Abstract
Bacteriophages represent rapid, readily targeted, and easily produced molecular probes for the detection of bacterial pathogens. Molecular biology techniques have allowed researchers to make significant advances in the bioengineering of bacteriophage to further improve speed and sensitivity of detection. Despite their host-specificity, bacteriophages have not been meaningfully leveraged in multiplex detection of bacterial pathogens. We propose a proof-of-principal phage-based scheme to enable multiplex detection. Our scheme involves bioengineering bacteriophage to carry a gene for a specific protease, which is expressed during infection of the target cell. Upon lysis, the protease is released to cleave a reporter peptide, and the signal detected. Here we demonstrate the successful: i) modification of T7 bacteriophage to carry TEV protease; ii) expression of TEV protease by E. coli following infection by our modified T7, an average of 2000 units of protease per phage are produced during infection; and iii) proof-of-principle detection of E. coli in 3 hours after a primary enrichment via TEV protease activity using a fluorescent peptide and using a designed target peptide for MALDI-TOF MS analysis. This proof-of-principle can be translated to other phage-protease-peptide combinations to enable multiplex bacterial detection, and readily adopted on multiple platforms, like MALDI-TOF MS or fluorescent readers, commonly found in labs.
Keywords: Biosensors, Bacteriophage, Protease, Multiplex
Introduction
There has been a renewed interest in the application of bacteriophages (phages) for the detection and control of bacterial pathogens. In the area of diagnostics, phages are attractive as detection elements due to their relative ease of production, host specificity, and potential for rapid signal amplification (Singh, et al., 2012). Furthermore, in comparison to immunological (Easter, 2003) and PCR-based (Wolffs, et al., 2005) detection methods, phage-based detection schemes can distinguish between viable and non-viable cells, as viable cells are required for phage proliferation (Denes and Wiedmann, 2014). This potentially means that phage-based diagnostics could have lower incidences of false positives. Several phage-based diagnostics have already entered the commercial market. Biomérieux (St. Louis, MO) has a product line called VIDAS® UP which leverages phage components for the detection of Salmonella, Listeria, and E. coli O157. Sample6 (Cambridge, MA) uses a phage-delivered luciferase reporter for the detection of environmental Listeria.
Individual testing for multiple pathogens requires large sample sizes, numerous selective media and reagents, and a significant amount of time and technician labor. Multiplex methods, which use a single reaction sample to test for multiple pathogens, are attractive due to their reduced complexity and time to result. The majority of multiplex systems leverage DNA (Settanni and Corsetti, 2007) or immunological (Dunbar, et al., 2003) based methods, and thus face the potential false positive issues due to the presence of dead cells. There has been limited research in applying phage in multiplex detection schemes. Work by Rees and Voorhees (2005) coupled phage amplification with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis (MALDI-TOF MS) for the simultaneous detection of E. coli and Salmonella, but the majority of bioengineered phage-based detection schemes have leveraged enzymatic reporters, like luciferase, that do not lend themselves readily to a single sample multiplex format (Schmelcher and Loessner, 2014, Singh, et al., 2012, Smartt, et al., 2012).
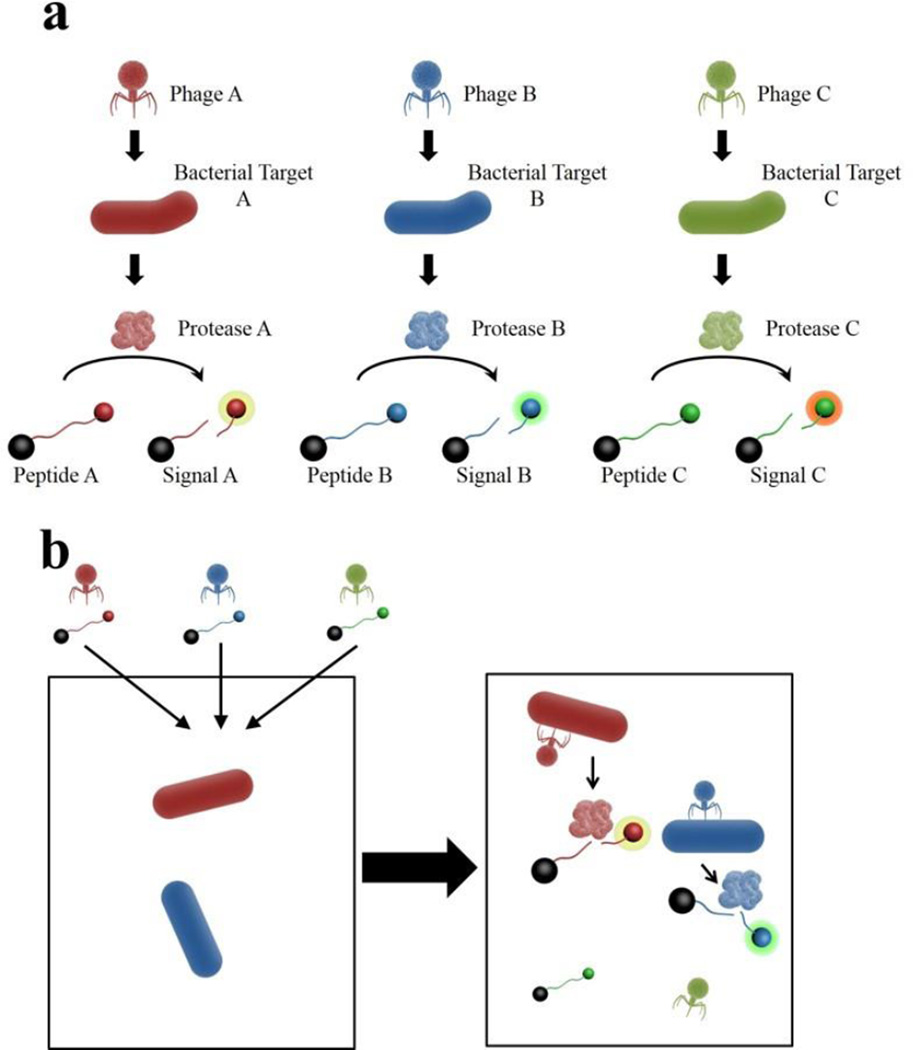
We propose a novel phage-based platform to enable multiplex detection of bacteria using phage bioengineered to carry genes encoding highly specific proteases, whose activity is detected by the cleavage of specific peptides (Fig. 1). The phage provides specificity for a viable bacterial target, and the protease-peptide pair provides further specificity and lays the groundwork for single sample multiplex detection. For our proof-of-principle, we genetically engineered T7, a phage with broad specificity for E. coli (Studier, 1972), to carry a gene for a protease from tobacco etch virus (TEV). TEV protease is a highly specific protease recognizing the amino acid sequence ENLYFQ(G/S), with cleavage between the Q and G or S amino acids (Kapust, et al., 2001). The cleavage event, indicative of the presence of TEV protease produced by a viable E. coli cells following infection with the engineered T7, can be detected using various platforms. Our work demonstrates the successful: i) modification of T7 bacteriophage to carry TEV protease gene; ii) expression of TEV protease by E. coli following T7 infection; and iii) detection of E. coli in sample via TEV protease activity. To show that this scheme was applicable to multiple platforms, we detected protease activity two ways. First, using a common fluorescent plate reader to detect the signal from a cleaved beacon peptide; and second, using MALDI-TOF MS to detect the production of a reporter peptide produced by the cleavage of a designed larger peptide. Both of these platforms are not only sensitive, but have the capability for high-throughput, multiplex screening of samples. We believe this scheme has the potential to be readily converted to other phage-protease-peptide combinations to enable multiplex detection of bacterial pathogens within a sample.
