAB | crystallin, alpha B (Cryab), mRNA. | NM_009964.1 | 0.5387105 | B |
| ASS1 | argininosuccinate synthetase 1 (Ass1), mRNA. | NM_007494.3 | 0.5595178 | |
| ESD | esterase D/formylglutathione hydrolase (Esd), mRNA. | NM_016903.2 | 0.5693265 | |
| INHBA | inhibin beta-A (Inhba), mRNA. | NM_008380.1 | 0.5792015 | B |
| CD44 | CD44 antigen (Cd44), transcript variant 2, mRNA | NM_001039150.1 | 0.5834 | B |
| GREM1 | gremlin1 (Grem1) mRNA. | NM_011824.1 | 0.5978743 | B |
| GDF15 | growth differentiation factor 15 (Gdf15), mRNA. | NM_011819.1 | 0.5985932 | T |
| MUSTN1 | musculoskeletal, embryonic nuclear protein 1 (Mustn1), mRNA. | NM_181390.1 | 0.6015734 | B |
| MDM2 | transformed mouse 3T3 cell double minute 2 (Mdm2), mRNA. | NM_010786.2 | 0.6034403 | |
| KCTD10 | potassium channel tetramerisation domain containing 10 (Kctd10), mRNA. | NM_026145.3 | 0.61132 | IC |
| EEF1A2 | eukaryotic translation elongation factor 1 alpha 2 (Eef1a2), mRNA. | NM_007906.2 | 0.6231256 | A |
| WHRN | whirlin (Whrn), transcript variant 3, mRNA | NM_001008792.1 | 0.6237 | |
| EPHX1 | epoxide hydrolase 1, microsomal (Ephx1), mRNA | NM_010145.2 | 0.6255 | |
| HIST1H4H | histone cluster 1, H4h (Hist1h4h), mRNA | NM_153173.2 | 0.635 | H |
| STMN2 | stathmin-like 2 (Stmn2), mRNA | NM_025285.2 | 0.645 | B |
| SCX | scleraxis (Scx), mRNA. | NM_198885.2 | 0.6553575 | B |
All -fold changes < 0.66, all p values < 0.05. Multiple entries for a gene indicate multiple probes. C: Clustering: A: apoptosis/survival, B: bone metabolism and bone morphogenic proteins, E: extracellular matrix, H: histone, HS: heat shock, IC: ion channel, P: prolactin, T: tumor growth factor, W: Wnt.
Table 4. Genes (50) downregulated by both Pthrp ablation and microgravity and upregulated by PTHrP1-36 treatment.
| Target ID | Definition (all probes: Mus musculus) | RefSeqID | Fold change PTHrP 1–36 treatment | C |
|---|---|---|---|---|
| RBM3 | PREDICTED: similar to RNA binding motif protein 3 (LOC100043257), mRNA. | XM_001480197.1 | 3.316729 | A |
| HIST1H2AD | histone cluster 1, H2ad (Hist1h2ad), mRNA. | NM_178188.3 | 3.070246 | H |
| HIST1H2AH | histone cluster 1, H2ah (Hist1h2ah), mRNA. | NM_175659.1 | 2.644405 | H |
| HIST1H2AN | histone cluster 1, H2an (Hist1h2an), mRNA. | NM_178184.1 | 2.629741 | H |
| HIST1H2AK | histone cluster 1, H2ak (Hist1h2ak), mRNA. | NM_178183.1 | 2.619065 | H |
| PTN | pleiotrophin (Ptn), mRNA. | NM_008973.2 | 2.610166 | B |
| HIST1H2AF | histone cluster 1, H2af (Hist1h2af), mRNA. | NM_175661.1 | 2.590586 | H |
| HIST1H2AO | histone cluster 1, H2ao (Hist1h2ao), mRNA. | NM_178185.1 | 2.363633 | H |
| 2310061N23RIK | interferon, alpha-inducible protein 27 (Ifi27), mRNA. | NM_029803.1 | 2.240007 | A |
| CENPA | centromere protein A (Cenpa), mRNA. | NM_007681.2 | 2.217246 | |
| HIST1H2AG | histone cluster 1, H2ag (Hist1h2ag), mRNA. | NM_178186.2 | 2.186001 | H |
| LUM | lumican (Lum), mRNA. | NM_008524.1 | 2.127828 | E |
| ITM2A | integral membrane protein 2A (Itm2a), mRNA. | NM_008409.2 | 2.072151 | B |
| IGFBP5 | insulin-like growth factor binding protein 5 (Igfbp5), mRNA. | NM_010518.2 | 2.068563 | I |
| DCN | decorin (Dcn), mRNA. | NM_007833.4 | 2.0663121.731693 | E |
| 1500015O10RIK | RIKEN cDNA 1500015O10 gene (1500015O10Rik), mRNA. | NM_024283.2 | 1.860714 | |
| MFAP2 | microfibrillar-associated protein 2 (Mfap2), mRNA. | NM_008546.2 | 1.8432941.5754411.501827 | B |
| TK1 | thymidine kinase 1 (Tk1), mRNA | NM_009387.1 | 1.8403 | |
| ALOX5AP | arachidonate 5-lipoxygenase activating protein (Alox5ap), mRNA | NM_009663.1 | 1.8329 | |
| CDKN1C | cyclin-dependent kinase inhibitor 1C (P57) (Cdkn1c), mRNA. | NM_009876.3 | 1.797543 | CC |
| GPR23 | G protein-coupled receptor 23 (Gpr23), mRNA | NM_175271.2 | 1.7145 | |
| CD52 | CD52 antigen (Cd52), mRNA. | NM_013706.1 | 1.688404 | |
| C1QA | complement component 1, q subcomponent, alpha polypeptide (C1qa), mRNA. | NM_007572.2 | 1.685787 | |
| CTSC | cathepsin C (Ctsc), mRNA. | NM_009982.2 | 1.68393 | E |
| ALDH3B1 | aldehyde dehydrogenase 3 family, member B1 (Aldh3b1), mRNA. | NM_026316.2 | 1.678022 | |
| TGFBI | transforming growth factor, beta induced (Tgfbi), mRNA. | NM_009369.1 | 1.664148 | T |
| FMO1 | flavin containing monooxygenase 1 (Fmo1), mRNA. | NM_010231.2 | 1.657767 | |
| IGF2 | insulin-like growth factor 2 (Igf2), mRNA. | NM_010514.2 | 1.656335 | I |
| CYP7B1 | cytochrome P450, family 7, subfamily b, polypeptide 1 (Cyp7b1), mRNA. | NM_007825.3 | 1.652679 | |
| CXCL12 | chemokine (C-X-C motif) ligand 12 (Cxcl12) mRNA. | NM_013655.2 | 1.637518 | CH |
| LDB2 | LIM domain-binding 2 (Ldb2) mRNA. | NM_010698.2 | 1.634978 | |
