Abstract
The risk of developing breast cancer is increased in women with family history of breast cancer and particularly in families with multiple cases of breast or ovarian cancer. Nevertheless, many women with a positive family history never develop the disease. Polygenic risk scores (PRSs) based on the risk effects of multiple common genetic variants have been proposed for individual risk assessment on a population level. We investigate the applicability of the PRS for risk prediction within breast cancer families. We studied the association between breast cancer risk and a PRS based on 75 common genetic variants in 52 Finnish breast cancer families including 427 genotyped women and pedigree information on ~4000 additional individuals by comparing the affected to healthy family members, as well as in a case–control dataset comprising 1272 healthy population controls and 1681 breast cancer cases with information on family history. Family structure was summarized using the BOADICEA risk prediction model. The PRS was associated with increased disease risk in women with family history of breast cancer as well as in women within the breast cancer families. The odds ratio (OR) for breast cancer within the family dataset was 1.55 [95 % CI 1.26–1.91] per unit increase in the PRS, similar to OR in unselected breast cancer cases of the case–control dataset (1.49 [1.38–1.62]). High PRS-values were informative for risk prediction in breast cancer families, whereas for the low PRS-categories the results were inconclusive. The PRS is informative in women with family history of breast cancer and should be incorporated within pedigree-based clinical risk assessment.
Electronic supplementary material
The online version of this article (doi:10.1007/s10549-016-3897-6) contains supplementary material, which is available to authorized users.
Keywords: Breast cancer, Common variants, PRS, Hereditary, Risk
Introduction
Individualized risk prediction is a major goal of clinical genetics. For prediction of breast cancer risk, family history is an important factor, women with a first-degree relative having twofold increased risk [1], while women from families with multiple breast/ovarian cancers or cancers diagnosed at younger age experience a much higher risk of developing the disease [2]. Rare mutations in high- and moderate-risk genes including BRCA1, BRCA2, TP53, PTEN,PALB2, and CHEK2 explain about 20 % of the familial relative risk for breast cancer [3]. A polygenic component comprising many variants of small effect contributes to the risk of developing the disease in the general population and may also modify the risk in cancer families [3–5].
Over the last few years, genome-wide association studies (GWAS) have been successful in identifying some of the common low-penetrance variants predisposing to breast cancer [6–8]. To date, more than seventy variants have been identified, which together explain about 14 % of the familial risk of breast cancer [5, 6]. Individually, the effect sizes associated with these common variants are small. However, their combined effect, summarized as a polygenic risk score (PRS), is more substantial [5]. In a recent population-based case–control study, eight percent of women at the high end of the PRS distribution were found to fall into a group of intermediate life-time risk (17–30 %) according to the UK NICE guidelines [9]. In recent studies, the PRS has been tested in combination with other risk prediction methods, such as BOADICEA and BRCAPRO [10], mammographic density (BI-RADS) [11], and a combination of family history and established risk factors (BCRAT and IBIS) [10].
The contribution of the PRS to disease risk for individuals with family history of breast cancer and within breast cancer families has not been studied extensively. Here, we investigate the association between a 75-variant PRS and disease status in individuals with and without family history in a large Finnish case–control study and 52 Finnish breast cancer families, which have an extensive pedigree information available and which have been well characterized in terms of their genetic and pathological characteristics. We use a family history score based on the BOADICEA risk prediction algorithm to evaluate whether the PRS predicts disease status among women sharing similar family history, and discuss clinical utility of the PRS for risk prediction in familial breast cancer.
Patients and methods
Study subjects
We included two separate sets of study subjects in the analyses. The case–control dataset consisted of
-
i:
three series of consecutive, unselected breast cancer patients (n = 1303) enrolled for their first primary breast cancer at the Helsinki University Central Hospital during 1997–1998, 2000, and 2001–2004 as described previously [12–14],
-
ii:
additional index cases (n = 378) with positive family history of breast cancer (one per family), tested negative for germ-line mutations in high penetrance susceptibility genes BRCA1 and BRCA2 from an ongoing collection started at 1995 at the Helsinki University Central Hospital, Department of Clinical Genetics [15, 16],
-
iii:
and healthy population controls (n = 1272, blood donors, Finnish Red Cross) (Supplementary Table 1).
