Introduction
Peanut allergy is the most prevalent food allergy in western countries. It affects 1-2 % of the population and is the leading cause of fatal food-induced anaphylaxis [1, 2]. Despite recent progress in desensitization based on early administration of peanuts, oral immunotherapy, and epicutaneous administration, a generally useful, clinically approved treatment is not available [3-6]. Currently, the only treatment is strict avoidance of peanut, but this is difficult to achieve because of widespread use of peanut in prepared foods. [7, 8].
Life-threatening peanut allergy is clearly an IgE mediated disease [9]. The initiation of an allergic reaction in a sensitized patient is due to interactions between specific peanut allergens and IgE bound to the high affinity receptor for IgE, FcεR1, on the surface of mast cells leading to cell activation, degranulation and release of histamine and inflammatory mediators that trigger allergic symptoms [10, 11]
Among the peanut allergens, Ara h 2 and Ara h 6 are the most potent allergens for IgE-mediated mast cell activation [12-16] and IgE binding to these proteins has higher diagnostic value than IgE binding to other peanut proteins [17-20]. Furthermore, Ara h 2 and Ara h 6 have highly redundant allergenic activity [21] that is most likely due to the very high homology within their IgE-binding domains [22].
Binding of IgE antibodies to specific regions of an allergen is a prerequisite for triggering of allergic reactions. Binding sites recognized by IgE antibodies are called IgE epitopes and are frequently categorized as linear or conformational. A linear epitope consists of continuous amino acids, while a conformational epitope contains amino acids that are distributed discontinuously over the protein sequence and come close to each other only when the protein is correctly folded. Therefore, conformational epitopes are dependent on the 3-dimensional structure (3D) of the protein [23-26].
Considerable advances have been made in identifying and characterizing linear IgE epitopes of peanut allergens but little is known about conformational IgE-binding epitopes. Linear IgE epitopes of Ara h 1, Ara h 2, Ara h 3, and Ara h 6 have been identified experimentally [22, 27-29] and linear and conformational epitopes of Ara h 6 and other peanut allergens have been predicted by computational methods[30]. In some studies, the diversity of linear IgE peptides is correlated with the severity of allergic reaction [27, 29], suggesting that linear epitopes are predominant [27, 28]. A study from our lab suggests that if peanut-specific IgE levels are normalized, binding to linear epitopes of Ara h 2 and Ara h 6 is inversely correlated with clinical severity of peanut allergy, suggesting a role for conformational epitopes [22].
Direct evidence to support the importance of conformational epitopes includes: 1) reduction of Ara h 2 by DTT disrupted its secondary structure and completely abrogated its IgE reactivity [31, 32], 2) a phase 1 study of recombinant modified peanut proteins on the basis of linear IgE epitope data did not show promising results [33], and 3) a study using hydroxyproline-containing epitopes of Ara h 2 to detect the relative contribution of linear and conformational epitopes to IgE binding showed that peanut allergic patients displayed variable levels of sensitization toward linear and conformational epitopes of Ara h 2 [34]. Taken together, these studies strongly suggest that conformational epitopes may play an important role in peanut allergy.
The most precise way to identify a conformational epitope is to determine the crystal structure of an antigen-antibody complex [16, 35-38], but this method is complicated, time-consuming, and is applicable only to monoclonal, and not polyclonal antibodies. An alternative approach is to screen a phage display library with polyclonal peanut allergen-specific IgE antibodies to identify mimics of allergenic IgE epitopes, called mimotopes [39]. A mimotope is defined as a molecule able to bind to the antigen binding site of an antibody molecule that is not necessarily identical with the original epitope, but an acceptable mimic of the essential features of the epitope [40]. Mimotopes and their corresponding epitopes are considered to have similar physicochemical properties and spatial organization [41-43]. Therefore, the phage display strategy is ideally suited for determination of individual polyclonal epitope recognition patterns. Phage display technology has been used to identify conformational epitopes in Ara h 1 and other allergens [42, 44-47].
In this study, we used affinity-purified polyclonal IgE antibodies from peanut–allergic subjects to screen a phage display library containing 12 amino acid random peptides as fusions to a coat protein, identified 41 unique peptide sequences, analyzed these mimotopes using EpiSearch [46, 48], and identified novel conformational IgE epitopes of the Ara h 2 and Ara h 6.
Materials and methods
Patients and sera
All adult patients and the parents or guardians of minors signed informed consent. Minors signed an assent. The University of Colorado Denver Institutional Review Board approved this study. Many of these subjects were previously described [22].
Sera were collected from peanut allergic patients who met the following criteria: 1) physician-diagnosed peanut allergy, 2) age >6 year, 3) peanut-specific IgE ≥ 10 KAU/L (ImmunoCap, ThermoFisher, Waltham, MA, USA), 4) no exposure to peanuts within 3 months, and 5) not receiving therapy with anti-IgE. Patients reported symptoms after naturally occurring exposure to peanuts were classified into grades of anaphylaxis according to criteria established by the World Health Organization for evaluation of allergic reactions in the context of allergen-specific immunotherapy [22]
Sera from four subjects with particularly high anti-peanut IgE (D44, D48, D64, D103) and known to bind a wide variety of linear IgE epitopes [22] were used to identify IgE mimotopes for Ara h 2 and Ara h 6 (Table 1). Sera from an additional 25 subjects (Supplementary Table 1) were used to determine the frequency of binding of specific mimotopes to IgE in a variety of sera.
Table 1.
Sera used for phage library screening
| Serum | Peanut- IgE (kU/L) | Ara h 2 epitopes* binding | Ara h 6 epitope* binding |
|---|---|---|---|
| D44 | 65 | 6/8 | 4/7 |
| D48 | 71 | 7/8 | 5/7 |
| D64 | 591 | 6/8 | 4/7 |
| D103 | 787 | 3/8 | 0/7 |
The numbers of Ara h 2 and Ara h 6 linear epitopes recognized by serum IgE compared with the total possible numbers of epitope in the allergens (22)
Allergens and antibodies
A chromatographic fraction of 2S albumins from raw peanuts was obtained as previously described [15], which consists of >97% Ara h 2 (79 %) and Ara h 6 (18 %). Other allergen detected (<3% by weight) are Ara h 7 and Ara h 8. Total IgE was affinity purified from peanut-allergic sera using affinity-purified goat anti-human IgE [49] coupled to AminoLink® resin (ThermoFisher, Waltham, MA, USA). IgE that bound to either Ara h 2 or Ara h 6 (Ara h 2/6-IgE) was then purified using Ara h 2/6 coupled to AminoLink® resin. Purification of Ara h 2 and Ara h 6 from raw peanut was as previously described [16]. Four individual rabbits were each immunized at monthly intervals five times SQ with 500μg either purified Ara h 2 (2 rabbits) or purified Ara h 6 (2 rabbits) with a mixture of complete Freund/incomplete Freund's adjuvant (YenZym, Inc., South San Francisco, CA). IgG from all sera were demonstrated to recognize the purified allergens in an ELISA assay.
