Abstract
Background
Single-stranded DNA aptamers are oligonucleotides of approximately 50 base pairs in length selected for their ability to bind proteins with high specificity and affinity. Emerging DNA aptamer-based technologies may address limitations of existing proteomic techniques, including low sample throughput, which have hindered proteomic analyses of large cohorts.
Methods
To identify early biomarkers of myocardial injury, we applied an aptamer-based proteomic platform that measures 1129 proteins to a clinically-relevant perturbational model of “planned” myocardial injury (PMI), patients undergoing septal ablation for hypertrophic cardiomyopathy. Blood samples obtained before, at 10 and 60 minutes after PMI, and protein changes were assessed by repeated measures ANOVA. The generalizability of our PMI findings was evaluated in a spontaneous MI (SMI) cohort (Wilcoxon rank-sum). We then tested the platform’s ability to detect associations between proteins and Framingham Risk Score (FRS) components in the Framingham Heart Study (FHS); performing regression analyses for each protein versus each clinical trait.
Results
We found 217 proteins that significantly changed in the peripheral vein blood after PMI in a derivation cohort (n=15; P < 5.70E-5). Seventy-nine of these proteins were validated in an independent PMI cohort (n=15; P < 2.30E-4); > 85% were directionally consistent and reached nominal significance. We detected many protein changes that are novel in the context of myocardial injury, including Dickkopf related protein 4, a WNT pathway inhibitor (peak increase 124%, P = 1.29E-15) and cripto, a growth factor important in cardiac development (peak increase 64%, P = 1.74E-4). Among the 40 validated proteins that increased within 1 hour after PMI, 23 were also elevated in patients with SMI (n=46; P < 0.05). Our FHS studies revealed 156 significant protein associations with the FRS (n=899), including aminoacylase 1 (β = 0.3386, P = 2.54E-22) and trigger factor 2 (β = 0.2846, P = 5.71E-17). Further, we developed a novel workflow integrating DNA-based immunoaffinity with mass spectrometry to analytically validate aptamer specificity.
Conclusions
Our results highlight an emerging proteomics tool capable of profiling over one thousand low abundance analytes with high sensitivity and high precision, applicable both to well-phenotyped perturbational studies as well as large human cohorts.
Keywords: protein, cardiovascular disease, myocardial infarction
Emerging proteomic technologies are beginning to permit the systematic characterization of human plasma samples. Protocols for “deep” (i.e., low abundance) unbiased biomarker discovery, predominantly focusing on liquid chromatography-tandem mass spectrometry (LC-MS/MS), have been applied to multiple disease states.1–8 However, formidable, inter-related obstacles remain. These include the wide dynamic range of plasma protein concentrations necessitating multiple “upfront” enrichment and separation techniques prior to mass spectrometry analysis and the paucity of rigorously characterized antibodies for follow-up studies. Together, these challenges contribute to diminished sample throughput and have precluded proteomic discovery profiling of low abundance analytes in large numbers of plasma samples.
Single-stranded DNA aptamers are oligonucleotides of approximately 50 base pairs in length that are selected for their ability to bind target proteins or peptides with high specificity and affinity. They can be immobilized to streptavidin-coated beads and incubated with samples to assay analytes in a highly multiplexed manner. Emerging DNA aptamer-based proteomic technologies may address important limitations of existing proteomic techniques,9–11 but data in humans are still limited, particularly studies integrating independent validation strategies. Applications to archived samples from population-based cohorts are also lacking. Here we applied an aptamer-based proteomic technology that measures 1129 proteins12 to identify potential novel biomarkers of cardiovascular disease.
As proof of principle, we first applied the platform to humans subjected to a robust, clinically-relevant perturbation. We studied patients undergoing alcohol septal ablation for hypertrophic cardiomyopathy, a model of “planned” myocardial injury in which each individual serves as his or her own biological control. The planned myocardial infarction (PMI) reproduces key clinical features of “spontaneous” MI, including chest pain, electrocardiographic changes and wall motion abnormalities, as well as the release of established markers of myocardial injury.13, 14 Leveraging the timed nature of the human model, we tested whether the platform can identify very early markers of myocardial injury.
We then applied the aptamer proteomics platform to samples from participants of the Framingham Heart Study (FHS) Offspring Study, enrolled between 1991–1995.15 In this community-based cohort, we tested whether the technology could identify novel proteins associated with cardiovascular risk factors in participants without overt cardiovascular disease. Finally, we developed a novel analytical workflow integrating DNA-based immunoaffinity and mass spectrometry to begin to examine the specificity of aptamer-based techniques. These studies document a reproducible aptamer-based protein profiling platform with approximately 20-fold better throughput than LC-MS/MS-based plasma proteomic techniques. Further, studies from both populations highlight many novel candidate biomarkers of cardiovascular disease.
METHODS
Study Patients
Patients with hypertrophic cardiomyopathy undergoing septal ablation
A total of thirty patients undergoing PMI using alcohol septal ablation for the treatment of symptomatic hypertrophic cardiomyopathy were included in this study (15 in the derivation cohort; 15 in the validation cohort). Inclusion criteria for this cohort and the procedure were performed as previously described.1, 2, 16 Blood was drawn at baseline, 10 minutes, 1 hour and 24 hrs after injury. An additional six patients consented to the placement of a catheter into the coronary sinus during the ablation, allowing for the simultaneous sampling of blood from the coronary sinus and femoral catheters at baseline, 10 minutes and 1 hour. Four patients undergoing elective, diagnostic cardiac catheterization for cardiovascular disease, but not acute myocardial ischemia, were used as controls for the PMI patients. Blood was drawn before the onset of cardiac catheterization and at 10 minutes and 1 hour after the procedure was begun.
Patients with spontaneous acute coronary syndromes and at-risk controls
We enrolled 23 patients undergoing emergent cardiac catheterization for acute ST-segment elevation, spontaneous MI within 8 hours of symptom onset. Femoral venous blood samples were obtained in the coronary catheterization suite upon initial presentation. Peripheral blood from 23 patients with negative standard Bruce protocol exercise tests was used as controls for spontaneous MI.17
Framingham Heart Study Participants
The FHS Offspring cohort was formed in 1971 with the enrollment of 5124 individuals in a community-based longitudinal cohort study.15 The analyses of proteins with cardiometabolic traits were performed on a case-cohort design sampling in which we selected baseline plasma samples from 899 participants (311 cases who developed incident cardiovascular disease (CVD) and 588 randomly selected controls who did not). Among the 899 participants, we also performed duplicate proteomic analyses on 94 individuals across the lowest and highest tertiles of the Framingham Risk Score (FRS) to assess platform reproducibility. Inclusion criteria as an incident case was based on cardiovascular disease events or diagnoses (including coronary heart disease, myocardial infarction, angina, coronary insufficiency, cerebrovascular accident, atherothrombotic infarction of the brain, transient ischemic attack, cerebrovascular disease, and intermittent claudication) occurring after exam 5. Participants with prevalent CVD as defined above at exam 5 were excluded.
Study Samples
Blood samples were collected in K2EDTA-treated tubes in PMI patients and citrate-treated tubes in FHS participants. Samples were centrifuged within 15 minutes at 2000g for 10 min to pellet cellular elements. The supernatant plasma was then aliquoted and frozen at −80 °C.
Proteomic assay overview
The SOMAscan™ proteomic profiling platform uses single-stranded DNA aptamers that target 1,129 proteins. All assays were performed using SOMAscan reagents according to the manufacturer’s detailed protocol.12 A detailed protocol is provided in Supplemental Methods and in Supplemental Figure 1. A complete list of the proteins is included in the Supplemental Table 1. Many of the proteins are either secreted or known to be shed from the cell surface, and thus the platform is particularly well-suited for plasma biomarker discovery.
