Abstract
Recent studies have detailed the genomic landscape of primary endometrial cancers, but their evolution into metastases has not been characterized. We performed whole-exome sequencing of 98 tumor biopsies including complex atypical hyperplasias, primary tumors, and paired abdominopelvic metastases to survey the evolutionary landscape of endometrial cancer. We expanded and reanalyzed TCGA-data, identifying novel recurrent alterations in primary tumors, including mutations in the estrogen receptor cofactor NRIP1 in 12% of patients. We found that likely driver events tended to be shared by primary and metastatic tissue-samples, with notable exceptions such as ARID1A mutations. Phylogenetic analyses indicated that the sampled metastases typically arose from a common ancestral subclone that was not detected in the primary tumor biopsy. These data demonstrate extensive genetic heterogeneity within endometrial cancers and relative homogeneity across metastatic sites.
Keywords: Cancer, Metastasis, Precursor, Endometrial cancer, Cancer genomics, Cancer evolution
Introduction
Endometrial cancer is the most common pelvic gynecologic malignancy in industrialized countries, partially due to the obesity epidemic1. Seventy-five percent of patients present with type I, endometrioid, tumors2, often with adjacent regions containing complex atypical hyperplasia (CAH), considered precursor lesions. Type I tumors are often estrogen responsive and portend a good prognosis. Type II tumors are the non-endometrioid subtypes, including the serous, carcinosarcoma, clear cell and undifferentiated histologies. These tend to occur in older, non-obese women, are rarely estrogen responsive, and carry a poor prognosis.
Recent large-scale sequencing studies of primary endometrioid and serous tumors have indicated that the difference in phenotype is reflected in distinct molecular subgroups, with further molecular subclustering3–5. While these studies detailed the patterns of somatic alterations across primary tumors, a comparative study of samples from endometrial CAH, primary tumors, and paired metastases has not been performed. It is not known whether metastases derive from the same or multiple lineages within the primary, or whether cancer cells require specific mutations that enable metastasis. The extent to which genetic events observed in a primary biopsy represent the full diversity of subclones across a metastatic cancer is also unknown. Such information would be helpful in understanding the biological underpinnings of endometrial cancer progression and to determine treatment strategies that target features that are homogenous throughout individual cancers6.
Here we address these questions using a collection of 98 extensively clinically annotated fresh-frozen samples ranging from precursor lesions to primary tumors and paired abdominopelvic metastases. We analyzed somatic mutations and allelic copy-number profiles between different biopsies from the same individual to reconstruct phylogenetic relationships and annotate putative cancer drivers across sites of disease. We also reanalyzed data from The Cancer Genome Atlas (TCGA) using updated methods, leading us to identify novel recurrent mutations in NRIP1 and patterns of heterogeneity within biopsies that mimic heterogeneity across multiple tumor sites.
Results
Patient and sample cohort
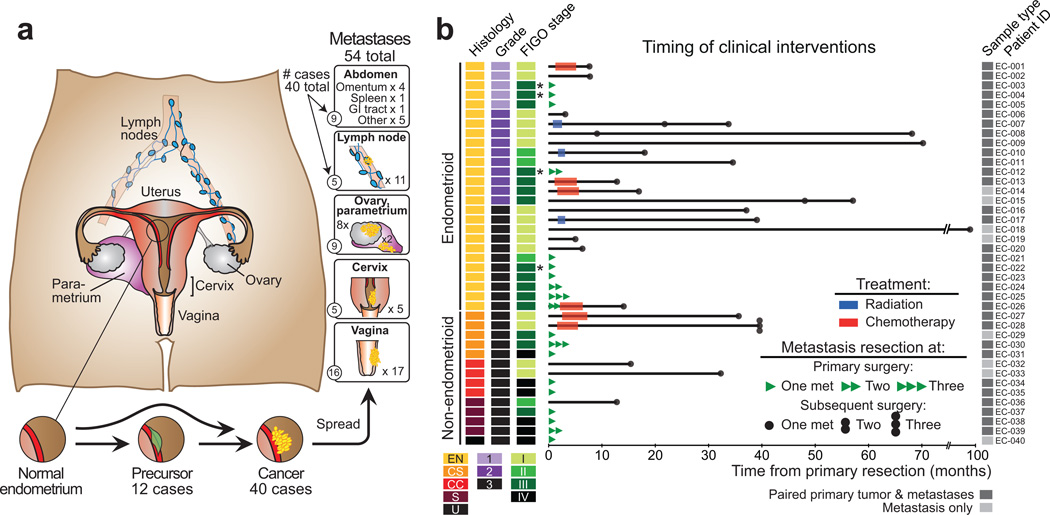
Our cohort consisted of a population-based patient series with extensive clinical annotation including complete follow-up information. We obtained fresh-frozen tumor tissue from 52 individuals: 12 with complex atypical endometrial hyperplasia (CAH) and 40 with metastatic endometrial cancer (EC). We analyzed 98 biopsies: 12 CAHs, 32 primary tumors, and 54 abdominopelvic metastases (Figure 1, Supplementary Table 1, and Supplementary Figures 1–2). Twenty-six primary tumors were associated with one or more paired metastases. Samples were subjected to whole-exome sequencing (WES) and/or Affymetrix SNP 6.0 arrays (Supplementary Figure 1). All samples undergoing WES were tested for microsatellite instability (MSI), enabling classification according to integrated molecular subgroups established by TCGA3 (Figure 2A, Supplementary Figure 2).
Figure 1. Samples assessed.
(a) Anatomic sites from which samples were obtained. (b) Histologic subtypes (EN: endometrioid, CC: clear cell, S: serous and U: undifferentiated carcinomas, and CS: carcinosarcoma), grade, FIGO 2009 stage at primary diagnosis, time of metastatic lesion sampling, and treatment after primary diagnosis. Asterisks indicate four cases that were clinically difficult to distinguish as metastatic or independent synchronous primary cancers at time of resection.
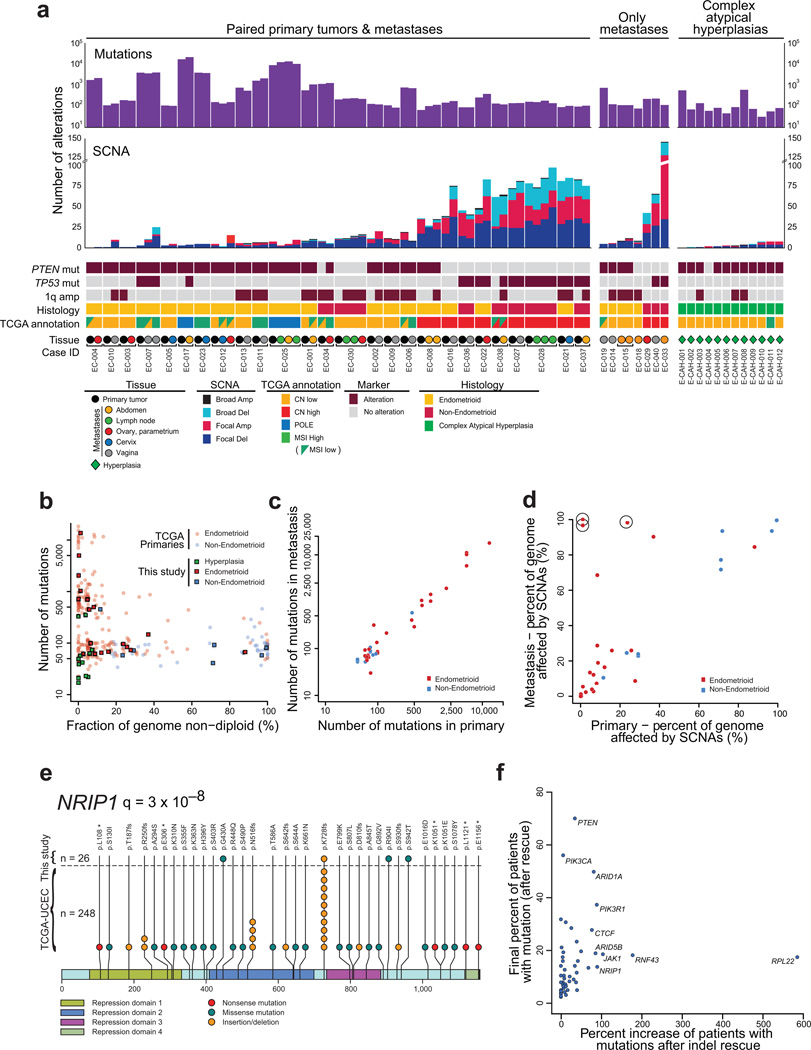
Figure 2. Somatic genetic alterations in complex atypical hyperplasias and primary and metastatic endometrial carcinomas.
