Abstract
Aromatic grasses of the genus Cymbopogon (Poaceae family) represent unique group of plants that produce diverse composition of monoterpene rich essential oils, which have great value in flavor, fragrance, cosmetic, and aromatherapy industries. Despite the commercial importance of these natural aromatic oils, their biosynthesis at the molecular level remains unexplored. As the first step toward understanding the essential oil biosynthesis, we performed de novo transcriptome assembly and analysis of C. flexuosus (lemongrass) by employing Illumina sequencing. Mining of transcriptome data and subsequent phylogenetic analysis led to identification of terpene synthases, pyrophosphatases, alcohol dehydrogenases, aldo-keto reductases, carotenoid cleavage dioxygenases, alcohol acetyltransferases, and aldehyde dehydrogenases, which are potentially involved in essential oil biosynthesis. Comparative essential oil profiling and mRNA expression analysis in three Cymbopogon species (C. flexuosus, aldehyde type; C. martinii, alcohol type; and C. winterianus, intermediate type) with varying essential oil composition indicated the involvement of identified candidate genes in the formation of alcohols, aldehydes, and acetates. Molecular modeling and docking further supported the role of identified protein sequences in aroma formation in Cymbopogon. Also, simple sequence repeats were found in the transcriptome with many linked to terpene pathway genes including the genes potentially involved in aroma biosynthesis. This work provides the first insights into the essential oil biosynthesis of aromatic grasses, and the identified candidate genes and markers can be a great resource for biotechnological and molecular breeding approaches to modulate the essential oil composition.
Keywords: Cymbopogon, aromatic grasses, transcriptome, essential oil, gene candidates, monoterpene biosynthesis
Introduction
Essential oils, also known as volatile or ethereal oils or essences, are the mixtures of highly fragrant compounds found in aromatic plants and flowers. The essential oil producing plants are distributed widely across the plant kingdom covering a large number of families including Lamiaceae (mint, basil, lavender), Rosaceae (roses), and Poaceae (aromatic grasses) (Nagegowda and Dudareva, 2006). Cymbopogon (aromatic grasses), the unique genus of Poaceae family known for its aromatic properties, comprises of about 180 species distributed across the world (Bertea and Maffei, 2010), of which 45 species have been reported in India (Padalia et al., 2011). These aromatic grasses are endowed with differential blend of several terpenoidal constituents and are large reserves of monoterpene rich essential oils (Devi et al., 2015). The major constituents of essential oils of these aromatic grasses comprise monoterpene alcohols geraniol (GOL) and citronellol (COL), aldehydes geranial (GAL), neral (NAL) and citronellal (CAL), and acetates such as citronellyl acetate (CA) and geranyl acetate (GA) (Khanuja et al., 2005). The essential oils of aromatic grasses have great importance in food flavors, fragrances, cosmetics, oral healthcare products, insect repellents, and aromatherapy. For instance, citral has wide industrial uses as raw material for perfumery, confectionery, Vitamin A and ionones (Moyler, 2010). Likewise, COL and its aldehyde CAL are used for manufacturing of flavor and fragrance agents. COL is also used as a raw material for the production of rose oxide (Pimentel et al., 2012). In addition, essential oils and their individual constituents from aromatic grasses possess potent pharmacological activities like cytotoxic, anti-inflammatory, antifungal, and antioxidant activities (Bayala et al., 2014).
Cymbopogon flexuosus (lemongrass), C. martinii (palmarosa), and C. winterianus (Java citronella), are three economically important species widely cultivated for extracting high value essential oils. These species are classified into three distinct groups based on their essential oil composition such as aldehyde type (lemongrass), alcohol type (palmarosa), and intermediate type (Java citronella) that accumulates both alcohol and aldehydes (Lavania et al., 2012). The growing global demand for Cymbopogon aromatic oils and their individual derivatives necessitates development of high yielding varieties that requires better understanding of genetic makeup and essential oil biosynthetic pathway in Cymbopogon. Breeding for essential oil improvement in Cymbopogon has been restricted to selection from natural populations because of problems associated with irregular flowering and seed setting. Moreover, attempts for mutation breeding have met with little success (Sharma and Ram, 2000). Hence, molecular breeding approaches using genomic resources could be promising for modulating the essential oil accumulation in Cymbopogon. Furthermore, understanding the biochemical and molecular mechanisms of essential oil biosynthesis could aid in metabolic engineering for enhanced essential oil production. Significant progress has been made in studying genetic analysis, chemodiversity and pharmacological effects of essential oils from Cymbopogon (Padalia et al., 2011; Lavania et al., 2012; Bayala et al., 2014; Rao et al., 2015). To date, reports on biochemical and molecular mechanisms of essential oil biosynthesis and molecular markers in this important genus remain very limited. As for the genomic resources, only 223 nucleotide and 180 protein sequences are reported in National Center for Biotechnology Information (NCBI) database, which provide little information on the genes responsible for the aroma formation in Cymbopogon species. Next generation sequencing (NGS) has emerged as a promising platform to discover novel genes, enzymes, transcription factors, and molecular markers from non-model plant species. In recent years, transcriptome approaches have been widely used for discovering and characterizing genes involved in secondary metabolic pathways (Yang et al., 2013; Rastogi et al., 2014; Niu et al., 2015) and also for identifying markers for molecular breeding (Shahin et al., 2012; Miah et al., 2013; Xu et al., 2013). Although leaf and root transcriptome analysis of citronella (C. winterianus) identified transcripts encoding MVA and MEP pathway genes, there was no investigation pertaining to downstream genes of essential oil biosynthetic pathway (Devi et al., 2016).
As the first step toward understanding the biosynthesis and regulation of essential oils and to generate genomic resources in aromatic grasses, here we report Illumina transcriptome sequencing, analysis and functional annotation of lemongrass (C. flexuosus) as a representative model for the genus Cymbopogon. In silico analysis of the transcriptome data identified several genes involved in essential oil biosynthesis, which included terpene synthases (TPS), pyrophosphatases (PPase), alcohol dehydrogenases (ADH), aldo-keto reductases (AKR), carotenoid cleavage dioxygenases (CCD), alcohol acetyltransferases (AAT) and aldehyde dehydrogenases (ALDH). Further, comparative essential oil profiling and gene expression analysis in different Cymbopogon sp. and homology modeling gave insights into their specific involvement in aroma biosynthesis. Also, SSR markers identified from the generated transcripotme will be useful resource for further genetic improvement by molecular breeding.
Materials and Methods
Plant Material, Library Preparation, and Sequencing
Mature leaves were collected from different plants of C. flexuosus cv. Krishna grown in field conditions (temperature 25°C ± 2°C and average humidity 60%) from CSIR-Central Institute of Medicinal and Aromatic Plants, Lucknow, India. The collected leaves were pooled and total RNA was isolated using Qiagen RNeasy mini kit (Qiagen, USA) following manufacturer’s protocol. The RNA integrity was assessed using Qubit 2.0 Fluorometer with Qubit RNA BR Assay kit (Life Technologies, USA) and on a 2100 Bioanalyzer using an Agilent RNA 6000 Pico kit (Agilent Technologies, USA). 4 μg of total RNA with an RNA Integrity Number (RIN) value of 7.0 was used for cDNA synthesis. cDNA library was prepared according to Illumina TruSeq RNA low throughput library protocol according to “TruSeq RNA Sample Preparation Guide” (Part # 15008136; Rev. A; November 2010). The library quality was assessed using 2100 Bioanalyzer using DNA 1000 kit (Agilent Technologies, USA), concentration measured using library quantification kit (Kapa Biosystems, USA) and sequencing was performed using the HiSeq2000 platform (Illumina Inc., USA) after indexing the sample and paired end library was prepared.
Assembly and Annotation
The generated raw reads were deposited in the NCBI Short Read Archive (SRA) (SRP066939). Raw reads obtained after sequencing were filtered to obtain processed reads by removing Illumina adapter and low quality bases (Q < 20). De novo assembly was done using Velvet-1.2.09 & Oases-0.2.8 with kmer size of 31 (Zerbino and Birney, 2008; Schulz et al., 2012). Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values were calculated by first aligning the trimmed reads to the assembled transcriptome using Bowtie2 program with upto 1-mismatch allowed in the seed region (length = 31bp). The assembled transcripts were annotated using BLASTX search against the NCBI Nr1 and UniProt2 database with an E-value cut-off of 10-5. GO terms were assigned to the annotated transcripts based on BLASTX hits against Universal Protein Resource (UniProt). Pathways were assigned by employing BLASTX search against Kyoto Encyclopaedia of Genes and Genomes (KEGG) using KEGG Automatic Annotation Server (KAAS)3. The transcription factor terms were assigned to each transcript by BLASTX search against Arabidopsis Gene Regulatory Information Server (AGRIS). The microsatellite program MISA Perl script4 was used for identification of SSRs. The parameters used were at least 12 repeats for mono-, 6 repeats for di-, 5 for each tri- and tetra-, 4 for each penta- and hexa- nucleotide sequences.
Identification of Genes Related to Essential Oil Biosynthesis
Candidate transcripts were identified on the basis of their KEGG, NCBI, and UniProt annotation. The transcripts were translated using ExPASy translate tool5. Amino acid sequence alignment was generated using MAFFT version 76 and BOXSHADE 3.217. Sequence relatedness and unrooted neighbor joining phylogenetic tree with 1000 bootstrap value was generated using Molecular Evolutionary Genetics Analysis tool version 6 (MEGA 6)8.
