Abstract
Staphylococcus lugdunensis is a major cause of aggressive endocarditis, but it is also responsible for a broad spectrum of infections. The differences in clinical and molecular characteristics between community-associated (CA) and health care-associated (HA) S. lugdunensis infections have remained unclear. We performed a retrospective study of S. lugdunensis infections between 2003 and 2014 to compare the clinical and molecular characteristics of CA and HA isolates. We collected 129 S. lugdunensis isolates in total: 81 (62.8%) HA isolates and 48 (37.2%) CA isolates. HA infections were more frequent than CA infections in children (16.0% versus 4.2%, respectively; P = 0.041) and the elderly (38.3% versus 14.6%, respectively; P = 0.004). The CA isolates were more likely to cause skin and soft tissue infections (85.4% versus 19.8%, respectively; P < 0.001). HA isolates were more frequently responsible for bacteremia of unknown origin (34.6% versus 4.2%, respectively; P < 0.001) and for catheter-related bacteremia (12.3% versus 0%, respectively; P = 0.011) than CA isolates. Fourteen-day mortality was higher for HA infections than for CA infections (11.1% versus 0%, respectively). A higher proportion of the HA isolates than of the CA isolates were resistant to penicillin (76.5% versus 52.1%, respectively; P = 0.004) and oxacillin (32.1% versus 2.1%, respectively; P < 0.001). Two major clonal complexes (CC1 and CC3) were identified. Sequence type 41 (ST41) was the most common sequence type identified (29.5%). The proportion of ST38 isolates was higher for HA than for CA infections (33.3% versus 12.5%, respectively; P = 0.009). These isolates were of staphylococcal cassette chromosome mec element (SCCmec)type IV, V, or Vt. HA and CA S. lugdunensis infections differ in terms of their clinical features, outcome, antibiotic susceptibilities, and molecular characteristics.
INTRODUCTION
Staphylococcus lugdunensis was first described by Freney et al. in 1988 (1). It has emerged as an important human pathogen in recent years, due to its ability to cause endocarditis and a broad spectrum of other infections, including skin and soft tissue infections, bloodstream infections, bone and joint infections, central nervous system infections, and intra-abdominal infections (2).
S. lugdunensis has a lower incidence than Staphylococcus aureus and Staphylococcus epidermidis (3–5). Many studies have focused on cases of bacteremia and endocarditis caused by S. lugdunensis and have shown that this bacterium can, like S. aureus, cause invasive infections but with higher rates of complications and mortality (4, 6–10). Endocarditis due to this bacterium has been reported to be mostly community associated (CA) (4, 7). Two retrospective studies showed that most CA S. lugdunensis infections were skin and soft tissue infections, with a total absence of oxacillin resistance (11, 12). We recently reported 48 cases of invasive S. lugdunensis infection. Most of these cases (43 of 48 [89.6%]) were health care-associated (HA) infections, and the frequency of oxacillin resistance was high (20%) (10). Staphylococcal cassette chromosome mec element (SCCmec) type V was the most common SCCmec type. We also found that the proportion of SCCmec type II isolates was higher for commensal isolates than for isolates causing disease in health care settings (13, 14).
Multilocus sequence typing (MLST) characterizes multilocus genetic variability on the basis of the sequences of multiple housekeeping genes (15). MLST has been used for population genetic analyses and studies of phylogenetic structure and global epidemiology in many microorganisms. MLST provides fully reproducible data that can be transferred between laboratories, a considerable advantage over pulsed-field gel electrophoresis (PFGE). It has been used to characterize the evolution and macroepidemiological features of Staphylococcus aureus (16) and Staphylococcus epidermidis (17, 18). An MLST method for S. lugdunensis was recently described by Chassain et al. (19). Twenty different sequence types (STs) and five clonal complexes (CCs) have been reported in France and Belgium. We have reported the presence of an oxacillin-resistant ST6 SCCmec type 2 clone endemic in Taiwan (13), but more detailed MLST typing data are required for S. lugdunensis.
The incidence of S. lugdunensis infection is low. Data concerning the clinical characteristics, outcome, antibiotic resistance, and SCCmec typing of CA and HA S. lugdunensis infections are therefore limited. The clonal complex (CC) and MLST typing data for CA and HA S. lugdunensis isolates are still lacking. In this study, we aimed to investigate the clinical features and molecular characteristics of CA and HA S. lugdunensis infections.
MATERIALS AND METHODS
Clinical setting.
This study was conducted at Chang Gung Memorial Hospital, a university hospital and tertiary referral center in northern Taiwan. This study was approved by the Chang Gung Medical Foundation institutional review board (approval no. 103-3231B). The study was divided into two time periods (study period I, 1 May 2003 to 31 May 2013; study period II, 1 July to 31 October 2014. In study period I, we collected S. lugdunensis isolates only from bacterial cultures established from normally sterile sites, including blood, ascites, synovial fluid, cerebrospinal fluid, pleural effusion, and amniotic fluid. In study period II, we collected S. lugdunensis isolates not only from cultures established from normally sterile sites but also from cultures of material from wounds, pus, and cervical swabs.