(a) Number of exonic mutations (top) and SCNAs (middle) detected in each tumor biopsy. PTEN, TP53, and 1q amplification status, histologic subtype, TCGA annotation, and tissue type are indicated on the bottom. (b) Number of mutations (y-axis) against fraction of the genome affected by SCNAs (x-axis) across complex atypical hyperplasias (green), primary lesions from endometrioid endometrial carcinoma (red) and non-endometrioid endometrial carcinoma (blue); primaries from our dataset (squares) and TCGA (dots). (c) Number of mutations detected in the primary tumor compared to their metastatic counterpart. (d) Fraction of the genome affected by SCNAs in metastases (y-axis) relative to paired primaries (x-axis). Circles indicate metastases that exhibit whole-genome doubling not observed in the primary biopsy. (e) Stick plot depicting detected mutations in NRIP1, a cofactor of the estrogen receptor. (f) Impact of indel rescue on the percentage of patients harboring mutations in known driver genes.
Novel significantly mutated genes and hotspots
The burden of somatic genetic alterations in our primary tumors was consistent with endometrial cancers profiled by TCGA. We observed similar rates of somatic mutation (minimum 40 to maximum 13,717) and SCNAs (Figure 2A and Supplementary Figure 3A–B), and an inverse correlation between both (P = 0.005; Figure 2B)7. However, mutation rates of some of the most frequently altered genes differed, with higher mutation rates for PPP2R1A, FGFR2, PIK3CA, and ARID1A, and lower rates for PIK3R1 in our dataset compared to TCGA. This may reflect different sample inclusion strategies (Supplementary Figure 4).
The burden of small insertions/deletions (indels) detected among primary endometrial cancers was higher than previously noted, particularly among MSI carcinomas. MSI, prevalent among endometrial carcinomas, leads to high rates of these indels. However, these events are often observed at low-allelic fractions in the paired normal samples due to sequencing error, and are typically discarded as non-somatic. We applied recently developed methods to “rescue” highly recurrent indels that are enriched in tumor samples (Supplementary Figure 5A)8. We identified an average of 156 and 21 indels per MSI and non-MSI tumor, respectively, compared to 16 and 4.4 in prior analyses9. We subjected the indels detected by our rescue strategy to additional technical validation using six approaches and achieved a 96% validation rate (see Methods).
We used these new calls and the combined dataset of our primary tumors and those of TCGA (274 primaries in total), to catalogue the significantly mutated genes in primary endometrial cancer. We identified 49 genes with significantly recurrent rates of mutation (Supplementary Table 2, Supplementary Figure 5B), including 21 that have not been previously described in endometrial cancer3,9. Of the 21 novel genes, four (NFE2L2, ERBB2, U2AF1, and ALPK2) have been found to be recurrently mutated in other primary cancer types using similar analytic methods to those used here9.
The other 17 novel significantly recurrently mutated genes included both ESR1, encoding the estrogen receptor alpha, and its binding partner NRIP1. Alterations in the estrogen pathway are considered risk factors for endometrioid endometrial cancer10, and recurrent rearrangements involving ESR1 have recently been identified in breast cancer11. However, significantly recurrent point mutations in the estrogen pathway have not been previously described in cancers that had not received anti-estrogen therapy.
We found NRIP1 mutations in 12.5% of patients, concentrated in two highly recurrent sites, p.Lys728fs (n = 11) and p.Asn516fs (n = 4) (Figure 2E, Supplementary Figure 5C–D). All but two of these indels were in MSI samples (20% of MSI samples). In addition, 14% of colorectal MSI samples analyzed by TCGA exhibited NRIP1 indels. NRIP1 binds to the AF2 domain of the estrogen receptor and is essential for its transcriptional activity12,13.
Mutations in ESR1 were detected in 4% of cancers and clustered in the ligand-binding domain (Supplementary Figure 5E–F). These included p.Tyr537(Cys/Asn/Ser) mutations (three patients) that have been shown to cause constitutive activation and resistance to tamoxifen therapy in breast cancer14,15. However, the only patient in our cohort with a p.Tyr537 mutation never received anti-estrogen treatment. Moreover, the endometrial cancers profiled by the TCGA were untreated, and prior malignancies were an exclusion criterion. These observations indicate that p.Tyr537(Cys/Asn/Ser) mutations can occur in endometrial cancers without prior anti-estrogen treatment.
Additional novel genes also included MAX, the binding partner of MYC family members. We identified two recurrently mutated sites in MAX: p.His28Arg (n = 5) and p.Arg60Gln (n = 2, Supplementary Figure 6A) that also occur in other cancers and appear to interface with DNA16,17. We also observed and validated significantly recurrent mutations in MYCN, as previously noted9, with a hotspot at p.Pro44Leu (n = 5; Supplementary Figure 6B–C). Mutations of MAX and MYCN never co-occurred with each other or with amplifications of MYC or MYCN (p = 0.36, Supplementary Figure 6D). MYCN is also recurrently amplified in endometrial cancers (Supplementary Figure 6E).
Even among genes previously noted to harbor significantly recurrent alterations, we often detected higher rates of alteration than previously noted, likely due to rescued indels (Supplementary Table 2). For instance, we observed ARID1A mutations in at least one biopsy from 49% of patients, a 40% increase over prior estimates. Genes in which polymerase slippage-associated indels have previously been identified, such as RPL2218,19, RNF438 and JAK120, showed even more dramatic gains (370%, 206%, and 163% increases respectively; Figure 2F). Conversely, the number of patients with biopsies exhibiting PIK3CA or CTNNB1 mutations increased by only 5.6% and 0% respectively. Overall, we called 39% more mutations (all indels) across all genes and 54% more mutations in recurrently altered genes (p = 0.28).
The higher rates of alteration detected in these recurrently mutated genes motivated us to reevaluate their relationship among TCGA data to clinical features of these cancers such as survival (Supplementary Table 3). No gene achieved statistical significance. Biases in sample selection and other factors impact the results of this analysis.
PI3K pathway alterations predominate in hyperplasias
Compared to primary tumors, CAHs exhibited few somatic mutations with two highly mutated exceptions (median 35 mutations per sample, range 17–348; Figure 2B; Supplementary Figure 3A). CAHs also exhibited less copy-number alteration (median 3.0% of genome altered vs. median of 11.9% in primaries, p = 0.005; Supplementary Figure 3B).
Mutations of at least one of PTEN and PIK3CA (usually PTEN) were present in all twelve samples (Supplementary Figure 7). Loss of heterozygosity of chromosome 10q, containing PTEN, and amplification of chromosome 1q were the only recurrent copy-number alterations, detected in four and three cases respectively. Other genes that were significantly mutated in primary endometrial cancers were also mutated at lower frequency in CAHs; mutations of ARID1A, CTCF, RNF43, ARHGAP35 and MYO10 were each mutated in two CAHs. Phosphatidylinositol 3-kinase (PI3K) pathway mutations have previously been shown to be prevalent in CAH21. Although it is possible that smaller fractions of CAH exhibit additional driver mutations, these results indicate that no other genes are mutated at a similar rate to PTEN and PIK3CA.
