Abstract
Background
Enterococci are normal commensals of human gut, but vancomycin-resistant enterococci (VRE) are a severe threat to human health. Antimicrobial-resistant enterococci have been reported previously from Indian surface waters. However, the presence of antimicrobial resistance and virulence markers in Enterococcus faecalis, the most dominant enterococci is yet to be investigated.
Objectives
The goal of this study was to analyse concentration of enterococci and distribution of antimicrobial resistance and virulence markers in E. faecalis isolates from river waters along an important north Indian city landscape.
Methods
We enumerated enterococci in river water samples (n = 60) collected from five sites across the Lucknow city landscape using the most probable number and membrane-filtration methods. The antimicrobial sensitivity profile of E. faecalis isolate was generated with the Kirby-Bauer antimicrobial disc diffusion assay. The multiplex PCR was used for genotypic characterization of vancomycin-resistance and virulence in E. faecalis isolates.
Results
Enterococci density (p < 0.0001) increased from up-to-down-stream sites. Multiplex PCR based genotypic characterization has shown a significant distribution of virulence-markers gelE, ace or efaA in the E. faecalis isolates (p < 0.05). The range of antimicrobial-resistance varied from 5 to 12 in the landscape with the frequency of vancomycin-resistant E. faecalis (VRE) ranging from 22 to 100 %.
Conclusion
The occurrence of pathogenic VRE in river Gomti surface water is an important health concern. The observed high background pool of resistance and virulence in E. faecalis in river waters has the potential to disseminate more alarming antimicrobial resistance in the environment and poses serious health risk in developing countries like India as VRE infections could lead to increased cost of healthcare.
Electronic supplementary material
The online version of this article (doi:10.1186/s40064-016-2870-5) contains supplementary material, which is available to authorized users.
Keywords: Mutiplex PCR, River water, Surface water, Vancomycin resistant E. faecalis
Background
Enterococci is considered to be a suitable ‘indicator’ of fecal contamination in aquatic environment as it survives longer compared to other fecal streptococci and coliforms (Cabelli et al. 1982; USEPA 2003). Enterococcus faecalis, most prevalent enterococci, is an enteric bacterium of mammalian intestinal tract. It is notorious as an opportunistic pathogen causing various infections (Thurlow et al. 2010; Parsek and Singh 2003). Majority of enterococcal infections (~80 %) are caused by E. faecalis due to its ability to acquire virulence traits and resistance to multiple antimicrobials (Gordon et al. 1992; Huycke et al. 1998; Ghoshal et al. 2006). The genome sequence of a clinical E. faecalis isolate V583 has revealed that more than a quarter of its genome consists of mobile or foreign DNA, including genes for vancomycin resistance and virulence markers (Paulsen et al. 2003). Vancomycin, once considered as the most powerful antimicrobial of “last resort,” is no longer efficacious in managing antimicrobial-resistant enterococci (Arias and Murray 2012). Further, the incidence of E. faecalis exhibiting antimicrobial-resistance viz. vancomycin resistance and virulence markers such as gelatinase (gelE), collagen binding protein (ace), endocarditis-associated antigen (efaA) and enterococcal surface protein (esp), have been explored mostly in clinical settings worldwide (Coque et al. 2002; Lowe et al. 1995; Eaton and Gasson 2001; Ghoshal et al. 2006; Mathur et al. 2003). However, very little is known about the distribution of antimicrobial-resistance and virulence-markers in E. faecalis isolates recovered from river waters (Novais et al. 2005; Moore et al. 2008; Lata et al. 2009b). River waters in developing countries are vulnerable to contamination due to discharge of untreated or partially treated municipal and industrial wastes, domestic and wild animal fecal waste and recreational activities on the banks (Ahmed et al. 2005; Ram et al. 2007). Such uninterrupted contamination of rivers lead to a bigger problem of nutrient enrichment or eutrophication of river waters. The eutrophication further leads to growth of aquatic macrophytes assisting the proliferation and prolonged survival of pathogens increasing public health risk (Whitman et al. 2003; Lata et al. 2009b).
River Gomti, a tributary of the river Ganga meets the major water requirement of more than 3.5 million populations of Lucknow city (Fig. 1). This river receives a discharge of 450 mLd untreated waste through different point sources due to the rapid industrialization and urbanization in the region (Singh et al. 2004). In India, river waters are used for drinking, bathing and carrying out various household chores. This demands determination of microbiological quality of river waters for remediation to protect public health. Reported prevalence of diarrheogenic and enterotoxigenic E. coli in the river waters of river Ganga and its tributary river Gomti has emphasized the necessity of information on recreational water quality ‘indicator’, E. faecalis in such riverine systems (Ram et al. 2007, 2008; Singh et al. 2010). This study investigates the prevalence and dissemination of vancomycin-resistant and pathogenic E. faecalis along the up-to-downstream landscape of Lucknow city (Fig. 1). The genotypic characterization is done by multiplex PCR assay panels comprising simultaneous amplification of four genes: sodA, vanB, gelE and either ace or efaA. In addition, the prevalence of esp virulence marker and multiple-antimicrobial-resistance was also examined amongst E. faecalis isolates from the river Gomti water.
Fig. 1.
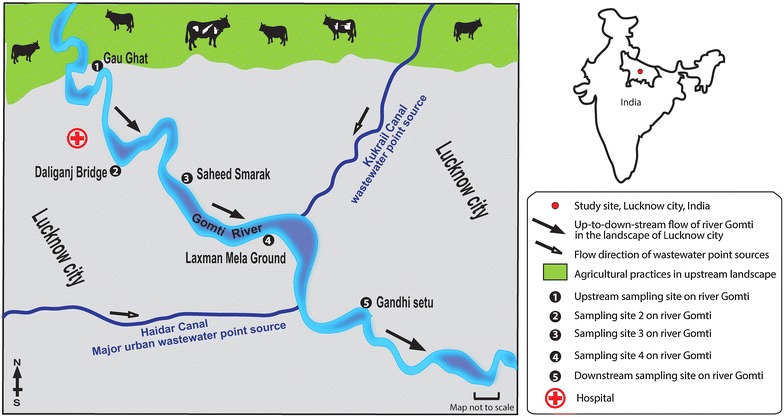
Map of study area/sampling sites in the landscape. Abbreviations: S#1, site 1: Gau-Ghat (the most upstream site), S#2, site 2: Daliganj Bridge, S#3, site 3: P Shaheed Smarak, S#4, site 4: Lakhshaman mela ground, S#5, site 5: Gandhi setu (the most downstream site). Black-and-white arrows indicate the direction of river water and wastewater flow in the up-to-downstream in the landscape respectively. Topographic data based upon Survey of the map of India
Adopted from www.ttkmaps.com
Results
Concentration of enterococci
Median concentrations of enterococci increased gradually and significantly (p < 0.0001) in the river Gomti waters from up-to-down-stream sites in the landscape. The most downstream site was found to be 123 and > 4500-fold more polluted than the most upstream site, using MPN test and membrane filtration method, respectively (Table 1; Additional file 1: Table S1). Concentrations of enterococci at downstream sites were about 10, 23 and 69-fold higher at site 2, 3 and 4, respectively when compared to the most upstream site, using MPN test (Additional file 1: Table S1). At the same time, median concentration of enterococci using membrane filtration method was 45, 296, 512-fold higher at site 2, 3 and 4, respectively (Additional file 1: Table S1).