| CORO1A | coronin, actin binding protein 1A (Coro1a), mRNA. | NM_009898.2 | 1.6294451.620518 | |
| EMR1 | EGF-like module containing, mucin-like, hormone receptor-like sequence 1 (Emr1), mRNA. | NM_010130.3 | 1.626247 | |
| KNG1 | kininogen 1 (Kng1), mRNA. | NM_023125.2 | 1.5999 | |
| OLFML1 | olfactomedin-like 1 (Olfml1), mRNA. | NM_172907.2 | 1.599738 | |
| AOC3 | amine oxidase, copper containing 3 (Aoc3), mRNA. | NM_009675.1 | 1.598282 | |
| GMFG | glia maturation factor, gamma (Gmfg), transcript variant 1, mRNA. | NM_022024.2 | 1.5965611.532455 | |
| NCF4 | neutrophil cytosolic factor 4 (Ncf4), mRNA. | NM_008677.1 | 1.593552 | |
| SFRP2 | secreted frizzled-related protein 2 (Sfrp2), mRNA. | NM_009144.1 | 1.589225 | W |
| SMARCA1 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 (Smarca1), mRNA. | NM_053123.3 | 1.5706 | |
| SIRPB1 | signal-regulatory protein beta 1 (Sirpb1), transcript variant 3, mRNA. | NM_001002898.1 | 1.559451 | |
| RASSF4 | Ras association (RalGDS/AF-6) domain family member 4 (Rassf4), mRNA. | NM_178045.3 | 1.558789 | |
| KCNAB2 | potassium voltage-gated channel, shaker-related subfamily, beta member 2 (Kcnab2), mRNA. | NM_010598.2 | 1.556591 | IC |
| LSP1 | PREDICTED: predicted gene, ENSMUSG00000043795 (ENSMUG00000043795), mRNA | XM_001480835.1 | 1.5468 | |
| COL3A1 | collagen, type III, alpha 1 (Col3a1), mRNA. | NM_009930.1 | 1.531741 | E |
| SRPX | sushi-repeat-containing protein (Srpx), mRNA. | NM_016911.4 | 1.522525 | |
| DAB2 | disabled 2 mitogen-responsive phosphoprotein (Dab2) mRNA | NM_023118.1 | 1.519832 | |
| EVI2A | ecotropic viral integration site 2a (Evi2a), transcript variant 2, mRNA. | NM_010161.3 | 1.517936 | |
| NID2 | nidogen 2 (Nid2), mRNA. | NM_008695.2 | 1.506611 | E |
| C1QB | complement component 1, q subcomponent, beta polypeptide (C1qb), mRNA. | NM_009777.2 | 1.503228 |
All -fold changes > 1.5, all p values < 0.05. C: Clustering: A: apoptosis/survival, B: bone metabolism and bone morphogenic proteins, CC: cell cycle, CH: chemokines, E: extracellular matrix, H: histone, I: Insulin-like growth factor, IC: ion channel, T: tumor growth factor, W: Wnt.
Discussion
Exposure to Space microgravity causes an important and rapid decrease in the BMD of astronauts’ weight-bearing bones similar to that observed in disuse osteoporosis on Earth [45]. Exercise in weightless conditions does not effectively counter spine and hip bone loss, and a better understanding of bone formation/resorption uncoupling mechanisms under these conditions is needed in order to provide effective countermeasures useful to flight personnel as well as to patients suffering from disuse-induced osteoporosis on Earth. Microgravity has been reported to reduce the expression of PTHrP in lung and bone cells, leading to speculation that this molecule can act as a gravisensor [33]. In view of the fact that bone loss appears to result in significant part from osteoblast dysfunction, we investigated the mechanistic implications of PTHrP metabolism in 0g-induced skeletal disorders using mouse wild-type trabecular osteoblasts (Pthrp+/+) which mimic astronauts’ TOs at the start of their mission. Comparison of viability of the wild-type cells with Pthrp+/- and -/- TOs confirmed the involvement of PTHrP in resistance to microgravity: a 6-day exposure to microgravity (actual and simulated) significantly decreased viability of Pthrp-/- trabecular osteoblasts while Pthrp+/+ cells appeared unaffected. An allelic dose-effect for PTHrP expression was even observed using heterozygous Pthrp+/- TOs in simulated 0g. Although Pthrp+/+ cells maintained viability after 6 days at 0g, a crucial observation made during exposure of TOs to longer periods of simulated microgravity revealed that the astronaut-mimicking Pthrp+/+ TOs start to lose viability at 2 weeks, suggesting early metabolic changes in the wild-type cells at 0g. Importantly, this sharp decline in viability could be countered by treatment with 10-8M PTHrP1-36 peptide which, when administered intermittently (in a pulsatile manner), has anabolic properties, in contrast with a continuous treatment which causes downregulation of osteogenic genes [36]. These observations confirmed the role of PTHrP in cell survival in microgravity in trabecular osteoblasts. In the present study, calvarial osteoblasts appeared much less sensitive to 0g conditions than trabecular osteoblasts, an observation which agrees with those made in human 6° head-down bedrest prolonged tests (which effectively mimic weightlessness) where weight-bearing bones incur substantial skeletal loss but cranium bones in fact appear to gain bone mass [46].
