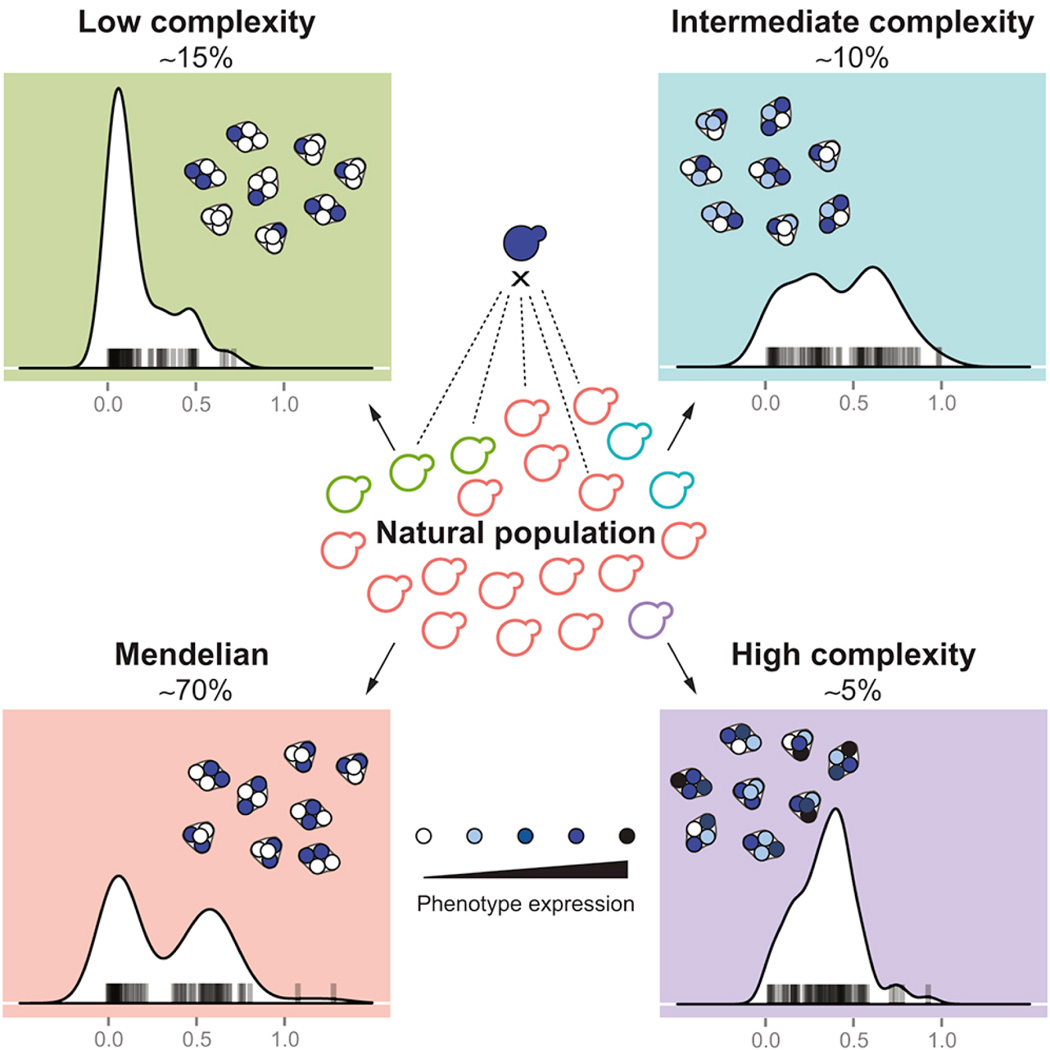
Summary
Mendelian traits are considered as the lower end of the complexity spectrum of heritable phenotypes. However, more than a century after the rediscovery of Mendel’s law, the global landscape of monogenic variants as well as their effects and inheritance patterns within natural populations is still not well understood. Using the yeast Saccharomyces cerevisiae, we performed a species-wide survey of Mendelian traits across a large population of isolates. We generated offspring from 41 unique parental pairs, and analyzed 1,105 cross/trait combinations. We found that 8.9% of the cases were Mendelian. Further tracing of causal variant revealed background-specific expressivity and modified inheritances, gradually transitioning from Mendelian to complex traits in 30% of the cases. In fact, when taking into account the natural population diversity, the hidden complexity of traits could be substantial, perplexing the phenotypic predictability even for simple Mendelian traits.
Graphical abstract
eTOC blurb
Mendelian traits are considered to be the least complex of all heritable phenotypes. By systematically identifying causal Mendelian variants and following their effects and inheritances across the S. cerevisiae species, Hou et al. uncovered the continuous complexity spectrum of monogenic mutations, where genotype is hardly predictive of a phenotype.
Introduction
Elucidating the genetic causes of the astonishing phenotypic diversity observed in natural populations is a major challenge in biology. Within a population, individuals display phenotypic variations in terms of morphology, growth, physiology, behavior, and disease susceptibility. The inheritance patterns of phenotypic traits can be classified as either monogenic or complex. While many traits are complex resulting from variation within multiple genes, their interaction and environmental factors (Mackay et al., 2009), some traits are primarily monogenic and conform to a simple Mendelian inheritance (Antonarakis and Beckmann, 2006). Nevertheless, while useful, this overly simplistic dichotomic view could potentially mask the continuous level of the underlying genetic complexity (Antonarakis et al., 2010; Badano and Katsanis, 2002; Dipple and McCabe, 2000). More than a century after the rediscovery of Mendel’s law, we still lack a global overview of the spectrum of genetic complexity of phenotypic variation within any natural population.
Complex traits can be predominantly controlled by variation in a single gene (Dipple and McCabe, 2000). Similarly, monogenic traits can be influenced by multiple genes in specific genetic backgrounds (Badano and Katsanis, 2002; Cooper et al., 2013; Dorfman, 2012; Nadeau, 2001; Thein, 2011). In fact, it is increasingly evident that monogenic mutations do not always strictly adhere to Mendelian inheritance (Cooper et al., 2013; Dorfman, 2012; Nadeau, 2001). For example, many human monogenic disorders, including sickle cell anemia and cystic fibrosis, could display significant clinical heterogeneity such as incomplete penetrance and variable levels of severity due to allelic interactions and background specific modifiers (Cooper et al., 2013). Recent genome-scale surveys of loss-of-function mutations have revealed considerable background effects in various model systems (Dowell et al., 2010; Hamilton and Yu, 2012; Paaby et al., 2015; Vu et al., 2015) and human cell lines (Blomen et al., 2015; Hart et al., 2015; Wang et al., 2015), where the mutant phenotypes could be highly variable even between closely related individuals.