Table 1.
Quantitative enumeration (log transformed) of enterococci collected from sites (n = 5) located on river Gomti in up-to-down-stream fashion
| Sampling site | CFU/100 mL water [Median 95 % CI (lower–upper)] |
MPN index/100 mLwater 95 % CI (lower–upper)a |
p valueb |
|---|---|---|---|
| Site 1 | 2.18 (2.16–2.21) | 1.11 (0.70–1.58) | <0.0001*** |
| Site 2 | 3.84 (3.83–3.84) | 2.15 (1.78–2.56) | |
| Site 3 | 4.66 (4.65–4.67) | 2.48 (2.00–3.11) | |
| Site 4 | 4.89 (4.89–4.90) | 2.95 (2.48–3.46) | |
| Site 5 | 5.84 (5.84–5.84) | 3.20 (2.78–3.72) | |
| Controlc | ND | ND |
ND not detected
*** Statistically significant at alpha < 0.05
aLower 95 % CI–upper 95 % CI is adopted from Table 9221.IV, Section 9221C. Estimation of bacterial density, APHA (1998); log transformed values shown in the Table 1; See Additional file 1: Table S1 for details
b p value was calculated using Chi square test for trend, χ2 = 1636; df = 1
cSterile Milli Q water used as control
Multiplex PCR for vancomycin resistant pathogenic E. faecalis
The multiplex PCR based genotypic characterization was carried out to determine the prevalence of vanB resistance and gelE, ace or efaA virulence markers in E. faecalis isolates from river Gomti (Tables 2 and 3; Fig. 2). The genotypic characterization has shown a significant distribution of virulence-markers along the landscape (p < 0.05). The median count investigated throughout the landscape was 3 (Table 4). According to genotypic characterization the frequency of vanB resistant E. faecalis is 27 % in the landscape. However, 100 % E. faecalis isolates recovered from river Gomti exhibit prevalence of gelE (gelatinase) and esp virulence markers. The esp gene was characterized as described earlier (Lata et al. 2009a). The ace gene was observed in ca. 58 % E. faecalis and the frequency of efaA virulence marker was ca. 97 % in the landscape. Further the frequency of various combinations of multiple-virulence-markers has been observed in the landscape (Table 3).
Table 2.
Multiplex PCR primers for vancomycin-resistance and virulence-markers detection in river water isolates of E. faecalis
| Multiplexed/singleplexed genes | Oligomer sequence (5′ → 3′) |
Oligo-mer conc.a (µM) | Product size (bp) |
Reference |
|---|---|---|---|---|
|
sodA
(E.faecalis) |
F: 5′-ACTTATGTGACTAACTTAACC-3′ R: 5′-TAATGGTGAATCTTGGTTTGG-3′ |
0.15 | 360 | Jackson et al. (2004) |
| vanB | F: 5′-CAAAGCTCCGCAGCTTGCATG-3′ R: 5′-TGCATCCAAGCACCCGATATAC-3′ |
0.20 | 484 | Dahl et al. (1999) |
| gelE | F: 5′-ACCCCGTATCATTGGTTT-3′ R: 5′-ACGCATTGCTTTTCCATC-3′ |
0.25 | 419 | Eaton and Gasson (2001) |
| ace b | F: 5′-GGAATGACCGAGAACGATGGC-3′ R: 5′-GCTTGATGTTGGCCTGCTTCCG-3′ |
0.29 | 616 | Creti et al. (2004) |
| efaA b | F: 5′-GCCAATTGGGACAGACCCTC-3′ R: 5′-CGCCTTCTGTTCCTTCTTTGGC-3′ |
0.30 | 688 | Creti et al. (2004) |
| esp c | F: 5′-TTGCTAATGCTAGTCCACGACC-3′ R: 5′-GCGTCAACACTTGCATTGCCGAA-3′ |
0.15 | 933 | Eaton and Gasson (2001) |
aOligomer concentration optimized for this study
bMultiplexed genes include either ace or efaA gene with sodA, vanB and gelE genes
cSingleplexed gene
Table 3.
Multiplex PCR based fingerprints of vancomycin-resistance and multiple-virulence-markers in E. faecalis isolates from river Gomti
|
E. faecalis
isolates |
Target genes | |||||
|---|---|---|---|---|---|---|
| Housekeeping gene | Antimicrobial resistance and Virulence genes | |||||
|
sodA
(E. faecalis) |
vanB | gelE | ace | efaA | Esp | |
| E. faecalis ATCC 51299 (ATCC, USA) (positive control) | + | + | +P | + | + | + |
| Site #1 | ||||||
| 2E | + | − | + | − | + | + |
| AE | + | − | +P | − | + | + |
| BE | + | − | +P | + | + | + |
| FE | + | + | + | − | + | + |
| GE | + | + | +P | − | + | + |
| Site #2 | ||||||
| 4E | + | + | +P | + | + | + |
| HE | + | + | +P | − | + | + |
| CE | + | + | +P | + | + | + |
| G2A | + | − | +P | − | + | + |
| G2B | + | − | + | + | + | + |
| Site #3 | ||||||
| 9E | + | − | +P | − | + | + |
| IE | + | − | + | − | + | + |
| 16E | + | − | +P | + | + | + |
| G3A | + | − | +P | + | + | + |
| G3B | + | − | +P | + | + | + |
| G3C | + | − | + | − | + | + |
| G3D | + | − | + | − | + | + |
| Site #4 | ||||||
| 11E | + | − | +P | + | + | + |
| 17E | + | − | + | + | + | + |
| 18E | + | + | +P | + | + | + |
| 19E | + | − | +P | − | + | + |
| 20E | + | + | +P | − | − | + |
| G4A | + | − | + | + | + | + |
| G4B | + | − | + | + | + | + |
| Site #5 | ||||||
| 15E | + | − | +P | − | + | + |
| 21E | + | − | + | + | + | + |
| 22E | + | + | +P | − | + | + |
| 23E | + | + | +P | + | + | + |
| 24E | + | − | + | + | + | + |
| G5A | + | − | +P | + | + | + |
| G5B | + | − | +P | + | + | + |
| G6A | + | − | + | + | + | + |
| G6B | + | − | +P | + | + | + |
ATCC American Type Culture Collection, USA
+P denotes E. faecalis isolates exhibiting gelatinase phenotype
Fig. 2.