In the case–control dataset, the index cases with positive family history of breast cancer were categorized into two groups based on whether they came from a family with two breast cancers in first-degree relatives (later referred to as “small families”) or from a family with three or more cases of breast or ovarian cancer in first- or second-degree relatives (later, “large families”).
The breast cancer family dataset consisted of 493 (427 women) genotyped study subjects and registry data for further 3992 family members from 52 Finnish families with multiple cases of breast cancer, collected systematically as described previously [16] (Supplementary Table 1). The criteria for including families in this study were the highest possible number of informative study subjects (women with breast cancer) with available DNA sample. The families were traced back to find the most recent common ancestors of the breast cancer patients and then traced forward including in the pedigree all the descendants of the most recent common ancestors according to records of church parish registries, Population Register Center, and Finnish Cancer Registry. The age and disease status were ascertained up to 31st December 2010. The index case of each family had previously been tested for germline mutations in BRCA1 and BRCA2 mutations and was found to be negative [16]. The number of family members varied between 22 and 356 (median 57.5) (Supplementary Table 2). Median proportion of affected women born between 1910 and 1970 was 22 % (Supplementary Table 2). The mean follow-up age of genotyped healthy women was 60.3 years, and the mean diagnosis age of genotyped breast cancer patients was 54.1 years. Seven pedigrees originated from two unrelated founder couples. One of the moderate-penetrance mutations, CHEK2:c.1100delC, PALB2:c.1592delT, or putative moderate penetrance mutation FANCM:c.5101C>T, was transmitted in seven families.
Genotyping
DNA was extracted from peripheral blood samples and genotyped at CNIO genotyping unit, Madrid, or Génome Québec Innovation Centre using a custom Illumina Infinium array, which was designed for the Collaborative Oncological Gene-environment Study (COGS). Data quality was monitored as described earlier [6]. Analyses were based on 75 variants reported to be associated (at P < 5 × 10-8) in the analysis of the COGS dataset or previous publications [5–8], with either overall breast cancer or ER-negative disease (Supplementary Table 3). All variants used in the risk score passed quality control filters. For the majority of variants, genotypes were missing in <0.1 % of individuals. Four variants used in the risk score (rs17879961, rs10941679, rs4973768, and rs13281615) had <0.4 % missing rate, and for one genotype, rs2943559, 1.06 % of individuals had missing genotypes. Twelve samples with missing genotype calling for over 10 % of the susceptibility variants were excluded from further analyses.
In the analyses, the genotype of each variant was represented by allele dose (the number of copies of the rare allele). Missing genotypes were imputed for controls using the mean of the known genotype doses, and for cases using the following formula:
where Ø is the effect size (per-allele Odds Ratio) for association between that variant and breast cancer risk in the case–control dataset, and p is the minor allele frequency in controls for the missing variant.
Three moderate-penetrance mutations, which are relatively frequent in the Finnish population, CHEK2:c.1100delC, PALB2:c.1592delT, and FANCM:c.5101C >T, had previously been genotyped locally using a custom made TaqMan assay (Applied Biosystems, Thermo Fisher Scientific Inc.), AmpliFluor fluorescent genotyping (KBiosciences), conformation sensitive gel electrophoresis heteroduplex conformation analysis and Sanger sequencing [17], or with the Sequenom MassARRAY system using iPLEX Gold assays, respectively, as described previously [18–20]. All study subjects of the case–control dataset were genotyped for all three moderate-penetrance mutations. However, in the breast cancer family dataset, the index case in each family was genotyped. If a mutation was found, all family members were genotyped for that mutation. If no mutation was found, all family members were considered as noncarriers.
