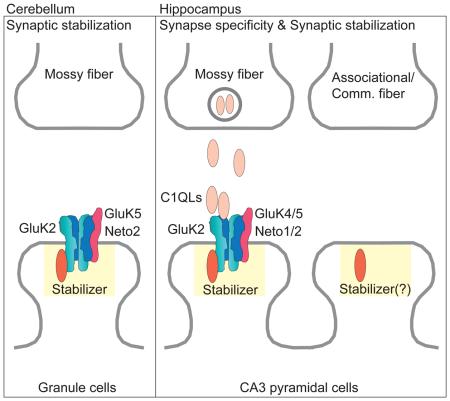
Abstract
Synaptic communication between neurons requires the precise localization of neurotransmitter receptors to the correct synapse type. Kainate-type glutamate receptors have restricted synaptic localization that is determined by the afferent presynaptic connection. The mechanisms that govern this input-specific synaptic localization remain unclear. Here we examine how subunit composition and specific subunit domains contribute to synaptic localization of kainate receptors. The cytoplasmic domain of the GluK2 low-affinity subunit stabilized kainate receptors at synapses. In contrast, the extracellular domain of the GluK4/5 high-affinity subunit synergistically controls the synaptic specificity of kainate receptors through interaction with C1q-like proteins. Thus the input-specific synaptic localization of the native kainate receptor complex involves two mechanisms that underlie specificity and stabilization of the receptor at synapses.
Graphical abstract
INTRODUCTION
Proper synaptic communication requires correct localization of neurotransmitter receptors to specific postsynaptic sites. Glutamate is the major excitatory transmitter in the vertebrate brain, and three classes of ionotropic glutamate receptors (kainate, AMPA, and NMDA) mediate the vast majority of synaptic transmission at excitatory synapses. Whereas most excitatory synapses contain AMPA- and NMDA-type receptors, kainate-type glutamate receptors (KARs) only localize to select synapses (Contractor et al., 2011; Darstein et al., 2003; Foster et al., 1981; Isaac et al., 2004; Monaghan and Cotman, 1982; Nicoll and Schmitz, 2005; Petralia et al., 1994). This restricted localization of KARs is apparent in the hippocampal stratum lucidum where mossy fiber axons projecting from dentate gyrus granule neurons form complex synapses with CA3 neurons (Castillo et al., 1997; Contractor et al., 2003; Darstein et al., 2003; Mulle et al., 1998; Petralia et al., 1994; Vignes and Collingridge, 1997). In contrast, KARs are found at all synapses in the cerebellum, where granule cells receive input from only one type of excitatory afferent, the mossy fiber (Yan et al., 2013). Mechanisms underlying these synaptic differences remain unclear.
KARs in the brain form a tripartite hetero-oligomeric complex consisting of the low-affinity GluK1-3 and high-affinity GluK4/5 KAR subunits along with Neto auxiliary subunits. Because KAR-mediated transmission is absent in primary cultured hippocampal neurons (Lerma et al., 1993), studying synapses in vivo using mouse gene-targeting approaches has been particularly useful in identifying KAR components required for synaptic localization and function. Knockout of the primary low-affinity subunit GluK2 abolishes KAR currents as well as localization of receptors (Mulle et al., 1998; Yan et al., 2013). In addition, GluK2 KO mice exhibit reduced expression of other components of the native KAR complexes, GluK4/5 and Neto1/2 (Christensen et al., 2004; Nasu-Nishimura et al., 2006; Ruiz et al., 2005; Straub et al., 2011; Zhang et al., 2009). In Neto1 KO mice, synaptic expression of KARs is unchanged at hippocampal mossy fiber–CA3 cell synapses while the decay kinetics of the current are dramatically faster (Straub et al., 2011). In mice in which both Neto1 and Neto2 are ablated (Neto1/2 DKO) or mice in which both high affinity subunits are knocked out (GluK4/5 DKO), KARs are reduced in the post-synaptic density (PSD) (Fernandes et al., 2009; Wyeth et al., 2014). In addition, GluK4/5 DKO mice lack KAR-mediated synaptic currents at mossy fiber synapses despite no obvious difference in the surface expression of the GluK2 subunit (Fernandes et al., 2009). Because dysregulation in multiple steps of receptor biogenesis, including protein expression, surface expression, synapse specific localization, and synaptic stabilization can affect synaptic activity of KARs, it remains unclear which components of the receptor complex contribute to synaptic stabilization and synapse-specific localization of KARs in the brain.
In this study, we used a gene-targeting approach to elucidate the mechanisms of synapse-specific localization of KARs by systematically examining the roles of the predominant subunits within the native receptor complex. The results demonstrate that the cytoplasmic domain of GluK2 plays specific roles in synaptic stabilization, but not in surface expression or protein levels in the brain. On the other hand, in CA3 pyramidal cells, the GluK4/5 high-affinity subunits localize KARs specifically to mossy fibers synapses, and the extracellular domain of GluK4/5 is required for this synapse specificity through an interaction with a member of the C1qL/nCLP protein family. Furthermore, the GluK2 cytoplasmic domain and the GluK4/5 extracellular domain synergistically control the synaptic abundance of KARs. These results imply that synapse-specific localization of KARs is mediated by two distinct mechanisms dependent on the constituent subunits of the heteromeric receptor complex.
RESULTS
GluK2, but not GluK5 or Neto2, is required for synaptic localization of KARs in the cerebellum
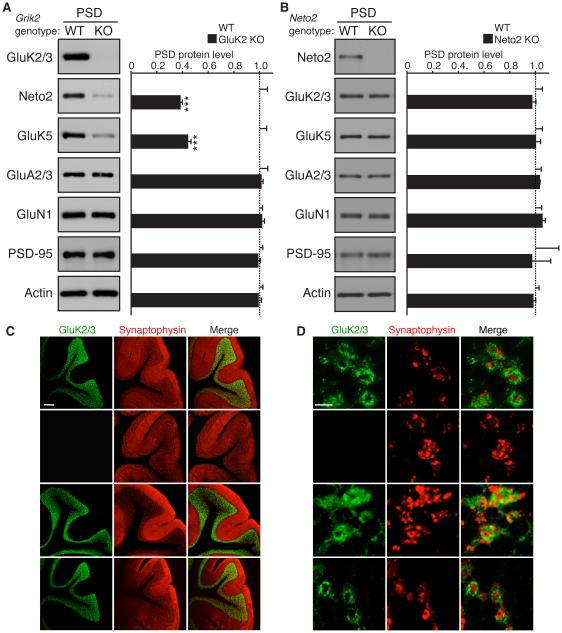
In cerebellar granule cells, the native KAR complex consists of GluK2, GluK5, and Neto2 (Yan et al., 2013). GluK2 and Neto2 are detected in the PSD fraction of both wild-type and GluK5 KO mice at similar levels (Yan et al., 2013). To identify responsible subunits for synaptic localization and protein expression of KARs, we evaluated protein levels in the cerebellar PSD fraction and total lysate from KO mice for each of the expressed subunits. We observed a substantial reduction in the amounts of GluK5 and Neto2 in both the PSD fraction and total lysate of GluK2 KO mice, but GluK5 expression was unaltered in preparations from Neto2 KO mice (Figure 1A and B, Figure S1). Levels of other synaptic proteins, including GluA2/3, GluN1, PSD-95, and actin were also unchanged in these mice (Figure 1A and B, Figure S1).
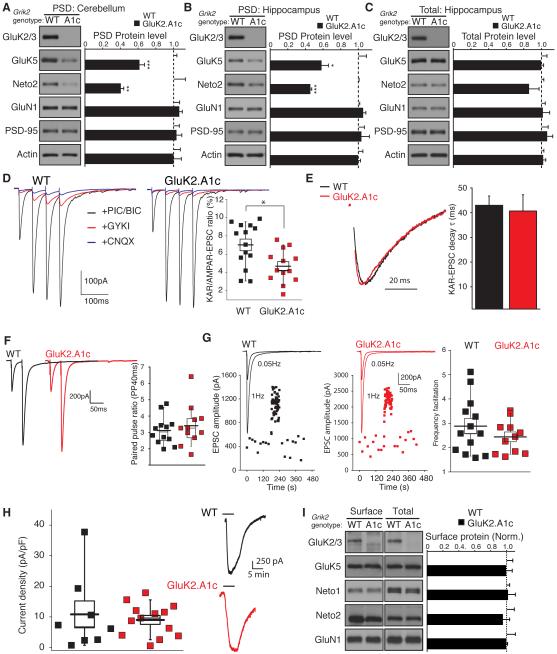
Figure 1. Synaptic localization of KARs is determined by GluK2, but not GluK5 or Neto2, in cerebellar granule cells.