However, although background effects on “monogenic” loss-of-function mutations are readily seen, such specific mutation type does not reflect the overall genetic diversity and complexity observed in natural populations (Auton et al., 2015; Cao et al., 2011; Strope et al., 2015). Specifically, the global landscape of natural genetic variants leading to Mendelian traits has never been thoroughly explored in any species, and their phenotypic effects and inheritance patterns within natural populations were largely unknown.
Here, we carried out a first species-wide identification of causal variants of Mendelian traits in the yeast S. cerevisiae to characterize in depth their phenotypic effects and transmission patterns across various genetic backgrounds. We generated a large number of crosses using natural isolates, and analyzed the fitness distribution and segregation patterns in the offspring for more than 1,100 cross/trait combinations. We found that 8.9% of the cases were Mendelian, among which most were caused by common variants and showed stable inheritances across the S. cerevisiae species. Interestingly, global phenotypic distribution patterns of multiple Mendelian traits across an extremely large population (~1000 isolates) were not necessarily correlated with patterns observed in the offspring from individual crosses. We further characterized a causal variant related to drug resistance and traced its effects across multiple genetic backgrounds. Significant deviations from the Mendelian expectation were observed with variable genetic complexities, illustrating the hidden complexity of a monogenic mutation across a yeast natural population.
Results
Global landscape of Mendelian traits in S. cerevisiae
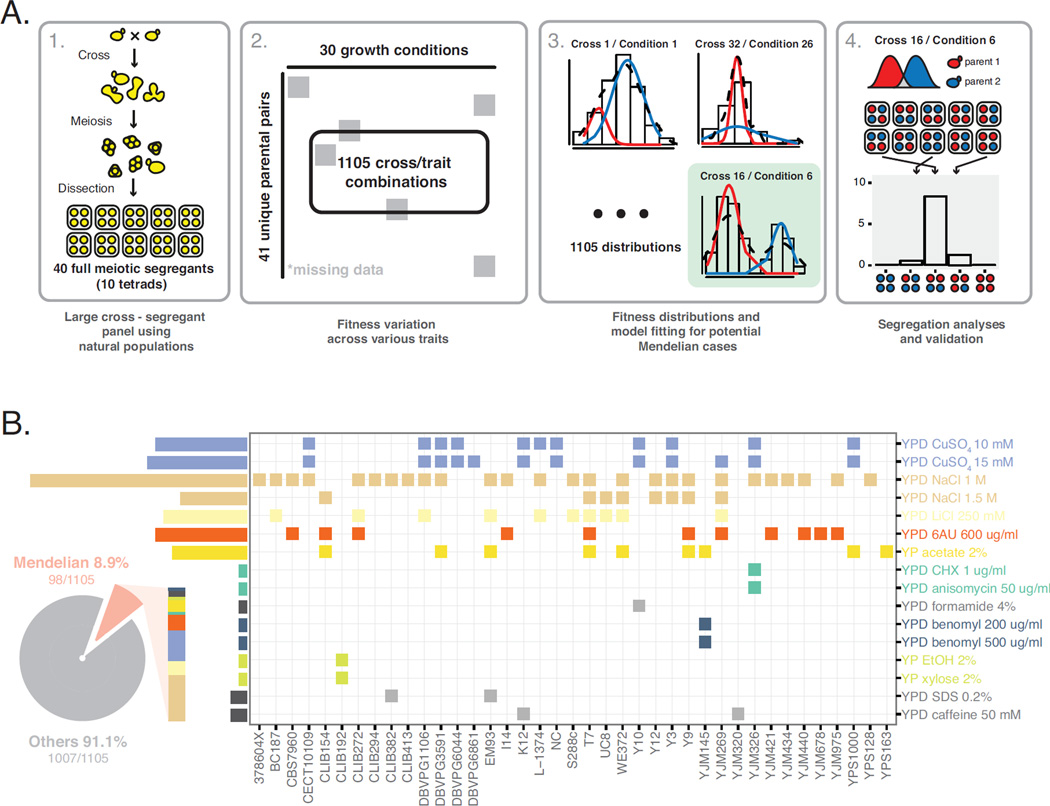
To obtain an overview of natural genetic variants leading to Mendelian traits in the S. cerevisiae species, we selected 41 diverse natural isolates spanning a wide range of ecological (tree exudates, drosophila, fruits, various fermentation and clinical isolates) and geographical sources (Europe, America, Africa and Asia) and performed systematic crosses with one strain Σ1278b (Supplemental Experimental Procedures). For each cross, we generated 40 offspring representing 10 individual meiosis (full tetrads), summing up to a panel of 1,640 full meiotic segregants from diverse parental origins (Figure 1A, panel 1). All segregants as well as the respective parental isolates were tested for 30 stress responsive traits related to various physiological and cellular processes, including different carbon sources, membrane and protein stability, signal transduction, sterol biosynthesis, transcription, translation, as well as osmotic and oxidative stress (Supplemental Experimental Procedures). In total, we tested 1,105 cross/trait combinations and analyzed the offspring fitness distribution patterns for each combination (Figure 1A, panel 2).
Fig 1. Comprehensive landscape of Mendelian traits in S. cerevisiae.
(A) Workflow of the detection of Mendelian traits. The workflow was defined as 4 steps, consisting with offspring generation, fitness measurements, model fitting and segregation analysis as indicated. (B) Distribution of all identified Mendelian traits spanning different crosses (x-axis) on conditions tested (y-axis). Each square represents any single Mendelian case and colors indicate different conditions. Pie chart represent the fraction of Mendelian cases relative to the entire dataset. See Figure S1 for method summary and Figure S2 for genomic mapping results for selected Mendelian cases.
For a Mendelian trait, contrasting phenotypes between the parental isolates were controlled by a single locus, therefore half of the offspring would inherit the causal allele and display a 2:2 segregation in any given tetrads. Consequently, the global offspring fitness distribution would follow a bimodal pattern with equal partitioning of segregants in either parental phenotype cluster. To detect such cases, we applied a bimodal distribution model with random latent variables for the observed fitness distributions for each cross/trait combination using an Expectation Maximization (EM) algorithm (Figure 1A, panel 3; Figure S1 and Supplemental Experimental Procedures). The principle of this method relies on unsupervised iterations of latent variables to fit any given set of data with maximum a posteriori (MAP) probability to a predefined model, which in our case, the presence of two normal distributions for the observed fitness values where the mean of each assigned cluster corresponds to either parental fitness value.
For each fitness distribution observed in a given cross/trait combination, the posterior probability that an individual belongs to either fitness cluster was computed (Experimental procedures), and the general features of the fitted bimodal model such as the means and standard deviations for both clusters as well as their relative proportions were extracted (Experimental procedures). To determine the cutoff values that allow for high confidence calling of bimodal cases and subsequent cluster assignments, we generated a simulated dataset of 1,000 fitness distributions with the same general features compared to the real data, and reapplied the model fitting procedure. Using the simulated data as a training set, we determined that a cutoff of posterior probability > 0.8 for cluster assignment while allowing less than 10% of overlapping between the clusters i.e. the percentage of individuals cannot be assigned at a given posterior probability cutoff, were the best parameters to maintain a high detection performance (area under the ROC = 0.824, Figure S1A) while minimizing case loss (Figure S1B).