The mechanisms by which PTHrP counters the signal of absent gravity were investigated through expression array analyses revealing that radical changes were already occurring in Pthrp+/+ TOs after 6 days in microgravity. Strikingly, the genes affected by 0g present an 87% overlap with genes affected by Pthrp ablation (comparison of Pthrp+/+ and Pthrp-/- TOs at 1g), and all changes proceed in the same direction for ablation and 0g exposure. This observation indicates that microgravity-induced metabolic changes in wild-type trabecular osteoblasts proceed mostly through PTHrP-controlled genes, affirming a prominent role of PTHrP in cell survival under microgravity. Among the most up- or downregulated genes common to 0g and Pthrp ablation are genes encoding members of the prolactin family, several genes involved in bone growth including Wnt signaling and insulin-like growth factor (IGF) signaling, genes encoding extracellular matrix (ECM)-related proteins and involved in BMP metabolism and, in mineralization, heat-shock proteins, chemokines, and genes encoding apoptosis/survival, cell cycle control as well as histones. A comparison of our microarray results with expression changes in wild-type mouse 2T3 pre-osteoblasts attached to growth container surface and exposed to simulated 0g for 3 days [47] confirms the loss of bone growth, mineralization, bone morphogenic and extra-cellular matrix proteins upon exposure to simulated microgravity by similar changes in expression of numerous common genes at 0g: osteoprotegerin, RAMP 2, lumican, decorin and various collagen proteins. Several cell signaling molecules are also similarly affected by microgravity in both studies: PDGFRα, IGF2, SFRP2, and members of the TNF and Fos family. The Wnt/βcatenin pathway is essential for bone formation in response to mechanical loading [48], and in our study, expression of mechanosensitive genes such as SFRP1 and 2 which regulate the Wnt pathway [49] was very strongly inhibited by exposure to 0g. This implicates the PTHrP pathway in this mechansensitive pathway in microgravity conditions, and agrees with results of Maycas et al [50] who report the involvement of PTH1R as a mechanosensor in osteocyte survival and Chen et al [51] who suggest PTHrP as a candidate mediator of the anabolic effects of mechanical force on bone.
Intermittent treatment with PTHrP1-36 of TOs not only reversed loss of viability (in short and long 0g exposures) but reversed expression changes in a significant proportion of the genes affected by microgravity. PTHrP1-36 also reversed many, but a lesser number, of the changes in gene expression modified by Pthrp ablation. In that respect, it must be noted that PTHrP1-36 will impact genes controlled by the N-terminus of PTHrP, and likely not genes controlled by the protein’s mid-region and C-terminus. Furthermore, reversal responses are likely to be dependent on PTHrP1-36 concentration, and a dose-response curve would need to be examined for its influence on genes not affected here which might be modified by higher concentrations of PTHrP1-36.
Of particular interest are the genes whose expression change in 0g was reversed by intermittent PTHrP1-36 treatment as their activity is clearly linked to the maintenance of viability obtained in wild-type TOs treated during long periods at 0g. The 74 genes (24 + 50 from central intersects in Fig 5C and 5D, common to ablation and 0g and reversed by PTHrP1-36) belong to similar clustering categories as above. Among the reversals caused by treatment, some strong candidates for microgravity-induced osteoporotic involvement appear. Two genes (Mrpplf3 and Plf2) that belong to the prolactin family were among the most highly-upregulated. Prolactins are hormones secreted by the pituitary gland and some extra-pituitary tissues. Mitogen-regulated protein/proliferin (MRP/PLF) proteins are members of the prolactin gene family involved in developmental processes. Prolactins have pleiotrophic actions in lactation and many other biological functions, and their expression is regulated by PTHrP [52, 53]. Exposure of pregnant rats to prolactins causes reduced ossification in pups [54], and abnormally high prolactin expression encountered in human conditions such as hyperprolactinemic hypogonadism, extended lactation, or as a consequence of the use of some anti-psychotic drugs, is associated with acute skeletal loss [55–57]. Our results indicate that exposure to 0g strikingly (by 12-fold) upregulates expression of prolactin genes Mrpplf3 and Plf2, with highly-efficient reversal by PTHrP1-36. Our observations agree with reports that Spaceflight increases prolactin levels in rats [58], and that exposure of healthy male volunteers to head-down bed rest causes significant elevation of prolactin levels [59]. Our results demonstrate expression of prolactin genes in osteoblastic cells and implicate their products in a major bone loss event that is PTHrP-controlled and unrelated to lactation.
