Abstract
Human infections with Salmonella enterica subspecies enterica serovar Senftenberg are often associated with exposure to poultry flocks, farm environments, or contaminated food. The recent emergence of multidrug-resistant isolates has raised public health concerns. In this study, comparative genomics and phenotypic analysis were used to characterize 14 Salmonella Senftenberg clinical isolates recovered from multiple outbreaks in Shenzhen and Shanghai, China, between 2002 and 2011. Single-nucleotide polymorphism analyses identified two phylogenetically distinct clades of S. Senftenberg, designated SC1 and SC2, harboring variations in Salmonella pathogenicity island 1 (SPI-1) and SPI-2 and exhibiting distinct biochemical and phenotypic signatures. Although the two variants shared the same serotype, the SC2 isolates of sequence type 14 (ST14) harbored intact SPI-1 and -2 and hence were characterized by possessing efficient invasion capabilities. In contrast, the SC1 isolates had structural deletion patterns in both SPI-1 and -2 that correlated with an impaired capacity to invade cultured human cells and also the year of their isolation. These atypical SC1 isolates also lacked the capacity to produce hydrogen sulfide. These findings highlight the emergence of atypical Salmonella Senftenberg variants in China and provide genetic validation that variants lacking SPI-1 and regions of SPI-2, which leads to impaired invasion capacity, can still cause clinical disease. These data have identified an emerging public health concern and highlight the need to strengthen surveillance to detect the prevalence and transmission of nontyphoidal Salmonella species.
INTRODUCTION
Nontyphoidal Salmonella (NTS) infections result in significant morbidity in both developed and developing countries, with ∼90 million gastroenteritis cases leading to ∼155,000 human deaths each year (1). In a recent study, the number of the deaths associated with NTS infection have been estimated at ∼680,000 in 2010 (2). In economically developed countries, NTS infection remains one of the leading causes of death among foodborne bacterial infections, with outbreaks common (3, 4). Traditionally, Salmonella isolates are typed into serovars that link O and H antigens with taxonomic assignments, host adaptation, and disease potential. However, and due to heterogeneity within serologically assigned serovars (5–7), multilocus sequence typing (MLST) has been recommended as an accurate typing method that could further recognize the evolutionary implications (7). A key step in the pathogenesis of Salmonella infections involves the invasion of eukaryotic cells and bacterial survival within phagocytes. These phenotypes are associated with the expression of virulence-associated determinants predominantly gained by the acquisition of horizontally transferred genetic elements inserted into the Salmonella genome that are collectively known as Salmonella pathogenicity islands (SPIs) (8). To date, 23 different SPIs have been identified (9). Two distinct type III secretion systems (T3SSs) encoded by SPI-1 and -2 are considered central to the pathogenicity of NTS. SPI-1 mediates the invasion of nonphagocytic cells and is necessary for enteropathogenesis. SPI-2 contributes to intracellular survival and replication. However, experimental evidence indicates that Salmonella can cause infection in the absence of an SPI-1 T3SS (10–13), as NTS harboring significant deletions within SPI-1 is capable of causing disease (14). In particular, strains of Salmonella enterica subsp. enterica serovar Senftenberg that lack SPI-1 have been linked to an outbreak of NTS in Guangdong, China, in 2002 (14). Moreover, Salmonella Senftenberg is a diverse serovar with isolates that have been assigned different sequence types (STs) that clustered into four distinct eBURST groups (eBGs), with some of these eBGs having been reported only from China (7).
S. Senftenberg isolates are often associated with colonization of hatcheries and are gradually eliminated during animal rearing. However, S. Senftenberg isolates that are able to persist throughout the rearing period into adulthood have recently emerged (15), with these same isolates displaying resistance to desiccation in soil (16). Clinically, infections caused by S. Senftenberg range from asymptomatic (17, 18) to severe infections resulting in large outbreaks (19, 20). The vehicles linked to outbreaks vary, with basil, fennel seeds (21), meat, and shellfish (22) implicated in different regions. Antimicrobial-resistant isolates of S. Senftenberg are associated mainly with animal sources (23). Recently, antimicrobial-resistant human isolates have been reported; for example, extremely drug-resistant strains of S. Senftenberg have been isolated from patients in Zambia (24). Moreover, illnesses caused by metallo-β-lactamase-producing S. Senftenberg that are associated with international travel have been identified recently in the United States (25). In addition, atypical S. Senftenberg isolates that lack the ability to produce hydrogen sulfide (H2S) have been identified in China (26). Thus, antimicrobial-resistant isolates of S. Senftenberg are emerging globally, and this, coupled with the persistence of S. Senftenberg in animals reared for food production and with an atypical biochemical signature, has potentially far-reaching public health implications.
Here, we explore the genetic and phenotypic differences between 14 clinical isolates of S. Senftenberg from China by using whole-genome sequencing and phenotyping. The studied isolates originated from multiple outbreaks that occurred in different locations between the years 2002 and 2011, with the majority of isolates lacking SPI-1 and characterized by various pulsed-field gel electrophoresis (PFGE) types. The analyses revealed the circulation of two distinct clades of S. Senftenberg in Shenzhen and Shanghai, China, that are distinguished by variations in their genomic architectures and phenotypes.
MATERIALS AND METHODS
Ethics statement.
All samples were collected for diagnostic purposes only. The ethics committee of the Shenzhen Center for Disease Control and Prevention approved the protocol (ethics committee approval number 2014009). The S. Senftenberg isolates described in this study were originally recovered during laboratory-based sentinel surveillance for diarrheal disease and outbreak detection of foodborne disease in Shenzhen City, Guangdong Province, and the Shanghai Municipality in China. The isolates were not collected for experimental purposes, and all the clinical data were anonymized and unlinked. Therefore, informed consent was not necessary.
Characterization of S. Senftenberg isolates.
