Abstract
Nontuberculous mycobacteria (NTM) are an important cause of pulmonary disease in patients with cystic fibrosis (CF). A new culture medium (RGM medium) for the isolation of rapidly growing mycobacteria from the sputum of cystic fibrosis patients has recently been reported. The aim of this study was to compare culture of sputum samples on RGM medium with culture using a standard automated liquid culture method. Sputum samples were obtained from 187 distinct patients with CF attending King's College Hospital, London, United Kingdom. Each sample was decontaminated with 3% oxalic acid and inoculated into a mycobacterial growth indicator tube (MGIT) that was monitored for 42 days using the Bactec MGIT 960 instrument. Each sample was also cultured, without decontamination, onto RGM medium, which was incubated for 10 days at 30°C. Mycobacteria were isolated from 28 patients (prevalence, 15%). Mycobacteria were detected in 24 samples (86%) using the MGIT and in 23 samples (82%) using RGM medium (P = 1.00). In this setting, RGM medium showed sensitivity equivalent to that of the MGIT for isolation of NTM from the sputum of patients with CF. RGM medium offers a simple, convenient tool that can be embedded into routine culture methods, allowing the culture of all sputum samples that are submitted from patients with CF.
INTRODUCTION
Nontuberculous mycobacteria (NTM) are an important cause of pulmonary disease in patients with cystic fibrosis (CF) (1). In the largest studies, the prevalence of NTM in sputum has been estimated to be 6 to 13% for patients with CF (2), and there is convincing evidence that prevalence is increasing in the CF population (3–6). NTM are detected by culture of sputum using methods that were designed primarily for the isolation of Mycobacterium tuberculosis. Consensus guidelines state that culture should be performed in liquid media using an automated growth detection system (such as the mycobacterial growth indicator tube [MGIT]) and note that concomitant culture on solid media (e.g., Lowenstein-Jensen medium) may increase the diagnostic yield (1).
Detection of NTM is frequently challenging due to the presence of other bacterial and fungal species in the lungs of CF patients. Competing species typically show a higher growth rate than mycobacteria and are frequently resistant to multiple antibiotics that might be used to select for growth of mycobacteria. To resolve this, sputum samples are subjected to treatment with chemicals in order to eliminate or reduce the burden of nonmycobacterial species. Chemical treatments include N-acetyl-l-cysteine (0.5%) plus sodium hydroxide (2%), 5% oxalic acid, or 1% chlorhexidine (1). However, such decontamination protocols are labor intensive, they may reduce the viability of NTM, and they may be ultimately unsuccessful if there is a high burden of nonmycobacterial species (7–9).
Culture on solid agar-based media is an attractive option as this can potentially allow recovery of NTM even in specimens that contain other viable species. For example, Esther et al. (10) demonstrated that extending the incubation of Burkholderia cepacia-selective agar (BCSA) from 5 to 14 days afforded an increase in the recovery rate of NTM from 0.7% to 2.8% using routine culture methods. However, not all NTM will grow on BCSA and overgrowth, particularly by fungi and Gram-negative bacteria, remains a problem (10). Preece et al. (11) recently described a new selective agar, RGM medium, to target rapidly growing species of mycobacteria, which represent the dominant species in many geographical areas (3, 4, 12). The sensitivity of RGM medium proved to be clearly superior to that of BCSA when evaluated with 502 sputum samples from patients with CF. The aim of this study was to evaluate RGM medium against the MGIT method for detection of mycobacteria in sputum samples from patients with CF.
MATERIALS AND METHODS
Patient samples.
Sputum samples that were routinely taken for culture for acid-fact bacilli (AFB) were prospectively collected from 187 distinct patients with cystic fibrosis attending King's College Hospital, London, United Kingdom, between August and December 2015. Both adults and children were sampled (age range, 7 to 56 years; mean age, 26 years).
Sample processing and culture methods.
