Abstract
We report the main characteristics of “Lachnoclostridium bouchesdurhonense” strain AT5T (=CSUR P2181), a new bacterial species isolated from the gut microbiota of an obese patient from Marseille.
Keywords: Culturomics, gut microbiota, “Lachnoclostridium bouchesdurhonense”, obese, taxonogenomics
In order to describe the bacterial flora of the gastrointestinal tract, a stool sample was collected from a 38-year-old obese patient from France. The patient, who provided informed oral consent, had a body mass index of 33 kg/m2. The study was approved by the ethics committee of the Institut Fédératif de Recherche 48, Marseille. The stool was cultivated using the culturomics approach [1], [2]. The culture of strain AT5T was achieved on 5% sheep’s blood–enriched Columbia agar (bioMérieux, Marcy l’Etoile, France). The colonies obtained were small, circular and smooth, with a mean diameter of 0.5 mm. Bacterial cells were Gram negative and rod shaped. Strain AT5T did not exhibit catalase and oxidase activities.
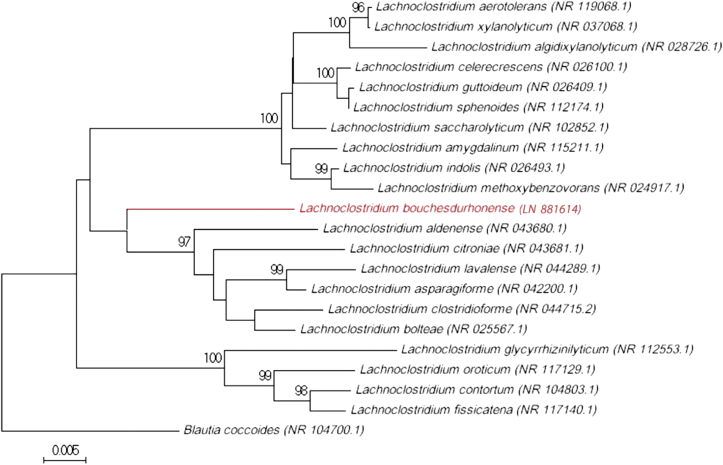
Colonies were not identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) using a Microflex spectrometer (Bruker Daltonics, Bremen, Germany) [3], [4]. Therefore, we sequenced the 16S rRNA gene of strain AT5T using the fD1-rP2 primers as previously described [5] and a 3130-XL sequencer (Applied Biosciences, Saint Aubin, France). The obtained sequence was 94.79% similar to the 16S rRNA gene sequence of Lachnoclostridium bolteae strain 16351 (GenBank accession number NR_025567) (Figure 1) [6]. According to the 16S rRNA gene sequence similarity for species demarcation of prokaryotes [7], [8], we propose that strain AT5T is representative of a new species within the recently described Lachnoclostridium genus [9] for which we propose the name “Lachnoclostridium bouchesdurhonense” sp. nov. (bouch.du.rhon.ense, L. fem. adj. bouchedurhonense, for Bouches-du-Rhône, the department where the city of Marseille is located, where strain AT5T was isolated).
Fig. 1.
Phylogenetic tree showing position of “Lachnoclostridium bouchesdurhonense” strain AT5T relative to other phylogenetically close species with validly published name. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values (>95%) obtained by repeating analysis 500 times to generate majority consensus tree. Blautia coccoides was used as outgroup. Scale bar indicates 5% nucleotide sequence divergence.
MALDI-TOF MS spectrum
The MALDI-TOF MS spectrum of “Lachnoclostridium bouchesdurhonense” is available online (http://www.mediterranee-infection.com/article.php?laref=256&titre=urms-database).
Nucleotide sequence accession number
The 16S rRNA gene sequence was deposited in GenBank under accession number LN881614.
Deposit in a culture collection
Strain AT5T was deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR) under number P2181.
Acknowledgement
This study was funded by the Fondation Méditerranée Infection.
Conflict of Interest
None declared.
References
- 1.Lagier J.C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 2.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.E., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 4.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore W.E.C., Johnson J.L., Holdeman L.V. Emendation of Bacteroidaceae and Butyrivibrio and descriptions of Desulfomonas gen. nov. and ten new species in the genera Desulfomonas, Butyrivibrio, Eubacterium, Clostridium, and Ruminococcus. Int J Syst Evol Microbiol. 1976;26:238–252. [Google Scholar]
- 7.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152. [Google Scholar]
- 8.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 9.Yutin N., Galperin M.Y. A genomic update on clostridial phylogeny: gram-negative spore formers and other misplaced clostridia. Environ Microbiol. 2013;15:2631–2641. doi: 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]