Statistical analyses
PMI studies
All protein values were log transformed because of their nonnormal distributions as determined by the Kolomogorov-Smirnov and Shapiro-Wilk normality tests. For the PMI and cardiac catheterization control studies, one-way repeated measures ANOVA was used to test differences in protein levels across time points (baseline, 10 minutes, 1 hour, 24 hours). The Greenhouse-Geisser adjustment was used if sphericity was violated. In analyses to identify proteins that might be altered by the cardiac catheterization procedure itself (without overt injury), a P < 0.05 was used to determine significant changes. Consequently, 253 proteins that were changed after heparin administration during cardiac catheterization were excluded from the PMI analyses. Bonferroni-corrected P threshold < 5.70E-05 (0.05/878 proteins on platform excluding 253 proteins on the SOMAscan that changed in catheterization heparin control patients) and < 2.30E-04 (0.05/217 proteins identified as significantly changed in derivation cohort) were used to determine significance in the derivation and validation PMI cohorts, respectively. A Wilcoxon rank-sum test was used in the spontaneous myocardial infarction (SMI) case-control analysis with a Bonferroni-corrected P threshold < 1.25E-03 (0.05/40 validated proteins shown to increase in first hour of myocardial injury) for significance. For the coronary sinus versus peripheral blood PMI analysis, two-way repeated measures ANOVA was performed to determine significant changes across time.
FHS studies
Protein measures were log transformed, then standardized (to mean =0 and SD=1). Age- and sex- adjusted regression analyses were performed for each protein (outcome) versus each clinical trait (FRS, age, female sex, total cholesterol, HDL cholesterol, SBP, diabetes, and smoking). Given the 1129 proteins analyzed, we used a Bonferroni-corrected P threshold of 5.54E-06 = [0.05/(1129 proteins x 8 traits)] to account for the number of statistical tests. Inverse probability sampling fractions for weights were applied in the analyses to account for the sampling algorithm. The sampling fractions were f1 = 311/324 cases and f0 = 588/2681 non-cases, so weights were w1 = 1/f1 and w0 = 1/f0 for cases and non-cases, respectively. We multiplied these weights by 899/3005 so weights sum to our sample size, 899. In the incidence analyses, we used proportional hazards regression models.
All analyses were performed with SAS Software version 9.4 (SAS Institute, Cary, NC). Figures were generated with Graph Pad Prism 5 (La Jolla, CA) and Tableau 9.1 (Seattle, WA).
Study Approval
All human study protocols were approved by the Institutional Review Boards of Massachusetts General Hospital and Boston University Medical Center. Informed consent was obtained from all participants.
RESULTS
Early protein changes in peripheral plasma of PMI patients
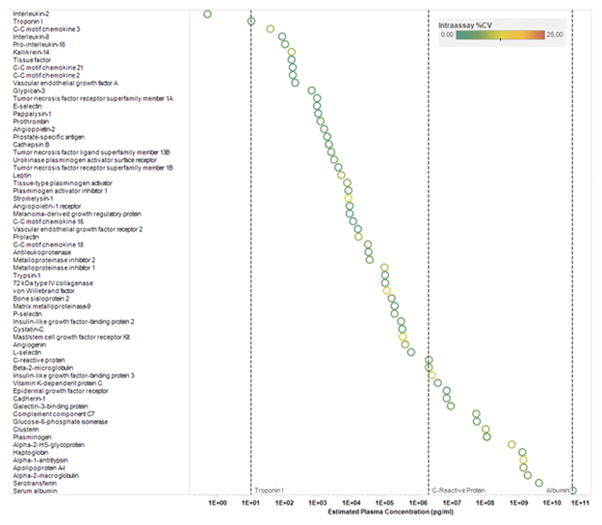
To assess reproducibility of the platform, we embedded pooled plasma control samples across a total of 43 individual plate assays and documented median intra-and inter-experimental coefficients of variance (CVs) of ≤ 8.2% for the 1129 proteins (see Supplemental Methods; a list of all proteins assayed are included in Supplemental Table 1). Figure 1 demonstrates the intra-assay CVs of representative proteins across a broad dynamic range of abundance. Using split sample analyses from 94 individuals, we also documented a median intraclass correlation of > 0.95 (Supplemental Figure 2).
Figure 1. Intra-assay CVs of selected proteins.
Data represent intra-assay CVs of selected proteins across a broad range of plasma concentrations. Proteins are sorted by increasing plasma concentration reference range (http://www.plasmaproteome.org/plasmaframes.htm). The color of the circle represents intra-assay CV as denoted in the key above. See Supplemental Table 1 for full protein names, Entrez Gene symbol/ID and UniProt ID.
To assess the linear dynamic response of the aptamer assay, we performed calibration curve experiments with standards for a subset of the novel proteins that were associated with PMI (see below). The calibration curves shown in Supplemental Figure 3 demonstrate a linear relationship to increasing concentrations of protein standards and provide estimated endogenous protein plasma concentration.
Clinical characteristics of the PMI study patients are detailed in Supplemental Table 2. We profiled peripheral blood samples across serial time points (baseline, 10 minutes, 1 hr and 24 hrs after injury) to characterize protein changes associated with myocardial injury. We determined that 253 proteins were nominally changed (P < 0.05) within 1 hour following the exposure of patients to intravenous heparin during cardiac catheterization (Supplemental Table 3), and therefore excluded these proteins from the analyses. We identified 217 proteins that were significantly changed within 24 hours post injury in a derivation cohort of 15 patients (Bonferroni-adjusted P < 5.70E-05, one-way ANOVA repeated measures). In a validation cohort of 15 patients, 87% of the 217 proteins had directionally consistent changes that reached nominal statistical significance (P < 0.05); changes in 79 proteins exceeded a repeat Bonferroni threshold (P < 2.30E-04, one-way ANOVA repeated measures). Table 1 details a subset of the 79 validated proteins that had changes of > 33% at any time point after PMI. All validated proteins are listed in Supplemental Table 4.
Table 1.
Protein changes in peripheral blood after myocardial injury
| Protein | % Change (IQR) 10 min vs. Baseline |
% Change (IQR) 1 hr vs. Baseline |
% Change (IQR) 24 hr vs. Baseline |
P |
|---|---|---|---|---|
| Angiogenin | 34.1 (29.3 to 61.8) | 24.5 (19.9 to 38.1) | −1.6 (−6.5 to 15.3) | 2.63 E-06 |
| Annexin VI | −46.0 (−75.6 to −32.3) | −34.9 (−76.9 to −30.7) | 33.9 (21.5 to 193.1) | 3.58E-06 |
| b-ECGF | 72.7 (32.3 to 102.4) | 103 (82.0 to 158.9) | −3.4 (−7.2 to 6.5) | 1.34E-10 |
| Cadherin-12 | 101 (34.9 to 168.7) | 213 (84.2 to 239.0) | 15.4 (−17.6 to 42.5) | 1.02E-04 |
| CDC37 | 45.0 (2.1 to 69.2) | 64.3 (22.6 to 81.9) | 7.4 (−6.5 to 18.3) | 2.67E-07 |
| Chymase | 41.3 (22.3 to 52.6) | 43.3 (30.5 to 67.5) | 19.1 (−1.7 to 28.9) | 1.91E-04 |