By applying these parameters, 318 cross/trait combinations were detected as bimodal with the parental isolates belonging to distinct clusters. All detected bimodal cases were robust against experimental noise, where the mean trait differences between the assigned clusters exceeded at least 2.5 times of the standard deviation estimated using technical replicates of the common parental strain Σ1278b (N ≥ 72) (Figure S1C). Considering that each segregant tested for a given cross/trait combination was genotypically distinct, bimodal cases detected using our method, while robust against noise, were rather conservative and therefore likely represented a lower-bound estimation.
For all bimodal cases, we further analyzed the phenotypic segregation patterns in the tetrads and identified 98 as Mendelian, displaying the characteristic 2:2 segregation (Figure 1A, panel 4). In total, identified Mendelian cases represented 8.9% (98/1105) across our sample, and were interspersed among various conditions including large number of instances related to NaCl (28 crosses), CuSO4 (13 crosses), 6-azauracil (11 crosses) and acetate (9 crosses) (Figure 1B). Other low frequency cases were found on conditions related to signal transduction (caffeine), carbon sources (ethanol and xylose) various other conditions (formamide, benomyl and SDS) and the antifungal drugs cycloheximide and anisomycin (Figure 1B). In addition, we observed co-segregation of unrelated traits (NaCl, acetate and 6-azauracil; Figure 1B), where the fitness variation patterns in the segregants were highly correlated (Pearson’s correlation ρ > 0.9). We further characterized cases with co-segregations, high frequency cases related to CuSO4 and the low frequency case related to resistance to the drugs cycloheximide and anisomycin in detail. For the selected cases, 80 additional full tetrads were tested and the 2:2 phenotypic segregation patterns were confirmed.
Molecular characterization of identified Mendelian traits
Using bulk segregant analysis followed by whole genome sequencing, we identified one locus for each case as expected. For all crosses displaying co-segregation with NaCl, the same ~60 kb region (480,000 – 540,000) on chromosome IV was mapped, spanning the ENA genes encoding for sodium and/or lithium efflux pumps (Figure S2A). While variations of the ENA genes were known to lead to osmotic stress tolerance (Ruiz and Arino, 2007), the phenotypic associations with other co-segregating traits (acetate and 6-azauracil) were previously unknown. Causal genes related to acetate and 6-azauracil were suspected to be in close genetic proximity with the ENA locus, however the precise identities of these genes remained unclear. For cases related to CuSO4, we mapped a 40 kb region on chromosome VIII (190,000 – 230,000; Figure S2C). We identified the CUP1 gene in this region, which encodes for a copper binding metallothionein (Figure S2C). In this case, the common parental strain Σ1278b was resistant to both concentrations of CuSO4 tested and the allelic version of CUP1 in Σ1278b led to stable Mendelian inheritance across multiple genetic backgrounds (Figure 1B).
Finally, the last characterized case involved two anti-fungal drugs cycloheximide and anisomycin, which was found in the cross between a clinical isolate YJM326 and Σ1278b (Figure 1B). Pooled segregants belonging to the higher fitness cluster showed allele frequency enrichment for the YJM326 parent across a ~100 kb region on chromosome VII (420,000 – 520,000; Figure S2B). Further analyses yielded PDR1 as the potential candidate, which encodes for a transcription factor involved in multidrug resistance. Using reciprocal hemizygosity analysis (Figure S3A) as well as plasmid-based complementation test (Figure S3B), we showed that the PDR1YJM326 allele was necessary and sufficient for the observed resistance.
Fitness distribution of identified Mendelian traits across large natural populations
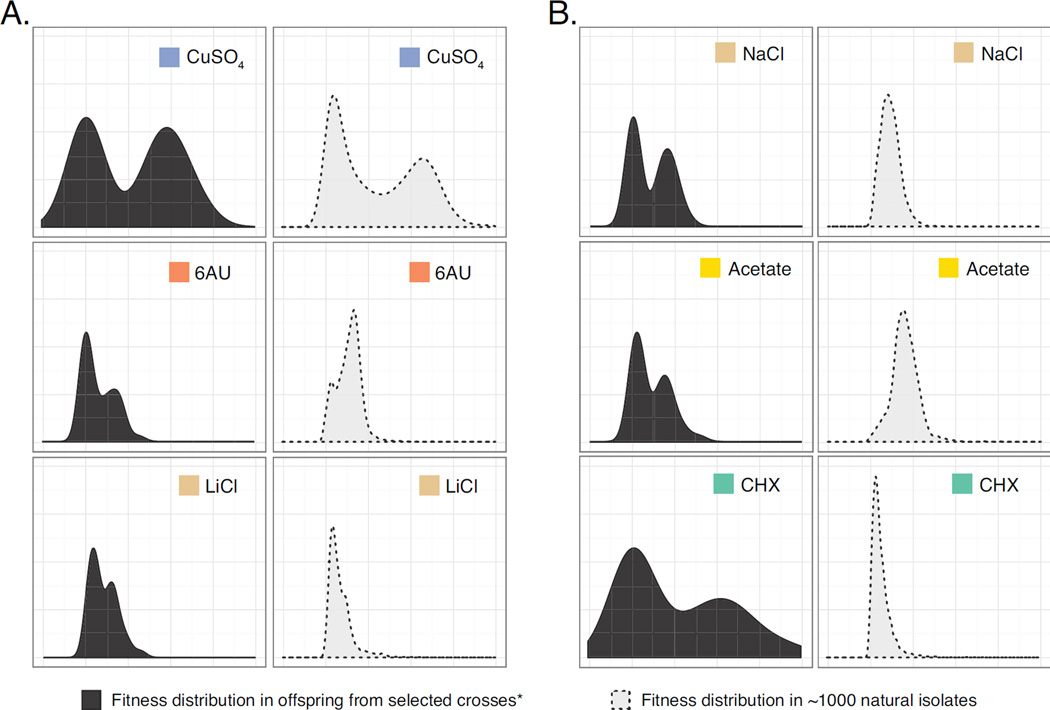
Although Mendelian traits could exhibit distinctive offspring distribution and segregation patterns in individual crosses, the general phenotypic distribution of such traits within a population was unclear. We measured the fitness distribution of an extremely large collection of ~1000 natural isolates of S. cerevisiae (the 1002 yeast genomes project, http://1002genomes.u-strasbg.fr/) on selected conditions related to identify Mendelian traits, including resistance to NaCl, LiCl, acetate, 6-azauracil, CuSO4 and cycloheximide (Figure 2). Interestingly, while some traits followed the same bimodal distribution model across the population as was observed in offspring from single crosses (Figure 2A), other traits with clear Mendelian inheritance pattern in crosses appeared to vary continuously at the population level (Figure 2B). This observation suggested that the phenotypic distribution within the population might not necessarily reflect the underlying genetic complexity of traits. Instead, the inheritance pattern for any given trait might largely be determined by specific combinations of parental genetic backgrounds.
