Abstract
Industrial enzyme market has been projected to reach US$ 6.2 billion by 2020. Major reasons for continuous rise in the global sales of microbial enzymes are because of increase in the demand for consumer goods and biofuels. Among major industrial enzymes that find applications in baking, alcohol, detergent, and textile industries are α-amylases. These are produced by a variety of microbes, which randomly cleave α-1,4-glycosidic linkages in starch leading to the formation of limit dextrins. α-Amylases from different microbial sources vary in their properties, thus, suit specific applications. This review focuses on the native and recombinant α-amylases from bacteria and archaea, their production and the advancements in the molecular biology, protein engineering and structural studies, which aid in ameliorating their properties to suit the targeted industrial applications.
Keywords: archaea, bacteria, dextrinizing α-amylases, saccharogenic α-amylases, protein engineering, native and recombinant α-amylases
Introduction
Starch is a glucose polymer, which is synthesized by a wide array of plant species. Starch granules contain of two types of α-glucans, amylose and amylopectin, overall representing 98–99% of its total dry weight. Amylose is a linear water insoluble polymer of glucose joined by α-1, 4 glycosidic bonds (99%), while amylopectin is branched water soluble polysaccharide with short α-1, 4 linked linear chains of 10–60 glucose units and α-1, 6 linked side chains with 15–45 glucose units that form the volume of starch molecule (Buleon et al., 1998; Tester et al., 2004). The ratio of the two polysaccharides depends on the botanical origin of the starch, but the representative levels of amylose to amylopectin are 25–28 and 72–75%, respectively.
Based on the mode of action, starch hydrolyzing enzymes may be endo-acting or exo-acting. α-Amylases (EC 3.2.1.1), categorized in GH-13 family of glyosyl hydrolases, are the extracellular endo-acting enzymes that hydrolyse α-1,4 glycosidic linkages of starch randomly, while bypassing the branch points and liberating α-limit dextrins as products (Antranikian, 1992; Gupta et al., 2003; Sivaramakrishnan et al., 2006). Amylolytic enzymes have been elegantly reviewed by several workers earlier (Vihinen and Mantasala, 1989; Henrissat, 1991; Antranikian, 1992; Svensson, 1994; Davies and Henrissat, 1995; Guzman-Maldonadao and Paredes-Lopez, 1995; Nigam and Singh, 1995; Crabb and Mitchinson, 1997; Janecek, 1997; Janecek et al., 1997, 1999; Sunna et al., 1997; Bisgaad-Frantzen et al., 1999; Pandey et al., 2000; Vieille and Zeikus, 2001; van der Maarel et al., 2002; Gupta et al., 2003; Rubin-Pitel and Zhao, 2006; Sivaramakrishnan et al., 2006). Another category belongs to carbohydrases that hydrolyze cyclodextrins. They are called maltogenic amylases (EC 3.2.1.133), which are catalytically versatile and can hydrolyze α-(1,4)- as well as α-(1,6)- linkages of the substrate molecule and transglycosylate the hydrolytic products. They possess an additional 130 residues at the N-terminus that are absent in the typical α-amylases. They can also hydrolyze acarbose, a potent inhibitor of α-amylases, to produce glucose and acarviosine-glucose [pseudotrisaccharide (PTS); (Li et al., 2011)]. β-Amylase (E.C.3.2.1.2) is an exoenzyme that hydrolyzes every alternate α-1,4 linkage from the non-reducing end, causing inversion of the anomeric configuration of the liberated maltose to its β-form, and hence, they are called β-amylases. This enzyme bypasses α–1,6-linkages of branched substrates producing maltose and high molecular weight β-limit dextrins (Vihinen and Mantasala, 1989). In comparison, glucoamylase (E.C.3.2.1.3), also known as amyloglucosidase or γ–amylase, slowly acts on α–1,4 linkages of α-glucans from the non-reducing ends and also α–1,6 linkages. It preferentially degrades polysaccharides with a high molecular weight. These enzymes hydrolyze starch to yield glucose in theoretically 100% yield (Kumar and Satyanarayana, 2004). CGTase (E.C.2.4.1.19) produces a series of α, β, γ cyclodextrins from starch, amylose, and other polysaccharides. In addition, CGTases also catalyze coupling reaction by which the rings are opened and transferred to co-substrates like glucose, maltose, or sucrose. The enzyme also catalyzes disproportionation reaction in which one or more glycosyl units are transferred between linear oligosaccharides. A hydrolysis reaction is also catalyzed by the enzyme that produces dextrins. Another starch hydrolyzing enzyme α–glucosidase (E.C.3.2.1.20) acts on α–1,4-and/or α–1,6-linkages of oligosaccharides from non-reducing end, which are formed by the action of other amylolytic enzymes, liberating α–D glucose units. Most enzymes show high affinity toward maltose and so they are called as maltases (Vihinen and Mantasala, 1989; Gupta et al., 2003; Sivaramakrishnan et al., 2006).
According to international starch market index, the total utilization of starch in the world in 2008 was 66 million tons. This increased to 75 million tons by 2012, which suggests an annual growth rate of 2–3%. Fructose syrups represent 72% of starch output, while ethanol alone 40% (http://starch.dk/isi/market/index.asp). The industrial enzyme market was valued at USD 4.2 billion in 2014 and is estimated to reach USD 6.2 billion by 2020. The food and beverage segment of industrial enzymes market is estimated to reach US$ 2.0 billion by 2020 (http://www.marketsandmarkets.com/PressReleases/industrial-enzymes.asp). Among industrial enzymes, starch hydrolyzing enzymes find applications in the production of ethanol, HFCS (high fructose corn syrup), and detergent and baking industries. Alcohol and starch enzyme market is estimated to be worth $2.24 billion by 2018 (http://www.marketsandmarkets.com/PressReleases/alcohol-starch-sugar-enzyme.asp).
Diversity of α-amylases
α-Amylases are divided into two categories according to the degree of hydrolysis. Saccharifying α-amylases produce free sugars and reduce the viscosity less rapidly in comparison with the amount of reducing sugars released. While liquefying α-amylases, on the other hand, break down the starch polymer, but do not produce free sugars and cause more rapid reduction in viscosity of starch pastes. α-Amylases have bacterial, archaeal, and eukaryotic origin. The bacterial α-amylases are quite distant from the eukaryotic enzymes evolutionarily. Among all α-amylases, the bacterial enzymes are the most diverse (Pandey et al., 2000). The temperature-activity optima for bacterial α-amylases range from ~25°C (Feller et al., 1992; AHA) to around 100°C (Rao and Satyanarayana, 2007b; Geobacillus thermoleovorans α-amylase). The pH optima of bacterial α-amylases vary from 1 to 11.5 (Vihinen and Mantasala, 1989; Pandey et al., 2000). α-Amylase from Bacillus sp. and Alicyclobacillus acidocaldarius showed an acidic pH optima of 1 and 3, respectively (Uchino, 1982; Schwermann et al., 1994). In contrast, alkaline amylase with optima of pH 9–10.5 had been reported from many alkaliphilic Bacillus spp. (Saito, 1973; Krishnan and Chandra, 1983; Lee et al., 1994; Shinke et al., 1996). An extremely alkaline α-amylase with pH optima of 11.5 was reported from Bacillus sp. GM8901 (Kim et al., 1995). Archaeal α-amylases, in general, are thermostable and acidic (pH 5–6) in nature. For example the highest temperature optimum has been reported as 100 and 130°C from archaea, Pyrococcus furiosus and P. woesei, respectively (Koch et al., 1991; Laderman et al., 1993). Molecular weight of α-amylases vary from about 10 to 210 kDa, 10 kDa enzyme from Bacillus caldolyticus (Grootegoed et al., 1973) and the highest molecular weight (210 kDa) from Chloroflexus aurantiacus has been reported (Ratanakhanokchai et al., 1992). Table 1 presents temperature and pH optima and molecular weights of α-amylases from bacteria and archaea.
Table 1.
Characteristics of bacterial and archaeal α-amylases.
| Source | Molecular weight | pH optimum | Temperature optimum (°C) | Km, Vmax | References |
|---|---|---|---|---|---|
| Acyclobacillus acidocaldarius | 160 | 3.0 | 75 | – | Matzke et al., 1997 |
| Alicyclobacillus sp. A4 | 64 | 4.2 | 75 | – | Bai et al., 2012 |
| Amphibacillus sp. NM-RA2 | – | 8 | 54 | – | Mesbah and Wiegel, 2014 |
| Bacillus acidicola | 66 | 4 | 60 | – | Sharma and Satyanarayana, 2010 |
| B. amyloliquefaciens | 52 | 6.0 | 55 | 1.92 mg ml−1, 351 Uml−1 | Demirkan et al., 2005 |
| B. amyloliquifaciens | 43 | 7 | 70 | – | Kikani and Singh, 2011 |
| B. circulans | 48 | 4.9 | 48 | 11.66 mg ml−1, 68.97U | Dey et al., 2002 |
| B. halodurans | 90, 85, 70, 65, 58 | 10.5–11 | 60–65 | – | Murakami et al., 2008 |
| B. licheniformis | 31 | 6.5 | 90 | – | Bozic et al., 2011 |
| B. licheniformis | 58 | 4–9 | 90 | – | Hmidet et al., 2008 |
| B. stearothermophilus | 59 | 5.6 | 80–82 | – | Ali et al., 2001 |
| B. stearothermophilus | 56 | 5.6 | 80 | – | Khemakhem et al., 2009 |
| B. subtilis AS-S01a | 21 | 9 | 55 | 1.9 mg ml−1, 198.2 U ml−1 | Roy et al., 2012 |
| B. subtilis KIBGE | 56 | 7.5 | 50 | 2.68 mg ml−1, 1773 U ml−1 | Bano et al., 2011 |
| Bacillus sp. | – | 8 | 110 | – | Pancha et al., 2010 |
| Bacillus sp. A3-15 | 86 | 8.5 | 60 | – | Burhan, 2008 |
| Bacillus sp. ANT-6 | 94.5 | 10.4 | 80 | – | Burhan et al., 2003 |
| Bacillus sp. KR8104 | 59 | 4.0–6.0 | 75–80 | – | Sajedi et al., 2005 |
| Bacillus sp. TS-23 | 65 | 9 | 60 | – | Lo et al., 2001 |
| Bacillus sp. WPD616 | 59 | 6 | 50 | – | Liu et al., 2006 |
| Bacillus sp. YX1 | 56 | 5.0 | 40–50 | – | Liu and Xu, 2008 |
| Bacillus sp. | 53 | 4.5 | 70 | – | Asoodeh et al., 2010 |
| Bacillus subtilis JS-2004 | – | 7 | 50 | – | Asgher et al., 2007 |
| Chromohalobacter sp. | 62, 72 | 9 | 65 | – | Prakash et al., 2009 |
| Chryseobacterium taeanense | 47 | 9 | 50 | – | Wang et al., 2011 |
| Desulfurococcus mucosus | – | 5.5 | 100 | – | Canganella et al., 1994 |
| Geobacillus caldoxylosilyticus TK4 | 70 | 7 | 50 | – | Kolcuoglu et al., 2010 |
| G. thermodenitrificans | 58 | 5.5 | 80 | 3.05 mg ml−1, 7.35 Uml−1 | Ezeji and Bahl, 2006 |
| G. thermoleovorans | 26 | 7 | 100 | – | Rao and Satyanarayana, 2007a |
| Geobacillus sp. IIPTN | 97 | 6.5 | 60 | 36 mg ml−1, 222 Uml−1 | Dheeran et al., 2010 |
| Geobacillus sp. LH8 | 52 | 5–7 | 80 | – | Khajeh et al., 2009 |
| Geobacillus sp. LH8 | 52 | 5–7 | 80 | 3 mg ml−1, 6.5 μ mol min−1 | Mollania et al., 2010 |
| Halomonas meridiana | – | 7.0 | 37 | – | Coronado et al., 2000a |
| Lactobacillus manihotivorans | 135 | 5.5 | 5 | – | Aguilar et al., 2000 |
| Marinobacter sp. EMB8 | 72 | 6–11 | 80 | 4.6 mg ml−1, 1.3 mg min−1ml−1 | Kumar and Khare, 2012 |
| Nesterenkonia sp. strain F | 100 | 7.5 | 45 | – | Shafiei et al., 2010 |
| Pyrococcus furiosus | 48 | 5.6 | 115 | – | Savchenko et al., 2002 |
| P. woesei | 45 | 5.5 | 95 | – | Frillingos et al., 2000 |
| Saccharopolyspora sp. A9 | 66 | 11 | 55 | – | Chakraborty et al., 2011 |
| Staphylothermus marinus | 82.5 | 5 | 100 | – | Li et al., 2010 |
| Sulfolobus solfataricus | 120 | 3 | 80 | – | Haseltine et al., 1996 |
| Thermococcus sp. CL1 | – | 6 | 98 | – | Jeon et al., 2014 |
| T. aggregans | – | 5.5 | 100 | – | Canganella et al., 1994 |
| T. celer | – | 5.5 | 90 | – | Canganella et al., 1994 |
| T. guaymagensis | – | 6.5 | 100 | – | Canganella et al., 1994 |
| T. hydrothermalis | – | 5.0–5.5 | 75–85 | – | Legin et al., 1998 |
| T. hydrothermalis | 53.5 | 5.5 | 85 | – | Horvathova et al., 2006 |
| T. profundus | 43 | 5.5 | 80 | – | Kwak et al., 1998 |
| Thermotoga maritima | 50 | 7 | 70 | – | Lim et al., 2003 |
Increased demands for α-amylases with desired properties for industrial applications also encouraged exploration from metagenomes from different habitats. The screening of clones from a soil metagenomic library constructed in pUC 19 vector led to find a putative amylase gene (amyM), which was over expressed and purified. This enzyme was optimally active at 42°C and pH 9.0 with transglycosylation activity and requirement for Ca2+ for its stability (Yun et al., 2004). In another attempt, a gene (pAMY) of 909 bp encoding an α-amylase was found when a soil-derived metagenomic library was screened. Phylogenetic analysis revealed that pAMY was closely related to amylases from uncultured bacteria. The enzyme with a molecular mass of 38 kDa exhibited optimum activity at 40°C and neutral pH (Sharma et al., 2010). A function-based screening of an acid mine drainage (AMD) derived metagenomic library led to the discovery two endo-acting amylases which shared no sequence similarity with any known amylase. They didn't have known amylolytic domains or could be assigned to any GH-family (Delavat et al., 2012). A thermostable α-amylase gene was isolated from a metagenomic library constructed from metagenome constructed from a pilot-plant biogas reactor. When the gene (1461 bp) was cloned in Escherichia coli, a very high titre was attained. Further characterization revealed that it has optimal activity at 80°C and highly salt tolerant [25% (w/v) NaCl]. This novel enzyme Amy13A showed 75% sequence identity to α-amylases from Petrotoga mobilis and Halothermothrix orenii (Jabbour et al., 2013). Vester et al. (2015) identified a cold-active α-amylase by a functional metagenomics approach. Sequence analysis revealed that the enzyme was similar to α-amylase from the class Clostridia. Its temperature and pH optima were 10–15°C and pH 8–9. α-Amylases have also been reported from metagenomic libraries from pygmy loris (Nycticebus pygmaeus) and cow dung (Xu et al., 2014; Sharma et al., 2015).
Production and characterization of α-amylases
Production of bacterial and archaeal α-amylases
The production of α-amylases has been studied extensively in submerged and/or solid state fermentations. In general, extracellular α-amylase production is growth associated (Murthy et al., 2009; Asoodeh et al., 2010; Abou Dobara et al., 2011). Among the physical parameters, the temperature and pH of the medium play an important role in α-amylase production. The influence of temperature on α-amylase production is related to the growth of the organism. Amylase production in bacteria has been studied in a wide range of temperatures. α-Amylase production has been reported from thermophilic and hyperthermophilic bacteria and archaea like Pyrococcus, Thermococcus, and Sulfolobus species (Leuschner and Antranikian, 1995; Sunna et al., 1997), G. thermoleovorans (Rao and Satyanarayana, 2007a), B. acidocaldarius (Buonocore et al., 1976), and Alicyclobacillus sp. A4 (Bai et al., 2012), from mesophiles B. amyloliquifaciens (Gangadharan et al., 2008), Halomonas meridiana (Coronado et al., 2000a) B. subtilis (Ravindar and Elangovan, 2013), Lactobacillus plantarum (Kanpiengjai et al., 2015) as well as from psychrotolerants and psychrophiles Microbacterium foliorum GA2 (Roohi and Kuddus, 2014) and Aeromonas veronii NS07 (Samie et al., 2012). Among different carbon sources used, starch, fructose, glucose, and rice flour, are known to support high enzyme production (Ezeji et al., 2005; Prakash et al., 2009). Carbon sources like glucose and maltose have been used for the production of α-amylase, but the use of starch remains ubiquitous (Mamo and Gessesse, 1999; Sajedi et al., 2005; Liu and Xu, 2008; Sharma and Satyanarayana, 2010). Industrially important enzymes have traditionally been produced in submerged fermentation, but these enzymes are also produced by solid state fermentation. Sodhi et al. (2005) and Hashemi et al. (2010) explained the use of wheat bran for the production of α-amylase. Agro-residues were used for cold-active α-amylase production from M. foliorum (Roohi and Kuddus, 2014). In general α-amylase production is inducible in nature (Aiyer, 2004; Ryan et al., 2006; Asoodeh et al., 2010; Abou Dobara et al., 2011), but in few cases α-amylase production is also constitutive (Rao and Satyanarayana, 2007a). Like most other inducible enzymes, α-amylase production is subjected to catabolite repression by maltose and glucose, starch hydrolytic products (Bhella and Altosaar, 1988; Morkeberg et al., 1995) with the exception of some Bacillus strains (Kalishwaralal et al., 2010). Nitrogen source is a major factor that affects α-amylase production. Many investigators had confirmed that organic nitrogen sources support maximum α-amylase yields (Saxena et al., 2007; Aqeel and Umar, 2010; Abou Dobara et al., 2011).