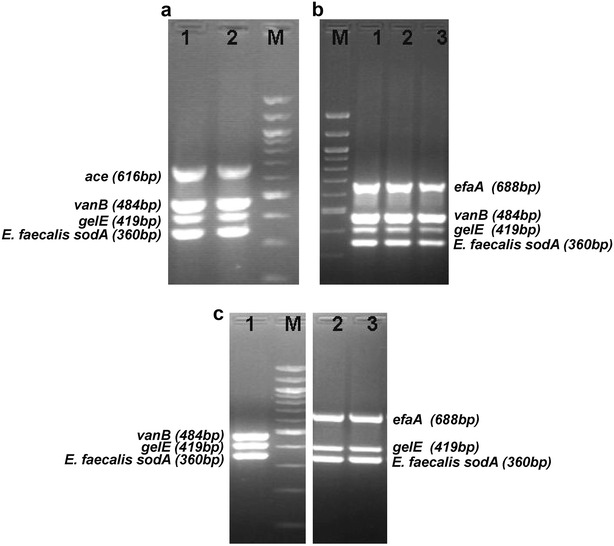
Multiplex PCR amplicons of E. faecalis isolates exhibiting vanB resistance and gelE, ace, efaA virulence-markers. a sodA + gelE + vanB + ace + genotype, lane 1: positive control (E. faecalis ATCC 51229), lane 2: 23E, lane M: 100 bp DNA ladder (New England Biolabs Inc.). b sodA + gelE + vanB + efA + genotype, lane M 100 bp DNA ladder (New England Biolabs Inc.), lane 1 positive control (E. faecalis ATCC 51229), lane 2 HE, lane 3 GE and c sodA + gelE + vanB + genotype, lane 1: 20E, lane M 100 bp DNA ladder (New England Biolabs Inc.), sodA + efaA + gelE + genotype, lane 2: G3D, lane 3 G3C
Table 4.
Multiple-virulence-markers observed in each E. faecalis isolate from the up-to-down-stream landscape of Lucknow city
| Sampling site | Virulence-markers characterized per isolate Median (range) |
p valuea |
|---|---|---|
| Site 1 | 3 (3–3) | 0.0313* |
| Site 2 | 4 (3–4) | 0.0313* |
| Site 3 | 3 (3–4) | 0.0078* |
| Site 4 | 4 (1–4) | 0.0078* |
| Site 5 | 4 (3–4) | 0.0020* |
a p value was calculated using Wilcoxon rank sum test
* Statistically significant at alpha = 0.05
Gelatinase exhibiting E. faecalis phenotypes
The phenotypic assay for determination of gelatinase activity shows high prevalence of gelatinase positive E. faecalis isolates in river Gomti throughout the landscape. Site 2 exhibits highest prevalence (80 %) of such isolates followed by the most downstream site 5 (67 %) and then most upstream site 1 (60 %) and 57 % for both site 3 and 4 (Table 3).
Antimicrobial-resistance in E. faecalis isolates
A high frequency of multiple-antimicrobial-resistance was recorded among E. faecalis tested. The range for the number of antimicrobials against which resistance was observed in each E. faecalis isolates increased significantly (p < 0.05) towards downstream in the landscape (Tables 5, 6). The median of antimicrobial-resistance per isolate was nine at Gau-Ghat, the most upstream site and increased to a value of eleven at site 3 and subsequently declined to nine at site 4 and 5. In general, E. faecalis isolates were resistant to at least five antimicrobials throughout the landscape. However, the range of antimicrobial-resistance varied from 5 to 12 in the landscape (Table 6). The frequency of vancomycin-resistant E. faecalis (VRE; both VanA and VanB phenotypes) is very high throughout the landscape; at site 1, 60 % E. faecalis isolates are VRE and 40 % are VanB phenotype, at site 2: 100 % VRE and 60 % VanB, at site 3: 100 % VanA phenotype VRE, at site 4: 71 % VRE and 28.5 % VanB phenotype, at site 5: 88 % VRE and 22 % VanB phenotype of E. faecalis were observed (Table 5).
Table 5.
Sitewise antimicrobial-resistance profile of E. faecalis isolates from River Gomti at Lucknow city
| Sampling site | E. faecalis isolates | Antimicrobial resistance |
|---|---|---|
| Site #1 | 2E | Ox,T,G,E,M |
| AE | A,T,P,G,E,Te,R,M,Va | |
| BE | A,S,Ox,T,G,P,E,R,M | |
| FE | Ox,T,G,R,M,Va | |
| GE | A,S,Ox,T,G,P,E,R,M,Va | |
| Site #2 | 4E | A,Ox,P,G,E,R,Va, |
| HE | S,Ox,T,K,G,E,R,M, Va | |
| CE | A,S,Ox,T,G,P,C,E,R,M,Va | |
| G2A | A,S,T,G,P,E,Te,R,M,Va | |
| G2B | A,S,Ox,T,G,P,E,Te,R,M,Va | |
| Site #3 | 9E | A,T,G,P,Te,R,M,Va |
| IE | A,S,Ox,T,P,C,G,E,Te,R,M,Va | |
| 16E | A,S,Ox,T,G,P,E,Te,R,Va | |
| G3A | A,S,Ox,T,G,P,E,Te,R,M,Va | |
| G3B | A,S,Ox,T,P,G,E,Te,R,M,Va | |
| G3C | A,S,Ox,T,E, P,G,Te,R,M,Va | |
| G3D | A,S,Ox,T,G,P,Te,R,M,Va | |
| Site #4 | 11E | S,G,E,R,M |
| 17E | A,S,OX,T,E,R,M | |
| 18E | A,S,Ox,T,G,P,C,E,R,M,Va | |
| 19E | A,S,Ox,T,P,C,E,Te,R,Va | |
| 20E | S,T,G,P,E,R,M,Va | |
| G4A | A,S,Ox,T,G,P,Te,R,M,Va | |
| G4B | A,S,T,G,P,Te,R,M,Va | |
| Site #5 | 15E | A,S,Ox,T,G,P,C,E,Te,R,M,Va |
| 21E | A,Ox,T,G,E,R | |
| 22E | A,S,T,G,P,C,E,Va | |
| 23E | A,S,Ox,G,P,C,E,Va | |
| 24E | A,P,E,Te,R,M,Va | |
| G5A | A,S,Ox,T,G,P,E,Te,R,M,Va | |
| G5B | A,S,Ox,T,G,P,Te,R,M,Va | |
| G6A | A,S,Ox,T,G,P,Te,R,M,Va | |
| G6B | A,S,Ox,T,G,P,Te,R,M,Va |
Antimicrobial abbreviation: Nx Norfloxacin, A ampicillin, Ox oxacillin, P penicillinG), M methicillin, G gentamycin, S streptomycin, T tetracycline, C chloramphenicol, E erythromycin, R rifampicin, Va vancomycin, Te teicoplanin
Table 6.