Case definition.
Clinical data, including the age and sex of the patient, chronic comorbidity, and sources of infection, were obtained retrospectively by reviewing the patients' medical records. The source of infection was identified, where possible, on the basis of the patient's medical history, symptoms, physical examination, laboratory test results, and radiographic findings. Only the first episode of S. lugdunensis infection in each given patient was included in the analysis. HA infections were defined as follows: (i) S. lugdunensis infection identified more than 48 h after admission to the hospital; (ii) S. lugdunensis infection in a patient with a history of hospitalization, surgery, dialysis, or residence in a long-term-care facility within the year preceding a positive culture; or (iii) S. lugdunensis infection in a patient fitted with an indwelling catheter or percutaneous medical device at the time of culture. Cases with none of the above-mentioned features were classified as CA infections.
Clinically significant S. lugdunensis bacteremia was defined according to the criteria proposed by Fadel et al. (8) and Souvenir et al. (20). Patients were considered to have clinically significant bacteremia if two consecutive blood cultures yielded positive results. Patients with a single positive blood culture were considered to have clinically significant bacteremia if they experienced one or more of the following symptoms or signs: prolonged fever, hypotension, leukocytosis, or neutropenia with a left-shifted differential, or disseminated intravascular coagulopathy combined with risk factors for infections caused by the skin flora, including long-term intravascular catheterization, peritoneal dialysis or hemodialysis, or extensive postsurgical infection with coagulase-negative staphylococci. Mortality rates were calculated from deaths within 14 days of the first culture positive for S. lugdunensis.
Microbiological methods.
The S. lugdunensis isolates obtained between May 2003 and May 2013 were first identified by Gram staining and biochemical methods (catalase-positive, coagulase-negative, pyrrolidonyl arylamidase-positive, and ornithine decarboxylase-positive results), with checking by PCR, as described by Noguchi et al. (21), and on a Bruker Biotyper (database 2.0) matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) system. Isolates obtained between July 2014 and October 2014 were identified on the basis of Gram staining and MALDI-TOF MS only, because the accuracy of MALDI-TOF MS had been well demonstrated by then (22).
Antimicrobial drug susceptibility testing.
We assessed the susceptibility of the isolates to penicillin, clindamycin, erythromycin, and trimethoprim-sulfamethoxazole by the disk diffusion method. We used cefoxitin disks rather than oxacillin disks to assess susceptibility to oxacillin, in accordance with Clinical and Laboratory Standards Institute guidelines (23).
SCCmec typing.
Isolates resistant to oxacillin were subjected to SCCmec typing with a multiplex PCR method detecting the ccr gene complex, the mec gene complex, and specific structures in adjacent regions, as described by Kondo et al. (24).
agr typing.
We carried out agr typing by multiplex PCR amplification of the conserved sequences of the agrC gene with two forward primers (5′-CTGTCATCCTTAGTGTAATTGCTG-3′ and 5′-GCCGGCATAATAGTCCCTTCTG-3′) and one reverse primer (5′-CGAACCTTTAGCTTATCTGTACC-3′), as previously described (14). We determined the agr specificity type of the isolates on the basis of the expected product sizes (586 bp for agr type I and 771 bp for agr type II).
Multilocus sequence typing.
The DNA sequences of seven housekeeping genes (aroE, dat, ddl, gmk, ldh, recA, and yqiL) were used for the multilocus sequence typing (MLST) of S. lugdunensis, as previously described by Chassain et al. (19). Each unique allelic profile was assigned to a sequence type (ST). The sequencing data obtained for the seven housekeeping genes were compared with those provided by the Institut Pasteur of Paris website (http://bigsdb.web.pasteur.fr/). Clonal complexes (CCs) were determined using the database of the Institut Pasteur of Paris. The sequences of the various alleles at each of the seven loci defined the allelic profile corresponding to the ST. We compared the STs of the isolates studied here with those available from the Institut Pasteur of Paris website with eBURST version 3 (http://eburst.mlst.net/; Department of Infectious Disease Epidemiology, Imperial College London, United Kingdom) software.
Hemolytic activity.
We assessed δ-hemolysin activity to evaluate agr function. This activity was analyzed by performing a cross-streaking test perpendicular to S. aureus RN4220, which produces only β-hemolysin, on a Columbia blood agar plate. We used S. aureus ATCC 25923 as the agr-positive control and S. aureus strain Mu 50 as the agr-negative control.
Statistical analysis.
The data were processed and statistical analyses performed with the Statistical Package for Social Sciences software version 18.0 (SPSS, Inc., Chicago, IL, USA). The data are expressed as absolute numbers and percentages. Differences in proportions between the two groups was analyzed in a two-tailed z-test for proportions, using an online statistics software (http://www.socscistatistics.com/tests/ztest/).