Fig. 1.
Phage-Protease-Peptide Multiplex Detection Scheme. (a) Phage specific to a bacterial target, is bioengineered to carry a gene for a specific protease. Protease acts on a specific peptide resulting in a unique fluorescent signal or peptide product. (b) Phage and corresponding peptides are added to a single sample. Signal is produced only if corresponding bacterial target is present in sample to produce the protease.
Materials and Methods
Bacterial strains, bacteriophage strains, media culture, and enumeration
Both the bacterial strain E. coli BL21 and the bacteriophage T7 (T7Select® 415-1) were purchased from EMD Millipore (Billerica, Massachusetts). All overnight cultures of E. coli BL21 were grown in 150 mL flasks containing 35 mL of Luria Broth (LB), pH 7.5, incubated at 37 °C with shaking at 200 RPM. The overnight cultures were serially diluted and spread plated on LB agar to confirm bacterial concentration for subsequent experiments. The titer of samples containing T7 bacteriophage were determined following the double agar overlay plaque assay on LB agar as commonly described (Kutter and Sulakvelidze, 2005).
Construction and isolation of engineered bacteriophage
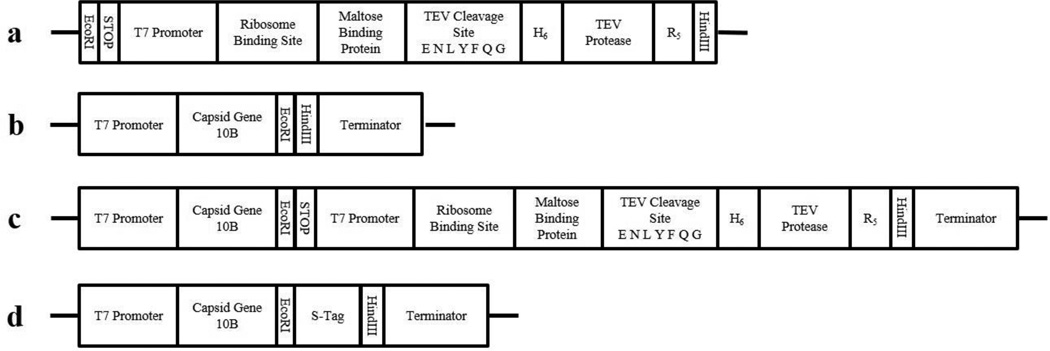
Using standard molecular techniques, we engineered T7 bacteriophage to carry the TEV protease gene for overexpression within E. coli. We designed a gene cassette (Fig. 2) to enable T7-induced expression of TEV protease in E. coli, and had the cassette synthesized within a pUC57 plasmid by GenScript USA Inc. (Piscataway, NJ). The TEV protease cassette was amplified with the iProof high-fidelity PCR kit (Bio-Rad Laboratories, Hercules, CA) using standard M13 forward and reverse primers. The PCR product was purified using QIAquick PCR Purification Kit (Qiagen, Valencia, CA), and then digested with restriction enzymes EcoRI and HindIII, both purchased from New England Biolabs (Ipswich, MA). The digested construct was then inserted into the T7 Select® 415-1 genome using T4 DNA ligase (Promega, Madison, WI) and packaged using the T7 Select® cloning kit (EMD Millipore) to create T7tev (Fig. 2). In parallel, we used the T7 Select® kit’s control DNA insert, which encodes for the S•Tag™, to create T7control (Fig. 2), a control phage which would not induce TEV protease expression. Both T7tev and T7control packaging reactions were propagated following the T7 Select® kit’s protocol and plated. Individual plaques were picked off plates using a sterile loop, dipped into 100 µL of LB, and stored at 4 °C. The isolated plaques were screened with the iProof PCR kit for the appropriate size insert using the T7Select® Up and Down primers included in the cloning kit. Isolated plaques containing the appropriate sized insert were propagated on E. coli BL21, and the subsequent lysates screened to confirm the presence of the insert. These lysates were passed through a 0.22 µm SCFA filter (Corning Life Science, Corning, NY), stored at 4 °C, titered, and used as the stock bacteriophage for subsequent experiments.
Fig. 2.
Diagram of DNA Constructs. (a) Our designed TEV protease gene cassette. (b) Genome of T7 Select® 415-1 indicating 10B capsid protein and insertion site. (c) Genome of T7tev with the insertion of our gene cassette. (d) Genome of T7control with insertion of the S•Tag™ control.
Fluorescent detection and quantification of TEV protease
Fluorescent detection of TEV protease activity was performed using the SensoLyte® 520 TEV Activity Assay Kit (AnaSpec, Inc., San Jose, CA). 50 µL of sample was mixed with 50 µL of substrate in a 96-well plate, incubated at 30 °C, and read every 10 minutes, over 90 minutes, using the Synergy 2 plate reader (BioTek Instruments, Inc., Winooski, VT) with Ex/Em filters 485/528 nm. For quantification of TEV protease in a sample, a standard curve was run using the TEV protease standards provided with the SensoLyte® kit. Our TEV samples were in LB and the provided standards were in the kit’s buffer, which meant the standards and our samples had a different base background noise. To account for this we compared Vmax, calculated using the Gen5 software (Biotek Instruments, Inc., Winooski, VT), of the fluorescence over 90 minutes between our samples and the standards to quantify the amount of TEV protease present.