Statistical analyses
Polygenic risk score
Statistical analyses were performed using R version 3.0.2 [21], including packages multiwayvcov [22] and lmtest [23]. A PRS summarizing the risk effects associated with 75 common breast cancer susceptibility variants [6–8] was calculated for each individual using the following formula: where n is the number of loci included in the model, a is the number of disease alleles at locus, i and OR is the corresponding per-allele odds ratio for breast cancer (Supplementary Table 3). The ORs for each variant, used as weights in the PRS, were taken from previously published estimates from the Breast Cancer Association Consortium (BCAC) [5]. The case–control component of the present study was included in this estimation, but it only comprised a small fraction (3 %) of the total BCAC data available for estimation of the ORs. To avoid ascertainment bias, studies oversampling for cases with family history were excluded from this estimation. The PRS-values were standardized by the mean (0.50) and standard deviation (0.45) of the PRS-values in healthy population controls, so that odds ratios could be reported as per unit standard deviation of the PRS. This corresponded well to a recently introduced PRS of 77 common variants [5], although we did not include data for two imputed variants on 11q13 (rs78540526 and rs75915166). When study subjects were categorized into centiles of the PRS distribution, the centile boundaries were defined on the basis of the PRS distribution in healthy population controls.
BOADICEA score
Familial history was quantified with a single quantity, which we term a BOADICEA score. This was calculated for all genotyped women (n = 427) in the 52 breast cancer families by applying the BOADICEA risk prediction algorithm [24]. The BOADICEA algorithm is implemented in a modified version of MENDEL [25], coded in FORTRAN 90. BOADICEA is a genetic model for breast and ovarian cancer that models risk in terms of BRCA1 and BRCA2 mutations and a polygene representing a large number of loci of small effect to capture the residual familial aggregation of breast cancer. The polygenotype is assumed to be normally distributed, but this is implemented using a binomial approximation, with transmission determined by a discrete hypergeometric polygenic model [26]. Thus, BOADICEA codes the polygenotype as a discrete variable (0–6). The polygenic score for each discrete polygenotype is the mean value from a standard normal distribution over the same probability interval. The BOADICEA algorithm can compute the posterior probability for each polygenotype, conditional on their family history. The overall BOADICEA score used in this analysis was calculated by summing the scores for the discrete polygenotypes weighted by the posterior probability. Hence, the BOADICEA score is an estimate of the mean polygenotype for the individual, given their family history (and hence would predict their future cancer risk, although it is not identical to the cancer risk output by the BOADICEA program, which also depends on age). The score was calculated assuming women were tested negative for the mutations in the BRCA1 and BRCA2 genes, with sensitivities of 0.7 and 0.8 for BRCA1 and BRCA2 testing, respectively. The study subject (target) was coded uninformative with regard to age and disease status (in effect, computing the mean polygenotype at birth, given the family history). Other available information on birth year, age, and cancer diagnoses of her relatives were included in the calculations as in the BOADICEA data form. For data standardizing purposes, only first-, second-, and third-degree relatives of the same and older generations were included in the input pedigrees.
Association analyses
Association between breast cancer risk and the PRS was modeled using logistic regression. Analysis performed within the breast cancer families (affected vs. healthy women) was adjusted for age (diagnosis age for cases, follow-up age for healthy women) and family history as summarized by the BOADICEA score, and additionally for moderate-penetrance mutations in CHEK2, PALB2, and FANCM (separate terms for each mutation, coded as 0, noncarriers; 1, heterozygotes; 2, homozygotes). P-values and 95 % confidence intervals (CI) were corrected using robust variance estimation, clustering the study subjects in families. Association between PRS and the tumor estrogen receptor (ER) expression was assessed with logistic regression after categorizing the study subjects into quintiles of the PRS distribution. Association between PRS and age at diagnosis was examined by linear regression. Pearson’s r was used as a measure of correlation in comparisons between PRS and BOADICEA scores.