Primaries and metastases have similar levels of alteration
Biopsies from primaries (PBs) and paired metastases (MBs) exhibited similar overall burdens of somatic genomic alteration (p = 0.81). Metastases exhibited a median of 94.5 mutations per biopsy vs. 93 in primaries (Supplementary Figure 3A). The number of mutations typically varied by 10.5% between a primary and its paired metastasis (Figure 2C). Metastases exhibited a median of 12 SCNAs, vs 11 per primary (p = 0.53); the number of SCNAs typically varied by 15.2% (Figure 2A). The fraction of genome altered was also similar between most primaries and paired metastases (Figure 2D).
Whole genome doubling events (“WGD”, detected using ABSOLUTE22,23) accounted for the largest differences in aneuploidy between paired MBs and PBs. Three PB-MB pairs exhibited WGD in only the MB; two of these MBs were from a single metastasis (Supplementary Figure 8). These MBs were therefore non-diploid (aneuploid) across most of their genomes. They also exhibited increased rates of localized SCNAs, a feature that has previously been associated with WGD in model systems24 and primary tumors23,25,26.
Primary biopsies lack half of alterations in metastases
While the overall somatic genomic alteration burden was similar between primaries and their matched metastases, only an average of 48% of specific mutations (Figure 3A) and 56% of SCNAs (Supplementary Figure 9) found in the MB were shared with the PB. Conversely, an average of 51% and 48% of mutations and SCNAs in the PB, respectively, were shared with each MB. The fraction of MB-specific mutations tended to increase with the anatomical distance of the metastasis site from the endometrium (ρ = 0.27; p = 0.13; Supplementary Figure 10A), consistent with data in prostate tumors27.
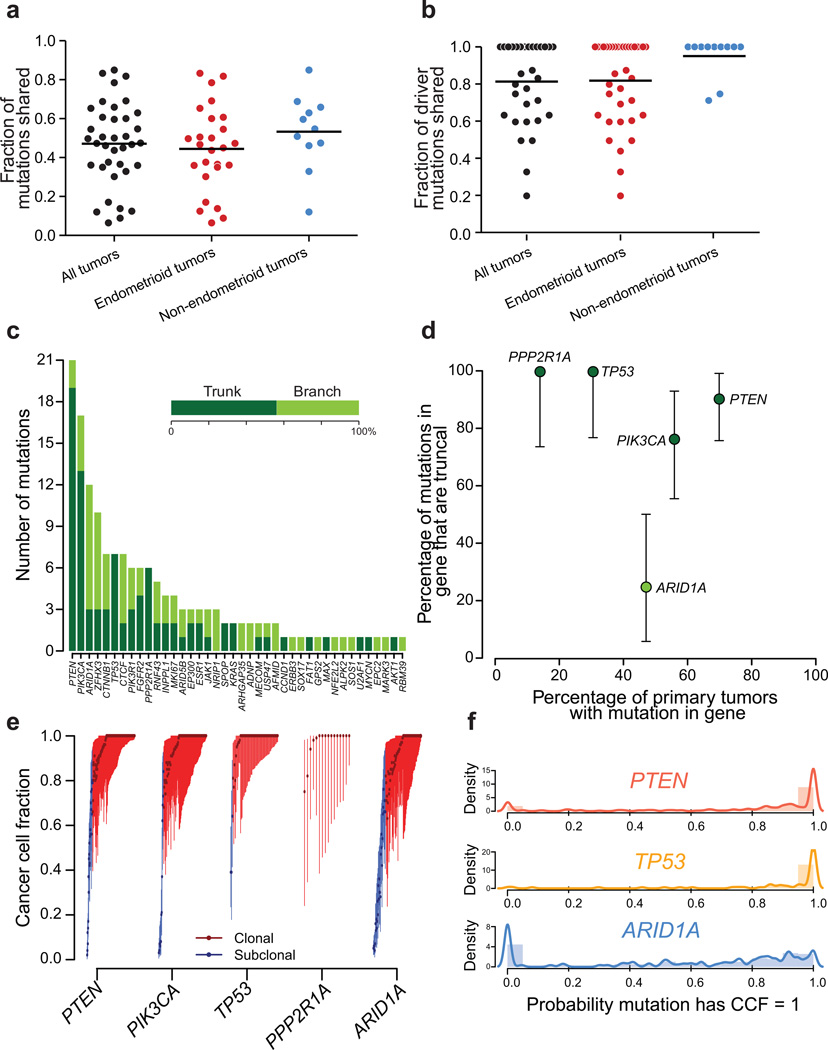
Figure 3. Heterogeneity among somatic mutations.
Fractions of (a) all mutations and (b) driver mutations detected in metastases that were truncal. Lines indicate the mean. (c) The number truncal and branch mutations involving the indicated driver genes. (d) Percentage of mutations that were detected in all paired biopsies in our dataset for frequently mutated driver genes. ARID1A is frequently mutated in the branches of phylogenies. (e) Distribution of cancer cell fraction (CCF) values for each detected mutation in TCGA endometrial samples for the indicated genes. Bars indicate 95% confidence intervals. (f) Distribution of the probability that each mutation detected in TCGA endometrial biopsies is clonal for the indicated genes. Hypermutated samples (> 1,000 mutations) were excluded from (e) and (f)
Distinguishing multiple clonally independent synchronous primaries from metastatic spread of a single primary has important implications for treatment28. We identified four cases (one in Supplementary Figure 10B) in which biopsies of different disease sites exhibited substantially different morphology, resulting in clinical calls of synchronous primaries. Sequencing revealed shared mutations, however, indicating a shared clonal origin. Conversely, prior analyses of other cancer types have revealed multiple clonally unrelated cancers in individual patients22,26. These results suggest that genetic evaluation is necessary to evaluate the clonal independence of cancer lesions.
Among the 186 arm-level SCNAs (comprising most of a chromosome arm) detected, 90 (48%) were heterogeneous across biopsies. Arm-level losses were more likely than gains to be shared (58% vs 40%; p = 0.02; Supplementary Figure 9A). Losses of 10q, harboring PTEN, and 17p, harboring TP53, were shared more often than other arm-level losses (p = 0.019 and 0.035). The most common arm-level gain, 1q, was truncal in only 6/12 cases (Supplementary Figure 9B). The heterogeneous events included both gains and losses of alternate homologous chromosomes in the PB and MBs (Supplementary Figures 9C–F), suggesting convergent evolution of these SCNAs. These observations imply that arm-level losses, at least some of which result in homozygous knockouts of tumor suppressor genes, tend to occur before arm-level gains in endometrial cancer evolution.
Rates of intratumoral heterogeneity among common drivers
An average PB shared 83% of its driver mutations with its paired MB (Figure 3B). We defined “driver mutations” as non-silent mutations of genes in Supplementary Table 2.2; we identified 1–27 (median 3) truncal driver mutations per patient. Among the 26 PBs, 15 (57.6%) contained driver mutations not detected in the paired MBs. The overlap among drivers exceeded the overlap in the overall number of mutations between primaries and metastases (mean 83% vs. 51%, p = 5.1×10−6). This suggests that the fraction of new mutations that have been identified as significantly recurrent decreases along the length of the evolutionary tree.