Phage screening
Phage library
The Ph.D. – 12 Phage Display Peptide library (New England BioLabs, Beverly, MA, USA), which displays 109 individual peptides was used in this study. Biotinylated mouse anti-human IgE (Invitrogen Life Technologies, Grand Island, NY, USA) was coupled to Dynabeads M-280 Streptavidin (Invitrogen Life Technologies, Grand Island, NY, USA) for two hours at room temperature and blocked with 2% skim milk in PBS. Peanut allergic sera (subjects D44, D48, D64, and D103; Table 1) were diluted 1:3 in 0.05% Tween 20 and 0.2% skim milk, and individually incubated with the beads overnight at 4°C.
First round of biopanning
Beads coated with human IgE were incubated with 10μl (~ 2 × 1011) of phages for overnight at 4°C, followed by extensive washing to remove unbound phages. Phages were eluted with elution buffer (0.2 M Glycine-HCI, pH 2.2, 1 mg/ml BSA) and neutralized with 1 M Tris-HCI, pH 9.0. The eluted phages are amplified in ER2738 E.coli for 4.5 hours, precipitated with 20%PEG/2.5 M NaCI and titrated.
Subsequent rounds of biopanning
The amplified phages from the first round were used in a second round of biopanning as described above except a negative selection was performed to remove the non-specifically-bound phages before the panning. After a third round of panning, the IgE-bound phages screened with the 4 individual sera were combined and eluted with elution buffer. The eluted phages were amplified in ER2738 E.coli, precipitated with 20%PEG/2.5 M NaCl and titrated. Single colonies were randomly selected from the titration plate and amplified.
Phage ELISA
Binding of IgE from peanut-allergic subjects
Microtiter plates were coated overnight at 4°C with goat anti-human IgE at 2 μg/ml in PBS. Subsequent steps were performed at room temperature for 1 hour. Between steps, plates were washed five times with 0.05% Tween 20 in PBS. Plates were first blocked with 2% skim milk in PBS. Then individual sera from peanut-allergic subjects were diluted 1:50, added to the plates and incubated with individual identified phage or wild type phage without peptide insert. IgE-bound phage colonies were detected with mouse anti-M13 phage conjugated HPR (GE Healthcare, Piscataway, NJ, USA). IgE positive colonies were further tested for their reactivity to affinity-purified anti-Ara h 2/6 IgE from four sera combined, using the same ELISA protocol. A optical density greater than OD + 3 × S.D. above the negative control (wild type phage) was regarded as a positive result. The Ara h 2/6 specific colonies were sequenced.
Binding of IgG from Ara h 2 or Ara h 6 immunized rabbits. Microtiter plates were coated with goat anti- rabbit IgG. Then rabbit anti- Ara h 2 or anti- Ara h 6 (1: 5000 dilution of sera pooled from 2 rabbits) was added, followed by individual phage, and mouse anti-M13 phage conjugated HPR. A optical density greater than OD + 3 × S.D. above the negative control (wild type phage) was regarded as a positive result.
Sequence alignments
Peptide sequences obtained from phage display experiments were aligned with Ara h 2 and Ara h 6 sequences using multiple sequence alignment program, ClustalW [50, 51], to find a consensus pattern of amino acids.
Mapping conformational epitopes
The EpiSearch method [46, 48] was used to map the potential epitope sites on the surface of Ara h 2 and Ara h 6. This approach uses patch analysis and solvent accessible surface area of amino acids to map peptides obtained from phage display experiments onto the 3D structure of an antigen protein. Despite the availability of high resolution structures of Ara h2 and Ara h6, we generated their 3D model structures because a highly disordered loop region is missing in the crystal structure of Ara h 2 (PDB ID: 3OB4) [52], and the orientation of loop regions is different in the NMR structure of Ara h 6 (PDB ID: 1W2Q) [14]. The model structure of Ara h 2 was generated using homology modeling technique wherein the sequence of Ara h 2 was submitted to a fold recognition server [53] and the best template structure was selected to generate a model structure of Ara h2 using MPACK [54-56]. Two additional model structures of Ara h 2 were generated using ROBETTA [57] and I-TASSER [58] to obtain additional information about the Ara h 2 disordered loop region. A comparison between the modeled and X-ray structures of Ara h 2 showed that the structures shared a similar protein fold but differed in the loop region. Hence, all 3D model structures of Ara h 2 were used as an input for the EpiSearch analysis (Supplement Fig. 1). We followed a similar strategy as described above for Ara h 2 to build model structures of Ara h 6. However, only the MPACK generated model structure of Ara h 6 was used for the EpiSearch analysis, since it shared a high structural similarity with its NMR structure (data not shown).
Statistics
GraphPad Prism 5.0c for the Macintosh (GraphPad; La Jolla, CA) was used to generate graphs and for statistical analysis. The following tests were used: Spearman rank order correlation coefficients for correlations and Fisher's exact test for comparing frequencies of two possible outcomes. All comparisons were two-tailed and a p value of <0.05 was considered to be statistically significant.