| CK-MB | 143 (46.6 to 305.1) | 506 (374.5 to 690.6) | 346 (138.7 to 1053.3) | 9.60E-08 |
| CK-MM | 42.0 (13.7 to 81.4) | 233 (146.1 to 287.7) | 234 (108.1 to 484.9) | 7.28E-07 |
| Cripto | 45.5 (14.0 to 66.1) | 63.6 (24.4 to 80.7) | 4.9 (−7.3 to 16.4) | 1.74E-04 |
| DKK-4 | 124 (101.9 to 157.4) | 56.7 (39.6 to 100.1) | −11.2 (−24.1 to 12.5) | 1.29E-15 |
| FABP | 178 (105.3 to 399.1) | 742 (551.8 to 1354.9) | 69.3 (5.3 to 235.0) | 1.28E-15 |
| FGF-18 | 134 (83.6 to 173.0) | 272 (189.9 to 359.2) | −8.4 (−27.3 to 98.2) | 6.31E-08 |
| FGF-8B | 29.4 (15.7 to 38.9) | 39.1 (26.1 to 56.9) | −0.1 (−10.1 to 8.8) | 1.54E-04 |
| GAS1 | 38.3 (18.8 to 51.7) | 65.3 (35.7 to 79.5) | 10.2 (1.0 to 15.1) | 4.73E-05 |
| GREM1 | 131 (62.2 to 154.3) | 176 (101.4 to 230.9) | 2.2 (−5.6 to 20.6) | 2.78E-07 |
| Histone H2A.z | −61.0 (−73.0 to −48.7) | −38.3 (−52.6 to −23.5) | 84.1 (31.4 to 257.0) | 5.41E-10 |
| IL-11 | 32.6 (15.8 to 48.0) | 44.4 (19.2 to 63.4) | 5.3 (−1.3 to 14.4) | 3.65E-08 |
| IL-5 | 31.2 (17.7 to 59.0) | 48.9 (30.5 to 71.8) | 23.8 (−5.3 to 50.8) | 3.98E-05 |
| IL-6 | −6.0 (−20.2 to 3.9) | 7.0 (−11.6 to 13.2) | 113 (35.9 to 173.7) | 1.20E-04 |
| KREM2 | 11.9 (4.6 to 27.8) | 36.1 (24.9 to 39.7) | −1.9 (−10.1 to 6.4) | 1.08E-04 |
| LBP | −28.9 (−40.5 to −26.6) | −30.5 (−36.9 to −23.1) | 78.5 (59.6 to 92.3) | 1.03E-07 |
| LDH-H1 | 6.7 (0.8 to 18.8) | 8.7 (1.7 to 20.3) | 73.4 (52.4 to 144.2) | 8.76E-14 |
| LIFsR | 196 (126.8 to 247.0) | 175 (114.2 to 217.0) | 19.2 (−3.4 to 27.7) | 2.68E-07 |
| MDHC | 50.3 (14.6 to 67.0) | 221 (149.3 to 279.4) | 143 (45.4 to 232.4) | 1.48E-08 |
| MMP-13 | 141 (102.7 to 237.8) | 64.8 (36.1 to 170.4) | 3.6 (−3.8 to 17.1) | 1.65E-14 |
| MMP-16 | 50.7 (20.9 to 68.5) | 62.2 (27.0 to 75.0) | 7.1 (4.2 to 36.9) | 7.33E-10 |
| Myoglobin | 88.3 (59.5 to 161.1) | 152 (105.7 to 231.0) | 16.6 (−2.2 to 32.3) | 8.53E-11 |
| NACA | 49.7 (16.6 to 70.4) | 43.3 (26.6 to 65.8) | 25.8 (11.2 to 46.6) | 4.72E-05 |
| NET4 | 115 (61.0 to 152.3) | 63.3 (20.1 to 82.5) | 1.7 (−9.9 to 23.4) | 1.11E-04 |
| PPIB | 42.7 (17.8 to 78.4) | 61.5 (38.3 to 95.5) | 22.9 (10.6 to 59.0) | 3.87E-05 |
| SCGF-alpha | 31.4 (19.0 to 55.7) | 48.0 (32.4 to 70.2) | 13.7 (2.3 to 34.2) | 5.89E-07 |
| SDF-1 | 120 (78.8 to 193.9) | 82.8 (45.3 to 119.4) | −6.7 (−17.3 to 3.5) | 4.41E-06 |
| ST4S6 | 43.6 (33.1 to 58.8) | 20.7 (12.9 to 34.5) | 7.2 (−5.4 to 33.3) | 9.94E-07 |
| TECK | 530 (264.8 to 634.9) | 213 (80.5 to 251.4) | −20.1 (−33.3 to 9.7) | 5.53E-18 |
| TPI | 45.1 (8.1 to 58.2) | 89.1 (59.3 to 145.7) | 2.4 (−9.1 to 37.5) | 4.21E-08 |
| Troponin I | 836 (452 to 1241) | 2692 (1756 to 5241) | 11400 (8435 to 20459) | 1.34E-26 |
| URB | 39.1 (23.7 to 51.9) | 18.9 (12.1 to 25.8) | −17.6 (−26.4 to −4.5) | 1.76E-04 |
Shown are proteins with ≥ 33% increase from baseline at any time point within validation cohort and P < 2.30E-04 (one-way ANOVA on log transformed RFU values), listed in alphabetical order. All proteins listed were significant in derivation cohort with P < 5.70E-05 (one-way ANOVA on log transformed RFU values). Change values denote median percent change (first and third quartiles). See Supplemental Table 1 for full protein names, Entrez Gene symbol/ID and UniProt ID.
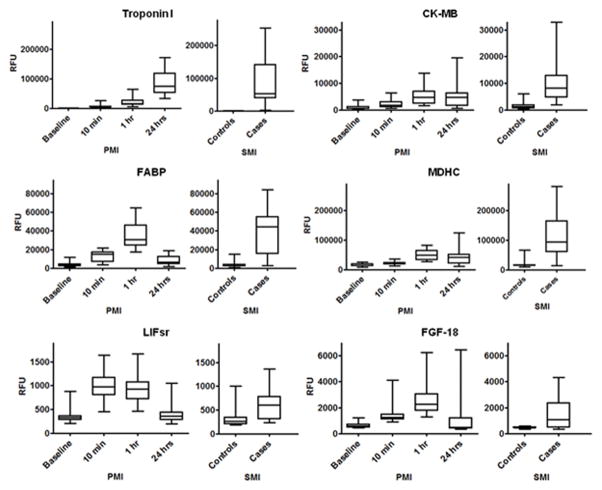
The dynamic changes in representative proteins are illustrated in Figure 2. We confirmed increases in well-established clinical markers of myocardial injury including troponin I and CK-MB, as well as other biomarkers previously identified by our group and others such as fatty acid binding protein (FABP)18, 19 and stromal derived factor-1 (SDF-1).20–22 Among the 79 proteins that changed in both derivation and validation cohorts, 31 increased by ≥ 33% within 1 hour of injury onset, and 25 proteins were increased within 10 minutes. Many of these protein changes were novel in the context of early myocardial injury (see Table 1), including fibroblast growth factor 18 (FGF 18), Dickkopf-related protein 4 (DKK-4), teratocarcinoma-derived growth factor 1 (Cripto-1) and phosphodiesterase 7A (PDE7A). We used immunoassays that were available for a subset of the novel proteins to confirm PMI findings (Supplemental Figure 4). Further, 12 of the protein changes identified on the aptamer platform were directionally consistent and significant in prior label free and isobaric tagging MS-based work on PMI samples.1, 2 Non-overlapping findings between the aptamer and prior MS-based studies are in part due to differences in experimental design and the targeted nature of the aptamer platform, though specific proteins were identified exclusively by each of the distinct proteomic approaches.
Figure 2. Protein markers that are increased early after the onset of myocardial injury in peripheral blood.
Data from selected proteins that increased in both derivation and validation cohorts [(P < 5.70E-05 in derivation cohort (n=15) and P < 2.30E-04 in validation cohort (n=15)]. P calculated by one-way ANOVA performed on log transformed RFU values. Edges of boxes denote 25th and 75th percentiles, lines denote median, whiskers denote minimum and maximum values. See Supplemental Table 1 for full protein names, Entrez Gene symbol/ID and UniProt ID. RFU: relative fluorescent units.
Changes in the coronary sinus during planned myocardial injury
In a small cohort of 6 PMI patients, we compared protein levels in samples obtained concurrently from peripheral blood and the coronary sinus (CS), the venous outflow from the heart. Higher protein levels in the coronary sinus samples as compared with the periphery suggest cardiac origin. Troponin-I and CK-MB, well-established markers of myocardial injury, were modestly enriched (~1.2-fold) in the coronary sinus at 1 hour after injury in this subset of individuals. Among the 79 validated proteins, 24 proteins were increased in the coronary sinus with ≥1.2 fold increase as compared to peripheral blood within 1 hour after myocardial injury. Supplemental Table 5 highlights the novel proteins found to be enriched in the coronary sinus within 1 hour of injury (nominal P < 0.05; 2-way ANOVA). Overall, there was concordance in the directionality of changes of the 79 proteins in the peripheral and CS samples (r > 0.88; P < 2.2E-16), again confirming the consistency of our findings across multiple sample time points and sampling sites.