Antimicrobial-resistance observed in each E. faecalis isolate in the up-to-down-stream landscape of Lucknow city
| Sampling site | Antimicrobial-resistance per isolate Median (range)a |
p valueb |
|---|---|---|
| Site 1 | 9 (5–10) | 0.0313* |
| Site 2 | 10 (7–11) | 0.0313* |
| Site 3 | 11 (8–12) | 0.0078* |
| Site 4 | 9 (5–11) | 0.0078* |
| Site 5 | 9 (6–12) | 0.0020* |
aMedian and range are reported to match more consistently with the nonparametric statistical tests performed
b p value was calculated using Wilcoxon rank sum test
* Statistically significant at alpha = 0.05
Discussion
This study was carried out to investigate the landscape dependent density and distribution of enterococci, prevalence of antimicrobial-resistance and virulence-markers among E. faecalis isolates recovered from river Gomti at Lucknow city. A multiplex PCR based genotypic characterization of vancomycin resistant pathogenic E. faecalis was carried out. The river Gomti exhibits high enterococci density at selected sites. The MPN index/100 mL values of enterococci recorded at all the sampling stations exceeded the standards set by regulatory authorities for river water reservoirs used for drinking and recreational purposes (US EPA 2003). The quantitative enumeration of enterococci by membrane filtration method has also shown a gradual increase in the concentration of enterococci towards the downstream sites in the landscape. These sites are highly polluted because of the recurring fecal pollution from nearby residential areas and cremation practices at local crematorium. Population pressure alongwith poor sanitation facilities might be responsible for the increased bacterial contamination in river waters of developing countries (Ram et al. 2007, 2008; Lata et al. 2009b). In the present study, an increasing trend of enterococci concentration was observed in the range of 1.5–1.6 × 102 CFU/100 mL, 6.8–6.9 × 103 CFU/100 mL, 4.4–4.6 × 104 CFU/100 mL, 7.7–7.8 × 104 CFU/100 mL and 6.8–6.9 × 105 CFU/100 mL at sites 1, 2, 3, 4 and 5 respectively. The most downstream site in the landscape, Gandhi setu receives contamination from city through two major wastewater point sources, including Kukrail nala and city’s largest crematorium located just upstream. Internationally, the single-sample advisory limit of enterococci for fresh water is 61 CFU/100 mL, and the 5-day geometric mean should not exceed 33 CFU/100 mL, while Indian standards have not defined the limit for enterococci in terms of CFU/100 mL (CPCB 2002; USEPA 2003).
A panel of multiplex PCR assay was developed in the current study for detection of vancomycin-resistant pathogenic E. faecalis in environmental waters (Table 2; Fig. 2). The frequency of E. faecalis isolates exhibiting multiple-virulence-markers and vanB genotype is 27 % in the landscape. However, 100 % E. faecalis isolates recovered from river Gomti exhibit prevalence of gelE and esp virulence markers. Gelatinase, a virulence marker widely reported in enterococci, hydrolyzes a broad range of bioactive substances and has been implicated in various infection models through biofilm formation (Parsek and Singh 2003; Nallapareddy et al. 2006). A recent study has shown the prevalence of gelatinase phenotype of enterococci with biofilm formation potential in agricultural environment and implicated it as a reservoir of clinically relevant strains (Macovei et al. 2009). The most frequent multiple-virulence-markers were gelE+esp+efaA+ and gelE+esp+efaA+ace+. Further the frequency of various combinations of multiple-antimicrobial-resistance and multiple-virulence-markers has been observed in the landscape. Among all, 39 % isolates were characterized with gelE+esp+efaA+ virulence markers and vancomycin resistance phenotype whereas 45 % were having methicillin resistance, VRE phenotype and gelE+esp+efaA+ace+ multiple-virulence-markers. Additionally 21 % of gelE+esp+efaA+ace+ isolates were observed to exhibit resistance to aminoglycoside–β-lactam–vancomycin–teicoplanin (G–Lactam–Va–Te); see Fig. 3 for details. The high frequency of VRE in the landscape, its association with other widely disseminated antimicrobials and virulence markers may lead to evolution of potential pathogenic VRE (Cetinkaya et al. 2000; Arias and Murray 2012). Additionally, 29 % isolates exhibit resistance to a macrolide, erythromycin, and rifampicin in association with tetracycline. A study from Denmark has reported carriage of coupled resistance of copper and antimicrobials (macrolide and glycopeptide) in pig-borne enterococci isolates contemplating persistence of the background pool of antimicrobial-resistance (BPAR) in that geographic region (Hasman and Aarestrup 2005). A set of other studies have reported the phenomenon of sustained BPAR in poultry and local population and its environmental carriage by plasmid maintenance systems (Sletvold et al. 2007b; Garcia-Migura et al. 2008). Moreover, previous study has implicated the possible transfer of linked virulence-traits and antimicrobial-resistance viz. vancomycin resistance (Heaton et al. 1996). The persistence of VRE in the environment even in the absence of antimicrobial selection pressure has been attributed to multiple types of plasmid maintenance systems that might play a pivotal role in persistence and dissemination of dangerous antimicrobial-resistant enterococci superbugs. (Pandey and Gerdes 2005; Sletvold et al. 2007a; Garcia-Migura et al. 2008). In general, E. faecalis isolates from the river Gomti exhibited resistance to 5–12 antimicrobials. The highest levels of multiple-antimicrobial-resistant E. faecalis were isolated from site 3 and 5 followed by sites 2, 4 and 1. Prevalence of such a high background pool of multiple-antimicrobial-resistance may be due to the lack of dilution of the contamination that is added to the river via various point and non-point sources. The river Gomti exhibits sluggish flow except for the monsoon period in the landscape. Although the specific sources of contaminants were not definitively determined, it is likely that the contaminants from nearby slums, cattle herds, poultries and the hospitals contributed to this contamination. Twenty-five city drains in the Lucknow area drain into the river Gomti (CPCB 2005). Heavy silting of the river reduces its carrying capacity. However, agricultural practices, poultry and dairy farming along site 1, the most upstream site might be a contributory factor for high prevalence of multiple-antimicrobial-resistance in the E. faecalis isolates recovered from this site. Although there is no data available from India in this context, the feed usage patterns in dairy farming or livestock operations and manure application in the agricultural practices are reported to be important contributing factors elsewhere (Aarestrup et al. 2000; Thurston-Enriquez et al. 2005). The VRE, both VanA and VanB phenotypes were ubiquitously present throughout the landscape. The increased frequency (100 %) of VRE at site 2 may be due to addition of contamination from the hospital located just upstream.