Briefly, fecal specimens were collected from outpatients with acute diarrhea or patients from suspected foodborne disease outbreaks; aliquots of stool samples were enriched in different enrichment broths as previously described (14). For each sample, a minimum of five typical colonies were selected and subjected to biochemical and serological tests for identification. The identified isolates were confirmed using API 20E biochemical test kits (bioMérieux SA, Marcy l'Etoile, France). Total nucleic acids were extracted from the stool samples for the detection of pathogens commonly associated with foodborne infections. Screening assessed the presence of Salmonella spp., Shigella spp., Vibrio spp., Staphylococcus aureus, Escherichia coli O157:H7, enterotoxigenic Escherichia coli (ETEC), enteropathogenic Escherichia coli (EPEC), enteroinvasive Escherichia coli (EIEC), enterohemorrhagic Escherichia coli (EHEC), Bacillus cereus, group A Streptococcus, Listeria monocytogenes, rotaviruses, noroviruses, adenoviruses, astroviruses, and sapoviruses (details of the primers and probes used in the screening are given in Table S1 in the supplemental material).
Following the discovery of SPI-1-negative Salmonella Senftenberg isolates in 2002, all S. Senftenberg isolates identified between 2002 and 2011 have been subjected to PCR testing for the SPI-1 region (14). Fourteen isolates were found to be SPI-1 negative. These isolates together with six SPI-1-positive isolates underwent PFGE typing and invasion assays as previously described (27). Briefly, Salmonella cells grown overnight at 37°C were diluted in LB and subcultured for 3 h. The cell pellets were recovered by centrifugation and resuspended in phosphate-buffered saline (PBS), and the cells were counted. Bacterial inocula were added directly to human cervical adenocarcinoma cells (HeLa cells; ATCC CCL2) and incubated for 10 min at 37°C in a 5% CO2 incubator. Extracellular bacteria were washed with PBS, and the HeLa cells were incubated in growth medium supplemented with gentamicin (50 μg/ml). The infected HeLa cells were solubilized in 1 ml of 1% Triton X-100–0.1% SDS in PBS for 5 min at room temperature. The solubilized cells were diluted in PBS and plated on LB agar to recover and count the invading bacteria. Salmonella serovar Typhimurium SL1344 was used as a positive control, and a mutant variant of SL1344 that lacked invA was used as a negative control. The experiment was repeated three times.
All antimicrobial susceptibility tests were performed using the disk diffusion method. Antibiotic discs were purchased from Oxoid (Oxoid Ltd., England). A panel of 18 antimicrobial agents covering 10 Clinical and Laboratory Standards Institute (CLSI) classes of antibiotics was used along with the zone diameter to determine if the isolates were resistant: ampicillin (10 μg, ≤13 mm), tetracycline (30 μg, ≤11 mm), streptomycin (10 μg, ≤11 mm), nalidixic acid (30 μg, ≤13 mm) trimethoprim (5 μg, ≤10 mm), trimethoprim-sulfamethoxazole (25 μg, ≤10 mm), gentamicin (10 μg, ≤12 mm), amoxicillin-clavulanic acid (30 μg, ≤13 mm), cephalothin (30 μg, ≤14 mm), ceftriaxone (30 μg, ≤19 mm), cefepime (30 μg, ≤19 mm), chloramphenicol (30 μg, ≤12 mm), kanamycin (30 μg, ≤13 mm), ciprofloxacin (5 μg, ≤15 mm), levofloxacin (5 μg, ≤13 mm), ceftazidime (30 μg, ≤17 mm), amikacin (30 μg, ≤14 mm), and cefoxitin (30 μg, ≤14 mm). Overnight cultures were spread on Mueller-Hinton (MH) agar, and the antibiotic discs were placed. The plates were incubated at 36°C for 24 h, and the breakpoints used for sensitive, intermediate, and resistant were defined by the Clinical and Laboratory Standards Institute (2012). E. coli ATCC 25922 was used as a quality control strain for antimicrobial susceptibility testing.
DNA extraction and genome sequencing.
All SPI-1-negative S. Senftenberg isolates together with one SPI-1-positive isolate that were representative of the PFGE patterns present in Guangdong Province and the Shanghai Municipality were selected for whole-genome sequencing. Isolates were cultured overnight in Luria broth medium at 37°C with aeration (vigorous shaking at 200 rpm). DNA was extracted using a Promega Wizard genomic DNA purification kit (Promega, Madison, WI, USA). Multiplexed paired-end Illumina libraries with 200-bp inserts were prepared for the S. Senftenberg isolates using the Nextera DNA library prep kit (Illumina, Inc.). The libraries were sequenced on an Illumina HiSeq2000 platform at the KAUST Bioscience Core Lab.
Construction of an SNP-based phylogenetic tree.
The 100-bp paired-end reads were mapped against 2,882 genes recently identified as the core genome of Salmonella enterica (28). SMALT (https://sanger.ac.uk/resources/software/smalt) was used to obtain the whole-genome alignments for all S. Senftenberg isolates sequenced in this study and publicly available reference genomes (EMBL accession numbers are given in Table S2 in the supplemental material). Single-nucleotide polymorphisms (SNPs) that had a quality score of more than 30 (Q30) and that were present in at least 75% of the mapped reads were concatenated and used to construct a maximum-likelihood phylogeny using the default settings of RAxML v.7.0.4 (29) in which the Salmonella core genome was used an outgroup to root the tree.
Construction of a decomposition network.
The SNPs that were identified in comparison to the S. enterica core genome when constructing the phylogenetic tree were used to construct a binary matrix. In the matrix, if an SNP was present in a locus, it was recorded as “0”; if an SNP was absent, it was recorded as “1.” The matrix was then analyzed using the SplitsTree software version 4.12.6 (30) using the binary −2 splits option to generate the split decomposition network.
Genome assembly and comparative genomics.