The rate at which driver mutations were shared across all biopsies varied by gene, ranging from 0%–100% (Figure 3C). For five genes, we had adequate power to determine whether mutations affecting them were truncal more or less often than the average rate among drivers (“trunk-biased” and “branch-biased” respectively, Figure 3D). Mutations of PTEN, TP53, and PPP2R1A were trunk-biased (Fisher’s two-tailed p = 0.006, p = 0.03, p = 0.04), suggesting they are early events and, in the cases of PTEN and PIK3CA, consistent with their prevalence among CAHs. PIK3CA did not exhibit significant bias in either direction. Mutations in ARID1A were only truncal in 25% of phylogenies, vs. 60% among other drivers, indicating significant branch-bias (p = 0.03). Immunohistochemical staining of ARID1A has also suggested subclonal loss29, and we confirmed across 54 samples that ARID1A mutations were associated with loss of ARID1A immunoreactivity (Supplementary Figure 11).
Analysis of heterogeneity within individual biopsies supports the finding of frequent heterogeneity of mutations in ARID1A. We calculated the likely fractions of sampled cancer cells (CCF) that carried each mutation as previously described23,30,31. Among TCGA data, mutations of PTEN and TP53 were almost exclusively clonal (81% and 92% of cases, respectively), whereas only two-thirds of ARID1A mutations were (dissimilar rates from PTEN; p = 1.6×10−11; Figure 3E–F).
Among seven phylogenies with PPP2R1A mutations, five also exhibited TP53 mutations (p = 0.02, Supplementary Figure 12A), and in all cases the PPP2R1A and TP53 mutations were truncal. We validated the association between PPP2R1A and TP53 mutations in the TCGA dataset using an approach that took into account varying degrees of genomic instability across cancers25,32 (Supplementary Figure 12B, Supplementary Table 5). The only gene whose mutation was positively correlated with TP53 was PPP2R1A (p = 0.0001; q = 0.08), and these were both enriched in non-endometrioid tumors. Combined, these two genes formed an isolated network corresponding to a subset of non-endometrioid tumors.
Mutations of PPP2R1A tended to cluster in two hotspots: p.Pro179Arg (n = 11) and p.Ser256Phe (n = 4, Supplementary Figure 12C), as previously noted33. The association between PPP2R1A and TP53 mutations among the 274 primaries was also primarily due to PPP2R1A hotspot mutations: 17 of 19 tumors with PPP2R1A hotspot mutations exhibited TP53 mutation (p = 5.8×10−8; Supplementary Figure 12D) whereas only three of 16 tumors with non-hotspot PPP2R1A mutations exhibited TP53 mutations (p = 0.56). The co-occurrence of PPP2R1A and TP53 mutations mirrors the dual mechanism of transformation by the SV40 oncovirus, wherein small-T antigen binds PPP2R1A to disrupt association with PP2A regulatory subunits and large-T antigen mediates TP53 inactivation (Supplementary Figure 12E).
Most metastases share an ancestor absent in the primary biopsy
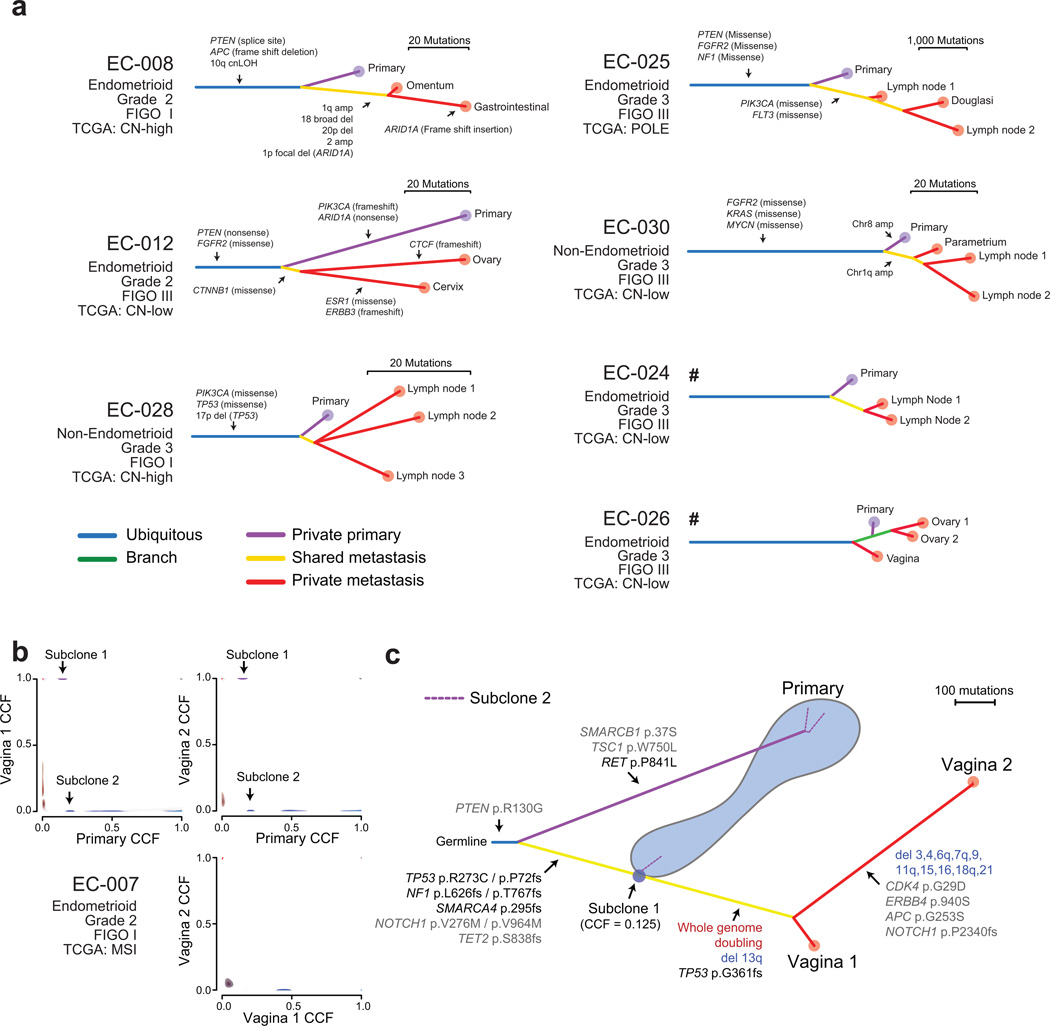
We determined phylogenetic relationships between tumor biopsies using both mutations and allelic copy-number alterations. In six of seven cases with multiple MBs, all MBs were more closely related to each other than to the PB (monophyly). This observation is consistent with these metastases having arisen from a limited fraction of these cancers (possibly even a single cell). In the seventh case, however, one of the MBs was more closely related to the PB than to the other MBs (polyphyly; Figure 4A, Supplementary Figures 13–14).
Figure 4. Phylogenetic trees for tumors with more than one metastasis.
(a) The labeled alterations constitute a subset of the alterations that distinguish between the indicated branches. Hash symbols indicate trees that were derived from SNP6.0 array data. (b) 2D phylogenetic illustration comparing subclonal structures between biopsies in patient EC-007. Cancer cell fraction (CCF) probability densities for the mutations supporting the metastasizing subclone (purple) are presented in Supplementary Figure 16. (c) Phylogenetic tree of patient EC-007. A subclone in the biopsy of the primary tumor shared a common ancestor with the two metastases, not detected in the dominant clone of the primary biopsy. Subclone 2 from (b) could be placed in three different phylogenetic locations as indicated by the dotted purple lines.
These results suggest that most metastases arise from one branched subclone of the cancer. If different metastases had no clonal relations beyond deriving from the same cancer (“independent branched subclones”), each sampled tissue would be equally likely to be most closely related to any of the other samples in a given patient. In that case, we would have expected one or two (expectation value 1.7) cases of monophyly in our data, a significantly different result from the six of seven cases of monophyly observed (p = 0.001; see Methods). Even the existence of two independent branched subclones, each the ancestor of half of the observable metastases in each patient, would have been expected to produce monophyly in only four of the seven phylogenies, a significantly different result from that observed (p = 0.018; see Methods).