Results
Identification and alignment of Ara h 2/6 IgE-mimotopes
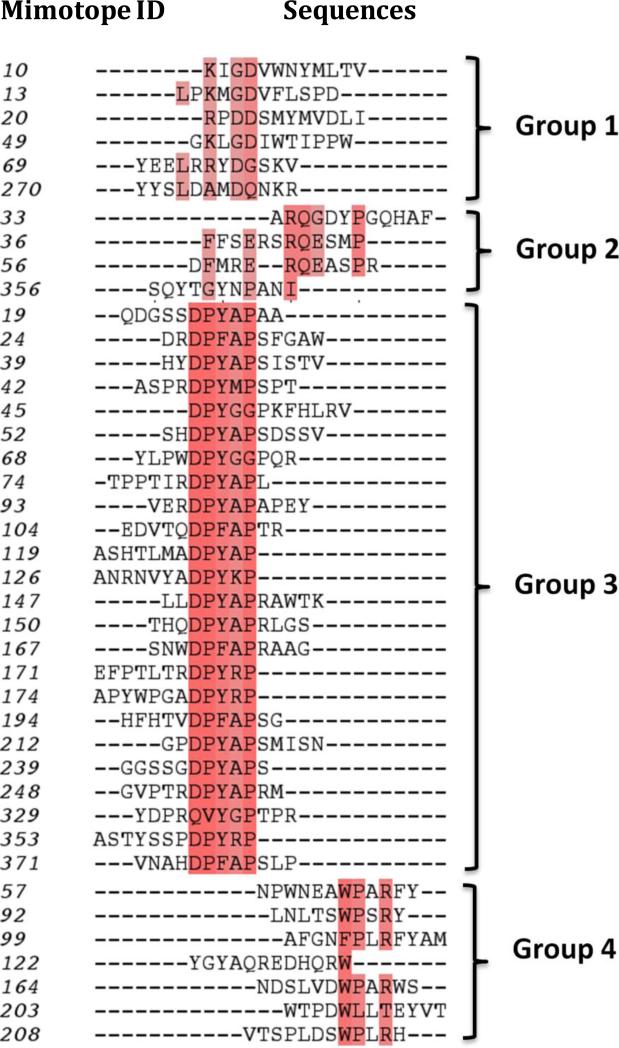
Forty-one individual peptide sequences were identified using affinity-purified anti-Ara h 2/6 IgE from four peanut allergic sera with relatively high levels of specific-IgE for peanut allergens (Table 1). The Ara h 2/6 mimotope sequences were then aligned to the primary sequences of Ara h 2 and Ara h 6. The sequence alignment results revealed the presence of four different patterns of peptides according to their amino acid composition and distribution (Fig. 1). The highly conserved residues for group 1- 4 are D; Q,P; DP(Y/F)XAP and (W/F) PXR. Of note, none of the mimotope sequences match a linear segment of >3 amino acids of the Ara h 2 or Ara h 6 sequences and most likely, mimic conformational epitopes of these allergens. All four peptide groups poorly aligned with different parts of Ara h 2 and Ara h 6 sequences. However, the peptides in group 3 showed the presence of a consensus sequence “DPY/F” that aligned with the linear sequence DPY in Ara h 2 and DSY in Ara h 6, respectively.
Fig. 1.
Sequence alignment of the mimotope. Consensus residues are colored in red.
The mimotopes bind IgE from a variety of peanut allergic sera
We tested reactivity of the identified Ara h 2/6 mimotopes to the four sera that were used to screen the phage library and to 25 additional sera from peanut allergic subjects (Table 2 and Supplementary Table 1). Of the 4 sera used to screen the phage library, serum D103 recognized the most mimotopes (98%), whereas D48 recognized only 44%. Among all sera assayed (n=29; Supplementary Table 1), D78, D80, and D103 recognized the largest numbers of mimotopes (98%) whereas D63 and D213 recognized the fewest (41%). Each serum had distinct IgE recognition patterns but the patterns were not correlated to the concentration of peanut specific IgE (data not shown).
Table 2.
Mimotopes from Group 1 are more frequently recognized by peanut-allergic sera (Summary of data shown in supplementary table 1)
| Group | Mimotopes | Sera | Positive | Negative | % |
|---|---|---|---|---|---|
| 1 | 6 | 29 | 137 | 37 | 78.7 |
| 2 | 4 | 29 | 71 | 45 | 45.5 |
| 3 | 25 | 29 | 415 | 310 | 57.2 |
| 4 | 7 | 29 | 135 | 68 | 66.5 |
Eight mimotopes (numbers: 10, 45, 49, 68, 69, 208, 353 and 371) were recognized by >90% of the sera and 4 of these (numbers: 49, 69, 208, 371) were recognized by 100% of the sera. Among the four groups of mimotopes, those from group 1 showed higher frequency of recognition by these sera (79%) compared to the other groups (46-67%) (p=<0.03) (Supplementary Table 1).
The mimotopes bind IgG from rabbits immunized with either Ara h 2 or Ara h 6
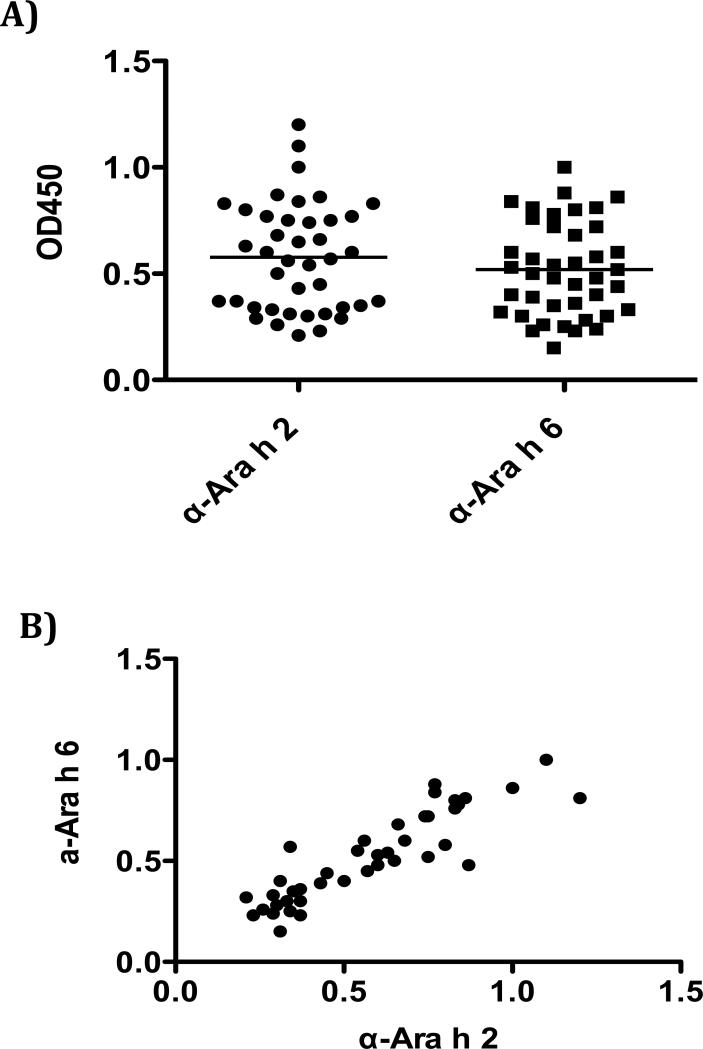
The mimotopes were tested for their ability to bind rabbit IgG that was raised against either Ara h 2 or Ara h 6 IgG. All mimotopes were reactive to anti-Ara h 2 and Ara h 6 IgG but not to pre-immune serum IgG. The mimotopes have similar binding intensity to both anti-Ara h 2 and anti-Ara h 6 IgG (Fig. 2A) and the binding intensities of the anti-Ara h 2 and anti-Ara h 6 IgG for the mimotopes were highly correlated (r= 0.8746;p< 0.0001, Fig. 2B). This further supports previous observations of high antigenic homology between Ara h 2 and Ara h 6.