Fig. 3.
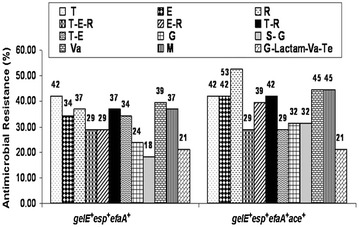
Frequency of various combinations of multiple-antimicrobial-resistance and multiple-virulence-markers among E. faecalis in the landscape. Abbreviations: T tetracycline, E erythromycin, R rifampicin, T–E–R tetracycline–macrolide–rifamycin, E–R (macrolide–rifamycin), T–R (tetracycline–rifamycin), T–E, (tetracycline–macrolide), G, gentamicin, S–G, streptomycin–gentamicin (aminoglycoside), Va, vancomycin (glycopeptide), M, methicillin (β-Lactam), G-Lactam-Va-Te (gentamicin-β-Lactam*-vancomycin-teicoplanin), *A–P–Ox–M ampicillin–penicillinG–oxacillin–methicillin (β-Lactam). All antimicrobial combinations derived from aforementioned antimicrobial abbreviations
The river Gomti identified as one of the most polluted rivers in India, is the lifeline of Lucknow city (population size ~3.5 million) as it meets about half of the total domestic water demand (about 550 mLd) of the city (Singh et al. 2004). In India, large population depends on untreated river water for carrying out daily needs (Hamner et al. 2006; Ram et al. 2007). The prevalence of vancomycin-resistant pathogenic E. faecalis isolates in river Gomti is a threat to public health as superbugs like VRE are among the most critical human health challenges of this century (Pruden et al. 2006). This study reports vancomycin-resistant pathogenic E. faecalis in river Gomti waters along an up to downstream city landscape. The emerging VRE and high background pool of multiple-antimicrobial-resistant and multi-virulent E. faecalis can contribute to the dissemination of resistance and virulence-markers in the diverse enterococci and other bacteria. Therefore, the presence of antimicrobial-resistant pathogenic E. faecalis in river waters of developing nations like India demands immediate environmental intelligence and surveillance for risk assessment and prevention strategies for protection of public health.
Conclusion
The occurrence of pathogenic VRE in river Gomti surface water is an important health concern. The observed high background pool of antimicrobial resistance and virulence in E. faecalis in river waters has the potential to disseminate more alarming antimicrobial resistance in the environment and poses serious health risk in developing countries like India as VRE gut inhabitation could lead to increased economic burden for healthcare.
Methods
Study site and Sample collection
The study was conducted along 10 km stretch of river Gomti in Lucknow city (Latitude: 25.55′N, Longitude: 80.58′E and Altitude: 123 m). River Gomti, flowing through the northern part of the country, is a major tributary of the Ganga River in India. Originating from a natural reservoir in the swampy and densely forested area near Madho-Tanda (altitude 200 m; latitude 28°34′N and E longitude 80°07′E) in the foothills of Himalayas, the river traverses a distance of about 730 km in the Indo-Gangetic alluvial region before its confluence with river Ganga. The river flowing through the Lucknow city (population: ~3.5 million), the State capital of Uttar Pradesh, meets about half of the total domestic water demand (about 550 mLd) of the town. The average flow of the river at Lucknow varies between 500 mLd in summers and 55,000 mLd during the monsoons. The river contains water throughout the year and exhibits sluggish flow except for the monsoon period. The Gomti River, identified as one of the most polluted tributary of river Ganga, receives 450 mLd of untreated domestic waste water in Lucknow city (Singh et al. 2004). In this study, five sites located in up-to-downstream on the river, namely Gau-ghat (site 1: located in upstream of river), Daliganj Bridge (site 2: bathing ghat and holy spot), Shaheed Smarak (site 3: bathing ghat and picnic spot), Lakshaman mela ground (site 4: recreational and picnic spots), Gandhi setu (site 5, most downstream location in the landscape) were selected based on various human activities on the banks of river Gomti. A cross-sectional approach was used to collect river water samples. Samples were collected in triplicate (n = 15) from five locations situated in up-to-down-stream fashion (Fig. 1). In brief, three transects were established randomly at each site and water samples (1 L) were collected 30 cm below the water surface from left, mid and right banks of the river along each transect. The samples of river water were stored in sterile glass bottles, labeled and transported on ice to the laboratory for analysis. Sample processing and analysis were conducted within 6 h after sample collection.
Isolation and enumeration of Enterococci
Quantitative enumeration of fecal streptococci or enterococci at selected sites was performed using the standard multiple tube fermentation technique (APHA 1998). In brief, tubes containing 20 mL Azide Dextrose Broth (ADB) were prepared. A series of tubes of ADB was inoculated with appropriate graduated quantities of sample (10, 1 and 0.1 mL). The inoculated tubes were incubated at 37 ± 0.5 °C and examined for turbidity after 24–48 h and calculated MPN index/100 mL surface water depending upon the number of positive tubes. Enterococci were recovered from each sample using standard membrane filtration method (APHA 1998). Briefly, tenfold dilutions of each water sample was prepared (100, 10−1, 10−2 and 10−3), and 10 mL of each dilution were filtered through 0.45-µm, 47-mm mixed cellulose ester filters (Millipore, Bedford, MA, USA) The filters were then placed on mE agar plates incubated at 41 ± 0.5 °C for 48 h for detection and enumeration of enterococci (Hi-Media, India). After 48 h, membrane filters from mE agar plates were placed on Bile Esculin Azide (BEA) agar plates and incubated at 41 ± 0.5 °C for 20 min. Colonies characteristic of enterococci, ranging from pink to dark red on mE agar and producing a brown to black precipitate on BEA agar, were considered presumptive enterococci (APHA 1998). Negative control filters and agar plates were included in each membrane filtration analysis. All colonies were counted, and concentration of enterococci per 100 mL water was determined from dilution plates containing 30–300 CFU. Presumptive enterococci recovered (n = 60) from each sample were identified by biochemical tests, including catalase test and PYR test. The growth of isolates was determined in 6.5 % NaCl, pH 9.6, and at 10°s and 45 °C, respectively. All confirmed enterococci isolates were archived in tryptic soy broth with 15 % (v/v) glycerol at -80 °C for further analyses.