The combination of low molecular weight dextran with Tween-80 increased 27-fold higher α-amylase production (Arnesen et al., 1998). Various metal ions like Ca2+, Fe2+, Mg2+, and K+ are added to the α-amylase production medium (Sajedi et al., 2005; Liu and Xu, 2008). Phosphate is a vital requirement for microbes as it regulates the synthesis of primary and secondary metabolites. Lower and higher levels of phosphate in the medium significantly affect the growth and enzyme production (Hillier et al., 1997; Sharma and Satyanarayana, 2010).
The production of amylases by microbes is affected considerably by physical and chemical parameters of the medium (Gigras et al., 2002; Rao and Satyanarayana, 2007a; Roohi and Kuddus, 2014; Sen et al., 2014). Traditionally “one-variable-at-a-time” approach has been used (Gokhale et al., 1991; Pham et al., 1998), but it is time consuming and does not permit understanding interactions among the process parameters. The statistical Plackett and Burman design, on the other hand, allows screening of critical culture variables (Sharma and Satyanarayana, 2006; Rao and Satyanarayana, 2007a; Kumar and Satyanarayana, 2008; Roohi and Kuddus, 2014), and response surface methodology (RSM) provides information about the optimum levels of each variable, interactions among them and their effects on the product yield (Gu et al., 2005; Rao and Satyanarayana, 2007a). The statistical approaches have been proved to be useful in optimizing medium components and cultural variables for maximizing amylase titres from many organisms like B. acidicola (Sharma and Satyanarayana, 2011), Bacillus sp. KR 8104 (Hashemi et al., 2010), and G. thermoleovorans (Rao and Satyanarayana, 2007a), Alcaligenes faecalis (Sen et al., 2014), and M. foliorum (Roohi and Kuddus, 2014).
Characterization of bacterial and archaeal α-amylases
Once produced and purified, an enzyme is biochemically characterized. α-Amylases show a wide range of substrate degradation. They degrade amylose, amylopectin, cyclodextrins, glycogen, and dextrins, but they possess highest specificity toward starch (Antranikian, 1992). Various metal ions influence activity of the enzymes (Vihinen and Mantasala, 1989). α-Amylase is a metal activated enzyme and has high affinity for Ca2+. In, general, Ca2+ enhances the activity and thermal stability of most of the α-amylases (Khajeh et al., 2001). The number of bound Ca2+ varies from 1 to 10. Dialysis against EDTA can remove the bound Ca2+. The Ca2+-free amylase can be reactivated by adding Ca2+. Although most of the α-amylases are Ca2+-dependent, there are reports of Ca2+-independent α-amylases too (Sajedi et al., 2005; Rao and Satyanarayana, 2007b; Hmidet et al., 2008; Asoodeh et al., 2010; Sharma and Satyanarayana, 2010). Besides Ca2+-independent α-amylases, there are also a few α-amylases, which are inhibited by Ca2+ ions (Babu and Satyanarayana, 1993; Tanaka and Hoshino, 2003; Mehta and Satyanarayana, 2013a). The metal ions, which inhibit α-amylase activity, include Hg2+ ions (Mamo and Gessesse, 1999; Asoodeh et al., 2010). Inhibition of α-amylase by Hg2+ ions indicates the presence of carboxyl groups in enzyme molecule (Dey et al., 2002). Furthermore, Hg2+ is also known to oxidize indole ring and to interact with aromatic ring present in tryptophan (Zhang et al., 2007; Liu et al., 2010). Various inhibitors such as dithiothreitol, β-mercaptoethanol, N-bromosuccinimide (NBS), p-hydroxy mercuribenzoic acid, iodoacetate, PMSF (phenylmethylsuphonyl fluoride), Woodward's reagent K, EDTA, and EGTA have been shown to inhibit α-amylases (Hamilton et al., 1999). Inhibition of enzyme activity by NBS demonstrates the role of tryptophan in maintaining the conformational stability of the enzyme (Rao and Satyanarayana, 2007b). Dithiothreitol and β-mercaptoethanol are the reducing agents, the effects of which suggest the role of—SH groups in the catalytic activity of enzyme. DTT may stimulate or inhibit the α-amylase (Ballschmiter et al., 2006; Rao and Satyanarayana, 2007b). In maltogenic α-amylase of Bacillus sp. WPD616, DTT had no effect on the α-amylase activity indicating that—SH groups have no role to play in the catalytic activity or these enzymes have no free and accessible—SH groups (Liu et al., 2006). The inhibition of α-amylase by PMSF indicates the role of seryl hydroxyl group in enzyme activity. Woodward's reagent K (WRK) has a role in the chemical modification of aspartic and glutamic acid residues (Paoli et al., 1997). The inactivation of amylase by WRK also indicates the involvement of acidic amino acids in the active site of the enzyme (Chauthaiwale and Rao, 1994; Komissarov et al., 1995; Sharma and Satyanarayana, 2010).
Gene cloning and over expression of recombinant α-amylases
Several attempts have been made on cloning α-amylase encoding genes from several bacteria and archaea in heterologous hosts such as E. coli. A thermostable α-amylase gene of 1203 bp encoding a 401-amino acid protein of Thermococcus profundus, was cloned and expressed in E. coli. Recombinant α-amylase production was 155.5-fold higher than that in the wild strain (Lee et al., 1996). Another α-amylase gene (1383 bp encoding 461 amino acid residues) from a hyperthermophilic archaeon, Pyrococcus sp. KODl, was cloned and expressed in E. coli. The molecular mass of this mature enzyme is 49.456 kDa with 435 amino acid residues which displays < 40% homology to other amylases. The optimum temperature and pH for the enzyme activity are 90°C and pH 6.5. Ca2+ (2.0 mM) enhanced the thermostability of this enzyme (Tachibana et al., 1996). The recombinant α-amylase from P. furiosus is Ca2+-independent. This α-amylase is a liquefying enzyme with a specific activity of 3900 U mg−1 at 98°C. It was optimally active at pH 5.5–6.0 and 100°C with a half-life of 13 h at 98°C (Dong et al., 1997). Another α-amylase encoding gene from the extremely thermophilic archaeon, T. hydrothermalis was cloned and expressed in E. coli. The recombinant α-amylase possesses molecular characteristics similar to Pyrococcus species encoding a protein of 457 amino acids with a 22 amino acid putative signal peptide and a 435 amino acid mature protein (molecular mass 49.236 kDa). This recombinant α-amylase is optimally active at 75–85°C and pH 5.0–5.5 (Lévêque et al., 2000; Horvathova et al., 2006). Hyperthermophilic archaeon, Pyrococcus woesei also produces hyperthermophilic α-amylase, the gene encoding which has been cloned and expressed in the moderate halophile Halomonas elongata. The 14 kb protein-coding sequence, and biochemical properties of the expressed protein are identical to the α-amylase gene of the closely related P. furiosus (Frillingos et al., 2000). A maltogenic α-amylase encoding gene with 588 amino acids from thermophilic Bacillus sp. WPD 616 was cloned and expressed in E. coli, which showed optimum activity at pH 6.0 and 50°C (Liu et al., 2006). Sajedi et al. (2005) reported cloning and expression of α-amylase gene (1328 bp) from Bacillus sp. KR-8104 that encodes 440 amino acids without 20 amino acids of N and C termini in E. coli. A gene encoding a thermostable α-amylase with the temperature optima of 75°C from G. thermoleovorans YN cloned in Bluescript® II KS(+) vector in E. coli has been sequenced. The corresponding amino acid sequence showed 99% sequence similarity with the known α-amylases from different Bacilli and Geobacilli (Berekaa et al., 2007). Another α-amylase gene, Amy N, from B. licheniformis NH1 was also cloned and expressed in E. coli using pDEST17 expression system. This recombinant α-amylase exhibited higher thermostability at 85°C (T1∕2 60 min) than the native amylase (8 min; Hmidet et al., 2008). In another study, the gene encoding α-amylase from B. subtilis PY22 was amplified by PCR and cloned into Pichia pastoris KM71H using the vector pPICZα, which allows methanol induced expression and secretion of the protein. The recombinant expression resulted in high levels of extracellular amylase production, as high as 22 mg/L in the shake flask culture supernatant (Karakas et al., 2010). A gene corresponding to thermo- and pH-stable maltogenic α-amylase of G. caldoxylosilyticus TK4 has been cloned into pET28a (+) vector and expressed in E. coli with 6xHis-tag at the N-terminus (Kolcuoglu et al., 2010). A gene encoding a hyperthermostable maltogenic α-amylase of Staphylothermus marinus (SMMA) was cloned and over expressed in E. coli. SMMA consisted of 696 amino acids with a predicted molecular weight of 82.5 kDa. The enzyme was active in acidic conditions (pH 3.5–5.0) with an optimal pH of 5.0, and was extremely thermostable with a temperature optimum of 100°C and a melting temperature of 109°C; both these favored starch conversion process (Li et al., 2010). The α-amylase encoding gene of an acidophile B. acidicola with N and C terminal truncation has been cloned in pET28a(+) and expressed in E. coli (Sharma and Satyanarayana, 2012). The 62 kDa recombinant α-amylase was optimally active at pH 4.0 and 60°C. Another raw starch digesting α-amylase gene (amyBS-I) with its signal peptide, from B. subtilis strain AS01a was cloned and expressed in E. coli. This recombinant enzyme was secreted extracellularly. The production was increased seven-fold by response surface optimization of culture conditions. This enzyme shows optimum activity at 70°C and pH 6.0. It is Ca2+ independent and was supplemented in bread dough for the amelioration of the bread quality as compared to the bread supplemented with commercial α-amylase (Roy et al., 2013). A gene encoding acidic, thermostable, and raw starch hydrolysing α-amylase was cloned from an extreme thermophile G. thermoleovorans and expressed (Mehta and Satyanarayana, 2013a). The ORF of 1650 bp encodes a 515 amino acid protein (Gt-amy) with a signal peptide of 34 amino acids at the N-terminus. The specific enzyme activity of recombinant Gt-amy is 1723 U mg−1 protein with a molecular weight of 59 kDa. It is optimally active at pH 5.0 and 80°C. Two other amylases with distinct properties from G. thermoleovorans have been cloned and expressed. One dimeric cyclodextrin-degrading maltogenic amylase with optimum temperature 80°C and pH activity between 5 and 9 (Mehta and Satyanarayana, 2013b), and the second raw starch hydrolysing thermostable α-amylase of 56 kDa with optimum activity at 60°C and pH 7.0 (Mehta and Satyanarayana, 2014). A gene encoding an α-amylase (Amy-E) from Exiguobacterium sp. SH3 was expressed in E. coli as a functional His-tagged protein of about 53 kDa with maximum activity at 30°C and pH 6.5. This enzyme retains 41% of its maximum activity at 0°C. This enzyme was biochemically characterized. It was halotolerant, and its activity was stimulated at high salt concentrations in the range of 1–5 M (Emampour et al., 2015). Another α-amylase gene from B. amyloliquifaciens was cloned into pMD18-T shuttle vector which was reconstructed to obtain vector pP43X for its heterologous expression in B. subtilis WB800. The recombinant purified enzyme had a specific activity of 5566 U/mg. Its productivity is 1.48-fold higher than the wild strain. The α-amylase gene encodes a protein of 514 amino acid residues with a predicted molecular weight of 58.4 kDa. The optimal conditions for its activity are pH 6.0 and 60°C. It was also biochemically characterized (Chen et al., 2015). The details of cloning and expression of α-amylase encoding genes are listed in Table 2.
Table 2.
Cloning and expression of α-amylase genes in heterologous hosts.
| Host | Gene length (bp) | Topt (°C) of recombinant α-amylase | pH optimum of recombinant α-amylase | References | |
|---|---|---|---|---|---|
| Bacillus amyloliquifaciens | B. subtilis WB800 | 1542 | 60 | 6.0 | Chen et al., 2015 |
| B. licheniformis NH1 | E. coli | 1539 | 90 | 5.0–10.0 | Hmidet et al., 2008 |
| B. subtilis WB800 | E. coli | 1545 | 60 | 6.0 | Chen et al., 2015 |
| B. acidicola | E. coli | 1479 | 60 | 4.0 | Sharma and Satyanarayana, 2012 |
| Bacillus sp. KR-8104 | E. coli | 1328 | 75–80 | 4.0–6.0 | Sajedi et al., 2005 |
| Bacillus sp. WPD 616 | E. coli | 1764 | 50 | 6.0 | Liu et al., 2006 |
| B. subtilis DR8806 | E. coli | 1545 | 70 | 5.0 | Emtenani et al., 2015 |
| B. subtilis PY22 | Pichia pastoris | 1960 | 60 | 7.0 | Karakas et al., 2010 |
| B. subtilis strain AS01a | E. coli | 1977 | 70 | 6.0 | Roy et al., 2013 |
| Exiguobacterium sp. SH3 | E. coli | 1443 | 30 | 6.5 | Emampour et al., 2015 |
| Geobacillus thermoleovorans | E. coli | 1650 | 80 | 5.0 | Mehta and Satyanarayana, 2013a |
| G. thermoleovorans | E. coli | 1767 | 80 | 5.0–9.0 | Mehta and Satyanarayana, 2013b |
| G. thermoleovorans | E. coli | 1542 | 60 | 7.0 | Mehta and Satyanarayana, 2014 |
| G. thermoleovorans YN | E. coli | 1649 | 75 | – | Berekaa et al., 2007 |
| G. caldoxylosilyticus TK4 | E. coli | 1740 | 50 | 7.0 | Kolcuoglu et al., 2010 |
| Halomonas meridiana | E. coli | 1371 | – | – | Coronado et al., 2000b |
| Pseudoalteromonas sp. MY-1 | E. coli | 2007 | 40 | 7.0 | Tao et al., 2008 |
| Pyrococcus furiosus | E. coli | 1380 | 100 | 5.5–6.0 | Dong et al., 1997 |
| Pyrococcus sp. KODl | E. coli | 1383 | 90 | 6.5 | Tachibana et al., 1996 |
| P. woesei | Halomonas elongata | 1400 | 90–100 | 5.5–6.0 | Frillingos et al., 2000 |
| Staphylothermus marinus | E. coli | 2088 | 100 | 5.0 | Li et al., 2010 |
| Streptomyces lividans TK24 | E. coli | 1719 | – | – | Yin et al., 1997 |
| Thermobifida fusca | Pichia pastoris | 1815 | 60 | 7.0 | Yang et al., 2010 |
| Thermococcus hydrothermalis | E. coli | 1371 | 75–85 | 5.0–5.5 | Lévêque et al., 2000; Horvathova et al., 2006 |
| T. profundus | E. coli | 1203 | 80 | 4.0–5.0 | Lee et al., 1996 |
| Thermoplasma volcanicum GSS1 | E. coli | 1872 | 80 | – | Kim et al., 2007 |
| Thermotoga maritima MSB8 | E. coli | 1269 | 70 | 7.0 | Lim et al., 2003 |
Protein engineering of α-amylases and molecular methods for their amelioration
Since several decades, a wide array of techniques have been developed allowing engineering of the enzyme properties. Directed evolution and site-directed mutagenesis are powerful tools for engineering enzymes, to improve their functions and to alter their properties like activity, selectivity, substrate specificity, stability, and solubility (Rubin-Pitel and Zhao, 2006). Thermostability, activity in acidic range and Ca2+-independence of α-amylases are desirable for their use in the starch saccharification process, and activity in alkaline range and oxidative stability is a prerequisite for its applicability in detergent industry. These and several other properties of α-amylases have been improved by various methods such as site-directed mutagenesis or directed evolution approach, which are described below.
Protein engineering of bacterial and archaeal α-amylases to increase thermostability
Many factors contribute to the stability of thermostable proteins including the presence of hydrogen bonds, electrostatic interactions, salt bridges, hydrophobic interactions, disulfide bonds, reduced entropy of unfolding, oligomerization, increased occurrence of proline residues, and others (Salminen et al., 1996; Russell et al., 1997; Vogt et al., 1997; Mehta and Satyanarayana, 2013b, 2015). Any of these properties of an enzyme can be modified by using methods such as site-directed mutagenesis, directed evolution, deletion mutation, and others for increasing its thermostability. Some examples of increasing thermostability of α-amylases are explained as follows. The first determinants of thermostability in bacterial α-amylases were identified by comparison of amino acid sequences of α-amylases of B. licheniformis (BLA) and B. amyloliquifaciens (BAA). Three stabilizing mutations in BAA were proposed, which caused a significant and additive thermostabilization of BAA; deletion of R176 and G177, and the substitution of K269A (Suzuki et al., 1989). This R176-G177 deletion showed similar effects on the thermostability of other α-amylases as well (Igarashi et al., 1998; Bisgaad-Frantzen et al., 1999). Conrad et al. (1995) separately determined four regions of particular importance for the thermostability of BLA, namely 34–76, 112–142, 174–179, and 263–276. Two of these coincide with the mutations identified earlier by Suzuki et al. (1989). BLA has been further mutated to withstand high temperature and acidic pH. Two positions 133 and 209 have been identified as important for its thermostability (Declerck et al., 1990; Joyet et al., 1992). Twelve different amino acids substitutions were made at H133 by a tRNA suppressor method, and H133Y substitution improved the stability the most (Declerck et al., 1990). Random mutagenesis and screening approach was used to create A209V substitution. The stabilizing effect of this mutation was an additive with the H133Y mutation, and the half-life of the double mutant was 10-fold higher at 90°C (Joyet et al., 1992). All naturally occurring amino acids were introduced in each of these two positions (Declerck et al., 1995). Of these, H133I substitution had stronger effect on thermostabilization than the previous substitution. Random mutagenesis and screening have also been used to identify M15T and N188S as stabilizing mutations of BLA (Shaw et al., 1999). A 23-fold enhancement in stability was achieved at pH 5.0 and 83°C in the presence of 5 mM CaCl2 by combining these mutations with H133Y/A209V. Shaw et al. (1999) also predicted a stabilizing effect for additional substitutions: A33S, A52S, N96Q, S148N, and A379S. Four rounds of DNA shuffling and subsequent recombination produced a variant from Thermus sp. IM6051 maltogenic amylase with a 15°C increase in optimum temperature as compared to wild-type (Kim et al., 2003). The half-life of this variant was 172 min at 80°C as compared to the wild type, which was completely inactivated at this temperature. After three rounds of DNA shuffling of B. thermalkalophilus ET2 maltogenic amylase, followed by recombination of selected mutations, variants with optimal reaction temperatures 10°C higher than the native enzyme was achieved. One of the variants carrying mutations displayed a 20-fold longer half-life than wild-type at 78°C (Tang et al., 2006). To improve the performance of a maltogenic amylase (Novamyl) as an antistaling agent for breads made at low pH, two epPCR libraries of Novamyl were constructed. These libraries were screened for improved thermal stability at 80°C and activity at pH 4.3 for 25 min. A triple mutant was better than the wild type for antistaling activity in bread made at pH 4.3 and exhibited a 10°C increase in melting temperature at pH 4.0 as compared to the wild type (Jones et al., 2008). The effect of deletion was investigated on the half-life of Bacillus sp. TS-23 α-amylase. An increased half-life at 70°C was observed for the mutant enzyme suggesting that Arg210-Ser211 deletion leads to a conformational change of the enzyme (Lin et al., 2008). Site-directed mutagenesis at Asn-75, Ser-76, and His-77 calcium binding sites of α-amylase from B. megaterium WHO resulted in increased thermostability. In the presence of calcium, conversion of His-77 to Glu resulted in a four-fold enhancement in enzyme half-life and a 9°C upward shift in T50, which was observed in comparison with the wild type. The H77E mutant was most stable with increased affinity for calcium ion and 5°C higher optimum temperature than the wild type (Ghollasi et al., 2013). The α-amylase (Ba-amy) of Bacillus acidicola was fused with gene fragments encoding partial N- and C-terminal region of thermostable α-amylase gene of G. thermoleovorans. The chimeric enzyme (Ba-Gt-amy) expressed in E. coli showed higher catalytic efficiency and thermostability than Ba-amy. The melting temperature (Tm) of Ba-Gt-amy (73.8°C) was also higher than Ba-amy (62°C). Circular dichroism (CD) spectra also revealed the stability of the chimeric enzyme (Parashar and Satyanarayana, 2016).