Fig. 2.
The mimotopes were reactive to anti-Ara h 2 and Ara h 6 IgG. A). The mimotopes have similar binding intensity to both anti-Ara h 2 and Ara h 6 sera. B). Correlation of the mimotopes binding to Ara h 2 and Ara h 6 IgG.
Mapping conformational epitopes in 3D model structures of Ara h 2 and Ara h 6
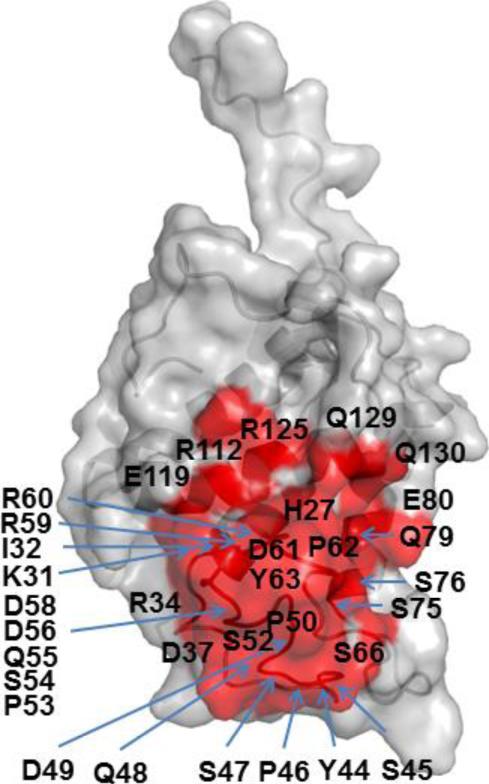
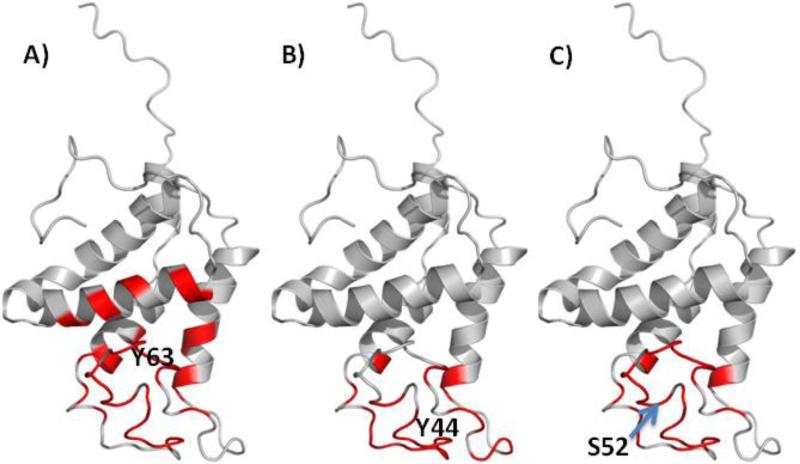
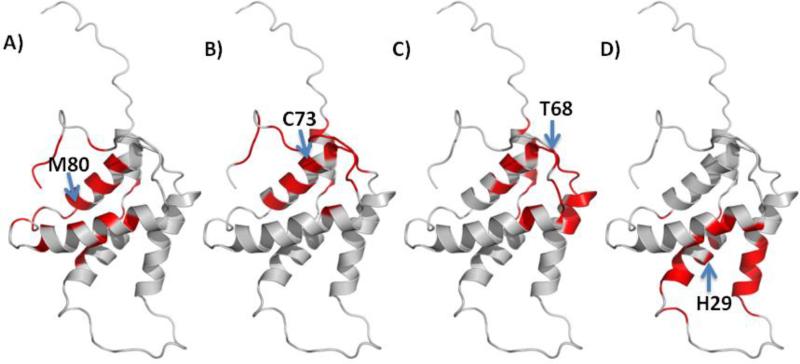
Potential epitope sites on the surface of Ara h 2 and Ara h 6 were mapped using EpiSearch. For Ara h 2, the mimotopes in all groups of peptides mapped to a surface patch centered on Y-63 (Fig. 3). In addition, mimotopes in group 1 and group 3 also mapped to surface patches centered on Y-44 and S-52 respectively (Fig. 4). For Ara h 6, the mimotopes in group 1 mapped to surface patches centered on M-80 and M-31, the mimotopes in group 2 mapped to surface patches centered on H-29, S-52 and Q-32, the mimtiopes in group 3 to surface patches centered on C-73 and those in group 4 mapped to a surface patch centered on T-68 (Fig. 5). All mimotopes represent conformational epitopes, as the surface areas consist of two or more sequential regions that are not neighboring in the primary sequences.
Fig. 3.
Molecular surface representation of Ara h 2 showing a consensus epitope site centered on Y63 (red).
Fig. 4.
Mapping mimotopes on Ara h 2 based on high scoring patch centered on: A) Y63, B) Y44 and C) S52.
Fig. 5.
Mapping conformational epitopes on Ara h 6 based on high scoring patch centered on: A) M80, B) C73, C) T68 and D) H29.