Distribution of components of KAR complex in the cerebellum of the indicated knockout (KO) mice. (A, B) Protein levels of GluK5 and Neto2 were reduced in the cerebellar PSD-enriched fraction of GluK2 KO mice (A), but unaltered in Neto2 KO mice (B). Protein levels of AMPAR (GluA2/3), NMDAR (GluN1), PSD-95, and actin were unaltered (n = 4 each). (C) GluK2/3 signal was not detected in the granular layer of GluK2 KO mice, but was detected in Neto2 and GluK5 KO mice. (D) High-magnification images of cerebellar glomeruli. No obvious change in GluK2 distribution was observed in Neto2 and GluK5 KO mice. Synaptophysin is a presynaptic marker. Scale bars: 200 µm (C), 5 µm (D). GL, PCL, and ML designate granular, Purkinje cell, and molecular layers, respectively. Data in A and B are given as mean ± s.e.m.; ***, P < 0.001 (Student’s t-test).
We next examined the distribution of GluK2 in the cerebellum using immunostaining. As expected, GluK2/3 signal was lost in the cerebellum of GluK2 KO mice (Figure 1C). In the cerebellar granular layer, GluK2 was distributed in ring-shapes around a presynaptic marker protein, synaptophysin, presumably indicating glomerular synapses between cerebellar mossy fibers and granule cells (Figure 1D). Consistent with our biochemical results (Figure 1A and B) and previous findings in GluK5 KO mice (Yan et al., 2013), we did not observe any obvious changes in GluK2 distribution in either Neto2 or GluK5 KO mice (Figure 1C and D). These data demonstrate that the signal for both synaptic localization and protein abundance of the native KAR complex in the cerebellum is not dependent on GluK5 or Neto2, but is inherent to the GluK2 subunit.
The GluK2 cytoplasmic domain is indispensible for receptor complex formation in the brain
We adopted a gene-targeting approach to determine GluK2 receptor subunit domain involvement in synaptic stabilization. Initially, we sought to generate a GluK2 mutant in which the receptor lacked synaptic stabilization, without altered expression. We focused on the cytoplasmic domain because altering the amino terminus of the receptor would likely result in disruption of heteromultimerization (Kumar et al., 2011; Meyerson et al., 2014; Sobolevsky et al., 2009).
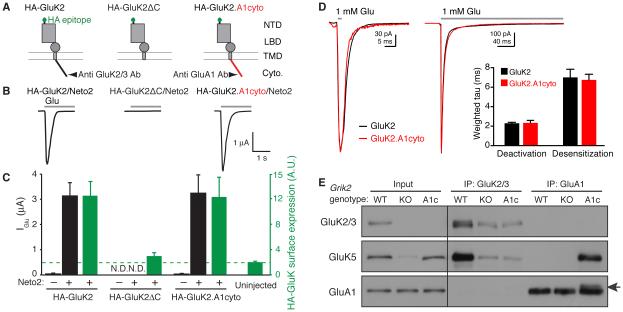
To evaluate the potential effects of GluK2 cytoplasmic mutations on KAR function, we injected cRNAs of extracellularly HA-epitope tagged GluK2 (HA-GluK2) and various mutants, along with Neto2, into Xenopus laevis oocytes (Figure 2A). We then measured glutamate-evoked currents by two-electrode voltage-clamp recording, and surface expression of HA-GluK2 with a chemiluminescence assay as described previously (Zhang et al., 2009). Neto2 co-expression significantly increased the peak amplitudes of glutamate-evoked currents from oocytes expressing HA-GluK2 (Figure 2B and C). Furthermore, surface expression of HA-GluK2 was significantly higher relative to the background level detected in uninjected oocytes (Figure 2C, green bar). On the other hand, deletion of the GluK2 cytoplasmic domain abolished both glutamate-evoked currents and surface expression (Figure 2A–C) as has been described previously (Yan et al., 2004). We then examined a chimera (GluK2.A1cyto) in which the cytoplasmic domain of GluK2 was replaced with that of the GluA1 AMPAR subunit, which shares only 9.9% sequence identity. Glutamate-evoked currents and surface expression of HA-GluK2.A1cyto were similar to those produced upon expression of wild-type HA-GluK2 (Figure 2B and C). To analyze receptor properties with a faster time resolution, we measured deactivation and desensitization of GluK2 and GluK2.A1cyto in outside-out patches from oocytes injected with cRNAs of Neto2, GluK5 and either GluK2 or GluK2.A1cyto. Outside-out membrane patches were exposed to brief (2 ms) and sustained (300 ms) rapid applications of 1 mM glutamate using a piezo-electric system. The deactivation and desensitization kinetics were similar with GluK2 and GluK2.A1cyto, and the weighted tau values obtained from bi-exponential fits to these decays were virtually identical for wild-type and mutant receptors (Figure 2D). Thus, GluK2.A1cyto can function as effectively as GluK2 in terms of channel activity and surface expression, at least in cRNA-injected oocytes.
Figure 2. Roles of the GluK2 cytoplasmic domain in formation of the KAR complex.
(A) Schematic diagram of each GluK2 mutant tested. GluK2ΔC indicates deletion of the C-terminal cytoplasmic domain, and GluK2.A1cyto indicates replacement of the cytoplasmic domain of GluK2 with that of the GluA1 AMPAR subunit. Epitopes for anti GluK2/3 or GluA1-antibodies (Ab) are indicated. NTD: N-terminal domain; LBD: ligand-binding domain; TMD: trans-membrane domain; cyto: cytoplasmic C-terminus. (B, C) Glutamate-evoked currents and surface expression were measured by two-electrode voltage-clamp recording and chemiluminescence assay in oocytes injected with various cRNAs, as indicated. (B) Representative traces are shown; gray bar indicates bath application of glutamate (1 mM). (C) Quantitation of peak amplitudes of glutamate-evoked currents (black) and surface expression of HA-tagged GluK2 (green) (n = 10 each). Deletion of the GluK2 cytoplasmic domain abolished surface expression, and replacing the cytoplasmic domain of GluK2 with that of GluA1 restored both surface expression and activity. Green dashed line indicates the background level, defined as the signal from un-injected oocytes. N.D.: not detectable. (D) Responses to 2 ms or 300 ms applications (bars) of 1 mM glutamate in outside-out oocyte membrane patches expressing GluK2 (black) or GluK2.A1cyto (red). Bar graph showing the mean weighted time constants of deactivation and desensitization from bi-exponential fits to the decay of currents. (E) GluK5 interaction with GluK2 or GluK2.A1cyto was analyzed by co-immunoprecipitation using cerebral cortical lysate from wild-type (WT), GluK2 knockout (KO), and GluK2.A1c KI mice (A1c), using antibodies shown in (A). GluK2.A1cyto was detected weakly at a slightly higher molecular weight than that of endogenous GluA1 (arrow). Total GluK5 expression was reduced in GluK2 KO, but not in GluK2A1c KI mice (Input). Data are shown as mean ± s.e.m.
Next, using gene-targeting techniques, we generated a GluK2 KI mouse in which the cytoplasmic domain of GluK2 was replaced with that of GluA1. The resultant KI mouse GluK2.A1c expresses GluK2.A1cyto instead of wild-type GluK2 (Figure S2). Grik2, the gene that encodes GluK2, encodes two alternative isoforms of the cytoplasmic domain, encoded by exon 17a/b. To ensure that all GluK2 proteins in the KI mice possessed a GluA1 cytoplasmic domain with appropriate stop codons, we inserted a 249-bp cassette encoding the GluA1 cytoplasmic domain with stop codons immediately after the GluK2 transmembrane domain in exon 16 of Grik2 (Figure S2A). Consequently, GluK2 containing the GluA1 cytoplasmic domain (GluK2.A1cyto) was expressed independently of GluK2 splicing. Proper targeting was confirmed by Southern blot and genomic PCR analysis (Figure S2B and C). GluK2.A1c KI mice were viable and fertile similar to GluK2 KO mice (Mulle et al., 1998).
Because of the positions of their epitopes, the anti-GluK2/3 antibody recognizes only GluK2 (but not GluK2.A1cyto), whereas the anti-GluA1 antibody recognizes GluK2.A1cyto and endogenous GluA1, but not GluK2 (Figure 2A). Thus, relative expression levels of GluK2.A1cyto to GluK2 in the brain cannot be directly evaluated. Therefore, to determine whether KAR expression is altered in GluK2.A1c mice, we first measured the GluK5 expression in GluK2.A1cyto and GluK2 KO mice. As previously demonstrated, GluK5 was reduced in GluK2 KO mice (Figure 2E, Input) (Christensen et al., 2004; Nasu-Nishimura et al., 2006; Ruiz et al., 2005). By contrast, GluK5 expression was unaltered in GluK2.A1c KI brains (Figure 2E, Input). Furthermore, in wild-type animals anti-GluK2/3 antibody immunoprecipitated GluK5 with GluK2/3 (Figure 2E) and Neto1 and Neto2 (Figure S2D). Given that this antibody recognizes both GluK2 and GluK3, the weak GluK5 bands observed in GluK2 KO and GluK2.A1c KI mice suggested that GluK3 was expressed at low levels. The anti-GluA1 antibody immunoprecipitated a specific band slightly higher than endogenous GluA1 (Figure 2E and S2D, arrow) whose molecular weight corresponded to the predicted molecular weight of GluK2.A1cyto; the low intensity of this band reflects the relative expression levels of GluA1 and GluK2.A1cyto in the brain. Importantly, GluK5, Neto1 and Neto2 were co-immunoprecipitated with comparable efficiency by anti-GluA1 antibody from GluK2.A1c KI brains and by anti-GluK2/3 antibody from wild-type brains (Figure 2E and S2D). These results suggest that the GluK2.A1cyto is expressed and forms a complex with GluK5 and Neto1/2 in brain as effectively as wild-type GluK2.