Protein engineering of bacterial and archaeal α-amylases for alteration of pH optima and stability
Site directed mutagenesis has also been employed for altering the optimum pH and increasing acid/basic stability of an enzyme. This method reduces the time and cost spent on screening libraries for improved variants. Saturated mutagenesis of the starch-binding domain of the α-amylase from B. licheniformis followed by selection for starch binding at low pH yielded a double mutant with an improved starch hydrolysis ratio at pH 4.5 as compared to the native protein (Verhaert et al., 2002). Site directed mutagenesis of soybean α-amylase led to the removal of hydrogen bond networks around the catalytic base residue (E380) of the enzyme (Hirata et al., 2004), raising the optimum from pH 5.4 to a more neutral pH range between 6.0 and 6.6. Richardson et al. (2002) used activity and sequence based screening to single and multi-organism DNA libraries from various environments to identify three different α-amylases with one or more aspects of the necessary phenotype: temperature stability, pH optimum, and lowered requirement of Ca2+. The genes encoding amylases with these properties were used as parental sequences for DNA shuffling in order to combine the best aspects of the three enzyme phenotypes. The two best chimeric α-amylases found by high throughput screening had 40-fold longer half-life for activity in the absence of Ca2+ ions at 90°C and pH 4.5 as compared to the most stable wild-type parent. The α-amylase from B. amyloliquefaciens with optimal activity at pH 6.0 was engineered by error prone PCR (epPCR) to produce a variant having optimal activity at alkaline pH for the detergent industry. The screening of an epPCR library for amylase activity at pH 7.0 and pH 10.0 yielded 26 variants with a very high activity. Sixteen of the selected variants were then randomly recombined by DNA shuffling and screened for increased activity at pH 10. The best mutant displayed a five-fold higher activity at pH 10 than the wild type (Bessler et al., 2003). Liu et al. (2008) observed that the mutations at two crucial positions Leu134 and Ser320 together affected the acid resistance of the α-amylase of B. licheniformis CICC 10181. Directed evolution was used to increase acid stability. In another example, protein stability and catalytic efficiency of α-amylase from B. subtilis was increased under acidic conditions by site-directed mutagenesis. Based on the three dimensional structure model analysis, four basic histidine (His) residues His222, His275, His293, and His310 in the catalytic domain were found to be important and single, double as well as triple mutants were constructed at these sites. The acidic stability of enzyme was significantly enhanced after mutation, and 45–92% of initial activity of mutants was retained after incubation at pH 4.5 and 25°C for 24 h as compared to the wild-type (39.5%). The catalytic efficiency for each active mutant was also much higher than that of wild-type at low pH. Due to increase in the hydrogen bonds and salt bridges after mutation, an obvious shift of the basic limb toward acidity was observed. These changes around the catalytic domain contributed to the significantly improved protein stability and catalytic efficiency at low pH (Yang et al., 2013).
Protein engineering of bacterial and archaeal α-amylases for increasing oxidative stability
Oxidation also has a demonstrated negative effect on the stability of α-amylases. Cysteine 362 was identified as the oxidation prone residue in BStA (Tomazic and Klibanov, 1988; Brosnan et al., 1992), and methionine 197, which is situated close to the active site, has been shown to be responsible for the inactivation of BLA (Borchert et al., 1995). The introduction of any non-sulfur-containing amino acid at position 197 was shown to greatly reduce the oxidation sensitivity of BLA. The specific activity was, however, very much dependent on the introduced side chain, with an apparent preference for the smaller side chains (Borchert et al., 1995). Bacillus sp. TS-23 α-amylase was truncated at both N and C termini, and further mutated to increase thermal and oxidative stability. Met231 was mutated to leucine and 483th codon of this enzyme was mutated to stop codon i.e., TAA by site-directed mutagenesis. The resultant engineered enzyme showed higher T1∕2 at 70°C and showed compatibility to detergents (oxidative stability; Chi et al., 2010). In another example, oxidative stability of α-amylase (AmyC) from Thermotoga maritima was improved by site-directed mutagenesis. In this, methionine residues at positions 43 and 44, 55, and 62 were mutated to alanine, which is oxidative-resistant. The M55A mutant showed 50% residual activity in the presence of H2O2, while the wild-type α-amylase was inactive (Ozturk et al., 2013).
Protein engineering of bacterial and archaeal α-amylases for the alteration of transglycosylation/hydrolysis ratio
Product re-hydrolyzation is a problem during maltooligosaccharide synthesis using maltogenic amylase. Maltogenic amylase (MAG1) from Bacillus lehensis G1 was mutated using a structure-guided protein engineering approach to decrease the hydrolysis activity of the enzyme. W359F, Y377F, and M375I mutations reduced steric interference, altered the subsite occupation and increased the internal flexibility to accommodate longer donor/acceptor molecules for transglycosylation, which resulted in an increase in the transglycosylation to hydrolysis ratio of up to 4.0-fold (Manas et al., 2015). Maltogenic amylases also use donor and acceptor sugar molecules to form α-1,4 or 1,6 glycosidic linkages between them. This process, called transglycosylation, is responsible for formation of undesirable maltotriose in the high maltose syrups. Maltogenic amylases with high hydrolytic activity than transglycosylation activity are used to decrease the maltotriose content of high maltose syrups. Maltogenic amylase from B. stearothermophilus was engineered by site-directed mutagenesis of W177 to decrease its transglycosylation activity. The mutant W177S exhibited notable increased maltose production with a minimum amount of maltotriose under industrial conditions (Sun et al., 2016). Role of His and Glu in the catalytic activity of B. licheniformis α-amylase (BLA) was determined by performing site-directed mutagenesis at His235. It was observed that the mutant enzyme (H235E) displayed high substrate transglycosylation activity, while the wild-type BLA exhibited high hydrolysis activity exclusively. This shows that Glu235 located on a wide open groove near subsite +1 is most probably involved in transglycosylation and may recognize and stabilize the non-reducing end glucose of the acceptor molecule (Tran et al., 2014)
Sequence and structural diversity of α-amylases
Conserved regions of GH-13 family
According to the Carbohydrate-Active enZYmes (CAZY) database, the α-amylases are grouped with different kinds of glycosyl hydrolases in GH-13 family, also known as α-amylase family (Henrissat, 1991). This family contains the enzymes with the following characteristics. They act on α-glycosidic bonds and hydrolyze it to produce α-anomeric mono-or oligosaccharides (hydrolysis) or form α-1,4 or 1,6 glycosidic linkages (transglycosylation), or show both activities. They possess a (β/α)8 or TIM barrel structure that contains the catalytic site residues (Figure 1). Their primary sequences have seven highly conserved regions with catalytic amino acids and some amino acids that are essential for the stability of the topology of conserved TIM barrel (Kuriki and Imanaka, 1999). The first four conserved regions (I to IV) are found in the TIM-barrel on β-strands 3, 4, and 5 and in the loop connecting β-strand 7 to α-helix 7. Amino acid residues corresponding to region I are found in the C-terminal end of the third β-strand of the TIM barrel (β3). This region includes the three conserved amino acids Asp 100, Asn104, and His105 (BLA numbering). Asp100 is essential for the active site integrity because it forms hydrogen bonding with Arg229. Arg229 is a completely conserved residue. This is present within hydrogen bond distance of the catalytic nucleophile Asp231 and the proton donor Glu26. Asn104 coordinates the conserved calcium ion between the A and B domains and does not directly stabilize the active site (Boel et al., 1990; MacHius et al., 1995, 1998). His105 stabilizes the interaction between the C-terminal of β3 and the rest of TIM barrel by hydrogen bonding to Asn104 and to the backbone oxygen of Tyr56 (BLA numbering), which is situated in the loop connecting β2 to α2. Region II is positioned in β4 and has the catalytic nucleophile Asp231 and the invariant residue Arg229. These two residues are indispensable for catalytic activity and are found in all α-amylases (Svensson, 1994). Lys234 and His235 are also present in this region, which form part of subsite +2. These amino acids bind the reducing end of the glucose chain in the substrate-binding cleft (Svensson, 1994). Region III includes the conserved residue and catalytic proton donor Glu261, which lies in the C-terminal part of the fifth L-strand of the TIM-barrel. The residues forming region IV are situated in the loop connecting L7 to K7 and protect the active site from the solvent. This region has fully conserved residue Asp328, which is postulated to be involved in substrate binding, substrate distortion and in elevating the pKa of Glu261. There is a strong preference for His, Phe, Val and Asn are often observed at positions 327, 323, 324, and 326, respectively (Klein et al., 1992; Knegtel et al., 1995; Strokopytov et al., 1995). The number of residues between the conserved Gly and Pro of the region VI differentiates between an α-amylase (7 residues) and a CGTase (8 residues; Janecek et al., 1995). Region VII does not contain conserved residues, but it usually starts with Gly followed by proline at i + 2 position.
Figure 1.

Schematic representation of the (β/α)8 barrel (TIM barrel).
GH-13 vs. GH-57 family: a comparison with respect to archaeal α-amylases
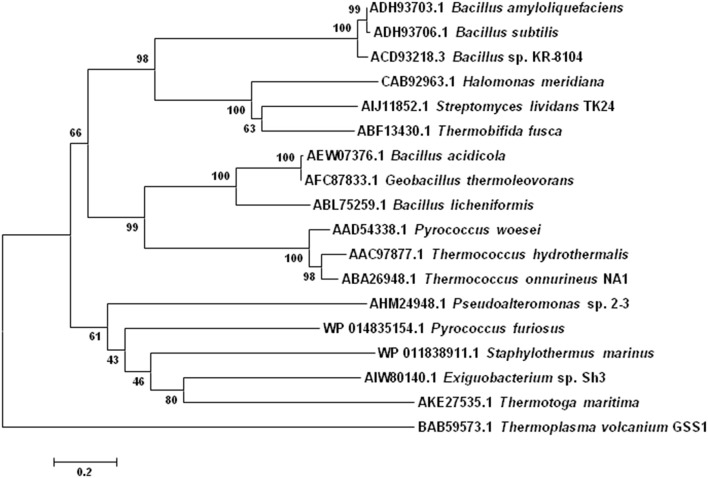
Comparison of the T. hydrothermalis α-amylase sequence with those of 21 other bacterial and archaeal α-amylases, which were representatives of more than 100 known α-amylase sequences, revealed that this enzyme as well as the other Thermococcales secreted α-amylases, contain most of the β-strands and the pentapeptide stretch located near the C-terminus of loop β3 → α3 that are present in family GH-13 (Janecek, 1997; Janecek et al., 1999). This indicates that α-amylase secreted by the members of Thermococcales belong to family GH-13. However, α-amylase from P. furiosus does not contain these conserved regions, so it belongs to GH-57 family instead. It was also showed that the α-amylase from the methanogenic archaeon Methanococcus jannaschii contains features of both glycosl hydrolase families, which indicates that these two families are either derived from a distant common ancestor or that one evolved from the other (Janecek, 1998). Also, the conserved regions of Thermococcale α-amylases show some characteristic sequence features (Janecek et al., 1999). A tryptophan is present in all the archaeal enzymes in the β4 strand. Moreover, a histidine, which has been suggested to be responsible for substrate binding, is replaced by glycine in archaeal enzymes (Svensson, 1994). It was suggested that the tryptophan found in β4 strand might be involved in catalysis instead of the missing histidine (Lee et al., 1996). Phylogenetic analysis revealed that archaeal α-amylases are closely related to plant α-amylases (Janecek et al., 1999). It also suggested that archaeal α-amylases are closer to the liquefying α-amylases, than to the saccharifying ones, which also agrees with the nature of the products of starch hydrolysis by the archaeal enzymes (Janecek et al., 1999). The phylogenetic analysis of various archaeal and bacterial α-amylases is shown in Figure 2.
Figure 2.
Phylogenetic tree showing relationships among various bacterial and archaeal α-amylases. The sequences of α-amylases from representative species of different bacterial and archaeal genera in FASTA format have been obtained from NCBI database. Phylogenetic tree was constructed using MEGA 4 software. The accession number of each bacterial and archaeal α-amylase sequence is mentioned that reveals the exact source of the sequence of representative species.
Domain organization in α-amylases
The structure of α-amylase, in general, consists of a polypeptide chain folded into three domains called A, B, and C (Figure 3). Domain A is the catalytic domain, which consists of N-terminal (β/α)8 barrel. This consists of a highly symmetrical fold which includes eight parallel β-strands organized in a barrel with a border of eight α-helices. This (β/α)8 barrel was first observed in chicken muscle triosephosphate isomerase (TIM), which (Banner et al., 1975), and hence, it is called TIM barrel (Figure 1). It is present in all the members of α-amylase family (Banner et al., 1975). The overhang between the third β-strand and the third α-helix of the TIM barrel forms Domain B. Domain B has an irregular β-sheet structure, forms a large part of the substrate binding cleft and varies considerably in structure among different α-amylases. This domain is presumed to play a role in the substrate specificity differences observed in the α-amylases (Janecek et al., 1997). Domain C constitutes the C-terminal part of the sequence and is composed of β-sandwich structure (MacGregor, 1988). Both domains B and C are located at opposite sides of the TIM barrel. Mehta and Satyanarayana (2014) have recently attributed the role of raw starch binding to the C domain of G. thermoleovorans α-amylase (Gt-amyII). Whether this is generalized for all α-amylases or only for this particular enzyme is not yet clear. In this enzyme, two tryptophan residues, W204 and W205, corresponding to W201/W202 of BaqA (B. aquimaris α-amylase) and W200/W201 of Anoxybacillus sp. α-amylase are present (Chai et al., 2012; Mok et al., 2013; Puspasari et al., 2013), which precedes β4 strand covering the catalytic nucleophile. These two tryptophan residues have been postulated to bind sugar residues of the raw starch in raw starch degrading α-amylase Baq via stacking interactions (Puspasari et al., 2013). Detailed structural analysis reveals that in GTA and Gt-amyII, W205 is not solvent accessible and is buried, in comparison to Baq, where both tryptophans are accessible to solvent (Mok et al., 2013). The domain C has also been shown to play a role in raw starch binding in barley α-amylase (Robert et al., 2003). In barley α-amylase, a “pair of sugar tongs” site in the domain C, formed by Ser378 and Tyr380, had been shown to function in starch recognition and binding (Bozonnet et al., 2007). It may be possible that a similar site is present in Gt-amyII (Mehta and Satyanarayana, 2014).
Figure 3.
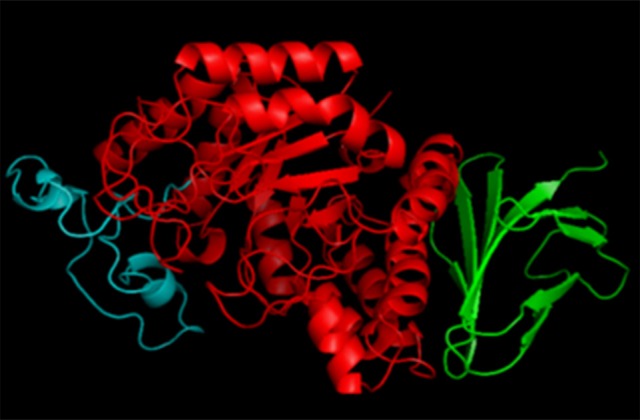
Domain organization in α-amylases. Domain A is shown in red, domain B in cyan, and domain C in green. The structure of this amylase is prepared using homology modeling. The sequence information for homology modeling is obtained from NCBI database for Gt-amyI amylase (Mehta and Satyanarayana, 2013a).
In the maltogenic amylases, the C-domain is followed by a D-domain, the function of which is also unknown (Nielsen and Borchert, 2000). A recent investigation by Tan et al. (2008) on the crystal structure of a thermostable α-amylase Amy B from H. orenii showed that it also has an additional N-terminal domain called N domain that forms a large groove, the N–C groove, in addition to the typical domain organization of GH-13 family (i.e., A, B, and C). It was shown that the N domain has been shown to increase the binding of enzyme to the raw starch. This N-domain is also found in maltogenic amylases. It has been shown to be responsible for thermostabilization via oligomerization and substrate affinity modifications in maltogenic amylases of G. thermoleovorans and Thermus sp. (Kim et al., 2001; Mehta and Satyanarayana, 2013b).