Fig 2. Fitness distribution patterns of identified Mendelian traits within large natural population.
Comparisons of the fitness distribution on 6 selected conditions in individual crosses (left panel, N=40) and across ~1000 natural isolates of S. cerevisiae (right panel, N=960) are shown. Conditions tested are color-coded. (A) Bimodal distribution patterns observed both in crosses and at the population level. (B) Bimodal distributions observed only in crosses but not within a population.
Hidden complexity of a rare Mendelian variant across different genetic backgrounds
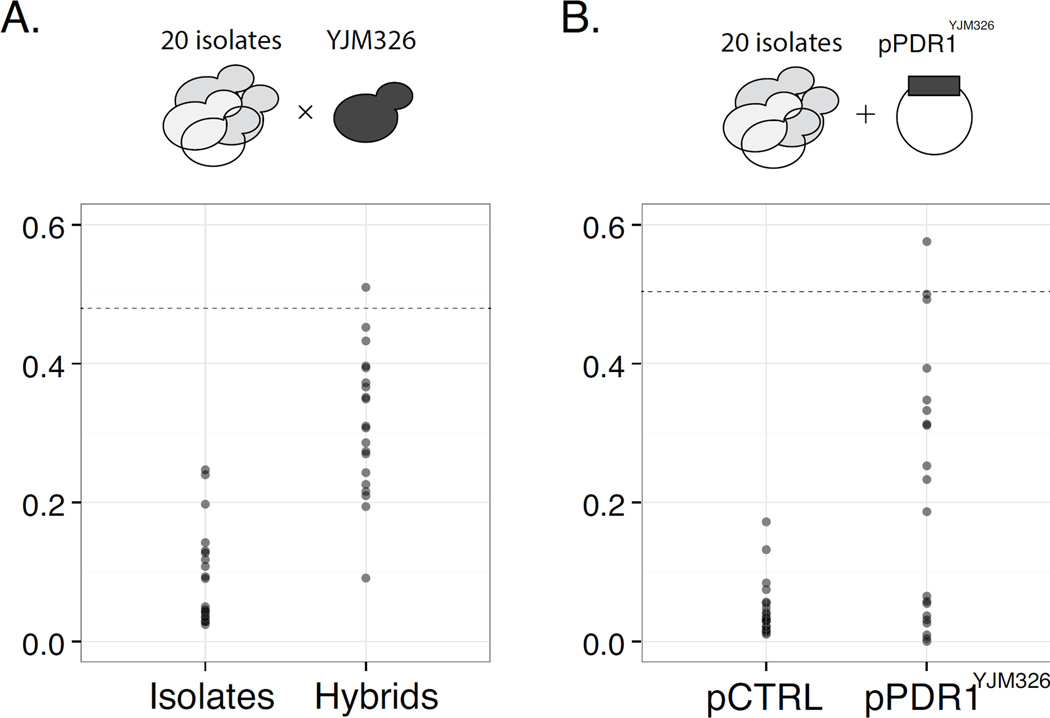
While focusing on highly frequent cases such as CuSO4 and NaCl provided indications about the transmission stability of common Mendelian variants and revealed previously unknown co-segregations, we were particularly interested in rare cases where the phenotypic effects and the general inheritance patterns across different genetic backgrounds were unknown. The identified Mendelian case related to the anti-fungal drugs cycloheximide and anisomycin could be considered as such. Across our panel, the parent YJM326 was the only highly fit isolate, and few isolates showed similar resistance level within the whole species (Figure 2B). To test the effect of the PDR1YJM326 allele in different backgrounds, we crossed the resistant isolate YJM326 with 20 diverse sensitive isolates. Counterintuitively, the resulting hybrids displayed continuous variation of the resistance in the presence of cycloheximide (Figure 3A). To test whether the resistance variation in the hybrids were due to allelic interactions at the PDR1 locus in different backgrounds, we introduced a plasmid carrying the PDR1YJM326 allele (pPDR1YJM326) into the same set of isolates, and quantified their fitness in the presence of cycloheximide (Figure 3B). Across all isolates tested, about half (11/20) expressed the resistant phenotype to various degrees (Figure 3B, Figure S3B). However, fitness between haploid isolates carrying pPDR1YJM326 and the corresponding hybrids were only weakly correlated (Pearson’s correlation ρ = 0.434), indicating that allelic interactions at the PDR1 locus only partly accounted for the observed variation (Figure S3B).
Fig 3. Effects of the PDR1YJM326 allele in different genetic backgrounds.
(A) Fitness variation of 20 isolates (left panel) in comparison with the same set of strains hybridized with YJM326 in the presence of drug. Fitness values (y-axis) correspond to the ratio between the growth in the presence of cycloheximide (YPD CHX 1µg/ml) and control media YPD. Dashed line indicates the fitness of the resistant strain YJM326. (B) Fitness variation of 20 isolates carrying empty control plasmid (pCTRL, left panel) or plasmid containing the PDR1YJM326 allele under its native promoter (pPDR1YJM326, right panel). Fitness values were measured in the presence of cycloheximide (YPD CHX 1µg/ml) with hygromycin to maintain plasmid stability. Dashed line indicates the fitness value of YJM326 carrying the plasmid pPDR1YJM326. See Figure S3 for detailed comparison for effect of hybrid and plasmid in individual genetic backgrounds.
The lack of correlation between hybrids and isolates carrying the plasmid with the PDR1YJM326 allele led us hypothesize the presence of potential modifiers in various hybrid backgrounds. To test this hypothesis, we evaluated the fitness distributions of the drug resistance in the offspring across the 20 hybrids generated previously. For each hybrid, 20 complete tetrads were tested in the presence of cycloheximide and the fitness distributions as well as the segregation patterns were assessed in the offspring (Figure S4A). In the absence of modifiers, haploid segregants are expected to have complete phenotypic penetrance, as the effects of intralocus interaction were eliminated. In this scenario, all crosses between any sensitive parental isolate and YJM326 should display a bimodal distribution in the offspring, with a 2:2 segregation of the phenotype.