The GluK2 cytoplasmic domain mediates synaptic stabilization, but not surface expression, of KARs in the cerebellum
We next examined protein localization in the cerebellum of GluK2.A1c KI mice. Immunostaining with the anti-GluK2/3 antibody demonstrated staining in the cerebellar granular layer in wild-type mice, but not in GluK2.A1c KI mice (Figure S3A), as predicted from the location of the epitope region (Figure 2A). On the other hand, due to expression of endogenous GluA1 in Bergmann glia and Purkinje cells, strong signal from anti-GluA1 antibody was evident in the cerebellar molecular layer of both wild-type and GluK2.A1c KI mice (Figure S3B). By increasing the laser power used for confocal microscopy, we were able to detect a specific anti-GluA1 antibody signal in the cerebellar granular layer of GluK2.A1c KI mice, but not in wild-type mice (Figure S3C). To eliminate endogenous GluA1 signal in the cerebellar molecular layer, we used GluK2.A1c KI; GluA1 KO double-mutant mice. The GluA1 signal in the molecular layer was abolished in GluA1 KO; GluK2.A1c KI double-mutant mice, whereas the signal in the granular layer was retained, indicating that the granular layer signal detected by anti-GluA1 antibody corresponds to GluK2.A1cyto protein (Figure S3C).
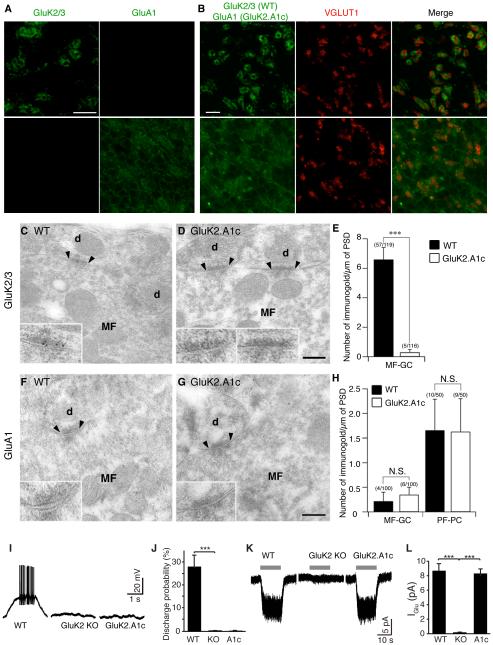
High-magnification images of the cerebellar granular layer revealed specific signals from the anti-GluK2/3 and GluA1 antibodies in wild-type and GluK2.A1cyto KI mice, respectively (Figure 3A). In wild-type mice, GluK2 was localized to cerebellar glomeruli as revealed by the GluK2 signal near the cerebellar mossy fiber presynaptic protein VGLUT1. In contrast, GluK2.A1cyto localized diffusely in the granular cell layer (Figure 3A and B). Using post-embedding immuno-electron microscopy, GluK2 signal was observed at the cerebellar mossy fiber–granule cell synapses in wild-type mice, but not in GluK2.A1c KI mice (Figure 3C–E). GluA1 signal was detected at parallel fiber– Purkinje cell synapses, but wild-type and GluK2.A1c KI mice had comparably low numbers of GluA1 particles at cerebellar mossy fiber–granule cell synapses (Figure 3F-H) consistent with a loss of synaptic GluK2.A1cyto.
Figure 3. The GluK2 cytoplasmic domain is required for synaptic KAR localization in cerebellar granule cells.
(A-H) Distribution of KARs in cerebellum of GluK2.A1c KI mice. (A) Immunohistochemical staining of the granular cell layer of mouse cerebellum. GluK2/3 signal was observed only wild-type (WT) mice, whereas GluA1 signal was observed only in GluK2.A1cyto KI mice. Because of no endogenous GluA1 in the granule cells, the GluA1 signal indicates specific expression of GluK2.A1cyto protein. (B) GluK2 was enriched at cerebellar glomeruli around the mossy fiber presynaptic marker VGLUT1, whereas GluK2.A1cyto was distributed diffusely. Scale bars: 10 µm. (C-H) Immuno-electron microscopic images of GluK2 and GluK2.A1cyto proteins. Inserts show high-magnification of labeled synapses. Scale bars: 200 nm. (C–E) GluK2 was detected at MF-GC synapses in WT mice, but not in GluK2.A1c KI mice. (F–H) No GluK2.A1cyto signal was detected at MF-GC synapses from both WT and GluK2.A1c KI mice with anti-GluA1 C-terminal antibody. By contrast, endogenous GluA1 was detected at similar levels in cerebellar parallel fiber (PF)-Purkinje cell (PC) synapses in both WT and GluK2.A1c KI mice. Numbers of immunogold-labeled synapses and total analyzed synapses are indicated in parentheses. (I-L) KAR activity was measured in cerebellar granule cells. To isolate KAR activity from other glutamate receptors, recordings were performed on the stargazer genetic background. (I, J) Mossy fiber-evoked responses were recorded under the whole-cell current-clamp configuration. KAR-dependent synaptic transmission at cerebellar mossy fiber–granule cell synapses was abolished in both GluK2 KO and GluK2A1c KI mice (WT n =5, KO and K2.A1c n = 4 each). (K, L) KAR activity at the cell surface was measured using 300 µM glutamate (gray bar) in the presence of 100 µM picrotoxin and 100 µM D-AP5. Surface KAR activity was detected at similar levels in WT and GluK2.A1cyto KI mice, but not in GluK2 KO (GluK2−/−) mice (WT n =5, KO and A1c n = 4 each). Data are given as mean ± s.e.m. ***, P < 0.001 (Student’s t-test).
Next, using whole-cell patch-clamp recordings, we measured KAR activity in cerebellar granule cells in acute cerebellar slices. Because KAR-mediated EPSCs are of small amplitude, but have a large effect on membrane depolarization during a train of stimulation, we evaluated the KAR contribution to EPSP–spike coupling using the AMPAR-deficient stargazer genetic background to clearly differentiate KAR activity from AMPAR activity (Yan et al., 2013). With NMDA activity blocked (100 µM APV), repetitive minimal stimulation of the cerebellar mossy fibers generated spikes in wild-type mice on the stargazer genetic background (Figure 3I and J). In contrast, no spikes were observed in GluK2 KO or GluK2.A1c KI mice (on the stargazer genetic background) (Figure 3I and J). These results suggest that synaptic KAR-dependent spike generation was abolished in the GluK2.A1c KI mice, as it is in GluK2 KO mice. Further supporting the critical role of the GluK2 cytoplasmic domain in synaptic transmission, GluK2.A1c KI; stargazer double-mutant mice exhibited severe locomotion deficits and died by postnatal day 30 similar to GluK2 KO; stargazer double-mutants reported previously (Yan et al., 2013).
We next investigated whether the total KAR current density was altered in granule cells in GluK2.A1cyto mice. To this end, we measured glutamate-evoked KAR-mediated currents in cerebellar granule cells from acute cerebellar slices under voltage-clamp configuration (Vh = −70 mV) with NMDA activity blocked (Yan et al., 2013). Saturating glutamate elicited KAR-mediated currents of similar amplitudes in both stargazer mice and GluK2.A1c; stargazer double-mutant mice, but not in GluK2 KO; stargazer double-mutant mice (Figure 3K and L), indicating that total surface KAR density remains the same in GluK2.A1c mice. These results suggest that the GluK2 cytoplasmic domain plays specific roles in synaptic stabilization without changes in surface activity in cerebellar granule cells.
The GluK2 cytoplasmic domain stabilizes receptors at synapses in hippocampus
We next investigated whether the GluK2 cytoplasmic domain also affects synaptic stabilization at hippocampal synapses. We measured protein levels in the PSD-enriched fractions from cerebellum and hippocampus. GluK5 protein levels in the cerebellar PSD fractions were reduced in GluK2.A1c KI mice with no changes in the levels of other synaptic proteins (Figure 4A). Similarly, in hippocampus, the protein levels of both GluK5 and Neto2 were specifically reduced in hippocampal PSD fractions (Figure 4B), without changes in total protein levels (Figure 4C), suggesting specific deficits in synaptic localization of KARs in hippocampus of GluK2.A1c KI mice.
Figure 4. Selective reduction in synaptic KARs in GluK2.A1c KI mice.