We performed similar calculations on phylogenies from prostate and pancreatic cancers for which genome-level sequencing had been performed on two to ten MBs and one to nine paired PBs27,34,35. Prostate and pancreatic cancers exhibited polyphyly in one of five (p = 0.05) and three of five cases (p = 0.98), respectively, consistent with the biopsied metastases arising from a limited fraction of the prostate cancers but perhaps not the pancreatic cancers.
No evidence of ubiquitous metastasis-specific mutations
We did not discover significantly recurrent metastasis-specific mutations. It is possible that metastasis-specific drivers remain undetected. To assess our power to detect metastasis specific drivers, we “spiked” hypothetical driver mutations into our dataset and then assessed whether we recovered them (Supplementary Figure 15). Our power exceeded 90% for genes mutated in at least 50% of metastases and remained greater than 50% for genes mutated in at least 20% of metastases. These results indicate with probability 0.9 that there are no metastasis-specific exomic mutations that recur in greater than 50% of abdominopelvic metastases. However, drivers of metastasis may include features not detectable through whole-exome sequencing, or combinations of mutations that we were not powered to detect.
We observed no significant excess of known driver mutations among metastases. Among our 26 phylogenies, 22 exhibited the same number of driver mutations in the primary and metastatic biopsies, three exhibited more drivers in the metastasis, and one exhibited more drivers in the primary (p = 0.63).
Detection of a metastasis-related subclone in the primary
We determined the cancer-cell fraction (CCF) carrying each mutation as described above. Different mutations tended to cluster around similar CCFs, indicating the presence of subclonal populations. Subclonal mutations were detected in every PB and MB. An average of 20% and 26% of mutations in MBs and PBs, respectively, had CCF < 1 (p = 0.26).
We focused on mutation clusters that were detected in more than one biopsy, but had CCF < 1 in at least one of them, as these may indicate seeding patterns from one biopsy to another22,30,36 (Figure 4B). In one patient, this analysis identified a subclone within the PB that was closely related to an ancestor of the MBs (Figure 4C, Supplementary Figures 16). We did not, however, find evidence of either oligoclonal seeding of metastases or re-seeding of either metastases or primaries. These results are consistent with a near-ubiquitous ‘branched-sibling’ relationship between primary-tumor samples and paired brain-metastases observed previously22.
Discussion
We present the first genome-wide analysis of genetic changes through endometrial cancer progression including complex atypical hyperplasias, primary tumors, and paired metastases. We observed striking heterogeneity between biopsies of paired primaries and metastases, with only half of mutations shared on average between any two biopsies. These biopsies did not fully sample these tumors, implying higher levels of heterogeneity than what we measured in both metastatic and primary tissue. As a result, some of the mutations detected only in metastases may have been present in unsampled regions of their paired primaries.
Across primary endometrial cancers, we identified 21 novel significantly mutated genes, owing in part to indel rescue in MSI tumors. Among these was NRIP1, which was mutated in 12.5% of tumors. NRIP1 is an obligate cofactor of the estrogen receptor13, and germline SNPs near NRIP1 have been associated with ER-positive breast cancer37. These data suggest that NRIP1 alterations are common drivers of endometrial cancer oncogenesis. However, variations in indel rates across the genome are not well-understood, and NRIP1 alterations were also seen in MSI colorectal cancers. Further characterization of the functional effects of NRIP1 alterations is necessary.
The varying rates in heterogeneity across mutations in different genes indicate the order in which these mutations are typically acquired during tumor evolution. In particular, likely drivers of primary oncogenesis tend to be more homogenous than likely passengers. Among the drivers, mutations in PIK3CA, PTEN, TP53, and PPP2R1A occurred earlier on average in tumor evolution. In the case of PIK3CA and PTEN, these findings from advanced cancers mirror the findings in CAHs, which almost exclusively exhibited PI3K pathway mutations21.
Conversely, mutations of ARID1A, a member of the BAF chromatin-remodeling complex, were frequently subclonal. This heterogeneity was mirrored by heterogeneity of mutations across BAF complex members in other cancers. Mutations of BAF complex members displayed the most phylogenetic heterogeneity of all known driver mutations in multi-region sequencing studies including mutations of PBRM1 in renal cell carcinomas38, SMARCA4 in gliomas39, and ARID1A/SMARCB1 in meningiomas40,41. An exception is malignant pediatric rhabdoid tumors, in which SMARCB1 mutation is typically the sole oncogenic driver42. These observations suggest that in many cancers, BAF complex perturbations may alter the epigenetic landscape of already established tumors rather than initiate tumor formation. The heterogeneity of ARID1A mutations also raises questions regarding the likely efficacy of ARID1A-directed therapies such as EZH2 inhibition43. However, we identified convergent evolution involving ARID1A mutations and other mechanisms of ARID1A inactivation might also converge to generate phenotypic homogeneity. Indeed, homogenous patterns of ARID1A loss were observed by immunostaining advanced lesions44,45.
We were unable to identify examples of tumor self-seeding, which have been observed in human prostate cancers and breast cancer models27,36,46. However, our data are insufficient to reject the hypothesis that these events occurred but only involved small fractions of cancer cells or cancer tissues that were not sampled.
Notably, we did not identify recurrent metastasis-specific driver mutations. Drivers of metastasis may be intergenic, epigenetic, or environmental events that are not well-assessed by whole-exome sequencing47. It is also possible that a great diversity of genetic events or combinations of events contribute to metastasis, each in a small subset of metastatic cancers, and that we had insufficient power to detect them. In this case, genomic analysis of metastases from many more patients will be required.
The observation of significantly recurrent monophyly is consistent with metastatic endometrial cancer cells sharing a feature that is associated with genetic ancestry. This may be a cryptic genetic event that enables metastasis, but it is also consistent with other explanations. For example, members of a lineage may happen to be located in an environment that is conducive to metastasis47. Alternatively, seeding the first metastasis may be a rate-limiting step, after which it is more likely that this metastasis will seed further metastases, a mode of spread that has been previously described in small-cell lung cancer mouse models36 and in human prostate cancer27.
Although our data suggest that large, clinically resected abdominopelvic metastases tend to arise from a limited fraction of endometrial cancer primaries, it is possible that metastases to other anatomic sites may exhibit different evolutionary relationships, and some histologic subtypes may be more likely to generate metastases from independent branched subclones. Only a single biopsy of the primary tumor was sampled in each case, and we could not infer how many independent metastatic lineages existed in the primary tumor, or what fraction of the tumor’s cells they comprised. Indeed, the single observed case of polyphyly might represent a cancer with more than one independent metastatic branched subclone, or a case in which the PB happened to sample descendants of the metastatic subclone within the primary tumor. Sampling more regions of primary tumors, in addition to multiple metastases from the same patient, should help resolve these issues.
Methods
Sample collection and description
The investigations within this study were approved by the Norwegian Social Science Data Services (15501), the local Institutional Review Board at Haukeland University Hospital, Bergen, Norway (REK-number 2009-2315) and at the Broad Institute (DFCI_12_049B), Cambridge MA, USA. All patients consented to inclusion in this study. Samples were collected from patients from western Norway from Sept 2002 until Sept 2012.
Biopsies were snap-frozen in liquid nitrogen and stored at −80°C. Tumor purity was assessed in hematoxylin-stained sections prior to DNA extraction. After sequencing, purity was also calculated by ABSOLUTE22,23,26; biopsies from primary tumors had equivalent purity to those from metastases (61.3% vs 59.9% respectively, p = 0.76). Blood samples were collected for reference as normal controls. Clinical information on all cases is presented in Supplementary Table 1.