In order to generate a multicontig draft genome for each of the 14 S. Senftenberg isolates, trimmed paired-end Illumina reads (>Q30) were assembled de novo using Velvet v0.7.03 (31). The parameters were optimized to give the best kmer size and at least 20× coverage of each kmer. S. Typhimurium strain SL1344 (FQ312003) was used as a reference to order the contigs using ABACAS (32). Twelve iterations of IMAGE were applied to close the gaps between the ordered contigs and improve the assembled genomes (33). The assembled genomes were annotated using Prokka (34). The comparisons between the draft genome assemblies of S. Senftenberg isolates sequenced in this study and the publicly available Salmonella reference genomes were performed using Mauve (35) with default parameters. Comparisons between individual genomes were performed using TBLASTX (36) and were viewed in the Artemis Comparison Tool (ACT) for manual comparison of the genomes (37). Regions of difference (RODs) were defined as insertions or replacements in the genomes of any of the studied S. Senftenberg isolates in comparison to S. Typhimurium strain SL1344.
MLST.
Multilocus sequence typing (MLST) sequence types were extracted from whole-genome data using the publicly available MLST server (https://cge.cbs.dtu.dk//services/MLST/) (38). STs were clustered into eBURST groups (eBGs) using software available on the MLST website (http://mlst.warwick.ac.uk/mlst/).
OrthoMCL and gene clustering.
All-versus-all BLAST (36) comparisons were generated for the predicted proteomes of S. Senftenberg isolates sequenced in this study along with the published S. enterica reference proteomes. OrthoMCL v2.0 (39) was used to generate the orthologous clusters with an inflation parameter, I, of 1.0. The protein families were checked manually through BLASTP with a cutoff of 1e−3 and a percentage identity greater than 50%. The hierarchical clustering (Euclidean distance, Ward method) and visualization were generated through the gplots package in R (http://www.r-project.org/).
Nucleotide sequence accession numbers.
All the sequencing data have been submitted to the European Nucleotide Archive (ENA) under accession numbers ERS626496 (isolate C02013), ERS626497 (C02014), ERS626498 (S09007), ERS626499 (S09008), ERS626500 (S09009), ERS626501 (S09010), ERS626502 (S09011), ERS626503 (S09012), ERS626504 (S09014), ERS626505 (S09015), ERS626506 (S09016), ERS626507 (S09017), ERS626508 (S10078), and ERS626509 (S11192).
RESULTS
Characterization of S. Senftenberg isolates.
The S. Senftenberg isolates characterized in this study were recovered from stool samples collected between 2002 and 2011 from patients (age range, 23 to 67 years) suffering from severe diarrheal infection and residing in Shenzhen or Shanghai, China (Table 1). In addition, the samples were screened at the molecular level for a panel of bacterial and viral agents associated with diarrhea. Apart from S. Senftenberg, no other pathogens were identified. All S. Senftenberg isolates were sensitive to the antimicrobial panel used in the study and exhibited only intermediate resistance to streptomycin.
TABLE 1.
Characterization of Salmonella Senftenberg isolates used in the study
| Isolate | Epidemiological data |
Clinical data |
Typing |
Invasion assay resultsa | |||||
|---|---|---|---|---|---|---|---|---|---|
| Location | Yr | Gender | Age (yr) | Symptomsb | MLST | eBG | Phenotypec | ||
| C02013 | Luohu, Shenzhen | 2002 | M | Fever, severe diarrhea, nausea, vomiting | ST217 | 30 | H2S− | 0.0049 ± 0.00003 | |
| C02014 | Luohu, Shenzhen | 2002 | M | Fever, severe diarrhea, nausea, vomiting | ST217 | 30 | H2S− | 0.176 ± 0.0225 | |
| S09007 | Changning,Shanghai | 2006 | F | 56 | Severe diarrhea, vomiting | ST1751 | 30 | H2S− | 0.0053 ± 0.0014 |
| S09008 | Changning,Shanghai | 2006 | M | 43 | Fever, severe diarrhea, vomiting | ST1751 | 30 | H2S− | 0.0046 ± 0.0006 |
| S09009 | Changning,Shanghai | 2006 | M | 38 | Severe diarrhea | ST1751 | 30 | H2S− | 0.0047 ± 0.0003 |
| S09010 | Changning,Shanghai | 2006 | F | 45 | Severe diarrhea, vomiting | ST1751 | 30 | H2S− | 0.0064 ± 0.0012 |
| S09011 | Changning,Shanghai | 2006 | M | 30 | Fever, severe diarrhea, vomiting | ST1751 | 30 | H2S− | 0.0065 ± 0.0011 |
| S09012 | Changning,Shanghai | 2006 | M | 67 | Severe diarrhea, vomiting | ST1751 | 30 | H2S− | 0.0070 ± 0.0020 |
| S09014 | Changning,Shanghai | 2006 | M | 54 | Severe diarrhea | ST1751 | 30 | H2S− | 0.0115 ± 0.0030 |
| S09015 | Changning,Shanghai | 2006 | M | 48 | Severe diarrhea | ST1751 | 30 | H2S− | 0.0099 ± 0.0039 |
| S09016 | Changning,Shanghai | 2006 | F | 46 | Fever, severe diarrhea | ST1751 | 30 | H2S− | 0.0180 ± 0.0035 |
| S09017 | Changning,Shanghai | 2006 | F | 50 | Fever, severe diarrhea | ST1751 | 30 | H2S− | 0.0031 ± 0.0012 |
| S10078 | Longgang, Shenzhen | 2010 | M | 23 | Fever, severe diarrhea | ST14 | 55 | H2S+ | 1.55 ± 0.0565 |
| S11192 | Shajing, Shenzhen | 2011 | M | 30 | Fever, severe diarrhea | ST185 | 30 | H2S− | 1.66 ± 0.0587 |
Invasion is expressed as the ratio of the number of gentamicin-resistant bacteria to the total number of bacteria in the inoculum. Values are the average ± standard deviation (SD) of the results for three experiments. S. Typhimurium SL1344 was used as a positive control (4.89 ± 0.3927), and the invA-lacking S. Typhimurium SL1344 mutant was used as a negative control (0.3860 ± 0.0839). The original data were analyzed by using SPSS software (13.0 version).
Severe diarrhea was defined as diarrheal cases that required hospitalization for 2 to 3 days.
H2S−; atypical non-hydrogen sulfide-producing isolates, H2S+; hydrogen sulfide-producing isolates.