(A) PSD-enriched fractions were purified from cerebella of wild-type (WT) and GluK2.A1c KI mice. GluK5 levels in the cerebellar PSD-enriched fraction was reduced in GluK2.A1c KI mice. (B and C) Protein levels in the PSD-enriched fraction (B) and total (C) were measured in hippocampi from WT and GluK2.A1c KI mice (n = 6). KAR components GluK5 and Neto2 were specifically reduced in the hippocampal PSD fraction, without changes in the total protein levels. Levels of other excitatory synaptic proteins (GluA1, GluN1 and PSD-95) were unaltered. (D–G) Synaptic activity at hippocampal mossy fiber–CA3 pyramidal cell synapses were measured under the whole-cell voltage-clamp configuration (Vh = −70 mV) in acute slices. (D) EPSCs were measured with combinations of various blockers following four consecutive stimulations of mossy fibers. EPSCs were isolated by addition of picrotoxin and bicuculline. KAR-mediated EPSCs were isolated as currents insensitive to 50 µ M GYKI53655 and sensitive to 10 µ M CNQX. The ratio of KAR-mediated to AMPAR-mediated EPSCs (the difference between total EPSCs and KAR-mediated EPSCs) was significantly reduced in GluK2.A1c KI mice (n = 13) relative to that in WT mice (n = 14). (E) No significant changes in the decay kinetics of KAR-mediated EPSCs were observed (WT n =13; K2.A1c n = 11). (F) Paired-pulse ratio of AMPAR-mediated EPSCs with a 40-ms interval did not differ (WT n =12; K2.A1c n = 11). (G) Frequency facilitation was unchanged (WT n =13; K2.A1c n = 11). (H) Kainate-evoked current density measured in CA3 pyramidal cells and two representative traces from mutant and wild-type mice (I) Surface expression of proteins in acute hippocampal slices was measured using cell-impermeable Sulfo-NHS-SS-biotin. No changes in GluK5, Neto1 and Neto2 as well as GluN1 were observed in the “Surface” and “Total” fractions between WT and GluK2.A1c KI mice (n = 4). Data are given as mean ± s.e.m.; * P < 0.05, ** P < 0.01, *** P < 0.005 (Student’s t-test).
We then examined KAR-mediated synaptic currents at hippocampal mossy fiber-CA3 pyramidal cell synapses in acute hippocampal slices. Mossy fiber EPSCs were evoked using a train (20 Hz) of four stimuli in the presence of GABAA (picrotoxin, 50 µM; bicuculline, 10 µM) and NMDA antagonists (D-APV, 50 µM) (Figure 4D). The AMPAR-mediated component of the mossy fiber EPSC was measured as the difference in synaptic response in the presence or absence of 50 µ M GYKI53655; while the KAR-mediated component of the EPSC was measured as the residual synaptic response in the presence of 50 µM GYKI53655, which could be subsequently blocked by 10 µ M CNQX (Figure 4D). The relative amplitude of the KAR-mediated component of the EPSC was 7.0 ± 0.6% of the total EPSC in wild-type mice, similar to that reported previously (Contractor et al., 2003). The relative contribution of the KAR to the amplitude of the EPSC was significantly reduced to 4.7 ± 0.4% in GluK2.A1c KI mice (Figure 4D). The decay kinetics of the KAR-mediated EPSCs were unchanged, suggesting that the subunit composition of synaptic KARs was preserved in GluK2.A1c KI neurons (Figure 4E). The absence of changes in the paired-pulse ratio (40 ms interval) and frequency facilitation of EPSCs suggested that mossy fiber synapses in GluK2.A1c mice did not have altered release probability and short-term plasticity (Figure 4F and G).
A reduction in synaptic KAR activity could be due to an overall reduction in KAR function at the cell surface. To examine this, we measured the total kainate current density. Application of the KAR agonist, kainic acid (10 µM; in the presence of AMPA, NMDA, and GABAA antagonists), to CA3 pyramidal cells demonstrated no difference in the total agonist-evoked KAR current density between recordings from wild-type and GluK2.A1c KI mice (Figure 4H). Furthermore, surface expression of KARs in acute hippocampal slices were examined using a cell-impermeable biotinylated reagent in acute hippocampal slices (Tomita et al., 2004), and no changes were observed in the surface expression of KAR components (GluK5, Neto1 and Neto2) or GluN1 in GluK2.A1c mice (Figure 4I). Under the same conditions, the intracellular protein tubulin was not detected at the surface (Figure S4). These results suggest that the GluK2 cytoplasmic domain is required for synaptic stabilization of the KAR complex at mossy fiber synapses.
Distinct mechanisms for synapse specificity and synaptic stabilization
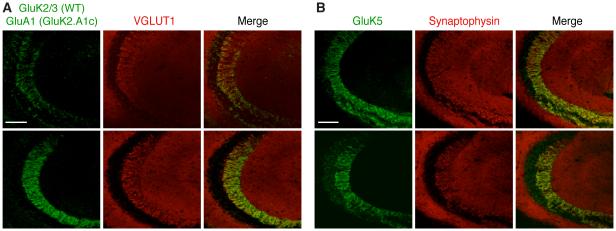
In hippocampal CA3 pyramidal cells, KARs are distributed with a restricted synapse-specific localization at the mossy fiber synapses in the stratum lucidum. Compared to the complete loss of the functional effects of synaptic KARs in cerebellar granule cells (Figure 3), the partial loss of KAR-mediated currents in the hippocampal CA3 pyramidal cells (Figure 4) (without a reduction in total and surface protein expression) prompted us to ask whether KARs redistributed to different synapses in the hippocampus of GluK2.A1c KI mice, thus losing synapse specificity to the mossy fiber-CA3 synapses. To examine this, we compared the distribution of endogenous GluK2 in wild-type hippocampus and GluK2.A1cyto in GluK2.A1c KI hippocampus. To avoid strong signals from endogenous GluA1 (Keinanen et al., 1990), we used GluK2.A1c KI; GluA1 KO double-mutant mice as in the cerebellum (Figure S3C). Both GluK2 and GluK2.A1cyto signals were observed at the stratum lucidum in both wild-type and GluK2.A1c; GluA1 double-mutant mice, respectively (Figure 5A). Similarly, endogenous GluK5 staining was also observed at the stratum lucidum of both mice (Figure 5B). Because KAR-mediated synaptic currents were reduced in GluK2.A1cyto KI hippocampus (Figure 4) but the localization of the KARs in the stratum lucidum is normal (Figure 5), this data suggests that the GluK2 cytoplasmic domain is critical for synaptic stabilization, but not for synapse specificity.
Figure 5. Distinct mechanisms for synaptic stabilization and synapse specificity of KARs in hippocampus.
KAR distribution was examined in hippocampus from wild-type (WT) and GluK2.A1c KI; GluA1 KO double-mutant mice. (A, B) GluK2 and GluK2.A1cyto (A) as well as GluK5 (B) were observed at the stratum lucidum in WT and GluK2.A1c KI; GluA1 KO double-mutant mice, respectively. Scale bar: 100 µ m.
Synapse specificity is mediated by the high affinity KAR subunits
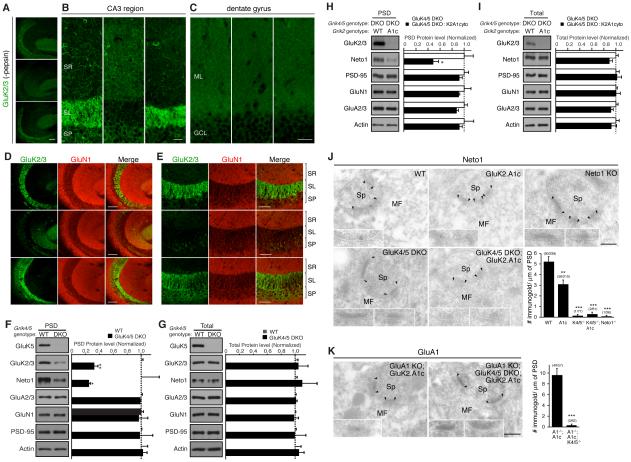
A major outstanding question is what determines synapse specificity of KARs. We examined this question in mice in which the components of the native KAR complex are disrupted. In hippocampal CA3 pyramidal cells the native KAR complex consists of at least five distinct subunits: one low-affinity subunit (GluK2), two high-affinity subunits (GluK4 and 5), and two auxiliary subunits (Neto1 and 2) (Fernandes et al., 2009; Mulle et al., 1998; Straub et al., 2011; Tang et al., 2011). Therefore, we systematically evaluated KAR localization in GluK4/5 DKO and Neto1/2 DKO mice. Staining for GluK2 in stratum lucidum was observed in wild-type and Neto1/2 DKO mice (Figure 6A). In contrast, GluK2 staining in the stratum lucidum was almost completely abolished in GluK4/5 DKO mice (Figure 6A). Surprisingly, the GluK2 signal in the stratum radiatum was increased in GluK4/5 DKO mice (Figure 6B) suggesting that GluK2 redistributed to the more distal dendrites of CA3. Analysis of a different hippocampal region, the dentate gyrus, found no similar redistribution of GluK2 staining in the GluK4/5 DKO mice (Figure 6C).