Results
Association between PRS and case–control status
The combined effect of 75 variants with confirmed associations with breast cancer risk was captured by constructing a PRS for each individual. The PRS was associated with the increased risk of breast cancer in unselected patient series and within breast cancer families (Table 1, Supplementary Fig. 1). When healthy population controls were used as a reference group, the odds ratio (OR) for association between the PRS and breast cancer risk was lower for sporadic cases (OR: 1.41, 95 % CI [1.30–1.54]) than for cases with positive family history, but did not differ between index cases of “small” (OR: 1.85 [1.63–2.11]) and “large” families (OR: 1.81 [1.59–2.06]). Similarly, when comparing affected women from the 52 breast cancer families to healthy population controls, the PRS was associated with the disease risk (OR: 1.82 [1.55–2.13]), but no difference was seen by the number of affected first-degree relatives (Table 1, Supplementary Fig. 1c). The PRS was significantly higher among healthy women in the 52 breast cancer families than among population controls (OR: 1.29 [1.12–1.48]), consistent with an association between PRS and positive family history of breast cancer (Table 1, Supplementary Fig. 1b).
Table 1.
Association between the PRS and breast cancer risk in case–control and breast cancer family datasets
| Sample group | N | PRS | p value | |
|---|---|---|---|---|
| OR [95 % CI] | ||||
| Case–control dataset | Healthy population controls | 1269 | Reference | |
| All BC cases | 1689 | 1.56 [1.45–1.68] | 9.2E−31 | |
| Unselected breast cancer cases | 1299 | 1.49 [1.38–1.62] | 3.5E−23 | |
| Sporadic breast cancer cases | 1020 | 1.41 [1.30–1.54] | 5.1E−16 | |
| Index cases from small families | 334 | 1.85 [1.63–2.11] | 8.8E−21 | |
| Index cases from large families | 305 | 1.81 [1.59–2.06] | 2.0E−19 | |
| 52 breast cancer families | Affected family members | 181 | 1.82 [1.55–2.13] | 1.8E−13 |
| Affected family members with no affected first-degree relatives | 50 | 1.62 [1.22–2.14] | 7.7E−4 | |
| Affected family members with one affected first-degree relative | 58 | 1.97 [1.52–2.57] | 4.0E−7 | |
| Affected family members with two affected first-degree relatives | 47 | 1.74 [1.30–2.33] | 1.8E−4 | |
| Affected family members with three or more affected first-degree relatives | 26 | 2.05 [1.40–3.02] | 2.5E−4 | |
| Healthy family members (52 breast cancer families) vs. healthy population controlsa | 246 vs. 1269 | 1.29 [1.12–1.48] | 3.4E−4 | |
aTest for association between PRS and positive family history of breast cancer comparing healthy women from the breast cancer families to healthy population controls
PRS as a risk predictor in breast cancer families
The correlation between PRS and BOADICEA score was 0.15 in affected and 0.0099 in healthy women. The association between the PRS and breast cancer risk was investigated in 427 women from 52 breast cancer families. Healthy female family members were considered as the reference group in this analysis. The OR for association between the PRS and breast cancer in these families was 1.55 [1.26–1.91] (P: 3.3E−5), adjusting for age and the family history (BOADICEA score). When the model was further adjusted for the moderate-penetrance mutations, the PRS OR was 1.59 [1.28–1.98] (P: 2.9E−5, Supplementary Table 4).