The ability of the 14 S. Senftenberg isolates to invade cultured human epithelial cells was assessed (Table 1). Generally, the S. Senftenberg isolates from China were less effective at invading cultured human epithelial cells than S. Typhimurium SL1344. However, the S. Senftenberg isolates S10078 and S11192 with intact SPI-1were significantly (P < 0.01) more effective in cell invasion than the other isolates tested in the study.
Phylogenomic analysis of S. Senftenberg isolates.
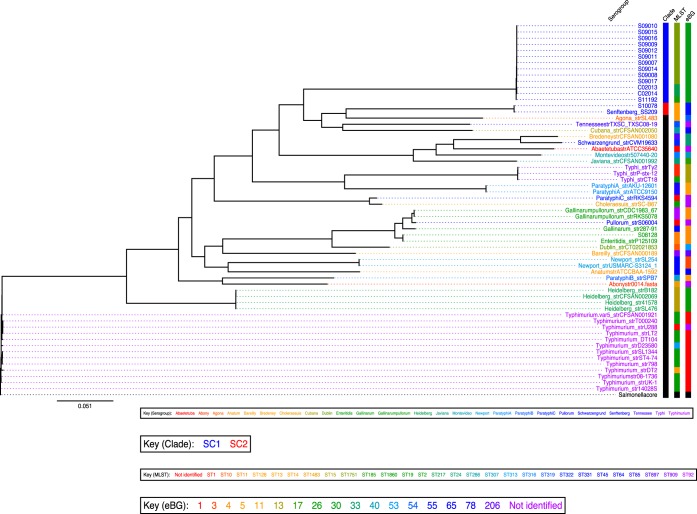
Initial PFGE analysis indicated that the 14 S. Senftenberg isolates represented six distinct PFGE patterns (see Fig. S1 in the supplemental material). Subsequently, whole-genome sequencing was applied to the S. Senftenberg isolates under study to assess the relationship between the isolates and to determine how they related phylogenetically to other NTS isolates. The S. Senftenberg sequences were compared to 44 S. enterica reference genomes available in the EMBL-EBI database, including the S. Senftenberg isolate SS209 (see Table S2 in the supplemental material), and they were also mapped against 2,882 genes recently defined as a core genome of S. enterica (28). The SNPs identified by sequencing were then used to construct a maximum-likelihood phylogenetic tree, in which the S. enterica core genome was used as an outgroup to root the tree (Fig. 1). S. Senftenberg isolates from Shenzhen and Shanghai clustered into two distinct clades, named here Senftenberg clades 1 (SC1) and 2 (SC2). All of the S. Senftenberg isolates from China, with the exception of S10078, grouped together into clade SC1. S10078 clustered in clade 2 with the poultry-persistent S. Senftenberg isolate SS209 (40) and formed a subbranch along with S. enterica serovar Agona strain SL483 (Fig. 1). SC1 isolates comprised three sequence types, ST185, ST217, and ST1751, that cluster together and form eBG30. Interestingly, Salmonella Senftenberg ST1751 has been reported only from China to date. In contrast, SC2 isolates included only ST14, which clusters within eBG55.
FIG 1.
An SNP-based maximum-likelihood phylogenetic tree of the S. enterica genomes. The phylogenetic tree shows the relation between S. Senftenberg isolates from China and the publicly available S. enterica reference genomes. The branch length corresponds to SNPs, and the scale given represents the number of substitutions per variable site. The Salmonella genomes are colored according to the serogroup. Color-coded information for each strain is shown on the right and includes phylogenetic clades, MLSTs, and eBGs. Black indicates not identified. SC1 isolates comprised three different STs (ST217, ST1751, and ST185) with a two-allele difference between ST1751 isolates and ST185 (S11192). Also, SC2 included isolates belonging to four different STs, i.e., ST14, ST13, ST78, and ST319, which belong to S. Senftenberg, S. Agona, S. Cubana, and S. Tennessee, respectively.
SNPs identified in the S. Senftenberg isolates sequenced in this study and the publicly available broiler chicken-persistent S. Senftenberg isolate SS209 were used to construct a split decomposition network based on the presence or absence of SNPs (see Fig. S2 in the supplemental material). Complementary to the phylogenetic tree, the split network helps to further dissect the differences between the sequences (30). A total of 12,695 SNPs (split 1; see Fig. S2) were found to distinguish between SC1 and SC2 S. Senftenberg isolates. However, only 30 SNPs (split 2; see Fig. S2) were unique to SC1 S. Senftenberg isolates (except isolate S11192). In addition, only 21 SNPs (split 3; see Fig. S2) were unique to the S. Senftenberg isolates from 2002 (C02013 and C02014), distinguishing them from the S. Senftenberg isolates recovered from the 2006 outbreak. A list of the SNPs for splits 2 and 3 and the potential impact on protein stability are shown in supplementary Tables S3 and S4, respectively.
Comparative genomic analysis of S. Senftenberg isolates.
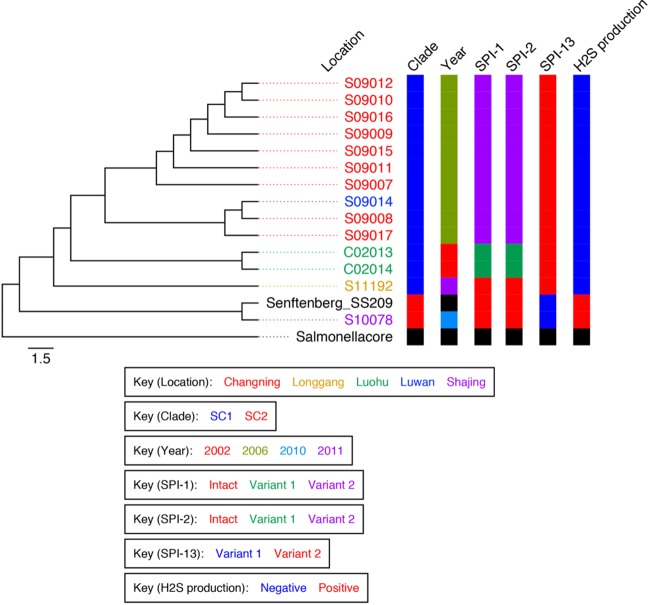
Structural variations among the key SPIs and sequence analyses correlated with the phylogenetic positions of these isolates on the SNP-based phylogenetic tree (Fig. 2). Clades 1 and 2 could be distinguished by clade-specific deletions in SPI-1, SPI-2, and SPI-13 (ROD 31; see Table S5 in the supplemental material).