Validation of candidate protein markers in spontaneous myocardial infarction
To begin to assess the generalizability of the findings from the planned MI cohort, we profiled a cohort of spontaneous myocardial infarction (SMI) patients presenting for acute coronary angiography (n=23 cases), as well as at-risk individuals without ischemia as determined by exercise stress testing (n=23 non-ischemic controls). Because the timing of sample collection relative to SMI onset was variable (7.0 ± 2.5 hrs), we focused on the PMI-derived candidate protein markers that were elevated at one hour, the closest available time point after the procedure. Among the 40 proteins that were increased after 1 hour of injury, twenty-three were elevated in SMI at nominal significance (P < 0.05). Fourteen of these proteins exceeded repeat Bonferroni threshold for statistical significance (Table 2) (P < 1.25E-03; Wilcoxon rank-sum). Several of these proteins are novel in the setting of acute myocardial infarction including malate dehydrogenase, cytoplasmic (MDHC), triosephosphate isomerase (TPI), leukocyte inhibitory factor soluble receptor (LIFsr), and fibroblast growth factor-18 (FGF18). Representative data for three known biomarkers and three novel proteins are demonstrated in Figure 3. Of note, twelve of the proteins that reached Bonferroni significance across all datasets were elevated as early as 10 minutes after myocardial injury.
Table 2.
Proteins increased in spontaneous and planned MI
| Protein | Fold Change (SMI vs. Control) | P |
|---|---|---|
| Troponin I | 177 | 2.43E-13 |
| CK-MB | 6.89 | 2.94E-10 |
| CK-MM | 6.05 | 3.88E-10 |
| FABP | 10.2 | 6.59E-10 |
| MDHC | 5.91 | 4.47E-09 |
| Myoglobin | 3.19 | 4.00E-07 |
| IL-6 | 1.63 | 4.21E-06 |
| LDH-H 1 | 1.70 | 1.73E-05 |
| TPI | 1.70 | 3.97E-05 |
| MMP-16 | 1.13 | 3.97E-05 |
| LIFsR | 1.89 | 4.45E-05 |
| Transketolase | 1.72 | 1.07E-04 |
| PDE7A | 1.22 | 4.76E-04 |
| FGF-18 | 3.18 | 5.73E-04 |
Shown are proteins increased in SMI cases vs. controls with P < 1.25E-03, listed in ascending P (Wilcoxon rank-sum on log transformed RFU values). All proteins listed were also increased in the PMI cohorts (derivation P < 5.70E-05 and validation P < 2.30E-04; one-way ANOVA on log transformed RFU values). See Supplemental Table 1 for full protein names, Entrez Gene symbol/ID and UniProt ID.
Figure 3. Proteins increased during planned and spontaneous myocardial injury.
Representative proteins that increased in PMI derivation and validation cohorts and an independent SMI cohort. PMI data shown from validation cohort (n=15; P < 2.30E-04; one-way ANOVA on log RFU values). SMI cohort (n=23 cases and 23 controls, P < 1.25E-03; Wilcoxon rank-sum on log RFU values). Edges of boxes denote 25th and 75th percentiles, lines denote median, whiskers minimum and maximum values. See Supplemental Table 1 for full protein names, Entrez Gene symbol/ID and UniProt ID. RFU: relative fluorescent units.
Proteins associated with cardiovascular risk traits in the Framingham Heart Study
We extended our initial perturbational studies to test whether the platform could identify novel protein associations with cardiac risk factors in a community-based sample. We studied 899 individuals from the Framingham Offspring Study (Supplemental Table 6) across a spectrum of the Framingham CHD Risk Score (FRS),23 which includes age, sex, total cholesterol, high-density lipoprotein (HDL) cholesterol, systolic blood pressure (SBP), smoking and diabetes status. The mean age for this cohort was 56 (± 12) years with a slight female predominance (52%) and mean FRS 3.43 (± 4.88). In the FHS sample, age- and sex-adjusted median absolute pairwise correlations between the proteins on the platform was 0.05, with over 93% of pairwise correlations < 0.2 and 99 % < 0.4.
Top protein correlations with established cardiovascular risk factors including sex, age, total cholesterol, HDL cholesterol, SBP, diabetes and smoking are shown in Supplemental Tables 7 – 13. As expected, female sex was positively associated with luteinizing and follicular stimulating hormones (P = 3.06E-149 and P = 2.72E-145; respectively) and negatively associated with PSA (P = 6.70E-52). In addition, we observed many novel relationships including smoking with circulating levels of the bone associated osteomodulin (OMD; P = 4.25E-25) and age with anti-angiogenic factor endostatin (P = 2.48E-10).
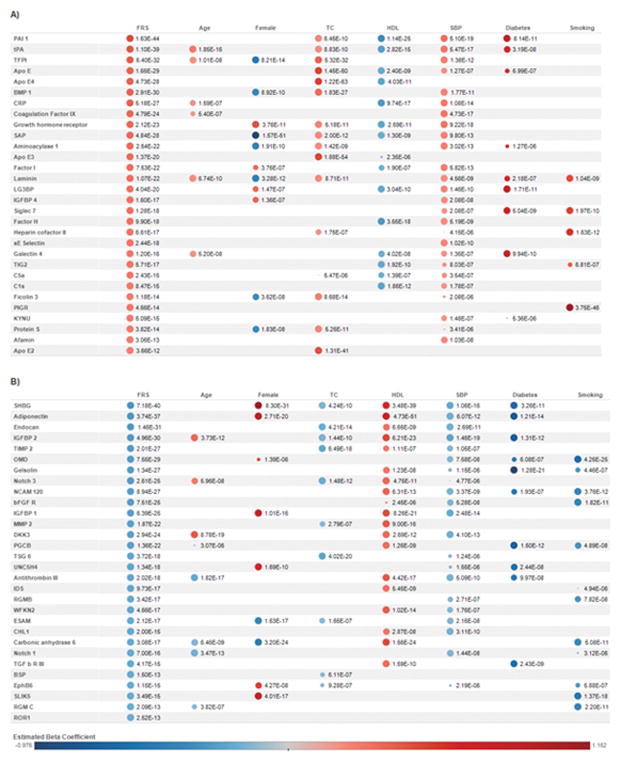
Age- and sex-adjusted regression analyses identified 156 proteins significantly associated with the FRS [(P < 5.54E-06; Bonferroni correction for (1129 proteins x 8 traits)]. Known biomarkers such as C-reactive protein (CRP, P = 5.18E-27) and apolipoprotein E subclasses (P < 3.66E-12) were positively associated with the FRS. There were numerous novel relationships identified as well, including aminoacylase 1 (P = 2.54E-22), laminin (P = 1.07E-22), trigger factor 2 (TIG2, P = 5.71E-17), and matrix metalloproteinase-12 (MMP-12, P = 3.93E-16). Figure 4 includes the top protein associations with FHS risk score along with the corresponding protein data with the clinical risk factors that comprise the FRS. All significant protein associations with FRS are listed in Supplemental Table 14.
Figure 4. Top protein associations with the Framingham Risk Score (FRS) and component clinical traits.
Shown are the top 30 proteins with positive (Panel A) and negative (Panel B) associations. Within groups of positive/negative associations, proteins are sorted by ascending P. Estimated beta coefficients and P were generated from age- and sex- adjusted regression analyses of each protein (log transformed then standardized) with FRS and clinical traits (standardized). The size of the circles corresponds to P (larger circles represent smaller P). Protein-trait associations with P > 5.54E-06 were not represented by circles. The color of the circles represents estimated beta coefficients as denoted in the key above. Abbreviations: FRS: Framingham Risk Score, TC: total cholesterol, HDL: high-density lipoprotein, SBP: systolic blood pressure. Diabetes and current smoking status were categorical variables. See Supplemental Table 1 for full protein names, Entrez Gene symbol/ID and UniProt ID.
In exploratory analyses, proportional hazards models were used to assess the association between baseline protein levels and future CVD, adjusting for age and sex (Table 3). One protein (tPA) reached Bonferroni adjusted significance. Proteins associated with incident disease with nominal P < 0.05 are presented in Supplemental Table 15. Following multivariable adjustment for all of the FHS components, cathepsin H was most strongly associated with incident disease, though this finding did not reach Bonferroni correction for multiple hypothesis testing (P = 0.002).
Table 3.