The most highly-downregulated gene by both 0g and Pthrp ablation with reversal by PTHrP1-36 is Rbm3 which encodes RNA-binding motif protein 3, an inhibitor of apoptosis [60, 61]. Its expression is upregulated by PTHrP1-36 which prevents TO death, suggesting Rbm3 involvement in loss of viability and cell death observed at 0g and after ablation. Several genes encoding proteins involved in bone growth, mineralization, BMP and extracellular matrix metabolism are significantly upregulated (Tnfrsf11b, Bdnf, Timp3, Cryab, Grem1, Inhba, Mustn1, Scx) or downregulated (Ptn, Itm2a, Lum, Dcn, Mfap2, Ctsc, Col3a1, Nid2) by both 0g and Pthrp ablation. Of interest among others, the highly-downregulated Ptn gene encodes pleitrophin, an osteoblast-secreted matrix-associated heparin-binding protein which enhances osteogenic differentiation of bone marrow cells [62]. Tnfrsf11b which encodes osteoprotegerin, the decoy to the receptor activator of nuclear factor kappa-B ligand (RANKL), regulates bone mass by inhibiting osteoclast differentiation and activation. It was found in our study to be strongly upregulated by both microgravity and Pthrp ablation in trabecular osteoblasts but downregulated by 0g in calvarial osteoblasts [63], suggesting a role in TO and CO different behaviors in microgravity. Also strongly upregulated is Timp3 (encoding tissue inhibitor of metalloprotease 3) which regulates ECM calcium deposition, and whose overexpression prevents osteoblast differentiation [64]. Gremlin overexpression is known to cause osteopenia and conditional deletion increases bone mass [65, 66], in agreement with our observation that 0g and ablation upregulate Grem1. Members of the IGF and chemokine families are downregulated by 0g and Pthrp ablation with reversal by PTHrP1-36. Igf2 increases bone formation and bone apposition rates [67, 68], and Igfbp5 stimulates osteoblast proliferation [69]. Chemokine ligand Cxcl12 expression accompanies osteoblast differentiation [70] and its downregulation suggests loss of osteoblast characteristics. All of the above changes point to rapid and profound modifications in trabecular osteoblast metabolism in 0g conditions.
The fact that bone loss in flight personnel is reversible to a certain extent upon return to Earth [5, 71, 72] indicates a major difference between Space-induced secondary osteoporosis and the primary aging-associated osteoporosis which is largely irreversible in absence of pharmacologic intervention. It is also interesting to note that in genome-wide association studies, genes associated with aging-related BMD loss do not overlap with those highlighted in our study except for PTHrP [73], osteoprotegerin [74], and Ptn [75, 76] suggesting mechanistic differences in bone loss control between these two types of osteoporosis.
Conclusions
In this study, we present in vitro evidence that naturally-expressed PTHrP in TOs has an important anti-apoptotic function in weightless conditions. Our results confirm a survival role for PTHrP in microgravity-exposed osteoblasts, and identify molecules of interest for further investigation in order to find efficient targets for prevention of bone loss due to skeletal unloading. In particular, the involvement of members of the prolactin family suggests a novel mechanistic hypothesis for transmission of the weightlessness stimulus. The role of PTHrP in maintaining low levels of prolactins may be crucial to prevent microgravity-induced osteoporosis and, by extension, disuse osteoporosis.
Supporting Information
The 4 genes downregulated by both 0g and PTHrP1-36 treatment are Fos, Zfp36, Pvrlz and JunB.
(TIF)
Pre-programmed valves allowed 5 ml (or less) volumes to be fed through the bioreactors. Each PTHrP1-36 treatment was preceded by a 1-min flush, and the 2-h treatment was followed by 2 growth medium rinses and an overnight growth medium feed.
(PDF)
(A) Genes (total 59) upregulated by 0g (6 days). Fold change > 2.0, p < 0.05. (B) Genes (total 129) downregulated by 0g (6 days). Fold change < 0.5, p value< 0.05. All probes: Mus musculus.
(PDF)
(A) Genes (total 270) upregulated by Pthrp ablation at 1 g. Fold change > 2.0, p < 0.05. (B) Genes (total 493) downregulated by Pthrp ablation at 1g. Fold change < 0.5, p value< 0.05. All probes: Mus musculus.
(PDF)
(A) Genes (total 102) upregulated by intermittent PTHrP1-36 treatment. Fold change > 1.5, p < 0.05. (B) Genes (total 61) downregulated by treatment. Fold changes < 0.66, p < 0.05.
(PDF)
Acknowledgments
This work was completed through the Canadian Space Agency (CSA) E/Osteo program contract # 9F-007-05-1657 and a grant to Dr. David Goltzman from the Canadian Institutes of Health Research (FRN-86703). We thank Dr Perry Johnson-Green (Senior Program Scientist, Life Sciences, Space Exploration, Canadian Space Agency) and Dr Nicole Buckley (Chief Scientist, Life Sciences and the International Space Station, Canadian Space Agency) for scientific help throughout the project, Engineers Bastien Dufour (Mission Manager) Luc Lefebvre (Head, International Space Station Projects and Payloads Development, Canadian Space Agency) Lowell Misener and Margaret Eberle (CALM Technologies) for excellent technical supervision, and Dr Richard Kremer (Department of Endocrinology, Faculty of Medicine, McGill University) for helpful scientific discussion.