RGM medium was prepared from its basic ingredients at the Microbiology Department, Freeman Hospital, Newcastle upon Tyne, United Kingdom, as previously described (13) and transferred by courier to Bedford Hospital where the culture of the sputum samples was performed. As the medium has been shown to have a shelf life of 12 weeks at 4°C (C. L. Preece and J. D. Perry, unpublished data), two batches of medium were used for the 5-month evaluation. Sputum samples were treated with an equal volume of Mucolyse sputum digestant (Pro-Lab Diagnostics), vortexed, and left at room temperature for 15 min. The samples were then centrifuged for 15 min at 3,000 rpm, and the supernatant was discarded to leave 2 to 3 ml of fluid. A 100-μl aliquot of this was cultured onto RGM medium, which was incubated at 30°C for 10 days. Cultures on RGM medium were examined for growth after 4, 7, and 10 days of incubation. A smear was prepared from any colonies recovered on the medium and stained using the auramine-phenol method for detection of AFB. AFB-positive smears were confirmed using Ziehl-Neelsen stain, while isolates that were not AFB were subjected to Gram staining.
The remaining sample was supplemented with 20 ml of 3% oxalic acid and was left for 2 h at room temperature, with vortexing every 15 min. After this decontamination, the sample was centrifuged at 3,000 rpm for 20 min. The supernatant was discarded, and the deposit was resuspended in 2 ml of phosphate-buffered water (E&O Laboratories). A 0.5-ml aliquot of the resuspended deposit was inoculated into a mycobacterial growth indicator tube (MGIT) (Becton Dickinson) along with 0.8 ml of growth supplement containing PANTA (polymyxin B-amphotericin B-nalidixic acid-trimethoprim-azlocillin) antibiotic supplement (Becton Dickinson) in accordance with the manufacturer's instructions. The tube was then loaded onto a Bactec MGIT 960 instrument (Becton Dickinson) and incubated for 42 days or until determined as positive by the instrument.
For positive MGIT cultures, the contents were removed to a sterile universal bottle and were centrifuged at 3,000 rpm for 30 min. Two smears of the deposit were prepared for staining by Gram stain and by the auramine-phenol method to detect AFB. AFB-positive smears were confirmed using Ziehl-Neelsen stain. Positive MGIT cultures with no evidence of bacteria by microscopy were reloaded onto the instrument within 5 h to complete their incubation period. For samples that contained non-acid-fast bacilli, the decontamination process was repeated on the original sample, and a new MGIT culture was initiated.
Confirmation of mycobacteria.
Isolates characterized as acid-fast bacilli by either method were referred to the National Mycobacterium Reference Laboratory, London, United Kingdom, for species identification using the Hain GenoType Mycobacterium CM kit (Hain Lifescience GmbH, Nehren, Germany) and/or sequencing of the 16S rRNA gene.
Statistical analysis.
The two culture methods were compared using McNemar's test with the continuity correction applied. Statistical significance was taken as a P value of <0.05.
RESULTS
Recovery of mycobacteria.