RESULTS
We collected 129 S. lugdunensis isolates in total: 67 (51.9%) during study period I (1 May 2003 to 31 May 2013) and 62 (48.1%) in study period II (1 July to 31 October 2014) (Table 1). During study period I, most of the isolates were obtained from blood cultures (56 of 67 [83.5%]) or from ascites (4 of 67 [6.0%]). In study period II, most of the isolates were obtained from pus cultures (39 of 62 [62.9%], 36 from skin and soft tissue infection, one from Bartholin's cyst infection, one from maxillary sinusitis, and one from deep neck infection) and wound culture (18 of 62 [29.0%]). Most of the isolates (60 of 67 [89.6%]) in study period I were HA. In contrast, more than half of the isolates (41 of 62 [66.1%]) in study period II were CA.
TABLE 1.
General data of isolates collected from study period I (May 2003 to May 2013) and study period II (July to October 2014)
| Characteristic | No. (%) for |
||
|---|---|---|---|
| Total (n = 129) | Study period I (n = 67) | Study period II (n = 62) | |
| Sample types | |||
| Blood | 58 (45.0) | 56 (83.6) | 2 (3.2) |
| Pus | 39 (30.2) | 0 | 39 (62.9) |
| Wound | 18 (14.0) | 0 | 18 (29.0) |
| Ascites | 4 (3.1) | 4 (6.0) | 0 |
| Body fluid | 3 (2.3) | 2 (3.0) | 1 (1.6) |
| Synovial fluid | 2 (1.6) | 2 (3.0) | 0 |
| Cerebrospinal fluid | 1 (0.8) | 1 (1.5) | 0 |
| Cervix | 1 (0.8) | 0 | 1 (1.6) |
| Pleural effusion | 1 (0.8) | 1 (1.5) | 0 |
| Tissue | 1 (0.8) | 0 | 1 (1.6) |
| Amniotic fluid | 1 (0.8) | 1 (1.5) | 0 |
| Type of infection | |||
| Community associated | 48 (37.2) | 7 (10.4) | 41 (66.1) |
| Health care associated | 81 (62.8) | 60 (89.6) | 21 (33.9) |
Overall, 55.8% of the 129 episodes of infection occurred in male patients, and the mean ± standard deviation age of the patients was 49.99 ± 24.90 years (Table 2). No difference in the sex ratio of patients was found between HA and CA infections. Despite the similar mean ages of patients with HA and CA infections, HA infections were found to be more frequent than CA infections in children (16.0% versus 4.2%, respectively; P = 0.041) and in the elderly (38.3% versus 14.6%, respectively; P = 0.004). HA infections were less frequent than CA infections in patients between the ages of 18 and 65 years (45.7% versus 81.3%, respectively; P < 0.001). Most of the episodes of HA infection in children concerned children under the age of 1 year (10 of 13 [76.9%]), and there were no episodes of CA infection in patients under the age of 1 year. Patients with HA infections had higher rates of comorbid conditions, including diabetes mellitus (39.5% versus 12.5, respectively; P = 0.001), end-stage renal disease (37.5% versus 0%, respectively; P < 0.001), and solid-organ cancers (22.2% versus 2.1%, respectively; P = 0.002), than patients with CA infections. The distribution of clinical infectious foci differed between HA and CA infections. CA infections were more likely to affect the skin and soft tissues (85.4% versus 19.8%, respectively; P < 0.001), mostly with abscesses and pus formation (31 of 42 [73.8%]), and were less likely to concern normally sterile sites. HA isolates accounted for a higher proportion of cases of bacteremia of unknown origin (34.6% versus 4.2%, respectively; P < 0.001) and catheter-related bacteremia (12.3% versus 0%, respectively; P = 0.011) than CA isolates. Five cases of infective endocarditis were identified. Three of these cases were HA, and the other two were CA. All the cases of HA endocarditis occurred in patients with end-stage renal disease treated by hemodialysis. We found that 14-day all-cause mortality was higher for HA than for CA infections (11.1% versus 0%, respectively; P = 0.017).
TABLE 2.