Characterizing TEV protease production
An overnight culture of E. coli BL21, prepared as previously described, was serially diluted in LB to achieve 109, 108, 107 and 106 CFU/mL. 900 µL of each dilution and a negative control of LB were placed in 15×100 mm test tubes, and mixed with 100 µL of 103 PFU/mL of T7tev in LB. Each combination was performed in triplicate. The samples were incubated at 37 °C with 200 rpm shaking for 3 hours. Samples were then passed through a 0.22 µm SCFA filter (Corning Life Science, Corning, NY), stored at 4 °C, tittered, and TEV protease activity fluorescently determined as previously described. The experiment was repeated with two separate overnight cultures to achieve biological triplicates.
Target peptide design and MALDI-TOF mass spectrometry analysis
We designed two 26 amino acid peptides, TEV-L and REF-L (Table 1), to detect TEV protease activity using MALDI-TOF MS. TEV-L was designed to contain the TEV protease cleavage site, and when cleaved by TEV protease to produce two identical 13 amino acid peptides, TEV-S (Table 1). The REF-L (Table 1) peptide was used as an internal reference. Samples of both TEV-L and REF-L were synthesized by Biopeptide Co., Inc. (San Diegeo, CA), at 98% purity. Samples were reconstituted in sterile water to make a 1 mM stock solution.
Table 1.
Peptides designed for MALDI-TOF detection of TEV protease activity. Bold letters designate the recognition sequence for TEV protease, and the hashtag indicates the cleavage site.
| Name | Amino acid sequence | Residue number | Monoisotopic mass |
|---|---|---|---|
| TEV-L | SHFLKKRENLYFQ#SHFLKKRENLYFQ | 26 | 3399.799 |
| TEV-S | SHFLKKRENLYFQ | 13 | 1708.905 |
| REF-L | GPHFLKKRETLFQGPHFLKKRETLFQ | 26 | 3181.767 |
Reagents for MALDI-TOF MS analysis are as follows: triflouroacetic acid (TFA), α -cyano-hydroxycinnamic acid (α-CHCA), and Tetrahydrofuran (THF). TFA and α-CHCA were purchased from Sigma-Aldrich (St. Louis, MO), and THF was purchased from Fisher Scientific (Pittsburgh, PA). For MALDI-TOF MS analysis, 5 µL of a matrix solution (0.16 M α-CHCA in 69: 30: 1% THF/H2O/TFA) was mixed with 5 µL of a sample. 2 µL of the matrix/sample mixture was then spotted onto a stainless steel target and the solvent was allowed to evaporate at room temperature. Analysis was performed using a Bruker Autoflex III smartbeam time-of-flight mass spectrometer (Bruker Daltonics, Inc., Billerica, MA). All spectra presented were obtained in reflectron mode and represent an average of 5000 shots acquired at 46% laser power.
Detection of E. coli through TEV protease activity using fluorescence and MALDI-TOF MS
Three colonies of E. coli BL21 streaked out onto an LB agar plate were selected, each put into 35 mL of LB in a 150 mL flask, and incubated at 37 °C with shaking at 200 rpm for 16 hours to simulate a primary enrichment. 150 µL of this primary enrichment was added to 35 mL of fresh LB in a 150 mL flask and incubated for 3 hours under the same conditions. Then 10 µL of T7tev phage stock (1.08×1011 PFU/mL) was added and the sample incubated for another two hours under the same conditions. LB inoculated with 10 µL of T7tev stock was used as a control. The resulting lysates were then filtered with a 0.22 µM filter to remove any cellular debris. Each lysate was analyzed in triplicate for TEV protease activity using the SensoLyte® fluorescent assay as previously described. For MALDI-TOF MS analysis 12.5 µL of each lysate was mixed with 12.5 µL of solution containing 10 µM of TEV-L and REF-L peptides in TEV reaction buffer from the SensoLyte® kit. Samples were incubated for 1 hour at 30 °C, heated to 80 °C for 10 minutes to deactivate any TEV present, and then cooled to 4 °C until MALDI-TOF MS analysis.
Results
Constructing a TEV protease producing bacteriophage
To create our proof-of-principle phage-based enzymatic multiplex detection of bacteria, we designed a gene cassette containing a gene for TEV protease (Fig. 2a) for insertion in the T7 genome (Fig. 2b), a phage with broad host range specificity for E. coli (Studier, 1972) We built upon a TEV protease mutant S219V which was shown to be more stable and catalytically efficient than the wild type when expressed in E. coli (Kapust, et al., 2001). The S219V TEV protease gene is linked to a maltose binding protein (MBP) at the N-terminus by a short amino acid sequence containing the TEV cleavage site sequence, ENLYFQG, followed by a 6X His-tag. At the C-terminus there is a 5X arginine tag. The MBP chaperone and ability of the protease to self-cleave from the chaperone was shown to increase solubility and activity of the protease when expressed in E. coli (Kapust and Waugh, 1999) Upstream of the start codon for MBP, we added the T7 promoter sequence and ribosome binding site sequence from the pET-3a plasmid (EMD Millipore), to allow for T7-mediated overexpression of the protease. The T7 Select® cloning kits are designed to enable phage-display, and the inserted peptide sequence is fused to 10B capsid protein of T7 (Novagen®, 2001) We wanted the protease to be free in solution, not attached to the phage capsid, so we followed the example of Lu and Collins (2007), and added a stop codon in all three reading frames upstream of our T7 promoter. Lastly, we added the restriction site for EcoRI at the 5’ terminus of our cassette and the HindIII site at the 3’ terminus. We removed a HindIII site found within the TEV protease gene. Our gene cassette was synthesized by Genscript, Inc., and the codon sequences optimized using their proprietary OptimumGene™ algorithm for expression within E. coli. The full sequence of our cassette can be found in Figure S1 or via Genbank, accession number KT183030. We cloned our cassette into the T7 Select®415-1 genome (Fig. 2c) and as a control cloned the T7 Select® kit’s control insert, encoding the S•Tag™ (Fig. 2d).