Essential Oil Extraction and GC- MS Analysis
Fresh leaf tissues (200 g) of C. flexuosus, C. winterianus and C. martinii, and inflorescence of C. martinii (200 g) were subjected to hydro-distillation (Li et al., 2014) using clevenger apparatus for 3h at 50°C and extracted oils were dried over Na2SO4. Gas Chromatography–Mass Spectrometry (GC–MS) analysis was performed in Agilent Technologies 7980A GC system coupled with 5977A MS detector (Agilent Technologies, USA). Essential oil was diluted 1000 times using pentane and toluene (0.0001%) was added as internal standard. One μl was injected in split mode in HP5-MS column (30 m × 250 μm with 0.25 μm film thickness). The oven temperature was adjusted to 40°C for 5 min followed by 150°C at the rate of 3°C/min, then with 5°C/min up to 200°C with a hold of 10 min and finally up to 300°C with a ramp rate of 10°C/min and a final hold for 10 min. The split ratio of 10:1 was maintained with Helium as the carrier gas at a flow rate of 1.0 ml/min. Compounds were identified using NIST/EPA/NIH MS library version 2.0g (Agilent Technologies, USA). The % composition of individual monoterpene components in different essential oils of Cymbopogon were determined after calibrating the peak area of internal standard toluene. The data shown is the average of three technical replicates.
qRT- PCR Analysis
RNA isolation, cDNA synthesis and qRT-PCR analysis were performed as reported previously (Kumar et al., 2015; Singh et al., 2015). Briefly, total RNA was extracted from 100 mg tissues of C. flexuosus (root, leaf, and inflorescence), C. winterianus (leaf and inflorescence), and C. martinii (leaf and inflorescence) using SpectrumTM Plant Total RNA Kit (Sigma–Aldrich, USA) according to the manufacturer’s instructions. In all cases, on-column DNase digestion was performed to remove trace amounts of DNA using DNase I (Sigma–Aldrich, USA). The DNA-free total RNA was quantified by UV-spectrophotometer (Kinetic Biospectrometer, Eppendorf, Germany). Two microgram of total RNA was used for first-strand cDNA synthesis with random hexamers using RevertAid H Minus Reverse Transcriptase (Thermo scientific Inc., Canada). Real-time qPCR was performed with a linear range of cDNA using Step One Real Time PCR System (Applied Biosystems, USA). For validating the stability of reference genes to be used for qPCR normalization, elongation factor 1α (EF1α), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and actin (ACT) were used (Supplementary Table S1). Only EF1α was further used for normalization because of its invariant expression under the tested conditions. qRT–PCR was performed with 5 μl of reaction volume containing 2.5 μl of 2X Maxima SYBR Green PCR master mix (Thermo Scientific, USA), 1:10 diluted cDNA and 2 μM gene-specific primers with following conditions, 94°C for 10 min for first cycle, followed by 40 cycles of 94°C for 15 s. 60°C for 15 s. Fold change differences in gene expression were analyzed using the comparative cycle threshold (Ct) method (Applied Biosystems, USA). All experiments were repeated using three technical replicates and data were analyzed statistically (±SD).
Homology Modeling and Docking
Homology models were built by comparative protein modeling with the help of MODELER 9.15 (Sali and Blundell, 1993), and validated using Ramachandran plot. The 3D coordinates of template structures were obtained from Protein Data Bank (PDB)9 by using BLASTP. The homology models of CfADH1 and CfADH2a were built using the X-ray structures of Populus tremuloides synapyl alcohol dehydrogenase (PDB ID: 1YQD), which shared 74% sequence identity. The 3D structure for AKR2b was generated using its suitable experimental structure homolog, perakine reductase from Rauvolfia serpentina (PDB ID: 3V0T), which shared 55% sequence identity. Hydroxycinnamoyl transferase (PDB ID: 4G0B; 30% sequence identity) structure from Coffea canephora and ALDH X-ray structure from Bos taurus (PDB ID: 1AG8; 62% sequence identity) were used to develop the homology models for CfAAT3 and CfALDH3, respectively. The models were visualized by Python Molecular Viewer (PMV) (Sanner, 1999). Homology models were modified by adding Kollman charges and polar hydrogen using ADT command. Docking studies were done using Autodock suite (Goodsell et al., 1996). The 3D substrate structures were created using Java molecular editor, and the structure topologies and energy minimization were carried out with GROMOS87 force fields with PRODRG suite (Schüttelkopf and van Aalten, 2004). Possible binding sites of substrates on candidate proteins were obtained by defining pregrids using autogrid command, and docking was done with Lamarckian genetic algorithm (LGA). The 50 LGA runs were performed in a defined grid map. Low RMSD and lowest ΔG of binding between receptor and the substrate compound was the criteria used for defining the possible binding site. The substrate bound complexes were visualized by PMV software.
Results and Discussion
De Novo Assembly and Functional Annotation
Aromatic grasses have been widely studied in terms of their essential oil composition but there are limited studies on its essential oil biosynthesis, which could be due to non-availability of genomic resources. Here, we have generated C. flexuosus leaf transcriptomic data using Illumina HiSeq2000 platform that has been used for sequencing many economically important monocots (Shahin et al., 2012; Davey et al., 2013). The transcript assembly details are provided in Table 1. Majority of the transcripts were between 150 and 1000 bases (75,261 transcripts) followed by 1000–2000 bases (14,565 transcripts) (Supplementary Figure S1A). The average length (635 bases), GC content (49.89%), and N50 values for the assembled transcripts were within the range of other monocot transcriptomes from Zea mays (Hansey et al., 2012), Lilium genus (Shahin et al., 2012; Du et al., 2015), and Musa balbisiana (Davey et al., 2013), which were assembled using the same sequencing platform. Of the total transcripts, 92,139 (99%) with FPKM of >1 (Supplementary Figure S1B) were subjected to BLASTX against different databases and the annotation summary is provided in Supplementary Table S2. The results showed that 82.80% (76,293) transcripts had at least one significant hit against NCBI non-redundant (Nr) protein database, with ∼ 98% having similarity of >60% at protein level, indicating high protein conservation (Supplementary Figures S1C,D).
Table 1.
RNA sequencing summary of C. flexuosus leaf transcriptome.
| Summary of RNA-Seq | C. flexuosus |
|---|---|
| Total Number of HQ Reads | 26936556 (26.93 Mb) |
| Number of paired-end reads after trimming | 23793466 (23.79 Mb) |
| Mean read quality (Phred score) | 33.85 |
| Number of bases (MB) | 2720.59 |
| Number of bases (GB) after trimming | 1.89 |
| Mean read length (bases) | 101 |
| kmer size | 31 |
| Number of assembled transcripts | 107,363 |
| Number of transcripts with length ≥ 150 bases | 92,937 |
| Maximum transcript length (bases) | 47,050 |
| Average transcript length (bases) | 635 |
| N50 value | 968 |
| Mean GC % of transcripts | 49.89 |
| Number of transcripts with FPKM ≥1.0 | 92,139 |
Organism distribution in NCBI annotations showed presence of 906 organisms that contained homologous genes for C. flexuosus transcripts with top 5 organisms belonging to Poaceae family. Oryza sativa ssp. japonica showed highest coverage with 39.6% (30,235) followed by Sorghum bicolor and Z. mays with 23.5% (17,909) and 14.36% (10,962), respectively (Supplementary Figure S2). In total, 69,984 transcripts represented 5,281 GO terms distributed among three main ontologies comprising molecular function (MF), cellular components (CC), and biological processes (BP) (Supplementary Figure S3). Various GO assignments of classified unigenes revealed the diversity of transcripts represented in lemongrass transcriptome. Since, trancription factors (TFs) play key role in regulating gene expression and metabolite accumulation in different metabolic pathways, a BLASTX search against AGRIS was performed that resulted in 5,867 transcripts (6.37%) with “transcription factor activity” belonging to 44 known TF families (Supplementary Figure S4). The top 5 hits of TFs were represented by Trihelix, C2H2, C3H, bHLH, and WRKY class. Further analysis and characterization of TFs from the transcriptome data could give insights into their role in regulating monoterpene biosynthesis in Cymbopogon. Overall, 76,409 (82.92%) transcripts significantly corresponded to known or unknown proteins present in public databases. A significant number of 15,730 (17.07%) transcripts remained unannotated, which may belong to untranslated regions, non-coding RNAs, intergenic spacers (Liu et al., 2012), or may be unique to C. flexuosus and can be a great resource for the discovery of novel genes.
Secondary Metabolic Pathways Identified in C. flexuosus Leaf Transcriptome
In order to find transcripts related to secondary metabolic pathways, assembled transcripts were annotated against KEGG, NCBI, and UniProt databases. About 26.21% (24,147) transcripts were assigned to 280 KEGG pathways among different categories that included metabolism, cellular processes, genetic information processing, environmental information processing, and others, with “metabolism” having the highest share of 36% (8,719 transcripts) (Supplementary Figure S5). Within the “metabolism” category, 38 metabolic pathways related to secondary metabolism (484 transcripts) were identified with terpenoid biosynthesis representing the largest group with 151 transcripts (Figure 1). This is in accordance with higher amounts of terpenes present in C. flexuosus essential oil. KEGG analyses of transcriptomes from terpene rich Ocimum basilicum and phenylpropanoid rich Ocimum sanctum also showed higher representation of respective pathway related transcripts (Rastogi et al., 2014), indicating that the distribution of transcripts involved in secondary metabolism correlates with the essential oil composition. Also, significant number of transcripts involved in the biosynthesis of secondary metabolites like phenylpropanoids (18%), alkaloids (12%), and flavonoids (10%) were represented. The total number of transcripts encoding enzymes involved in major metabolic pathways obtained from KEGG, NCBI, and UniProt annotations are represented in Supplementary Tables S3 and S4.
FIGURE 1.
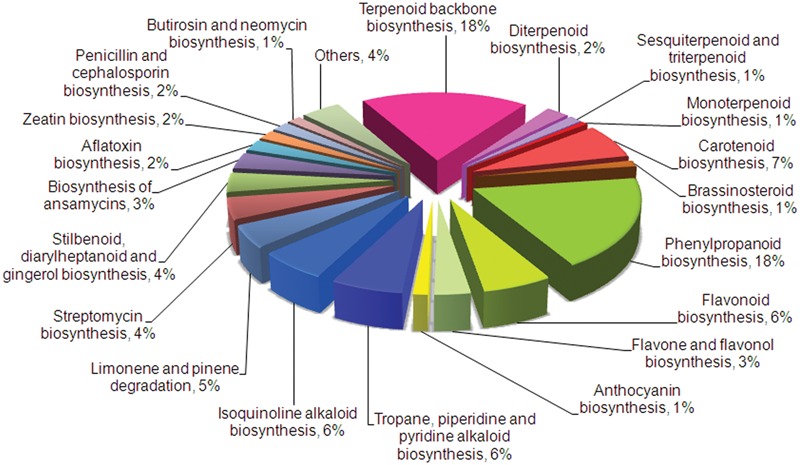
KEGG classifications of genes involved in secondary metabolism in Cymbopogon flexuosus leaf transcriptome. Relative percentage of transcripts involved in various secondary metabolic pathways is shown.