Top FHS protein associations with incident cardiovascular disease (age- and sex-adjusted)
| Protein | HR per s.d. (CI) | P |
|---|---|---|
| tPA | 1.65 (1.31–2.07) | 2.1E-05 |
| Cathepsin H | 1.47 (1.21–1.79) | 1.3E-04 |
| NCAM 120 | 0.68 (0.55–0.83) | 2.5E-04 |
| IDS | 0.69 (0.56–0.85) | 3.9E-04 |
| C1s | 1.42 (1.17–1.73)) | 5.1E-04 |
| IGFBP4 | 1.48 (1.18–1.86) | 7.8E-04 |
Shown are proteins with P < 0.001, listed by ascending P. Values are hazard ratios per standard deviation (95% confidence intervals) for incident CVD from proportional hazards regression models. See Supplemental Table 1 for full protein names, Entrez Gene symbol/ID and UniProt ID.
Technical validation of aptamer findings
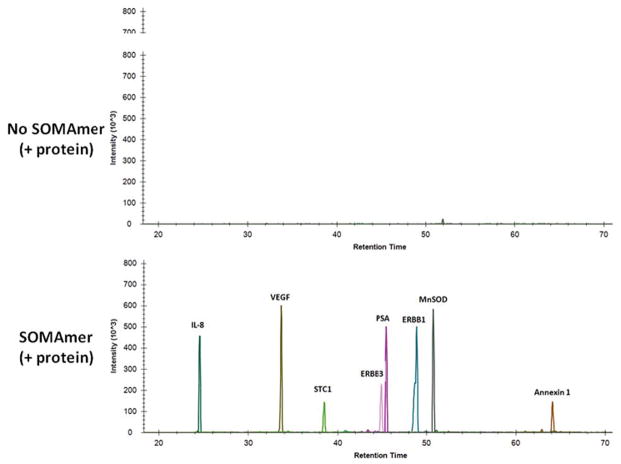
To begin to systematically assess platform specificity, we developed protocols using aptamers for immunoaffinity pulldown of intact proteins followed by digestion to peptides and analysis by targeted mass spectrometry. We acquired 29 aptamers and their cognate proteins, across a range of sizes and functions, including both intracellular and secreted proteins. We first added a mixture of the protein standards into plasma with the corresponding mixture of aptamer-coated beads. After overnight incubation, the beads were boiled and analyzed by SDS-PAGE. All of the proteins were characterized by one predominant band at the anticipated molecular weight (data not shown). Conditions for eluting bound proteins from aptamers were then optimized and eluates were digested with trypsin and analyzed by LC-MS/MS (see Supplemental Methods). Figure 5a demonstrates confirmatory ion chromatograms for 8 of the 29 proteins that were also found to be statistically significant in our FHS analyses (FRS, age or sex). All 29 aptamers demonstrated enrichment of their corresponding spiked-in protein relative to the plasma proteome background. A total of 532 peptides from the 29 proteins were identified by LC-MS/MS following aptamer affinity enrichment (Supplemental Tables 16 and 17). In addition to the proteins targeted for enrichment and analysis, we also detected many high abundance plasma proteins by LC-MS/MS (Supplemental Table 18). This is a common finding in all bead pull down experiments. Response curves for aptamer-enriched proteins were linear over a wide dynamic range of spiked protein concentrations (Figure 5b and Supplemental Tables 19 and 20). The median LOD (limit of detection) was 0.41 fmol/uL for 19 peptides (corresponding to 9 proteins) using heavy isotope peptides quantified by amino acid analysis (AAA; median R2 = 0.96). A similar median LOD for 106 peptides (corresponding to 29 proteins) was derived using light peak areas only (median R2 = 0.93). These LODs are comparable to other antibody-based enrichment methods analyzed by LC-MRM-MS using heavy isotope peptide standards. These results indicate that the presence of non-specific proteins of even high abundance does not substantially affect quantification in a targeted MS mode of analysis and that the reproducibility of the aptamer-based enrichment process offers sufficient interference-free precision to allow for comparable peptide (and therefore protein) quantification from matrices as complex as plasma.
Figure 5.
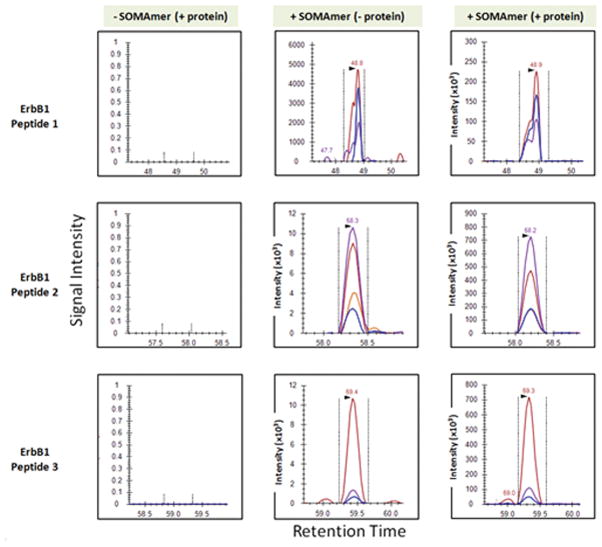
Figure 5a. Aptamers bind their cognate proteins spiked into plasma
Proteins were spiked into plasma in the absence (top panel) or presence (bottom panel) of biotinylated aptamers bound to streptavidin bead. Following elution and digestion of the affinity enriched sample, LC-MRM-MS analysis was performed. Shown are MS signal intensities for peptides unique to eight proteins in the study that were extracted from the total ion chromatogram.
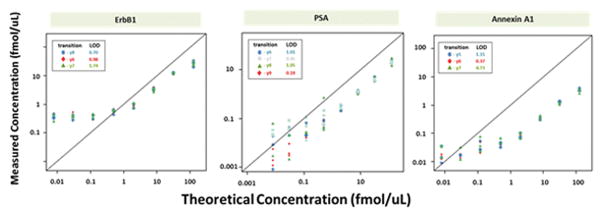
Figure 5b. Response curves for ERBB1, PSA and Annexin A1 spiked into plasma.
Measured protein concentrations are compared to the expected concentrations based on the observed ratio of analyte peptide from spiked protein standard to heavy isotope labeled standard. Limits of detection (LODs) were determined for each of the three selected transitions for the proteins. Calibration curves were prepared by method of standard addition in plasma, enriched using a multiplex of 29 aptamers, digested with trypsin and quantified by LC-MRM-MS using heavy isotope labeled standards for selected peptides as described in Supplemental Methods. Regression analysis was performed and the slope and the x-intercept were used to estimate the relative recovery and lower limit of detection (LLOD) respectively (see Supplemental Methods and Supplemental Tables 19 and 20). Response was linear over 3 orders of concentration for all peptides, and LODs were in the range of 10’s to 100’s of amol/uL except in cases where the endogenous protein was present at sufficiently high levels and caused an earlier plateau in response as was observed for ERBB1.
Figure 5c. Demonstration of aptamers binding endogenous protein (ERBB1)
MS response of fragment-ion transitions monitored for three peptides unique to ERBB1 are plotted for plasma samples containing protein standards enriched using SA beads without aptamers (column 1), and with aptamers in the absence (column 2) or presence (column 3) of the added protein standard. The peak areas of co-eluting fragment-ion transitions in the same retention time window are designated by color (Supplemental Table 17). The relative intensities of the fragment ions for each peptide observed in the positive control (protein standards and aptamer added), and the patient plasma samples without exogenous protein added, confirms the identification and detection of these peptides and of endogenous ERBB1 in patient plasma using aptamer affinity enrichment. See Supplemental Methods for details of sample handling, affinity enrichment, elution, digestion and MS analysis.
We then tested the limits of detection of this system for endogenous proteins. Aptamer enriched samples were digested and then analyzed by LC-MRM-MS for proteotypic peptides for the 8 proteins shown in Figure 5a (and Supplemental Table 17). Figure 5c details findings for the ERBB1 protein in control versus aptamer containing samples. As expected, we confirmed the detection of peptides unique for ERBB1 protein in samples containing both the aptamer and the added protein standard. However, even in samples that did not have protein standards added, the same peptides from endogenous ERBB1 protein were detected by LC-MRM-MS. None was observed in the absence of aptamer. Approximately one quarter of the proteins could be detected at endogenous levels under these conditions (Supplemental Tables 19 and 20).