Discussion
Characterization of conformational IgE-epitopes of important peanut allergens is of fundamental importance for understanding mechanisms underlying allergic reactions to foods. In this study, we screened a phage peptide display library that displays 12-mer peptides and identified IgE mimotopes of the most potent peanut allergens, Ara h 2 and Ara h 6. We used this phage library, because it has been demonstrated that the majority of binding interfaces of protein heterodimers is larger than 600 A2, which suggests that the minimum length an epitope should be 8 amino acids [59, 60]. Thus, compared to other libraries that use 7-mer peptides, use of 12-mer peptides ensures a higher affinity interaction and increases the ability to detect important conformational epitopes. Because we are interested in the specificity and diversity of the IgE response to Ara h 2 and Ara h 6, a specific challenge is the identification of epitopes to which the concentration of cognate IgE is low in sera. We have optimized the methods by using avidin-biotin system to increase the detection sensitivity so that signal intensity. Also in this study, we performed acidic elution instead of competitive elution as described by Bogh and colleagues [61], because in our preliminary experiments (not shown), we identified more allergen-specific mimotopes with acidic elution than with competitive elution. By screening this phage peptide library with affinity-purified IgE from 4 peanut allergic patients, we identified 41 individual mimotopes of native Ara h 2 and Ara h 6.
The 3-dimensional structures of Ara h 2 and Ara h 6 have been determined by X- ray crystallography of an Ara h 2 - maltose binding protein fusion protein [52] and by nuclear magnetic resonance for Ara h 6 [14]. Ara h 2 and Ara h 6 share a compact conformation characterized by five α-helical structures and are stabilized by a network of four (Ara h 2) or five (Ara h 6) conserved disulfide bridges [62]. The fifth disulfide bond in Ara h 6 links the C-terminus to the core structure whereas the equivalent region in Ara h 2 is flexible and without regular secondary structure elements[14, 52]. The compact structure of both Ara h 2 and Ara h 6[14] contributes to the high resistance of these peanut allergens to proteolytic cleavage and their thermodynamic stability [14], and possibly leads to the importance of conformational epitopes [22].
In addition to their folding pattern, Ara h 2 and Ara h 6 share 59% of sequence homology and 75% α-helical structural identity [14]. Furthermore, 5 of 7 IgE-binding linear epitopes of Ara h 2 are highly homologous (70-93%) to similar regions of Ara h 6 and binding of IgE to each of these 5 linear epitope pairs is highly correlated [22]. In assays of IgE/FcεR1 cross-linking, Ara h 2 and Ara h 6 have similar potency and do not show either additivity or synergy, suggesting that their allergenic function is highly redundant [63].
The 41 mimotopes that we identified segregated into 4 groups. Mimotopes from group 1 were recognized by more sera compared to each of the other 3 groups (p<0.03). Four mimotopes, numbers 49 and 69, (group 1) and numbers 371 and 208 from groups 3 and 4 respectively, were recognized by all the tested sera. An additional 4 mimopes (numbers: 10, 45, 68, and 150) were recognized by 90-93% of the sera, suggesting immunodominance. However, we found that recognition of these mimotopes was not related to either peanut-specific IgE levels or to clinical histories. Nonetheless, it is likely that these mimotopes are the mimics of epitopes that are important for the allergenicity of Ara h 2 and Ara h 6.
Because of the sequence homology and structural identify of these two allergens, the mimotopes were screened with anti-Ara h 2/6 IgE that was purified from patient sera. We then tested rabbit anti-Ara h 2 or anti-Ara h 6 sera to determine if there is significant cross-reactivity of the identified mimotopes. The mimotopes were recognized by IgG from both anti-Ara h 2 and anti-Ara h 6 sera and this binding was highly correlated, further confirming that these mimotopes are mimics of both Ara h 2 and Ara h 6 epitopes. The cross-reactivity of the mimotopes is consistent with the findings from the structural analysis that all mimotope sequences mapped to overlapping surface patches on both Ara h 2 and Ara h 6.
Although several mimotopes were recognized by ≥90% of the sera tested, each serum had a distinct recognition pattern indicating a broad variation in IgE conformational epitopes among patients suffering from peanut allergy. Similarly, the analysis of IgE binding to linear epitopes of Ara h 2 and Ara h 6 demonstrated a high degree of heterogeneity of the IgE binding patterns among peanut allergic patients [22, 27]. Such results likely reflect the individual development of antibody repertoires, the diverse polyclonal nature of specific antibody responses, and individual progression of affinity maturation in peanut allergic patients.
Finally, we used EpiSearch [46, 48] to analyze the best matches between the amino acid composition of the mimotopes and surface-exposed patches on the 3D model structures of Ara h 2 and Ara h 6. (Figs. 4 and 5). In the case of Ara h2, the EpiSearch analysis using four different input peptide groups predicted high scoring patches in the vicinity of a patch centered on Y63 (Fig. 3). In case of Ara h 6, the EpiSearch analysis predicted four top scoring patches centered on M80, T68, C73 and H29, three of which, the patches centered on M80, T68, and C73 share several overlapping residues. Further, structural alignment between Ara h 2 and Ara h 6 showed that the patch centered on H29 in Ara h 6 partially overlaps with the high scoring patch centered on Y63 in Ara h 2 (Figs. 3 and 5). Of note, the mimotopes in group 3 have a consensus sequence DPY/F that aligns with the linear sequence DPY in Ara h 2 and DSY in Ara h 6 also reported using computational analysis[30]. These unique consensus sequences were also predicted by EpiSearch analysis and are likely part of conformational epitopes on Ara h 2 and Ara h 6, respectively.
In conclusion, the mimotopes selected by the phage-display technology are most likely to be conformational. This argument is supported by the finding that the mimotopes map to the surface exposed areas of Ara h 2/ 6 and the finding that linear sequences were not observed. This study provides a new approach to identify epitopes that are potentially important for initiation of allergic reactions.
Supplementary Material
Acknowledgements
This work was supported by RO1-AI052164 (SCD), a supplemental ARRA grant (SCD), R01-AI099029 (SCD), R21-AI109090(WB) and R21-AI112792(XC) from the National Institute of Allergy and Infectious Diseases and divisional funds. Also, this work was supported by NIH/NCRR Colorado CTSI Grant Number UL1 RR025780. We thank all of our subjects who donated serum and Spodra Eglite, our study coordinator. Contents are the authors’ sole responsibility and do not necessarily represent official NIH views.