Identification of Transcripts Related to Terpenoid Metabolism
The essential oil of Cymbopogon comprises mainly of monoterpene alcohols (GOL, COL), aldehydes (GAL, NAL, CAL), and acetates (GA, CA). The schematic representation of the steps involved in biosynthesis of these compounds is shown in Figure 2. Terpenoids are derived from two five-carbon precursors, isopentenyl diphosphate (IPP) and its isomer, dimethylallyl diphosphate (DMAPP) via the cytosolic mevalonic acid (MVA) pathway and plastidial methylerythritol phosphate (MEP) pathway (Nagegowda, 2010). In C. flexuosus transcriptome, 56 and 77 transcripts were annotated for 7 genes of MEP pathway and 6 genes of MVA pathway, respectively (Supplementary Table S3). IPP and DMAPP are further converted to geranyl diphosphate (GPP), farnesyl diphosphate (FPP), and geranylgeranyl diphosphate (GGPP), by GPP synthase (GPPS), FPP synthase (FPPS), and GGPP synthase (GGPPS), respectively (Nagegowda, 2010). In this analysis, we identified 8 transcripts for GPPS, 9 transcripts each for FPPS and GGPPS (Supplementary Table S3). In general, plants produce monoterpenes from GPP, which is formed by GPPS. Both homomeric and heteromeric forms of GPPS have been reported in plants (Rai et al., 2013), however, mostly heteromeric GPPS have been shown to be involved in monoterpene bisoynthesis (Wang and Dixon, 2009). Although C. flexuosus contained transcripts for both homomeric and heteromeric GPPS (Supplementary Table S3), only heteromeric GPPS may be involved in monoterpene biosynthesis similar to dicots (Rai et al., 2013).
FIGURE 2.

Biosynthetic pathway of essential oil biosynthesis in C. flexuosus. Broken arrows indicate multiple steps. Green arrows indicate alternate route of biosynthesis or already characterized steps in other plants. Double headed arrow indicates bidirectional reaction. Sequences analyzed in this study is shown in red color. AAT, alcohol acetyltransferase; ADH, alcohol dehydrogenase; AKR, aldo-keto reductase; ALDH, aldehyde dehydrogenase; CCD, carotenoid cleavage dioxygenase, DMAPP, dimethylallyl diphosphate; FPPS, Farnesyl diphosphate synthase; GES, geraniol synthase; GPP, Geranyl diphosphate; GP, Geranyl monophosphate; GPPS, GPP synthase; GGPP, Geranylgeranyl diphosphate; GGPPS, GGPP synthase; IPP, isopentenyl diphosphate; NUDX, nudix hydroxylase; PPase, pyrophosphatase.
Geranyl diphosphate is further converted to GOL either by geraniol synthase (GES) in plastids (Simkin et al., 2013) or through recently proposed NUDX1 of Nudix hydrolase superfamily in the cytosol (Magnard et al., 2015) or by yet to be identified PPase (Nah et al., 2001) (Figure 2). We identified 16, 26, and 17 transcripts for TPS, NUDX, and PPase, respectively, which could play a role in GOL biosynthesis (Supplementary Table S3). It is also proposed that GOL could be formed from GAL through the action of ADH/AKR (Iijima et al., 2014; Sato-Masumoto and Ito, 2014). Genes involved in downstream conversion of GOL into its aldehyde, acetate or acid derivatives involve ADH/AKR, AAT, and ALDH at their respective steps of synthesis (Figure 2). Our search resulted in 92 ADHs, 38 AKRs, 35 AATs, and 88 ALDHs (Supplementary Table S3). Aldehydes (GAL and NAL) could also be synthesized by a novel route utilizing carotenoids as substrate through the action of CCDs as reported in rice and tomato (Ilg et al., 2009, 2014). C. flexuosus transcriptome contained 11 transcripts annotated as CCDs (Supplementary Table S3).
Mining of Genes Related to Essential Oil Biosynthesis in C. flexuosus
For mining of candidate genes, only those transcripts encoding full length proteins were considered for further analyses. Search for candidates involved in GOL formation yielded 1, 6, and 8 genes encoding TPS (Supplementary Figure S6), PPase, and NUDX, respectively. BLAST analysis of C. flexuosus putative NUDX candidates showed no significant homology to the recently characterized NUDX from Rosa hybrida (RhNUDX1) (Magnard et al., 2015). Also, when RhNUDX1 was used to search homologous candidates against NCBI monocot and other databases including Oryzabase10, Rice Genome Annotation Project11, Phytozome v10.312 and PlantGDB13, it did not yield any homologous NUDX candidates except in Zostera marina (seagrass) (52% identity) of Zosteraceae family. The obtained results could possibly be due to the reason that Z. marina is more close to dicots, further suggesting that divergence of monocots and dicots occurred after Z. marina got established as a marine plant (Kong et al., 2014). Since there were no homologs for RhNUDX1 in C. flexuosus, only TPS and PPase were considered for further analyses. Although GES enzymes involved in the formation of GOL have been characterized in dicots (Iijima et al., 2004; Dong et al., 2013; Simkin et al., 2013), they are yet to be identified in monocots. The identified CfTPS1 in this study had the closest similarity (63% identity) with putative LIS/NES from rice. Among the characterized GES from other plants, Vitis vinifera GES exhibited a low similarity (36% identity) to CfTPS1 (Supplementary Figure S6; Supplementary Table S5). As for the PPase, it has been reported that in rice seedlings FPPase and GGPPase activities are involved in formation of farnesol and geranylgeraniol from FPP and GGPP, respectively (Nah et al., 2001). A similar phosphatase activity could be possibly involved in GOL formation from GPP in monocots. However, the molecular evidence for such activity still needs to be determined.
Members of MDR superfamily (medium chain dehydro genases/reductases) such as cinnamyl alcohol dehydrogenases (CAD) and ADH families have been reported in citral formation. In this study, among 46 transcripts encoding ADH proteins, 9 sequences (denoted as CfADH1, CfADH2a-b, CfADH3a-e, and CfADH4) showing high homology to already characterized geraniol dehydrogenase (GeDH) and CAD (with GeDH activity) were considered for their relatedness through phylogeny (Figure 3A and Supplementary Table S5). CfADH1, CfADH2a, and CfADH2b were clustered in the CAD class-II having specific GeDH activity catalyzing conversion of GOL/nerol (NOL) to GAL/NAL (Figure 3A and Supplementary Table S5). CfADH1 exhibited highest amino acid identity (59%) with Zingiber officinale (ZoGeDH1) that catalyzes bidirectional inter-conversion of GOL to GAL (Iijima et al., 2014). CfADH2a and CfADH2b exhibited highest identity of 65 and 62% identity, respectively, to ZoGeDH1 (Iijima et al., 2014) (Figure 3A and Supplementary Table S5). CfADH3a-e formed a different clade with multifunctional CAD class-I members, sharing 77% identity with O. basilicum ObCAD1 (Iijima et al., 2006) and 74% identity with Artemisia annua AaCAD (Li et al., 2012) that exhibited catalytic ability toward different aliphatic and aromatic aldehydes/alcohols including GAL/GOL. CfADH4 formed a separate clade with members of benzyl alcohol denydrogenase (benzyl/aryl ADHs) family exhibiting GeDH activity. CfADH4 shared 35 and 31% identity to GeDH from astigmatid mite Carpoglyphus lactis (Noge et al., 2008) and Castellaniella defragrans (Lüddeke et al., 2012), respectively. Search for AKRs (members of ADH family) yielded 3 candidates showing maximum homology to the characterized AKRs from Perilla (Sato-Masumoto and Ito, 2014) involving GOL to GAL conversion with CfAKR1 sharing 62% identity; CfAKR2a and CfAKR2b having 68% identity (Figure 3B and Supplementary Table S5). From 7 transcripts encoding CCD proteins, one sequence named as CfCCD1 exhibited very high homology with rice (99%) and tomato (74–78%) CCDs that have been recently characterized to produce GAL and NAL by breaking C7–C8/C7 = C8 bonds (Ilg et al., 2009, 2014) (Figure 3C, Supplementary Figure S12; Supplementary Table S5).
FIGURE 3.