Approximately 10% of the amylolytic enzymes possess a separate domain for binding to raw starch. Besides α-amylases, the starch binding function has been reported from some glycoside hydrolases, cyclodextrin glucanotransferases, and acarviose transferases from glycoside hydrolase family GH13, β-amylases from GH14, and glucoamylases from GH15. SBDs are classified under CBMs (carbohydrate binding modules), which have been classified into 67 families (http://www.cazy.org/Carbohydrate-Binding-Modules.html) on the basis of amino acid sequence similarity. Starch-binding domain belongs to seven different families of CBMs (CBM20, CBM21, CBM 25, CBM26, CBM34, CBM41, and CBM48). α-Amylase SBDs are majorly classified in CBM20 and CBM25 families. Although large number of bacterial, archaeal as well as fungal SBD sequences are available at CAZy database; only a few α-amylase SBDs have been characterized and well described (Iefuji et al., 1996; Sumitani et al., 2000; Guillén et al., 2007; Yamaguchi et al., 2012). Florencio et al. (2000) and Morlon-Guyot et al. (2001) reported the presence of starch binding domains (SBDs) in three α-amylases from Lactobacilli. The genes encoding the α-amylases have been sequenced (Giraud and Cuny, 1997; Morlon-Guyot et al., 2001), and the amino acid sequence analysis revealed that the C-terminal possesses SBD belonging to CBM 26 family formed by direct tandem repeat units; four modules have been reported in Lactobacillus plantarum and L. manihotivorans α-amylases and five in that from L. amylovorus enzyme. The enhanced affinity in the SBD of the maltohexaose-forming amylase from B. halodurans is due to the simultaneous interaction of the two tandem CBMs present in the enzyme (one from family CBM 25 and the other from family CBM 26). Structural features of SBDs indicate that they belong to type B glycan chain-binding CBMs. The members of this class comprise several subsites that are able to accommodate individual sugar units of the polymeric ligands. These CBMs show higher affinity toward oligosaccharides and higher polymers, and show negligible interaction with oligosaccharides with a degree of polymerization (DP) of three or less. These are called as “chain binders.” According to the Boraston's classification of CBMs based on the kind of fold (Boraston et al., 2004; Hashimoto, 2006), most of the SBDs are classified in this fold family 1 of CBMs.
Ca2+ and CL− binding sites in α-amylase structure
The most intensively studied bacterial and archaeal α-amylases for which 3D structures have been resolved (Kim et al., 1999) include: Alteromonas haloplanctis (AHA), B. subtilis (BSUA), B. amyloliquefaciens (BAA), B. licheniformis (BLA), P. woesei/furiosus (PWA/PFA; Koradi et al., 1996). These α-amylases vary considerably with respect to the properties. In general, all known α-amylases contain a conserved calcium ion, which is present at the interface between domains A and B (Boel et al., 1990; MacHius et al., 1995; Linden et al., 2003). This calcium ion is indispensable for its stability and activity of α-amylases (MacHius et al., 1998). If the calcium is removed experimentally, the α-amylase loses its catalytic properties, whereas its restoration recovers the activity. Calcium ion is positioned far away from the active site to contribute directly in catalysis. Therefore, the role of calcium ion has been suggested to be structural (Hsiu et al., 1964; Larson et al., 1994). Calcium ion also plays a role in the stabilization of thermostability of α-amylases. This can be explained by the fact that salting out of hydrophobic residues by calcium ion in the protein structure leads to the formation of a compact structure (Buisson et al., 1987). Many α-amylases contain a single calcium ion, while α-amylases from Bacillus spp. contain three or four calcium ions and one sodium ion and a calcium—sodium—calcium metal triad, which bridges between domains A and B (Feller et al., 1999; Linden et al., 2003). This metal triad contributes to maintaining the conformational stability of protein as well as for resisting thermal inactivation of the enzyme (MacGregor, 1988; Khajeh et al., 2001; Goyal et al., 2005). There are conserved Ca2+ binding sites to which calcium ion bind in most of the α-amylases. A new type of Bacillus α-amylase (Amy K38), a calcium-free amylase has been reported, in which two sodium ions retain the structure and function of the enzyme, instead of calcium ions (Lee et al., 2006). Besides this, there are other α-amylases that are Ca2+-independent. These include enzymes from P. furiosus, Thermus sp., G. thermoleovorans etc (Koch et al., 1990; Malhotra et al., 2000; Nonaka et al., 2003; Prakash and Jaiswal, 2010; Mehta and Satyanarayana, 2013b, 2014). Ca2+, which stimulates most of the amylases, inhibits another α-amylase in G. thermoleovorans (Mehta and Satyanarayana, 2013a), and in B. coagulans B 49 (Babu and Satyanarayana, 1993). Besides the primary Ca2+ binding sites, calcium ions also bind to the secondary sites, which involves catalytic residues (Asp and Glu) located at the bottom of substrate binding cleft. The presence of these secondary Ca2+ binding sites at catalytic sites explains inhibition of α-amylase by Ca2+ at higher concentration (MacHius et al., 1995; Mehta and Satyanarayana, 2013a). Many α-amylases also contain a chloride ion in the active site, which enhances the catalytic efficiency of the enzyme (Levitsky and Steer, 1974; Feller et al., 1996). Although majorly found in mammalian α-amylases (Larson et al., 1994; Brayer et al., 1995; Ramasubbu et al., 1996), a chloride ion has also been reported in a α-amylase (AHA) from the psychrophilic bacterium, A. haloplanctis (Aghajari et al., 1998). Chloride binding increases the affinity of the enzyme for the calcium ion, therefore it is much likely that binding of chloride ion also induces conformational changes around the active site (Levitsky and Steer, 1974).
Active site cleft of α-amylases
In total, three steps are involved in the catalytic mechanism for retaining glycosyl hydrolases (Davies and Henrissat, 1995). Firstly, the glycosidic oxygen is protonated by the proton donor (Glu261). This is followed by a nucleophilic attack on the C1 of the sugar residue in subsite-1 by Asp231 (BLA numbering; Nielsen et al., 1999). Once the glycon part of the substrate leaves, a water molecule is activated presumably by the deprotonated Glu261. This water molecule hydrolyses the covalent bond between the nucleophilic oxygen and the Cl of the sugar residue in subsite-1, thereby completing the catalytic cycle (Nielsen et al., 1999).
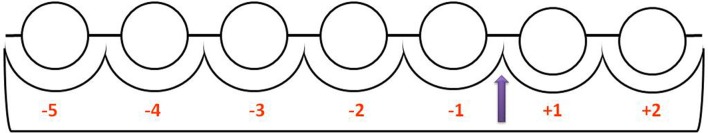
At the C-terminus of the β-strands in the TIM barrel, in the interface between domain A and domain B lies the active site cleft. The substrate binding cleft can accommodate from four to ten glucose units as revealed by the X-ray structures of α-amylases complexed with the α-amylase inhibitor acarbose. Certain of the amino acid residues of α-amylase binds to each glucose unit, which is said to constitute the binding subsite for that glucose unit. Subsite nomenclature was defined by Davies et al. (1997). The location of scissile bond defines the numbering of the subsites, with negative subsite numbers on the non-reducing side of the scissile bond, and positive subsite numbers on the reducing end (Figure 4). In the α-amylases the number of subsites present on the reducing end of the scissile bond are two or three (subsites +1, +2, and +3), while those on the non-reducing side of scissile bond varies between two and seven (MacGregor, 1988; Brzozowski et al., 2000). The X-ray structures of α-amylases complexed with acarbose shows that acarbose occupies subsites −1 to +4. Density for more than one sugar units is also observed in these X-ray structures in addition to the density for the four sugar units from acarbose (Brzozowski and Davies, 1997; Dauter et al., 1999). A study on a chimaeric Bacillus α-amylase (BA2) revealed that the extra density originates from a transglycosylation event that produces longer chained sugar molecules, which span the subsites −7 to +3, in comparison to an active site mutant of BA2, in which acarbose molecule spans the subsites −7 to −4. This observation suggests that the catalytic activity of the α-amylases is necessary for converting acarbose into a more potent longer-chained inhibitor by transglycosylation (Brzozowski et al., 2000).
Figure 4.
Active site subsite nomenclature for glycosyl hydrolases.
Applications of α-amylase
α-Amylases can be used at industrial level in sugar, textile, baking, paper, and brewing industries (Figure 5). Every application requires an α-amylase with specific characteristics (Figure 6). Different commercially available α-amylases with varying properties are used for different applications. A list of commercially available α-amylases is presented in Table 3. The foremost applicability of α-amylases in sugar industry is in the formation of high fructose corn syrups (HFCS), which are used in huge quantities in the beverage industry as sweeteners for soft drinks. Starch saccharification is required for the production of high fructose corn syrups (HFCS; Guzman-Maldonadao and Paredes-Lopez, 1995; Crabb and Mitchinson, 1997). During liquefaction, starch granules are gelatinized in a jet cooker at 105–110°C for 5 min (pH 5.8–6.5) and then partially hydrolyzed using thermostable α-amylase at 95°C for 2–3 h (Vieille and Zeikus, 2001). After liquefaction, the pH is adjusted to 4.2–5.0 and the temperature is lowered to 55–60°C for the saccharification step, which produces dextrins. The amylolytic enzymes that produce specific malto-oligosaccharides in high yields from starch have gained significant attention. Such enzymes are widely used in the food, chemical and pharmaceutical industries (Nigam and Singh, 1995). Although maltogenic α-amylases that yield 53–80% maltose have been reported, their industrial potential is limited because of their moderate thermostability, Ca2+ requirement and transglycosylation property. The use of Ca2+-independent enzymes in starch hydrolysis eliminates the addition of Ca2+ in starch liquefaction and its subsequent removal by ion exchangers from the product streams (van der Maarel et al., 2002). α-Amylases from several microbes such as G. thermoleovorans, Bacillus megaterium VUMB109, P. furiosus, and others have been reported to possess most of the desired properties, which enable them to produce high yields of maltooligosaccharides and maltose (Rao and Satyanarayana, 2007b; Jana et al., 2013; Cuong et al., 2016). Some raw starch degrading α-amylases have also been reported which can act at the native pH of starch, thereby avoiding the pH adjustment step in the starch saccharification process. Hydrolysis rates of corn and wheat starches at 60°C were 65.5 and 70.3% with 10.0% starch slurry (Mehta and Satyanarayana, 2013a). Hydrolysis rates of 60 and 37% of 1% raw wheat and corn starches in 4 h has been recorded with 0.07 U mg−1 starch of B. licheniformis ATCC 9945a α-amylase (Bozic et al., 2011). The α-amylase of Alicyclobacillus at 0.5 U mg−1 starch hydrolyzed 52.0% of 1.0% corn starch in 2 h (Bai et al., 2012). While G. thermoleovorans subsp. stromboliensis α- amylase at 500 U g−1 starch, hydrolyzed 50.0% of the corn starch (20%) in 12 h (Finore et al., 2011).
Figure 5.
Applications of α-amylases.
Figure 6.
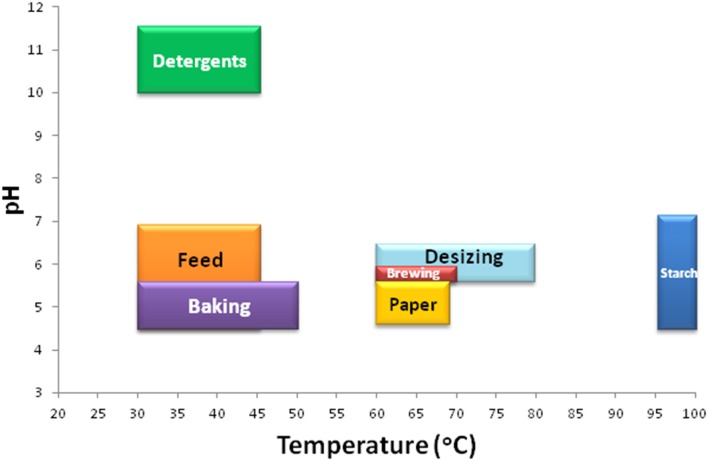
Characteristics of α-amylases for specific applications. A bar graph is plotted using the range of temperature and pH requirements of α-amylases to be used for different industrial applications. The temperature/pH range of α-amylases to be used in detergent industry, feed, baking, desizing, brewing, paper industry, and starch saccharification are 30–45°C/10.0–11.5, 30–45°C/4.5–7.0, 30–50°C/4.5–5.5, 60–80°C/5.5–6.5, 60–70°C/5.5–6.0, 60–70°C/4.5–5.5, 95–100°C/4.5–7.0, respectively.
Table 3.
Commercially available bacterial α-amylases.
| Commercial name of α-amylase | Manufacturer | Producer microorganism | Application |
|---|---|---|---|
| Amzyme TX | Parchem7 | Bacillus amyloliquifaciens | Foods and feeds |
| Aquazym 120l | Novo Nordisk, Denmark5 | – | Desizing of textiles |
| Aquazym Ultra 250l | Novo Nordisk, Denmark5 | – | Desizing of textiles |
| BANTM | Novozymes | B. amyloliquifaciens | Foods and feeds, paper industry |
| Enzymex (Cocktail), | Exotic Biosolutions Pvt. Ltd.4 | B. amyloliquifaciens | Foods and feeds |
| Fructamyl® FHT | ERBSLOEH3 | – | Starch saccharification |
| Liquozyme® SC DC | Novozymes6 | Genetically engineered from B. licheniformis | Starch saccharification |
| Natalase® | Novozymes6 | – | Detergent industry |
| Stainzyme® plus | Novozymes6 | Genetically engineered | Detergent industry |
| Thermamyl®, Takaterm | Novo Nordisk, Denmark5 | B. licheniformis | Detergent industry, paper industry |
| Validase BAA | DSM Valley Research2 | B. amyloliquifaciens | Food industry |
| VERON® XTENDER | AB enzymes1 | – | Baking industry |
Another application of α-amylases is in textile desizing. During the weaving process the warp (chain) threads are exposed to considerable mechanical strain. In order to prevent breakage, they are usually reinforced by coating (sizing) with a gelatinous substance (size) like starch. Small amounts of fats or oils may be also added to the size, with the aim of lubricating the warp coat surface. As a consequence of the sizing, the warp threads of the fabric are not able to absorb water or finishing agents to a sufficient degree. For that, size must be removed (desizing) before finishing. The complete removal of starch containing size without fiber damage is best obtained by using enzymatic desizing agents. The enzymatic desizing process has three stages. First stage is impregnation. In this stage, enzyme solution is absorbed by the fabric. This stage involves thorough wetting of fabric with enzyme solution at 70°C or higher. An amylase enzyme for desizing must be active at 70°C or higher and optimum pH 5.5–6.5, although efficient desizing have been reported at lower temperatures as well (Cavaco-Paulo and Gübitz, 2003; Chand et al., 2012, 2014). Maximal textile desizing was achieved at 45°C (Chand et al., 2014) and at 60°C (Chand et al., 2012), respectively at pH 4–5. During this stage, gelatinization of the size (starch) is to the highest possible extent. After this, the cloth is incubated under optimum conditions so that the size is broken down by the enzyme. Then, an after wash is performed in which the breakdown products from the size are removed from the fabric. This is best obtained by a subsequent detergent wash (with NaOH) at the highest possible temperature. The enzymes Aquazym 120l, Aquazym Ultra 250l, and Termamyl 60l are available commercially for desizing. The enzymes are commercially available from Novo Nordisk. The advantage of using enzymes in this process is that they are specific for starch, removing it without damage to the support fabric (http://www.novonordisk.com).
Another major application of these enzymes is in baking industry. To prevent the staling of bread and other baked goods, and to improve its texture and shelf-life, the dough is supplemented with various additives (Pritchard, 1992). The bacterial maltogenic α-amylases with intermediate thermostability are known to act as antistaling agents, thereby reducing the crumb firmness during storage (Kumar and Satyanarayana, 2008) by production of malto-oligosaccharides (DP 2–12) and allowing the yeast to act continuously during dough fermentation and early stages of baking. Several studies have proved that supplementation of α-amylases to the dough improves the crumb grain, volume, texture, flavor, and shelf-life of the bread (Van Dam and Hille, 1992; Rao and Satyanarayana, 2007b; Sharma and Satyanarayana, 2010; Amigo et al., 2016). Wild type α-amylase from G. thermoleovorans is a high maltose forming amylase which improved crumb structure, texture, and shelf life (Rao and Satyanarayana, 2007b). The bread prepared by supplementing the dough with α-amylase of B. acidicola had a higher moisture content, reducing sugars and soluble protein than the bread made by using commercial enzyme. It also had high shelf-life of 3 days at room temperature and showed amelioration in the texture and softness (Sharma and Satyanarayana, 2010). Besides these applications, α-amylases are also used for the clarification of haze formed in beer or fruit juices, in the animal feeds for improving the digestibility, in the fields of laundry and dish washing detergents (van der Maarel et al., 2002; Roy et al., 2012; Roy and Mukherjee, 2013).
α-Amylases are also being investigated currently for several other applications such as biodegradation of n-alkanes, synthesis of nanoparticles and others. α-Amylase from Bacillus subtilis TB1 was studied for its applicability in the biodegradation of alkanes. The efficiency of biodegradation was better in the presence of starch and the obtained residual hydrocarbons in the system were 53% less than the samples without starch. In silico docking of α-amylase with different molecular weight n-alkanes also supported this (Karimi and Biria, 2016). In a study, bioreductive potential of Micrococcus luteus for the synthesis of gold nanoparticles (GNPs) was investigated. Extracellular α-amylase and cell wall teichuronic acid (TUA) of M. luteus was used in the synthesis of gold nanoparticles. The synthesized GNPs were characterized by UV–VIS spectrometry, transmission electron microscopy (TEM), Fourier transform infrared spectroscopy (FTIR), and dynamic light scattering (DLS; Arunkumar et al., 2013).