Figure 6. High-affinity GluK4/5 subunits mediate synapse specificity of KARs in the hippocampus.
The distribution of KAR components in the hippocampus was examined by immunohistochemistry and biochemical fractionation. (A and B) Immunostaining of hippocampal sections without pepsin treatment (see Experimental Procedures) revealed reduction in the GluK2/3 signal at the stratum lucidum (SL) in GluK4/5 double-knockout (DKO) mice. On the other hand, GluK2/3 signal at the stratum radiatum (SM) was elevated in GluK4/5 DKO mice. (C) GluK2/3 distribution in the dentate gyrus was unaltered (G; ML = molecular layer, GCL = granular layer). (D, E) Immunostaining of hippocampal sections after pepsin treatment. In wild-type mice (WT), a strong GluK2/3 signal was detected at the stratum lucidum, but not at the stratum radiatum or stratum pyramidale (SP). This highly compartmentalized pattern was abolished in GluK4/5 DKO mice, but was preserved in Neto1/2 DKO with a slight increase in the GluK2/3 signal at the SP. Images represent GluK2/3 localization at lower (D) and higher (E) magnifications. Scale bars: 100 μm (A, D), 50 μm (E), 25 μm (B, C). (F, G) Protein levels in the PSD fraction (F) and total (G) were measured in hippocampus (n = 5). Protein levels of KAR components (GluK2/3 and Neto1) were significantly reduced in the PSD fraction of GluK4/5 DKO, but total expression was unaltered. (H, I) Protein levels in the PSD fraction (H) and total (I) were measured in hippocampus (n = 3-4). Protein levels of KAR component (Neto1) were further reduced in the PSD fraction of GluK2.A1c KI; GluK4/5 DKO triple-mutant mice, but total expression was unaltered. (J, K) Immuno-electron microscopic images of Neto1 protein. PSDs are indicated by arrowheads. Inserts show high-magnification of labeled synapses. Scale bars = 200 nm (J) Neto1 was detected at hippocampal MF-CA3 synapses in WT mice, but not in Neto1 KO mice. On the other hand, Neto1 was reduced in GluK2.A1c KI mice. No Neto1 signal was detected in GluK4/5 DKO and GluK2.A1c KI; GluK4/5 DKO triple-mutant mice. (K) GluK2.A1cyto signal detected by anti GluA1 antibody was detected in GluK2.A1c KI; GluA1 KO double-mutant mice, but not in GluK2.A1c KI; GluK4/5 DKO, GluA1 KO quadruple-mutant mice. Numbers of immunogold-labeled synapses and total analyzed synapses are indicated in parentheses. Data are given as mean ± s.e.m. *, P < 0.05; ** P < 0.01; ***, P < 0.001 (Student’s t-test).
Epitopes of postsynaptic proteins such as PSD-95 are sometimes masked by high-density protein networks at the PSD; consequently, pepsin treatment can often enhance the signal from synaptic proteins, presumably by exposing buried epitopes (Fukaya and Watanabe, 2000). We therefore further examined protein distribution in hippocampal sections treated with pepsin. In pepsin-treated sections, strong GluK2 and GluK5 signal at the stratum lucidum were detected in wild-type and Neto1/2 DKO mice, with a slight increase in the GluK2 signal at the stratum pyramidale in Neto1/2 DKOs (Figure 6D and E, S5A and B). Consistent with our initial observations (Figure 6A and B), GluK2 and GluK5 signal was substantially reduced at the stratum lucidum of GluK4/5 DKO mice (Figure 6D and E, S5A and B), whereas no alterations in GluK2 were observed in the cerebellum of GluK4/5 DKO (Figure S5C and D). These results indicate that synapse specificity of KARs in hippocampal CA3 pyramidal cells is mediated by the GluK4/5 high-affinity subunits.
Previous work in GluK4/5 DKO mice revealed the abolishment of KAR-mediated EPSCs, and a reduced number of GluK2 immuno-electron gold particles at the hippocampal mossy fiber–CA3 pyramidal cell synapses (Fernandes et al., 2009). Our complementary biochemical assay revealed that protein levels of the KAR components GluK2/3 and Neto1 were reduced in the hippocampal PSD fraction of GluK4/5 DKO mice (Figure 6F), with no changes in total protein levels (Figure 6G). However, approximately 30% of the KARs were still detected in the PSD fraction of GluK4/5 DKO mice (Figure 6F). In GluK2A1c: GluK4: GluK5 triple-mutant mice we saw a further reduction of Neto1 in the PSD fraction (Figure 6H) without changes in total protein levels (Figure 6I), suggesting that the GluK2 cytoplasmic domain and GluK4/5 have mutually exclusive roles in localizing KARs to mossy fiber synapses and stabilizing the receptors in the PSD.
Since the biochemical fraction does not provide spatial information, we further analyzed the localization of Neto1 in GluK2.A1c; GluK4; GluK5 triple-mutant mice by post-embedding immuno-electron microscopy. Consistent with reduced synaptic activity and expression of KARs (Figure 4B and D), Neto1 expression in hippocampal mossy fiber-CA3 cell synapses was decreased in GluK2.A1c KI mice, and almost completely disappeared in Neto1 KO mice (Figure 6J). Under the same conditions, Neto1 signal was not detected in either GluK4/5 DKO or GluK4/5:GluK2.A1c triple mutant mice (Figure 6J). Specific reduction of GluK2.A1cyto was also confirmed by comparing GluK2.A1 signal in GluK2.A1c; GluA1 KO double-mutant mice and GluK2.A1c; GluA1; GluK4/5 quadruple-mutant mice (Figure 6K). Together with the observed changes in GluK2 distribution (Figure 6A and B), these data indicate that GluK4/5 primarily determines synapse specificity while there is an independent role in stabilization of KARs at synapses by the GluK2 cytoplasmic domain in CA3 pyramidal cells in the hippocampus.
The extracellular domain of GluK5 determines synapse specificity
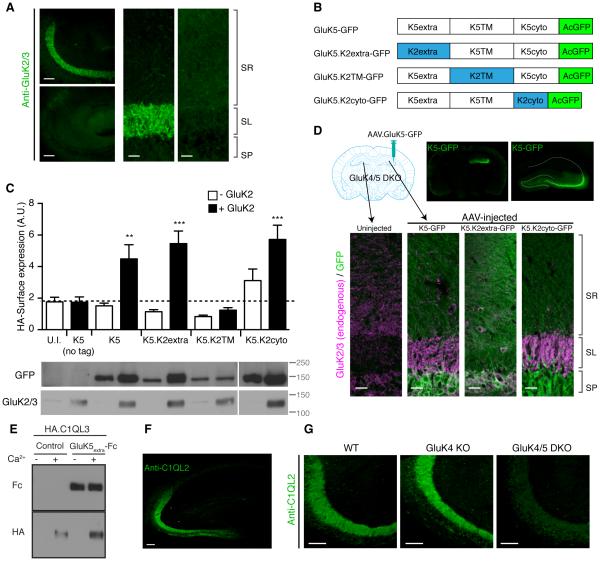
We next examined which domain of GluK4/5 is required for synapse specificity. To this end, we took an approach of rescuing synapse specificity in GluK4/5 DKO by re-introducing GluK4/5 mutants. We confirmed that loss of GluK5 expression in GluK4 KO mice further reduced GluK2 signal in the stratum lucidum (Figure 7A). Thus, we generated GFP-tagged chimeric constructs by swapping domains between GluK5 and GluK2 (Figure 7B). We specifically sought to identify a domain substitution that results in loss of synaptic specificity without deficits in surface expression. Thus, we first examined surface expression of the chimeric receptor in cRNA-injected oocytes. Extracellularly HA-tagged GluK5-GFP was expressed at the cell surface when co-expressed with GluK2 (Figure 7C). Similarly, chimeric HA-GluK5 with the extracellular domain of GluK2 (HA-K5.K2extra) or with the cytoplasmic domain of GluK2 without the epitope of anti GluK2/3 antibody (HA-K5.K2cyto) showed normal surface expression when co-expressed with GluK2. In contrast, the HA-GluK5 chimera with the transmembrane domains of GluK2 (HA-K5.K2TM) failed to express at the surface despite significant protein expression (Figure 7C). We concluded from this that HA-K5.K2TM has deficits in complex assembly and/or forward trafficking, and concentrated on examining the roles of the other two chimeras HA-K5.K2extra and HA-K5.K2cyto in vivo.
Figure 7. The GluK5 extracellular domain mediates synapse specificity.