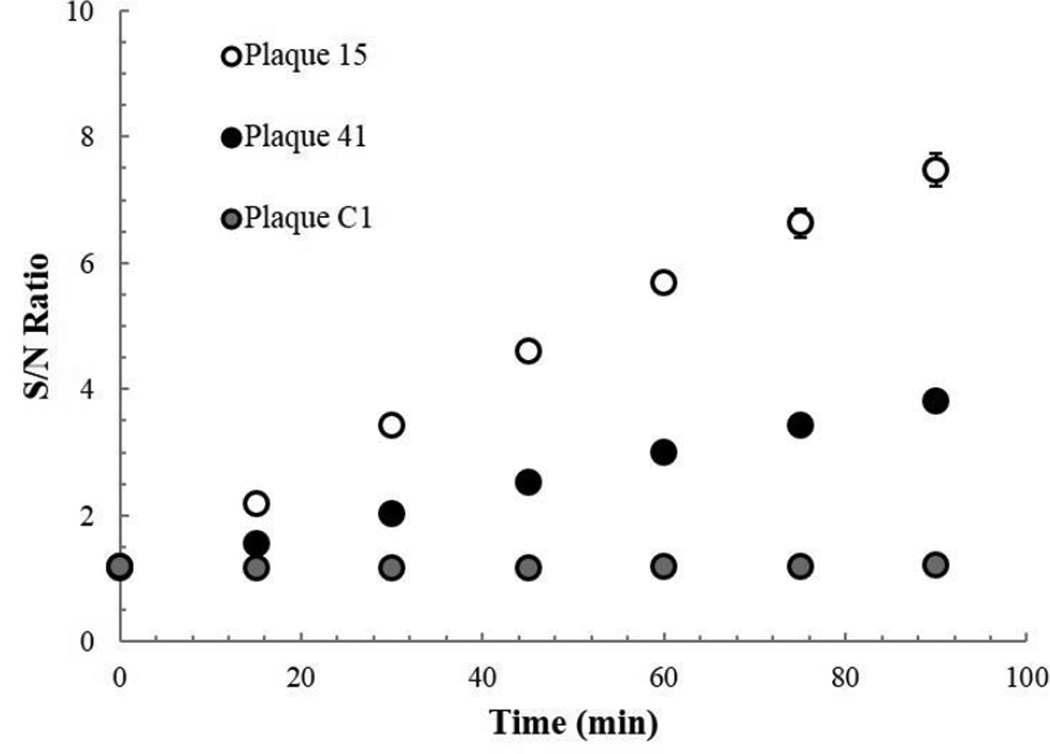
After cloning both inserts, we used PCR to screen plaques for phage that contained either our gene cassette or the control insert. We choose two plaques, 15 and 41, carrying our gene cassette and one plaque carrying the control insert, to look for TEV protease activity. We propagated the plaques in a broth of E. coli, and after incubation filtered the lysates to remove any remaining cells. We took samples of each of the three lysates and tested for TEV protease activity using the SensoLyte® fluorescent TEV assay. We monitored fluorescence over 90 minutes, and divided the signal by the signal of a known negative control, a sample containing only LB and the fluorescent peptide substrate (Fig. 3). After 90 minutes, we could clearly observe an increase in fluorescence vs. the negative control in both phage samples carrying the TEV gene cassette and did not observe an increase in fluorescence in the phage sample carrying the control insert. We thus show that not only have we successfully introduced our TEV protease gene cassette in T7, but that the protease is produced in an active form by E. coli following infection by our T7tev. For all further experiments, we used T7tev phage derived from our plaque 15 lysate.
Fig. 3.
Confirming TEV protease product. Ratio of fluorescent signal in lysate samples to that in an LB negative control over time. Plaque 15 and 41 are phage confirmed by PCR to carry the TEV protease gene cassette, Plaque C1 carries the S•Tag™ insert.
Characterizing TEV protease production
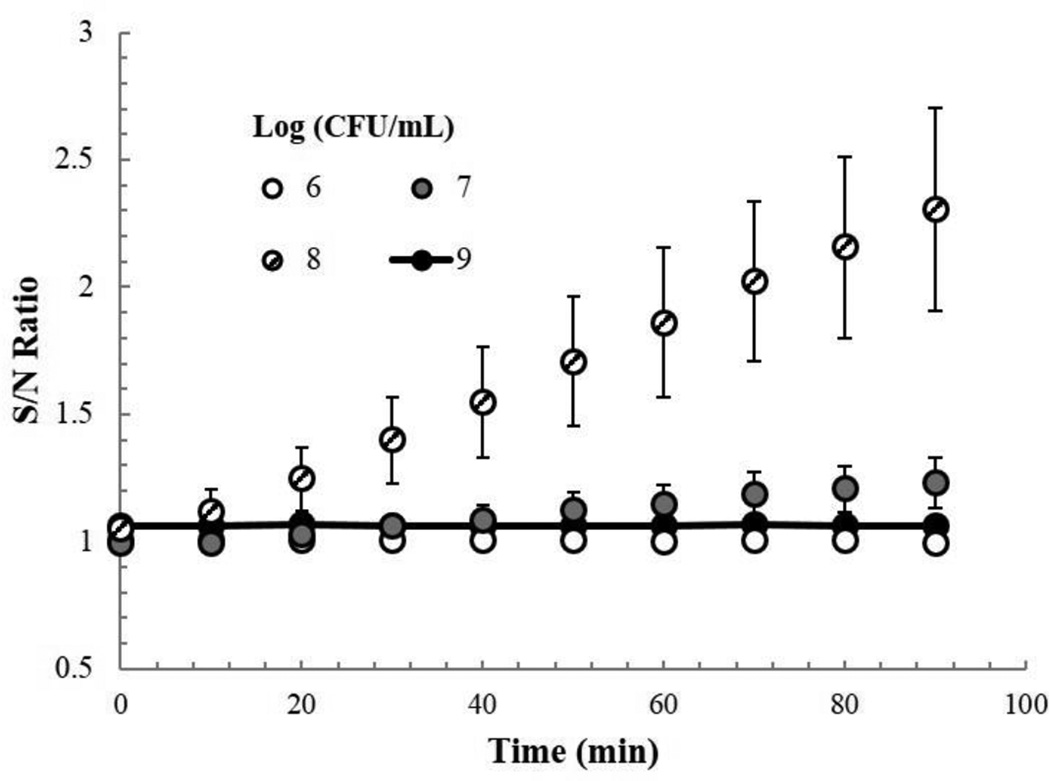
In order to understand how much TEV protease was being produced and how many E. coli cells were needed to obtain a signal, we incubated several concentrations of E. coli BLS21, 109 to 106 cfu/mL, and a negative control of LB, with 102 PFU of T7tev, for three hours at 37 °C. We then filtered the samples and monitored fluorescence over 90 minutes, and normalized the signal to the negative control (Fig. 4). We observed an increase in fluorescence vs. the negative control after 90 minutes in samples with a starting CFU/mL of 107 and 108, but observed no increase in the others.