Abbreviations used
- Ara h 2/6
Ara h 2 and Ara h 6
References
- 1.Sicherer SH, Munoz-Furlong A, Godbold JH, Sampson HA. US prevalence of self-reported peanut, tree nut, and sesame allergy: 11-year follow-up. J Allergy Clin Immunol. 2010;125:1322–6. doi: 10.1016/j.jaci.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 2.Venter C, Arshad SH. Epidemiology of food allergy. Pediatr Clin North Am. 2011;58:327–49. ix. doi: 10.1016/j.pcl.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Du Toit G, Roberts G, Sayre PH, Bahnson HT, Radulovic S, Santos AF, Brough HA, Phippard D, Basting M, Feeney M, Turcanu V, Sever ML, Gomez Lorenzo M, Plaut M, Lack G, Team LS. Randomized trial of peanut consumption in infants at risk for peanut allergy. N Engl J Med. 2015;372:803–13. doi: 10.1056/NEJMoa1414850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fleischer DM, Burks AW, Vickery BP, Scurlock AM, Wood RA, Jones SM, Sicherer SH, Liu AH, Stablein D, Henning AK, Mayer L, Lindblad R, Plaut M, Sampson HA, Consortium of Food Allergy R Sublingual immunotherapy for peanut allergy: a randomized, double-blind, placebo-controlled multicenter trial. J Allergy Clin Immunol. 2013;131:119–27. e1–7. doi: 10.1016/j.jaci.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nurmatov U, Venderbosch I, Devereux G, Simons FE, Sheikh A. Allergen-specific oral immunotherapy for peanut allergy. Cochrane Database Syst Rev. 2012;9:CD009014. doi: 10.1002/14651858.CD009014.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vickery BP, Lin J, Kulis M, Fu Z, Steele PH, Jones SM, Scurlock AM, Gimenez G, Bardina L, Sampson HA, Burks AW. Peanut oral immunotherapy modifies IgE and IgG4 responses to major peanut allergens. J Allergy Clin Immunol. 2013;131:128–34. e1–3. doi: 10.1016/j.jaci.2012.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King RM, Knibb RC, Hourihane JO. Impact of peanut allergy on quality of life, stress and anxiety in the family. Allergy. 2009;64:461–8. doi: 10.1111/j.1398-9995.2008.01843.x. [DOI] [PubMed] [Google Scholar]
- 8.King JC, Blumberg J, Ingwersen L, Jenab M, Tucker KL. Tree nuts and peanuts as components of a healthy diet. J Nutr. 2008;138:1736S–40S. doi: 10.1093/jn/138.9.1736S. [DOI] [PubMed] [Google Scholar]
- 9.Sicherer SH, Sampson HA. Food allergy: Epidemiology, pathogenesis, diagnosis, and treatment. J Allergy Clin Immunol. 2014;133:291–307. doi: 10.1016/j.jaci.2013.11.020. quiz 08. [DOI] [PubMed] [Google Scholar]
- 10.Soares-Weiser K, Takwoingi Y, Panesar SS, Muraro A, Werfel T, Hoffmann-Sommergruber K, Roberts G, Halken S, Poulsen L, van Ree R, Vlieg-Boerstra BJ, Sheikh A, Allergy EF. Anaphylaxis Guidelines G, The diagnosis of food allergy: a systematic review and meta-analysis. Allergy. 2014;69:76–86. doi: 10.1111/all.12333. [DOI] [PubMed] [Google Scholar]
- 11.Sicherer SH, Sampson HA. Peanut allergy: emerging concepts and approaches for an apparent epidemic. J Allergy Clin Immunol. 2007;120:491–503. doi: 10.1016/j.jaci.2007.07.015. quiz 04-5. [DOI] [PubMed] [Google Scholar]
- 12.Koppelman SJ, de Jong GA, Laaper-Ertmann M, Peeters KA, Knulst AC, Hefle SL, Knol EF. Purification and immunoglobulin E-binding properties of peanut allergen Ara h 6: evidence for cross-reactivity with Ara h 2. Clin Exp Allergy. 2005;35:490–7. doi: 10.1111/j.1365-2222.2005.02204.x. [DOI] [PubMed] [Google Scholar]
- 13.Kulis M, Chen X, Lew J, Wang Q, Patel OP, Zhuang Y, Murray KS, Duncan MW, Porterfield HS, A WB, Dreskin SC. The 2S albumin allergens of Arachis hypogaea, Ara h 2 and Ara h 6, are the major elicitors of anaphylaxis and can effectively desensitize peanut-allergic mice. Clin Exp Allergy. 2012;42:326–36. doi: 10.1111/j.1365-2222.2011.03934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehmann K, Schweimer K, Reese G, Randow S, Suhr M, Becker WM, Vieths S, Rosch P. Structure and stability of 2S albumin-type peanut allergens: implications for the severity of peanut allergic reactions. Biochem J. 2006;395:463–72. doi: 10.1042/BJ20051728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porterfield HS, Murray KS, Schlichting DG, Chen X, Hansen KC, Duncan MW, Dreskin SC. Effector activity of peanut allergens: a critical role for Ara h 2, Ara h 6, and their variants. Clin Exp Allergy. 2009;39:1099–108. doi: 10.1111/j.1365-2222.2009.03273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Zhuang Y, Wang Q, Moutsoglou D, Ruiz G, Yen SE, Dreskin SC. Analysis of the effector activity of Ara h 2 and Ara h 6 by selective depletion from a crude peanut extract. J Immunol Methods. 2011;372:65–70. doi: 10.1016/j.jim.2011.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klemans RJ, Broekman HC, Knol EF, Bruijnzeel-Koomen CA, Otten HG, Pasmans SG, Knulst AC. Ara h 2 is the best predictor for peanut allergy in adults. J Allergy Clin Immunol Pract. 2013;1:632–8. e1. doi: 10.1016/j.jaip.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 18.Klemans RJ, Knol EF, Bruijnzeel-Koomen CA, Knulst AC. The diagnostic accuracy of specific IgE to Ara h 6 in adults is as good as Ara h 2. Allergy. 2014;69:1112–4. doi: 10.1111/all.12424. [DOI] [PubMed] [Google Scholar]
- 19.Kukkonen AK, Pelkonen AS, Makinen-Kiljunen S, Voutilainen H, Makela MJ. Ara h 2 and Ara 6 are the best predictors of severe peanut allergy: a double-blind placebo-controlled study. Allergy. 2015;70:1239–45. doi: 10.1111/all.12671. [DOI] [PubMed] [Google Scholar]