(A) Immunostaining of hippocampal sections revealed substantial reduction of GluK2/3 signal in the stratum lucidum layer in GluK4/5 DKO compared to GluK4 KO mice. (B) Schematic diagram of chimeras of GluK5 and GluK2 with GFP at their C-terminus. (C) Surface expression of the extracellularly HA-tagged GluK5-GFP chimeras in cRNA-injected oocytes was measured using chemiluminescence assay. HA-K5-GFP alone did not express at the cell surface. On the other hand, GluK2 co-expression enhanced surface expression of HA-GluK5-GFP, HA-GluK5.K2extra-GFP and HA-GluK5.K2cyto-GFP, but not HA-GluK5.K2TM-GFP (n = 6-8). Expression of chimeric proteins was confirmed by western blotting. (D) Upon stereotaxic injection of AAV carrying GluK5-GFP, GluK5-GFP signal was observed in AAV-injected hemispheres in GluK4/5 DKO hippocampus (top). Re-introducing GluK5-GFP and GluK5.K2cyto-GFP into GluK4/5 DKO restored the stratum lucidum localization of endogenous GluK2 (Magenta), whereas GluK5.K2extra-GFP failed. Composite images were shown. (E) HA-tagged C1QL3/nCLP3 bound to the GluK5 extracellular domain tagged with human Fc domain (GluK5extra-Fc). Two proteins expressed independently were mixed and pulled down with protein A-sepharose. HA-C1QL3 was pulled down with GluK5extra-Fc strongly, but not with bovine serum albumin (control). Addition of calcium (Ca2+) was required for their interaction. (F) Immunostaining of C1QL2/nCLP2 in the hippocampus resulted in a selective distribution at the stratum lucidum, mimicking the distribution pattern of KARs. (G) The stratum lucidum distribution of C1QL2 was markedly reduced in GluK4/5 DKO mice. Data in C are given as mean ± s.e.m. ** P < 0.01; ***, P < 0.001. Scale bars: 100 μm (A, left panels; F, G), 20 μm (A, right panel; D, bottom).
We generated adeno-associated virus (AAV) carrying each GluK5 chimera tagged with GFP under the synapsin promoter, and injected the viral particles stereotaxically into the hippocampus of GluK4/5 DKO mice (Figure 7D). 1-2 weeks following AAV delivery, clear GFP signal was observed at the CA3 region of the injected hemisphere (Figure 7D, top). Loss of GluK2/3 signal at the stratum lucidum in GluK4/5 DKO mice was rescued by re-introducing wild-type GluK5-GFP and the GluK5.K2cyto-GFP (Figure 7D). On the other hand, expression of the GluK5.K2extra-GFP failed to enrich endogenous GluK2 to the stratum lucidum, despite diffuse GFP signal through all the layers (Figure 7D). These results strongly suggest that the extracellular domain of GluK5 is required for synapse specificity, and the differential KAR localization at mossy fiber – CA3 synapses in the stratum lucidum.
This finding lead us to speculate that a molecule that interacts with the extracellular domain of GluK5, which is localized to mossy fiber terminals, acts to recruit KARs to this synapse. Previous reports have demonstrated that another member of the glutamate receptor family, the delta2 receptor, is clustered by binding to the secreted protein Cbln1 through its extracellular domain localizing these receptors to cerebellar PF-PC synapses (Matsuda et al., 2010; Uemura et al., 2010). Similar members of the C1QL family protein, C1QL2/nCLP2 and C1QL3/nCLP3, are expressed in hippocampal dentate granule cells and specifically localized to the stratum lucidum (Iijima et al., 2010; Shimono et al., 2010). We therefore asked whether this mossy fiber enriched protein might play a role in localizing KARs to mossy fiber synapses.
We first examined an interaction between C1QL3/nCLP3 and the extracellular domain of GluK5. We individually expressed either HA-tagged C1QL3/nCLP3 or the GluK5 extracellular domain tagged with human Fc in FreeStyle HEK cells. The media containing the secreted proteins were mixed, followed by pull-down with protein A-beads. We found that the GluK5extra-Fc pull-downed HA-C1QL3/nCLP3 compared to control protein (bovine serum albumin) in a calcium dependent manner (Figure 7E). Similar calcium–dependent oligomerization and interaction of C1QL family proteins with other potential receptors has previously been reported (Bolliger et al., 2011; Kakegawa et al., 2015; Ressl et al., 2015). We next confirmed that C1QL2/nCLP2 signal is enriched in the stratum lucidum of wild-type mice using anti C1QL2 antibody (Figure 7F). Interestingly, we found that enrichment of the C1QL2/nCLP2 at the stratum lucidum was significantly reduced in GluK4/5 DKO compared to GluK4 KO mice (Figure 7G) suggesting that the loss of the high-affinity subunits disrupts the localization of C1QL2/nCLP2. Together, our results support the model that the extracellular domain of GluK5 binds to C1QLs secreted from mossy fiber terminals, and this interaction acts to promote synapse-specific localization of KARs at mossy fiber synapses in the stratum lucidum (Figure S6).
DISCUSSION
We found that mechanisms of synaptic stabilization and synapse specificity in the cerebellum and hippocampus are controlled by distinct complex components and receptor subunit domains (Figure S6). The intracellular domain of the low-affinity GluK2 subunit stabilizes receptors at synapses without changing total and surface expression levels. On the other hand, synapse-specific distribution of KARs in the hippocampal stratum lucidum was disrupted in the GluK4/5 DKO, and is dependent upon the extracellular domain of GluK5 that binds to the mossy fiber-enriched, secreted C1QL family of proteins.
Mechanisms of stabilization of KARs at synapses
Our systematic analysis of the roles of KAR components in the brain revealed that the cytoplasmic domain of the GluK2 low-affinity subunit is the domain responsible for stabilizing KARs at synapses. Several of the KAR subunits and auxiliary subunits have been implicated in the synaptic localization of KARs (Copits et al., 2011; Hirbec et al., 2003; Palacios-Filardo et al., 2014; Tang et al., 2012; Yan et al., 2004), but defining a specific domain involved in synaptic stabilization especially at anatomically defined synapses has not been possible. Generation of mice expressing a modified GluK2 subunit demonstrated that the carboxyl terminal cytoplasmic domain of GluK2 does not affect the surface (or total) expression of KARs and has no effect on the total current density of neuronal KARs. These results suggest that the loss or reduction of synaptic localization and activity of KARs in the cerebellum and hippocampus of GluK2.A1c KI mice is due to loss of synaptic stabilization.
A general concern with the approach of germline manipulation is that there may be unknown or uncontrolled developmental compensation in these mice. While this is still a possibility, it is important to take this approach to validate pioneering studies in expression systems, and our results highlight some differences with what has been observed in culture. For instance, it has been demonstrated in heterologous cells and primary cultured neurons that phosphorylation sites on the C-terminal cytoplasmic domain regulate surface expression of GluK2 (Nasu-Nishimura et al., 2010). However, we found no changes in surface expression and current density of KARs in neurons in GluK2.A1c KI mice (Figure 4H and I). The cytoplasmic domain of GluA1 that was selected for the chimeric construct may itself have a trafficking signal for surface expression. Generation of chimeric constructs with other cytoplasmic domains may provide further insight.
Mechanisms for synapse specificity of KAR in the hippocampus
KARs in hippocampal CA3 pyramidal neurons localize to the stratum lucidum, where mossy fiber axons form glutamatergic synapses. However, the mechanisms that underlie this restricted synapse-specific localization have remained unknown. Here, we demonstrate that the extracellular domain of the high-affinity GluK5 subunit contributes to the synapse specific localization of KARs through an interaction with a secreted, mossy fiber-enriched protein, C1QL. Indeed, GluK4/5 DKO mice exhibit no kainate-mediated synaptic currents at MF-CA3 synapses (Fernandes et al., 2009). The absence of kainate-mediated transmission in GluK4/5 DKO mice could be due to loss of synaptic KARs or a change in the functional properties of the KAR complex. At cerebellar mossy fiber–granule cell synapses in GluK5 KO mice, there is a loss of KAR-mediated transmission, although the synaptic localization of GluK2 is unaltered (Yan et al., 2013). At these synapses, GluK5 forms a complex with GluK2 and Neto2, yielding a KAR complex that is functional (Yan et al., 2013). Combined with these observations, loss of KAR-mediated signaling in GluK4/5 DKO mice may be due to altered function as well as reduction in the localization of synaptic receptors.
It has long been established that lesioning granule cells with colchinine and subsequent loss of the mossy fiber axons results in the loss of high affinity [3H] kainate labeling, presumably reflecting loss of KARs in the stratum lucidum (Represa et al., 1987). Though it was not clear if this reflected redistribution of postsynaptic KARs, our data would suggest that the presynaptic axons and possibly associated factors are required for the localization of KARs to the stratum lucidum. Here we demonstrate that one potential factor associated with mossy fiber axons, the secreted C1QL/nCLP protein family interacts with GluK4/5-containing receptors at the stratum lucidum in CA3 pyramidal cells.