Future perspectives and conclusions
There has been a marked increase in the research and development in the fields pertaining to the enzyme applications in industrial and medical sectors. α-Amylases are being used in several industries for a variety of applications. Cloning, expression, structural studies, and protein engineering of α-amylases from different bacterial and archaeal sources have been carried out for evolving enzymes with the desired characteristics for specific industrial applications. Using newer technologies and approaches, exploration of α-amylases from novel sources must be continued. It is also essential to bring down the cost of enzyme production and meeting consumer demands in all the sectors where this enzyme finds application.
Author contributions
Both the authors DM and TS have contributed to the data collection and writing this review in its final form.
Funding
Waiver request has been accepted.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Abou Dobara M. I, El-Sayed A. K., El-Fallal A. A., Omar N. F. (2011). Production and partial characterization of high molecular weight extracellular α-amylase from Thermoactinomyces vulgaris isolated from Egyptian soil. Pol. J. Microbiol. 60, 65–71. [PubMed] [Google Scholar]
- Aghajari N., Feller G., Gerday C., Haser R. (1998). Crystal structures of the psychrophilic alpha-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor. Protein Sci. 7, 564–572. 10.1002/pro.5560070304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar G. J., Morlon-Guyot B., Trejo-Aguilar C., Guyot J. P. (2000). Purification and characterization of an extracellular α-amylase produced by Lactobacillus manihotivorans LMG 18010T an amylolytic lactic acid bacterium. Enzyme Microb. Technol. 27, 406–413. 10.1016/S0141-0229(00)00230-1 [DOI] [PubMed] [Google Scholar]
- Aiyer P. (2004). Effect of C: N ratio on alpha amylase production by Bacillus licheniformis SPT 27. Afr. J. Biotechnol. 3, 519–522. [Google Scholar]
- Ali M. B., Mhiri S., Mezghani M., Bejar S. (2001). Purification and sequence analysis of the atypical maltohexaose-forming α-amylase of the B. stearothermophilus US100. Enzyme Microb. Technol. 28, 537–542. 10.1016/S0141-0229(01)00294-0 [DOI] [PubMed] [Google Scholar]
- Amigo J. M., Del Olmo Alvarez A., Engelsen M. M., Lundkvist H., Engelsen S. B. (2016). Staling of white wheat bread crumb and effect of maltogenic alpha-amylases. Part 1: spatial distribution and kinetic modeling of hardness and resilience. Food Chem. 208, 318–325. 10.1016/j.foodchem.2016.02.162 [DOI] [PubMed] [Google Scholar]
- Antranikian G. (1992). Microbial degradation of starch, in Microbial Degradation of Natural Products, ed Winkelmann A. (Germany: Weinheim VCH; ), 27–56. [Google Scholar]
- Aqeel B., Umar D. (2010). Effect of alternative carbon and nitrogen sources on production of α-amylase by Bacillus megaterium. World Appl. Sci. J. 8, 85–90. 10.1016/j.jbiotec.2008.07.1864 [DOI] [Google Scholar]
- Arnesen S., Eriksen S. H., Olsen J., Jensen B. (1998). Increased production of α-amylase from Thermomyces lanuginosus by the addition of Tween 80. Enzyme Microb. Technol. 23, 249–252. 10.1016/S0141-0229(98)00040-4 [DOI] [Google Scholar]
- Arunkumar P., Thanalakshmi M., Kumar P., Premkumar K. (2013). Micrococcus luteus mediated dual mode synthesis of gold nanoparticles: involvement of extracellular alpha-amylase and cell wall teichuronic acid. Colloids Surf. B Biointerfaces 103, 517–522. 10.1016/j.colsurfb.2012.10.051 [DOI] [PubMed] [Google Scholar]
- Asgher M., Asad M. J., Rahman S. U., Legge R. L. (2007). A thermostable α-amylase from a moderately thermophilic Bacillus subtilis strain for starch processing. J. Food Eng. 79, 950–995. 10.1016/j.jfoodeng.2005.12.053 [DOI] [Google Scholar]
- Asoodeh A., Chamanic J., Lagzian M. (2010). A novel thermostable, acidophilic α-amylase from a new thermophilic “Bacillus sp. Ferdowsicous” isolated from Ferdows hot mineral spring in Iran: Purification and biochemical characterization. Int. J. Biol. Macromol. 46, 289–297. 10.1016/j.ijbiomac.2010.01.013 [DOI] [PubMed] [Google Scholar]
- Babu K. R., Satyanarayana T. (1993). Extracellular calcium-inhibited α-amylase of Bacillus coagulans B 49. Enzyme Microb. Technol. 15, 1066–1069. 10.1016/0141-0229(93)90056-8 [DOI] [Google Scholar]
- Bai Y., Huang H., Meng K., Shi P., Yang P., Luo H., et al. (2012). Identification of an acidic α-amylase from Alicyclobacillus sp. A4 and assessment of its application in the starch industry. Food Chem. 131, 1473–1478. 10.1016/j.foodchem.2011.10.036 [DOI] [Google Scholar]
- Ballschmiter M., Fütterer O., Liebl W. (2006). Identification and characterization of a novel intracellular alkaline α-amylase from the hyperthermophilic bacterium Thermotoga maritima MSB8. Appl. Environ. Microbiol. 72, 2206–2211. 10.1128/AEM.72.3.2206-2211.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banner D. W., Bloomer A. C., Petsko G. A., Phillips D. C., Pogson C. I., Wilson I. A., et al. (1975). Structure of chicken muscle triose phosphate isomerase determined crystallographically at 2.5 angstrom resolution using amino acid sequence data. Nature 255, 609–614. [DOI] [PubMed] [Google Scholar]
- Bano S., Qader S. A. U., Aman A., Syed M. N., Azhar A. (2011). Purification and characterization of Novel α-Amylase from Bacillus subtilis. AAPS PharmSciTech 12, 2–22. 10.1208/s12249-011-9586-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berekaa M. M., Soliman N. A., Fattah A. (2007). Production, partial characterization and cloning of thermostable α-amylase of a thermophiles Geobacillus thermoleovorans. Biotechnology 6, 175–183. 10.3923/biotech.2007.175.183 [DOI] [Google Scholar]
- Bessler C., Schmitt J., Maurer K. H., Schmid R. D. (2003). Directed evolution of a bacterial alpha-amylase: toward enhanced pH-performance and higher specific activity. Protein Sci. 12, 2141–2149. 10.1110/ps.0384403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhella R. S., Altosaar I. (1988). Role of CAMP in the mediation of glucose catabolite repression of glucoamylase synthesis in Aspergillus awamori. Curr. Genet. 14, 247–252. 10.1007/BF00376745 [DOI] [PubMed] [Google Scholar]
- Bisgaad-Frantzen H., Svendsen A., Norman B., Pedersen S., Kjruli S., Outtrup H., et al. (1999). Development of industrially important α-amylases. J. Appl. Glycosci. 46, 199–206. 10.5458/jag.46.199 [DOI] [Google Scholar]
- Boel E., Brady L., Brzozowski A. M., Derewenda Z., Dodson G. G., Jensen V. J., et al. (1990). Calcium binding in alpha-amylases: an X-ray diffraction study at 2.1-A resolution of two enzymes from Aspergillus. Biochemistry 29, 6244–6249. [DOI] [PubMed] [Google Scholar]
- Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004). Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781. 10.1042/BJ20040892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert T. V., Lassen S. F., Svendsen A., Frantzen H. B. (1995). Oxidation stable amylases for detergents, in Carbohydrate Bioengineering, eds Petersen S. B., Svensson B., Pedersen S. (Amsterdam: Elsevier Science; ), 175–179. [Google Scholar]
- Bozic N., Ruiz J., Lopez-Santin J., Vujcic V. (2011). Production and properties of the highly efficient raw starch digesting α-amylase from a Bacillus licheniformis ATCC 9945a. Biochem. Eng. J. 53, 203–209. 10.1016/j.bej.2010.10.014 [DOI] [Google Scholar]
- Bozonnet S., Jensen M. T., Nielsen M. M., Aghajari N., Jensen M. H., Kramhoft B., et al. (2007). The “pair of sugar tongs” site on the non-catalytic domain C of barley alpha-amylase participates in substrate binding and activity. FEBS J. 274, 5055–5067. 10.1111/j.1742-4658.2007.06024.x [DOI] [PubMed] [Google Scholar]
- Brayer G. D., Luo Y., Withers S. G. (1995). The structure of human pancreatic alpha-amylase at 1.8 A resolution and comparisons with related enzymes. Protein Sci. 4, 1730–1742. 10.1002/pro.5560040908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnan M. P., Kelly C. T., Fogarty W. M. (1992). Investigation of the mechanisms of irreversible thermoinactivation of Bacillus stearothermophilus alpha-amylase. Eur. J. Biochem. 203, 225–231. [DOI] [PubMed] [Google Scholar]
- Brzozowski A. M., Davies G. J. (1997). Structure of the Aspergillus oryzae alpha-amylase complexed with the inhibitor acarbose at 2.0 A resolution. Biochemistry 36, 10837–10845. 10.1021/bi970539i [DOI] [PubMed] [Google Scholar]
- Brzozowski A. M., Lawson D. M., Turkenburg J. P., Bisgaard-Frantzen H., Svendsen A., Borchert T. V., et al. (2000). Structural analysis of a chimeric bacterial alpha-amylase. High-resolution analysis of native and ligand complexes. Biochemistry 39, 9099–9107. 10.1021/bi0000317 [DOI] [PubMed] [Google Scholar]
- Buisson G., Duee E., Haser R., Payan F. (1987). Three dimensional structure of porcine pancreatic alpha-amylase at 2.9 A° resolution. Role of calcium in structure and activity. EMBO J. 6, 3909–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buleon A., Colonna P., Planchot V., Ball S. (1998). Starch granules: structure and biosynthesis. Int. J. Biol. Macromol. 23, 85–112. 10.1016/S0141-8130(98)00040-3 [DOI] [PubMed] [Google Scholar]
- Buonocore V., Caporale C., De Rosa M., Gambacorta A. (1976). Stable, inducible thermoacidophilic α-amylase from Bacillus acidocaldarius. J. Bacteriol. 128, 515–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhan A. (2008). Highly thermostable, thermophilic, alkaline, SDS and chelator resistant amylase from a thermophilic Bacillus sp. isolate A3-15. Bioresour. Technol. 99, 3071–3076. 10.1016/j.biortech.2007.06.019 [DOI] [PubMed] [Google Scholar]
- Burhan A., Nisa U., Gokhan C., Omer C., Ashabil A., Osman G. (2003). Enzymatic properties of a novel thermostable, thermophilic, alkaline and chelator resistant amylase from an alkaliphilic Bacillus sp. isolate ANT-6. Process Biochem. 38, 1397–1403. 10.1016/S0032-9592(03)00037-2 [DOI] [Google Scholar]
- Canganella F., Andrade C. M., Antranikian G. (1994). Characterization of amylolytic and pullulytic enzymes from thermophilic archaea and from a new Fervidobacterium species. Appl. Microbiol. Biotechnol. 42, 239–245. 10.1007/BF00902723 [DOI] [Google Scholar]
- Cavaco-Paulo A., Gübitz G. M. (2003). Textile Processing with Enzymes. Sawston; Cambridge: Woodhead Publishing Ltd. [Google Scholar]
- Chai Y. Y., Rahman R. N., Illias R. M., Goh K. M. (2012). Cloning and characterization of two new thermostable and alkalitolerant alpha-amylases from the Anoxybacillus species that produce high levels of maltose. J. Ind. Microbiol. Biotechnol. 39, 731–741. 10.1007/s10295-011-1074-9 [DOI] [PubMed] [Google Scholar]
- Chakraborty S., Khopade A., Biao R., Jian W., Liu X. Y., et al. (2011). Characterization and stability studies on surfactant, detergent and oxidant stable α-amylase from marine haloalkaliphilic Saccharopolyspora sp. A9. J. Mol. Catal. B Enzym 68, 52–58. 10.1016/j.molcatb.2010.09.009 [DOI] [Google Scholar]
- Chand N., Nateri A. S., Sajedi R. H., Mahdavi A., Rassa M. (2012). Enzymatic desizing of cotton fabric using a Ca2+-independent α-amylase with acidic pH profile. J. Mol. Catal. B Enzym. 83, 46–50. 10.1016/j.molcatb.2012.07.003 [DOI] [Google Scholar]
- Chand N., Sajedi R. H., Nateri A. S., Khajeh K., Rassa M. (2014). Fermentative desizing of cotton fabric using an α-amylase-producing Bacillus strain: optimization of simultaneous enzyme production and desizing. Process Biochem. 49, 1884–1888. 10.1016/j.procbio.2014.07.007 [DOI] [Google Scholar]
- Chauthaiwale J., Rao M. (1994). Chemical modification of xylanase from alkalothermophilic Bacillus species: evidence for essential carboxyl group. Biochem. Biophys. Acta 1204, 164–168. 10.1016/0167-4838(94)90004-3 [DOI] [PubMed] [Google Scholar]
- Chen J., Chen X., Dai J., Xie G., Yan L., Lu L., et al. (2015). Cloning, enhanced expression and characterization of an alpha-amylase gene from a wild strain in B. subtilis WB800. Int. J. Biol. Macromol. 80, 200–207. 10.1016/j.ijbiomac.2015.06.018 [DOI] [PubMed] [Google Scholar]
- Chi M. C., Chen Y. H., Wu T. J., Lo H. F., Lin L. L. (2010). Engineering of a truncated alpha-amylase of Bacillus sp. strain TS-23 for the simultaneous improvement of thermal and oxidative stabilities. J. Biosci. Bioeng. 109, 531–538. 10.1016/j.jbiosc.2009.11.012 [DOI] [PubMed] [Google Scholar]
- Conrad B., Hoang V., Polley A., Hofemeister J. (1995). Hybrid Bacillus amyloliquefaciens and Bacillus licheniformis α-amylases. Construction, properties and sequence determinants. Eur. J. Biochem. 230, 481–490. 10.1111/j.1432-1033.1995.0481h.x [DOI] [PubMed] [Google Scholar]
- Coronado M. J., Vargas C., Hofemeister J., Ventosa A., Nieto J. J. (2000a). Production and biochemical characterization of an α-amylase from the moderate halophile Halomonas meridiana. FEMS Microbiol. Lett. 183, 67–71. 10.1016/S0378-1097(99)00628-X [DOI] [PubMed] [Google Scholar]
- Coronado M. J., Vargas C., Mellado E., Tegos G., Drainas C., Nieto J. J., et al. (2000b). The alpha-amylase gene amyH of the moderate halophile Halomonas meridiana: cloning and molecular characterization. Microbiology 146, 861–868. 10.1099/00221287-146-4-861 [DOI] [PubMed] [Google Scholar]
- Crabb W. D., Mitchinson C. (1997). Enzymes involved in the processing of starch to sugars. Trends Biotech. 15, 349–352. [Google Scholar]
- Cuong N. P., Lee W. H., Oh I. N., Thuy N. M., Kim D. G., Park J. T., et al. (2016). Continuous production of pure maltodextrin from cyclodextrin using immobilized Pyrococcus furiosus thermostable amylase. Process Biochem. 51, 282–287. 10.1016/j.procbio.2015.11.022 [DOI] [Google Scholar]
- Dauter Z., Dauter M., Brzozowski A. M., Christensen S., Borchert T. V., Beier L., et al. (1999). X-ray structure of Novamyl, the five-domain “maltogenic” alpha-amylase from Bacillus stearothermophilus: maltose and acarbose complexes at 1.7A resolution. Biochemistry 38, 8385–8392. 10.1021/bi990256l [DOI] [PubMed] [Google Scholar]
- Davies G., Henrissat B. (1995). Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859. 10.1016/S0969-2126(01)00220-9 [DOI] [PubMed] [Google Scholar]
- Davies G. J., Wilson K. S., Henrissat B. (1997). Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 321, 557–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Declerck N., Joyet P., Gaillardin C., Masson J. M. (1990). Use of amber suppressors to investigate the thermostability of Bacillus licheniformis alpha-amylase. Amino acid replacements at 6 histidine residues reveal a critical position at His-133. J. Biol. Chem. 265, 15481–15488. [PubMed] [Google Scholar]
- Declerck N., Joyet P., Trosset J. Y., Garnier J., Gaillardin C. (1995). Hyperthermostable mutants of Bacillus licheniformis alpha-amylase: multiple amino acid replacements and molecular modelling. Protein Eng. 8, 1029–1037. [DOI] [PubMed] [Google Scholar]
- Delavat F., Phalip V., Forster A., Plewniak F., Lett M. C., Lievremont D. (2012). Amylases without known homologues discovered in an acid mine drainage: significance and impact. Sci. Rep. 2:354. 10.1038/srep00354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirkan E. S., Mikami B., Adachi M., Higasa T., Shigeru U. (2005). α-Amylase from B. amyloliquefaciens: purification, characterization, raw starch degradation and expression in E. coli. Process Biochem. 40, 2629–2636. 10.1016/j.procbio.2004.08.015 [DOI] [Google Scholar]
- Dey G., Palit S., Banerjee R., Maiti B. R. (2002). Purification and characterization of maltooligosaccharide-forming amylase from Bacillus circulans GRS 313. J. Ind. Microbiol. Biotechnol. 28, 193–200. 10.1038/sj/jim/7000220 [DOI] [PubMed] [Google Scholar]