- 20.Keet CA, Johnson K, Savage JH, Hamilton RG, Wood RA. Evaluation of Ara h2 IgE thresholds in the diagnosis of peanut allergy in a clinical population. J Allergy Clin Immunol Pract. 2013;1:101–3. doi: 10.1016/j.jaip.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Wang Q, El-Mezayen R, Zhuang Y, Dreskin SC. Ara h 2 and Ara h 6 have similar allergenic activity and are substantially redundant. Int Arch Allergy Immunol. 2013;160:251–8. doi: 10.1159/000341642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otsu K, Guo R, Dreskin SC. Epitope analysis of Ara h 2 and Ara h 6: characteristic patterns of IgE-binding fingerprints among individuals with similar clinical histories. Clin Exp Allergy. 2015;45:471–84. doi: 10.1111/cea.12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pomes A. Relevant B cell epitopes in allergic disease. Int Arch Allergy Immunol. 2010;152:1–11. doi: 10.1159/000260078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varshney S, Goldblum RM, Kearney C, Watanabe M, Midoro-Horiuti T. Major mountain cedar allergen, Jun a 1, contains conformational as well as linear IgE epitopes. Mol Immunol. 2007;44:2781–5. doi: 10.1016/j.molimm.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schein CH, Ivanciuc O, Midoro-Horiuti T, Goldblum RM, Braun W. An Allergen Portrait Gallery: Representative Structures and an Overview of IgE Binding Surfaces. Bioinform Biol Insights. 2010;4:113–25. doi: 10.4137/BBI.S5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Power TD, Ivanciuc O, Schein CH, Braun W. Assessment of 3D models for allergen research. Proteins. 2013;81:545–54. doi: 10.1002/prot.24239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shreffler WG, Beyer K, Chu TH, Burks AW, Sampson HA. Microarray immunoassay: association of clinical history, in vitro IgE function, and heterogeneity of allergenic peanut epitopes. J Allergy Clin Immunol. 2004;113:776–82. doi: 10.1016/j.jaci.2003.12.588. [DOI] [PubMed] [Google Scholar]
- 28.Shreffler WG, Lencer DA, Bardina L, Sampson HA. IgE and IgG4 epitope mapping by microarray immunoassay reveals the diversity of immune response to the peanut allergen, Ara h 2. J Allergy Clin Immunol. 2005;116:893–9. doi: 10.1016/j.jaci.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 29.Flinterman AE, Knol EF, Lencer DA, Bardina L, den Hartog Jager CF, Lin J, Pasmans SG, Bruijnzeel-Koomen CA, Sampson HA, van Hoffen E, Shreffler WG. Peanut epitopes for IgE and IgG4 in peanut-sensitized children in relation to severity of peanut allergy. J Allergy Clin Immunol. 2008;121:737–43. e10. doi: 10.1016/j.jaci.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 30.Mishra A, Jain A, Arora N. Mapping B-cell epitopes of major and minor peanut allergens and identifying residues contributing to IgE binding. J Sci Food Agric. 2016;96:539–47. doi: 10.1002/jsfa.7121. [DOI] [PubMed] [Google Scholar]
- 31.Albrecht M, Kuhne Y, Ballmer-Weber BK, Becker WM, Holzhauser T, Lauer I, Reuter A, Randow S, Falk S, Wangorsch A, Lidholm J, Reese G, Vieths S. Relevance of IgE binding to short peptides for the allergenic activity of food allergens. J Allergy Clin Immunol. 2009;124:328–36. 36, e1–6. doi: 10.1016/j.jaci.2009.05.031. [DOI] [PubMed] [Google Scholar]
- 32.Maleki SJ, Viquez O, Jacks T, Dodo H, Champagne ET, Chung SY, Landry SJ. The major peanut allergen, Ara h 2, functions as a trypsin inhibitor, and roasting enhances this function. J Allergy Clin Immunol. 2003;112:190–5. doi: 10.1067/mai.2003.1551. [DOI] [PubMed] [Google Scholar]
- 33.Wood RASS, Burks AW, Grishin A, Henning AK, Lindblad R, Stablein D, Sampson HA. A phase 1 study of heat/phenol-killed, E. coli-encapsulated, recombinant modified peanut proteins Ara h 1, Ara h 2, and Ara h 3 (EMP-123) for the treatment of peanut allergy. Allergy. 2013;68:803–08. doi: 10.1111/all.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernard H, Guillon B, Drumare MF, Paty E, Dreskin SC, Wal JM, Adel-Patient K, Hazebrouck S. Allergenicity of peanut component Ara h 2: Contribution of conformational versus linear hydroxyproline-containing epitopes. J Allergy Clin Immunol. 2015;135:1267–74. e1–8. doi: 10.1016/j.jaci.2014.10.025. [DOI] [PubMed] [Google Scholar]
- 35.Seiskari T, Kondrashova A, Viskari H, Kaila M, Haapala AM, Aittoniemi J, Virta M, Hurme M, Uibo R, Knip M, Hyoty H. Allergic sensitization and microbial load--a comparison between Finland and Russian Karelia. Clin Exp Immunol. 2007;148:47–52. doi: 10.1111/j.1365-2249.2007.03333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Padavattan S, Schirmer T, Schmidt M, Akdis C, Valenta R, Mittermann I, Soldatova L, Slater J, Mueller U, Markovic-Housley Z. Identification of a B-cell epitope of hyaluronidase, a major bee venom allergen, from its crystal structure in complex with a specific Fab. J Mol Biol. 2007;368:742–52. doi: 10.1016/j.jmb.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 37.Padavattan S, Flicker S, Schirmer T, Madritsch C, Randow S, Reese G, Vieths S, Lupinek C, Ebner C, Valenta R, Markovic-Housley Z. High-affinity IgE recognition of a conformational epitope of the major respiratory allergen Phl p 2 as revealed by X-ray crystallography. J Immunol. 2009;182:2141–51. doi: 10.4049/jimmunol.0803018. [DOI] [PubMed] [Google Scholar]
- 38.Amar SaD, S.C. Urticaria and Angioedema. Primary Care: Clinics in Office Practice. 2008 In press. [Google Scholar]
- 39.Luzzago A, Felici F, Tramontano A, Pessi A, Cortese R. Mimicking of discontinuous epitopes by phage-displayed peptides, I. Epitope mapping of human H ferritin using a phage library of constrained peptides. Gene. 1993;128:51–7. doi: 10.1016/0378-1119(93)90152-s. [DOI] [PubMed] [Google Scholar]