Our cohort included 12 patients with complex atypical hyperplasia (CAH) and 40 patients with metastatic endometrial cancer. Of this latter group, 26 had endometrioid endometrial carcinoma (EEC) and 14 had non-endometrioid endometrial carcinomas (NEEC). Time interval and treatment given between resection of the primary tumor and metastatic lesions are detailed in Figure 1 and Supplementary Table 1. All histopathologic diagnoses were subjected to formal histopathologic revision and/or established in a tumor board setting as previously reported4,48,49.
We performed genomic characterization of 98 biopsies from these 52 individuals. We performed whole-exome sequencing on 81 biopsies from 45 individuals, including twenty-six with paired primary and metastatic lesions, five with more than one metastasis, and eight metastases without paired primaries, along with DNA from paired blood in all cases. We also analyzed SCNAs in 76 samples from 37 patients using Affymetrix SNP 6.0 arrays (Supplementary Figure 2). These included 59 samples from 30 patients that had also undergone WES, 10 additional metastases with paired primary tumors (from six cases, including three cases with more than one metastasis) and one unpaired metastasis (Supplementary Figure 1).
Assessment of microsatellite instability (MSI) status
MSI testing was performed on all samples subjected to whole-exome sequencing using the marker set employed by TCGA3. DNA was whole-genome amplified using the GenomePlex Complete Whole Genome Amplification kit (Sigma Aldrich). The probe set consisted of BAT25, BAT26, BAT40, TGFBRII, D2S123, D5S346 and D17S250. No markers were positive in the normal controls (blood). None of the patients in this study were diagnosed with hereditary nonpolyposis colorectal cancer (HNPCC).
Exome Sequencing and SNP Array profiling
Genomic DNA was isolated from frozen tissues using the Qiagen DNAamp kit or a standard proteinase K protocol. Samples were sequenced on an Illumina HiSeq-2000 to an average of 77× depth (85.6% targeted reads > 20× coverage). The average rate of read alignment was 98.6%. Affymetrix SNP 6.0 arrays were used for a subset of samples. Seven CAHs, out of 19 profiled, were found to have both an exceptionally low purity (less than 25% per ABSOLUTE analysis23) and low burden of mutations. Upon manual review, the mutations whose allelic fractions were higher than 10% were enriched in regions with low mapping quality. These seven samples were therefore excluded from further analyses.
Validation sequencing
We subjected the indels detected by our rescue strategy to additional technical validation using six approaches. First, we tried calling indels in the normal samples using the paired tumor as the control, resulting in a mean of 0.16 indels called per normal, as opposed to a mean of 62 indels per tumor. Second, we applied an independent indel caller, Strelka50, which detected 86.5% of the indels we had rescued. Third, we conducted Sanger sequencing to validate rescued indels across 88 genomic loci, including 77 loci across a variety of genes and 11 loci in genes with recurrent indels: NRIP1, ARID1A, and RPL22 (Supplementary Table 4). We sequenced a total of 127 indel events from the 77 loci, 123 (97%) of which validated. Among the 11 sites in NRIP1, ARID1A, and RPL22, we sequenced a total of 25 indels, of which 23 (92%) validated. The exceptions were two samples with low allelic-fraction (0.02 and 0.05) events. None of the indels were detected in the paired normals. Fourth, we performed Illumina amplicon sequencing across these 25 indel events to 10,000X depth, and detected all 25, including the two low allelic-fraction events. Fifth, we performed Sanger sequencing from subcloned PCR products for the sample with the NRIP1 mutation with an allelic fraction of 0.05 and validated the mutation in the tumor DNA but not in its normal DNA. Sixth, we detected two NRIP1 p.Lys728fs indels by Sanger sequencing across an independent cohort of 37 endometrial cancer samples (Supplementary Figure 5D). Across the 78 rescued indels subjected to validation sequencing, we validated 75 (96%) in at least one assay.
Sanger sequencing was done by PCR with subsequent sequencing in both directions using forward and reverse M13 primers (Supplementary Table 4) and whole-genome amplified (as described above) or original DNA, and analyzed on an Applied Biosystem 3730XL Instrument as previously described50. Chromatograms were analyzed with FinchTV (1) or Sequence Scanner (2). For selected NRIP1 mutations with low allelic fraction, the PCR product was subcloned into the pGEM T-easy vector (Promega) for sequencing of single colonies.
A selection of indel variants were also amplicon sequenced on a MiSeq instrument (Illumina) to a coverage of >10,000× (Supplementary Table 4). DNA that had been subjected to WES and not to whole genome amplification underwent PCR to amplify the relevant sites for DNA input. All mutations detected by WES of non-TCGA samples and presented in any stick-plot figure were subject to validation sequencing.
Immunohistochemistry
Immunohistochemistry was performed on full sections of formalin-fixed paraffin embedded tissue (FFPE) and three representative cylinders mounted in tissue microarrays (TMA) as described previously44, employing anti-ARID1A rabbit monoclonal antibody (Abcam 182560, clone number EPR13501, dilution 1:2000). The staining index was assessed in TMA samples, as a product of staining intensity (0–3) and area of positive tumor cells (1: ≤10%, 2: 10–50% and 3: ≥50%), providing a scale from 0 (negative staining) to 9 (full positive staining)46.
Somatic mutation calling
Somatic mutations were called with MuTect51. One source of false-positive somatic mutations is contamination with foreign DNA. We estimated this by CONTEST and found the mean level of contamination to be 0.4%. This information was used by MUTECT to determine the floor for accepting potential low-variant allele frequency mutations. OxoG artifacts were removed using the Broad Institute OxoG3 filter52. Insertions and deletions (indels) were called with Indelocator. Additional indels were rescued according to the following previously established criteria: at least 50 reads in both the tumor and normal, > 0.2 allelic fraction for the variant read in the tumor, and < 0.05 allelic fraction for the variant read in the normal8. These indels often occurred in microsatellites: among 4,752 microsatellites covered by exome sequencing, 1,196 exhibited indels in at least one sample, and these accounted for 1,882 of the 30,096 indels we detected. All of these recurrent-site indels were supported by small numbers of supporting reads in the paired normal samples; this accounts for their absence in prior analyses. To ensure the fidelity of this approach, we swapped tumor and normal labels to determine the false positive rate of indel calls. A median of 0 and maximum of 2 indels were falsely called exome-wide in this approach (Supplementary Figure 5A). In contrast, we rescued more than 100 mutations in 13 tumor samples using this approach. The variance of the number of mutations in PBs and MBs was approximately equal (p = 0.22, F-test).
Copy-number analysis
Relative copy-number profiles from Affymetrix SNP 6.0 arrays were determined as previously described25. Relative copy-number profiles from exome sequencing data were determined by normalizing exome coverage data to values from blood controls and generating segmented copy-number profiles. These were paired with germline heterozygous sites to obtain allele-specific relative copy-number profiles, as previously described22,26. The relative allele-specific copy-number profiles were paired with exome mutation data for each tumor sample as input to ABSOLUTE22,23,26. for final determination of discrete allele-specific copy-number profiles. The sequence of events that led to each allelic copy-number profile was inferred using a maximum parsimony approach25.
Mutation correlations analysis
The mutations detected in primary cancers from this cohort were combined with mutations detected in endometrial cancers profiled by TCGA3 to detect correlations and anticorrelations between mutated genes, using a previously described approach that maintains the marginal counts of both the number of mutations within each sample and the number of events within each gene32. Ultramutated samples and rescued indels in MSI tumors were excluded from this analysis. P-values were calculated using 10,000 permutations of the observed data. The network of correlated interactions was plotted using Cytoscape where the negative-log of the q value for positive correlation is proportional to the spring constant of an edge between two nodes.
Associations with survival were determined by a Kaplan-Meier analysis using the R package survival. P-values were computed by the log-rank test.