FIG 2.
An SNP-based maximum-likelihood phylogram of S. Senftenberg. A zoom-in view of the phylogenetic tree of S. Senftenberg isolates from China and poultry-persistent isolate SS209 is shown. The scale given represents the substitutions per variable site. The colors of the isolate designations indicates the locations from which the isolates were recovered. Color-coded information for each isolate is shown on the right and includes phylogenetic clade, year of isolation, structural patterns of SPI-1, SPI-2, and SPI-13, and the biochemical feature. The SC1 clade includes the newly emerged atypical non-hydrogen sulfide (H2S)-producing isolates and, except for S11192, shared deletions in SPI-1 and SPI-2. The SC2 clade includes H2S-producing isolates and is characterized by intact SPI-1 and SPI-2.
SC2 S. Senftenberg isolates S10078 and SS209, together with S11192, harbored intact SPI-1 and SPI-2 regions similarly to the S. Typhimurium reference strain SL1344. In contrast, all of the SC1 S. Senftenberg isolates sequenced in this study except S11192 harbored deletions in both SPI-1 and SPI-2. However, the deletion pattern separated the isolates into two distinct subgroups. For SPI-1, S. Senftenberg isolates C02013 and C02014, recovered in 2002, were characterized by the deletion of 37 protein-coding genes (CDSs), beginning with locus SL1344_2846 (sprB) through to SL1344_2883. S. Senftenberg isolates from the 2006 outbreak in Shanghai shared identical deletions in 33 loci from SL1344_2850 (orgA) through to SL1344_2883 but retained SL1344_2846 through SL1344_2849 (see Fig. S3 in the supplemental material). Similarly, for SPI-2, the 2002 isolates (C02013 and C02014) shared identical deletions of 11 CDSs from SL1344_1328 to SL1344_1339, including the genes ssaD, ssaE, sseA, sseBa, sscA, sseC, sseD, sseF, sscB, sseF, and sseG, while the 2006 isolates shared deletions in only four CDSs, from SL1344_1325 through SL1344_1328 (ssrB, ssrA, ssaB, and ssaC).
Interestingly, all S. Senftenberg isolates sequenced in this study and isolate SS209 shared an insertion of ∼9,616 bp between yfbK and nuoN (ROD 23; see Fig. S4 in the supplemental material) that comprised one CDS encoding outer membrane protein autotransporter precursor IcsA. icsA shows identical homology with shdA in S. Cubana and S. Agona and hence may play a key role in intestinal colonization.
All isolates in both clades shared an intact T3SS autotransporter gene encoding the protein MisL that was previously identified as an extracellular matrix adhesin involved in intestinal colonization (41, 42). They also shared a CS54 island identical to the pattern seen in S. Cubana and S. Agona. All identified RODs are listed in Table S5 in the supplemental material.
Proteins associated with Salmonella Senftenberg isolates.
The presence and absence of genes encoding different protein families were determined across the studied S. Senftenberg isolates in comparison with the published Salmonella enterica reference genomes. The protein families that show patterns associated with S. Senftenberg were selected and are shown in Fig. 3. All S. Senftenberg isolates shared an O-antigen structure similar to that seen in S. Anatum (Fig. 3, profile A). All S. Senftenberg isolates are characterized by the deletion of the O-antigen loci rfbU, rfbV, rfbX, rfbJ, rfbH, rfbG, rfbF, and rfbI, with an insertion of 4,753 bases (ROD 65; see Fig. S5 in the supplemental material) consisting of two CDSs, one encoding a mannosyl transferase enzyme which catalyzes the assembly of a mannose sugar substituted into the O subunit and one encoding WZY proteins that are involved in O-antigen polymerization, which is the linkage between O-antigen repeat and the adhesion of branched sugar.
FIG 3.
Conservation of protein families across S. Senftenberg isolates. A heat map shows the presence (red) or absence (white) of different protein families across S. Senftenberg isolates along with the published S. enterica reference genomes. Protein clusters were generated using OrthoMCL 2.0 with inflation parameter 1 and checked manually using BLASTP. The annotation was added to the right of the heat map and is highlighted by color based on the function. Hypothetical proteins were included in the analysis if they were part of a conserved sequence block containing other proteins with functional annotation.
All S. Senftenberg isolates shared five CDSs (ROD 63; see Table S5 in the supplemental material) encoding arsenic resistance proteins (Fig. 3, profile B) that have been identified in only a few Salmonella serovars (Newport, Tennessee, Thompson, Agona, Montevideo, and Javiana).
All S. Senftenberg isolates are characterized by novel variants of CDSs (ROD 18; see Table S5 in the supplemental material) that are involved in sugar metabolism, membrane transport, and anaerobic growth (Fig. 3, profile C). Also, orthologs for the aphD gene encoding alkylhydroperoxidase AhpD and a CDS encoding a LysR family transcriptional regulator were identified only in S. Senftenberg, S. Cubana, and S. Tennessee. AhpD is an NADH-dependent peroxidase and peroxynitrite reductase system that has been identified as a key element of the Mycobacterium tuberculosis defense system against oxidative stress (43).
SC1-specific protein clusters associated with virulence and environmental persistence.
All of the SC1 S. Senftenberg isolates were characterized by novel variants of CDSs (ROD 26b; see Table S5 in the supplemental material) encoding proteins involved in carbohydrate transport and metabolism and metal homeostasis (Fig. 3, profile D).