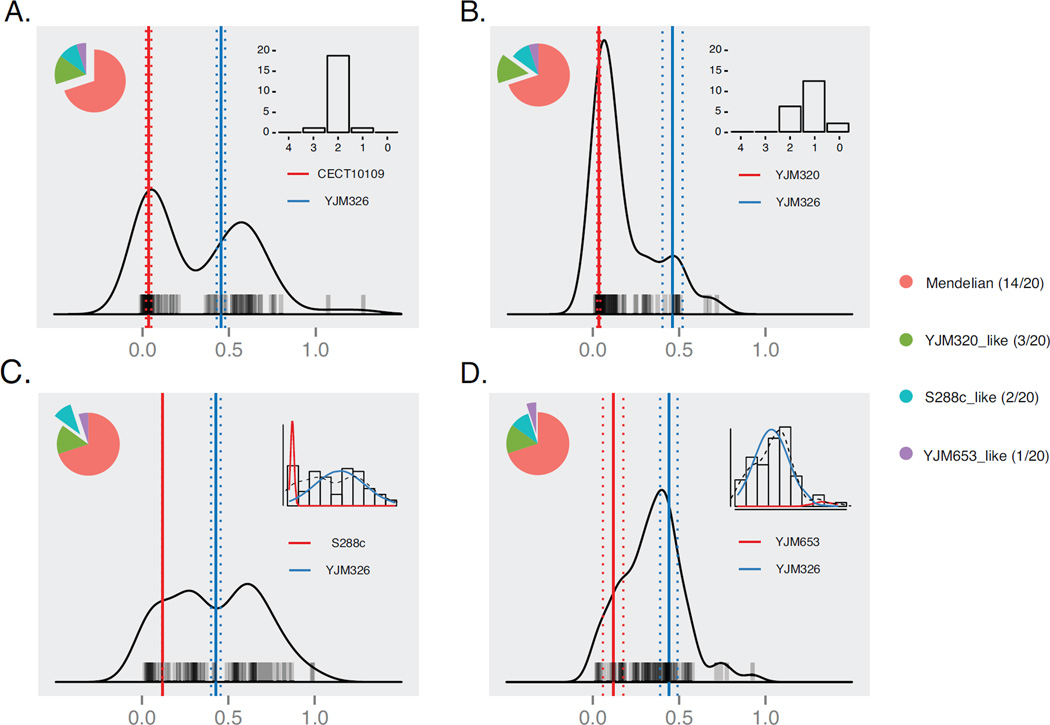
Interestingly, while most of the tested crosses (14/20) displayed Mendelian segregation as was observed in the cross between YJM326 and Σ1278b, several crosses showed clear deviation of the expected phenotypic distribution (Figure 4, Figure S4). In addition to Mendelian cases (Figure 4A), 3 other types of distribution were observed (Figure 4B–D). In total, such cases represent ~30% of all crosses tested. Of these crosses, 15% (3/20, between YJM320, Y3, Y9 and YJM326) showed incomplete penetrance, indicating possible suppressors of the PDR1YJM326 allele (Figure 4B). We observed a 1:4:1 ratio between tetrads containing 2, 1 and 0 resistant segregants, possibly indicating that two independent loci, including PDR1, were involved (Figure 4B, Figure S4). Furthermore, 10% of the crosses (2/20, between S288c, YJM440 and YJM326) showed enriched high fitness offspring, with an intermediate peak between the sensitive and resistant clusters. This observation suggests the presence of epistatic interactions from these specific genetic backgrounds, resulting as a transitional resistant phenotype cluster with higher genetic complexity (Figure 4C). The levels of genetic complexity in these cases are suspected to be low, but the precise number of genes involved remained unclear.
Fig 4. Post-Mendelian inheritance patterns of drug resistance in different hybrid contexts.
(A–D) Offspring fitness distribution patterns observed in hybrids originated from 20 sensitive isolates and YJM326 in the presence of cycloheximide (YPD CHX 1µg/ml). 80 offspring were tested for each case, and examples of Mendelian (A) and non-Mendedian (C–D) inheritance patterns are shown. Phenotypic segregation is indicated at the upper right side for cases with biased bimodal distributions (A and B). For cases with more complex patterns, maximum likelihood fittings of a bimodal model were shown instead (C and D). For nonbimodal cases the model fitting results were shown instead. Parental origins for each cross are shown, and the fitness values of the sensitive (red) or resistant (blue) parental strains are presented as vertical bars, with dotted bars corresponding to ± standard variation (N = 4). See Figure S4 for offspring fitness distributions for each of the 20 crosses, and additional offspring distributions for selected crosses with biased Mendelian patterns.
In addition to cases with low level of deviations from Mendelian expectations, we also found one cross (between YJM653 and YJM326) with largely biased offspring fitness distribution, for which a bimodal distribution with distinctive parental clusters was rejected by our model. In this case, the resistant phenotype was no longer caused by a single Mendelian factor, and the underlying genetic determinants were undoubtedly complex (Figure 4D). For selected crosses in each type of biased distributions, additional offspring (160 from 40 complete tetrads) were generated and the fitness distribution patterns were further confirmed (Figure S4B). Contrasting to other identified Mendelian traits with a stable inheritance patterns across the population, the PDR1 case represented a perfect example illustrating the hidden complexity of a simple Mendelian trait within natural population of the yeast S. cerevisiae.
Discussion
By performing a species-wide survey of monogenic variants in S. cerevisiae, we obtained a first lower-bound estimation of the proportion of Mendelian traits within a natural population. We showed that genes and alleles underlying the onset of Mendelian traits are variable in terms of their type, frequency and genomic distribution at the population level. Remarkably, by tracing the effect of one causal Mendelian variant PDR1YJM326 across the population, we demonstrated that the genetic complexity of traits could be dynamic, transitioning from clear Mendelian to diverse complex inheritance patterns depending on various genetic backgrounds.
Yeasts and more particularly S. cerevisiae have been extensively used as a model for dissecting many complex traits that were of medical, industrial and evolutionary interests (Bloom et al., 2013; Ehrenreich et al., 2012; Mukherjee et al., 2014; Steinmetz et al., 2002; Treusch et al., 2015). A trend emerging from studying complex traits in this species was that causal variants do not distribute randomly across the genome, and several hotspots have been identified (Fay, 2013). As a result, a low number of loci were found to be involved in high numbers of unrelated phenotypes, despite the fact that underlying causal genes could be different. Interestingly, causal variants in Mendelian traits seemed to follow the same trend as supported by our data. In fact, we observed phenotypic co-segregation of unrelated conditions such as resistance to acetate, 6-azauracil and osmotic stress, and showed that only a single region on chromosome IV was involved (Figure S2). In addition, the observed co-segregations showed relatively high population frequencies, with more than 15% of the crosses co-segregating on at least two different conditions (Figure 1B). This effect of linkage could possibly lead to biased phenotype assortments across the population, although the underlying evolutionary origin is unknown.