- Dheeran P., Kumar S., Jaiswal Y. K., Adhikari D. K. (2010). Characterization of hyperthermostable α-amylase from Geobacillus sp. IIPTN. Appl. Microbiol. Biotechnol. 86, 1857–1866. 10.1007/s00253-009-2430-9 [DOI] [PubMed] [Google Scholar]
- Dong G., Vieille C., Savchenko A., Zeikus J. G. (1997). Cloning, sequencing, and expression of the gene encoding extracellular α-amylase from Pyrococcus furiosus and biochemical characterization of the recombinant enzyme. Appl. Environ. Microbiol. 63, 3569–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emampour M., Noghabi K. A., Zahiri H. S. (2015). Molecular cloning and biochemical characterization of a novel cold-adapted α-amylase with multiple extremozyme characteristics. J. Mol. Catal. B Enzym. 111, 79–86. 10.1016/j.molcatb.2014.10.012 [DOI] [Google Scholar]
- Emtenani S., Asoodeh A., Emtenani S. (2015). Gene cloning and characterization of a thermostable organic-tolerant alpha-amylase from Bacillus subtilis DR8806. Int. J. Biol. Macromol. 72, 290–298. 10.1016/j.ijbiomac.2014.08.023 [DOI] [PubMed] [Google Scholar]
- Ezeji T. C., Bahl H. (2006). Purification, characterization, and synergistic action of phytate-resistant α-amylase and α-glucosidase from Geobacillus thermodenitrificans HRO10. J. Biotechnol. 125, 27–38. 10.1016/j.jbiotec.2006.02.006 [DOI] [PubMed] [Google Scholar]
- Ezeji T. C., Wolf A., Bahl H. (2005). Isolation, characterization, and identification of Geobacillus thermodenitrificans HRO10, an α-amylase and α-glucosidase producing thermophile. Can. J. Microbiol. 51, 685–693. 10.1139/w05-054 [DOI] [PubMed] [Google Scholar]
- Feller G., Bussy O., Houssier C., Gerday C. (1996). Structural and functional aspects of chloride binding to Alteromonas haloplanctis alpha-amylase. J. Biol. Chem. 271, 23836–23841. [DOI] [PubMed] [Google Scholar]
- Feller G., d'Amico D., Gerday C. (1999). Thermodynamic stability of a cold-active alpha-amylase from the Antarctic bacterium Alteromonas haloplanctis. Biochemistry 38, 4613–4619. [DOI] [PubMed] [Google Scholar]
- Feller G., Lonhienne T., Deroanne C., Libioulle C., Van Beeumen J., Gerday C. (1992). Purification, characterization, and nucleotide sequence of the thermolabile α-amylase from the antarctic psychrotroph Alteromonas haloplanctis A23. J. Biol. Chem. 267, 5217–5221. [PubMed] [Google Scholar]
- Finore I., Kasavi C., Poli A., Romano I., Oner E. T. (2011). Purification, biochemical characterization and gene sequencing of a thermostable raw starch digesting α-amylase from Geobacillus thermoleovorans subsp. stromboliensis subsp. nov. World J. Microbiol. Biotechnol. 27, 2425–2433. 10.1007/s11274-011-0715-5 [DOI] [Google Scholar]
- Florencio J. A., Eiras-Stofella D. R., Soccol C. R., Raimbault M., Guyot J. P., Fontana J. D. (2000). Lactobacillus plantarum amylase acting on crude starch granules. Native isoforms and activity changes after limited proteolysis. Appl. Biochem. Biotechnol. 84–86, 721–730. 10.1385/ABAB:84-86:1-9:721 [DOI] [PubMed] [Google Scholar]
- Frillingos S., Linden A., Niehaus F., Vargas C., Nieto J. J., Ventosa A., et al. (2000). Cloning and expression of α-amylase from the hyperthermophilic archaeon Pyrococcus woesei in the moderately halophilic bacterium Halomonas elongata. J. Appl. Microbiol. 88, 495–503. 10.1046/j.1365-2672.2000.00988.x [DOI] [PubMed] [Google Scholar]
- Gangadharan D., Sivaramakrishnan S., Nampoothiri K. M., Sukumaran R. K., Pandey A. (2008). Response surface methodology for the optimization of alpha amylase production by Bacillus amyloliquefaciens. Bioresour. Technol. 99, 4597–4602. 10.1016/j.biortech.2007.07.028 [DOI] [PubMed] [Google Scholar]
- Ghollasi M., Ghanbari-Safari M., Khajeh K. (2013). Improvement of thermal stability of a mutagenised alpha-amylase by manipulation of the calcium-binding site. Enzyme Microb. Technol. 53, 406–413. 10.1016/j.enzmictec.2013.09.001 [DOI] [PubMed] [Google Scholar]
- Gigras P., Sahai V., Gupta R. (2002). Statistical media optimization and production of its ITS α-amylase from Aspergillus oryzae in a bioreactor. Curr. Microbiol. 45, 203–208. 10.1007/s00284-001-0107-4 [DOI] [PubMed] [Google Scholar]
- Giraud E., Cuny G. (1997). Molecular characterization of the alpha-amylase genes of Lactobacillus plantarum A6 and Lactobacillus amylovorus reveals an unusual 3′ end structure with direct tandem repeats and suggests a common evolutionary origin. Gene 198, 149–157. [DOI] [PubMed] [Google Scholar]
- Gokhale D. V., Patil S. G., Bastawde K. B. (1991). Optimization of cellulase production by Aspergillus niger NCIM 1207. Appl. Biochem. Biotechnol. 30, 99–109. [DOI] [PubMed] [Google Scholar]
- Goyal N., Gupta J. K., Soni S. K. (2005). A novel raw starch digesting thermostable α-amylase from Bacillus sp. I-3 and its use in the direct hydrolysis of raw potato starch. Enzyme Microb. Technol. 37, 723–734. 10.1016/j.enzmictec.2005.04.017 [DOI] [Google Scholar]
- Grootegoed J. A., Lauwers A. M., Heinen W. (1973). Separation and partial purification of extracellular amylase and protease from Bacillus caldolyticus. Arch. Microbiol. 90, 223. [DOI] [PubMed] [Google Scholar]
- Gu X.-B., Zheng Z.-M., Yu H.-Q., Wang J., Liang F.-L., Liu R.-L. (2005). Optimization of medium constituents for a novel lipopeptide production by Bacillus subtilis MO-01 by a response surface method. Process Biochem. 40, 3196–3201. 10.1016/j.procbio.2005.02.011 [DOI] [Google Scholar]
- Guillén D., Santiago M., Linares L., Perez R., Morlon J., Ruiz B., et al. (2007). Alpha-amylase starch binding domains: cooperative effects of binding to starch granules of multiple tandemly arranged domains. Appl. Environ. Microbiol. 73, 3833–3837. 10.1128/AEM.02628-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Gigras P., Mohapatra H., Goswami V. K., Chauhan B. (2003). Microbial α-amylases: a biotechnological perspective. Process Biochem. 38, 1599–1616. 10.1016/S0032-9592(03)00053-0 [DOI] [Google Scholar]
- Guzman-Maldonadao H., Paredes-Lopez O. (1995). Amylolytic enzymes and products derived from starch: a review. Crit. Rev. Food Sci. Nutr. 35, 373–403. [DOI] [PubMed] [Google Scholar]
- Hamilton L. M., Kelly C. T., Fogarty W. M. (1999). Purification and properties of the raw starch-degrading α-amylase of Bacillus sp. IMD 434. Biotechnol. Lett. 21, 111–115. [Google Scholar]
- Haseltine C., Rolfsmeier M., Blum P. (1996). The glucose effect and regulation of α-amylase synthesis in the hyperthermophilic archaeaon Sulfolobus solfataricus. J. Bacteriol. 178, 945–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi M., Razavi S. H., Shojaosadati S. A., Mousavi S. M., Khajeh K., Safari M. (2010). Development of a solid-state fermentation process for production of an alpha amylase with potentially interesting properties. J. Biosci. Bioeng. 110, 333–337. 10.1016/j.jbiosc.2010.03.005 [DOI] [PubMed] [Google Scholar]
- Hashimoto H. (2006). Recent structural studies of carbohydrate-binding modules. Cell. Mol. Life Sci. 63, 2954–2967. 10.1007/s00018-006-6195-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B. (1991). A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier P., Wase D. A. J., Emery A. N., Solomons G. L. (1997). Instability of α-amylase production and morphological variation in continuous culture of Bacillus amyloliquefaciens is associated with plasmid loss. Process Biochem. 32, 51–59. [Google Scholar]
- Hirata A., Adachi M., Sekine A., Kang Y. N., Utsumi S., Mikami B. (2004). Structural and enzymatic analysis of soybean beta-amylase mutants with increased pH optimum. J. Biol. Chem. 279, 7287–7295. 10.1074/jbc.M309411200 [DOI] [PubMed] [Google Scholar]
- Hmidet N., Bayoudh A., Berrin J. G., Kanoun S., Juge N., Nasri M. (2008). Purification and biochemical characterization of a novel α-amylase from Bacillus licheniformis NH1 Cloning, nucleotide sequence and expression of amyN gene in Escherichia coli. Process Biochem. 43, 499–510. 10.1016/j.procbio.2008.01.017 [DOI] [Google Scholar]
- Horvathova V., Godany A., Sturdik E., Janecek S. (2006). α-Amylase from Thermococcus hydrothermalis: re-cloning aimed at the improved expression and hydrolysis of corn starch. Enzyme Microb. Technol. 39, 1300–1305. 10.1016/j.enzmictec.2006.03.016 [DOI] [Google Scholar]
- Hsiu J., Fischer E. H., Stein E. A. (1964). Alpha-amylases as calcium-metalloenzymes. II. Calcium and the catalytic activity. Biochemistry 3, 61–66. [DOI] [PubMed] [Google Scholar]
- Iefuji H., Chino M., Kato M., Iimura Y. (1996). Raw-starch-digesting and thermostable alpha-amylase from the yeast Cryptococcus sp. S-2: purification, characterization, cloning and sequencing. Biochem. J. 318, 989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K., Hatada Y., Ikawa K., Araki H., Ozawa T., Kobayashi T., et al. (1998). Improved thermostability of a Bacillus alpha-amylase by deletion of an arginine-glycine residue is caused by enhanced calcium binding. Biochem. Biophys. Res. Commun. 248, 372–377. 10.1006/bbrc.1998.8970 [DOI] [PubMed] [Google Scholar]
- Jabbour D., Sorger A., Sahm K., Antranikian G. (2013). A highly thermoactive and salt-tolerant alpha-amylase isolated from a pilot-plant biogas reactor. Appl. Microbiol. Biotechnol. 97, 2971–2978. 10.1007/s00253-012-4194-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana M., Maity C., Samanta S., Pati B. R., Islam S. S., Mohapatra P. K. D., et al. (2013). Salt-independent thermophilic α-amylase from Bacillus megaterium VUMB109: an efficacy testing for preparation of maltooligosaccharides. Ind. Crop Prod. 41, 386–391. [Google Scholar]
- Janecek S. (1997). alpha-Amylase family: molecular biology and evolution. Prog. Biophys. Mol. Biol. 67, 67–97. [DOI] [PubMed] [Google Scholar]
- Janecek S. (1998). Sequence of archaeal Methanococcus jannaschii alpha-amylase contains features of families 13 and 57 of glycosyl hydrolases: a trace of their common ancestor? Folia Microbiol. (Praha) 43, 123–128. [DOI] [PubMed] [Google Scholar]
- Janecek S., Leveque E., Belarbi A., Haye B. (1999). Close evolutionary relatedness of alpha-amylases from Archaea and plants. J. Mol. Evol. 48, 421–426. [DOI] [PubMed] [Google Scholar]
- Janecek S., MacGregor E. A., Svensson B. (1995). Characteristic differences in the primary structure allow discrimination of cyclodextrin glucanotransferases from α-amylases. Biochem. J. 305, 685–686. 10.1042/bj3050685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janecek S., Svensson B., Henrissat B. (1997). Domain evolution in the alpha-amylase family. J. Mol. Evol. 45, 322–331. [DOI] [PubMed] [Google Scholar]
- Jeon E. J., Jung J. H., Seo D. H., Jung D. H., Holden J. F., Park C. S. (2014). Bioinformatic and biochemical analysis of a novel maltose-forming α-amylase of the GH57 family in the hyperthermophilic archaeon Thermococcus sp. CL1. Enzyme Microb. Technol. 60, 9–15. 10.1016/j.enzmictec.2014.03.009 [DOI] [PubMed] [Google Scholar]
- Jones A., Lamsa M., Frandsen T. P., Spendler T., Harris P., Sloma A., et al. (2008). Directed evolution of a maltogenic alpha-amylase from Bacillus sp. TS-25. J. Biotechnol. 134, 325–333. 10.1016/j.jbiotec.2008.01.016 [DOI] [PubMed] [Google Scholar]
- Joyet P., Declerck N., Gaillardin C. (1992). Hyperthermostable variants of a highly thermostable alpha-amylase. Biotechnology 10, 1579–1583. [DOI] [PubMed] [Google Scholar]
- Kalishwaralal K., Gopalram S., Vaidyanathan R., Deepak V., Pandian S., Gurunathan S. (2010). Optimization of α-amylase production for the green synthesis of gold nanoparticles. Colloids Surf. B Biointerfaces 77, 174–180. 10.1016/j.colsurfb.2010.01.018 [DOI] [PubMed] [Google Scholar]
- Kanpiengjai A., Lumyong S., Nguyen T.-H., Haltrich D., Khanongnuch C. (2015). Characterization of a maltose-forming α-amylase from an amylolytic lactic acid bacterium Lactobacillus plantarum S21. J. Mol. Catal. B Enzym. 120, 1–8. 10.1016/j.molcatb.2015.06.010 [DOI] [Google Scholar]
- Karakas B., Inana M., Certel M. (2010). Expression and characterization of Bacillus subtilis PY22 α-amylase in Pichia pastoris. J. Mol. Catal. B Enzym. 64, 129–134. 10.1016/j.molcatb.2009.07.006 [DOI] [Google Scholar]
- Karimi M., Biria D. (2016). The synergetic effect of starch and alpha amylase on the biodegradation of n-alkanes. Chemosphere 152, 166–172. 10.1016/j.chemosphere.2016.02.120 [DOI] [PubMed] [Google Scholar]
- Khajeh K., Hosseinkhani S., Mollania N. (2009). Purification, biochemical characterization and sequence analysis of a phytate-resistant and thermostable α-amylase from Geobacillus spp. LH8. New Biotechnol. 25, S87. [DOI] [PubMed] [Google Scholar]
- Khajeh K., Ranjbar B., Naderi-Manesh H., Habibi A. E., Nemat-Goegani M. (2001). Chemical modification of lysine residues in Bacillus α-amylases: effect on activity and stability. Biochim. Biophys. Acta 1548, 229–237. 10.1016/S0141-0229(01)00296-4 [DOI] [PubMed] [Google Scholar]
- Khemakhem B., Ali M. B., Aghajari N., Juy M., Haser R., Bejar S. (2009). Engineering of the α-amylase from Geobacillus stearothermophilus US100 for detergent incorporation. Biotechnol. Bioeng. 102, 380–390. 10.1002/bit.22083 [DOI] [PubMed] [Google Scholar]
- Kikani B. A., Singh S. P. (2011). Single step purification and characterization of a thermostable and calcium independent α-amylase from Bacillus amyloliquifaciens TSWK1-1 isolated from Tulsi Shyam hot spring reservoir, Gujarat (India). Int. J. Biol. Macromol. 48, 676–681. 10.1016/j.ijbiomac.2011.02.010 [DOI] [PubMed] [Google Scholar]
- Kim J. W., Kim Y. H., Lee H. S., Yang S. J., Kim Y. W., Lee M. H., et al. (2007). Molecular cloning and biochemical characterization of the first archaeal maltogenic amylase from the hyperthermophilic archaeon Thermoplasma volcanium GSS1. Biochim. Biophys. Acta 1774, 661–669. 10.1016/j.bbapap.2007.03.010 [DOI] [PubMed] [Google Scholar]
- Kim J.-S., Cha S.-S., Kim H.-J., Kim T.-J., Ha N.-C. (1999). Crystal structure of a maltogenic amylase provides insights into a catalytic versatility. J. Biol. Chem. 274, 26279–26286. [DOI] [PubMed] [Google Scholar]
- Kim T. J., Nguyen V. D., Lee H. S., Kim M. J., Cho H. Y., Kim Y. W., et al. (2001). Modulation of the multisubstrate specificity of Thermus maltogenic amylase by truncation of the N-terminal domain and by a salt-induced shift of the monomer/dimer equilibrium. Biochemistry 40, 14182–14190. 10.1021/bi015531u [DOI] [PubMed] [Google Scholar]
- Kim T. U., Gu B. G., Jeong J. Y., Byun S. M., Shin Y. C. (1995). Purification and characterization of a maltotetraose forming alkaline aamylase from an alkalophilic Bacillus sp. GM8901. Appl. Environ. Microbiol. 61, 3105–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. W., Choi J. H., Kim J. W., Park C., Kim J. W., Cha H., et al. (2003). Directed evolution of Thermus maltogenic amylase toward enhanced thermal resistance. Appl. Environ. Microbiol. 69, 4866–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C., Hollender J., Bender H., Schulz G. E. (1992). Catalytic center of cyclodextrin glycosyltransferase derived from X-ray structure analysis combined with site-directed mutagenesis. Biochemistry 31, 8740–8746. [DOI] [PubMed] [Google Scholar]
- Knegtel R. M., Strokopytov B., Penninga D., Faber O. G., Rozeboom H. J., Kalk K. H., et al. (1995). Crystallographic studies of the interaction of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 with natural substrates and products. J. Biol. Chem. 270, 29256–29264. [DOI] [PubMed] [Google Scholar]
- Koch R., Spreinat A., Lemke K., Antranikian G. (1991). Purification and properties of a hyperthermoactive α-amylase from the archaeobacterium Pyrococcus woesei. Arch. Microbiol. 155, 572–578. [Google Scholar]