Data Availability
Microarray data was deposited in the NCBI GEO database and is available under accession number GSE 78980. All other relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was completed through the Canadian Space Agency (CSA) E/Osteo program contract # 9F-007-05-1657. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Goltzman D. Discoveries, drugs and skeletal disorders. Nat Rev Drug Discov. 2002;1(10):784–96. [DOI] [PubMed] [Google Scholar]
- 2.Nagaraja MP, Risin D. The current state of bone loss research: data from spaceflight and microgravity simulators. Journal of cellular biochemistry. 2013;114(5):1001–8. 10.1002/jcb.24454 [DOI] [PubMed] [Google Scholar]
- 3.Carmeliet G, Vico L, Bouillon R. Space flight: a challenge for notmal bone homeostasis. Critical Reviews in Eukaryotic Gene Expression. 2001;11(1–3):131–44. [PubMed] [Google Scholar]
- 4.Payne MWC, Williams DR, Trudel G. Space flight rehabilitation. American journal of physical medicine & rehabilitation. 2007;86(7):583–91. [DOI] [PubMed] [Google Scholar]
- 5.Vernikos J, Schneider VS. Space, gravity and the physiology of aging: parallel or convergent disciplines? A mini-review. Gerontology. 2010;56(2):157–66. 10.1159/000252852 [DOI] [PubMed] [Google Scholar]
- 6.Cavanagh PR, Licata AA, Rice AJ. Exercise and pharmacological countermeasures for bone loss during long-duration space flight. Gravitational and Space Biology. 2005;18:39–58. [PubMed] [Google Scholar]
- 7.LeBlanc A, Schneider V, Shackelford L, West S, Oganov V, Bakulin A, et al. Bone mineral and lean tissue loss after long duration spaceflight. Journal of Bone and Mineral Research; American Society of Bone and Mineral Research 18th Annual Meeting1996. p. S323.
- 8.LeBlanc A, Shackelford L, Schneider V. Future Human Bone Research in Space. Bone. 1998;22(5, Supplement 1):113S–6S. [DOI] [PubMed] [Google Scholar]
- 9.Shackelford LC, LeBlanc AD, Driscoll TB, Evans HJ, Rianon NJ, Smith SM, et al. Resistance exercise as a countermeasure to disuse-induced bone loss. Journal of Applied Physiology. 2004;97(1):119–29. [DOI] [PubMed] [Google Scholar]
- 10.Vico L, Collet P, Guignandon A, Lafage-Proust M-Hln, Thomas T, Rehailia M, et al. Effects of long-term microgravity exposure on cancellous and cortical weight-bearing bones of cosmonauts. The Lancet. 2000;355(9215):1607–11. [DOI] [PubMed] [Google Scholar]
- 11.Kaderka J, Young LR, Paloski WH. A critical benefit analysis of artificial gravity as a microgravity countermeasure. Acta Astronautica. 2010;67(9–10):1090–102. [Google Scholar]
- 12.Buckey J. Bone loss: managing calcium and bone loss in space. Space Physiology. 2006;5:3–25. [Google Scholar]
- 13.Lang T, LeBlanc A, Evans H, Lu Y, Genant H, Yu A. Cortical and Trabecular Bone Mineral Loss From the Spine and Hip in Long-Duration Spaceflight. Journal of Bone and Mineral Research. 2004;19(6):1006–12. [DOI] [PubMed] [Google Scholar]
- 14.LeBlanc A, Schneider V, Shackelford L, West S, Oganov V, Bakulin A, et al. Bone mineral and lean tissue loss after long duration space flight. Journal of Musculoskeletal and Neuronal Interactions. 2000;1(2):157–60. [PubMed] [Google Scholar]
- 15.Nabavi N, Khandani A, Camirand A, Harrison RE. Effects of microgravity on osteoclast bone resorption and osteoblast cytoskeletal organization and adhesion. Bone. 2011;49(5):965–74. 10.1016/j.bone.2011.07.036 [DOI] [PubMed] [Google Scholar]
- 16.Moore ST, MacDougall HG. Journey to Mars: Physiological Effects and Operational Consequences of Long-Duration Microgravity Exposure. Journal of Cosmology. 2010;12:3781–93. [Google Scholar]
- 17.LeBlanc A, Driscoll TB, Shackelford L, Evans HJ, Rianon NJ, Smith SM, et al. Alendronate as an effective countermeasure to disuse induced bone loss. Journal of Musculoskeletal and Neuronal Interactions. 2002;2(4):335–43. [PubMed] [Google Scholar]
- 18.Watanabe Y, Ohshima H, Mizuno K, Sekiguchi C, Fukunaga M, Kohri K, et al. Intravenous Pamidronate Prevents Femoral Bone Loss and Renal Stone Formation During 90-Day Bed Rest. Journal of Bone and Mineral Research. 2004;19(11):1771–8. [DOI] [PubMed] [Google Scholar]
- 19.Mashiba T, Turner CH, Hirano T, Forwood MR, Jacob DS, Johnston CC, et al. Effects of high-dose etidronate treatment on microdamage accumulation and biomechanical properties in beagle bone before occurrence of spontaneous fractures. Bone. 2001;29(3):271–8. [DOI] [PubMed] [Google Scholar]
- 20.Lloyd SAJ, Travis ND, Lu T, Bateman TA. Development of a low-dose anti-resorptive drug regimen reveals synergistic suppression of bone formation when coupled with disuse. Journal of Applied Physiology. 2008;104(3):729–38. 10.1152/japplphysiol.00632.2007 [DOI] [PubMed] [Google Scholar]
- 21.Esbrit P, Alcaraz MJ. Current perspectives on parathyroid hormone (PTH) and PTH-related protein (PTHrP) as bone anabolic therapies. Biochemical pharmacology. 2013;85(10):1417–23. 10.1016/j.bcp.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 22.Esbrit P, Herrera S, Portal-Núñez S, Nogués X, Díez-Pérez A. Parathyroid Hormone-Related Protein Analogs as Osteoporosis Therapies. Calcified tissue international. 2015:1–11. [DOI] [PubMed] [Google Scholar]