Fig. 4.
TEV protease activity from varying starting cell concentrations. Ratio of fluorescent signal in T7tev lysates (with starting cell concentrations of 106, 107, 108, and 109 CFU/mL) to that of a control with no cells, over time.
Using the TEV protease standards provided with SensoLyte® assay we determined the amount of protease produced in the 107 and 108 CFU/mL samples to be 400 ± 300 ng of TEV/mL and 1500 ± 500 ng of TEV/mL, respectively (data not shown). TEV protease is a 25 kDa protein (Kapust, et al., 2001), and using its molecular weight we can calculate the number of TEV protease molecules present per mL in these two samples to be 1013.0 and 1013.6, respectively. The final phage titer of the 107 and 108 samples was determined to be 109.86 ± .06 and 1010.17 ± .13 PFU/mL, respectively. Dividing the average number of molecules of TEV protease produced by the average amount of phage produced, results in approximate values of 1380 and 2754 in the 107 and 108 samples, indicating that roughly 2000 molecules of TEV protease are produced for every phage produced by an infected E. coli BL21 cell.
Designing Peptides for MALDI-TOF
MALDI-TOF mass spectrometry can detect small peptides with a relative molecular mass of ~1000–3000 at sample concentrations as low as 1 nM, though as the complexity of the sample matrix increases the sensitivity typically decreases to > 1 µM of target peptide (Hortin, 2006). We designed two peptides, TEV-L and REF-L (Table 1), for detection by MALDI-TOF MS. TEV-L was designed so that when cleaved by TEV protease it would produce two identical peptides, TEV-S (Table 1). This provided us with two advantages: first, a doubling in signal would be expected as for every cleavage of TEV-L we would get two TEV-S peptides; and second, increased specificity would be present since cleavage of TEV-L by a protease other than TEV would likely not result in the production of two identical smaller peptides.
Detection of E. coli through TEV protease activity using fluorescence and MALDI-TOF
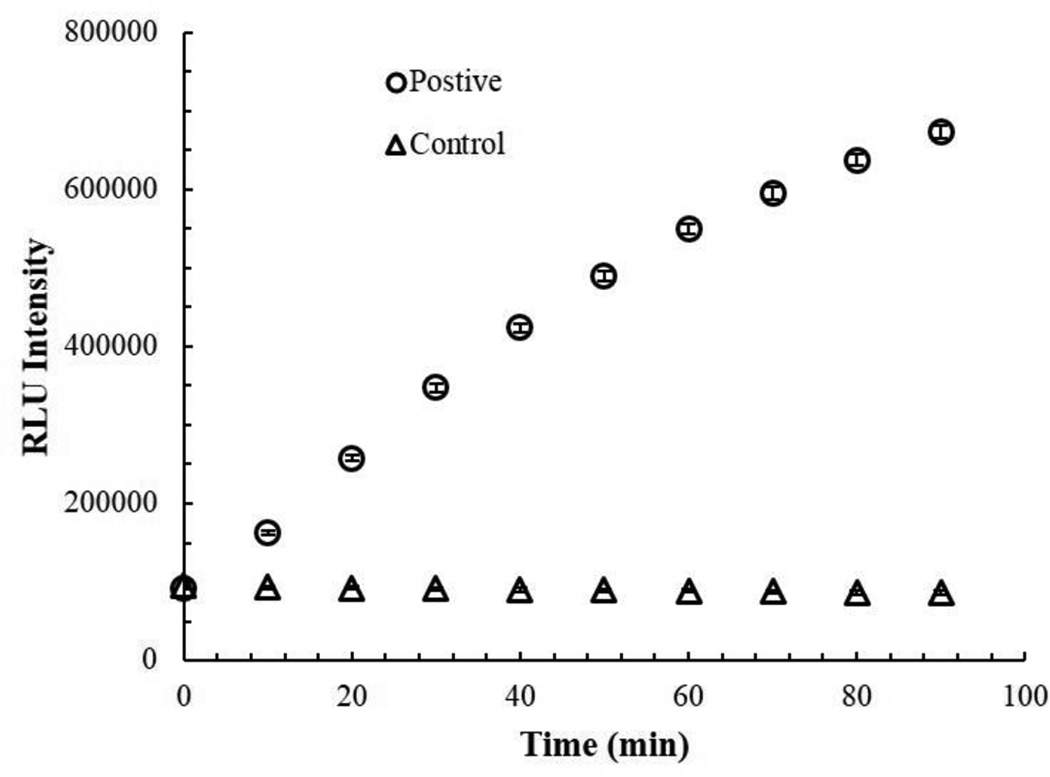
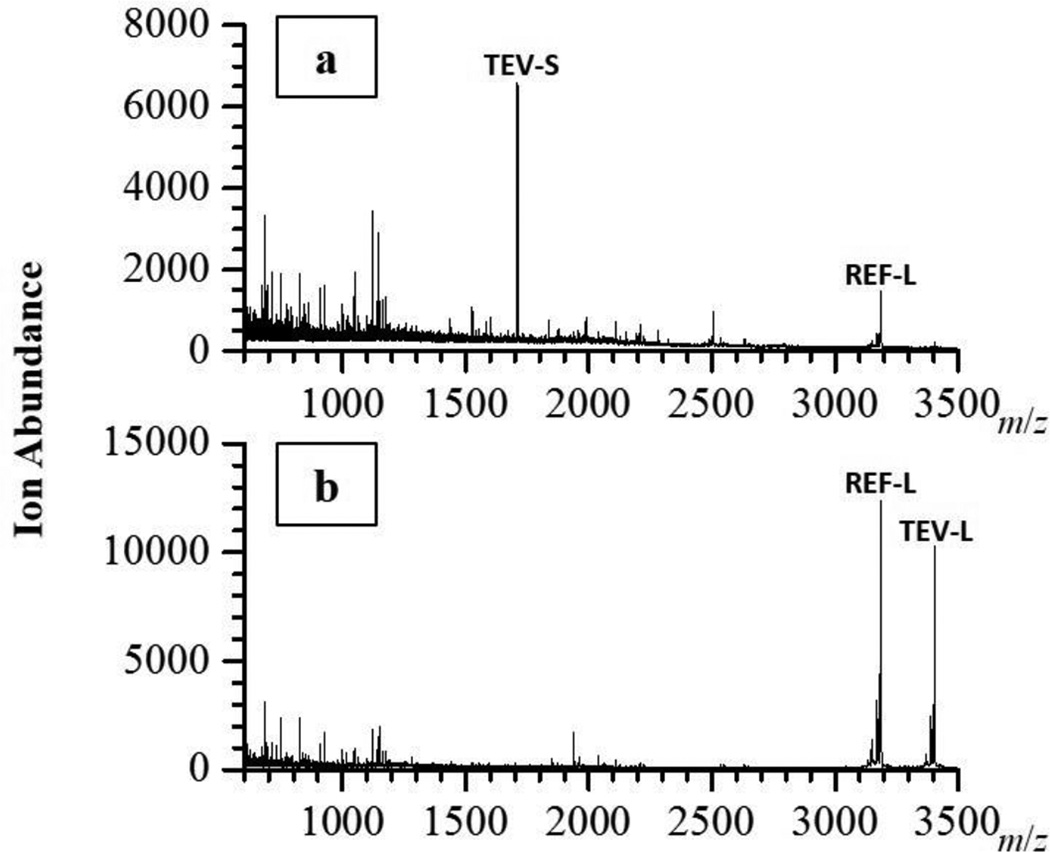
For a proof-of-principle, we used a culture of E. coli BL21grown from a single colony for 16 hours in LB as a stand in for a non-specific primary enrichment typically used for the detection of food borne pathogens, like E. coli O157:H7 and Salmonella spp. (FDA, 1998). We took a portion of this primary enrichment and inoculated fresh LB, incubated for 3 hours, then added our T7tev, and incubated another 2 hours. The samples were filtered to remove any remaining bacterial cells, and then analyzed for TEV protease activity using the fluorescent kit (Fig. 5) and MALDI-TOF MS (Fig. 6). Both methods were able to detect TEV protease activity in the samples containing E. coli. In the majority of the E. coli positive samples, MALDI-TOF MS analysis could not detect any remaining TEV-L, whereas a TEV-S peak was always observed.
Fig. 5.
Fluorescent detection of E. coli via T7-induced TEV protease activity. Fluorescent signal intensity of E.coli positive samples and a negative (Control) over time.
Fig. 6.
MALDI-TOF detection of E. coli via T7-induced TEV protease activity. Spectra of representative E.coli positive sample (a) and negative control (b). The peaks corresponding to TEV-L, TEV-S, and REF-L are labeled.
Discussion