Distribution of gender, age, comorbidity, source of infection, and outcome between health care- and community-associated infectionsa
| Characteristic | Total (n = 129) | Health care associated (n = 81) | Community associated (n = 48) | P value |
|---|---|---|---|---|
| Gender | ||||
| Male | 73 (56.6) | 44 (54.3) | 29 (60.4) | 0.496 |
| Age (yr) | ||||
| Mean ± SD | 49.99 ± 24.90 | 51.35 ± 28.13 | 47.70 ± 18.26 | 0.423 |
| 0–17 | 15 (11.6) | 13 (16.0) | 2 (4.2) | 0.041b |
| <1 | 10 (7.8) | 10 (12.3) | 0 | 0.011b |
| 18–64 | 76 (58.9) | 37 (45.7) | 39 (81.3) | <0.001c |
| ≥65 | 38 (29.5) | 31 (38.3) | 7 (14.6) | 0.004d |
| Comorbidity | ||||
| Diabetes mellitus | 38 (29.5) | 32 (39.5) | 6 (12.5) | 0.001d |
| End-stage renal disease | 32 (24.8) | 32 (37.5) | 0 | <0.001c |
| Liver cirrhosis | 6 (4.7) | 6 (7.4) | 0 | 0.054 |
| Solid-organ malignancy | 19 (14.7) | 18 (22.2) | 1 (2.1) | 0.002d |
| Hematologic malignancy | 2 (1.6) | 2 (2.5) | 0 | 0.271 |
| Cerebrovascular accident | 18 (14.0) | 14 (17.3) | 4 (8.3) | 0.156 |
| Source of infection | ||||
| Skin and soft tissue | 57 (44.2) | 16 (19.8) | 41 (85.4) | <0.001c |
| Unknown source of bacteremia | 30 (23.3) | 28 (34.6) | 2 (4.2) | <0.001c |
| Catheter-related bloodstream infection | 10 (7.8) | 10 (12.3) | 0 | 0.011b |
| Bone and joints | 8 (6.2) | 7 (8.6) | 1 (2.1) | 0.136 |
| Arteriovenous fistula/graft | 6 (4.7) | 6 (7.4) | 0 | 0.053 |
| Infective endocarditis | 5 (3.9) | 3 (3.7) | 2 (4.2) | 0.986 |
| Intra-abdomen | 4 (3.1) | 4 (4.9) | 0 | 0.119 |
| Genital system | 3 (2.3) | 2 (2.5) | 1 (2.1) | 0.889 |
| Peritoneal dialysis-related peritonitis | 2 (1.6) | 2 (2.5) | 0 | 0.271 |
| Lung | 1 (0.8) | 1 (1.2) | 0 | 0.441 |
| Central nervous system | 1 (0.8) | 1 (1.2) | 0 | 0.441 |
| Deep neck | 1 (0.8) | 1 (1.2) | 0 | 0.441 |
| Sinusitis | 1 (0.8) | 1 (1.2) | 0 | 0.441 |
| 14-day all-cause mortality | 9 (7.0) | 9 (11.1) | 0 | 0.017b |
All data are presented as the no. (%), unless otherwise stated.
P < 0.05.
P < 0.001.
P < 0.01.
Higher proportions of HA than of CA isolates were resistant to penicillin (76.5% versus 52.1%, respectively; P = 0.004) and oxacillin (32.1% versus 2.1%, respectively) (Table 3). Rates of resistance to clindamycin and erythromycin were similar in HA and CA isolates (clindamycin resistance: 23.5% versus 18.8%, respectively, P = 0.529; erythromycin resistance: 27.2% versus 20.8%, respectively, P = 0.424). Rates of resistance to trimethoprim-sulfamethoxazole were low in both groups. SCCmec typing of the 27 oxacillin-resistant isolates was successful for 25 isolates, with the other two isolates being nontypeable (NT) (Table 3). The oxacillin-resistant isolates of SCCmec types II (5 of 81 [6.2%]), IV (1 of 81 [1.2%]), V (17 of 81 [21%]), and Vt (2 of 81 [2.5%]) were obtained from patients with HA infections. Only one oxacillin-resistant isolate were identified in a case of CA infection, and this isolate was nontypeable for SCCmec. No statistical difference in agr typing results was observed between the HA and CA isolates. The percentage of isolates displaying agr dysfunction was similar in the two groups.
TABLE 3.
Antibiotic resistance, distribution of SCCmec typing, agr group, and δ-hemolysin, clonal complex, and multilocus sequence typing among Staphylococcus lugdunensis isolates that cause health care- and community-associated infection
| Characteristica | No. (%) for |
P value | ||
|---|---|---|---|---|
| Total (n = 129) | Health care associated (n = 81) | Community associated (n = 48) | ||
| Antibiotic resistance | ||||
| Penicillin | 87 (67.4) | 62 (76.5) | 25 (52.1) | 0.004b |
| Oxacillin | 27 (20.9) | 26 (32.1) | 1 (2.1) | <0.001c |
| Clindamycin | 28 (21.7) | 19 (23.5) | 9 (18.8) | 0.529 |
| Erythromycin | 32 (24.8) | 22 (27.2) | 10 (20.8) | 0.424 |
| TMP-SMX | 3 (2.3) | 1 (1.2) | 2 (4.2) | 0.285 |
| SCCmec type | ||||
| II | 5 (3.9) | 5 (6.2) | 0 | 0.078 |
| IV | 1 (0.8) | 1 (1.2) | 0 | 0.441 |
| V | 17 (13.2) | 17 (21.0) | 0 | <0.001c |
| Vt | 2 (1.6) | 2 (2.5) | 0 | 0.271 |
| NT | 2 (1.6) | 1 (1.2) | 1 (2.1) | 0.703 |
| agr type | ||||
| I | 57 (44.2) | 31 (38.3) | 26 (54.2) | 0.078 |
| II | 72 (55.8) | 50 (61.7) | 22 (45.8) | 0.078 |
| δ-Hemolysin | ||||
| Negative | 10 (7.8) | 7 (8.6) | 3 (6.3) | 0.624 |
| Clonal complex and MLST typing | ||||
| CC1 | 56 (43.4) | |||
| ST6 | 2 (1.6) | 2 (2.5) | 0 | 0.271 |
| ST25 | 11 (8.5) | 5 (6.2) | 6 (12.5) | 0.215 |
| ST41 | 38 (29.5) | 24 (29.6) | 14 (29.2) | 0.841 |
| ST44 | 3 (2.3) | 0 | 3 (6.3) | 0.002b |
| ST45 | 2 (1.6) | 0 | 2 (4.2) | 0.064 |
| CC2 | 2 (1.6) | |||
| ST29 | 2 (1.6) | 1 (1.2) | 1 (1.2) | 0.703 |
| CC3 | 33 (25.6) | |||
| ST38 | 33 (25.6) | 27 (33.3) | 6 (12.5) | 0.009b |
| CC4 | 12 (9.3) | |||
| ST33 | 12 (9.3) | 6 (7.4) | 6 (12.5) | 0.337 |
| CC7 | 1 (0.8) | |||
| ST27 | 1 (0.8) | 0 | 1 (2.1) | 0.194 |
| CC8 | 22 (17.1) | |||
| ST36 | 15 (11.6) | 12 (14.8) | 3 (6.3) | 0.141 |
| ST39 | 7 (5.4) | 2 (2.5) | 5 (10.4) | 0.054 |
| Singletons | ||||
| ST43 | 3 (2.3) | 2 (2.5) | 1 (2.1) | 0.889 |
TMP-SMX, trimethoprim-sulfamethoxazole; NT, nontypeable; MLST, multilocus typing sequence typing.