- 23.McCauley LK, Martin TJ. Twenty-five years of PTHrP progress: From cancer hormone to multifunctional cytokine. Journal of Bone and Mineral Research. 2012;27(6):1231–9. 10.1002/jbmr.1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wysolmerski JJ. Parathyroid Hormone-Related Protein: An Update. Journal of Clinical Endocrinology & Metabolism. 2012;97(9):2947–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amizuka N, Warshawsky H, Henderson JE, Goltzman D, Karaplis AC. Parathyroid hormone-related peptide-depleted mice show abnormal epiphyseal cartilage development and altered endochondral bone formation. The Journal of Cell Biology. 1994;126(6):1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karaplis AC, Luz A, Glowacki J, Bronson RT, Tybulewicz VL, Kronenberg HM, et al. Lethal skeletal dysplasia from targeted disruption of the parathyroid hormone-related peptide gene. Genes & Development. 1994;8(3):277–89. [DOI] [PubMed] [Google Scholar]
- 27.Miao D, He B, Karaplis AC, Goltzman D. Parathyroid hormone is essential for normal fetal bone formation. Journal of Clinical Investigation. 2002;109(9):1173–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amizuka N, Karaplis AC, Henderson JE, Warshawsky H, Lipman ML, Matsuki Y, et al. Haploinsufficiency of parathyroid hormone-related peptide (PTHrP) results in abnormal postnatal bone development. Developmental biology. 1996;175(1):166–76. [DOI] [PubMed] [Google Scholar]
- 29.Miao D, He B, Jiang Y, Kobayashi T, Soroc xE, et al. Osteoblast-derived PTHrP is a potent endogenous bone anabolic agent that modifies the therapeutic efficacy of administered PTH 1–34. The Journal of Clinical Investigation. 2005;115(9):2402–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horwitz MJ, Tedesco MB, Garcia-Ocana A, Sereika SM, Prebehala L, Bisello A, et al. Parathyroid Hormone-Related Protein for the Treatment of Postmenopausal Osteoporosis: Defining the Maximal Tolerable Dose. Journal of Clinical Endocrinology & Metabolism. 2010;95(3):1279–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardwick T, Kalia P, Baawa-Ameyaw J, Lozano D, Sinha J, Di Silvio L. Continued vs Intermittent Cell Exposure to Parathyroid Hormone-Related Peptide (1–36) on Osteogenic and Angiogenic Gene Expression in Human Osteoblasts and Bone Marrow Stromal Cells. Bone & Joint Journal Orthopaedic Proceedings Supplement. 2013;95(SUPP 13):24-. [Google Scholar]
- 32.De Gortázar A, Alonso V, Alvarez-Arroyo M, Esbrit P. Transient exposure to PTHrP (107–139) exerts anabolic effects through vascular endothelial growth factor receptor 2 in human osteoblastic cells in vitro. Calcified tissue international. 2006;79(5):360–9. [DOI] [PubMed] [Google Scholar]
- 33.Torday JS. Parathyroid hormone-related protein is a gravisensor in lung and bone cell biology. Advances in Space Research. 2003;32(8):1569–76. 10.1016/S0273-1177(03)90397-8 [DOI] [PubMed] [Google Scholar]
- 34.Maniatopoulos C, Sodek J, Melcher A. Bone formation in vitro by stromal cells obtained from bone marrow of young adult rats. Cell and tissue research. 1988;254(2):317–30. [DOI] [PubMed] [Google Scholar]
- 35.Zayzafoon M, Gathings WE, McDonald JM. Modeled Microgravity Inhibits Osteogenic Differentiation of Human Mesenchymal Stem Cells and Increases Adipogenesis. Endocrinology. 2004;145(5):2421–32. [DOI] [PubMed] [Google Scholar]
- 36.Horwitz MJ, Tedesco MB, Sereika SM, Syed MA, Garcia-Ocana A, Bisello A, et al. Continuous PTH and PTHrP infusion causes suppression of bone formation and discordant effects on 1, 25 (OH) 2Vitamin D. Journal of Bone and Mineral Research. 2005;20(10):1792–803. [DOI] [PubMed] [Google Scholar]
- 37.Blaise SA, Nédélec E, Schroeder H, Alberto J-M, Bossenmeyer-Pourié C, Guéant J-L, et al. Gestational Vitamin B Deficiency Leads to Homocysteine-Associated Brain Apoptosis and Alters Neurobehavioral Development in Rats. The American Journal of Pathology. 2007;170(2):667–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frisch S, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. The Journal of Cell Biology. 1994;124(4):619–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ontiveros C, McCabe LR. Simulated microgravity suppresses osteoblast phenotype, Runx2 levels and AP-1 transactivation. Journal of Cellular Biochemistry. 2003;88(3):427–37. [DOI] [PubMed] [Google Scholar]
- 40.Sarkar D, Nagaya T, Koga K, Nomura Y, Gruener R, Seo H. Culture in Vector-Averaged Gravity Under Clinostat Rotation Results in Apoptosis of Osteoblastic ROS 17/2.8 Cells. Journal of Bone and Mineral Research. 2000;15(3):489–98. [DOI] [PubMed] [Google Scholar]
- 41.Blazejczyk M, Miron M, Nadon R. FlexArray: a statistical data analysis software for gene expression microarrays. Montréal, QC2007. Available from: http://genomequebec.mcgill.ca/FlexArray.