In general, Mendelian traits were considered as rare especially in human disorders, however, no directly estimation of the proportion of Mendelian relative to complex traits was available at the population level, and what types of genes were more susceptible to cause Mendelian inheritance were unknown. Our data showed that across a yeast natural population, causal alleles involved in direct response to stress, such as transporters (ENA) or metal-binding genes (CUP1) were more likely to follow Mendelian inheritance. In fact, a large number of Mendelian traits identified in our sample were related to these two loci, and the inheritance patterns were extremely stable, displaying 2:2 segregations with little influence of the genetic backgrounds. Similar pattern was found in a Mendelian trait related ammonium resistance in natural isolates of S. cerevisiae, where a transporter gene TRK1 was involved (Reisser et al., 2013). The stable inheritance patterns of traits caused by alleles with direct phenotypic effect could potentially due to the lack of regulatory complexity. As was supported by laboratory evolution experiments, amplifications of this type of genes were frequent, conferring to rapid acquisition of resistances in stress conditions such as salt (Anderson et al., 2010), copper (Fogel and Welch, 1982; Gerstein et al., 2015), sulfate (Gresham et al., 2008) and glucose limitations (Dunham et al., 2002).
By contrast, depending on the gene involved, a given Mendelian trait could lead to complex inheritance patterns across different genetic backgrounds, as evidence by the causal allele PDR1 related to resistance to cycloheximide and anisomycin. By crossing the strain YJM326 carrying the resistant allele PDR1YJM326 with diverse natural isolates, we showed that although most crosses retained stable 2:2 segregations, the inheritance pattern of the resistance phenotype in some cases displayed various deviations from Mendelian expectation, including reduced penetrance (3/20), increased genetic complexity (2/20) and in one extreme case, transition from monogenic to complex trait. We propose that the observed post-Mendelian inheritance patterns are due to the functional nature of the PDR1 gene. In fact, as PDR1 encodes for a transcriptional factor with complex regulatory networks and impact multiple downstream effector genes (Moye-Rowley, 2003), the resulting phenotypic expression would possibly be influenced by variations of a large number of genes that are involved in the same network in different genetic backgrounds.
Overall, our data provided a first comprehensive view of natural genetic variants that lead to the onset of Mendelian traits in a yeast population. We showed that monogenic mutations could exhibit post-Mendelian modifications such as pleiotropy, incomplete dominance as well as variations in expressivity and penetrance due to differences in specific genetic backgrounds. Depending on the parental combination, the inheritance might display a Mendelian, intermediate or complex pattern, showing the continuum of the complexity spectrum related to a monogenic mutation, as illustrated by the example of the drug resistance involving PDR1YJM. However, while Mendelian traits could be related to common or rare variants, we found that the overall fitness distribution patterns of such traits at the population level, for some instances if not all, were not informative regarding their genetic complexity. Collectively, phenotypic prediction even for simple Mendelian variants may not be an easy task, in part due to the lack of prediction power using population data and the scarcity of large-scale family transmission information, such as the case for diseases in human. Future studies using pairwise crosses covering a larger panel of conditions in yeasts, or in other model organisms, may provide general trends and a more complete picture regarding the phenotypic predictability of monogenic traits.
Experimental Procedures
Strains
Isolates from diverse ecological and geographical sources used in this study are detailed in Supplemental Experimental Procedures. All strains are stable haploids with deletion of the HO gene (Liti et al., 2009; Schacherer et al., 2009). Laboratory strains FY4, FY5 (isogenic to S288c) and Σ1278b were used. Deletion mutants in the Σ1278b background were obtained from the gene deletion collection kindly provided by Dr. Charles Boone (Dowell et al., 2010). YJM326 Δpdr1 strain was generated by insertion of hygromycin resistance cassette HygMX using homologous recombination.
Media and culture conditions
Detailed media compositions for phenotyping of the segregant panel are listed in Supplemental Experimental Procedures. Growth and maintenance of the strains are carried on standard rich media YPD (1% yeast extract, 2% peptone and 2% glucose). A final concentration of 200 µg/ml hygromycin (Euromedex) was supplemented to maintain the plasmids carrying a resistance marker gene HygMX. Sporulation was induced on potassium acetate plates (1% potassium acetate, 2% agar). All procedures are performed at 30°C unless otherwise indicated.
Crosses and generation of the offspring
For the construction of the segregant panel, 41 diverse isolates (MATα) were crossed with the lab strain Σ1278b (MATa) on YPD (Supplemental Experimental Procedures). Resulting diploids were sporulated for 2–3 days on sporulation medium (10 g/L potassium acetate, 20 g/L agar) at 30°C. Tetrad dissections were performed using the MSM 400 dissection microscope (Singer instrument) on YPD agar after digestion of the tetrad asci with zymolyase (MP Biomedicals MT ImmunO 20T). A total of 10 tetrads containing four viable spores were retained per cross. Same protocol was used for the validation of Mendelian cases with 2:2 segregation as well as the crosses between 20 isolates with the drug resistant strain YJM326 (Supplemental Experimental Procedures), in these cases 80 and 20 full tetrads were tested for each cross, respectively.
High-throughput phenotyping and growth quantification
Quantitative phenotyping was performed using end point colony growth on solid media. Strains were pregrown in liquid YPD medium and pinned onto a solid YPD matrix plate to a 384 density format using a replicating robot RoTor (Singer instruments). At least two replicates of each parental strain were present on the corresponding matrix, and 16 replicates for the common parent Σ1278b. The matrix plates were incubated overnight to allow sufficient growth, which were then replicated on 30 media conditions including YPD as a pinning control (see detailed compositions of the media in Supplemental Experimental Procedures). The plates were incubated for 48 hours at 30°C and were scanned at the 24, 40, 48 hour time points with a resolution of 600 dpi at 16-bit grayscale. Quantification of the colony size was performed using the Colony Area plugin in ImageJ, and the fitness of each strain on the corresponding condition was measured by calculating the normalized growth ratio between stress media and YPD using the software package ScreenMill (Dittmar et al., 2010). Phenotyping data related to crosses with YJM326 were analyzed using the R package Gitter (Wagih and Parts, 2014).