Multiplex PCR assay
All enterococci isolates confirmed by biochemical tests were subjected to genotypic characterization by multiplex Polymerase Chain Reaction (PCR) technique. The multiplex PCR assay format was designed comprising four genes; either ace or efaA gene with sodA (specific for detection of E. faecalis), vanB and gelE gene. All the primers used in this assay have been reported earlier in singleplex PCR format for detection of respective genes in clinical settings (Table 2; supplementary information). Genomic DNA was extracted from overnight grown cells of enterococci isolates (n = 60) using Genelute bacterial genomic DNA kit (Sigma, USA). PCR assay was performed in reaction volume of 50 µl. In brief, The mastermix consisted of 1× hot start Taq buffer, 3.0 mM MgCl2, 0.4 mM deoxynucleotide triphosphate mix, 1.5 U of Hotstart Taq polymerase (MBI Fermentas), 300 pg of template DNA, sterile molecular grade water, forward and reverse primer (Metabion, Germany) concentration optimized for this study has been described in (Table 2; supplementary information). The PCR protocol employed an initial denaturation at 95 °C for 3 min followed by 30 cycles of denaturation at 95 °C; 30 s, annealing at 51 °C; 30 s, elongation at 72 °C; 50 s and final extension at 72 °C for 8 min. The amplicons were electrophoresed on 1.5 % agarose gel in Tris–acetate-EDTA buffer supplemented with 0.5 µg/mL of ethidium bromide. Standard DNA molecular weight markers used were 100 bp DNA ladder (MBI Fermentas, USA). The presence of gene esp encoding enterococcal surface protein in different E. faecalis isolates was also examined (Table 5.2). E. faecalis ATCC 51229, E. faecium ATCC 35667, 27270, E. durans ATCC 49470, E. hirae ATCC 9790, E. coli MTCC 723, Vibrio cholerae ATCC 51394, Salmonella typhimurium ATCC 13311 and Salmonella typhimurium ATCC 14028 were used throughout the study as reference/standard strains.
Gelatinase phenotype determination
Gelatinase production was detected by spotting mid-log phase grown pure culture of individual E. faecalis isolates (n = 33) on freshly prepared tryptic soy agar plates (Himedia, India) containing 1.5 % of skimmed milk. Plates were incubated overnight at 37 °C and then cooled to ambient temperature for 2 h. The appearance of a transparent halo around the colonies was considered to be a positive indication of gelatinase production (Gilmore et al. 2002).
Antimicrobial susceptibility testing
A panel of thirteen antimicrobials (antimicrobial abbreviation:mcg/disc) impregnated on paper discs (Himedia Ltd., India) belonging to eight different group of antimicrobials as Fluoroquinolone: Norfloxacin (Nx:10mcg), β-lactam: Ampicillin (A:10mcg), Oxacillin (Ox:1mcg), PenicillinG (P:10 units), Methicillin (M:5mcg), Aminoglycoside: Gentamycin (G:10mcg), Streptomycin (S:10mcg), Tetracycline: Tetracycline (T:30mcg), Phenicol: Chloramphenicol (C:30mcg), Macrolide: Erythromycin (E:15mcg), Rifamycin: Rifampicin (R:5mcg), Glycopeptides: Vancomycin (Va:30mcg), Teicoplanin (Te:30mcg) were used for testing the sensitivity of isolated organisms by Kirby-Bauer disc diffusion method as described by Clinical Laboratory Standards Institute (CLSI 2002). In brief, pure E. faecalis isolate culture colonies (3–4) were transferred to tubes containing 5 mL brain heart infusion broth and incubated in rotary shaker (Innova 4230, New Brunswick, USA) at 150 rpm and 37 + 1 °C for 4–6 h to yield a suspension of 106 cells/mL. The inoculum was uniformally spread on the sterile Muller-Hinton agar plates (90 mm diameter) using the sterile cotton swab. Four to six antimicrobial discs were applied aseptically, at least 24 mm apart on Mueller–Hinton agar plates. The plates were incubated immediately at 35 + 1 °C; 16–18 h for all enlisted antimicrobials and 24 h for vancomycin. The test was performed in triplicate for each E. faecalis isolate. The diameter of zones showing inhibitions were measured to the nearest mm and recorded. A zone size interpretive chart was used to determine sensitivity/resistance of antimicrobials (CLSI 2002).
Determination of Vancomycin-resistant VanA and VanB phenotype of E. faecalis
VanA phenotyping
Antimicrobials: Vancomycin (Van) and Teicoplanin (Te) were used for this study (Sigma-aldrich, USA). After Biochemical and microbiological confirmation, E.faecalis isolates from each sampling site were subjected to tube macrodilution method of antimicrobial susceptibility testing (Van: ≥ 64 µg/mlL, Te: ≥ 16 µg/mL) with slight modification. In brief, well characterized E. faecalis isolates were grown in tubes containing 5 mL Brain Heart Infusion broth and incubated in rotary shaker (INNOVA 4230, New Brunswick, USA) at 150 rpm and 37 + 1 °C for 4–6 h to yield a suspension of 106 cells/mL. The inoculum of 106 cells was added to sterile Muller-Hinton broth tubes containing Van: ≥ 64 µg/mL, Te: ≥ 16 µg/mL. The tubes were incubated immediately at 35 + 1 °C for 24 h. The tubes showing consistent growth were recorded as VanA positive. E. faecium ATCC 51559 was used as VanA positive and sterile water as negative control.