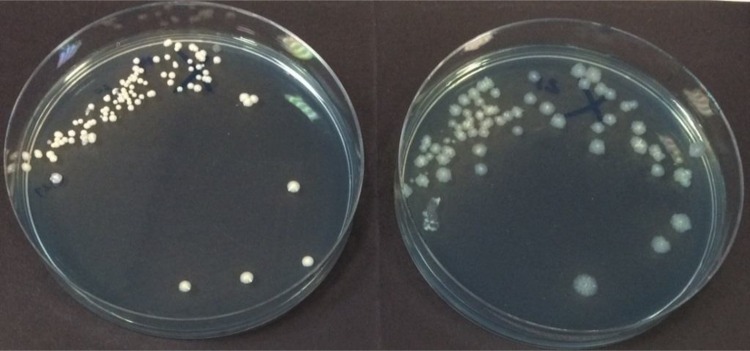
A total of 28 isolates of mycobacteria were recovered from the sputum samples of 28 distinct patients using a combination of both methods (prevalence, 15%). Mycobacterium abscessus complex was clearly dominant and accounted for 90% of all isolates of mycobacteria. Twenty-four isolates of mycobacteria were recovered using the MGIT (Table 1). Of the four isolates that were not recovered by the MGIT, cultures from two sputum samples containing M. abscessus complex were contaminated with nonmycobacterial species (yeasts and Gram-positive cocci), and these were not eradicated using a repeat round of decontamination. The two other cultures were negative after 42 days of incubation. Twenty-three isolates of mycobacteria were recovered using RGM medium. For two sputum samples containing M. abscessus complex that were not recovered by RGM medium, cultures on RGM medium were contaminated with nonmycobacterial species (Gram-negative bacilli and Gram-positive cocci). For the other three mycobacteria that were not detected by RGM medium (one Mycobacterium chelonae and two Mycobacterium mucogenicum), MGIT cultures required >23 days to generate a positive signal, suggesting that the isolates were slow growing and/or present at a very low inoculum. There was no statistical difference between the two methods for recovery of mycobacteria (P = 1.00). Only 9 of the 28 sputum samples that yielded mycobacteria (32%) were AFB positive when the sputum samples were stained using the auramine-phenol method (8 yielded M. abscessus complex and 1 yielded Mycobacterium avium complex). For these 9 samples, mycobacteria were recovered from all samples by both methods. Figure 1 shows the appearance of M. abscessus complex on RGM medium.
TABLE 1.
Numbers of mycobacteria recovered from 187 sputum samples using the MGIT and culture on RGM mediuma
| Species | No. of mycobacteria recovered |
||
|---|---|---|---|
| Total | MGIT | RGM | |
| M. abscessus complex | 21 | 19 | 19 |
| M. avium complex | 1 | 1 | 1 |
| M. chelonae | 3 | 2 | 2 |
| M. mucogenicum | 3 | 2 | 1 |
| Total | 28 | 24 | 23 |
The sensitivities for mycobacteria were as follows: mycobacteria growth indicator tube (MGIT), 86%; RGM medium, 82%.
FIG 1.
Appearance of Mycobacterium abscessus subsp. abscessus on RGM medium showing smooth (left) and rough (right) colony types after 7 days of incubation at 30°C.
Time to results for both methods.
The average time taken for the 24 isolates of mycobacteria to generate a positive signal using the MGIT was 6 days (range, 2 h to 25 days), whereas the average time to generate colonies on RGM medium was 5.7 days (range, 4 to 10 days). Cultures on RGM medium were only examined after 4, 7, and 10 days, and it is plausible that daily examination of cultures may have allowed a faster time to detection, but this would need to be demonstrated. The MGIT declared a negative result for 151 samples. The average time to achieve a negative result was 44 days (range, 42 to 63 days). For 12 samples, no result was available due to successive contamination events. Negative results for RGM cultures were always available after 10 days.
Recovery of nonmycobacterial species.
Of 187 MGIT cultures, 64 (34%) grew nonmycobacterial species and required a further round of decontamination treatment. Ultimately, 12 MGIT cultures (6.4%) had to be abandoned due to contamination. The microorganisms implicated included Gram-negative bacilli (n = 21), Gram-positive cocci (n = 19), and yeasts (n = 31) with mixtures of these occasionally encountered. For RGM medium, nonmycobacterial species were recovered from 31 cultures (16.6%). The vast majority of these (n = 27) were Gram-negative bacilli, but two fungal isolates and one Gram-positive coccus were also recovered. A strain of Segniliparus rugosus was isolated from an AFB smear-positive sputum sample on RGM medium only (the corresponding MGIT culture was contaminated with Gram-negative bacilli and Gram-positive cocci). This acid-fast species has been implicated as an opportunistic pathogen in patients with CF, and, in a small case series, was associated with a rapid decline in lung function (14).