P < 0.01.
P < 0.001.
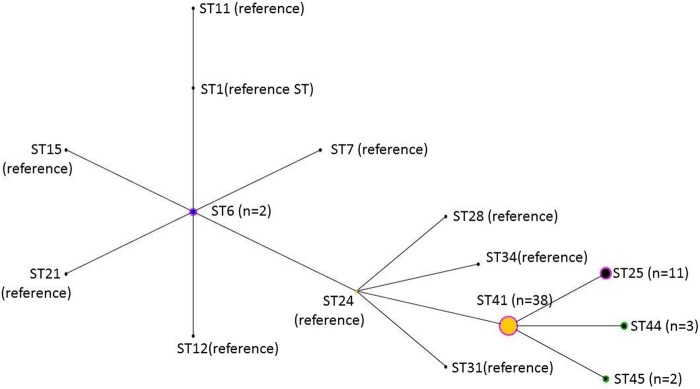
Six CCs were identified by MLST typing. CC1 was the most frequently isolated clonal complex (n = 56 [43%]), followed by CC3 (n = 33 [25.6%]) and CC8 (n = 22 [17.1%]) (Table 3). We identified 12 STs in total. The most frequently encountered STs were ST41 (belonging to CC1, n = 38 [29.8%]), ST38 (belonging to CC3, n = 33 [25.6%]), and ST36 (belonging to CC2, n = 15 [11.6%]). We identified three new STs not present in the database, which we named ST43, ST44, and ST45. The allelic profile of ST43 was 1-4-3-1-5-7-4 for aroE, dat, ddl, gmk, ldh, recA, and yqiL. This ST is most closely related to ST33 (6-4-3-1-5-7-3) and ST9 (1-4-3-1-1-3-4), from which it differs by the alleles at two loci. The allelic profile of ST44 was 1-1-7-1-5-9-2, and this ST is most similar to ST41 (1-1-1-1-5-9-2), from which it differs by the allele at one locus. The allelic profile of ST45 was 1-2-1-1-5-9-2, and this ST is most closely related to ST41, from which it differs by the allele at one locus. Some genetic evolution was observed in clonal complex 1. The ancestral genotype was ST41 in our study, and eBURST identified ST25, ST44, and ST45 as being derived from ST41 (Fig. 1). ST41 isolates were similarly distributed between HA (29.6%) and CA (29.2%) infections. ST38 isolates were more frequent among HA than among CA isolates (33.3% versus 12.5%, respectively; P = 0.009).
FIG 1.
eBURST diagram showing the relatedness among the major cluster (clonal complex 1) of S. lugdunensis isolates in this study and compared with the reference sequence types (STs) in the database of the Institut Pasteur (http://bigsdb.web.pasteur.fr/). Each circle represents an ST. The size of the circle represents the proportion of the number of isolates in the study.
Most of the oxacillin-resistant isolates (n = 27) belonged to ST38 (n = 21), with ST41 (n = 4) and ST6 (n = 2) being the next most frequent sequence types for these isolates (Table 4). The ST38 oxacillin-resistant isolates were mostly of SCCmec type V (17 of 21), followed by SCCmec type Vt (2 of 21) and SCCmec type IV (1 of 21). The ST6 (two isolates) and ST41 oxacillin-resistant isolates (three isolates) were SCCmec type II. Two oxacillin-resistant isolates were SCCmec type NT: one belonged to ST38 and the other to ST41. The ST38 oxacillin-resistant isolates of SCCmec type V were obtained at our hospital in 2009, the type Vt isolates were obtained in 2011, and the type IV isolates were obtained in 2012. The ST6 oxacillin-resistant isolates of SCCmec type II were obtained in 2010, and the ST41 oxacillin-resistant isolates of SCCmec type II were obtained in 2012. The only oxacillin-resistant isolate from a CA infection belonged to ST38 and SCCmec type NT and was obtained in 2014.