Detection of cancer subclones within biopsies
For each mutation, cancer-cell fractions (CCFs) were calculated by ABSOLUTE23,30 by integrating information from local allelic copy-number, biopsy purity and variant allele counts. Posterior distributions over CCF values across samples cancer tissues from a given case were then subjected to a clustering procedure in order to identify subpopulations of cells and reduce the uncertainty over CCF estimates22,26,36.
We determined from this analysis that most detected subclones with CCF < 1 were restricted to individual tissue samples (data not shown). This enabled us to construct biopsy-level phylogenetic trees without further consideration of cancer-tissue heterogeneity. One exception occurred where a subclone was detected in the primary-tumor sample of case EC-007 that was an ancestor of the paired metastases.
For the analysis of cancer cell fraction of mutations in TCGA data, we performed ABSOLUTE across all tumor samples in the TCGA endometrial dataset. ABSOLUTE computes a probability distribution of the CCF of each mutation and includes a probability that each mutation is subclonal. To exclude the possibility that passenger mutations in hypermutated samples could confound our analysis, we excluded hypermutated samples (>1000 detected mutations) from this analysis.
Phylogenetic tree reconstruction
To improve the sensitivity of mutation calls in each biopsy, we utilized a previously described ‘forced-calling’ strategy22,26. This procedure effectively rescues mutations that failed to reach the evidence threshold of Mutect in a given biopsy, provided that they were confidently detected in another sample from the same case.
Phylogenetic trees were constructed using an implementation of clonal ordering. Force-called mutations were converted into a binary incidence matrix depending on their absence/presence in a set of paired biopsies. We calculated the power to detect each mutation in each biopsy based on local allelic copy-number and purity23. Where a mutation was not detected in one biopsy, but power to detect it was less than 0.95, the mutation was excluded from the incidence matrix and separately annotated. A distance matrix was computed from the final incidence matrix using the following distance metric:
where ma corresponds to the binary vector of mutations in biopsy a and mb is the vector describing biopsy b. Hierarchical clustering of this distance matrix was performed using the complete linkage method in R.
Homologous chromosome tracking across tumor biopsies
Germline heterozygous sites were determined from exome sequencing of the normal blood control sample. The allelic fraction of these sites was determined at each site in the exome in all paired tumor samples (primaries and metastases). Purity estimates (p) from ABSOLUTE were used to generate purity-adjusted minor allelic fractions (mAF) at each site.
These allelic fractions were multiplied by the local total copy-number (CNT) by ABSOLUTE to graph a point estimate for each SNP of the major and minor tumor alleles. A point estimate for the minor allelic copy-number (mACN) at each site was calculated as follows:
The major allele in the reference tumor for each site was defined as whichever allele count was greater (variant vs. reference). The expected major allele at each SNP was colored red in the resulting plot. In the test tumor, the same major and minor alleles estimated from the reference (primary) tumor are used and colored accordingly. Homologous chromosome tracking was performed across every pair-wise comparison in the cohort. Resulting plots were manually reviewed for discordant tumor haplotype alterations.
Instances in which the chromosome undergoing copy loss/gain in the test tumor is opposite the homologous chromosome undergoing loss/gain in the reference tumor indicate separate events in the genetic history of the tumor. Raw plots for selected chromosomes for case EC-022 are shown in Supplementary Figure 9.
Significance analysis of phylogenies
Under the null hypothesis that evolutionary distances are randomly distributed among pairs of cancer tissues from a given patient, we expect all configurations of phylogenetic trees involving biopsies of the primary tumor and metastases to be equally probable. In cases of two MBs and one PB, the phylogeny could have three configurations: either the PB is the most distantly related biopsy, or either of the MBs is the most distantly related biopsy. Therefore, monophyly would be observed in one-third of cases. In cases of three MBs and one PB, the phylogenetic tree could include two clades with two members each (with a one-third probability53) or clades with one and three members, respectively (with a two-thirds probability53). Only the latter is consistent with monophyly, and only if the PB is in the clade by itself (with a probability of one-quarter), for a one-sixth probability of monophyly overall. We used these probabilities to calculate p-values indicating the likelihood of obtaining the observed or a greater rate of monophyly.
If two independent subclones each gave rise to half of the observed metastases, any two metastases would have a 50% chance of deriving from different subclones, with a probability of monophyly of one-third (as above), and a 50% chance of deriving from the same subclone. We assume that if the MBs derive from the same subclone, they necessarily exhibit monophyly (a conservative assumption: it is possible that the PB would by chance represent that same subclone within the primary tumor, in which case polyphyly would still be possible). We used these and similar considerations for phylogenies with three MBs to calculate p-values indicating the likelihood of obtaining the observed or a greater rate of monophyly, assuming an equivalent number of patients and PB and MB samples in each patient.
Significance analysis of mutations in primary endometrial cancers
We combined the force-called mutation lists (without indel rescue) from our primary tumors with the mutations from the TCGA9. We applied MUTSIG2CV on this list of mutations. Genes with q-values less than 0.1 were considered significantly mutated. Separately, we combined the force-called mutation lists (with indel rescue) from our primary tumors with the mutations from TCGA that included indel rescue8. We considered any genes that were mutated in greater than 10% of samples and whose q-value was less than 10−5 as significant.
Significance analysis of metastasis-associated driver mutations
For each phylogeny in our dataset, we selected the set of mutations that were detected in every paired metastasis biopsy that were not detected in the biopsy of the primary tumor. We applied MUTSIG2CV to this set of mutations and considered any mutations whose q-value was less than 0.25 as significant.
Power to detect metastasis-associated drivers
We used an empirical approach to determine our power to detect mutations that conferred the ability to metastasize. We used the list of metastasis-specific mutations that we previously constructed as a pool into which we “spiked” hypothetical driver gene mutations at decreasing frequencies. We then assessed the rate at which these hypothetical driver genes were recovered as significant (q < 0.25).
The spiking procedure used non-silent mutations randomly selected from 585,491 exomic mutations detected in TCGA tumors to ensure consistency with mutational background rates and genomic covariates observed in human tumors. For every gene, the number of patients selected for spiking was randomly drawn for the binomial distribution with probability of a success equal to the proposed frequency of driver gene mutation in metastases. When each hypothetical driver gene was proposed, the gene was set to exhibit hotspot mutations with probability 1/3 such that the same mutation was spiked in for each patient selected. After a mutation was spiked into the genome of a given patient, a randomly selected mutation previously observed in the patient was removed to preserve the total number of mutations.
Percentage of mutations in driver genes found in all biopsies
To calculate the percentage of mutations in each driver gene that were truncal, we used force-calling mutation lists annotated with detection power from ABSOLUTE22,23,26. We then determined whether the mutation was detected in all biopsies from the same patient. If the mutation was present in all biopsies, then the number of trunk mutations was incremented by one. If a mutation was not detected in a given biopsy, and power to detect the mutation was greater than 0.8, the number of branch mutations was incremented by one. If there was not sufficient power to detect the mutation in one or more biopsies lacking the mutation, then the mutation was not counted towards the trunk or branch counts. To exclude that possibility that passenger mutations in driver genes could confound our analysis, two phylogenies with POLE exonuclease mutations and ultramutated genomes (15,095 and 30,601 mutations detected) were excluded from this analysis.
Supplementary Material
Acknowledgments
We thank Ellen Valen, Britt Edvardsen, Kadri Madissoo, Reidun Kopperud and Bendik Nordanger for excellent technical assistance.
We would like to dedicate this manuscript to Helga Birgitte Salvesen, who unexpectedly passed away prior to its publication. She was a tremendously generous and insightful colleague and a dear friend. We will all miss her dearly.