DISCUSSION
Automated liquid culture is a laborious, time-consuming, and relatively expensive method for recovery of mycobacteria. It is particularly problematic in the processing of sputum samples from patients with CF as a large amount of competing flora may require eradication to avoid the subsequent contamination of liquid cultures by nonmycobacterial species. In this study, one-third of the MGIT cultures required a repeat round of decontamination due to growth of nonmycobacterial species, and 6.4% of cultures had to be abandoned. Despite these limitations and in the absence of adequate alternative methods, consensus guidelines by international experts recommend automated liquid culture as the principal method for the detection of mycobacteria from patients with CF (1). Perhaps mindful of the burden that such methods place on diagnostic laboratories, international guidelines recommend that cultures only be submitted annually for investigation of NTM in spontaneously expectorating individuals with a stable clinical course.
In the first published evaluation of the use of RGM medium, Preece et al. demonstrated performance superior to the use of BCSA for recovery of mycobacteria (11). The study examined 502 consecutive sputum samples from patients with CF in the United Kingdom and reported that 54 isolates of NTM were recovered on RGM medium versus 17 recovered on BCSA (P ≤ 0.0001). A recent study in Germany (13) also demonstrated a greater yield of mycobacteria on RGM medium than on a more selective formulation of BCSA in an evaluation with 224 sputum samples (P = 0.023). Due to the very high selectivity of RGM medium, an increased inoculum of 100 μl of digested sputum was used in the German study in an attempt to maximize recovery of mycobacteria, and this approach has been adopted in this study.
These studies prompted us to investigate how RGM medium might compare with formal AFB culture. We report here equivalent performance of RGM medium and the MGIT method (P = 1.00), suggesting that culture using RGM medium might have the potential to replace automated liquid culture for routine surveillance of NTM in patients with CF.
It should be emphasized that RGM medium was primarily designed for isolation of rapidly growing mycobacteria such as M. abscessus complex. There are insufficient available data to assess the ability of RGM medium to recover slower-growing isolates of NTM such as M. avium complex. We would therefore encourage further trials of RGM medium, particularly in locations where slower-growing species predominate in the CF population.
In conclusion, the use of RGM medium is an attractive alternative method for culture of sputum samples for isolation of NTM. No decontamination of the sample is required, making processing of samples extremely straightforward; consequently, the method can be easily embedded into routine culture methods. This means that all samples referred for routine culture can be conveniently screened for NTM.
ACKNOWLEDGMENTS
We are grateful to the staff of the National Mycobacterium Reference Laboratory, London, United Kingdom, for species identification of mycobacteria.
The Freeman Hospital Microbiology Department (represented by C.L.P. and J.D.P.) receives funding from bioMérieux for the development and evaluation of culture media, and J.D.P. has performed consultancy work for the same company. The other authors declare no conflicts of interest.
Funding Statement
This study used internal funding only.
REFERENCES