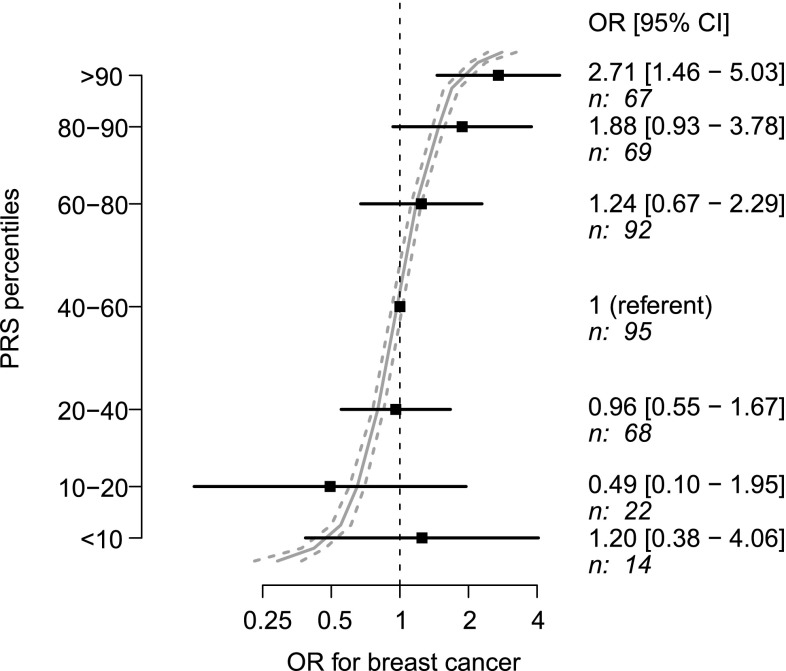
Previously, the relevance of the PRS in risk prediction has been evaluated by deriving the relative and absolute risk estimates for women at specific percentiles of the PRS distribution [5]. We also assigned the affected and healthy women from the breast cancer families into centiles of the PRS distribution based on the healthy population controls. The ORs for breast cancer estimated in the 52 families were similar in all categories to those obtained in the BCAC dataset of population-based case–control studies [5], except for the lowest ten percent, where the number of study subjects was quite low and the confidence interval included the published estimate (Fig. 1). The OR for women in the highest ten percent was 2.71 [1.46–5.03] (P = 0.0015).
Fig. 1.
Estimated effect sizes (odds ratios: OR with confidence intervals: CI) by percentile of the PRS in the 52 breast cancer families
Estrogen receptor status and age at diagnosis
Among cases, lower PRS-values were associated with higher proportion of ER-negative disease in both nonfamilial (P = 0.00082) and familial index cases (P = 0.023), but this trend was not observed within breast cancer families (P = 0.22)(Supplementary Table 5). No significant association between PRS and age at diagnosis was seen in our case–control or breast cancer family dataset.
Discussion
In this paper, we have investigated the potential of the PRS to improve risk stratification in the setting of familial breast cancer. The PRS we constructed used information from 75 variants, almost all breast cancer susceptibility variants known to date. We tested for association of this PRS with breast cancer risk among different groups according to their family history of breast cancer. The PRS was on average higher in patients with family history of breast cancer in a first-degree relative than in sporadic cases in the case–control dataset (Table 1). We neither saw a difference in the PRS between the “large” and “small” families, nor an increasing trend with the number of affected first-degree relatives within the family dataset. Epidemiological studies have reported life-time risk for women with two affected first-degree relatives to be higher than for women with only one affected relative (21.1 vs. 13.3 %) [1]. This difference was not reflected in a change in PRS, although power for this comparison was limited in our dataset (Table 1, Supplementary Fig. 1). The PRS was significantly associated with increased risk of breast cancer within the breast cancer families, when comparing affected to healthy women. Furthermore, the PRS was higher among the healthy women of the breast cancer families compared with healthy controls from the population (Table 1, Supplementary Fig. 1), supporting the notion that common genetic risk variants cluster in breast cancer families.