- Koch R., Zablowski P., Sprienat A., Antranikian G. (1990). Extremely thermostable amylolytic enzyme from the archaebacterium Pyrococcus furiosus. FEMS Microbiol. Lett. 71, 21–26. [Google Scholar]
- Kolcuoglu Y., Colak A., Faiz O., Ali O. B. (2010). Cloning, expression and characterization of highly thermo- and pH-stable maltogenic amylase from a thermophilic bacterium Geobacillus caldoxylosilyticus TK4. Process Biochem. 45, 821–828. 10.1016/j.procbio.2010.02.001 [DOI] [Google Scholar]
- Komissarov A. A., Romanova D. V., Debabov V. G. (1995). Complete inactivation of Escherichia coli uridine phosphorylase by modification of Asp with Woodward's reagent K. J. Biol. Chem. 270, 10050–10055. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter M., Wuthrich K. (1996). MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55, 29–32. [DOI] [PubMed] [Google Scholar]
- Krishnan T., Chandra A. K. (1983). Purification and characterization of α-amylase from Bacillus licheniformis CUMC 305. Appl. Environ. Microbiol. 46, 430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Satyanarayana T. (2004). Biotechnological aspects of thermophilic fungal glucoamylases, in Emerging Trends in Mycology, Plant Pathology and Microbial Biotechnology, eds Bagyanarayana G., Bhadraiah B., Kunwar I. K. (Hyderabad: B.S. Publications; ), 539–563. [Google Scholar]
- Kumar P., Satyanarayana T. (2008). Optimization of culture variables for improving glucoamylase production by alginate-trapped Thermomucor indicae-seudaticae using statistical methods. Bioresour. Technol. 98, 1252–1259. 10.1016/j.biortech.2006.05.019 [DOI] [PubMed] [Google Scholar]
- Kumar S., Khare S. K. (2012). Purification and characterization of maltooligosaccharide-forming α-amylase from moderately halophilic Marinobacter sp. EMB8. Bioresour. Technol. 116, 247–251. 10.1016/j.biortech.2011.11.109 [DOI] [PubMed] [Google Scholar]
- Kuriki T., Imanaka T. (1999). The concept of the alpha-amylase family: structural similarity and common catalytic mechanism. J. Biosci. Bioeng. 87, 557–565. [DOI] [PubMed] [Google Scholar]
- Kwak Y. S., Akiba T., Kudo T. (1998). Purification and characterization of α-amylase from hyperthermophilic archaeon Thermococcus profundus, which hydrolyzes both α-1,4 and α-1,6 glucosidic linkages. J. Fermen. Bioeng. 86, 363–367. [Google Scholar]
- Laderman K. A., Davis B. R., Krutzsch H. C., Lewis M. S., Griko Y. V., Privalov P. L., et al. (1993). The purification and characterization of an extremely thermostable α-amylase from hypothermophilic archaebacterium Pyrococcus furiosus. J. Biol. Chem. 268, 24394–24401. [PubMed] [Google Scholar]
- Larson S. B., Greenwood A., Cascio D., Day J., McPherson A. (1994). Refined molecular structure of pig pancreatic alpha-amylase at 2.1 A resolution. J. Mol. Biol. 235, 1560–1584. 10.1006/jmbi.1994.1107 [DOI] [PubMed] [Google Scholar]
- Lee J. T., Kanai H., Kobayashi T., Akiba T., Kudo T. (1996). Cloning, nucleotide sequence, and hyperexpression of α-amylase gene from an archaeon Thermococcus profundus. J. Fermen. Bioeng. 82, 432–438. [Google Scholar]
- Lee S., Mouri Y., Minoda M., Oneda H., Inouye K. (2006). Comparison of the wild-type α-amylase and its variants enzymes in Bacillus amyloliquefaciens in activity and thermal stability, and insights into engineering the thermal stability of Bacillus α-amylase. J. Biochem. 139, 1007–1015. 10.1093/jb/mvj107 [DOI] [PubMed] [Google Scholar]
- Lee S. P., Morikawa M., Takagi M., Imanaka T. (1994). Cloning of the aapT gene and characterization of its product, α-amylase pullulanase (AapT), from thermophilic and alkaliphilic Bacillus sp. Strain XAL601. Appl. Environ. Microbiol. 60, 3764–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legin E., Copinet A., Duchiron F. (1998). Production of thermostable amylolytic enzymes by Thermococcus hydrothermalis. Biotech. Lett. 20, 363–367. [Google Scholar]
- Leuschner C., Antranikian G. (1995). Heat-stable enzymes from extremely thermophilic and hyperthermophilic microorganisms. World J. Microbiol. Biotechnol. 11, 95–114. [DOI] [PubMed] [Google Scholar]
- Lévêque E., Haye B., Belarbi A. (2000). Cloning and expression of an α-amylase encoding gene from the hyperthermophilic archaebacterium Thermococcus hydrothermalis and biochemical characterisation of the recombinant enzyme. FEMS Microbiol. Lett. 186, 67–71. 10.1111/j.1574-6968.2000.tb09083.x [DOI] [PubMed] [Google Scholar]
- Levitsky A., Steer M. L. (1974). The allosteric activation of mammalian α-amylase by chloride. Eur. J. Biochem. 41, 171–180. [DOI] [PubMed] [Google Scholar]
- Li D., Park J. T., Li X., Kim S., Lee S., Shim J. H., et al. (2010). Overexpression and characterization of an extremely thermostable maltogenic amylase, with an optimal temperature of 100 degrees C, from the hyperthermophilic archaeon Staphylothermus marinus. N. Biotechnol. 27, 300–307. 10.1016/j.nbt.2010.04.001 [DOI] [PubMed] [Google Scholar]
- Li F., Zhu X., Li Y., Cao H., Zhang Y. (2011). Functional characterization of a special thermophilic multifunctional amylase OPMA-N and its N-terminal domain. Acta Biochim. Biophys. Sin. 43, 324–334. 10.1093/abbs/gmr013 [DOI] [PubMed] [Google Scholar]
- Lim W. J., Park S. R., An C. L., Lee J. Y., Hong S. Y., Shin E. C., et al. (2003). Cloning and characterization of a thermostable intracellular alpha-amylase gene from the hyperthermophilic bacterium Thermotoga maritima MSB8. Res. Microbiol. 154, 681–687. 10.1016/j.resmic.2003.09.005 [DOI] [PubMed] [Google Scholar]
- Lin L.-L., Huang C.-C., Lo H.-F. (2008). Impact of Arg210-Ser211 deletion on thermostability of a truncated Bacillus sp. strain TS-23 α-amylase. Process Biochem. 43, 559–565. 10.1016/j.procbio.2008.01.020 [DOI] [Google Scholar]
- Linden A., Mayans O., Meyer-Claucke W., Antranikian G., Wilmanns M. (2003). Differential regulation of a hyperthermophilic α-amylase with a novel (Ca, Zn) two metal center by zinc. J. Biol. Chem. 278, 9875–9884. 10.1074/jbc.M211339200 [DOI] [PubMed] [Google Scholar]
- Liu B., Wang Y., Zhang X. (2006). Characterization of a recombinant maltogenic amylase from deep sea thermophilic Bacillus sp. WPD616. Enzyme Microb. Technol. 39, 805–810. 10.1016/j.enzmictec.2006.01.003 [DOI] [Google Scholar]
- Liu W., Shi P., Chen Q., Yang P., Wang G., Wang Y., et al. (2010). Gene cloning, overexpression, and characterization of a xylanase from Penicillium sp. CGMCC 1699. Appl. Biochem. Biotechnol. 162, 1–12. 10.1016/j.enzmictec.2006.01.003 [DOI] [PubMed] [Google Scholar]
- Liu X. D., Xu Y. (2008). A novel raw starch digesting α-amylase from a newly isolated Bacillus sp. YX-1: Purification and characterization. Bioresour. Technol. 99, 4315–4320. 10.1016/j.biortech.2007.08.040 [DOI] [PubMed] [Google Scholar]
- Liu Y.-H., Lu F.-P., Li Y., Wang J. L., Gao C. (2008). Acid stabilization of Bacillus licheniformis alpha amylase through introduction of mutations. Appl. Microbiol. Biotechnol. 80, 795–803. 10.1007/s00253-008-1580-5 [DOI] [PubMed] [Google Scholar]
- Lo H.-F., Lin L.-L., Chen H.-L., Hsu W.-H., Chang C.-T. (2001). Enzymic properties of a SDS-resistant Bacillus sp. TS-23 α-amylase produced by recombinant Escherichia coli. Process Biochem. 36, 743–750. 10.1016/S0032-9592(00)00273-9 [DOI] [Google Scholar]
- MacGregor E. A. (1988). Alpha-amylase structure and activity. J. Protein Chem. 7, 399–415. [DOI] [PubMed] [Google Scholar]
- MacHius M., Declerck N., Huber R., Wiegand G. (1998). Activation of Bacillus licheniformis alpha-amylase through a disorder–>order transition of the substrate-binding site mediated by a calcium-sodium-calcium metal triad. Structure 6, 281–292. [DOI] [PubMed] [Google Scholar]
- MacHius M., Wiegand G., Huber R. (1995). Crystal structure of calcium-depleted Bacillus licheniformis alpha-amylase at 2.2 A resolution. J. Mol. Biol. 246, 545–559. 10.1006/jmbi.1994.0106 [DOI] [PubMed] [Google Scholar]
- Malhotra R., Noorwez S. M., Satyanarayana T. (2000). Production and partial characterization of thermostable and calcium-independent α-amylase of an extreme thermophile Bacillus thermooleovorans NP54. Lett. Appl. Microbiol. 31, 378–384. 10.1046/j.1472-765x.2000.00830.x [DOI] [PubMed] [Google Scholar]
- Mamo G., Gessesse A. (1999). Purification and characterization of two raw-starch–digesting thermostable α-amylase from a thermophilic Bacillus. Enzyme Microb. Technol. 25, 433–438. [Google Scholar]
- Manas N. H. A., Jonet M. A., Murad A. M. A., Mahadi N. M., Illias R. M. (2015). Modulation of transglycosylation and improved malto-oligosaccharide synthesis by protein engineering of maltogenic amylase from Bacillus lehensis G1. Process Biochem. 50, 1572–1580. 10.1016/j.procbio.2015.06.005 [DOI] [Google Scholar]
- Matzke J., Schwermann B., Baker E. P. (1997). Acidostable and acidophilic proteins: the example of the α-amylase from Alicyclobacillus acidocaldarius. Comp. Biochem. Physiol. A Physiol. 118, 475–479. [DOI] [PubMed] [Google Scholar]
- Mehta D., Satyanarayana T. (2013a). Biochemical and molecular characterization of recombinant acidic and thermostable raw starch hydrolysing α-amylase from an extreme thermophile Geobacillus thermoleovorans. J. Mol. Catal. B Enzym. 85–86, 229–238. 10.1016/j.molcatb.2012.08.017 [DOI] [Google Scholar]
- Mehta D., Satyanarayana T. (2013b). Dimerization mediates thermo-adaptation, substrate affinity and transglycosylation in a highly thermostable maltogenic amylase of Geobacillus thermoleovorans. PLoS ONE 8:e73612. 10.1371/journal.pone.0073612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D., Satyanarayana T. (2014). Domain C of thermostable alpha-amylase of Geobacillus thermoleovorans mediates raw starch adsorption. Appl. Microbiol. Biotechnol. 98, 4503–4519. 10.1007/s00253-013-5459-8 [DOI] [PubMed] [Google Scholar]
- Mehta D., Satyanarayana T. (2015). Structural elements of thermostability in the maltogenic amylase of Geobacillus thermoleovorans. Int. J. Biol. Macromol. 79, 570–576. 10.1016/j.ijbiomac.2015.04.011 [DOI] [PubMed] [Google Scholar]
- Mesbah N. M., Wiegel J. (2014). Halophilic alkali- and thermostable amylase from a novel polyextremophilic Amphibacillus sp. NM-Ra2. Int. J. Biol. Macromol. 70, 222–229. 10.1016/j.enzmictec.2013.03.009 [DOI] [PubMed] [Google Scholar]
- Mok S. C., Teh A. H., Saito J. A., Najimudin N., Alam M. (2013). Crystal structure of a compact alpha-amylase from Geobacillus thermoleovorans. Enzyme Microb. Technol. 53, 46–54. 10.1016/j.enzmictec.2013.03.009 [DOI] [PubMed] [Google Scholar]
- Mollania N., Khajeh K., Hosseinkhani S., Dabirmanesh B. (2010). Purification and characterization of a thermostable phytate resistant α-amylase from Geobacillus sp. LH8. Int. J. Biol. Macromol. 46, 27–36. 10.1016/j.ijbiomac.2009.10.010 [DOI] [PubMed] [Google Scholar]
- Morkeberg R., Carlsen M., Nielsen J. (1995). Induction and repression of α-amylase production in batch and continuous cultures of Aspergillus oryzae. Microbiology 141, 2449–2454. [DOI] [PubMed] [Google Scholar]
- Morlon-Guyot J., Mucciolo-Roux F., Rodriguez Sanoja R., Guyot J. P. (2001). Characterization of the L. manihotivorans alpha-amylase gene. DNA Seq. 12, 27–37. 10.3109/10425170109042048 [DOI] [PubMed] [Google Scholar]
- Murakami S., Nagasaki K., Nishimoto H., Shigematu R., Umesaki J., et al. (2008). Purification and characterization of five alkaline, thermotolerant, and maltotetraose-producing α-amylases from Bacillus halodurans MS-2-5, and production of recombinant enzymes in Escherichia coli. Enzyme Microb. Technol. 43, 321–328. 10.1016/j.enzmictec.2008.05.006 [DOI] [Google Scholar]
- Murthy P., Naidu M., Srinivas P. (2009). Production of α-amylase under solid-state fermentation utilizing coffee waste. J. Chem. Technol. Biotechnol. 84, 1246–1249. 10.1002/jctb.2142 [DOI] [Google Scholar]
- Nielsen J. E., Borchert T. V. (2000). Protein engineering of bacterial α-amylases. Biochim. Biophys. Acta 1543, 253–274. 10.1016/S0167-4838(00)00240-5 [DOI] [PubMed] [Google Scholar]
- Nielsen J. E., Beier L., Otzen D., Borchert T. V., Frantzen H. B., Andersen K. V., et al. (1999). Electrostatics in the active site of an alpha-amylase. Eur. J. Biochem. 264, 816–824. [DOI] [PubMed] [Google Scholar]
- Nigam P., Singh D. (1995). Enzyme and microbial systems involved in starch processing. Enzyme Microb. Technol. 17, 770–778. [Google Scholar]
- Nonaka T., Fujihashi M., Kita A., Hagihara H., Ozaki K., et al. (2003). Crystal structure of calcium-free α -amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites. J. Biol. Chem. 278, 24818–24824. 10.1074/jbc.M212763200 [DOI] [PubMed] [Google Scholar]
- Ozturk H., Ece S., Gundeger E., Evran S. (2013). Site-directed mutagenesis of methionine residues for improving the oxidative stability of alpha-amylase from Thermotoga maritima. J. Biosci. Bioeng. 116, 449–451. 10.1016/j.jbiosc.2013.04.018 [DOI] [PubMed] [Google Scholar]
- Pancha I., Jain D., Shrivastav A., Mishra S. K., Shethia B., et al. (2010). A thermoactive α-amylase from a Bacillus sp. isolated from CSMCRI salt farm. Int. J. Biol. Macromol. 47, 288–291. 10.1016/j.ijbiomac.2010.04.006 [DOI] [PubMed] [Google Scholar]
- Pandey A., Nigam P., Soccol C. R., Soccol V. T., Singh D., Mohan R. (2000). Advances in microbial amylases. Appl. Biochem. Biotechnol. 31, 135–152. [DOI] [PubMed] [Google Scholar]
- Paoli P., Fiaschi T., Cirri P., Camici G., Manao G., Cappugi G., et al. (1997). Mechanism of acylphosphatase inactivation by Woodward's reagent K. Biochem. J. 328, 855–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar D., Satyanarayana T. (2016). A chimeric alpha-amylase engineered from Bacillus acidicola and Geobacillus thermoleovorans with improved thermostability and catalytic efficiency. J. Ind. Microbiol. Biotechnol. 43, 473–484. 10.1007/s10295-015-1721-7 [DOI] [PubMed] [Google Scholar]
- Pham P. L., Taillandier P., Delmas M., Strehaiano P. (1998). Optimization of the culture medium for xylanase production by Bacillus sp. using statistical experimental designs. World J. Microbiol. Biotechnol. 14, 185–190. [Google Scholar]
- Prakash B., Vidyasagar M., Madhukumar M. S., Muralikrishna G., Sreeramulu K. (2009). Production, purification, and characterization of two extremely halotolerant, thermostable, and alkali-stable α-amylases from Chromohalobacter sp. TVSP 101. Process Biochem. 44, 210–215. 10.1016/j.procbio.2008.10.013 [DOI] [Google Scholar]
- Prakash O., Jaiswal N. (2010). α-Amylase: an ideal representative of thermostable enzymes. Appl. Biochem. Biotechnol. 160, 2401–2414. 10.1007/s12010-009-8735-8744 [DOI] [PubMed] [Google Scholar]
- Pritchard P. A. (1992). Studies on the bread improving mechanisms of fungal α-amylase. J. Biol. Educ. 26, 12–18. [Google Scholar]
- Puspasari F., Radjasa O. K., Noer A. S., Nurachman Z., Syah Y. M., van der Maarel M., et al. (2013). Raw starch-degrading alpha-amylase from Bacillus aquimaris MKSC 6.2: isolation and expression of the gene, bioinformatics and biochemical characterization of the recombinant enzyme. J. Appl. Microbiol. 114, 108–120. 10.1111/jam.12025 [DOI] [PubMed] [Google Scholar]
- Ramasubbu N., Paloth V., Luo Y., Brayer G. D., Levine M. J. (1996). Structure of human salivary alpha-amylase at 1.6 A resolution: implications for its role in the oral cavity. Acta Crystallogr. D Biol. Crystallogr. 52, 435–446. 10.1107/S0907444995014119 [DOI] [PubMed] [Google Scholar]
- Rao J. L. U. M., Satyanarayana T. (2007a). Improving production of hyperthermostable and high maltose-forming α-amylase by an extreme thermophile Geobacillus thermoleovorans using response surface methodology and its applications. Bioresour. Technol. 98, 345–352. 10.1016/j.biortech.2005.12.022 [DOI] [PubMed] [Google Scholar]