The SC1 S. Senftenberg isolates are also characterized by variants of CDSs (ROD 31; see Table S5 in the supplemental material) that seem to be associated with bacterial persistence in nonhuman environments and share homology only with S. Cubana. These proteins are involved in the degradation of organic substances and tellurite resistance (Fig. 3, profile E).
All of the S. Senftenberg isolates also have a unique insertion at phoN (ROD 55; see Table S5 in the supplemental material). SC1 isolates were characterized by an insertion of ∼13,000 bases comprised of eight CDSs that encode fimbrial proteins (Fig. 3, profile F1). Orthologs of these genes were identified in S. Bredeney, S. Anatum, S. Montevideo, S. Schwarzengrund, S. Cubana, and S. Bareilly. SC2 S. Senftenberg isolates shared a different insertion of ∼9,938 bases that comprised nine CDSs encoding enzymes involved in transportation and phosphorylation of a broad range of carbohydrates and amino acids (Fig. 3, profile F2).
DISCUSSION
This study has demonstrated through phylogenomic analysis that S. Senftenberg isolates (with identical O-antigen loci and hence sharing a common serology pattern) are characterized by not one but at least two different bacterial clades that correlate with eBGs, have distinct structural variations in SPI-1 and -2, and exhibit differing capacities for invading human cultured cells and biochemical signatures. Interestingly, there is some evidence that the two S. Senftenberg clades cover different geographical regions. The atypical isolates were identified for the first time in Luohu (2002) before being detected in Changning (2006) and more recently in Shajing (2011). Moreover, SNP analysis indicates that the SPI-1-negative isolates (all SC1 isolates except S1192) uniquely share 30 SNPs that distinguish them from the other Salmonella Senftenberg isolates. The 2002 SPI-1-negative isolates also uniquely share a further 21 SNPs that distinguish them from the 2006 isolates.
The SC1 clade includes the newly emerged atypical non-hydrogen sulfide (H2S)-producing isolates characterized by significant deletions in SPI-1 and -2, while SC2 includes the poultry-persistent isolates of ST14. The atypical SC1 isolates, assigned to eBG30, comprise three distinct STs, with ST1751 having been reported only from China and being associated with recent outbreaks (26). Interestingly, the majority of Salmonella Senftenberg ST1751 isolates are non-H2S producers. However, both H2S producer and nonproducer strains have previously been isolated from the same patient (26). There is some evidence supporting the possibility that non-H2S-producing Salmonella isolates might have an increased ability to survive in the host intestine (44). The non-H2S producers lack the metabolic activity to utilize thiosulfate, which enhances anaerobic respiration through tetrathionate metabolism (45, 46).
The structural variation in SPI-1 might explain the differences in the invasion capacities observed between SC1 (with the exception of S11192) and SC2 isolates. The SPI-1-intact S. Senftenberg isolates S10078 and S11192 were significantly more efficient in cell invasion than the other isolates tested in the study.
Comparative genomic analysis indicated that both of the Senftenberg clades shared identical genomic regions encoding key T3SSs that have been associated with intestinal colonization and host persistence. For instance, all of the studied isolates shared a gain of an ∼9-kb DNA fragment that included the homolog of the autotransporter precursor shdA. This is in addition to identical copies of misL and the CS54 island. Although S. Senftenberg isolates were characterized by a genomic gain at phoN, both clades had distinct insertions that differ in size and CDS content and hence might lead to different pathogenic potentials. The gain within SC1 isolates included fimbria-encoded CDSs that could contribute to pathogenicity (host colonization), while SC2 gains included CDSs involved in carbohydrate and amino acid metabolic pathways. These findings, coupled with the observation that SPI-1-negative S. Senftenberg isolates contain additional copies of the host colonizer shdA, might offer a plausible explanation for how SPI-1-negative Senftenberg isolates can still be pathogenic and cause disease in humans.
Our findings provide comprehensive genetic evidence for the emergence of atypical variants of S. Senftenberg in China that probably evolved through independent deletion events in SPI-1 and -2 and are characterized by distinct pathogenic potentials. Both variants have the same metalloid-resistant phenotype through the acquisition of arsenic resistance operons that have been shown to confer resistance to arsenite and antimonite in E. coli (47). More importantly, arsenic-resistant bacteria are metabolically adapted to arsenic-induced osmotic and oxidative stress (48). These variants probably evolved in arsenic-contaminated environments, such as poultry hatcheries (as arsenic-based compounds are added to most chicken feed to promote growth and to kill parasites that cause diarrhea [49]) or contaminated groundwater (40). However, the SC1 isolates seem to have further capacities to persist in different environments through the acquisition of genes that have been associated with metabolic and degradation pathways. For instance, the SC1 isolates (all except S09008) share the genes encoding the YeeV-YeeU toxin-antitoxin system that has been identified in many prokaryotic genomes and is associated with the enhanced persistence of bacteria in response to environmental stress (50, 51). Recently, this toxin-antitoxin module has been shown to promote the colonization and survival of Salmonella in mouse mesenteric lymph nodes (52).
Our findings highlight the heterogeneity of Salmonella serovar Senftenberg and provide comprehensive genetic evidence for the emergence of atypical Senftenberg variants with distinct biochemical signatures and pathogenic potentials. These variants are difficult to characterize using serology and other molecular subtyping approaches, and therefore this study highlights the need to strengthen whole-genome surveillance to detect the prevalence and transmission of such atypical Salmonella variants.
Supplementary Material
ACKNOWLEDGMENTS
We thank the KAUST Bioscience Core Laboratory team for generating the whole-genome sequencing data.
This work was supported by the National Natural Science Foundation of China (grant no. 81071433 to Q.H.), the China National Science and Technology Major Projects Foundation (grant no. 2012ZX10004215-003-005 and 2016ZX10004215-005-005 to Q.H.), and the Shenzhen Municipal Science and Technology Program (CXZZ20140411105636301) and Faculty Baseline Research Funds (BRF) from KAUST (to A.P.).
The funders had no role in study design, data collection, and interpretation or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00052-16.