TABLE 4.
MLST and year of isolation of oxacillin-resistant Staphylococcus lugdunensis (n = 27)
| MLST typing (n) | Clonal complex | SCCmec typing (no. of isolates) | No. of isolates by yr of isolation |
|||||
|---|---|---|---|---|---|---|---|---|
| 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | |||
| ST6 (2) | CC1 | II (2) | 1 | 1 | ||||
| ST38 (21) | CC3 | IV (1) | 1 | |||||
| V (17) | 2 | 5 | 4 | 1 | 2 | 3 | ||
| Vt (2) | 1 | 1 | ||||||
| NT (1) | 1a | |||||||
| ST41 (4) | CC1 | II (3) | 2 | 1 | ||||
| NT (1) | 1 | |||||||
The isolates belonged to community-associated infections. Other isolates belonged to health care-associated infections.
DISCUSSION
This is the first study to compare of HA and CA S. lugdunensis infections directly, in a total of 129 cases. Overall, HA isolates were found to be more likely than CA isolates to cause bloodstream infection. In contrast, CA isolates mostly caused skin and soft tissue infections. The age distributions of the patients presenting HA and CA infections was also different. HA infections were more frequent than CA infections in children and the elderly. HA infections had a higher 14-day all-cause mortality than CA infections. Oxacillin resistance was almost exclusively limited to HA isolates carrying SCCmec types II, IV, V, and Vt. The emergence of oxacillin-resistant isolates of ST38, ST41, and ST6 in health care settings was noted.
CA and HA S. lugdunensis infections were found to have different clinical presentations in this study. CA infections were less invasive than HA infections and were more likely to cause skin and soft tissue infections with abscess formation. The presentation of CA S. lugdunensis infection was similar to that previously reported (11, 12). In contrast, HA isolates were more frequent than CA isolates in cases of bacteremia of unknown origin and catheter-related bloodstream infection. S. lugdunensis endocarditis has been widely reported in many studies and is associated with high mortality, at about 70% (2, 6, 25). S. lugdunensis is thought to behave similarly to Staphylococcus aureus in cases of endocarditis, but the overall mortality for S. lugdunensis infections is lower than that for S. aureus infections, especially for CA infections. We identified three cases of HA endocarditis and two of CA endocarditis in this study. S. lugdunensis endocarditis may, therefore, occur in both HA and CA settings. All the HA cases of endocarditis occurred in patients on hemodialysis. These findings are consistent with those of a previous study by Ho et al. showing high rates (54.6%) of S. lugdunensis carriage in patients on long-term renal replacement therapy (26). These high rates of carriage in patients with end-stage renal disease suggest that there may be a risk of endocarditis in patients on hemodialysis.
We identified only one oxacillin-resistant isolate obtained from a CA infection, and in this case, the isolate was nontypeable by SCCmec. SCCmec types IV, V, and Vt were considered to be CA in oxacillin-resistant S. aureus and S. epidermidis isolates, but we found oxacillin-resistant isolates of SCCmec types IV, V, and Vt only in cases of HA infections. No SCCmec type IV, V, or Vt isolate was identified in cases of CA S. lugdunensis infection. Only one case of CA oxacillin-resistant S. lugdunensis SCCmec type V infection has been reported to date, in Brazil (27). This finding suggests that the rate of oxacillin resistance in S. lugdunensis remains low in the community. The origin of the oxacillin-resistant S. lugdunensis isolates of SCCmec type V remains unclear, but in this study, they did not appear to originate from the community.
The MLST method for S. lugdunensis was developed in 2012, and previous reports identified three major CCs (CC1, CC2, and CC3) and showed that the most frequent STs were ST3 (belonging to CC3), ST2 (belonging to CC2), and ST1 (belonging to CC1) in France (19). Ho et al. (26) reported that ST3 was also the most common ST colonizing patients with end-stage renal disease in Hong Kong, and 25.3% of the ST3 isolates identified were found to be of SCCmec type V. In our study in Taiwan, the most common CCs were CC1 and CC3, as in France and Hong Kong, but the most common STs were different. In our hospital, the most common ST was ST41, belonging to CC1, and ST38, belonging to CC3. These differences between different countries highlight the genetic diversity of S. lugdunensis in different regions. We also identified a clone of oxacillin-resistant S. lugdunensis ST6 of SCCmec type II in our hospital environment, and most of the isolates of this clone did not cause infection (13). ST41 was the most common ST in both HA and CA infections, but ST38 has recently emerged as a cause of HA infections, probably because the increasing numbers of oxacillin-resistant ST38 isolates are of SCCmec type IV, V, or Vt. In addition to the ST6 oxacillin-resistant isolates of SCCmec type II identified in 2010, we also identified ST41 oxacillin-resistant isolates of SCCmec type II in 2012, but there were fewer of these isolates than ST38 oxacillin-resistant isolates. High levels of antibiotic use may favor the emergence of oxacillin-resistant isolates in health care settings.