- 40.Geysen HM, Rodda SJ, Mason TJ. A priori delineation of a peptide which mimics a discontinuous antigenic determinant. Mol Immunol. 1986;23:709–15. doi: 10.1016/0161-5890(86)90081-7. [DOI] [PubMed] [Google Scholar]
- 41.Folgori A, Tafi R, Meola A, Felici F, Galfre G, Cortese R, Monaci P, Nicosia A. A general strategy to identify mimotopes of pathological antigens using only random peptide libraries and human sera. EMBO J. 1994;13:2236–43. doi: 10.1002/j.1460-2075.1994.tb06501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mittag D, Batori V, Neudecker P, Wiche R, Friis EP, Ballmer-Weber BK, Vieths S, Roggen EL. A novel approach for investigation of specific and cross-reactive IgE epitopes on Bet v 1 and homologous food allergens in individual patients. Mol Immunol. 2006;43:268–78. doi: 10.1016/j.molimm.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 43.Garcia-Casado G, Pacios LF, Diaz-Perales A, Sanchez-Monge R, Lombardero M, Garcia-Selles FJ, Polo F, Barber D, Salcedo G. Identification of IgE-binding epitopes of the major peach allergen Pru p 3. J Allergy Clin Immunol. 2003;112:599–605. doi: 10.1016/s0091-6749(03)01605-1. [DOI] [PubMed] [Google Scholar]
- 44.Hantusch B, Krieger S, Untersmayr E, Scholl I, Knittelfelder R, Flicker S, Spitzauer S, Valenta R, Boltz-Nitulescu G, Scheiner O, Jensen-Jarolim E. Mapping of conformational IgE epitopes on Phl p 5a by using mimotopes from a phage display library. J Allergy Clin Immunol. 2004;114:1294–300. doi: 10.1016/j.jaci.2004.06.048. [DOI] [PubMed] [Google Scholar]
- 45.Bogh KL, Nielsen H, Madsen CB, Mills EN, Rigby N, Eiwegger T, Szepfalusi Z, Roggen EL. IgE epitopes of intact and digested Ara h 1: a comparative study in humans and rats. Mol Immunol. 2012;51:337–46. doi: 10.1016/j.molimm.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 46.Tiwari R, Negi SS, Braun B, Braun W, Pomes A, Chapman MD, Goldblum RM, Midoro-Horiuti T. Validation of a phage display and computational algorithm by mapping a conformational epitope of Bla g 2. Int Arch Allergy Immunol. 2012;157:323–30. doi: 10.1159/000330108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Untersmayr E, Szalai K, Riemer AB, Hemmer W, Swoboda I, Hantusch B, Scholl I, Spitzauer S, Scheiner O, Jarisch R, Boltz-Nitulescu G, Jensen-Jarolim E. Mimotopes identify conformational epitopes on parvalbumin, the major fish allergen. Mol Immunol. 2006;43:1454–61. doi: 10.1016/j.molimm.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 48.Negi SS, Braun W. Automated detection of conformational epitopes using phage display Peptide sequences. Bioinform Biol Insights. 2009;3:71–81. doi: 10.4137/bbi.s2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dreskin SC, Goldsmith PK, Strober W, Zech LA, Gallin JI. Metabolism of immunoglobulin E in patients with markedly elevated serum immunoglobulin E levels. J Clin Invest. 1987;79:1764–72. doi: 10.1172/JCI113017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beitzel K, Mazzocca AD, Obopilwe E, Boyle JW, McWilliam J, Rincon L, Dhar Y, Arciero RA, Amendola A. Biomechanical properties of double- and single-row suture anchor repair for surgical treatment of insertional Achilles tendinopathy. Am J Sports Med. 2013;41:1642–8. doi: 10.1177/0363546513487061. [DOI] [PubMed] [Google Scholar]
- 51.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 52.Mueller GA, Gosavi RA, Pomes A, Wunschmann S, Moon AF, London RE, Pedersen LC. Ara h 2: crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity. Allergy. 2011;66:878–85. doi: 10.1111/j.1398-9995.2010.02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanner M, Widmer A, Senn H, Braun W. GEOM: a new tool for molecular modelling based on distance geometry calculations with NMR data. J Comput Aided Mol Des. 1989;3:195–210. doi: 10.1007/BF01533068. [DOI] [PubMed] [Google Scholar]
- 55.Mumenthaler C, Braun W. Automated assignment of simulated and experimental NOESY spectra of proteins by feedback filtering and self-correcting distance geometry. J Mol Biol. 1995;254:465–80. doi: 10.1006/jmbi.1995.0631. [DOI] [PubMed] [Google Scholar]
- 56.Oezguen N, Zhou B, Negi SS, Ivanciuc O, Schein CH, Labesse G, Braun W. Comprehensive 3D-modeling of allergenic proteins and amino acid composition of potential conformational IgE epitopes. Mol Immunol. 2008;45:3740–7. doi: 10.1016/j.molimm.2008.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Helm RM, Burks AW. Sensitization and allergic response and intervention therapy in animal models. J AOAC Int. 2004;87:1441–7. [PubMed] [Google Scholar]
- 58.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones S, Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci U S A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meloen RH, Puijk WC, Slootstra JW. Mimotopes: realization of an unlikely concept. J Mol Recognit. 2000;13:352–9. doi: 10.1002/1099-1352(200011/12)13:6<352::AID-JMR509>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 61.Bogh KL, Nielsen H, Eiwegger T, Madsen CB, Mills EN, Rigby NM, Szepfalusi Z, Roggen EL. IgE versus IgG4 epitopes of the peanut allergen Ara h 1 in patients with severe allergy. Mol Immunol. 2014;58:169–76. doi: 10.1016/j.molimm.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 62.Moreno FJ, Clemente A. 2S Albumin Storage Proteins: What Makes them Food Allergens? Open Biochem J. 2008;2:16–28. doi: 10.2174/1874091X00802010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen X, Wang Q, EI-Mezayen R, Zhuang Y, Dreskin SC. Ara h2 and Ara h 6 have similar allergenic activity and are substantially redudent. Int Arch Allergy Immunol. 2012;160:251–58. doi: 10.1159/000341642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.