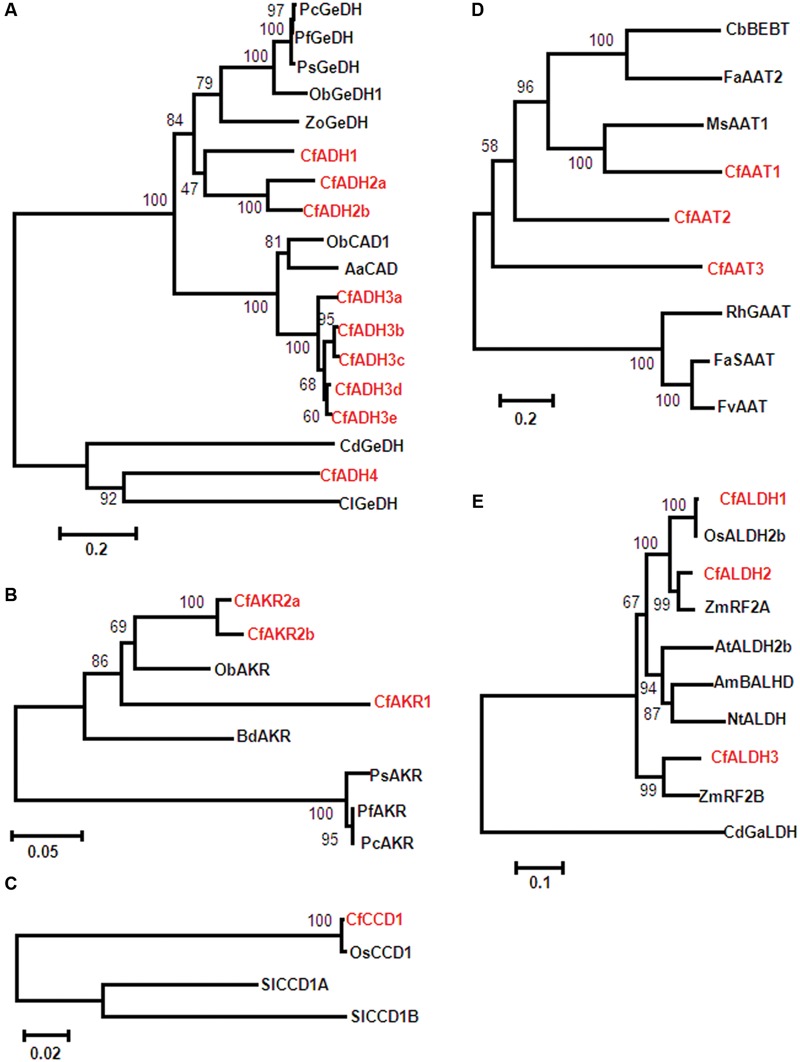
Phylogenetic relationship of C. flexuosus candidate sequences. The evolutionary relationship was analyzed by unrooted Neighbor–Joining (NJ) using MEGA6 (Tamura et al., 2013). The branch length of the line indicates evolutionary distance and numbers represent confidence of the phylogenetic tree calculated by bootstrap analysis from 1000 replicates. Phylogenetic analysis of ADH (A), AKR (B), CCD (C), AAT (D), and ALDH (E) candidates. The abbreviation and accession numbers are: (A) CAD, cinnamayl alcohol dehydrogenase; GeDH, geraniol dehydrogenase; Aa, Artemisia annua (ACB54931); Cd, Castellaniella defragrans (WP_043683915); Cl, Carpoglyphus lactis (B2N193); Cf, C. flexuosus; Ob, Ocimum basilicum (ObCAD1_Q2KNL5, ObGeDH1_AAX831C7); Pc, Perilla citriodora (AFY63473); Pf, Perilla frutescens (AFY63472); Ps, Perilla setoyensis (AFY63474); Zo, Zingiber officinale (BAR42579). (B) Bd, Brachypodium distachyon (XP_003575318); Cf, C. flexuosus; Ob, Oryza brachyantha (XP_006652179); Pc, P. citriodora (AFV99149); Pf, P. frutescens (AFV99148); Ps, P. setoyensis (AFV99150). (C) Cf, C. flexuosus; Os, Oryza sativa (AK066766); Sl, Solanum lycopersicum (SlCCD1A_AAT68187 and SlCCD1B_AAT68188). (D) Ms, Musa sapientum (CAC09063); CbBEBT, Clarkia breweri benzyl alcohol O-benzoyltransferase (AAN09796), FaSAAT- Faragaria ananassa alcohol acyltransferase (AAG13130); FvAAT2, Fragaria vesca alcohol acyltransferase (AAN07090); RhGAAT, Rosa hybrida acetyl CoA geraniol/citronellol acetyltransferase (AAW31948). (E) AmBALHD, Antirrhinum majus benzaldehyde dehydrogenase (ACM89738); At, Arabidopsis thaliana (Q8S528); CdGaLDH, C. defragrans geranial dehydrogenase (CCF55023); Cf, C. flexuosus; Nt, Nicotiana tabacum (CAA71003), Os, O. sativa (AAF73828); ZmRF, Zea mays restoration factor (ZmRF2A_AAC49371, ZmRF2B_AAL99613).
With respect to AAT enzymes involved in GA/CA formation, 6 AATs have been identified from plants including banana AAT1(Beekwilder et al., 2004), Clarkia breweri benzyl alcohol O-benzoyltransferase (Dudareva et al., 1998), Fragaria ananassa AAT (Cumplido-Laso et al., 2012), F. chiloenisis AAT (González et al., 2009), and R. hybrida acetyl CoA geraniol/citronellol acetyltransferase (RhGAAT1) (Shalit et al., 2003). Out of 8 AATs mined, CfAAT1, CfAAT2, and CfAAT3 showing 28, 40, and 23% identity, respectively, to banana AAT1, and RhGAAT1 were considered for further analyses (Figure 3D and Supplementary Table S5).
Geranic acid (GAc), an oxygenated monoterpene present in trace amounts in Cymbopogon sp. has anticancer/tyrosinase inhibitor and antifungal activity (Yang et al., 2011). GAc has been reported to be formed from GAL by ALDH enzyme. ALDH acting on GAL has so far only been reported from C. defragrans that catalyzes the NAD+ dependent oxidation of GAL to GAc (Lüddeke et al., 2012). Of 18 candidates encoding ALDH proteins, 3 CfALDH sequences were closely related to previously characterized ALDH from other plants (Figure 3E and Supplementary Table S5). While CfALDH1 shared highest homology with uncharacterized OsALDH2b (99%) from rice, CfALDH2 and CfALDH3 had close relationship with maize fertility-restorer (RF) genes ZmRF2B (84%) and ZmRF2A (95%), respectively (Cui et al., 1996; Liu et al., 2001) (Figure 3E and Supplementary Table S5). All CfALDH proteins showed ∼37% identity with characterized C. defragrans geranial dehydrogenase (GALDH) (Figure 3E and Supplementary Table S5).
Comparative Essential Oil Profiling and Gene Expression Analysis
The chemical diversity in essential oil imparts different fragrances to aromatic grasses, which could be due to the differential expression of genes involved in conversion of basic substrate into their derivatives. Also, it has been reported in many plant species that the level of terpenoid volatiles correlates with the expression of corresponding genes (Nagegowda, 2010). Hence, comparative gene expression and metabolite analyses could further facilitate narrowing down the possible gene candidates involved in essential oil formation. Since strong homology of gene sequences exists among the closely related species within the same genus, gene specific primers, designed based on lemongrass transcriptome, were used for determining the expression levels in all three Cymbopogon species that accumulate varying composition of monoterpenes (Lavania et al., 2012). First, leaf essential oils of C. flexuosus, C. winterianus, C. martinii, and inflorescence of C. martinii were analyzed (Figure 4A and Supplementary Figure S7). C. flexuosus (group I - aldehyde type) was rich in GAL (42%) and NAL (33%) which are together called as citral with relatively trace amounts of GOL and GA (Figure 4B). Essential oil of C. winterinus (group II- intermediate type) was well represented by different levels of various monoterpene derivatives with CAL (39%) representing the major constituent followed by 21% of alcohols (GOL and COL) and 11% of acetates (GA and CA) (Figure 4B). Essential oils from leaf and inflorescence of C. martinii were dominated by GOL (71 and 57%) and GA (5 and 15%) (Figure 4B and Supplementary Figure S7).
FIGURE 4.
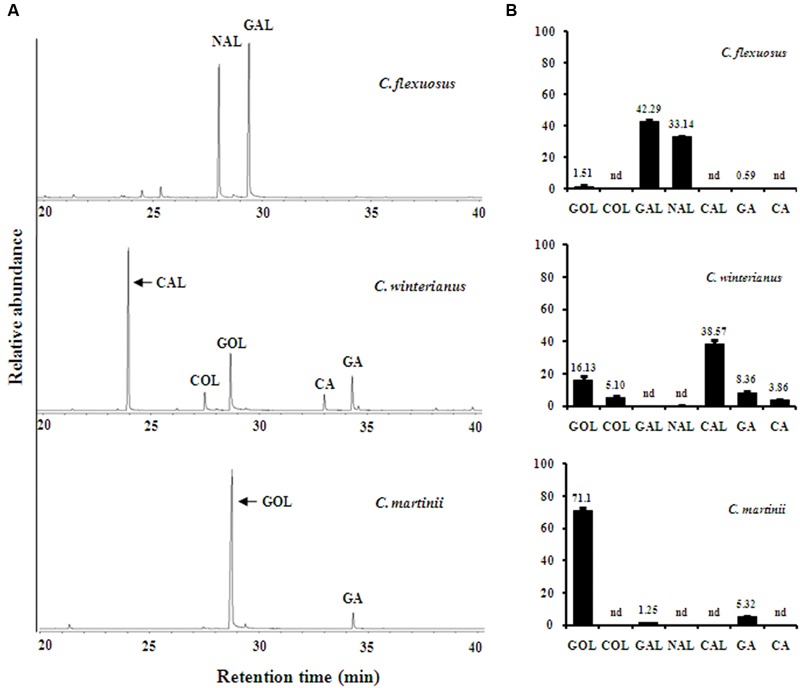
Gas Chromatography–Mass Spectrometry (GC–MS) profile representing relative abundance of essential oil components in Cymbopogon. Essential oils were hydrodistilled from 200 g fresh leaf tissue of C. flexuosus, C. winterianus, and C. martinii. One micro litre of diluted oil (1:1000 in pentane) was analyzed by GC–MS using HP5-MS column (A). The compounds were identified using NIST/EPA/NIH mass spectral library version 2.0 g and relative abundance of individual components was determined from pooled leaf samples with three technical replicates (B). Abbreviations: CA, citronellyl acetate; CAL, citronellal; COL, citronellol; GA, geranyl acetate; GAL, geranial; GOL, geraniol; NAL, neral; nd, not detected.
Next, the expression of identified candidate genes was compared in Cymbopogon that were used for essential oil analysis. Before proceeding for qPCR analysis of candidate genes, endogenous reference genes were selected and validated for transcript normalization. Among the selected reference genes, EF1α exhibited highest stability across different Cymbopogon species and in different tissues (Supplementary Figure S8A). Hence, EF1α was used for normalization in all subsequent qPCR analysis. For analysis, the species/gene having the least Ct was set to 100% to determine the abundance of transcripts relative to other species/genes. The expression of TPS1 was highest in C. winterianus followed by C. flexuosus (24%) and negligible in C. martinii (Figure 5). Tissue specific expression of CfTPS1 in C. flexuosus indicated minimal expression in root as compared to leaf (Supplementary Figure S8C). PPase1 exhibited highest expression in C. martinii followed by C. winterianus (15%) with least in C. flexuosus (5%). Although PPase2 followed similar trend as that of PPase1, the expression of PPase1 was ∼ three to eightfold higher compared to PPase2 in all three species. The GOL content in C. flexuosus has been reported to be 4 and 0.4% in leaf and root, respectively (Rao et al., 2015). Hence, to determine the involvement of PPases, tissue specific expression was studied, which showed a differential trend of PPase expression. While PPase1 showed fivefold higher expression in leaf compared to root, PPase2 exhibited sevenfold higher abundance in root (Figure 5 and Supplementary Figure S8C). Based on the GOL content, comparative and tissue specific expression, we implicate the involvement of PPase1 in GOL formation in aromatic grasses, similar to involvement of FPPase and GGPPase in farnesol and geranylgeraniol formation in rice (Nah et al., 2001).