Funding sources
This study was supported by The Research Council of Norway, The Norwegian Cancer Society, Helse Vest, The University of Bergen, Bergen Research Foundation, the National Institutes of Health (award numbers T32GM007753, 5R01CA188228, and 1F30CA192725), and Novartis Institutes for Biomedical Research.
Footnotes
Accession Codes
Pending dbGAP approval
Author Contributions
E.A.H. and H.B.S. initiated and W.J.G., E.A.H., S.L.C., R.B., and H.B.S. designed the study. E.A.H., M.K.H., A.B., H.M.J.W., I.M.S., K.K.M., J.T., K.W., L.B., and H.B.S. performed sample collection, annotation, and curation. W.J.G., E.A.H., A.T.-W., A.D.C., F.H., T.I.Z., K.M.S., K.K., J.A.W., M.S.L., S.L.C., R.B., and H.B.S. performed the data analyses. E.A.H., K.M.S., and C.K. performed validation, MSI, and immunohistochemistry experiments. W.J.G., E.A.H., M.R., A.C., K.K.M., J.T., C.K., M.G., E.H., O.K.V., M.S.L., G.G., S.L.C., R.B., and H.B.S. contributed reagents and algorithms. All authors critically revised the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Code availability
Software packages used for sequence analysis are publicly available at the following URLs:
Mutect (3)
Indelocator (4)
Recapseg (5)
ABSOLUTEv1.2 (6)
References
- 1.Bhaskaran K, et al. Body-mass index and risk of 22 specific cancers: a population-based cohort study of 5.24 million UK adults. Lancet. 2014;384:755–765. doi: 10.1016/S0140-6736(14)60892-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15:10–17. doi: 10.1016/0090-8258(83)90111-7. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research, N. et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salvesen HB, et al. Integrated genomic profiling of endometrial carcinoma associates aggressive tumors with indicators of PI3 kinase activation. Proc Natl Acad Sci U S A. 2009;106:4834–4839. doi: 10.1073/pnas.0806514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dutt A, et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl Acad Sci U S A. 2008;105:8713–8717. doi: 10.1073/pnas.0803379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salvesen HB, Haldorsen IS, Trovik J. Markers for individualised therapy in endometrial carcinoma. Lancet Oncol. 2012;13:e353–e361. doi: 10.1016/S1470-2045(12)70213-9. [DOI] [PubMed] [Google Scholar]
- 7.Ciriello G, et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giannakis M, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet. 2014;46:1264–1266. doi: 10.1038/ng.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodruff JD, Pickar JH. Incidence of endometrial hyperplasia in postmenopausal women taking conjugated estrogens (Premarin) with medroxyprogesterone acetate or conjugated estrogens alone. The Menopause Study Group. Am J Obstet Gynecol. 1994;170:1213–1223. doi: 10.1016/s0002-9378(94)70129-6. [DOI] [PubMed] [Google Scholar]
- 11.Veeraraghavan J, et al. Recurrent ESR1-CCDC170 rearrangements in an aggressive subset of oestrogen receptor-positive breast cancers. Nat Commun. 2014;5:4577. doi: 10.1038/ncomms5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavailles V, et al. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosell M, et al. Complex formation and function of estrogen receptor alpha in transcription requires RIP140. Cancer Res. 2014;74:5469–5479. doi: 10.1158/0008-5472.CAN-13-3429. [DOI] [PubMed] [Google Scholar]
- 14.Toy W, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45:1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson DR, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45:1446–1451. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamburov A, et al. Comprehensive assessment of cancer missense mutation clustering in protein structures. Proc Natl Acad Sci U S A. 2015;112:E5486–E5495. doi: 10.1073/pnas.1516373112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang MT, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2015 doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novetsky AP, et al. Frequent mutations in the RPL22 gene and its clinical and functional implications. Gynecol Oncol. 2013;128:470–474. doi: 10.1016/j.ygyno.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong B, Le Gallo M, Bell DW. The mutational landscape of endometrial cancer. Curr Opin Genet Dev. 2015;30:25–31. doi: 10.1016/j.gde.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren Y, et al. JAK1 truncating mutations in gynecologic cancer define new role of cancer-associated protein tyrosine kinase aberrations. Sci Rep. 2013;3:3042. doi: 10.1038/srep03042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayes MP, et al. PIK3CA and PTEN mutations in uterine endometrioid carcinoma and complex atypical hyperplasia. Clin Cancer Res. 2006;12:5932–5935. doi: 10.1158/1078-0432.CCR-06-1375. [DOI] [PubMed] [Google Scholar]
- 22.Brastianos P, et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discovery. 2015 doi: 10.1158/2159-8290.CD-15-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nature biotechnology. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stachler M, et al. Paired Exome Analysis of Barrett’s Esophagus and Adenocarcinoma. Nature Genetics. 2015 doi: 10.1038/ng.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gundem G, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015 doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright JD, Barrena Medel NI, Sehouli J, Fujiwara K, Herzog TJ. Contemporary management of endometrial cancer. Lancet. 2012;379:1352–1360. doi: 10.1016/S0140-6736(12)60442-5. [DOI] [PubMed] [Google Scholar]
- 29.Mao TL, et al. Loss of ARID1A expression correlates with stages of tumor progression in uterine endometrioid carcinoma. Am J Surg Pathol. 2013;37:1342–1348. doi: 10.1097/PAS.0b013e3182889dc3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landau, Dan A, et al. Evolution and Impact of Subclonal Mutations in Chronic Lymphocytic Leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohr JG, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25:91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao S, et al. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc Natl Acad Sci U S A. 2013;110:2916–2921. doi: 10.1073/pnas.1222577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McFadden DG, et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell. 2014;156:1298–1311. doi: 10.1016/j.cell.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghoussaini M, et al. Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nat Genet. 2012;44:312–318. doi: 10.1038/ng.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerlinger M, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson BE, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abedalthagafi MS, et al. ARID1A and TERT promoter mutations in dedifferentiated meningioma. Cancer Genet. 2015 doi: 10.1016/j.cancergen.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torres-Martin M, et al. Whole exome sequencing in a case of sporadic multiple meningioma reveals shared NF2, FAM109B, and TPRXL mutations, together with unique SMARCB1 alterations in a subset of tumor nodules. Cancer Genet. 2015 doi: 10.1016/j.cancergen.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 42.Lee RS, et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest. 2012;122:2983–2988. doi: 10.1172/JCI64400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bitler BG, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21:231–238. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Werner HM, et al. ARID1A loss is prevalent in endometrial hyperplasia with atypia and low-grade endometrioid carcinomas. Mod Pathol. 2013;26:428–434. doi: 10.1038/modpathol.2012.174. [DOI] [PubMed] [Google Scholar]
- 45.Wiegand KC, et al. Loss of BAF250a (ARID1A) is frequent in high-grade endometrial carcinomas. J Pathol. 2011;224:328–333. doi: 10.1002/path.2911. [DOI] [PubMed] [Google Scholar]
- 46.Kim MY, et al. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 48.Berg A, et al. Molecular profiling of endometrial carcinoma precursor, primary and metastatic lesions suggests different targets for treatment in obese compared to non-obese patients. Oncotarget. 2015;6:1327–1339. doi: 10.18632/oncotarget.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wik E, et al. Endometrial Carcinoma Recurrence Score (ECARS) validates to identify aggressive disease and associates with markers of epithelial-mesenchymal transition and PI3K alterations. Gynecol Oncol. 2014;134:599–606. doi: 10.1016/j.ygyno.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 50.Saunders CT, et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 51.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Costello M, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013;41:e67. doi: 10.1093/nar/gks1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu S, Degnan JH, Steel M. Clades, clans, and reciprocal monophyly under neutral evolutionary models. Theor Popul Biol. 2011;79:220–227. doi: 10.1016/j.tpb.2011.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.