Input-specific synaptic stabilization of KARs
In this work, we show that input-specific synaptic localization of KARs observed in the hippocampus, but not in cerebellum, is mediated by two distinct mechanisms: synaptic stabilization through the GluK2 cytoplasmic domain and targeting to mossy fiber through the GluK5 extracellular domain. It is possible that the GluK2 cytoplasmic domain is required for stable synaptic localization, analogous to the roles of the TARP PDZ-binding domain of synaptic AMPAR (Bats et al., 2007; Chen et al., 2000; Schnell et al., 2002; Sumioka et al., 2011), whereas GluK4/5 accumulates at high levels around synapses at the stratum lucidum and is inserted into synapses by diffusion, rather than being captured at the synapse. In support of this idea, CaMKII phosphorylation of GluK5 modulates lateral diffusion of KARs during plasticity (Carta et al., 2013). In GluK2.A1c KI mice there are no changes in both the surface expression of the other KAR subunits and total current density, but the synaptic KAR currents are reduced, supporting the idea that extrasynaptic KARs at the neuron surface are normal but may not be captured efficiently at the PSD.
In order to sustain proper neuronal function, synaptic proteins must localize to the appropriate types of synapses. Our dissection of the KAR complex uncovered processes involved in synapse-specific localization of KARs and revealed mechanisms underlying synaptic stabilization. Further investigations, and in particular further delineation of the interacting proteins that mediate each mechanism, and whether there exists a general model for KAR synapse localization, will reveal fundamental features of synaptic receptor proteins and their regulation within multiple types of synaptic connections.
EXPERIMENTAL PROCEDURES
Animals
All animal experiments were carried out in accordance with protocols approved by the Institutional Animal Care and Use Committees of both Yale University and Northwestern University, following guidelines described, in accordance with National Institutes of Health guidelines. GluK2.A1cyto knock-in mice was generated with standard procedures using homologous recombinations of ES cells, and details are described in Supplementary Experimental Procedures.
Biochemical analysis
PSD fraction and co-immunoprecipitation were performed as described (Zhang et al., 2009, Straub et al., 2011). Fractions were analyzed by SDS-PAGE and western blotting, and samples were adjusted by protein amount. Antibodies used are listed in Supplementary Experimental Procedures.
Immunostaining
Immunostaining and immunoelectron microscopy were done as described previously (Straub et al., 2011). Step-by-step procedures are detailed in Supplementary Experimental Procedures.
Electrophysiology
Whole cell recording of acute cerebellar slices was performed as described (Yan et al, 2013). Horizontal slices from the ventral hippocampus (350 µm) were prepared from mice aged P19 – P27 as previously described (Fernandes et al., 2009). Patch-clamp recordings of glutamate-evoked currents in outside-out patches were done at room temperature with an EPC-9 amplifier (HEKA) and Patch Master acquisition software, essentially as described (Robert and Howe, 2003). Measurements of surface expression and activity of receptors expressed in oocytes were performed as described (Zhang et al., 2009, Straub et al., 2011). Further details are described in Supplementary Experimental Procedures.
Statistical analysis
All data are given as the mean ± standard error of the mean (s.e.m.). Statistical significance was calculated using the unpaired Student’s t-test or one-way ANOVA, as indicated.
Supplementary Material
HIGHLIGHTS.
The GluK2 cytoplasmic domain mediates synaptic stabilization.
Surface kainate receptor activity depends on GluK2, but not its cytoplasmic domain.
The extracellular domain of high-affinity GluK subunits mediates synaptic specificity.
Input-specific synaptic localization of kainate receptors is mediated by two mechanisms.
eTOC Blurb.
Synaptic communication between neurons requires neurotransmitter receptors to be localized precisely to the correct synapse type. Straub et al. identify two distinct mechanisms that lead to input-specific synaptic localization of the kainate receptor complex in the brain.
ACKNOWLEDGEMENTS
The authors thank the members of the Tomita lab and the Contractor Lab for helpful discussions. We thank Dr. Megumi Morimoto-Tomita for generating various reagents, Dr. S.F. Heinemann (Salk Institute), Dr. Peter Seeburg (Max Planck Institute), Deltagen, Genentech, KOMP for generating each of GluK2/4/5 knockouts, GluA1 knockout, Neto2 knockout, Neto1 knockout mice, and Jackson laboratory and MMRRC for maintaining GluK2, GluK5, Neto2 knockout and stargazer mice. Parts of this study were supported by NIH/NIMH R01 MH085080 and the Yale fund (S.T.) and NIH/NIMH R01 MH099114 (A.C.). M.W. is supported by Grants-in-Aid for Scientific Research provided by the Ministry of Education, Culture, Sports, Science, and Technology of Japan. C.S. was supported by a Boehringer-Ingelheim Fonds PhD fellowship, and Y.N was supported by NIH/NINDS NRSA post-doctoral fellowship (F32 NS093952).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
S.T. conceived the project and S.T. and A.C. wrote the manuscript. C.S. and Y.N. performed and analyzed biochemistry and immunohistochemistry. C.S. and S.T. generated GluK2 knockin mice. Y.N. generated and analyzed GluK5 chimeras and AAV-injected animals. D.Y. performed and analyzed cerebellar slice physiology. T.N. H.B.F. and A.C. performed and analyzed hippocampal slice physiology. M.Y. and M.W. performed and analyzed immuno-electron microscopy. P.Z. and J.R.H. performed the fast application experiments in outside-out patches. All authors contributed to the final version of the manuscript.
REFERENCES
- Bats C, Groc L, Choquet D. The interaction between Stargazin and PSD-95 regulates AMPA receptor surface trafficking. Neuron. 2007;53:719–734. doi: 10.1016/j.neuron.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Bolliger MF, Martinelli DC, Sudhof TC. The cell-adhesion G protein-coupled receptor BAI3 is a high-affinity receptor for C1q-like proteins. Proc Natl Acad Sci U S A. 2011;108:2534–2539. doi: 10.1073/pnas.1019577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carta M, Opazo P, Veran J, Athane A, Choquet D, Coussen F, Mulle C. CaMKII-dependent phosphorylation of GluK5 mediates plasticity of kainate receptors. EMBO J. 2013;32:496–510. doi: 10.1038/emboj.2012.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo PE, Malenka RC, Nicoll RA. Kainate receptors mediate a slow postsynaptic current in hippocampal CA3 neurons. Nature. 1997;388:182–186. doi: 10.1038/40645. [DOI] [PubMed] [Google Scholar]
- Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408:936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- Christensen JK, Paternain AV, Selak S, Ahring PK, Lerma J. A mosaic of functional kainate receptors in hippocampal interneurons. J Neurosci. 2004;24:8986–8993. doi: 10.1523/JNEUROSCI.2156-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contractor A, Mulle C, Swanson GT. Kainate receptors coming of age: milestones of two decades of research. Trends Neurosci. 2011;34:154–163. doi: 10.1016/j.tins.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contractor A, Sailer AW, Darstein M, Maron C, Xu J, Swanson GT, Heinemann SF. Loss of kainate receptor-mediated heterosynaptic facilitation of mossy-fiber synapses in KA2−/− mice. J Neurosci. 2003;23:422–429. doi: 10.1523/JNEUROSCI.23-02-00422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copits BA, Robbins JS, Frausto S, Swanson GT. Synaptic Targeting and Functional Modulation of GluK1 Kainate Receptors by the Auxiliary Neuropilin and Tolloid-Like (NETO) Proteins. J Neurosci. 2011;31:7334–7340. doi: 10.1523/JNEUROSCI.0100-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darstein M, Petralia RS, Swanson GT, Wenthold RJ, Heinemann SF. Distribution of kainate receptor subunits at hippocampal mossy fiber synapses. J Neurosci. 2003;23:8013–8019. doi: 10.1523/JNEUROSCI.23-22-08013.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes HB, Catches JS, Petralia RS, Copits BA, Xu J, Russell TA, Swanson GT, Contractor A. High-affinity kainate receptor subunits are necessary for ionotropic but not metabotropic signaling. Neuron. 2009;63:818–829. doi: 10.1016/j.neuron.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster AC, Mena EE, Monaghan DT, Cotman CW. Synaptic localization of kainic acid binding sites. Nature. 1981;289:73–75. doi: 10.1038/289073a0. [DOI] [PubMed] [Google Scholar]