FIGURE 5.
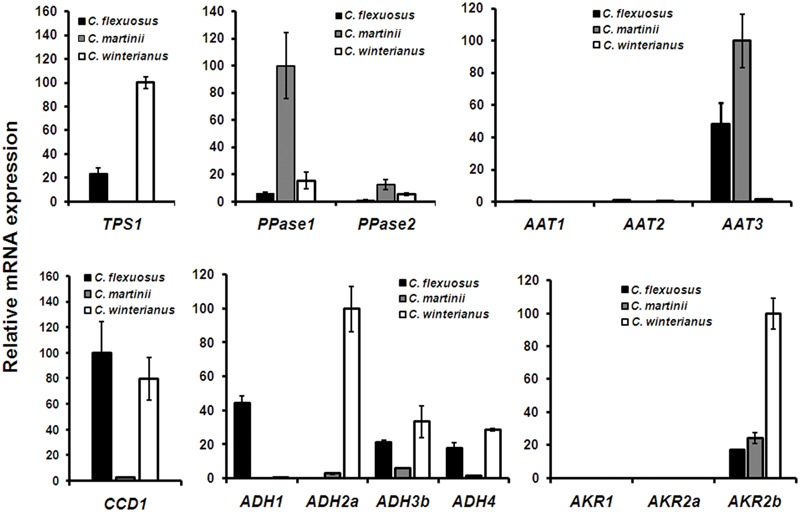
Comparative expression of candidate genes in Cymbopogon. Real-time qPCR analysis of TPS, PPase, CCD, ADH, AKR, and AAT candidates in C. flexuosus, C. martinii, and C. winterianus. While the expression of all candidates except AAT was compared in leaf tissue, the expression of AAT was analyzed in inflorescence. In each graph, the species/gene having the least Ct was set to 100 to determine the relative abundance of transcripts in other species. CfEF1α was used as endogenous control for normalization. Each data point represents the mean ± standard error (SE).
As aldehydes (GAL, NAL, and CAL) are reported to be formed via different routes involving ADH/AKR (Iijima et al., 2006; Sato-Masumoto and Ito, 2014) and CCD (Ilg et al., 2009, 2014), the expression of transcripts encoding the respective enzymes were analyzed. Of 9 ADH candidates used for phylogeny (Figure 3A), ADH1, ADH2a, ADH3b, and ADH4 having highest FPKM were used for expression analysis. In addition, all 3 AKR candidates were used for expression analyses. All 4 ADH candidates showed differential expression with varying levels among the three species analyzed (Figure 5). While expression of ADH2a, ADH3b, ADH4, and AKR2b was highest in C. winterianus, ADH1 exhibited highest expression in C. flexuosus. AKR1 and AKR2a showed basal level of expression in all species as compared to AKR2b (Figure 5). The expression of all four ADH candidates was least in C. martinii and AKR2b was least in C. flexuosus (Figure 5). In C. flexuosus, the expression of ADH1, ADH2a, and AKR2b, and ADH3b and ADH4 was higher in leaf and root, respectively (Figure 6 and Supplementary Figure S8C). The species and tissue specific expression of ADH1, and ADH2a and AKR2b was in agreement with citral and CAL content, respectively (Rao et al., 2015) (Figures 4B and 5). A similar trend in ZoGeDH expression and GAL content was reported in ginger tissues (Iijima et al., 2014). The higher expression of ADH3b and ADH4 in roots compared to leaf implied that they may not be involved in monoterpene aldehyde formation (Supplementary Figure S8C). The transcript encoding CCD1 exhibited highest expression in C. flexuosus, followed by C. winterianus (80%) with negligible expression in C. martinii, which corroborated with the aldehyde (citral and CAL) content in respective species (Figures 4B and 5). Similar to CfADH1, CfADH2a and CfAKR2b, expression of CfCCD1 was consistent with the aldehyde content in leaf of C. flexuosus (Figure 6). As suggested for aldehyde formation in O. basilicum (Iijima et al., 2006), multiple enzymes (ADH1, ADH2a, AKR2b, and CCD1) could be responsible for differential accumulation of different aldehydes in Cymbopogon species.
FIGURE 6.
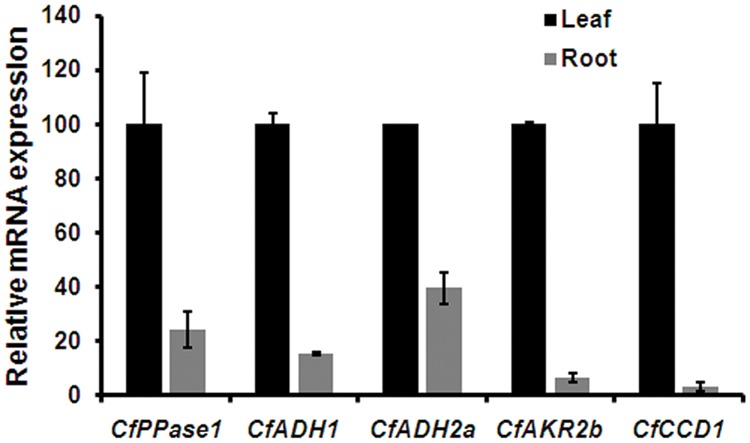
Tissue specific expression of candidate genes in C. flexuosus. Real-time qPCR analysis of candidate genes in leaf and root of C. flexuosus. The expression in leaf was set to 100 to determine the relative abundances of mRNA in root. CfEF1α was used as endogenous control for normalization. Each data point represents the mean ± standard error (SE).
Esterification of alcohol to corresponding acetates by AATs is an important conjugative step in essential oil biosynthetic pathway. These acetates form one of the most predominant volatile esters in essential oils of different aromatic grasses. The essential oil profile of C. martinii revealed ∼21% of GA in inflorescence (Supplementary Figure S7B). Among the three AATs analyzed, only AAT3 exhibited significant expression in inflorescence and was high in C. martinii followed by C. flexuosus (∼48%) with negligible expression in C. winterianus (Figure 5). Also, the tissue specific expression (which accumulates high GA) indicated proportional expression with GA content (Supplementary Figures S7B,C). These results are consistent with previous reports of correlation of expression of acetyl-CoA:benzylalcohol acetyltransferase in C. breweri and GAAT in R. hybrida with floral scent acetates (Dudareva et al., 1998; Shalit et al., 2003). Hence, AAT3 may be the possible candidate involved in GA formation in Cymbopogon sp. Among the 3 ALDH candidates, while ALDH3 showed higher expression in all three species, ALDH1 and ALDH2 expression was negligible, implicating the possible role of ALDH3 in geranic/citronellic acid formation in Cymbopogon (Supplementary Figure S8B).
Molecular Modeling and Docking
Analysis of gene expression and essential oil profiling indicated the involvement of CfADH1, CfADH2a, CfAKR2b, CfAAT3, and CfALDH3 in Cymbopogon aroma biosynthesis. To further support the role of these candidates, in silico studies were performed to know the possible 3D structure and substrate interactions. The conversion of hydroxyl group to aldehyde requires a co-enzyme NAD+ and co-factor zinc (Zn++), and conserved catalytic motifs. While conserved glycine rich “GXGXXG” and catalytic “GHEXXGXXXXXGV” motifs are reported for ADH activity (Lüddeke et al., 2012; Iijima et al., 2014), AKR requires conserved “DXXXXY” motif having catalytic aspartic acid (D) and tyrosine (Y) for its enzyme function (Kavanagh et al., 2008). Sequence analysis revealed the presence of these conserved motifs in CfADH1, CfADH2a, and CfAKR2b (Supplementary Figures S10A,B and S11), suggesting their NAD-dependent dehydrogenase and aldo-keto reductase activity, respectively. Indeed, molecular docking data clearly demonstrated that the GOL binds very proximal to the conserved catalytic motifs of CfADH1, CfADH2a, and CfAKR2b proteins (Figures 7A–C; Supplementary Figure S10A–C and S11; Supplementary Table S6). Among the tested aliphatic and aromatic alcohols, GOL exhibited the lowest ΔG energy for all three proteins (Supplementary Table S6). Further, the docking revealed that the active site topology of CfADH1 and CfADH2b contains glutamic acid (E) and histidine (H) residues, where GOL binds and gets converted to GAL in presence of NAD+ (Figures 7A,B). The conserved “E” plays a vital role in ADH function, as it facilitates substrate binding and coordination of Zn++ ion during catalysis (Ryde, 1995). The GOL bound complex of CfAKR2b exhibited the ΔG energy of -6.96 kcal/mol (Figure 7C; Supplementary Table S6). It is possible that the co-factor NAD+ binds to the conserved sites in ADH and AKR where the reduction of NAD+ occurs by accepting hydrogen from GOL (Strommer, 2011). In the case of CfAAT3 also, docking studies demonstrated higher affinity of CfAAT3 for GOL (ΔG energy of -5.67 kcal/mol) along with farnesol (ΔG energy of -5.97 kcal/mol) among the tested aliphatic and aromatic alcohols (Figure 7D and Supplementary Table S6). The GOL-bound CfAAT3 complex revealed that the conserved histidine (H) and aspartic acid (D) of catalytic HXXXD motif are closely located in the active site (Figure 7D and Supplementary Figure S13), where GOL is converted to GA by acetyltransferase activity (Aharoni et al., 2000; D’Auria, 2006). Similarly for CfALDH3, the molecular modeling and docking studies revealed greater affinity (ΔG energy of -6.16 kcal/mol) for GAL-bound CfALDH3 complex (Supplementary Figures S9 and S14; Supplementary Table S6). Overall, the results from molecular modeling supported the gene expression data, further indicating the involvement of these candidates in essential oil formation. Nevertheless, further experimental evidences (i.e., site directed mutagenesis and in vitro assays) are needed to confirm their precise role in Cymbopogon aroma formation.
FIGURE 7.