Our 52 breast cancer families comprised 427 genotyped and ~4000 nongenotyped individuals. The proportion of affected women in the breast cancer families ranged between 6 and 67 % per family (median 22 %) (Supplementary Table 2). Two-thirds of the healthy women of the dataset were first-degree relatives of the breast cancer cases, and the remainder second- or third-degree relatives. The BOADICEA risk prediction algorithm is able to capture complex family structure, including information on more distant relatives as well as information on ages of diagnosis or interview, and is more informative than simpler measures of family history. We therefore calculated a ‘BOADICEA score’ as an estimate of the mean polygenotype for each individual, given their family history. The correlation between our BOADICEA score and the PRS in affected women from the 52 breast cancer families was 0.15, consistent with the relatively small fraction of the heritability explained by these variants. As expected, the correlation among the healthy women was lower (Pearson’s r = 0.0099). These estimates are somewhat higher than that reported recently between PRS and BOADICEA risk prediction output (0.01 [−0.05–0.07]) by an Australian study [10]. This could be explained partly by the different study designs, as the Australian study included a large number of sporadic cases, and any pedigree data were collected by interview, whereas we collected family data systematically from population and cancer registries. Furthermore, they included in the correlation both breast cancer cases and healthy controls, which may have masked an existing correlation in cases.
The PRS was significantly associated with breast cancer risk in a logistic regression model adjusted for age and the BOADICEA score (OR: 1.55 [1.26–1.91]). The magnitude of the effect size associated with the PRS was consistent with the estimate made in the unselected series of the case–control dataset (OR: 1.49 [1.38–1.62]) and with the estimate reported in a recent population-based study (OR: 1.55 [1.52–1.58]) [5], and provides further support for using the PRS in risk prediction. Furthermore, when the model was adjusted for the moderate-penetrance mutations in CHEK2, PALB2, and FANCM, the OR associated with the PRS was very similar (1.59 [1.28–1.98]) supporting a multiplicative mode of interaction between the low- and moderate-penetrance genetic variants. This is been demonstrated explicitly for CHEK2:c.1100delC and the PRS (Muranen et al. in review), but further analyses in larger datasets would be needed to evaluate such interactions for the FANCM and PALB2 variants.
A recent study examined the utility of the PRS for risk prediction in breast cancer families in the BCFR [27]; our results are broadly in agreement with the conclusions of this prospective study. This study used only 24 variants rather than the full complement of 75 (or 77 as in [5]), so we would have expected to see a larger effect size in our study. However, we were not able to directly compare the effect sizes reported, as the BCFR-study was prospective in design, used Cox regression for modeling, and the effect of the PRS was studied in the context of 10-year BOADICEA risk estimates. In addition, individuals with moderate-penetrance mutations were excluded from their analyses, while these individuals were included in our analyses.
Our results on subtype-specific associations are consistent with previous observations that most common genetic variants are more strongly associated with ER-positive disease, while fewer ER-negative specific variants have been identified. Although pathology information in the familial study was limited, we noted a higher proportion of ER-negative cancers among women with the lowest PRS in this study. Noteworthy, none of these cancers were from carriers of PALB2 or FANCM mutations, which have been previously associated with ER-negative disease [19, 20]. It would be of interest to study the pathology of breast cancers within families and evaluate any correlations in the context of the PRS.
The effect sizes associated with the PRS can be used to derive estimates of the absolute risk of breast cancer according to PRS [5]. In women with a family history of breast cancer, the baseline absolute risk of developing breast cancer is higher. Our observations indicate that the relative risks associated with the PRS are similar to those in the general population, consistent with a model in which the PRS multiplies the effects of other familial risk factors. Hence, the effect of the PRS on the absolute risk of disease will be much greater than in the general population. For example, if the pedigree-based familial risk for a woman was about 17 % [9], and her PRS was in the highest 20 centile, the combined risk would be 30.9 %, moving her from intermediate- to high-risk category according to UK NICE guidelines [9]. By comparison, women with no data on family history would have to be in the top centile of the PRS to have the same absolute risk (~30 %). We did not find support for the protective effect associated with low PRS-values (Fig. 1). This may reflect low power, as few women have very low PRS in the breast cancer families. However, it might be explained by the presence of yet-unidentified moderate/high-risk genetic or shared environmental factors [28].