- Fukaya M, Watanabe M. Improved immunohistochemical detection of postsynaptically located PSD-95/SAP90 protein family by protease section pretreatment: a study in the adult mouse brain. J Comp Neurol. 2000;426:572–586. [PubMed] [Google Scholar]
- Hashimoto K, Fukaya M, Qiao X, Sakimura K, Watanabe M, Kano M. Impairment of AMPA receptor function in cerebellar granule cells of ataxic mutant mouse stargazer. J Neurosci. 1999;19:6027–6036. doi: 10.1523/JNEUROSCI.19-14-06027.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirbec H, Francis JC, Lauri SE, Braithwaite SP, Coussen F, Mulle C, Dev KK, Coutinho V, Meyer G, Isaac JT, et al. Rapid and differential regulation of AMPA and kainate receptors at hippocampal mossy fibre synapses by PICK1 and GRIP. Neuron. 2003;37:625–638. doi: 10.1016/s0896-6273(02)01191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima T, Miura E, Watanabe M, Yuzaki M. Distinct expression of C1q-like family mRNAs in mouse brain and biochemical characterization of their encoded proteins. Eur J Neurosci. 2010;31:1606–1615. doi: 10.1111/j.1460-9568.2010.07202.x. [DOI] [PubMed] [Google Scholar]
- Isaac JT, Mellor J, Hurtado D, Roche KW. Kainate receptor trafficking: physiological roles and molecular mechanisms. Pharmacol Ther. 2004;104:163–172. doi: 10.1016/j.pharmthera.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Kakegawa W, Mitakidis N, Miura E, Abe M, Matsuda K, Takeo YH, Kohda K, Motohashi J, Takahashi A, Nagao S, et al. Anterograde C1ql1 signaling is required in order to determine and maintain a single-winner climbing fiber in the mouse cerebellum. Neuron. 2015;85:316–329. doi: 10.1016/j.neuron.2014.12.020. [DOI] [PubMed] [Google Scholar]
- Keinanen K, Wisden W, Sommer B, Werner P, Herb A, Verdoorn TA, Sakmann B, Seeburg PH. A family of AMPA-selective glutamate receptors. Science. 1990;249:556–560. doi: 10.1126/science.2166337. [DOI] [PubMed] [Google Scholar]
- Kumar J, Schuck P, Mayer ML. Structure and assembly mechanism for heteromeric kainate receptors. Neuron. 2011;71:319–331. doi: 10.1016/j.neuron.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerma J, Paternain AV, Naranjo JR, Mellstrom B. Functional kainate-selective glutamate receptors in cultured hippocampal neurons. Proc Natl Acad Sci U S A. 1993;90:11688–11692. doi: 10.1073/pnas.90.24.11688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Miura E, Miyazaki T, Kakegawa W, Emi K, Narumi S, Fukazawa Y, Ito-Ishida A, Kondo T, Shigemoto R, et al. Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science. 2010;328:363–368. doi: 10.1126/science.1185152. [DOI] [PubMed] [Google Scholar]
- Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S. Structural mechanism of glutamate receptor activation and desensitization. Nature. 2014;514:328–334. doi: 10.1038/nature13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan DT, Cotman CW. The distribution of [3H]kainic acid binding sites in rat CNS as determined by autoradiography. Brain Res. 1982;252:91–100. doi: 10.1016/0006-8993(82)90981-7. [DOI] [PubMed] [Google Scholar]
- Mulle C, Sailer A, Perez-Otano I, Dickinson-Anson H, Castillo PE, Bureau I, Maron C, Gage FH, Mann JR, Bettler B, et al. Altered synaptic physiology and reduced susceptibility to kainate-induced seizures in GluR6-deficient mice. Nature. 1998;392:601–605. doi: 10.1038/33408. [DOI] [PubMed] [Google Scholar]
- Nasu-Nishimura Y, Hurtado D, Braud S, Tang TT, Isaac JT, Roche KW. Identification of an endoplasmic reticulum-retention motif in an intracellular loop of the kainate receptor subunit KA2. J Neurosci. 2006;26:7014–7021. doi: 10.1523/JNEUROSCI.0573-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasu-Nishimura Y, Jaffe H, Isaac JT, Roche KW. Differential regulation of kainate receptor trafficking by phosphorylation of distinct sites on GluR6. J Biol Chem. 2010;285:2847–2856. doi: 10.1074/jbc.M109.081141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll RA, Schmitz D. Synaptic plasticity at hippocampal mossy fibre synapses. Nat Rev Neurosci. 2005;6:863–876. doi: 10.1038/nrn1786. [DOI] [PubMed] [Google Scholar]
- Palacios-Filardo J, Aller MI, Lerma J. Synaptic Targeting of Kainate Receptors. Cerebral cortex. 2014 doi: 10.1093/cercor/bhu244. [DOI] [PubMed] [Google Scholar]
- Petralia RS, Wang YX, Wenthold RJ. Histological and ultrastructural localization of the kainate receptor subunits, KA2 and GluR6/7, in the rat nervous system using selective antipeptide antibodies. J Comp Neurol. 1994;349:85–110. doi: 10.1002/cne.903490107. [DOI] [PubMed] [Google Scholar]
- Represa A, Tremblay E, Ben-Ari Y. Kainate binding sites in the hippocampal mossy fibers: localization and plasticity. Neuroscience. 1987;20:739–748. doi: 10.1016/0306-4522(87)90237-5. [DOI] [PubMed] [Google Scholar]
- Ressl S, Vu BK, Vivona S, Martinelli DC, Sudhof TC, Brunger AT. Structures of C1q-like proteins reveal unique features among the C1q/TNF superfamily. Structure. 2015;23:688–699. doi: 10.1016/j.str.2015.01.019. [DOI] [PubMed] [Google Scholar]
- Robert A, Howe JR. How AMPA receptor desensitization depends on receptor occupancy. J Neurosci. 2003;23:847–858. doi: 10.1523/JNEUROSCI.23-03-00847.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Sachidhanandam S, Utvik JK, Coussen F, Mulle C. Distinct subunits in heteromeric kainate receptors mediate ionotropic and metabotropic function at hippocampal mossy fiber synapses. J Neurosci. 2005;25:11710–11718. doi: 10.1523/JNEUROSCI.4041-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell E, Sizemore M, Karimzadegan S, Chen L, Bredt DS, Nicoll RA. Direct interactions between PSD-95 and stargazin control synaptic AMPA receptor number. Proc Natl Acad Sci U S A. 2002;99:13902–13907. doi: 10.1073/pnas.172511199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono C, Manabe R, Yamada T, Fukuda S, Kawai J, Furutani Y, Tsutsui K, Ikenaka K, Hayashizaki Y, Sekiguchi K. Identification and characterization of nCLP2, a novel C1q family protein expressed in the central nervous system. Journal of biochemistry. 2010;147:565–579. doi: 10.1093/jb/mvp203. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub C, Hunt DL, Yamasaki M, Kim KS, Watanabe M, Castillo PE, Tomita S. Distinct functions of kainate receptors in the brain are determined by the auxiliary subunit Neto1. Nat Neurosci. 2011;14:866–873. doi: 10.1038/nn.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumioka A, Brown TE, Kato AS, Bredt DS, Kauer JA, Tomita S. PDZ binding of TARPgamma-8 controls synaptic transmission but not synaptic plasticity. Nat Neurosci. 2011;14:1410–1412. doi: 10.1038/nn.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Ivakine E, Mahadevan V, Salter MW, McInnes RR. Neto2 interacts with the scaffolding protein GRIP and regulates synaptic abundance of kainate receptors. PloS one 7, e51433. 2012 doi: 10.1371/journal.pone.0051433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Pelkey KA, Ng D, Ivakine E, McBain CJ, Salter MW, McInnes RR. Neto1 Is an Auxiliary Subunit of Native Synaptic Kainate Receptors. J Neurosci. 2011;31:10009–10018. doi: 10.1523/JNEUROSCI.6617-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Fukata M, Nicoll RA, Bredt DS. Dynamic interaction of stargazin-like TARPs with cycling AMPA receptors at synapses. Science. 2004;303:1508–1511. doi: 10.1126/science.1090262. [DOI] [PubMed] [Google Scholar]
- Uemura T, Lee SJ, Yasumura M, Takeuchi T, Yoshida T, Ra M, Taguchi R, Sakimura K, Mishina M. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- Vignes M, Collingridge GL. The synaptic activation of kainate receptors. Nature. 1997;388:179–182. doi: 10.1038/40639. [DOI] [PubMed] [Google Scholar]
- Wyeth MS, Pelkey KA, Petralia RS, Salter MW, McInnes RR, McBain CJ. Neto auxiliary protein interactions regulate kainate and NMDA receptor subunit localization at mossy fiber-CA3 pyramidal cell synapses. J Neurosci. 2014;34:622–628. doi: 10.1523/JNEUROSCI.3098-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Yamasaki M, Straub C, Watanabe M, Tomita S. Homeostatic control of synaptic transmission by distinct glutamate receptors. Neuron. 2013;78:687–699. doi: 10.1016/j.neuron.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Sanders JM, Xu J, Zhu Y, Contractor A, Swanson GT. A C-terminal determinant of GluR6 kainate receptor trafficking. J Neurosci. 2004;24:679–691. doi: 10.1523/JNEUROSCI.4985-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, St-Gelais F, Grabner CP, Trinidad JC, Sumioka A, Morimoto-Tomita M, Kim KS, Straub C, Burlingame AL, Howe JR, et al. A transmembrane accessory subunit that modulates kainate-type glutamate receptors. Neuron. 2009;61:385–396. doi: 10.1016/j.neuron.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.