In this work we demonstrate the production of a highly specific protease delivered by an engineered phage for the detection of E. coli using fluorescent and MALDI-TOF MS platforms. While the bioengineering of reporter phage is not new (Schmelcher and Loessner, 2014, Singh, et al., 2012, van der Merwe, et al., 2014), to our knowledge we are the first to demonstrate the use of a protease as a reporter with application in multiple detection platforms. While our proof-of-principle is specific for E. coli, we believe our scheme can be readily modified for the detection of other bacterial species in combination with other specific proteases. Successful bioengineering of phage specific for other species, like Salmonella and Listeria, has been well demonstrated (van der Merwe, et al., 2014). The successful expression of viral proteases in gram negative bacteria, like E. coli, has also been demonstrated (Graves, et al., 1988, Liu, et al., 1999, Raran-Kurussi, et al., 2013). This phage-protease-peptide combination lays the groundwork for a novel multiplex detection scheme of various bacteria within a sample.
Of the two platforms we leveraged for the detection of E. coli via phage-induced TEV protease activity, fluorescence is likely the most widely adoptable. Fluorescent/luminescent readers, both high-throughput, like Synergy™ 2 (BioTek) or GloMax® (Promega), and smaller mobile readers, like the Quantus™ Fluorometer (Promega) are commercially available and commonly used. There is also a wide range of fluorescent/quencher markers for substrate-based protease probes (Sanman and Bogyo, 2014, Xia, et al., 2008) suggesting that a mature phage-protease-fluorescent peptide multiplex scheme could be easily implemented by many laboratories.
The upfront equipment cost necessary for a MALDI-TOF MS system may be higher than that of a fluorescent system, but the cost is lowering to that point that when combined with the low cost and high speed of sample analysis, MALDI-TOF systems are beginning to be adopted in high and low resource countries (Fall, et al., 2015, Martiny, et al., 2014, Neville, et al., 2011). MALDI-TOF MS analysis offers several advantages for our detection scheme. The first is the higher resolving power of MALDI-TOF MS over fluorescence. MALDI-TOF MS differentiates small peptides based on their mass-to-charge (m/z) ratios, and they need only differ by a couple m/z units to be easily distinguished. One could readily design a very wide range of peptides for detection, and thus the breadth of the multiplexing capability is much greater with MALDI-TOF MS. To expand the breadth of multiplexing with a fluorescent readout, one would need to incorporate more and more fluorescent molecules whose emission spectra may not be resolved well. The second advantage MALDI-TOF MS offers is specificity. There is the possibility that a given sample may contain an extraneous protease that cleaves our peptide, producing a signal in our fluorescent scheme, resulting in a false positive. The TEV-L peptide is designed to be cleaved into two identical peptides, TEV-S, by TEV protease. It is unlikely that the activity of another protease would result in peptides whose m/z ratio would be identical to TEV-S. Thus, using the MALDI-TOF MS platform we can be further assured that the signal we measure is only due to TEV protease activity, which would only be present if viable E. coli, infected by T7tev, were present in the original sample. A third advantage to the MALDI-TOF MS platform is the signal amplification inherent in our scheme, where TEV protease cleavage of the TEV-L peptide results in two TEV-S peptides. This can be observed by comparing the signal TEV-L to REF-L in the control to the signal of TEV-S to REF-L in the E. coli positive samples (Fig. 6). There is the potential that this signal amplification and the sensitivity of the MALDI-TOF MS could improve the bacterial limit of detection beyond that of the fluorescent scheme, which is approximately 107 CFU/mL (Fig. 4). Further work determining the limit of detection for the peptides, optimizing the sample matrix for both TEV activity and background signal, and the use of sample enrichment tools that can be readily coupled with MALDI, like Surface-Enhanced Laser Desorption/Ionization (Tang, et al., 2004), need to be explored before understanding if this benefit can be realized.