Conclusions
Our results suggest that it would be valuable to combine the PRS with pedigree-based risk estimation in the context of familial disease.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank all the individuals taking part in this study; Drs. Kirsimari Aaltonen, Tuomas Heikkinen, Päivi Heikkilä and Karl von Smitten for their help with the patient samples and data; and research nurse Irja Erkkilä for the assistance in the data collection and management. This work was supported by the Helsinki University Hospital Research Fund, the Finnish Cancer Society, the Nordic Cancer Union, the Academy of Finland [266528] and the Sigrid Juselius Foundation. The work of TAM has been supported by Finnish Cultural Foundation and Orion-Farmos Research Foundation. Genotyping of the iCOGS array, and the Breast Cancer Association Consortium, was supported by Cancer Research UK (C1287/A10118, C1287/A12014, C1287/A10710) and by the European Community's Seventh Framework Programme under grant agreement n° 223175 (HEALTH-F2-2009-223175) (COGS). NM was supported by Cancer Research UK (C1287/ C1287/A16563) and the PERSPECTIVE project, funded from the Government of Canada through Genome Canada and the Canadian Institutes of Health Research. AL was supported by Cancer Research UK.
Abbreviations
- CI
Confidence interval
- ER
Estrogen receptor
- OR
Odds ratio
- PRS
Polygenic risk score
Compliance with ethical standards
The study was carried out with permissions from Helsinki University Hospital Ethics Committee and the Ministry of Social Affairs and Health, Finland. Written informed consent was obtained from all participants.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Taru A. Muranen and Nasim Mavaddat have been contributed equally to this work.
References
- 1.Collaborative Group on Hormonal Factors in Breast Cancer Familial breast cancer: collaborative reanalysis of individual data from 52 epidemiological studies including 58,209 women with breast cancer and 101,986 women without the disease. Lancet. 2001;358:1389–1399. doi: 10.1016/S0140-6736(01)06524-2. [DOI] [PubMed] [Google Scholar]
- 2.Metcalfe KA, Finch A, Poll A, Horsman D, Kim-Sing C, Scott J, Royer R, Sun P, Narod SA. Breast cancer risks in women with a family history of breast or ovarian cancer who have tested negative for a BRCA1 or BRCA2 mutation. Br J Cancer. 2009;100:421–425. doi: 10.1038/sj.bjc.6604830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antoniou AC, Easton DF. Models of genetic susceptibility to breast cancer. Oncogene. 2006;25:5898–5905. doi: 10.1038/sj.onc.1209879. [DOI] [PubMed] [Google Scholar]
- 4.Antoniou AC, Pharoah PD, McMullan G, Day NE, Stratton MR, Peto J, Ponder BJ, Easton DF. A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Br J Cancer. 2002;86:76–83. doi: 10.1038/sj.bjc.6600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mavaddat N, Pharoah PD, Michailidou K, et al. Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst. 2015 doi: 10.1093/jnci/djv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michailidou K, Hall P, Gonzalez-Neira A, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Closas M, Couch FJ, Lindstrom S, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45:392–398. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.French JD, Ghoussaini M, Edwards SL, et al. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am J Hum Genet. 2013 doi: 10.1016/j.ajhg.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Collaborating Centre for Cancer (UK) (2013). DOI:NBK247567 [bookaccession] [PubMed]