Model fitting procedure and detection of traits with Mendelian inheritance
For each cross/trait combination, a bimodal distribution was fitted using the R package “mixtools” (https://cran.r-project.org/web/packages/mixtools/index.html) with k = 2 and maxit = 500. Mean (μ), standard deviation (σ), proportion of each cluster (λ) and posterior probability of each cluster for each individual were extracted from the output file. To determine cutoff values of posterior probability for cluster assignment, a simulated dataset was generated, by simulating two normal distributions with n*λ and n*(1−λ) individuals for each cluster, respectively, with mean and standard deviation randomly sampled from observations in real data. For each simulated set, the two normal distributions generated were combined, and the procedure was repeated for 1000 times to generate a training set with 1000 distributions (Supplemental Experimental Procedures). The training set was then subjected to model fitting with the same parameters (Supplemental Experimental Procedures). The mean (μ), standard deviation (σ), proportion of each cluster (λ) and posterior probability of each cluster for each individual were extracted again, and the training dataset was evaluated against the real data (Supplemental Experimental Procedures). In this case, as the prior probability of cluster assignment was known for each simulated individual, it is possible to test for the detection sensitivity and specificity (Receiver Operating Characteristic or ROC) using varied cutoff parameters. A sequence starting from 0.5 to 0.95 (increment 0.05) for posterior probability and a sequence from 0 to 0.9 (increment 0.1) for percentage of non overlapping individuals were tested. The ROC curves and area under the curve (AUC) were calculated for each combination of cutoff parameters using R package “ROCR”(Sing et al., 2005). Cutoffs of 0.8 for posterior probability and 0.9 for percentage of non-overlapping were retained to ensure confident detections (Figure S1A–B). In this case, a maximum of 10% of individuals are allowed to have a posterior probability of less than 0.8, adjusting for the internal phenotypic stochasticity and noise. The defined parameters were applied on real data and cases passed the filter were preceded to cluster assignment. For bimodal cases with parental pairs that belong to either phenotype cluster, the segregation patterns were determined. As each cross/trait combination consists of 40 individual, a cutoff of 0.9 for percentage of non-overlapping would allow maximum of 4 individuals that could not be assigned with confidence. In theory, 4 tetrads maybe impacted. To adjust for this effect, a trait is considered as Mendelian when at least 7/10 of the tetrads display a 2:2 segregation with posterior probability > 0.8. All analyses were performed in R.
Evaluation of detection robustness against experimental noise
The levels of experimental noise were estimated using technical replicates of the Σ1278b parental strain. For each condition, at least 72 replicates were tested, and the noise level for a given condition was calculated as the standard deviation. To evaluate the robustness of the detected bimodal distributions, we compared the mean fitness differences between the assigned clusters to the observed noise related to the corresponding condition (Figure S1C). For all 318 detected bimodal cases, the mean fitness differences between the assigned clusters were higher than at least 2.5 times of the standard deviation. Among which, the identified Mendelian cases displayed mean cluster fitness differences higher than 3 times standard deviation (Z scores = ± 3, Figure S1C). These observations suggest that the Mendelian cases identified here were of high confidence and were unlikely to be due to distribution stochasticity or experimental noise.
Bulk segregant analysis
In total, 6 crosses representative of the identified Mendelian traits were subjected to bulk segregant analyses in order to identify the genomic regions involved. Crosses involved were between Σ1278b and EM93, I14, YJM269 and CLIB272 in presence of salt and a second stressor, and between Σ1278b and YJM326 for antifungal drugs and copper sulfate. For each case, 50 independent viable spores tetrads exhibiting 2:2 segregation on the corresponding conditions were separately grown overnight at 30 °C in liquid YPD and were pooled by equal optical density readings at 600 nm. Pooled segregants were subjected to whole genome sequencing and the genomic regions involved in each trait were determined by looking at allele frequency variation.
Genotyping strategy and data treatment
Genomic DNA from the pool was extracted using Genomic-tips 100/G columns and Genomic DNA buffers (QIAGEN) as described previously (Friedrich et al., 2015). Sequencing of the samples was performed using Illumina Hiseq 2000 except for the cycloheximide pool, for which we used MiSeq technology. Reads were mapped to the Σ1278b genome with the Burrows-Wheeler Aligner (BWA, version 0.7.4) allowing 5 mismatches and 1 gap (Li et al., 2009). The ‘-I’ flag has been added for the MiSeq Pool because reads were encoded in Illumina 1.9 format. Single nucleotide polymorphism (SNP) calling has been performed using GATK v3.3-0 (McKenna et al., 2010), with default parameters. The allele frequency of Σ1278b was calculated for each polymorphic position by adding the allele balance ratio, with the “VariantAnnotator” command of GATK.
Reciprocal hemizygosity test
To perform reciprocal hemizygosity test on drug resistance, the wild type strains Σ1278b and YJM326 were crossed with each other and with deletion mutant strains of PDR1 in both genetic backgrounds. Individual zygotes were isolated using the MSM 400 dissection microscope (Singer instrument) on YPD plate and the ploidy of the hybrids was checked on sporulation media after 2–3 days at 30°C. Phenotypic effects of the sensitive and resistance alleles were evaluated using drop test on selective media YPD CHX 1µg/ml and YPD as a growth control. Plates were scanned after 48 hours of incubation at 30°C.
Plasmids construction and phenotyping
Centromeric plasmid was constructed to test the allelic effect of the drug resistance allele PDR1YJM326 in different genetic backgrounds using Gateway cloning technology (Invitrogen). Fragment containing PDR1 and its native promoter and terminator regions flanked by attB1/attB2 recombination sites was amplified from the genomic DNA of YJM326 and Σ1278b, and was cloned into an empty centromeric plasmid with HphMX resistance marker (pCTRL, Treusch et al., 2015), according to instructions. The resulting plasmids, pPDR1YJM326 and pPDR1Σ1278b were verified using restriction enzymes and PCR amplification with internal primers of PDR1 gene. 20 diverse natural isolates were transformed with pPDR1YJM326 as well as the empty control plasmid pCTRL using EZ transformation kit (MP biomedicals). Transformants were selected on YPD media containing 200 µg/ml hygromycin. Growth quantification of isolates carrying the pPDR1YJM326 or pCTRL plasmids in the presence of drug was performed as described previously, and 200 µg/ml hygromycin were supplemented to all media to maintain the selection pressure during the phenotyping procedure.
Supplementary Material
Highlights.
In a natural population, at least ~9% of traits follow a Mendelian inheritance
Non-random distribution of causal Mendelian loci leads to co-segregating phenotypes
Monogenic variants may have a continuous expressivity spectrum in natural population
Background could lead to the transition from Mendelian to complex traits
Acknowledgments
We thank Matt Rockman, David Botstein and Bernard Dujon for comments on the manuscript. The authors thank the National Institutes of Health (NIH Grant R01 GM101091-01) and the Agence Nationale de la Recherche (ANR Grant 2011-JSV6-004-01) for financial support. M.J.D. is a Rita Allen Foundation Scholar and a Senior Fellow in the Genetic Networks program at the Canadian Institute for Advanced Research. A.S. is supported by a grant from Région Alsace. J.H. is supported in part by a grant from the Ministère de l’Enseignement Supérieur et de la Recherche and in part by a fellowship from the medical association La Ligue contre le Cancer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
All Illumina sequencing reads generated in this study have been deposited in the European Nucleotide Archive under the study accession number PRJEB11500.