PCR-based detections methods have clearly demonstrated their sensitivity, though DNA contamination remains a challenge. Coupling PCR with a sample pre-treatment of ethidium bromide monoazide or propidium monoazide, dyes that irreversible bind DNA, has been shown to enable PCR-based differentiation of viable and non-viable cells (Kobayashi, et al., 2009, Rudi, et al., 2005, Wang and Mustapha, 2010). Studies, however, suggest more research is needed to optimize the successful use of these dyes. When used at low concentrations, interfering compounds or high levels of non-viable cells, both of which may be found in environmental or foods samples, can still result in false positive (Seinige, et al., 2014, Taylor, et al., 2014). At high concentrations, these dyes are toxic, expensive, and have been shown to have a negative effect on the detection of DNA from viable cells of clinical important pathogens (Kobayashi, et al., 2009, Nocker, et al., 2006, Taylor, et al., 2014). It therefore seems prudent to further probe the capabilities and potential applications of phage-based detection of viable bacterial cells.
A challenge for phage-based bacterial detection is the condition of the target cell. Environmental conditions, temperature, bacterial growth rate, membrane composition, and cell injury are all factors that have been shown to impact the success of phage infection and subsequent productivity of infection (Denes and Wiedmann, 2014). This is a key reason we focused on bacterial detection following primary enrichment, to ensure the cells were in a state favorable for infection. In fact, when we looked for TEV activity in samples containing 109 CFU/mL, we could detect none (Fig. 4). This likely occurs because at a concentration of 109 CFU/mL the majority of E. coli cells are in stationary rather than log phase, thus significantly limiting cell growth and phage replication. To address this upper limit, we added a step where a portion of the primary enrichment sample was added to fresh both and incubated to allow the cells to re-enter log phase before the addition of phage. Another reason for a primary enrichment is that the level of a pathogenic bacteria in a sample like food can be quite low, and the sample sizes needed to detect their presence are large enough, 25 g to 375 g (FDA, 1998), to make detection without an enrichment step unfeasible. A study by Zhao and Doyle (2001) found that injured pathogenic bacteria only grew to levels between 104.0 to 108.3 CFU/mL when revived in pre-enrichment broth after 24 hours. A study by Suo and Wang (2013), using SEL broth (Kim and Bhunia, 2008), found that injured pathogenic cells of E. coli, Salmonella, and Listeria reached levels > 109 CFU/mL after a 20 hour enrichment. These studies further highlight the need for the appropriate enrichment conditions to ensure bacterial and successful detection. In future work we intended to implement a concentration step, either filtration based or using phage-based magnetic bead separation (Chen, et al., 2015) following appropriate primary enrichment conditions to ensure that necessary cell concentrations are consistently achieved.
In summary, bioengineering of bacteriophage offers novel platforms for the specific and sensitive multiplex detection of bacterial pathogens within a sample. By utilizing molecular reporters with multiple modes of detection, one can adapt a base technology, in this case phage-based enzymatic reporters, to meet the varying capabilities of laboratories around the world. To further explore this multi-platform approach to detection, our lab is also designing a paper-fluidic, immuno-lateral flow assay for maltose binding protein, a by-product of our T7-mediated overexpression of TEV protease, as an alternative detection method for the presence of E. col that can be performed in resource-limited environments, like on-farm. Continued research and novel phage constructs will further expand the application and use of phage-based diagnostics for bacterial detection in the real world.
Acknowledgments
Funding: This material is based upon work supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture, and the Food Science department of the University of Massachusetts. S.D.A. would like to acknowledge the USDA NIFA National Needs Fellowship (2011-38420-200044), USDA NIFA Pre-Doctoral Award (2015-67011-22805), and the Northeast Alliance for Graduate Education and the Professoriate and NIH IMSD grant (GM099649) for their support. R.W.V acknowledges support from the National Institutes of Health R01 CA169140. M.A.C.S. acknowledges support from a National Institutes of Health training grant T32 GM008515.
Footnotes
Conflict of Interest: Authors declare that no conflict of interest exists.
Ethical Approval: This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Chen J, Duncan B, Wang L, Rotello VM, Nugen SR. Bacteriophage-based Nanoprobes for Rapid Bacterial Separation. Personal Communication. 2015 doi: 10.1039/c5nr03779d. [DOI] [PubMed] [Google Scholar]
- Denes T, Wiedmann M. Environmental responses and phage susceptibility in foodborne pathogens: implications for improving applications in food safety. Curr Opin Biotechnol. 2014:45–49. doi: 10.1016/j.copbio.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Dunbar SA, Vander Zee CA, Oliver KG, Karem KL, Jacobson JW. Quantitative, multiplexed detection of bacterial pathogens: DNA and protein applications of the Luminex LabMAP system. J Microbiol Methods. 2003:245–252. doi: 10.1016/s0167-7012(03)00028-9. [DOI] [PubMed] [Google Scholar]
- Easter MC. Rapid microbiological methods in the pharmaceutical industry. Boca Raton, Fla: Interpharm/CRC; 2003. [Google Scholar]
- Fall B, Lo CI, Samb-Ba B, Perrot N, Diawara S, Gueye MW, Sow K, Aubadie-Ladrix M, Mediannikov O, Sokhna C, Dieme Y, Chatellier S, Wade B, Raoult D, Fenollar F. The ongoing revolution of MALDI-TOF mass spectrometry for microbiology reaches tropical Africa. Am J Trop Med Hyg. 2015:641–647. doi: 10.4269/ajtmh.14-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA. Bacteriological Analytical Manual. 8th Edition, Revision A. FDA; 1998. [Google Scholar]
- Graves MC, Lim JJ, Heimer EP, Kramer RA. An 11-kDa form of human immunodeficiency virus protease expressed in Escherichia coli is sufficient for enzymatic activity. Proc Natl Acad Sci U S A. 1988:2449–2453. doi: 10.1073/pnas.85.8.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortin GL. The MALDI-TOF mass spectrometric view of the plasma proteome and peptidome. Clin Chem. 2006:1223–1237. doi: 10.1373/clinchem.2006.069252. [DOI] [PubMed] [Google Scholar]
- Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- Kapust RB, Waugh DS. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 1999:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Bhunia AK. SEL, a selective enrichment broth for simultaneous growth of Salmonella enterica, Escherichia coli O157:H7, and Listeria monocytogenes. Appl Environ Microbiol. 2008:4853–4866. doi: 10.1128/AEM.02756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Oethinger M, Tuohy MJ, Hall GS, Bauer TW. Improving clinical significance of PCR: use of propidium monoazide to distinguish viable from dead Staphylococcus aureus and Staphylococcus epidermidis. J Orthop Res. 2009:1243–1247. doi: 10.1002/jor.20872. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Oethinger M, Tuohy MJ, Hall GS, Bauer TW. Unsuitable distinction between viable and dead Staphylococcus aureus and Staphylococcus epidermidis by ethidium bromide monoazide. Lett Appl Microbiol. 2009:633–638. doi: 10.1111/j.1472-765X.2009.02585.x. [DOI] [PubMed] [Google Scholar]
- Kutter E, Sulakvelidze A. Bacteriophages : biology and applications. Boca Raton, FL: CRC Press; 2005. [Google Scholar]
- Liu BL, Viljoen GJ, Clarke IN, Lambden PR. Identification of further proteolytic cleavage sites in the Southampton calicivirus polyprotein by expression of the viral protease in E. coli. J Gen Virol. 1999:291–296. doi: 10.1099/0022-1317-80-2-291. [DOI] [PubMed] [Google Scholar]
- Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci U S A. 2007:11197–11202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny D, Cremagnani P, Gaillard A, Miendje Deyi VY, Mascart G, Ebraert A, Attalibi S, Dediste A, Vandenberg O. Feasibility of matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS) networking in university hospitals in Brussels. Eur J Clin Microbiol Infect Dis. 2014:745–754. doi: 10.1007/s10096-013-2006-6. [DOI] [PubMed] [Google Scholar]
- Neville SA, Lecordier A, Ziochos H, Chater MJ, Gosbell IB, Maley MW, van Hal SJ. Utility of matrix-assisted laser desorption ionization-time of flight mass spectrometry following introduction for routine laboratory bacterial identification. J Clin Microbiol. 2011:2980–2984. doi: 10.1128/JCM.00431-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocker A, Cheung CY, Camper AK. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J Microbiol Methods. 2006:310–320. doi: 10.1016/j.mimet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Novagen®. T7Select® System Manual User Protocol TB178 Rev. D 0311JN. Darmstadt, Germany: EMD Chemicals Inc; 2001. [Google Scholar]
- Raran-Kurussi S, Tozser J, Cherry S, Tropea JE, Waugh DS. Differential temperature dependence of tobacco etch virus and rhinovirus 3C proteases. Anal Biochem. 2013:142–144. doi: 10.1016/j.ab.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees JC, Voorhees KJ. Simultaneous detection of two bacterial pathogens using bacteriophage amplification coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2005:2757–2761. doi: 10.1002/rcm.2107. [DOI] [PubMed] [Google Scholar]
- Rudi K, Moen B, Dromtorp SM, Holck AL. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl Environ Microbiol. 2005:1018–1024. doi: 10.1128/AEM.71.2.1018-1024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanman LE, Bogyo M. Activity-based profiling of proteases. Annu Rev Biochem. 2014:249–273. doi: 10.1146/annurev-biochem-060713-035352. [DOI] [PubMed] [Google Scholar]
- Schmelcher M, Loessner MJ. Application of bacteriophages for detection of foodborne pathogens. Bacteriophage. 2014:e28137. doi: 10.4161/bact.28137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seinige D, von Kockritz-Blickwede M, Krischek C, Klein G, Kehrenberg C. Influencing factors and applicability of the viability EMA-qPCR for a detection and quantification of Campylobacter cells from water samples. PLoS One. 2014:e113812. doi: 10.1371/journal.pone.0113812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settanni L, Corsetti A. The use of multiplex PCR to detect and differentiate food- and beverage-associated microorganisms: a review. J Microbiol Methods. 2007:1–22. doi: 10.1016/j.mimet.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Singh A, Arutyunov D, Szymanski CM, Evoy S. Bacteriophage based probes for pathogen detection. Analyst. 2012:3405–3421. doi: 10.1039/c2an35371g. [DOI] [PubMed] [Google Scholar]
- Smartt AE, Xu TT, Jegier P, Carswell JJ, Blount SA, Sayler GS, Ripp S. Pathogen detection using engineered bacteriophages. Analytical and Bioanalytical Chemistry. 2012:3127–3146. doi: 10.1007/s00216-011-5555-5. [DOI] [PubMed] [Google Scholar]
- Studier FW. Bacteriophage T7. Science. 1972:367–376. doi: 10.1126/science.176.4033.367. [DOI] [PubMed] [Google Scholar]
- Suo B, Wang Y. Evaluation of a multiplex selective enrichment broth SEL for simultaneous detection of injured Salmonella, Escherichia coli O157:H7 and Listeria monocytogenes. Braz J Microbiol. 2013:737–742. doi: 10.1590/s1517-83822013000300011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N, Tornatore P, Weinberger SR. Current developments in SELDI affinity technology. Mass Spectrom Rev. 2004:34–44. doi: 10.1002/mas.10066. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Bentham RH, Ross KE. Limitations of Using Propidium Monoazide with qPCR to Discriminate between Live and Dead Legionella in Biofilm Samples. Microbiol Insights. 2014:15–24. doi: 10.4137/MBI.S17723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe RG, van Helden PD, Warren RM, Sampson SL, Gey van Pittius NC. Phage-based detection of bacterial pathogens. Analyst. 2014:2617–2626. doi: 10.1039/c4an00208c. [DOI] [PubMed] [Google Scholar]
- Wang L, Mustapha A. EMA-real-time PCR as a reliable method for detection of viable Salmonella in chicken and eggs. J Food Sci. 2010:M134–M139. doi: 10.1111/j.1750-3841.2010.01525.x. [DOI] [PubMed] [Google Scholar]
- Wolffs P, Norling B, Radstrom P. Risk assessment of false-positive quantitative real-time PCR results in food, due to detection of DNA originating from dead cells. J Microbiol Methods. 2005:315–323. doi: 10.1016/j.mimet.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Xia Z, Xing Y, So MK, Koh AL, Sinclair R, Rao J. Multiplex detection of protease activity with quantum dot nanosensors prepared by intein-mediated specific bioconjugation. Anal Chem. 2008:8649–8655. doi: 10.1021/ac801562f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Doyle MP. Evaluation of universal preenrichment broth for growth of heat-injured pathogens. J Food Prot. 2001:1751–1755. doi: 10.4315/0362-028x-64.11.1751. [DOI] [PubMed] [Google Scholar]