We did not observe any difference in the distribution of agr types and δ-hemolysin activity between CA and HA isolates. The agr system is important in the pathogenesis of staphylococci. The δ-hemolysin activity represents the function of agr gene, which is correlated with toxin expression profiles and biofilm formation of S. aureus (28). Different agr types are associated with CA and HA S. aureus infections (29). Although the role of the agr gene in S. aureus infection was well documented, we did not found any statistic difference between agr types or agr gene function and CA or HA S. lugdunensis infection. The role of the agr gene in S. lugdunensis infections still needs further investigation.
Our study is subject to several limitations. First, the clinical data were collected retrospectively by chart review. In cases of bloodstream infection, the origin of the infection was determined on the basis of clinical judgment. Second, the inclusion criteria and durations of the two periods were different, making it difficult to balance the numbers of CA and HA infections, and the distributions of CA and HA infections differed between the two study periods. It remains unclear whether changes in the molecular epidemiological features of these infections occurred between the two time periods, with potential effects on our results. Third, no healthy volunteers with S. lugdunensis carriage were enrolled in our study for further comparisons of the molecular characteristics of isolates of community origin. Finally, our study was conducted at a single center and therefore may not be representative of the national situation. Further multicenter data should be collected to evaluate the genetic evolution of S. lugdunensis in Taiwan.
In conclusion, our data show that HA and CA S. lugdunensis infections differ in terms of their clinical features and antibiotic resistance profiles. Most STs were similarly distributed between HA and CA infections, but ST38 isolates predominated in the HA setting. The emergence of resistant strains of ST6, ST38, and ST41 in cases of HA infections was noted.
ACKNOWLEDGMENTS
This work was supported by grants from Chang Gung Memorial Hospital (grant CMRPG3D1771) and the Ministry of Science and Technology of Republic of China (Taiwan) (grant MOST104-2320-B-182A-005-MY3).
REFERENCES
- 1.Freney J, Brun Y, Bes M, Meugnier H, Grimont F, Grimont PA, Nervi C, Fleurette J. 1988. Staphylococcus lugdunensis sp. nov. and Staphylococcus schleiferi sp. nov., two species from human clinical specimens. Int J Syst Bacteriol 38:168–172. doi: 10.1099/00207713-38-2-168. [DOI] [Google Scholar]
- 2.Frank KL, Del Pozo JL, Patel R. 2008. From clinical microbiology to infection pathogenesis: how daring to be different works for Staphylococcus lugdunensis. Clin Microbiol Rev 21:111–133. doi: 10.1128/CMR.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shin JH, Jung HJ, Lee HR, Kim JH, Kim HR, Lee JN. 2007. Prevalence, identification, and antimicrobial susceptibility of Staphylococcus lugdunensis from various clinical specimens in Korea. Jpn J Infect Dis 60:312–313. [PubMed] [Google Scholar]
- 4.Choi SH, Chung JW, Lee EJ, Kim TH, Lee MS, Kang JM, Song EH, Jun JB, Kim MN, Kim YS, Woo JH, Choi SH. 2010. Incidence, characteristics, and outcomes of Staphylococcus lugdunensis bacteremia. J Clin Microbiol 48:3346–3349. doi: 10.1128/JCM.00609-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu CJ, Shen DX, Guo J, Wang KF, Wang H, Yan ZQ, Chen R, Ye LY. 2012. Clinical and microbiological characterization of Staphylococcus lugdunensis isolates obtained from clinical specimens in a hospital in China. BMC Microbiol 12:168. doi: 10.1186/1471-2180-12-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vandenesch F, Etienne J, Reverdy ME, Eykyn SJ. 1993. Endocarditis due to Staphylococcus lugdunensis: report of 11 cases and review. Clin Infect Dis 17:871–876. doi: 10.1093/clinids/17.5.871. [DOI] [PubMed] [Google Scholar]
- 7.Zinkernagel AS, Zinkernagel MS, Elzi MV, Genoni M, Gubler J, Zbinden R, Mueller NJ. 2008. Significance of Staphylococcus lugdunensis bacteremia: report of 28 cases and review of the literature. Infection 36:314–321. doi: 10.1007/s15010-008-7287-9. [DOI] [PubMed] [Google Scholar]