Finally, we performed orthogonal analyses to assess quantitation of proteins leveraging anti-peptide antibodies, as we have done previously.2 Supplemental Figure 5 demonstrates concordance between the aptamer-based scan and anti-peptide immunoaffinity enrichment MS for two representative proteins, troponin I and spondin-1 in PMI patients (n=11 and 12, respectively). Troponin serves as a model “positive control”. Spondin-1 increased in myocardial injury, although it is also significantly changed in heparinized catheterization control patients. Taken together, these findings highlight a new workflow leveraging the strength of LC-MS to verify the specificity and quantitation of the higher throughput aptamer method.
DISCUSSION
Recent advances in aptamer-based proteomic technologies offer the prospect of proteomic analyses with breadth and throughput not previously possible. However, there are limited data available in studies integrating replication studies or in archived population-based samples. To assess the applicability of this technology in human cardiovascular disease, we used a clinical model of MI that produced robust clinical and biochemical changes and allowed precise kinetic analysis in patients who served as their own controls. Using an aptamer-based platform, we confirmed established biomarkers of myocardial injury such as CK-MB and troponin and extended prior work by identifying many new proteins not previously associated with MI. Many of the protein changes occurred within 10 minutes of myocardial injury, which might ultimately provide additional information on top of established biomarkers and electrocardiographic changes. We next applied the platform to investigate proteins associated with subtle cardiovascular risk factors in samples from FHS participants. We found many novel protein associations with cardiovascular risk traits in samples archived for over 20 years.
Techniques that stochastically sample proteins or peptides, such as mass spectrometry, face formidable challenges when applied to the human plasma proteome, including the sheer number of proteins and their extensive range of concentrations. Depletion strategies targeting a subset of high abundance proteins mitigate this problem somewhat, but at the cost of throughput and reproducibility. We and others have integrated protocols for “deep” (i.e., low abundance) biomarker discovery, coupling chemical isobaric mass tag labeling as well as “label free” approaches with liquid chromatography-tandem mass spectrometry (LC-MS/MS).1–8, 24, 25 Recent reports have suggested the identification of greater than 3,000 proteins in plasma, but these analyses take approximately 24 hours per sample when accounting for depletion strategies and multiple types of chromatographic separation prior to MS analysis.1 Others have reported higher throughput mass spectrometry-based approaches using alternative multiplexing techniques and minimal fractionation that can identify ~150–250 plasma proteins at a faster rate of 2.4 hours per sample.26 The aptamer-based technique we employed here is greater than 20-fold faster than deep proteomic analyses1 and 6-fold more rapid than the higher throughput approach reported by Cominetti et al., which detects less than a quarter of the number of analytes.26 While the number of proteins presently assayed is less than can be obtained by deep mass spectrometry, this trade-off is reasonable if the goal is to interrogate large cohorts. It is anticipated that additional reagents will become available as an increasing number of aptamers are generated against recombinant proteins. The experimental design of our PMI model addressed some of the study design challenges that have limited proteomic biomarker studies. By allowing each individual to serve as his/her own biological control in a perturbation experiment with serial sampling, we constrained inter-individual variability and enhanced power to identify statistically meaningful changes. In addition to the known markers of myocardial injury, we were able to identify several candidate biomarkers that may also highlight specific pathways in response to myocardial injury, including myocardial cyto-protection and regeneration. Proteins implicated in angiogenesis, such as angiogenin and FGF1,27–30 are increased in PMI. We identified an increase in WNT pathway inhibitor, Dickkopf-related protein 4 (DKK-4) and DKK receptor KREM2, highlighting possible WNT pathway involvement in myocardial injury.31 Changes in the leukocyte inhibitor factor (LIF) pathway, including decreases in cardiotrophin 1 and increases in LIF soluble receptor (LIFsr), may suggest a cell differentiation response to injury.32, 33 Other proteins important in cardiac development and regeneration also increased early after myocardial injury including SDF-1,34 Cripto,35 and FGF-8β.36, 37 Unfortunately the small samples size of the coronary sinus studies does not allow for definitive differentiation between cardiac and non-cardiac origin of protein changes.
With several layers of validation and potentially interesting biological findings from our PMI studies, we investigated the more subtle phenotype of cardiovascular risk in archived samples from FHS participants (n=899). We examined the protein associations with the clinical risk factors of cardiovascular disease including age, gender, total cholesterol, HDL, SBP, diabetes and smoking. We identified circulating proteins not previously associated with these clinical traits, highlighting potentially novel pathways. For example, osteomodulin, a protein involved in biomineralization of bones, demonstrates a strongly negative association with smoking, an established risk factor for osteoporosis.38 Thus, lower levels may reflect (or be participating in) decreased bone density. We then evaluated protein associations with the composite of these risk factors in the Framingham Risk Score (FRS). As might be expected, CRP5–7 and apolipoprotein E39, 40 were confirmed as top associations with FRS. Novel associations with FRS included MMP 12 and laminin as well as numerous other proteins. Interestingly, a common variant in the MMP-12 locus was recently found to be associated with atherosclerotic stroke, raising the possibility of a causal pathway.41 Laminin chain isotype composition was found to be different in the atherosclerotic plaque tissue as compared to intima media from both APO E-deficient mice and humans.42
Further, we developed a new method incorporating DNA-aptamers into an immunoaffinity LC-MS based workflow for orthogonal validation of findings. Our data confirm that the aptamers enriched for their targeted proteins in spike-in studies. In the conditions tested to date, LC-MS/MS analyses also provided unambiguous identification of endogenous peptides from targeted proteins as well, though sensitivity was not sufficient for all proteins studied. Additional studies modifying key analytical parameters such as the amount of starting material or aptamer, as well as LC-MS conditions, are ongoing.
We recognize limitations of our studies. While this aptamer-based proteomics platform provides very broad coverage with high throughput, its coverage of the global human proteome remains limited and may therefore be agnostic to changes of analytes not targeted. In our PMI studies, we excluded over 200 proteins changed by heparin administration and/or the cardiac catheterization procedure itself (nominal P < 0.05). This very conservative effort to limit potential confounding may result in a bias toward the null where we fail to identify significant protein changes. Although our PMI studies included derivation and validation cohorts, the total number of patients remains relatively small. While our FHS study was well powered to test the analytical aspects of the platform and cross-sectional associations, we were underpowered for incident CVD analyses. Future analyses must be performed in larger, ethnically diverse populations to confirm and extend our findings. Finally, because groups of proteins cluster within biological pathways, the assumption of independent statistical tests is overly stringent. Thus, it is important to acknowledge the possibility of false negative results from using Bonferroni-corrected P values.
In summary, we demonstrated the application of a novel aptamer-based proteomics platform for the discovery of blood markers of myocardial injury and to highlight potential pathways associated with cardiovascular risk traits. Further, we developed a novel DNA aptamer-based immunoaffinity MS-based workflow for the technical validation of these findings. Beyond the potential diagnostic utility we demonstrate here in our PMI studies, findings in the archived FHS samples highlight a broad spectrum of proteins associated with cardiometabolic risk for exploration in functional studies.
Supplementary Material
CLINICAL PERSPECTIVE.
What is new?
Proteomic technologies hold considerable promise for biomarker and pathway discovery in cardiovascular disease; however, existing techniques have significant drawbacks, including variable reproducibility and low sample throughput, which together have hindered proteomic analyses of large numbers of human samples.
Here we applied a novel technology that uses single- stranded DNA aptamers to measure over 1100 proteins in a single blood sample.
What are the clinical implications?
We found very early markers of myocardial injury in a robust, clinically relevant model of “planned” myocardial injury, patients undergoing septal ablation for hypertrophic cardiomyopathy; we then tested whether the platform could detect novel associations between proteins and cardiovascular risk factors in individuals without overt cardiovascular disease using archived samples from the Framingham Heart Study.
Our results highlight an emerging proteomics tool that captures a large number of low abundance analytes with high sensitivity and high precision, and we provide important proof-of-principle for clinical applications.
Acknowledgments
SOURCES OF FUNDING: This work was supported by NIH Contract N01HC25195 to the FHS and HHSN268201000033C to REG and National Cancer Institute (NCI) CPTAC award U24CA160034 to SAC.