- 42.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols. 2008;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 43.Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, et al. Technical brief funrich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–601. [DOI] [PubMed] [Google Scholar]
- 44.Unsworth BR, Lelkes PI. Growing tissues in microgravity. Nature Medicine. 1998;4(8):901–7. [DOI] [PubMed] [Google Scholar]
- 45.LeBlanc A, Spector E, Evans H, Sibonga J. Skeletal responses to space flight and the bed rest analog: a review. Journal of Musculoskeletal and Neuronal Interactions. 2007;7(1):33 [PubMed] [Google Scholar]
- 46.Robling AG, Castillo AB, Turner CH. Biomechanical and molecular regulation of bone remodeling. Annu Rev Biomed Eng. 2006;8:455–98. [DOI] [PubMed] [Google Scholar]
- 47.Pardo SJ, Patel MJ, Sykes MC, Platt MO, Boyd NL, Sorescu GP, et al. Simulated microgravity using the Random Positioning Machine inhibits differentiation and alters gene expression profiles of 2T3 preosteoblasts. American Journal of Physiology—Cell Physiology. 2005;288(6):C1211–C21. [DOI] [PubMed] [Google Scholar]
- 48.Robinson JA, Chatterjee-Kishore M, Yaworsky PJ, Cullen DM, Zhao W, Li C, et al. Wnt/β-catenin signaling is a normal physiological response to mechanical loading in bone. Journal of Biological Chemistry. 2006;281(42):31720–8. [DOI] [PubMed] [Google Scholar]
- 49.Rolfe RA, Nowlan NC, Kenny EM, Cormican P, Morris DW, Prendergast PJ, et al. Identification of mechanosensitive genes during skeletal development: alteration of genes associated with cytoskeletal rearrangement and cell signalling pathways. BMC genomics. 2014;15(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maycas M, Ardura JA, de Castro LF, Bravo B, Gortázar AR, Esbrit P. Role of the parathyroid hormone type 1 receptor (PTH1R) as a mechanosensor in osteocyte survival. Journal of Bone and Mineral Research. 2015;30(7):1231–44. 10.1002/jbmr.2439 [DOI] [PubMed] [Google Scholar]
- 51.Chen X, Macica CM, Ng KW, Broadus AE. Stretch-Induced PTH-Related Protein Gene Expression in Osteoblasts. Journal of bone and mineral research. 2005;20(8):1454–61. [DOI] [PubMed] [Google Scholar]
- 52.Sowers M. Pregnancy and lactation as risk factors for subsequent bone loss and osteoporosis. Journal of Bone and Mineral Research. 1996;11(8):1052–60. [DOI] [PubMed] [Google Scholar]
- 53.VanHouten JN, Wysolmerski JJ. Low estrogen and high parathyroid hormone-related peptide levels contribute to accelerated bone resorption and bone loss in lactating mice. Endocrinology. 2003;144(12):5521–9. [DOI] [PubMed] [Google Scholar]
- 54.Coss D, Yang L, Kuo CB, Xu X, Luben RA, Walker AM. Effects of prolactin on osteoblast alkaline phosphatase and bone formation in the developing rat. American Journal of Physiology-Endocrinology And Metabolism. 2000;279(6):E1216–E25. [DOI] [PubMed] [Google Scholar]
- 55.Greenspan SL, Neer RM, Ridgway EC, Klibanski A. Osteoporosis in men with hyperprolactinemic hypogonadism. Annals of internal medicine. 1986;104(6):777–82. [DOI] [PubMed] [Google Scholar]
- 56.O’sullivan S, Grey A, Singh R, Reid I. Bisphosphonates in pregnancy and lactation-associated osteoporosis. Osteoporosis international. 2006;17(7):1008–12. [DOI] [PubMed] [Google Scholar]
- 57.O'Keane V, Meaney AM. Antipsychotic drugs: a new risk factor for osteoporosis in young women with schizophrenia? Journal of clinical psychopharmacology. 2005;25(1):26–31. [DOI] [PubMed] [Google Scholar]
- 58.Macho L, Jezova D, Jurcovicova J, Kvetnanský R, Vigas M, Serova L. Effect of space flight on the development of endocrine functions in rats. Endocrine regulations. 1993;27(1):17–22. [PubMed] [Google Scholar]
- 59.Cagnati P, Puppo F. Multiparametricendocrine evaluation of healthy subjects undergoing prolonged head down tilt bed rest anti-gravitary posture. Stress. 2013;1:3. [Google Scholar]
- 60.Kita H, Carmichael J, Swartz J, Muro S, Wyttenbach A, Matsubara K, et al. Modulation of polyglutamine-induced cell death by genes identified by expression profiling. Human Molecular Genetics. 2002;11(19):2279–87. [DOI] [PubMed] [Google Scholar]
- 61.Sutherland LC, Rintala-Maki ND, White RD, Morin CD. RNA binding motif (RBM) proteins: A novel family of apoptosis modulators? Journal of Cellular Biochemistry. 2005;94(1):5–24. [DOI] [PubMed] [Google Scholar]
- 62.Tare RS, Oreffo ROC, Clarke NMP, Roach HI. Pleiotrophin/Osteoblast-Stimulating Factor 1: Dissecting Its Diverse Functions in Bone Formation. Journal of Bone and Mineral Research. 2002;17(11):2009–20. [DOI] [PubMed] [Google Scholar]