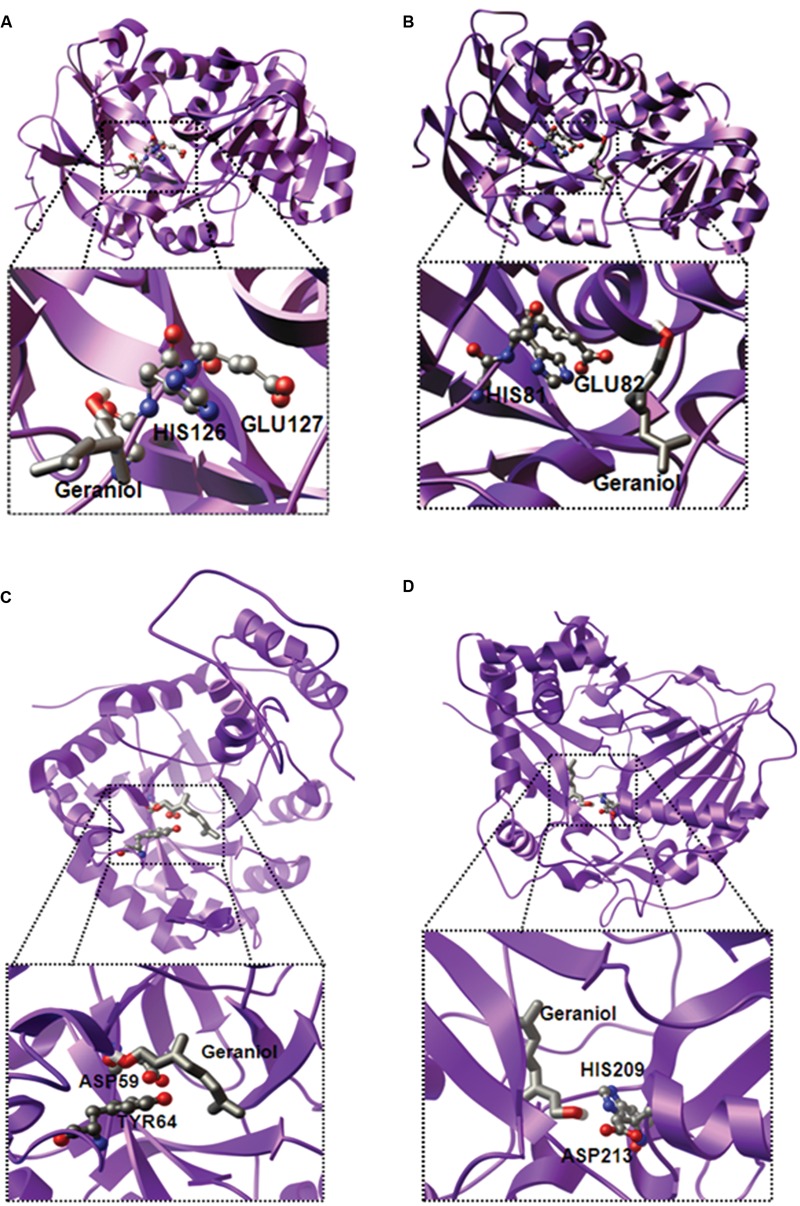
Three-dimensional ribbon model of the complete structure of CfADH1 (A) CfADH2a (B), CfAKR2b (C), and CfAAT3 (D). The homology models of CfADH1 and 2a, CfAKR2b, CfAAT3, and CfALDH3 were built using the X-ray structures of Populus tremuloides synapyl alcohol dehydrogenase (PDB ID: 1YQD), Rauvolfia serpentina perakine reductase (PDB ID: 3V0T), and Coffea canephora hydroxycinnamoyl transferase (PDB ID: 4G0B), respectively. The substrate bound complexes were visualized by PMV software (http://mgltools.scripps.edu). The active site residues are represented as ball and stick, and substrates are shown as stick (colored by atom type).
SSR Mining and Distribution Analysis
Simple sequence repeats (SSRs) are tandem repeats of DNA sequences present in abundance and are distributed throughout the genome (Liu et al., 2013). They are highly versatile PCR-based markers, which are successfully used in marker-assisted selection (MAS), comparative genomics, genetic diversity, and evolutionary studies (Liu et al., 2013). Transcriptome SSR markers, also called as genic-SSRs or EST-SSRs, exhibit high inter-specific transferability as they are located in the coding region of the gene in contrast to genomic SSRs (Liu et al., 2013). Although several molecular markers consisting of RAPD, ISSR, and genomic SSRs have been developed (Khanuja et al., 2005; Kumar et al., 2007; Adhikari et al., 2015; Bishoyi et al., 2016), so far no genic-SSRs are available for Cymbopogon. In recent years, NGS technologies has massively increased the number of SSR markers discovered for both major crops and non-model plant species including Lilium (Shahin et al., 2012), O. sativa (Miah et al., 2013), and Setaria viridis (Xu et al., 2013). Here, for the first time we have mined genic/EST SSRs from lemongrass transcriptome that can be utilized for further crop improvement. Mining of assembled transcripts from C. flexuosus leaf transcriptome resulted in 10,715 (11.5%) transcripts containing 12,968 promising SSRs, of which 1,805 (16.8%) sequences contained >1 SSRs (Supplementary Table S7). A total of 966 SSRs were found to be present in compound formation (Supplementary Table S7). For the motif type, tri-nucleotide (59.8%) repeats were most abundant, followed by mono- (24.2%) and di- nucleotide repeats (13.1%) (Figure 8 and Supplementary Table S7), which was consistent with earlier reports on other monocot species (Davey et al., 2013; Miah et al., 2013). The most common tri-nucleotide repeats were CCG/CGG in lemongrass (Figure 8 and Supplementary Table S8), which was also previously reported for Cymbopogon jwarancusa using genomic library (Kumar et al., 2007). The abundance of CCG repeats has been observed as a special feature of monocot genomes attributed to their higher GC content (Morgante et al., 2002). The most common mono- and di-nucleotide repeats were A/T and AG/CT, respectively (Figure 8 and Supplementary Table S8). Several SSR motifs were linked to transcripts of several secondary metabolic pathways including terpenoid pathways with 6 and 9 SSR motifs linked to MEP pathway and MVA pathway, respectively (Supplementary Tables S3 and S4). In addition, SSR motifs linked to genes involved in other pathways such as steroid, brassinosteroid, alkaloid and phenylpropanoid were also found (Supplementary Table S4). Among the short listed candidates possibly involved in essential oil biosynthesis, transcripts encoding CfPPase1, CfADH1, and CfADH2a contained (AG)6 and (GCC)5ggccgatccgccgcccggcgatgatg(CGT)5 at 5′ UTR, (A)19 at 5′ UTR, and (GGC)5 within the ORF, respectively. Similar to our observation, it has been previously reported that dinucleotide SSR motifs are located in the 5′ UTRs or in the introns of the genes, which are known to regulate promoter activity (Morgante and Olivieri, 1993; Debrauwere et al., 1997). The presence of trinucleotide motifs in the ORF of the genes can be attributed to the tolerance for frame shift mutations in coding regions (Richard and Dujon, 1997; Varshney et al., 2005). The genic/EST-SSRs generated in this study hold great potential for identifying functional markers and will aid in genetic and genomic studies in Cymbopogon as they are better tools than other markers because of their co-dominant inheritance, multi-allelic nature, and high reproducibility (Zhao et al., 2012). The role of these motifs in pathway genes containing SSRs needs to be further investigated in Cymbopogon.
FIGURE 8.
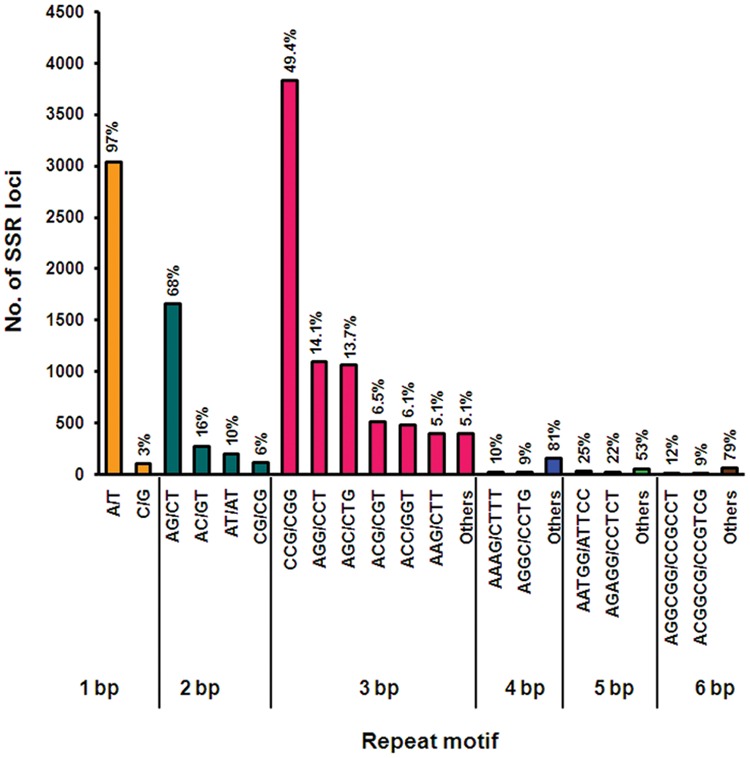
Summary of SSR types identified in C. flexuosus transcriptome. Total number of SSR loci in each repeat motif is represented in Y-axis. Percentage value in the graph indicates the share of individual repeat motif for each repeat type considering the total number in each repeat type as 100%.
Conclusion
Cymbopogon is the most important essential oil producing aromatic grass of Poaceae family. The transcriptome resource for this economically important genus remains unexplored to date. Here, we have generated a gene catalog for lemongrass (C. flexuosus) using de novo transcriptome assembly, which led to the discovery of potential genes (PPase, ADH, AKR, CCD, AAT, and ALDH) involved in essential oil biosynthesis. Notably, comparative and tissue specific expression of identified genes correlated with the essential oil profiles of different Cymbopogon species, supporting their possible involvement in essential oil biosynthesis. In addition, molecular modeling of identified proteins supported the gene expression, thereby validating their role in essential oil biosynthesis. Biochemical and in planta functional characterization of identified candidates could unravel different steps of essential oil biosynthesis and regulation in Cymbopogon, which would be the subject of our future investigations. The putative SSR markers generated in this study could be used for association mapping and molecular breeding programs to modulate/improve essential oil profile/yield in aromatic grasses. This report, for the first time, provides transcriptomic insights into the essential oil biosynthesis of aromatic grass species. We anticipate that this work will take the research on Cymbopogon to the next level, facilitating characterization of genes, regulators and functional markers, and also engineering of essential oil biosynthetic pathway in aromatic grasses or through synthetic biology approaches.