Footnotes
AUTHOR CONTRIBUTIONS:
DN designed the study, analyzed and interpreted data and wrote the manuscript. SS, DS and LF designed and performed the aptamer-based proteomic scan experiments. MK, under the direction of MGL, performed statistical analyses. EK, HK under the direction of SAC designed, performed and analyzed the MS-based proteomic experiments, and SAC contributed to the writing of the manuscript. XS, MB, JOS contributed to data analyses and manuscript generation. MAF and RSV contributed to experimental design. MS contributed to interpretation of data and manuscript revision. TJW conceived the study, designed the experiments, analyzed and interpreted the data. REG conceived the study, designed the experiments, analyzed and interpreted the data and wrote the manuscript.
DISCLOSURES: None
References
- 1.Keshishian H, Burgess MW, Gillette MA, Mertins P, Clauser KR, Mani DR, Kuhn EW, Farrell LA, Gerszten RE, Carr SA. Multiplexed, quantitative workflow for sensitive biomarker discovery in plasma yields novel candidates for early myocardial injury. Molecular & cellular proteomics: MCP. 2015;14:2375–2393. doi: 10.1074/mcp.M114.046813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addona TA, Shi X, Keshishian H, Mani DR, Burgess M, Gillette MA, Clauser KR, Shen D, Lewis GD, Farrell LA, Fifer MA, Sabatine MS, Gerszten RE, Carr SA. A pipeline that integrates the discovery and verification of plasma protein biomarkers reveals candidate markers for cardiovascular disease. Nature biotechnology. 2011;29:635–643. doi: 10.1038/nbt.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang H, Hanash S. Mass spectrometry based proteomics for absolute quantification of proteins from tumor cells. Methods. 2015;81:34–40. doi: 10.1016/j.ymeth.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang S, Mercado-Uribe I, Hanash S, Liu J. Itraq-based proteomic analysis of polyploid giant cancer cells and budding progeny cells reveals several distinct pathways for ovarian cancer development. PloS one. 2013;8:e80120. doi: 10.1371/journal.pone.0080120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin X, Subramanian S, Hwang SJ, O’Donnell CJ, Fox CS, Courchesne P, Muntendam P, Gordon N, Adourian A, Juhasz P, Larson MG, Levy D. Protein biomarkers of new-onset cardiovascular disease: Prospective study from the systems approach to biomarker research in cardiovascular disease initiative. Arteriosclerosis, thrombosis, and vascular biology. 2014;34:939–945. doi: 10.1161/ATVBAHA.113.302918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prentice RL, Zhao S, Johnson M, Aragaki A, Hsia J, Jackson RD, Rossouw JE, Manson JE, Hanash SM. Proteomic risk markers for coronary heart disease and stroke: Validation and mediation of randomized trial hormone therapy effects on these diseases. Genome medicine. 2013;5:112. doi: 10.1186/gm517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halim SA, Neely ML, Pieper KS, Shah SH, Kraus WE, Hauser ER, Califf RM, Granger CB, Newby LK. Simultaneous consideration of multiple candidate protein biomarkers for long-term risk for cardiovascular events. Circulation Cardiovascular genetics. 2015;8:168–177. doi: 10.1161/CIRCGENETICS.113.000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ping P, Vondriska TM, Creighton CJ, Gandhi TK, Yang Z, Menon R, Kwon MS, Cho SY, Drwal G, Kellmann M, Peri S, Suresh S, Gronborg M, Molina H, Chaerkady R, Rekha B, Shet AS, Gerszten RE, Wu H, Raftery M, Wasinger V, Schulz-Knappe P, Hanash SM, Paik YK, Hancock WS, States DJ, Omenn GS, Pandey A. A functional annotation of subproteomes in human plasma. Proteomics. 2005;5:3506–3519. doi: 10.1002/pmic.200500140. [DOI] [PubMed] [Google Scholar]
- 9.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, Carter J, Dalby AB, Eaton BE, Fitzwater T, Flather D, Forbes A, Foreman T, Fowler C, Gawande B, Goss M, Gunn M, Gupta S, Halladay D, Heil J, Heilig J, Hicke B, Husar G, Janjic N, Jarvis T, Jennings S, Katilius E, Keeney TR, Kim N, Koch TH, Kraemer S, Kroiss L, Le N, Levine D, Lindsey W, Lollo B, Mayfield W, Mehan M, Mehler R, Nelson SK, Nelson M, Nieuwlandt D, Nikrad M, Ochsner U, Ostroff RM, Otis M, Parker T, Pietrasiewicz S, Resnicow DI, Rohloff J, Sanders G, Sattin S, Schneider D, Singer B, Stanton M, Sterkel A, Stewart A, Stratford S, Vaught JD, Vrkljan M, Walker JJ, Watrobka M, Waugh S, Weiss A, Wilcox SK, Wolfson A, Wolk SK, Zhang C, Zichi D. Aptamer-based multiplexed proteomic technology for biomarker discovery. PloS one. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies DR, Gelinas AD, Zhang C, Rohloff JC, Carter JD, O’Connell D, Waugh SM, Wolk SK, Mayfield WS, Burgin AB, Edwards TE, Stewart LJ, Gold L, Janjic N, Jarvis TC. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19971–19976. doi: 10.1073/pnas.1213933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vant-Hull B, Gold L, Zichi DA. Theoretical principles of in vitro selection using combinatorial nucleic acid libraries. In: Beaucage Serge L, et al., editors. Current protocols in nucleic acid chemistry. Unit 9 1. Chapter 9. 2000. [DOI] [PubMed] [Google Scholar]
- 12.Hathout Y, Brody E, Clemens PR, Cripe L, DeLisle RK, Furlong P, Gordish-Dressman H, Hache L, Henricson E, Hoffman EP, Kobayashi YM, Lorts A, Mah JK, McDonald C, Mehler B, Nelson S, Nikrad M, Singer B, Steele F, Sterling D, Sweeney HL, Williams S, Gold L. Large-scale serum protein biomarker discovery in duchenne muscular dystrophy. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:7153–7158. doi: 10.1073/pnas.1507719112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lakkis NM, Nagueh SF, Dunn JK, Killip D, Spencer WH., 3rd Nonsurgical septal reduction therapy for hypertrophic obstructive cardiomyopathy: One-year follow-up. J Am Coll Cardiol. 2000;36:852–855. doi: 10.1016/s0735-1097(00)00767-1. [DOI] [PubMed] [Google Scholar]
- 14.Lakkis NM, Nagueh SF, Kleiman NS, Killip D, He ZX, Verani MS, Roberts R, Spencer WH., 3rd Echocardiography-guided ethanol septal reduction for hypertrophic obstructive cardiomyopathy. Circulation. 1998;98:1750–1755. doi: 10.1161/01.cir.98.17.1750. [DOI] [PubMed] [Google Scholar]
- 15.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The framingham offspring study. Design and preliminary data. Preventive medicine. 1975;4:518–525. doi: 10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- 16.Keshishian H, Addona T, Burgess M, Mani DR, Shi X, Kuhn E, Sabatine MS, Gerszten RE, Carr SA. Quantification of cardiovascular biomarkers in patient plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2009;8:2339–2349. doi: 10.1074/mcp.M900140-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis GD, Farrell L, Wood MJ, Martinovic M, Arany Z, Rowe GC, Souza A, Cheng S, McCabe EL, Yang E, Shi X, Deo R, Roth FP, Asnani A, Rhee EP, Systrom DM, Semigran MJ, Vasan RS, Carr SA, Wang TJ, Sabatine MS, Clish CB, Gerszten RE. Metabolic signatures of exercise in human plasma. Science translational medicine. 2010;2:33ra37. doi: 10.1126/scitranslmed.3001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okamoto F, Sohmiya K, Ohkaru Y, Kawamura K, Asayama K, Kimura H, Nishimura S, Ishii H, Sunahara N, Tanaka T. Human heart-type cytoplasmic fatty acid-binding protein (h-fabp) for the diagnosis of acute myocardial infarction. Clinical evaluation of h-fabp in comparison with myoglobin and creatine kinase isoenzyme mb. Clinical chemistry and laboratory medicine: CCLM/FESCC. 2000;38:231–238. doi: 10.1515/CCLM.2000.034. [DOI] [PubMed] [Google Scholar]