- 63.Rucci N, Rufo A, Alamanou M, Teti A. Modeled microgravity stimulates osteoclastogenesis and bone resorption by increasing osteoblast RANKL/OPG ratio. Journal of Cellular Biochemistry. 2007;100(2):464–73. [DOI] [PubMed] [Google Scholar]
- 64.Suzuki H, Nezaki Y, Kuno E, Sugiyama I, Mizutani A, Tsukagoshi N. Functional roles of the tissue inhibitor of metalloproteinase 3 (TIMP-3) during ascorbate-induced differentiation of osteoblastic MC3T3-E1 cells. Bioscience, biotechnology, and biochemistry. 2003;67(8):1737–43. [DOI] [PubMed] [Google Scholar]
- 65.Gazzerro E, Pereira RC, Jorgetti V, Olson S, Economides AN, Canalis E. Skeletal Overexpression of Gremlin Impairs Bone Formation and Causes Osteopenia. Endocrinology. 2005;146(2):655–65. [DOI] [PubMed] [Google Scholar]
- 66.Gazzerro E, Smerdel-Ramoya A, Zanotti S, Stadmeyer L, Durant D, Economides AN, et al. Conditional Deletion of Gremlin Causes a Transient Increase in Bone Formation and Bone Mass. Journal of Biological Chemistry. 2007;282(43):31549–57. [DOI] [PubMed] [Google Scholar]
- 67.Martel-Pelletier J, Di Battista JA, Lajeunesse D, Pelletier JP. IGF/IGFBP axis in cartilage and bone in osteoarthritis pathogenesis. Inflammation Research. 1998;47(3):90–100. [DOI] [PubMed] [Google Scholar]
- 68.Nannuru K, Singh R. Tumor-Stromal Interactions in Bone Metastasis. Current Osteoporosis Reports. 2010;8(2):105–13. 10.1007/s11914-010-0011-6 [DOI] [PubMed] [Google Scholar]
- 69.Amaar YG, Thompson GR, Linkhart TA, Chen S-T, Baylink DJ, Mohan S. Insulin-like Growth Factor-binding Protein 5 (IGFBP-5) Interacts with a Four and a Half LIM Protein 2 (FHL2). Journal of Biological Chemistry. 2002;277(14):12053–60. [DOI] [PubMed] [Google Scholar]
- 70.Mishima S, Nagai A, Abdullah S, Matsuda C, Taketani T, Kumakura S, et al. Effective ex vivo expansion of hematopoietic stem cells using osteoblast-differentiated mesenchymal stem cells is CXCL12 dependent. European Journal of Haematology. 2010;84(6):538–46. 10.1111/j.1600-0609.2010.01419.x [DOI] [PubMed] [Google Scholar]
- 71.Sibonga J, Evans H, Sung H, Spector E, Lang T, Oganov V, et al. Recovery of spaceflight-induced bone loss: bone mineral density after long-duration missions as fitted with an exponential function. Bone. 2007;41(6):973–8. [DOI] [PubMed] [Google Scholar]
- 72.Tilton FE, Degioanni J, Schneider V. Long-term follow-up of Skylab bone demineralization. Aviation, space, and environmental medicine. 1980;51(11):1209 [PubMed] [Google Scholar]
- 73.Estrada K, Styrkarsdottir U, Evangelou E, Hsu Y-H, Duncan EL, Ntzani EE, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nature genetics. 2012;44(5):491–501. 10.1038/ng.2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Y-J, Zhang L, Papasian CJ, Deng H-W. Genome-wide association studies for osteoporosis: a 2013 update. Journal of bone metabolism. 2014;21(2):99–116. 10.11005/jbm.2014.21.2.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benisch P, Schilling T, Klein-Hitpass L, Frey SP, Seefried L, Raaijmakers N, et al. The Transcriptional Profile of Mesenchymal Stem Cell Populations in Primary Osteoporosis Is Distinct and Shows Overexpression of Osteogenic Inhibitors. PLoS ONE. 2012;7(9):e45142 10.1371/journal.pone.0045142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Trost Z, Trebse R, Prezelj J, Komadina R, Logar DB, Marc J. A microarray based identification of osteoporosis-related genes in primary culture of human osteoblasts. Bone. 2010;46(1):72–80. 10.1016/j.bone.2009.09.015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The 4 genes downregulated by both 0g and PTHrP1-36 treatment are Fos, Zfp36, Pvrlz and JunB.
(TIF)
Pre-programmed valves allowed 5 ml (or less) volumes to be fed through the bioreactors. Each PTHrP1-36 treatment was preceded by a 1-min flush, and the 2-h treatment was followed by 2 growth medium rinses and an overnight growth medium feed.
(PDF)
(A) Genes (total 59) upregulated by 0g (6 days). Fold change > 2.0, p < 0.05. (B) Genes (total 129) downregulated by 0g (6 days). Fold change < 0.5, p value< 0.05. All probes: Mus musculus.
(PDF)
(A) Genes (total 270) upregulated by Pthrp ablation at 1 g. Fold change > 2.0, p < 0.05. (B) Genes (total 493) downregulated by Pthrp ablation at 1g. Fold change < 0.5, p value< 0.05. All probes: Mus musculus.
(PDF)
(A) Genes (total 102) upregulated by intermittent PTHrP1-36 treatment. Fold change > 1.5, p < 0.05. (B) Genes (total 61) downregulated by treatment. Fold changes < 0.66, p < 0.05.
(PDF)
Data Availability Statement
Microarray data was deposited in the NCBI GEO database and is available under accession number GSE 78980. All other relevant data are within the paper and its Supporting Information files.