- 1.Floto RA, Olivier KN, Saiman L, Daley CL, Herrmann JL, Nick JA, Noone PG, Bilton D, Corris P, Gibson RL, Hempstead SE, Koetz K, Sabadosa KA, Sermet-Gaudelus I, Smyth AR, van Ingen J, Wallace RJ, Winthrop KL, Marshall BC, Haworth CS. 2016. US Cystic Fibrosis Foundation and European Cystic Fibrosis Society consensus recommendations for the management of non-tuberculous mycobacteria in individuals with cystic fibrosis: executive summary. Thorax 71:88–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martiniano SL, Nick JA, Daley CL. 2016. Nontuberculous mycobacterial infections in cystic fibrosis. Clin Chest Med 37:83–96. doi: 10.1016/j.ccm.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Qvist T, Gilljam M, Jönsson B, Taylor-Robinson D, Jensen-Fangel S, Wang M, Svahn A, Kötz K, Hansson L, Hollsing A, Hansen CR, Finstad PL, Pressler T, Høiby N, Katzenstein TL, Scandinavian Cystic Fibrosis Study Consortium (SCFSC). 2015. Epidemiology of nontuberculous mycobacteria among patients with cystic fibrosis in Scandinavia. J Cyst Fibros 14:46–52. doi: 10.1016/j.jcf.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seddon P, Fidler K, Raman S, Wyatt H, Ruiz G, Elston C, Perrin F, Gyi K, Bilton D, Drobniewski F, Newport M. 2013. Prevalence of nontuberculous mycobacteria in cystic fibrosis clinics, United Kingdom, 2009. Emerg Infect Dis 19:1128–1130. doi: 10.3201/eid/1907.120615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bar-On O, Mussaffi H, Mei-Zahav M, Prais D, Steuer G, Stafler P, Hananya S, Blau H. 2015. Increasing nontuberculous mycobacteria infection in cystic fibrosis. J Cyst Fibros 14:53–62. doi: 10.1016/j.jcf.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Raidt L, Idelevich EA, Dübbers A, Küster P, Drevinek P, Peters G, Kahl BC. 2015. Increased prevalence and resistance of important pathogens recovered from respiratory specimens of cystic fibrosis patients during a decade. Pediatr Infect Dis J 34:700–705. doi: 10.1097/INF.0000000000000714. [DOI] [PubMed] [Google Scholar]
- 7.Bange FC, Böttger EC. 2002. Improved decontamination method for recovering mycobacteria from patients with cystic fibrosis. Eur J Clin Microbiol Infect Dis 21:546–548. doi: 10.1007/s10096-002-0760-y. [DOI] [PubMed] [Google Scholar]
- 8.Ferroni A, Vu-Thien H, Lanotte P, Le Bourgeois M, Sermet-Gaudelus I, Fauroux B, Marchand S, Varaigne F, Berche P, Gaillard JL, Offredo C. 2006. Value of the chlorhexidine decontamination method for recovery of nontuberculous mycobacteria from sputum samples of patients with cystic fibrosis. J Clin Microbiol 44:2237–2239. doi: 10.1128/JCM.00285-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buijtels PC, Petit PL. 2005. Comparison of NaOH-N-acetyl cysteine and sulfuric acid decontamination methods for recovery of mycobacteria from clinical specimens. J Microbiol Methods 62:83–88. doi: 10.1016/j.mimet.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Esther CR Jr, Hoberman S, Fine J, Allen S, Culbreath K, Rodino K, Kerr A, Gilligan P. 2011. Detection of rapidly growing mycobacteria in routine cultures of samples from patients with cystic fibrosis. J Clin Microbiol 49:1421–1425. doi: 10.1128/JCM.02379-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Preece CL, Perry A, Gray B, Kenna DT, Jones AL, Cummings SP, Robb A, Thomas MF, Brodlie M, O'Brien CJ, Bourke SJ, Perry JD. 2016. A novel culture medium for isolation of rapidly-growing mycobacteria from the sputum of patients with cystic fibrosis. J Cyst Fibros 15:186−191. doi: 10.1016/j.jcf.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Roux AL, Catherinot E, Ripoll F, Soismier N, Macheras E, Ravilly S, Bellis G, Vibet MA, Le Roux E, Lemonnier L, Gutierrez C, Vincent V, Fauroux B, Rottman M, Guillemot D, Gaillard JL, Jean-Louis Herrmann Group OMA . 2009. Multicenter study of prevalence of nontuberculous mycobacteria in patients with cystic fibrosis in France. J Clin Microbiol 47:4124–4128. doi: 10.1128/JCM.01257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Preece CL, Wichelhaus TA, Perry A, Jones AL, Cummings SP, Perry JD, Hogardt M. Evaluation of various culture media for detection of rapidly-growing mycobacteria from patients with cystic fibrosis. J Clin Microbiol, in press. doi: 10.1128/JCM.00471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler WR, Sheils CA, Brown-Elliott BA, Charles N, Colin AA, Gant MJ, Goodill J, Hindman D, Toney SR, Wallace RJ Jr, Yakrus MA. 2007. First isolations of Segniliparus rugosus from patients with cystic fibrosis. J Clin Microbiol 45:3449–3452. doi: 10.1128/JCM.00765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]