Author Contributions
Conceived and designed the experiment: DN and DR. Performed the experiments: SM, SK, DR, VD, HS, and SR. Contributed reagents/materials/analysis tools: AS. Wrote the paper: SM, SK, DR, and DN.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors express their sincere gratitude to Prof. AK. Tripathi, Director, CSIR-CIMAP. NGS service by SciGenom Labs, Cochin, India is also acknowledged.
Funding. This work was supported by the GAP-210 (BT/HRD/35/24/2006) as part of Ramalingaswami Fellowship (DBT, Govt. of India) to DN and TFYP project (BSC0203) of CSIR-CIMAP. DN thanks the Department of Biotechnology, Government of India, for the Ramalingaswami Fellowship. SM and VD are the recipients of a Research Fellowship from UGC, and CSIR, respectively.
UniProtKB, http://www.uniprot.org/blast/
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01129
References
- Adhikari S., Saha S., Bandyopadhyay T. K., Ghosh P. (2015). Efficiency of ISSR marker for characterization of Cymbopogon germplasms and their suitability in molecular barcoding. Plant Syst. Evol. 301 439–450. 10.1007/s00606-014-1084-y [DOI] [Google Scholar]
- Aharoni A., Keizer L. C. P., Bouwmeester H. J., Sun Z., Alvarez-Huerta M., Verhoeven H. A., et al. (2000). Identification of the SAAT gene involved in strawberry flavour biogenesis by use of DNA microarrays. Plant Cell 12 647–661. 10.1105/tpc.12.5.647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayala B., Bassole I. H., Scifo R., Gnoula C., Morel L., Lobaccaro J. M., et al. (2014). Anticancer activity of essential oils and their chemical components – a review. Am. J. Cancer Res. 4 591–607. [PMC free article] [PubMed] [Google Scholar]
- Beekwilder J., Alvarez-Huerta M., Neef E., Verstappen F. W., Bouwmeester H. J., Aharoni A. (2004). Functional characterization of enzymes forming volatile esters from strawberry and banana. Plant Physiol. 135 1865–1878. 10.1104/pp.104.042580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertea C. M., Maffei M. E. (2010). “The genus Cymbopogon: botany, including anatomy, physiology, biochemistry, and molecular biology,” in Essential Oil- Bearing Grasses ed. Akhila A. (Boca Raton, FL: CRC Press; ) 1–24. [Google Scholar]
- Bishoyi A. K., Sharma A., Kavane A., Geetha K. A. (2016). Varietal discrimination and genetic variability analysis of Cymbopogon using RAPD and ISSR markers analysis. Appl. Biochem. Biotechnol. 179 659–670. 10.1007/s12010-016-2022-y [DOI] [PubMed] [Google Scholar]
- Cui X., Wise R. P., Schnable P. S. (1996). The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science 272 1334–1336. 10.1126/science.272.5266.1334 [DOI] [PubMed] [Google Scholar]
- Cumplido-Laso G., Medina-Puche L., Moyano E., Hoffmann T., Sinz Q., Ring L., et al. (2012). The fruit ripening-related gene FaAAT2 encodes an acyl transferase involved in strawberry aroma biogenesis. J. Exp. Bot. 63 4275–4290. 10.1093/jxb/ers120 [DOI] [PubMed] [Google Scholar]
- D’Auria J. C. (2006). Acyltransferases in plants: a good time to be BAHD. Curr. Opin. Plant Biol. 9 331–340. 10.1016/j.pbi.2006.03.016 [DOI] [PubMed] [Google Scholar]
- Davey M. W., Gudimella R., Harikrishna J. A., Sin L. W., Khalid N., Keulemans J. (2013). A draft Musa balbisiana genome sequence for molecular genetics in polyploid, inter- and intra-specific Musa hybrids. BMC Genomics 14:683 10.1186/1471-2164-14-683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debrauwere H., Gendrel C. G., Lechat S., Dutreix M. (1997). Differences and similarities between various tandem repeat sequences: minisatellites and microsatellites. Biochimie 79 577–586. 10.1016/S0300-9084(97)82006-8 [DOI] [PubMed] [Google Scholar]
- Devi K., Dehury B., Phukon M., Modi M. K., Sen P. (2015). Novel insights into structure-function mechanism and tissue-specific expression profiling of full-length dxr gene from Cymbopogon winterianus. FEBS Open Bio 5 325–334. 10.1016/j.fob.2015.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi K., Mishra S. K., Sahu J., Panda D., Modi M. K., Sen P. (2016). Genome wide transcriptome profiling reveals differential gene expression in secondary metabolite pathway of Cymbopogon winterianus. Sci. Rep. 6:21026 10.1038/srep21026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Miettinen K., Goedbloed M., Verstappen F. W., Voster A., Jongsma M. A., et al. (2013). Characterization of two geraniol synthases from Valeriana officinalis and Lippia dulcis: similar activity but difference in subcellular localization. Metab. Eng. 20 198–211. 10.1016/j.ymben.2013.09.002 [DOI] [PubMed] [Google Scholar]
- Du F., Wu Y., Zhang L., Li X.-W., Zhao X.-Y., Wang W.-H., et al. (2015). De novo assembled transcriptome analysis and SSR marker development of a mixture of six tissues from Lilium oriental hybrid ‘Sorbonne.’ Plant Mol. Biol. Rep. 33 281–293. 10.1007/s11105-014-0746-9 [DOI] [Google Scholar]
- Dudareva N., D’Auria J. C., Nam K. H., Raguso R. A., Pichersky E. (1998). Acetyl-CoA:benzylalcohol acetyltransferase – an enzyme involved in floral scent production in Clarkia breweri. Plant J. 14 297–304. 10.1046/j.1365-313X.1998.00121.x [DOI] [PubMed] [Google Scholar]
- González M., Gaete-Eastman C., Valdenegro M., Figueroa C. R., Fuentes L., Herrera R., et al. (2009). Aroma development during ripening of Fragaria chiloensis fruit and participation of an alcohol acyltransferase (FcAAT1) gene. J. Agric. Food Chem. 14 9123–9132. 10.1021/jf901693j [DOI] [PubMed] [Google Scholar]
- Goodsell D. S., Morris G. M., Olson A. J. (1996). Automated docking of flexible ligands: applications of AutoDock. J. Mol. Recognit. 9 1–5. [DOI] [PubMed] [Google Scholar]
- Hansey C. N., Vaillancourt B., Sekhon R. S., de Leon N., Kaeppler S. M., Buell C. R. (2012). Maize (Zea mays L.) genome diversity as revealed by RNA-sequencing. PLoS ONE 7:e33071 10.1371/journal.pone.0033071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima Y., Gang D. R., Fridman E., Lewinsohn E., Pichersky E. (2004). Characterization of geraniol synthase from the peltate glands of sweet basil. Plant Physiol. 134 370–379. 10.1104/pp.103.032946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima Y., Koeduka T., Suzuki H., Kubota K. (2014). Biosynthesis of geranial, a potent aroma compound in ginger rhizome (Zingiber officinale): molecular cloning and characterization of geraniol dehydrogenase. Plant Biotechnol. 31 525–534. 10.5511/plantbiotechnology.14.1020a [DOI] [Google Scholar]
- Iijima Y., Wang G., Fridman E., Pichersky E. (2006). Analysis of the enzymatic formation of citral in the glands of sweet basil. Arch. Biochem. Biophys. 448 141–149. 10.1016/j.abb.2005.07.026 [DOI] [PubMed] [Google Scholar]
- Ilg A., Beyer P., Babili S. A. (2009). Characterization of the rice carotenoid cleavage dioxygenase 1 reveals a novel route for geranial biosynthesis. FEBS J. 276 736–747. 10.1111/j.1742-4658.2008.06820.x [DOI] [PubMed] [Google Scholar]
- Ilg A., Bruno M., Beyer P., Babili S. A. (2014). Tomato carotenoid cleavage dioxygenases 1A and 1B: relaxed double bond specificity leads to a plenitude of dialdehydes, mono-apocarotenoids and isoprenoid volatiles. FEBS Open Bio 4 584–593. 10.1016/j.fob.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh K. L., Jörnvall H., Persson B., Oppermann U. (2008). Medium- and short-chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 65 3895–3906. 10.1007/s00018-008-8588-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanuja S. P. S., Shasany A. K., Anubha P., Lal R. K., Darokar M. P., Naqvi A. A., et al. (2005). Essential oil constituents and RAPD markers to establish species relationship in Cymbopogon Spreng (Poaceae). Biochem. Syst. Ecol. 33 171–186. 10.1016/j.bse.2004.06.011 [DOI] [Google Scholar]
- Kong F., Li H., Sun P., Zhou Y., Mao Y. (2014). De novo assembly and characterization of the transcriptome of seagrass Zostera marina using Illumina paired-end sequencing. PLoS ONE 9:e112245 10.1371/journal.pone.0112245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J., Verma V., Shahi A. K., Qazi G. N., Balyan H. S. (2007). Development of simple sequence repeat markers in cymbopogon species. Planta Med. 73 262–266. 10.1055/s-2007-967121 [DOI] [PubMed] [Google Scholar]
- Kumar K., Kumar S. R., Dwivedi V., Rai A., Shukla A. K., Shanker K., et al. (2015). Precursor feeding studies and molecular characterization of geraniol synthase establish the limiting role of geraniol in monoterpene indole alkaloid biosynthesis in Catharanthus roseus leaves. Plant Sci. 239 56–66. 10.1016/j.plantsci.2015.07.007 [DOI] [PubMed] [Google Scholar]
- Lavania U. C., Srivastava S., Lavania S., Basu S., Misra N. K., Mukai Y. (2012). Autopolyploidy differentially influences body size in plants, but facilitates enhanced accumulation of secondary metabolites, causing increased cytosine methylation. Plant J. 71 539–549. 10.1111/j.1365-313X.2012.05006.x [DOI] [PubMed] [Google Scholar]
- Li X., Ma D., Chen J., Pu G., Ji Y., Lei C., et al. (2012). Biochemical characterization and identification of a cinnamyl alcohol dehydrogenase from Artemisia annua. Plant Sci. 193 85–95. 10.1016/j.plantsci.2012.05.011 [DOI] [PubMed] [Google Scholar]
- Li Y., Fabiano-Tixier A-S., Chemat F. (2014). “Essential oils: from conventional to green extraction”, in Essential Oils as Reagents in Green Chemistry ed. Sharma S. K. (Berlin: Springer; ) 9–20. 10.1007/978-3-319-08449-7_2 [DOI] [Google Scholar]