REFERENCES
- 1.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease ‘Burden of Illness’ Studies. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 2.Ao TT, Feasey NA, Gordon MA, Keddy KH, Angulo FJ, Crump JA. 2015. Global burden of invasive nontyphoidal Salmonella disease, 2010(1). Emerg Infect Dis doi: 10.3201/eid2106.140999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scharff RL. 2012. Economic burden from health losses due to foodborne illness in the United States. J Food Prot 75:123–131. doi: 10.4315/0362-028X.JFP-11-058. [DOI] [PubMed] [Google Scholar]
- 4.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rabsch W, Andrews HL, Kingsley RA, Prager R, Tschape H, Adams LG, Baumler AJ. 2002. Salmonella enterica serotype Typhimurium and its host-adapted variants. Infect Immun 70:2249–2255. doi: 10.1128/IAI.70.5.2249-2255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prager R, Rabsch W, Streckel W, Voigt W, Tietze E, Tschape H. 2003. Molecular properties of Salmonella enterica serotype paratyphi B distinguish between its systemic and its enteric pathovars. J Clin Microbiol 41:4270–4278. doi: 10.1128/JCM.41.9.4270-4278.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, Krauland MG, Hale JL, Harbottle H, Uesbeck A, Dougan G, Harrison LH, Brisse S, S. enterica MLST Study Group. 2012. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog 8:e1002776. doi: 10.1371/journal.ppat.1002776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groisman EA, Ochman H. 1996. Pathogenicity islands: bacterial evolution in quantum leaps. Cell 87:791–794. doi: 10.1016/S0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 9.Hayward MR, Jansen V, Woodward MJ. 2013. Comparative genomics of Salmonella enterica serovars Derby and Mbandaka, two prevalent serovars associated with different livestock species in the UK. BMC Genomics 14:365. doi: 10.1186/1471-2164-14-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aiastui A, Pucciarelli MG, Garcia-del Portillo F. 2010. Salmonella enterica serovar Typhimurium invades fibroblasts by multiple routes differing from the entry into epithelial cells. Infect Immun 78:2700–2713. doi: 10.1128/IAI.01389-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Radtke AL, Wilson JW, Sarker S, Nickerson CA. 2010. Analysis of interactions of Salmonella type three secretion mutants with 3-D intestinal epithelial cells. PLoS One 5:e15750. doi: 10.1371/journal.pone.0015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosselin M, Abed N, Virlogeux-Payant I, Bottreau E, Sizaret PY, Velge P, Wiedemann A. 2011. Heterogeneity of type III secretion system (T3SS)-1-independent entry mechanisms used by Salmonella Enteritidis to invade different cell types. Microbiology 157:839–847. doi: 10.1099/mic.0.044941-0. [DOI] [PubMed] [Google Scholar]
- 13.Rosselin M, Virlogeux-Payant I, Roy C, Bottreau E, Sizaret PY, Mijouin L, Germon P, Caron E, Velge P, Wiedemann A. 2010. Rck of Salmonella enterica, subspecies enterica serovar enteritidis, mediates zipper-like internalization. Cell Res 20:647–664. doi: 10.1038/cr.2010.45. [DOI] [PubMed] [Google Scholar]
- 14.Hu Q, Coburn B, Deng W, Li Y, Shi X, Lan Q, Wang B, Coombes BK, Finlay BB. 2008. Salmonella enterica serovar Senftenberg human clinical isolates lacking SPI-1. J Clin Microbiol 46:1330–1336. doi: 10.1128/JCM.01255-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boumart Z, Roche SM, Lalande F, Virlogeux-Payant I, Hennequet-Antier C, Menanteau P, Gabriel I, Weill FX, Velge P, Chemaly M. 2012. Heterogeneity of persistence of Salmonella enterica serotype Senftenberg strains could explain the emergence of this serotype in poultry flocks. PLoS One 7:e35782. doi: 10.1371/journal.pone.0035782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedersen TB, Olsen JE, Bisgaard M. 2008. Persistence of Salmonella Senftenberg in poultry production environments and investigation of its resistance to desiccation. Avian Pathol 37:421–427. doi: 10.1080/03079450802216561. [DOI] [PubMed] [Google Scholar]
- 17.Mehta G, Malik A, Singh S, Kumari S. 1992. Asymptomatic Salmonella senftenberg carriage in a neonatal ward. J Hosp Infect 22:317–322. doi: 10.1016/0195-6701(92)90017-G. [DOI] [PubMed] [Google Scholar]
- 18.L'Ecuyer PB, Diego J, Murphy D, Trovillion E, Jones M, Sahm DF, Fraser VJ. 1996. Nosocomial outbreak of gastroenteritis due to Salmonella senftenberg. Clin Infect Dis 23:734–742. doi: 10.1093/clinids/23.4.734. [DOI] [PubMed] [Google Scholar]
- 19.Rushdy AA, Stuart JM, Ward LR, Bruce J, Threlfall EJ, Punia P, Bailey JR. 1998. National outbreak of Salmonella senftenberg associated with infant food. Epidemiol Infect 120:125–128. doi: 10.1017/S0950268897008546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pezzoli L, Elson R, Little C, Fisher I, Yip H, Peters T, Hampton M, De Pinna E, Coia JE, Mather HA, Brown DJ, Nielsen EM, Ethelberg S, Heck M, de Jager C, Threlfall J. 2007. International outbreak of Salmonella Senftenberg in 2007. Euro Surveill 12:E070614.3. [DOI] [PubMed] [Google Scholar]
- 21.Ilic S, Duric P, Grego E. 2010. Salmonella Senftenberg infections and fennel seed tea, Serbia. Emerg Infect Dis 16:893–895. doi: 10.3201/eid1605.091555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu XB, Yuan ZA, Jin HM, Xiao WJ, Gu BK, Chen M, Ran L, Diao BW, Cui ZG, Hu QH, Kan B. 2009. Study on the epidemiological characteristics and molecular typing of Salmonella enterica subsp. enterica serovar Senftenberg in Shanghai. Zhonghua Liu Xing Bing Xue Za Zhi 30:933–937. (In Chinese.) [PubMed] [Google Scholar]