- 8.Fadel HJ, Patel R, Vetter EA, Baddour LM. 2011. Clinical significance of a single Staphylococcus lugdunensis-positive blood culture. J Clin Microbiol 49:1697–1699. doi: 10.1128/JCM.02058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tseng SP, Lin YT, Tsai JC, Hung WC, Chen HJ, Chen PF, Hsueh PR, Teng LJ. 2015. Genotypes and phenotypes of Staphylococcus lugdunensis isolates recovered from bacteremia. J Microbiol Immunol Infect 48:397–405. doi: 10.1016/j.jmii.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Lin JF, Cheng CW, Kuo AJ, Liu TP, Yang CC, Huang CT, Lee MH, Lu JJ. 2015. Clinical experience and microbiologic characteristics of invasive Staphylococcus lugdunensis infection in a tertiary center in northern Taiwan. J Microbiol Immunol Infect 48:406–412. doi: 10.1016/j.jmii.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 11.Wu AB, Wang MC, Tseng CC, Lin WH, Teng CH, Huang AH, Hung KH, Chiang-Ni C, Wu JJ. 2011. Clinical and microbiological characteristics of community-acquired Staphylococcus lugdunensis infections in southern Taiwan. J Clin Microbiol 49:3015–3018. doi: 10.1128/JCM.01138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bocher S, Tonning B, Skov RL, Prag J. 2009. Staphylococcus lugdunensis, a common cause of skin and soft tissue infections in the community. J Clin Microbiol 47:946–950. doi: 10.1128/JCM.01024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng CW, Liu TP, Yeh CF, Lee MH, Chang SC, Lu JJ. 2015. Persistence of a major endemic clone of oxacillin-resistant Staphylococcus lugdunensis sequence type 6 at a tertiary medical centre in northern Taiwan. Int J Infect Dis 36:72–77. doi: 10.1016/j.ijid.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Yeh CF, Liu TP, Cheng CW, Chang SC, Lee MH, Lu JJ. 2015. Molecular characteristics of disease-causing and commensal Staphylococcus lugdunensis isolates from 2003 to 2013 at a tertiary hospital in Taiwan. PLoS One 10:e0134859. doi: 10.1371/journal.pone.0134859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maiden MCJ, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou JJ, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas JC, Vargas MR, Miragaia M, Peacock SJ, Archer GL, Enright MC. 2007. Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J Clin Microbiol 45:616–619. doi: 10.1128/JCM.01934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozitskaya S, Olson ME, Fey PD, Witte W, Ohlsen K, Ziebuhr W. 2005. Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J Clin Microbiol 43:4751–4757. doi: 10.1128/JCM.43.9.4751-4757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chassain B, Lemee L, Didi J, Thiberge JM, Brisse S, Pons JL, Pestel-Caron M. 2012. Multilocus sequence typing analysis of Staphylococcus lugdunensis implies a clonal population structure. J Clin Microbiol 50:3003–3009. doi: 10.1128/JCM.00988-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Souvenir D, Anderson DE Jr, Palpant S, Mroch H, Askin S, Anderson J, Claridge J, Eiland J, Malone C, Garrison MW, Watson P, Campbell DM. 1998. Blood cultures positive for coagulase-negative staphylococci: antisepsis, pseudobacteremia, and therapy of patients. J Clin Microbiol 36:1923–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noguchi N, Goto K, Ro T, Narui K, Ko M, Nasu Y, Utsumi K, Takazawa K, Moriyasu F, Sasatsu M. 2010. Using the tannase gene to rapidly and simply identify Staphylococcus lugdunensis. Diagn Microbiol Infect Dis 66:120–123. doi: 10.1016/j.diagmicrobio.2009.03.028. [DOI] [PubMed] [Google Scholar]
- 22.Carbonnelle E, Beretti JL, Cottyn S, Quesne G, Berche P, Nassif X, Ferroni A. 2007. Rapid identification of staphylococci isolated in clinical microbiology laboratories by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 45:2156–2161. doi: 10.1128/JCM.02405-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement CLSI document M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 24.Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother 51:264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu PY, Huang YF, Tang CW, Chen YY, Hsieh KS, Ger LP, Chen YS, Liu YC. 2010. Staphylococcus lugdunensis infective endocarditis: a literature review and analysis of risk factors. J Microbiol Immunol Infect 43:478–484. doi: 10.1016/S1684-1182(10)60074-6. [DOI] [PubMed] [Google Scholar]
- 26.Ho PL, Leung SMH, Chow KH, Tse CWS, Cheng VCC, Tse H, Mak SK, Lo WK. 2015. Carriage niches and molecular epidemiology of Staphylococcus lugdunensis and methicillin-resistant S. lugdunensis among patients undergoing long-term renal replacement therapy. Diagn Microbiol Infect Dis 81:141–144. doi: 10.1016/j.diagmicrobio.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Pereira EM, Schuenck RP, Nouer SA, dos Santos KRN. 2011. Methicillin-resistant Staphylococcus lugdunensis carrying SCCmec type V misidentified as MRSA. Braz J Infect Dis 15:293–295. doi: 10.1016/S1413-8670(11)70192-1. [DOI] [PubMed] [Google Scholar]
- 28.Cheung GY, Wang R, Khan BA, Sturdevant DE, Otto M. 2011. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect Immun 79:1927–1935. doi: 10.1128/IAI.00046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naimi TS, LeDell KH, Como-Sabetti K, Borchardt SM, Boxrud DJ, Etienne J, Johnson SK, Vandenesch F, Fridkin S, O'Boyle C, Danila RN, Lynfield R. 2003. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 290:2976–2984. doi: 10.1001/jama.290.22.2976. [DOI] [PubMed] [Google Scholar]