- Liu F., Cui X., Horner H. T., Weiner H., Schnable P. S. (2001). Mitochondrial aldehyde dehydrogenase activity is required for male fertility in maize. Plant Cell 13 1063–1078. 10.1105/tpc.13.5.1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Jung C., Xu J., Wang H., Deng S., Bernad L., et al. (2012). Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24 4333–4345. 10.1105/tpc.112.102855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.-R., Li W.-Y., Long D., Hu C.-G., Zhang J.-Z. (2013). Development and characterization of genomic and expressed SSRs in Citrus by genome-wide analysis. PLoS ONE 8:e75149 10.1371/journal.pone.0075149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüddeke F., Wülfing A., Timke M., Germer F., Weber J., Dikfidan A., et al. (2012). Geraniol and geranial dehydrogenases induced in anaerobic monoterpene degradation by Castellaniella defragrans. Appl. Environ. Microbiol. 78 2128–2136. 10.1128/AEM.07226-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnard J.-L., Roccia A., Caissard J.-C., Vergne P., Sun P., Hecquet R., et al. (2015). Biosynthesis of monoterpene scent compounds in roses. Science 349 81–83. 10.1126/science.aab0696 [DOI] [PubMed] [Google Scholar]
- Miah G., Rafii M. Y., Ismail M. R., Puteh A. B., Rahim H. A., Islam KhN., et al. (2013). A review of microsatellite markers and their applications in rice breeding programs to improve blast disease resistance. Int. J. Mol. Sci. 14 22499–22528. 10.3390/ijms141122499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M., Hanafey M., Powell W. (2002). Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 30 194–200. 10.1038/ng822 [DOI] [PubMed] [Google Scholar]
- Morgante M., Olivieri A. M. (1993). PCR-amplified microsatellites as markers in plant genetics. Plant J. 3 175–182. 10.1111/j.1365-313X.1993.tb00020.x [DOI] [PubMed] [Google Scholar]
- Moyler D. A. (2010). “Citral from Lemongrass and other natural sources: its toxicology and Legislation” in Essential Oil- Bearing Grasses ed. Akhila A. (Boca Raton, FL: CRC Press; ) 223–238. [Google Scholar]
- Nagegowda D. A. (2010). Plant volatile terpenoid metabolism: biosynthetic genes, transcriptional regulation and subcellular compartmentation. FEBS Lett. 584 2965–2973. 10.1016/j.febslet.2010.05.045 [DOI] [PubMed] [Google Scholar]
- Nagegowda D. A., Dudareva N. (2006). “Plant biochemistry and biotechnology of flavor compounds and essential oils” in Medicinal Plant Biotechnology: From Basic Research to Industrial Applications ed. Kayser O., Quax W. J. (Weinheim: Wiley-VCH Verlag GmbH; ). [Google Scholar]
- Nah J., Song S. J., Back K. (2001). Partial characterization of farnesyl and geranylgeranyl diphosphatases induced in rice seedlings by UV-C irradiation. Plant Cell Physiol. 42 864–867. 10.1093/pcp/pce102 [DOI] [PubMed] [Google Scholar]
- Niu J., Hou X., Fang C., An J., Ha D., Qiu L., et al. (2015). Transcriptome analysis of distinct Lindera glauca tissues revealed the differences in the unigenes related to terpenoid biosynthesis. Gene 559 22–30. 10.1016/j.gene.2015.01.002 [DOI] [PubMed] [Google Scholar]
- Noge K., Kato M., Mori N., Kataoka M., Tanaka C., Yamasue Y., et al. (2008). Geraniol dehydrogenase, the key enzyme in biosynthesis of the alarm pheromone, from the astigmatid mite Carpoglyphus lactis (Acari: Carpoglyphidae). FEBS J. 275 2807–2817. 10.1111/j.1742-4658.2008.06421.x [DOI] [PubMed] [Google Scholar]
- Padalia R. C., Verma R. S., Chanotiya C. S., Yadav A. (2011). Chemical fingerprinting of the fragrant volatiles of nineteen indian cultivars of Cymbopogon spreng (Poaceae). Rec. Nat. Prod. 5 290–299. [Google Scholar]
- Pimentel M., Molina G., Bertucci T. C. P., Pastore G. (2012). Biotransformation of citronellol in rose oxide by Pseudomonas spp. Chem. Eng. Trans. 27 295–300. [Google Scholar]
- Rai A., Smita S. S., Singh A. K., Shanker K., Nagegowda D. A. (2013). Heteromeric and homomeric geranyl diphosphate synthases from Catharanthus roseus and their role in monoterpene indole alkaloid biosynthesis. Mol. Plant 6 1531–1549. 10.1093/mp/sst058 [DOI] [PubMed] [Google Scholar]
- Rao B. R. R., Adinarayana G., Rajput D. K., Kumar A. N., Syamasundar K. V. (2015). Essential oil profiles of different parts of East Indian lemongrass {Cymbopogon flexuosus (Nees ex Steud.) Wats.}. J. Essent. Oil Res. 27 225–231. 10.1080/10412905.2015.1007218 [DOI] [Google Scholar]
- Rastogi S., Meena S., Bhattacharya A., Ghosh S., Shukla R. K., Sangwan N. S., et al. (2014). De novo sequencing and comparative analysis of holy and sweet basil transcriptomes. BMC Genomics 15:588 10.1186/1471-2164-15-588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard G. F., Dujon B. (1997). Trinucleotide repeats in yeast. Res. Microbiol. 148 731–744. 10.1016/S0923-2508(97)82449-7 [DOI] [PubMed] [Google Scholar]
- Ryde U. (1995). On the role of Glu-68 in alcohol dehydrogenase. Protein Sci. 4 1124–1132. 10.1002/pro.5560040611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A., Blundell T. L. (1993). Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234 779–815. 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- Sanner M. F. (1999). Python: a programming language for software integration and development. J. Mol. Graph. Mod. 17 7–61. [PubMed] [Google Scholar]
- Sato-Masumoto N., Ito M. (2014). Two types of alcohol dehydrogenase from Perilla can form citral and perillaldehyde. Phytochemistry 104 12–20. 10.1016/j.phytochem.2014.04.019 [DOI] [PubMed] [Google Scholar]
- Schulz M. H., Zerbino D. R., Vingron M., Birney E. (2012). Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28 1086–1092. 10.1093/bioinformatics/bts094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüttelkopf A. W., van Aalten D. M. (2004). PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60 1355–1363. 10.1107/S0907444904011679 [DOI] [PubMed] [Google Scholar]
- Shahin A., van Kaauwen V., Esselink D., Bargsten J. W., van Tuyl J. M., Visser R. G. F., et al. (2012). Generation and analysis of expressed sequence tags in the extreme large genomes Lilium and Tulipa. BMC Genomics 13:640 10.1186/1471-2164-13-640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalit M., Guterman I., Volpin H., Bar E., Tamari T., Menda N., et al. (2003). Volatile ester formation in roses. Identification of an acetyl-coenzyme A. Geraniol/citronellol acetyltransferase in developing rose petals. Plant Physiol. 131 1868–1876. 10.1104/pp.102.018572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma J. R., Ram R. S. (2000). “Genetics and genotype improvement of Cymbopogon species,” in Cymbopogon: The Aromatic Grass: A Monograph eds Kumar S., Dwivedi S., Kukreja A. K., Sharma J. R., Bagchi G. D. (Lucknow: CIMAP- CSIR Publication; ) 85–128. [Google Scholar]
- Simkin A. J., Miettinen K., Claudel P., Burlat V., Guirimand G., Courdavault V., et al. (2013). Characterization of the plastidial geraniol synthase from Madagascar periwinkle which initiates the monoterpenoid branch of the alkaloid pathway in internal phloem associated parenchyma. Phytochemistry 85 36–43. 10.1016/j.phytochem.2012.09.014 [DOI] [PubMed] [Google Scholar]
- Singh A. K., Dwivedi V., Rai A., Pal S., Reddy S. G., Rao D. K., et al. (2015). Virus-induced gene silencing of Withania somnifera squalene synthase negatively regulates sterol and defence-related genes resulting in reduced withanolides and biotic stress tolerance. Plant Biotechnol. J. 13 1287–1299. 10.1111/pbi.12347 [DOI] [PubMed] [Google Scholar]
- Strommer J. (2011). The plant ADH gene family. Plant J. 66 128–142. 10.1111/j.1365-313X.2010.04458.x [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney R. K., Graner A., Sorrells M. E. (2005). Genic microsatellite markers in plants: features and applications. Trends Biotechnol. 23 48–55. 10.1016/j.tibtech.2004.11.005 [DOI] [PubMed] [Google Scholar]
- Wang G., Dixon R. A. (2009). Heterodimeric geranyl(geranyl) diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 106 9914–9919. 10.1073/pnas.0904069106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Li Y., Ma X., Ding J., Wang K., Wang S., et al. (2013). Whole transcriptome analysis using next-generation sequencing of model species Setaria viridis to support C4 photosynthesis research. Plant Mol. Biol. 83 77–87. 10.1007/s11103-013-0025-4 [DOI] [PubMed] [Google Scholar]
- Yang L., Ding G., Lin H., Cheng H., Kong Y., Wei Y., et al. (2013). Transcriptome analysis of medicinal plant Salvia miltiorrhiza and identification of genes related to tanshinone biosynthesis. PLoS ONE 8:e80464 10.1371/journal.pone.0080464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Stoopena G., Yalpanib N., Vervoortc J., de Vosa R., Vostera A., et al. (2011). Metabolic engineering of geranic acid in maize to achieve fungal resistance is compromised by novel glycosylation patterns. Metab. Eng. 13 414–425. 10.1016/j.ymben.2011.01.011 [DOI] [PubMed] [Google Scholar]
- Zerbino D. R., Birney E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18 821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Williams R., Prakash C. S., He G. (2012). Identification and characterization of gene-based SSR markers in date palm (Phoenix dactylifera L.). BMC Plant Biol. 12:237 10.1186/1471-2229-12-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