- 10.Dite GS, MacInnis RJ, Bickerstaffe A, et al. Breast cancer risk prediction using clinical models and 77 independent risk-associated snps for women aged under 50 years: Australian Breast Cancer Family Registry. Cancer Epidemiol Biomarkers Prev. 2015;25:359–365. doi: 10.1158/1055-9965.EPI-15-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vachon CM, Pankratz VS, Scott CG, et al. The contributions of breast density and common genetic variation to breast cancer risk. J Natl Cancer Inst. 2015 doi: 10.1093/jnci/dju397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Syrjakoski K, Vahteristo P, Eerola H, Tamminen A, Kivinummi K, Sarantaus L, Holli K, Blomqvist C, Kallioniemi OP, Kainu T, Nevanlinna H. Population-based study of BRCA1 and BRCA2 mutations in 1035 unselected Finnish breast cancer patients. J Natl Cancer Inst. 2000;92:1529–1531. doi: 10.1093/jnci/92.18.1529. [DOI] [PubMed] [Google Scholar]
- 13.Kilpivaara O, Bartkova J, Eerola H, et al. Correlation of CHEK2 protein expression and c.1100delC mutation status with tumor characteristics among unselected breast cancer patients. Int J Cancer. 2005;113:575–580. doi: 10.1002/ijc.20638. [DOI] [PubMed] [Google Scholar]
- 14.Fagerholm R, Hofstetter B, Tommiska J, et al. NAD(P)H:quinone oxidoreductase 1 NQO1*2 genotype (P187S) is a strong prognostic and predictive factor in breast cancer. Nat Genet. 2008;40:844–853. doi: 10.1038/ng.155. [DOI] [PubMed] [Google Scholar]
- 15.Vahteristo P, Bartkova J, Eerola H, et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet. 2002;71:432–438. doi: 10.1086/341943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eerola H, Blomqvist C, Pukkala E, Pyrhonen S, Nevanlinna H. Familial breast cancer in southern Finland: how prevalent are breast cancer families and can we trust the family history reported by patients? Eur J Cancer. 2000;36:1143–1148. doi: 10.1016/S0959-8049(00)00093-9. [DOI] [PubMed] [Google Scholar]
- 17.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weischer M, Nordestgaard BG, Pharoah P, et al. CHEK2*1100delC heterozygosity in women with breast cancer associated with early death, breast cancer-specific death, and increased risk of a second breast cancer. J Clin Oncol. 2012;30:4308–4316. doi: 10.1200/JCO.2012.42.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiiski JI, Pelttari LM, Khan S, et al. Exome sequencing identifies FANCM as a susceptibility gene for triple-negative breast cancer. Proc Natl Acad Sci USA. 2014;111:15172–15177. doi: 10.1073/pnas.1407909111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heikkinen T, Karkkainen H, Aaltonen K, Milne RL, Heikkila P, Aittomaki K, Blomqvist C, Nevanlinna H. The breast cancer susceptibility mutation PALB2 1592delT is associated with an aggressive tumor phenotype. Clin Cancer Res. 2009;15:3214–3222. doi: 10.1158/1078-0432.CCR-08-3128. [DOI] [PubMed] [Google Scholar]
- 21.R Development Core Team . R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2012. [Google Scholar]
- 22.Graham N, Mahmood A, Hagströmer B (2015) multiwayvcov: Multi-way Standard Error Clustering, R package version 1.2.2
- 23.Zeileis A, Hothorn T. Diagnostic checking in regression relationships. R News. 2002;2:7–10. [Google Scholar]
- 24.Lee AJ, Cunningham AP, Kuchenbaecker KB, Mavaddat N, Easton DF, Antoniou AC. BOADICEA breast cancer risk prediction model: updates to cancer incidences, tumour pathology and web interface. Br J Cancer. 2014;110:535–545. doi: 10.1038/bjc.2013.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lange K, Weeks D, Boehnke M. Programs for pedigree analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol. 1988;5:471–472. doi: 10.1002/gepi.1370050611. [DOI] [PubMed] [Google Scholar]
- 26.Lange K. An approximate model of polygenic inheritance. Genetics. 1997;147:1423–1430. doi: 10.1093/genetics/147.3.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Feng B, Miron A, et al. Breast cancer risk prediction using a polygenic risk score in the familial setting: a prospective study from the Breast Cancer Family Registry and kConFab. Genet Med. 2016 doi: 10.1038/gim.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.