- Rao J. L. U. M., Satyanarayana T. (2007b). Purification and characterization of a hyperthermostable and high maltogenic α-amylase of an extreme thermophile Geobacillus thermoleovorans. Appl. Biochem. Biotechnol. 142, 179–193. 10.1007/s12010-007-0017-4 [DOI] [PubMed] [Google Scholar]
- Ratanakhanokchai K., Kaneko J., Kamio Y., Izaki K. (1992). Purification and properties of a maltotetraose and maltotriose producing amylase from Chloroflexus aurantiacus. Appl. Environ. Microbiol. 58, 2490–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindar D. J., Elangovan N. (2013). Molecular identification of amylase producing Bacillus subtilis and detection of optimal conditions. J. Pharm. Res. 64, 426–430. 10.1016/j.jopr.2013.04.001 [DOI] [Google Scholar]
- Richardson T. H., Tan X., Frey G., Callen W., Cabell M., Lam D., et al. (2002). A novel, high performance enzyme for starch liquefaction. Discovery and optimization of a low pH, thermostable alpha-amylase. J. Biol. Chem. 277, 26501–26507. 10.1074/jbc.M203183200 [DOI] [PubMed] [Google Scholar]
- Robert X., Haser R., Gottschalk T. E., Ratajczak F., Driguez H., Svensson B., et al. (2003). The structure of barley alpha-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: a pair of sugar tongs. Structure 11, 973–984. 10.1016/S0969-2126(03)00151-5 [DOI] [PubMed] [Google Scholar]
- Roohi Kuddus M. (2014). Bio-statistical approach for optimization of cold-active α-amylase production by novel psychrotolerant M. foliorum GA2 in solid state fermentation. Biocatal. Agric. Biotechnol. 3, 175–181. 10.1016/j.bcab.2013.09.007 [DOI] [Google Scholar]
- Roy J. K., Borah A., Mahanta C. L., Mukherjee A. K. (2013). Cloning and overexpression of raw starch digesting α-amylase gene from Bacillus subtilis strain AS01a in Escherichia coli and application of the purified recombinant α-amylase (AmyBS-I) in raw starch digestion and baking industry. J. Mol. Catal. B Enzym. 97, 118–129. 10.1016/j.molcatb.2013.07.019 [DOI] [Google Scholar]
- Roy J. K., Mukherjee A. K. (2013). Applications of a high maltose forming, thermo-stable α-amylase from an extremely alkalophilic Bacillus licheniformisstrain AS08E in food and laundry detergent industries. Biochem. Eng. J. 77, 220–230. 10.1016/j.bej.2013.06.012 [DOI] [Google Scholar]
- Roy J. K., Rai S. K., Mukherjee A. K. (2012). Characterization and application of a detergent-stable alkaline alpha-amylase from Bacillus subtilis strain AS-S01a. Int. J. Biol. Macromol. 50, 219–229. 10.1016/j.ijbiomac.2011.10.026 [DOI] [PubMed] [Google Scholar]
- Rubin-Pitel S. B., Zhao H. (2006). Recent advances in biocatalysis by directed enzyme evolution. Comb. Chem. High Throughput Screen. 9, 247–257. [DOI] [PubMed] [Google Scholar]
- Russell R. J. M., Ferguson J. M., Hough D. W., Danson M. J., Taylor G. L. (1997). The crystal structure of citrate synthase from the hyperthermophilic Archaeon Pyrococcus furiosus at 19 angstrom resolution. Biochemistry 36, 9983–9994. [DOI] [PubMed] [Google Scholar]
- Ryan S. M., Fitzgerald G. F., Van Sinderen D. (2006). Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in Bifidobacterial strains. Appl. Environ. Microbiol. 72, 5289–5296. 10.1128/AEM.00257-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito N. A. (1973). Thermophilic extracellular α-amylase from Bacillus licheniformis. Arch. Biochem. Biophys. 155, 290–298. [DOI] [PubMed] [Google Scholar]
- Sajedi R. H., Naderi-Mahesh H., Khajeh K., Ahmadvand R., Ranjbar B. A., Asoodeh A., et al. (2005). A calcium independent α-amylase that is active and stable at low pH from the Bacillus sp. KR-8104. Enzyme Microb. Technol. 36, 666–671. 10.1016/j.enzmictec.2004.11.003 [DOI] [Google Scholar]
- Salminen T., Teplyakov A., Kankare J., Cooperman B. S., Lahti R., et al. (1996). An unusual route to thermostability disclosed by the comparison of Thermus thermophilus and Escherichia coli inorganic pyrophosphatases. Protein Sci. 5, 1014–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samie N., Noghabi K. A., Gharegozloo Z., Zahiri H. S., Ahmadian G., Sharafi H., et al. (2012). Psychrophilic α-amylase from Aeromonas veronii NS07 isolated from farm soils. Process Biochem. 47, 1381–1387. 10.1016/j.procbio.2012.05.007 [DOI] [Google Scholar]
- Savchenko A., Vieille C., Kang S., Zeikus G. (2002). Pyrococcus furiosus α-amylase is stabilized by calcium and zinc. Biochemistry 41, 6193–6201. 10.1021/bi012106s [DOI] [PubMed] [Google Scholar]
- Saxena R. K., Dutt K., Agarwal L., Nayyar P. (2007). A highly thermostable and alkaline amylase from a Bacillus sp. PNS. Bioresour. Technol. 98, 260–265. 10.1016/j.biortech.2006.01.016 [DOI] [PubMed] [Google Scholar]
- Schwermann B., Pfau K., Liliensiek B., Schleyer M., Fischer T., Bakker E. P. (1994). Purification, properties and structural aspects of a thermoacidophilic α-amylase from Alicyclobacillus acidocaldarius ATCC 27009: insight into acidostability of proteins. Eur. J. Biochem. 226, 981–991. [DOI] [PubMed] [Google Scholar]
- Sen S. K., Dora T. K., Bandyopadhyay B., Mohapatra P. K. D., Raut S. (2014). Thermostable alphα-amylase enzyme production from hot spring isolates Alcaligenes faecalis SSB17 – statistical optimization. Biocatal. Agric. Biotechnol. 3, 218–226. 10.1016/j.bcab.2014.03.005 [DOI] [Google Scholar]
- Shafiei M., Ziaee A. A., Amoozegar M. A. (2010). Purification and biochemical characterization of a novel SDS and surfactant stable, raw starch digesting, and halophilic α-amylase from a moderately halophilic bacterium, Nesterenkonia sp. strain F. Process Biochem. 45, 694–699. 10.1016/j.procbio.2010.01.003 [DOI] [Google Scholar]
- Sharma A., Satyanarayana T. (2010). High maltose-forming, Ca2+-independent and acid stable α-amylase from a novel acidophilic bacterium, Bacillus acidicola. Biotechnol. Lett. 32, 1503–1507. 10.1007/s10529-010-0322-9 [DOI] [PubMed] [Google Scholar]
- Sharma A., Satyanarayana T. (2011). Optimization of medium components and cultural variables for enhanced production of acidic high maltose-forming and Ca2+-independent α-amylase by Bacillus acidicola. J. Biosci. Bioeng. 111, 550–553. 10.1016/j.jbiosc.2011.01.004 [DOI] [PubMed] [Google Scholar]
- Sharma A., Satyanarayana T. (2012). Cloning and expression of acidstable, high maltose-forming, Ca2+-independent a-amylase from an acidophile Bacillus acidicola and its applicability in starch hydrolysis. Extremophiles 16, 515–522. 10.1007/s00792-012-0451-2 [DOI] [PubMed] [Google Scholar]
- Sharma D. C., Satyanarayana T. (2006). A marked enhancement in the production of a highly alkaline and thermostable pectinase by Bacillus pumilus dcsr1 in submerged fermentation by using statistical methods. Bioresour. Technol. 97, 727–733. 10.1016/j.biortech.2005.04.012 [DOI] [PubMed] [Google Scholar]
- Sharma P., Pushpanathan M., Jayashree S., Gunasekaran P., Rajendhran J. (2015). Identification of periplasmic α-amlyase from cow dung metagenome by product induced gene expression profiling (pigex). Ind. J. Microbiol. 55, 57–65. 10.1007/s12088-014-0487-3 [DOI] [Google Scholar]
- Sharma S., Khan F. G., Qazi G. N. (2010). Molecular cloning and characterization of amylase from soil metagenomic library derived from Northwestern Himalayas. Appl. Microbiol. Biotechnol. 86, 1821–1828. 10.1007/s00253-009-2404-y [DOI] [PubMed] [Google Scholar]
- Shaw A., Bott R., Day A. G. (1999). Protein engineering of alpha-amylase for low pH performance. Curr. Opin. Biotechnol. 10, 349–352. 10.1016/S0958-1669(99)80063-9 [DOI] [PubMed] [Google Scholar]
- Shinke R., Aoki K., Murokam S., Inoue T., Babat T. (1996). Alkaline and thermophilic amylases of industrial use, in Enzyme Engineering XIII, Vol. 799, eds Dordick J. S., Russell A. J. (New York, NY: Annals of the New York Academy of Sciences; ), 332–340. [Google Scholar]
- Sivaramakrishnan S., Gangadharan D., Nampoothiri K. M., Soccol C. R., Pandey A. (2006). α-amylases from microbial sources – an overview on recent developments. Food Technol. Biotechnol. 44, 173–184. [Google Scholar]
- Sodhi H. K., Sharma K., Gupta J. K., Soni S. K. (2005). Production of a thermostable α-amylase from Bacillus sp. PS-7 by solid state fermentation and its synergistic use in the hydrolysis of malt starch for alcohol production. Process Biochem. 40, 525–534. 10.1016/j.procbio.2003.10.008 [DOI] [Google Scholar]
- Strokopytov B., Penninga D., Rozeboom H. J., Kalk K. H., Dijkhuizen L., Dijkstra B. W. (1995). X-ray structure of cyclodextrin glycosyltransferase complexed with acarbose. Implications for the catalytic mechanism of glycosidases. Biochemistry 34, 2234–2240. [DOI] [PubMed] [Google Scholar]
- Sumitani J., Tottori T., Kawaguchi T., Arai M. (2000). New type of starch-binding domain: the direct repeat motif in the C-terminal region of Bacillus sp. no. 195 alpha-amylase contributes to starch binding and raw starch degrading. Biochem. J. 350, 477–484. 10.1042/bj3500477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Duan X., Wang L., Wu J. (2016). Enhanced maltose production through mutagenesis of acceptor binding subsite +2 in Bacillus stearothermophilus maltogenic amylase. J. Biotechnol. 217, 53–61. 10.1016/j.jbiotec.2015.11.007 [DOI] [PubMed] [Google Scholar]
- Sunna A., Moracci M., Rossi M., Antranikian G. (1997). Glycosyl hydrolases from hyperthermophiles. Extremophiles 1, 2–13. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Ito N., Yuuki T., Yamagata H., Udaka S. (1989). Amino acid residues stabilizing a Bacillus alpha-amylase against irreversible thermoinactivation. J. Biol. Chem. 264, 18933–18938. [PubMed] [Google Scholar]
- Svensson B. (1994). Protein engineering in the alpha-amylase family: catalytic mechanism, substrate specificity, and stability. Plant Mol. Biol. 25, 141–157. [DOI] [PubMed] [Google Scholar]
- Tachibana Y., Leclere M. M., Fujiwara S., Takag M., Imanaka T. (1996). Cloning and Expression of the α-Amylase Gene from the Hyperthermophilic archaeon Pyrococcus sp. KOD 1, and characterization of the enzyme. J. Fermen. Bioeng. 82, 224–232. [Google Scholar]
- Tan T. C., Mijts B. N., Swaminathan K., Patel B. K., Divne C. (2008). Crystal structure of the polyextremophilic alpha-amylase AmyB from Halothermothrix orenii: details of a productive enzyme-substrate complex and an N domain with a role in binding raw starch. J. Mol. Biol. 378, 852–870. 10.1016/j.jmb.2008.02.041 [DOI] [PubMed] [Google Scholar]
- Tanaka A., Hoshino E. (2003). Secondary calcium binding parameter of Bacillus amyloliquifaciens α-amylase obtained from inhibition studies. J. Biosci. Bioeng. 96, 262–267. 10.1016/S1389-1723(03)80191-3 [DOI] [PubMed] [Google Scholar]
- Tang S. Y., Le Q. T., Shim J. H., Yang S. J., Auh J. H., Park C., et al. (2006). Enhancing thermostability of maltogenic amylase from Bacillus thermoalkalophilus ET2 by DNA shuffling. FEBS J. 273, 3335–3345. 10.1111/j.1742-4658.2006.05337.x [DOI] [PubMed] [Google Scholar]
- Tao X., Jang M. S., Kim K. S., Yu Z., Lee Y. C. (2008). Molecular cloning, expression and characterization of alpha-amylase gene from a marine bacterium Pseudoalteromonas sp. MY-1. Indian J. Biochem. Biophys. 45, 305–309. [PubMed] [Google Scholar]
- Tester R. F., Karkalas J., Qi X. (2004). Starch—composition, fine structure and architecture. J. Cereal Sci. 39, 151–165. 10.1016/j.jcs.2003.12.001 [DOI] [Google Scholar]
- Tomazic S. J., Klibanov A. M. (1988). Mechanisms of irreversible thermal inactivation of Bacillus α-amylases. J. Biol. Chem. 263, 3086–3091. [PubMed] [Google Scholar]
- Tran P. L., Cha H. J., Lee J. S., Park S. H., Woo E. J., Park K. H. (2014). Introducing transglycosylation activity in Bacillus licheniformis alpha-amylase by replacement of His235 with Glu. Biochem. Biophys. Res. Commun. 451, 541–547. 10.1016/j.bbrc.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Uchino F. (1982). A thermophilic and unusually acidophilic amylase produced by a thermophilic acidophilic Bacillus sp. Agric. Biol. Chem. 46, 7–13. [Google Scholar]
- Van Dam H. W., Hille J. D. R. (1992). Yeast and enzymes in bread making. Cereal Foods World 37, 245–252. [Google Scholar]
- van der Maarel M. J., van der Veen B., Uitdehaag J. C., Leemhuis H., Dijkhuizen L. (2002). Properties and applications of starch-converting enzymes of the alpha-amylase family. J. Biotechnol. 94, 137–155. 10.1016/S0168-1656(01)00407-2 [DOI] [PubMed] [Google Scholar]
- Verhaert R. M., Beekwilder J., Olsthoorn R., van Duin J., Quax W. J. (2002). Phage display selects for amylases with improved low pH starch-binding. J. Biotechnol. 96, 103–118. 10.1016/S0168-1656(02)00041-X [DOI] [PubMed] [Google Scholar]
- Vester J. K., Glaring M. A., Stougaard P. (2015). An exceptionally cold-adapted alpha-amylase from a metagenomic library of a cold and alkaline environment. Appl. Microbiol. Biotechnol. 99, 717–727. 10.1007/s00253-014-5931-0 [DOI] [PubMed] [Google Scholar]
- Vieille C., Zeikus G. J. (2001). Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65, 1–43. 10.1128/MMBR.65.1.1-43.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihinen M., Mantasala P. (1989). Microbial amylolytic enzymes. Crit. Rev. Biochem. Mol. Biol. 24, 329–418. [DOI] [PubMed] [Google Scholar]
- Vogt G., Woell S., Argos P. (1997). Protein thermal stability, hydrogen bonds, and ion pairs. J. Mol. Biol. 269, 631–643. [DOI] [PubMed] [Google Scholar]
- Wang S. L., Liang Y. C., Liang T. W. (2011). Purification and characterization of a novel alkali-stable α-amylase from Chryseobacterium taeanense TKU001, and application in antioxidant and prebiotic. Process Biochem. 46, 745–750. 10.1016/j.procbio.2010.11.022 [DOI] [Google Scholar]
- Xu B., Yang F., Xiong C., Li J., Tang X., Zhou J., et al. (2014). Cloning and characterization of a novel alpha-amylase from a fecal microbial metagenome. J. Microbiol. Biotechnol. 24, 447–452. 10.4014/jmb.1310.10121 [DOI] [PubMed] [Google Scholar]
- Yamaguchi R., Inoue Y., Tokunaga H., Ishibashi M., Arakawa T., Sumitani J., et al. (2012). Halophilic characterization of starch-binding domain from Kocuria varians alpha-amylase. Int. J. Biol. Macromol. 50, 95–102. 10.1016/j.ijbiomac.2011.10.007 [DOI] [PubMed] [Google Scholar]
- Yang C. H., Huang Y. C., Chen C. Y., Wen C. Y. (2010). Expression of Thermobifida fusca thermostable raw starch digesting alpha-amylase in Pichia pastoris and its application in raw sago starch hydrolysis. J. Ind. Microbiol. Biotechnol. 37, 401–406. 10.1007/s10295-009-0686-9 [DOI] [PubMed] [Google Scholar]
- Yang H., Liu L., Shin H. D., Chen R. R., Li J., Du G., et al. (2013). Structure-based engineering of histidine residues in the catalytic domain of alpha-amylase from Bacillus subtilis for improved protein stability and catalytic efficiency under acidic conditions. J. Biotechnol. 164, 59–66. 10.1016/j.jbiotec.2012.12.007 [DOI] [PubMed] [Google Scholar]
- Yin X. H., Gagnat J., Gerbaud C., Guerineau M., Virolle M. J. (1997). Cloning and characterization of a new alpha-amylase gene from Streptomyces lividans TK24. Gene 197, 37–45. [DOI] [PubMed] [Google Scholar]
- Yun J., Kang S., Park S., Yoon H., Kim M. J., Heu S., et al. (2004). Characterization of a novel amylolytic enzyme encoded by a gene from a soil-derived metagenomic library. Appl. Environ. Microbiol. 70, 7229–7235. 10.1128/AEM.70.12.7229-7235.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G. M., Huang J., Huang G. R., Ma L. X., Zhang X. E. (2007). Molecular cloning and heterologous expression of a new xylanase gene from Plectosphaerella cucumerina. Appl. Microbiol. Biotechnol. 74, 339–346. 10.1007/s00253-006-0648-3 [DOI] [PubMed] [Google Scholar]