- 23.Stepan RM, Sherwood JS, Petermann SR, Logue CM. 2011. Molecular and comparative analysis of Salmonella enterica Senftenberg from humans and animals using PFGE, MLST and NARMS. BMC Microbiol 11:153. doi: 10.1186/1471-2180-11-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendriksen RS, Joensen KG, Lukwesa-Musyani C, Kalondaa A, Leekitcharoenphon P, Nakazwe R, Aarestrup FM, Hasman H, Mwansa JC. 2013. Extremely drug-resistant Salmonella enterica serovar Senftenberg infections in patients in Zambia. J Clin Microbiol 51:284–286. doi: 10.1128/JCM.02227-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasheed JK, Kitchel B, Zhu W, Anderson KF, Clark NC, Ferraro MJ, Savard P, Humphries RM, Kallen AJ, Limbago BM. 2013. New Delhi metallo-beta-lactamase-producing Enterobacteriaceae, United States. Emerg Infect Dis 19:870–878. doi: 10.3201/eid1906.121515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi S, Xie J, Liu N, Li P, Xu X, Li H, Sun J, Wang J, Liang B, Yang C, Wang X, Hao R, Wang L, Wu Z, Zhang J, Wang Y, Huang L, Sun Y, Klena JD, Meng J, Qiu S, Song H. 2014. Emergence and prevalence of non-H2S-producing Salmonella enterica serovar Senftenberg isolates belonging to novel sequence type 1751 in China. J Clin Microbiol 52:2557–2565. doi: 10.1128/JCM.00377-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steele-Mortimer O, Brumell JH, Knodler LA, Meresse S, Lopez A, Finlay BB. 2002. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell Microbiol 4:43–54. doi: 10.1046/j.1462-5822.2002.00170.x. [DOI] [PubMed] [Google Scholar]
- 28.Leekitcharoenphon P, Lukjancenko O, Friis C, Aarestrup FM, Ussery DW. 2012. Genomic variation in Salmonella enterica core genes for epidemiological typing. BMC Genomics 13:88. doi: 10.1186/1471-2164-13-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 30.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. [DOI] [PubMed] [Google Scholar]
- 31.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Assefa S, Keane TM, Otto TD, Newbold C, Berriman M. 2009. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics 25:1968–1969. doi: 10.1093/bioinformatics/btp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai IJ, Otto TD, Berriman M. 2010. Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps. Genome Biol 11:R41. doi: 10.1186/gb-2010-11-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 35.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Carver T, Berriman M, Tivey A, Patel C, Bohme U, Barrell BG, Parkhill J, Rajandream MA. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L, Stoeckert CJ Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grepinet O, Boumart Z, Virlogeux-Payant I, Loux V, Chiapello H, Gendrault A, Gibrat JF, Chemaly M, Velge P. 2012. Genome sequence of the persistent Salmonella enterica subsp. enterica serotype Senftenberg strain SS209. J Bacteriol 194:2385–2386. doi: 10.1128/JB.00255-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dorsey CW, Laarakker MC, Humphries AD, Weening EH, Baumler AJ. 2005. Salmonella enterica serotype Typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol Microbiol 57:196–211. doi: 10.1111/j.1365-2958.2005.04666.x. [DOI] [PubMed] [Google Scholar]
- 42.Abd El Ghany M, Jansen A, Clare S, Hall L, Pickard D, Kingsley RA, Dougan G. 2007. Candidate live, attenuated Salmonella enterica serotype Typhimurium vaccines with reduced fecal shedding are immunogenic and effective oral vaccines. Infect Immun 75:1835–1842. doi: 10.1128/IAI.01655-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guimarães BG, Souchon H, Honore N, Saint-Joanis B, Brosch R, Shepard W, Cole ST, Alzari PM. 2005. Structure and mechanism of the alkyl hydroperoxidase AhpC, a key element of the Mycobacterium tuberculosis defense system against oxidative stress. J Biol Chem 280:25735–25742. doi: 10.1074/jbc.M503076200. [DOI] [PubMed] [Google Scholar]
- 44.Sakano C, Kuroda M, Sekizuka T, Ishioka T, Morita Y, Ryo A, Tsukagoshi H, Kawai Y, Inoue N, Takada H, Ogaswara Y, Nishina A, Shimoda MA, Kozawa K, Oishi K, Kimura H. 2013. Genetic analysis of non-hydrogen sulfide-producing Salmonella enterica serovar Typhimurium and S. enterica serovar infantis isolates in Japan. J Clin Microbiol 51:328–330. doi: 10.1128/JCM.02225-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Baumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Baumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carlin A, Shi W, Dey S, Rosen BP. 1995. The ars operon of Escherichia coli confers arsenical and antimonial resistance. J Bacteriol 177:981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang A, Teplitski M, Rathinasabapathi B, Ma L. 2010. Characterization of arsenic-resistant bacteria from the rhizosphere of arsenic hyperaccumulator Pteris vittata. Can J Microbiol 56:236–246. doi: 10.1139/W10-005. [DOI] [PubMed] [Google Scholar]
- 49.Christen K. 2001. Chickens, manure, and arsenic. Environ Sci Technol 35:184A–185A. doi: 10.1021/es012337m. [DOI] [PubMed] [Google Scholar]
- 50.Gerdes K, Maisonneuve E. 2012. Bacterial persistence and toxin-antitoxin loci. Annu Rev Microbiol 66:103–123. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- 51.Yamaguchi Y, Inouye M. 2011. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat Rev Microbiol 9:779–790. doi: 10.1038/nrmicro2651. [DOI] [PubMed] [Google Scholar]
- 52.De la Cruz MA, Zhao W, Farenc C, Gimenez G, Raoult D, Cambillau C, Gorvel JP, Meresse S. 2013. A toxin-antitoxin module of Salmonella promotes virulence in mice. PLoS Pathog 9:e1003827. doi: 10.1371/journal.ppat.1003827. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.