- 19.Glatz JF, van der Vusse GJ, Simoons ML, Kragten JA, van Dieijen-Visser MP, Hermens WT. Fatty acid-binding protein and the early detection of acute myocardial infarction. Clinica chimica acta; international journal of clinical chemistry. 1998;272:87–92. doi: 10.1016/s0009-8981(97)00255-6. [DOI] [PubMed] [Google Scholar]
- 20.Abbott JD, Huang Y, Liu D, Hickey R, Krause DS, Giordano FJ. Stromal cell-derived factor-1alpha plays a critical role in stem cell recruitment to the heart after myocardial infarction but is not sufficient to induce homing in the absence of injury. Circulation. 2004;110:3300–3305. doi: 10.1161/01.CIR.0000147780.30124.CF. [DOI] [PubMed] [Google Scholar]
- 21.Ma J, Ge J, Zhang S, Sun A, Shen J, Chen L, Wang K, Zou Y. Time course of myocardial stromal cell-derived factor 1 expression and beneficial effects of intravenously administered bone marrow stem cells in rats with experimental myocardial infarction. Basic research in cardiology. 2005;100:217–223. doi: 10.1007/s00395-005-0521-z. [DOI] [PubMed] [Google Scholar]
- 22.Pillarisetti K, Gupta SK. Cloning and relative expression analysis of rat stromal cell derived factor-1 (sdf-1)1: Sdf-1 alpha mrna is selectively induced in rat model of myocardial infarction. Inflammation. 2001;25:293–300. doi: 10.1023/a:1012808525370. [DOI] [PubMed] [Google Scholar]
- 23.Wilson PW, D’Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–1847. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 24.Addona TA, Shi X, Keshishian H, Mani DR, Burgess M, Gillette MA, Clauser KR, Shen D, Lewis GD, Farrell LA, Fifer MA, Sabatine MS, Gerszten RE, Carr SA. A pipeline that integrates the discovery and verification of plasma protein biomarkers reveals candidate markers for cardiovascular disease. Nat Biotechnol. 2011;29:635–643. doi: 10.1038/nbt.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keshishian H, Burgess MW, Gillette MA, Mertins P, Clauser KR, Mani DR, Kuhn EW, Farrell LA, Gerszten RE, Carr SA. Multiplexed, quantitative workflow for sensitive biomarker discovery in plasma yields novel candidates for early myocardial injury. Mol Cell Proteomics. 2015;14:2375–93. doi: 10.1074/mcp.M114.046813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cominetti O, Nunez Galindo A, Corthesy J, Oller Moreno S, Irincheeva I, Valsesia A, Astrup A, Saris WH, Hager J, Kussmann M, Dayon L. Proteomic biomarker discovery in 1000 human plasma samples with mass spectrometry. Journal of proteome research. 2016;15:389–399. doi: 10.1021/acs.jproteome.5b00901. [DOI] [PubMed] [Google Scholar]
- 27.Xue L, Greisler HP. Angiogenic effect of fibroblast growth factor-1 and vascular endothelial growth factor and their synergism in a novel in vitro quantitative fibrin-based 3-dimensional angiogenesis system. Surgery. 2002;132:259–267. doi: 10.1067/msy.2002.125720. [DOI] [PubMed] [Google Scholar]
- 28.Pandit AS, Feldman DS, Caulfield J, Thompson A. Stimulation of angiogenesis by fgf-1 delivered through a modified fibrin scaffold. Growth factors. 1998;15:113–123. doi: 10.3109/08977199809117187. [DOI] [PubMed] [Google Scholar]
- 29.Gao X, Xu Z. Mechanisms of action of angiogenin. Acta biochimica et biophysica Sinica. 2008;40:619–624. doi: 10.1111/j.1745-7270.2008.00442.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Gao X, Weng C, Xu Z. Interaction between angiogenin and fibulin 1: Evidence and implication. Acta biochimica et biophysica Sinica. 2008;40:375–380. doi: 10.1111/j.1745-7270.2008.00420.x. [DOI] [PubMed] [Google Scholar]
- 31.Monaghan AP, Kioschis P, Wu W, Zuniga A, Bock D, Poustka A, Delius H, Niehrs C. Dickkopf genes are co-ordinately expressed in mesodermal lineages. Mechanisms of development. 1999;87:45–56. doi: 10.1016/s0925-4773(99)00138-0. [DOI] [PubMed] [Google Scholar]
- 32.Kawahara Y, Manabe T, Matsumoto M, Kajiume T, Matsumoto M, Yuge L. Lif-free embryonic stem cell culture in simulated microgravity. PloS one. 2009;4:e6343. doi: 10.1371/journal.pone.0006343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pennica D, Shaw KJ, Swanson TA, Moore MW, Shelton DL, Zioncheck KA, Rosenthal A, Taga T, Paoni NF, Wood WI. Cardiotrophin-1. Biological activities and binding to the leukemia inhibitory factor receptor/gp130 signaling complex. The Journal of biological chemistry. 1995;270:10915–10922. doi: 10.1074/jbc.270.18.10915. [DOI] [PubMed] [Google Scholar]
- 34.Penn MS. Importance of the sdf-1:Cxcr4 axis in myocardial repair. Circulation research. 2009;104:1133–1135. doi: 10.1161/CIRCRESAHA.109.198929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bianco C, Cotten C, Lonardo E, Strizzi L, Baraty C, Mancino M, Gonzales M, Watanabe K, Nagaoka T, Berry C, Arai AE, Minchiotti G, Salomon DS. Cripto-1 is required for hypoxia to induce cardiac differentiation of mouse embryonic stem cells. The American journal of pathology. 2009;175:2146–2158. doi: 10.2353/ajpath.2009.090218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reifers F, Walsh EC, Leger S, Stainier DY, Brand M. Induction and differentiation of the zebrafish heart requires fibroblast growth factor 8 (fgf8/acerebellar) Development. 2000;127:225–235. doi: 10.1242/dev.127.2.225. [DOI] [PubMed] [Google Scholar]
- 37.Ilagan R, Abu-Issa R, Brown D, Yang YP, Jiao K, Schwartz RJ, Klingensmith J, Meyers EN. Fgf8 is required for anterior heart field development. Development. 2006;133:2435–2445. doi: 10.1242/dev.02408. [DOI] [PubMed] [Google Scholar]
- 38.Law MR, Hackshaw AK. A meta-analysis of cigarette smoking, bone mineral density and risk of hip fracture: Recognition of a major effect. Bmj. 1997;315:841–846. doi: 10.1136/bmj.315.7112.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson PW, Schaefer EJ, Larson MG, Ordovas JM. Apolipoprotein e alleles and risk of coronary disease. A meta-analysis. Arteriosclerosis, thrombosis, and vascular biology. 1996;16:1250–1255. doi: 10.1161/01.atv.16.10.1250. [DOI] [PubMed] [Google Scholar]
- 40.Eichner JE, Dunn ST, Perveen G, Thompson DM, Stewart KE, Stroehla BC. Apolipoprotein e polymorphism and cardiovascular disease: A huge review. American journal of epidemiology. 2002;155:487–495. doi: 10.1093/aje/155.6.487. [DOI] [PubMed] [Google Scholar]
- 41.Traylor M, Makela KM, Kilarski LL, Holliday EG, Devan WJ, Nalls MA, Wiggins KL, Zhao W, Cheng YC, Achterberg S, Malik R, Sudlow C, Bevan S, Raitoharju E, Oksala N, Thijs V, Lemmens R, Lindgren A, Slowik A, Maguire JM, Walters M, Algra A, Sharma P, Attia JR, Boncoraglio GB, Rothwell PM, de Bakker PI, Bis JC, Saleheen D, Kittner SJ, Mitchell BD, Rosand J, Meschia JF, Levi C, Dichgans M, Lehtimaki T, Lewis CM, Markus HS Metastroke ISGCWTCC. A novel mmp12 locus is associated with large artery atherosclerotic stroke using a genome-wide age-at-onset informed approach. PLoS genetics. 2014;10:e1004469. doi: 10.1371/journal.pgen.1004469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rauch U, Saxena A, Lorkowski S, Rauterberg J, Bjorkbacka H, Durbeej M, Hultgardh-Nilsson A. Laminin isoforms in atherosclerotic arteries from mice and man. Histology and histopathology. 2011;26:711–724. doi: 10.14670/HH-26.711. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.