VanB phenotyping
Antimicrobial Vancomycin (Van) was used for this study (Sigma-aldrich, USA). The E. faecalis isolates from each sampling site were subjected to tube macrodilution method of antimicrobial susceptibility testing (Van: 4–1024 µg/mL) with slight modification. The tube macrodilution method CLSI (2005) for antimicrobial susceptibility testing (Van: 4–1024 µg/mL) was adopted with slight modification. In brief, well characterized E. faecalis isolates were grown in tubes containing 5 mL Brain Heart Infusion broth and incubated in rotary shaker (INNOVA 4230, New Brunswick, USA) at 150 rpm and 37 + 1 °C for 4–6 h to yield a suspension of 106 cells/mL. The inoculum of 106 cells was added to sterile Muller-Hinton broth tubes containing Van: 4–1024 µg/mL in twofold dilution series. The tubes were incubated immediately at 35 + 1 °C for 24 h. The tubes showing consistent growth were recorded as VanB positive. E. faecalis ATCC 51299 served as VanB positive and sterile water as negative control.
Statistical analyses
The concentrations of enterococci obtained using MPN analysis test and membrane filtration method from up-to-downstream river water samples was compared, Chi square test for trend was applied for the purpose. The prevalence and distribution of antimicrobial-resistance and virulence-markers among isolates from up-to-downstream landscape was assessed using wilcoxon rank-sum tests. All statistical tests were performed using GraphPad Prism version 6.0 for Windows (GraphPad Software, San Diego, CA, USA, www.graphpad.com).
Authors’ contributions
PL carried out the phenotypic antimicrobial susceptibility profiling, determination of gelatinase activity, multiplex PCR based characterization for virulence markers and vancomycin resistance in E. faecalis isolates, analysed the data, wrote the manuscript and made illustration for study site. SR contributed to sample collection and microbiological analysis. RS conceived the study, analysed the data and wrote the manuscript. All authors contributed to the general content and structure of the final manuscript.
Acknowledgements
We thank Director, IITR, Lucknow, for providing the necessary facilities for this study. This work was supported by CSIR (Council of Scientific and Industrial Research) Network Project SMM-05. P.L. and S.R. received financial support (Senior Research Fellowship) from the CSIR.
Competing interests
The authors declare they have no competing interests.
Abbreviations
- VRE
vancomycin resistant enterococci
- PCR
polymerase chain reaction
- BPAR
background pool of antimicrobial resistance
- MPN
maximum probable number
- CFU
colony forming unit
Additional file
10.1186/s40064-016-2870-5 Quantitative enumeration of enterococci collected from sites (n=5) located on river Gomti in up-to-down-stream fashion (See Table 1 for log transformed values).
Contributor Information
Pushpa Lata, Email: pushpalata0@gmail.com.
Siya Ram, Email: siyaram1@rediffmail.com.
Rishi Shanker, Phone: +91-79-26302414-18, Email: rishishanker@gmail.com, Email: rishi.shanker@ahduni.edu.in.
References
- Aarestrup F, Kruse H, Tast E, Hammerum A, Jensen L. Associations between the use of antimicrobial agents for growth promotion and the occurrence of resistance among E. faecium from broilers and pigs in Denmark, Finland, and Norway. Microb Drug Resist. 2000;6:63–70. doi: 10.1089/mdr.2000.6.63. [DOI] [PubMed] [Google Scholar]
- Ahmed W, Neller R, Katouli M. Host species-specific metabolic fingerprint database for Enterococci and Escherichia coli and its application to identify sources of fecal contamination in surface waters. Appl Environ Microbiol. 2005;71:4461–4468. doi: 10.1128/AEM.71.8.4461-4468.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APHA (1998) Standard methods for the examination of water and wastewater
- Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10(4):266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabelli VJ, Dufour AP, McCabe LJ, Levin MA. Swimming-associated gastroenteritis and water quality. Am J Epidemiol. 1982;115(4):606–616. doi: 10.1093/oxfordjournals.aje.a113342. [DOI] [PubMed] [Google Scholar]
- Cetinkaya Y, Falk P, Mayhall C. Vancomycin-resistant enterococci. Clin Microbiol Rev. 2000;13:686–707. doi: 10.1128/CMR.13.4.686-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2002) National Committee for Clinical Laboratory Standards, Wayne [PubMed]
- Coque T, Willems R, Canton R, Del Campo R, Baquero F. High occurrence of esp among ampicillin-resistant and vancomycin-susceptible Enterococcus faecium clones from hospitalized patients. J Antimicrob Chemother. 2002;50:1035–1038. doi: 10.1093/jac/dkf229. [DOI] [PubMed] [Google Scholar]
- CPCB (2002) Central Pollution Control Board, New Delhi
- CPCB . Central Pollution Control Board. India: New Delhi; 2005. [Google Scholar]
- Creti R, Imperi M, Bertuccini L, Fabretti F, Orefici G, Di Rosa R, Baldassarri L. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J Med Microbiol. 2004;53(Pt 1):13–20. doi: 10.1099/jmm.0.05353-0. [DOI] [PubMed] [Google Scholar]
- Dahl KH, Simonsen GS, Olsvik O, Sundsfjord A. Heterogeneity in the vanB gene cluster of genomically diverse clinical strains of vancomycin-resistant enterococci. Antimicrob Agents Chemother. 1999;43(5):1105–1110. doi: 10.1128/aac.43.5.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton T, Gasson M. Molecular screening of enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol. 2001;67:1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Migura L, Hasman H, Svendsen C, Jensen L. Relevance of hot spots in the evolution and transmission of Tn1546 in glycopeptide-resistant Enterococcus faecium (GREF) from broiler origin. J Antimicrob Chemother. 2008;62:681–687. doi: 10.1093/jac/dkn265. [DOI] [PubMed] [Google Scholar]
- Ghoshal U, Garg A, Tiwari D, Ayyagiri A. Emerging vancomycin resistance in enterococci in India. Indian J Pathol Microbiol. 2006;49:620–622. [PubMed] [Google Scholar]
- Gilmore M, Coburn P, Nallapareddy S, Murray B. Enterococcal virulence. In: Gilmore M, Clewell DB, Courvalin P, Dunny GM, Murray BE, Rice LB, editors. The enterococci: pathogenesis, molecular biology and antibiotic resistance. Washington, DC: American Society for Microbiology Press; 2002. p. 317. [Google Scholar]