References
- Anderson JB, Funt J, Thompson DA, Prabhu S, Socha A, Sirjusingh C, Dettman JR, Parreiras L, Guttman DS, Regev A, et al. Determinants of divergent adaptation and Dobzhansky-Muller interaction in experimental yeast populations. Current biology : CB. 2010;20:1383–1388. doi: 10.1016/j.cub.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonarakis SE, Beckmann JS. Mendelian disorders deserve more attention. Nature reviews Genetics. 2006;7:277–282. doi: 10.1038/nrg1826. [DOI] [PubMed] [Google Scholar]
- Antonarakis SE, Chakravarti A, Cohen JC, Hardy J. Mendelian disorders and multifactorial traits: the big divide or one for all? Nature reviews Genetics. 2010;11:380–384. doi: 10.1038/nrg2793. [DOI] [PubMed] [Google Scholar]
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano JL, Katsanis N. Beyond Mendel: an evolving view of human genetic disease transmission. Nature reviews Genetics. 2002;3:779–789. doi: 10.1038/nrg910. [DOI] [PubMed] [Google Scholar]
- Blomen VA, Majek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, Sacco R, van Diemen FR, Olk N, Stukalov A, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- Bloom JS, Ehrenreich IM, Loo WT, Lite TL, Kruglyak L. Finding the sources of missing heritability in a yeast cross. Nature. 2013;494:234–237. doi: 10.1038/nature11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Gunther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nature genetics. 2011;43:956–963. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C, Kehrer-Sawatzki H. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Human genetics. 2013;132:1077–1130. doi: 10.1007/s00439-013-1331-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipple KM, McCabe ER. Modifier genes convert "simple" Mendelian disorders to complex traits. Molecular genetics and metabolism. 2000;71:43–50. doi: 10.1006/mgme.2000.3052. [DOI] [PubMed] [Google Scholar]
- Dittmar JC, Reid RJ, Rothstein R. ScreenMill: a freely available software suite for growth measurement, analysis and visualization of high-throughput screen data. BMC bioinformatics. 2010;11:353. doi: 10.1186/1471-2105-11-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman R. Modifier gene studies to identify new therapeutic targets in cystic fibrosis. Current pharmaceutical design. 2012;18:674–682. doi: 10.2174/138161212799315920. [DOI] [PubMed] [Google Scholar]
- Dowell RD, Ryan O, Jansen A, Cheung D, Agarwala S, Danford T, Bernstein DA, Rolfe PA, Heisler LE, Chin B, et al. Genotype to phenotype: a complex problem. Science. 2010;328:469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, Botstein D. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich IM, Bloom J, Torabi N, Wang X, Jia Y, Kruglyak L. Genetic architecture of highly complex chemical resistance traits across four yeast strains. PLoS genetics. 2012;8:e1002570. doi: 10.1371/journal.pgen.1002570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC. The molecular basis of phenotypic variation in yeast. Current opinion in genetics & development. 2013;23:672–677. doi: 10.1016/j.gde.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel S, Welch JW. Tandem gene amplification mediates copper resistance in yeast. Proc Natl Acad Sci U S A. 1982;79:5342–5346. doi: 10.1073/pnas.79.17.5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich A, Jung P, Reisser C, Fischer G, Schacherer J. Population genomics reveals chromosome-scale heterogeneous evolution in a protoploid yeast. Molecular biology and evolution. 2015;32:184–192. doi: 10.1093/molbev/msu295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein AC, Ono J, Lo DS, Campbell ML, Kuzmin A, Otto SP. Too much of a good thing: the unique and repeated paths toward copper adaptation. Genetics. 2015;199:555–571. doi: 10.1534/genetics.114.171124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, DeSevo CG, Botstein D, Dunham MJ. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS genetics. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton BA, Yu BD. Modifier genes and the plasticity of genetic networks in mice. PLoS genetics. 2012;8:e1002644. doi: 10.1371/journal.pgen.1002644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nature reviews Genetics. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moye-Rowley WS. Transcriptional control of multidrug resistance in the yeast Saccharomyces. Progress in nucleic acid research and molecular biology. 2003;73:251–279. doi: 10.1016/s0079-6603(03)01008-0. [DOI] [PubMed] [Google Scholar]
- Mukherjee V, Steensels J, Lievens B, Van de Voorde I, Verplaetse A, Aerts G, Willems KA, Thevelein JM, Verstrepen KJ, Ruyters S. Phenotypic evaluation of natural and industrial Saccharomyces yeasts for different traits desirable in industrial bioethanol production. Applied microbiology and biotechnology. 2014;98:9483–9498. doi: 10.1007/s00253-014-6090-z. [DOI] [PubMed] [Google Scholar]
- Nadeau JH. Modifier genes in mice and humans. Nature reviews Genetics. 2001;2:165–174. doi: 10.1038/35056009. [DOI] [PubMed] [Google Scholar]
- Paaby AB, White AG, Riccardi DD, Gunsalus KC, Piano F, Rockman MV. Wild worm embryogenesis harbors ubiquitous polygenic modifier variation. eLife. 2015;4 doi: 10.7554/eLife.09178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisser C, Dick C, Kruglyak L, Botstein D, Schacherer J, Hess D. Genetic Basis of Ammonium Toxicity Resistance in a Sake Strain of Yeast: A Mendelian Case. G3. 2013 doi: 10.1534/g3.113.005884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Arino J. Function and regulation of the Saccharomyces cerevisiae ENA sodium ATPase system. Eukaryotic cell. 2007;6:2175–2183. doi: 10.1128/EC.00337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- Steinmetz LM, Sinha H, Richards DR, Spiegelman JI, Oefner PJ, McCusker JH, Davis RW. Dissecting the architecture of a quantitative trait locus in yeast. Nature. 2002;416:326–330. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- Strope PK, Skelly DA, Kozmin SG, Mahadevan G, Stone EA, Magwene PM, Dietrich FS, McCusker JH. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome research. 2015;25:762–774. doi: 10.1101/gr.185538.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein SL. Genetic modifiers of sickle cell disease. Hemoglobin. 2011;35:589–606. doi: 10.3109/03630269.2011.615876. [DOI] [PubMed] [Google Scholar]
- Treusch S, Albert FW, Bloom JS, Kotenko IE, Kruglyak L. Genetic Mapping of MAPK-Mediated Complex Traits Across S. cerevisiae. PLoS genetics. 2015;11:e1004913. doi: 10.1371/journal.pgen.1004913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu V, Verster AJ, Schertzberg M, Chuluunbaatar T, Spensley M, Pajkic D, Hart GT, Moffat J, Fraser AG. Natural Variation in Gene Expression Modulates the Severity of Mutant Phenotypes. Cell. 2015;162:391–402. doi: 10.1016/j.cell.2015.06.037. [DOI] [PubMed] [Google Scholar]
- Wagih O, Parts L. gitter: a robust and accurate method for quantification of colony sizes from plate images. G3. 2014;4:547–552. doi: 10.1534/g3.113.009431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.