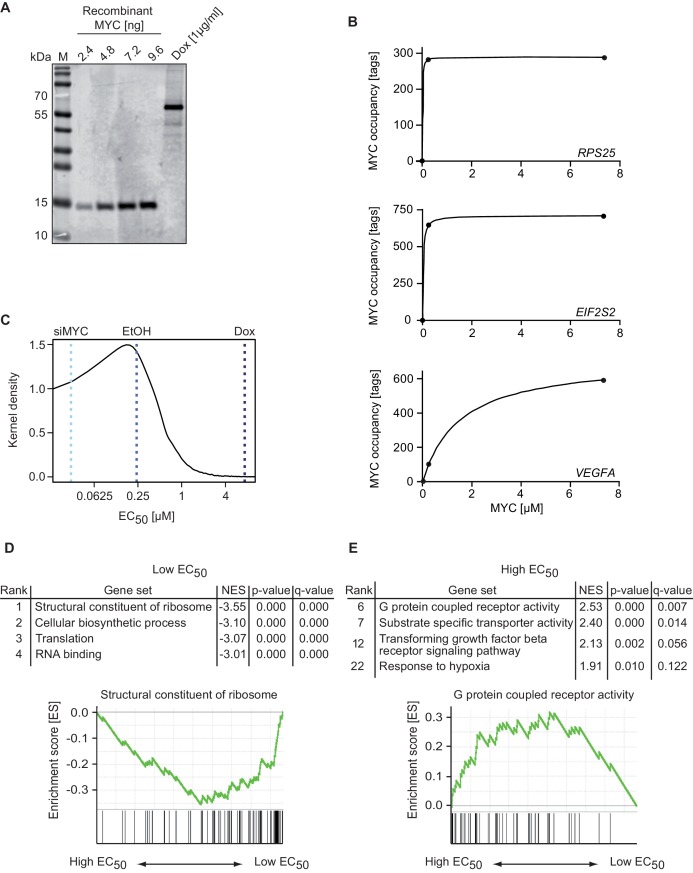
Figure 2. MYC binds with a wide range of affinity (EC50 values) to target genes.
(A) Immunoblot of MYC in U2OSTet-On cells treated with 1 µg/ml doxycycline and a recombinant MYC protein fragment, which was used for absolute quantification of cellular MYC levels (M: marker). Absolute quantification is based on biological triplicates shown in Figure 2—figure supplement 1D–F. (B) Diagram of MYC occupancy calculated in ChIP-sequencing experiments of EtOH- or doxycycline-treated U2OSTet-On cells (y-axis) versus the cellular MYC concentration (x-axis). The line was fitted using a Michaelis-Menten model and non-linear regression. (C) Density plot of the distribution of the EC50 values calculated for all MYC-bound genes. Dashed lines indicate the cellular MYC concentration in uninduced (EtOH, blue line), 1 µg/ml doxycycline treated (Dox, dark blue line), or MYC-depleted (siMYC, light blue) U2OSTet-On cells. (D, E) GSE analysis using the MSigDB C5 (GO gene sets) collection, of genes sorted according to EC50 values. Enrichment plots of two gene sets enriched in the GSE analysis are shown as examples. NES: normalized enrichment score. Both, gene sets with very low (D) and very high (E) EC50 values are shown.