- Gordon S, Swenson J, Hill B, Pigott N, Facklam R, Cooksey R, Thornsberry C, Jarvis W, Tenover F. Antimicrobial susceptibility patterns of common and unusual species of enterococci causing infections in the United States. J Clin Microbiol. 1992;30:2373–2378. doi: 10.1128/jcm.30.9.2373-2378.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamner S, Tripathi A, Mishra RK, Bouskill N, Broadaway SC, Pyle BH, Ford TE. The role of water use patterns and sewage pollution in incidence of water-borne/enteric diseases along the Ganges river in Varanasi, India. Int J Environ Health Res. 2006;16(2):113–132. doi: 10.1080/09603120500538226. [DOI] [PubMed] [Google Scholar]
- Hasman H, Aarestrup F. Relationship between copper, glycopeptide, and macrolide resistance among Enterococcus faecium strains isolated from pigs in Denmark between 1997 and 2003. Antimicrob Agents Chemother. 2005;49:454–456. doi: 10.1128/AAC.49.1.454-456.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton M, Discotto L, Pucci M, Handwerger S. Mobilization of vancomycin resistance by transposon-mediated fusion of a VanA plasmid with an Enterococcus faecium sex pheromone-response plasmid. Gene. 1996;171:9–17. doi: 10.1016/0378-1119(96)00022-4. [DOI] [PubMed] [Google Scholar]
- Huycke M, Sahm D, Gilmore M. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis. 1998;4:239–249. doi: 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C, Fedorka-Cray P, Barrett J. Use of a genus and species-specific multiplex PCR for identification of enterococci. J Clin Microbiol. 2004;42:3558–3565. doi: 10.1128/JCM.42.8.3558-3565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata P, Ram S, Agrawal M, Shanker R. Enterococci in river Ganga surface waters: propensity of species distribution, dissemination of antimicrobial-resistance and virulence-markers among species along landscape. BMC Microbiol. 2009;9(1):140. doi: 10.1186/1471-2180-9-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata P, Ram S, Agrawal M, Shanker R. Real time PCR for the rapid detection of vanA Gene in surface waters and aquatic macrophyte by molecular beacon probe. Environ Sci Technol. 2009;43(9):3343–3348. doi: 10.1021/es803635y. [DOI] [PubMed] [Google Scholar]
- Lowe A, Lambert P, Smith A. Cloning of an Enterococcus faecalis endocarditis antigen: homology with adhesins from some oral streptococci. Infect Immun. 1995;63:703–706. doi: 10.1128/iai.63.2.703-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macovei L, Ghosh A, Thomas V, Hancock L, Mahmood S, Zurek L. Enterococcus faecalis with the gelatinase phenotype regulated by the fsr operon and with biofilm-forming capacity are common in the agricultural environment. Environ Microbiol. 2009;11:1540–1547. doi: 10.1111/j.1462-2920.2009.01881.x. [DOI] [PubMed] [Google Scholar]
- Mathur P, Kapil A, Chandra R, Sharma P, Das B. Antimicrobial resistance in Enterococcus faecalis at a tertiary care centre of northern India. Indian J Med Res. 2003;118:25–28. [PubMed] [Google Scholar]
- Moore D, Guzman J, McGee C. Species distribution and antimicrobial resistance of enterococci isolated from surface and ocean water. J Appl Microbiol. 2008;105:1017–1025. doi: 10.1111/j.1365-2672.2008.03828.x. [DOI] [PubMed] [Google Scholar]
- Nallapareddy SR, Singh KV, Sillanpaa J, Garsin DA, Hook M, Erlandsen SL, Murray BE. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116(10):2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novais C, Coque T, Ferreira H, Sousa J, Peixe L. Environmental contamination with vancomycin-resistant enterococci from hospital sewage in Portugal. Appl Environ Microbiol. 2005;71:3364–3368. doi: 10.1128/AEM.71.6.3364-3368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey D, Gerdes K. Toxin–antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299(5615):2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- Pruden A, Pei R, Storteboom H, Carlson KH. Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ Sci Technol. 2006;40:7445–7450. doi: 10.1021/es060413l. [DOI] [PubMed] [Google Scholar]
- Ram S, Vajpayee P, Shanker R. Prevalence of multi-antimicrobial-agent resistant, shiga toxin and enterotoxin producing Escherichia coli in surface waters of river Ganga. Environ Sci Technol. 2007;41(21):7383–7388. doi: 10.1021/es0712266. [DOI] [PubMed] [Google Scholar]
- Ram S, Vajpayee P, Shanker R. Rapid culture-independent quantitative detection of enterotoxigenic Escherichia coli in surface waters by real-time PCR with molecular beacon. Environ Sci Technol. 2008;42(12):4577–4582. doi: 10.1021/es703033u. [DOI] [PubMed] [Google Scholar]
- Singh KP, Malik A, Mohan D, Sinha S. Multivariate statistical techniques for the evaluation of spatial and temporal variations in water quality of Gomti River (India)—a case study. Water Res. 2004;38(18):3980–3992. doi: 10.1016/j.watres.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Singh G, Vajpayee P, Ram S, Shanker R. Environmental reservoirs for enterotoxigenic Escherichia coli in south Asian Gangetic riverine system. Environ Sci Technol. 2010;44(16):6475–6480. doi: 10.1021/es1004208. [DOI] [PubMed] [Google Scholar]
- Sletvold H, Johnsen P, Simonsen G, Aasnaes B, Sundsfjord A, Nielsen K. Comparative DNA analysis of two vanA plasmids from Enterococcus faecium strains isolated from poultry and a poultry farmer in Norway. Antimicrob Agents Chemother. 2007;51:736–739. doi: 10.1128/AAC.00557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sletvold H, Johnsen PJ, Simonsen GS, Aasnæs B, Sundsfjord A, Nielsen KM. Comparative DNA analysis of two vanA plasmids from Enterococcus faecium strains isolated from poultry and a poultry farmer in Norway. Antimicrob Agents Chemother. 2007;51:736–739. doi: 10.1128/AAC.00557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow LR, Thomas VC, Narayanan S, Olson S, Fleming SD, Hancock LE. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect Immun. 2010;78(11):4936–4943. doi: 10.1128/IAI.01118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston-Enriquez J, Gilley J, Eghball B. Microbial quality of runoff following land application of cattle manure and swine slurry. J Water Health. 2005;3:157–171. [PubMed] [Google Scholar]
- USEPA (2003) Bacterial water quality standards for recreational waters (freshwater and marine waters). EPA-823-R-03-008
- Whitman RL, Shively DA, Pawlik H, Nevers MB, Byappanahalli MN. Occurrence of Escherichia coli and enterococci in Cladophora (chlorophyta) in nearshore water and beach sand of Lake Michigan. Appl Environ Microbiol. 2003;69:4714–4719. doi: 10.1128/AEM.69.8.4714-4719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


