Abstract
Background
A previous genome-wide association study deduced that one (ARS-BFGL-NGS-39328), two (Hapmap26001-BTC-038813 and Hapmap31284-BTC-039204), two (Hapmap26001-BTC-038813 and BTB-00246150), and one (Hapmap50366-BTA-46960) genome-wide significant single nucleotide polymorphisms (SNPs) associated with milk fatty acids were close to or within the fatty acid synthase (FASN), peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (PPARGC1A), ATP-binding cassette, sub-family G, member 2 (ABCG2) and insulin-like growth factor 1 (IGF1) genes. To further confirm the linkage and reveal the genetic effects of these four candidate genes on milk fatty acid composition, genetic polymorphisms were identified and genotype-phenotype associations were performed in a Chinese Holstein cattle population.
Results
Nine SNPs were identified in FASN, among which SNP rs41919985 was predicted to result in an amino acid substitution from threonine (ACC) to alanine (GCC), five SNPs (rs136947640, rs134340637, rs41919992, rs41919984 and rs41919986) were synonymous mutations, and the remaining three (rs41919999, rs132865003 and rs133498277) were found in FASN introns. Only one SNP each was identified for PPARGC1A, ABCG2 and IGF1.
Association studies revealed that FASN, PPARGC1A, ABCG2 and IGF1 were mainly associated with medium-chain saturated fatty acids and long-chain unsaturated fatty acids, especially FASN for C10:0, C12:0 and C14:0. Strong linkage disequilibrium was observed among ARS-BFGL-NGS-39328 and rs132865003 and rs134340637 in FASN (D´ > 0.9), and among Hapmap26001-BTC-038813 and Hapmap31284-BTC-039204 and rs109579682 in PPARGC1A (D´ > 0.9). Subsequently, haplotype-based analysis revealed significant associations of the haplotypes encompassing eight FASN SNPs (rs41919999, rs132865003, rs134340637, rs41919992, rs133498277, rs41919984, rs41919985 and rs41919986) with C10:0, C12:0, C14:0, C18:1n9c, saturated fatty acids (SFA) and unsaturated fatty acids (UFA) (P = 0.0204 to P < 0.0001).
Conclusion
Our study confirmed the linkage between the significant SNPs in our previous genome-wide association study and variants in FASN and PPARGC1A. SNPs within FASN, PPARGC1A, ABCG2 and IGF1 showed significant genetic effects on milk fatty acid composition in dairy cattle, indicating their potential functions in milk fatty acids synthesis and metabolism. The findings presented here provide evidence for the selection of dairy cows with healthier milk fatty acid composition by marker-assisted breeding or genomic selection schemes, as well as furthering our understanding of technological processing aspects of cows’ milk.
Electronic supplementary material
The online version of this article (doi:10.1186/s12863-016-0418-x) contains supplementary material, which is available to authorized users.
Keywords: Association analysis, Candidate gene, Haplotype, Milk fatty acids, Single nucleotide polymorphism
Background
Recently, an increasing number of genes have been reported as associated with milk production for dairy cattle breeding, and great improvements have been obtained. Many quantitative trait locus (QTL) analysis and association studies revealed the DGAT1, GHR, FASN and PPARGC1A genes as promising candidate genes for milk production traits [1–12]. Nevertheless, there have been few reports [13–22] of association studies involving milk fatty acid traits, which should be considered because of their close relation with milk flavor and nutritional properties. High concentrations of saturated fatty acids (SFAs) such as C12:0, C14:0 and C16:0 increase the risks of coronary artery disease (CAD) by promoting the concentrations of blood low density lipoprotein (LDL) cholesterol [23], while polyunsaturated fatty acids (PUFAs) have the ability to reduce blood fat and cholesterol levels by inhibiting fat formation and enzyme activities acting on fat [24, 25]. Thus, increasing the ratio of PUFAs to SFAs would be beneficial to human health. A previous genome-wide association study (GWAS) revealed that several significant single nucleotide polymorphisms (SNPs) close to or within the FASN, PPARGC1A, ABCG2 and IGF1 genes were associated with milk fatty acids in Chinese Holstein dairy cattle [26]. In addition, the FASN, PPARGC1A, ABCG2 and IGF1 genes were observed to be associated significantly with milk production traits in our previous candidate genes analysis in Chinese Holstein cattle [27–30]. Therefore, we deduced that the significant SNPs might be linked with the causative mutations in these four genes. The purpose of the present study was to identify the genetic effects of the FASN, PPARGC1A, ABCG2 and IGF1 genes on traits of milk fatty acids in a Chinese Holstein cattle population. In addition, linkage disequilibrium (LD) analyses were conducted among the SNPs identified in our previous GWAS and in this study.
Methods
Phenotypic data and traits
Complete details of the milk sample collection and the detection method for milk fatty acids have been reported previously [26]. Briefly, fat was extracted from 2 mL of milk and then methyl esterification of fats was performed. One milliliter of methyl esters of fatty acids were prepared and determined by gas chromatography using a gas chromatograph (6890 N, Agilent) equipped with a flame-ionization detector and a high polar fused silica capillary column (SPTM-2560, 100 m × 0.25 mm ID, 0.20 μm film; Cat. No. 24056). About 1 μL of the sample was injected under the specific gas chromatography conditions. Finally, individual fatty acids were identified and quantified by comparing the methyl ester chromatograms of the milk fat samples with the chromatograms of pure fatty acids methyl ester standards (SupelcoTM 37 Component FAME Mix), and were measured as the weight proportion of total fat weight (wt/wt%). Phenotypic values of 10 main milk fatty acids were tested directly using gas chromatography, which included SFAs of C10:0, C12:0, C14:0, C16:0, C18:0, monounsaturated fatty acids (MUFAs) of C14:1, C16:1, C18:1n9c, and PUFAs of CLA (cis-9, trans-11 C18:2), C18:2n6c. Based on the phenotypes of 10 tested milk fatty acids, six additional traits were obtained including SFA, UFA, SFA/UFA (the ratio of SFA to UFA), C14 index, C16 index and C18 index. The three indices were calculated as , [31].
The population in this study comprised 346 Chinese Holstein cows, which were the daughters of 13 sire families from 13 farms of the Beijing Sanyuan Dairy Farm Center. Sixteen main milk fatty acid traits were considered in this association study.
Genomic DNA extraction
The whole blood samples corresponding to the 346 Chinese Holstein cows with phenotypic values were collected. Genomic DNA was extracted from blood samples of the cows using a TIANamp Genomic DNA kit (TianGen, Beijing, China) according to the manufacturer’s instructions and frozen semen of the sires using a standard phenol-chloroform procedure. The quantity and quality of the extracted DNA were measured using a NanoDrop™ ND-2000c Spectrophotometer (Thermo Scientific, Inc.) and by gel electrophoresis.
SNP identification and genotyping
A DNA pool was constructed from aforementioned 13 Holstein bulls (50 ng/μL for each individual) whose daughters were used for the association analysis to identify potential SNPs in the FASN, PPARGC1A, ABCG2 and IGF1 genes. For FASN, a total of 30 pairs of PCR primers (Additional file 1, Table S1) were designed to amplify all the exons and their partial flanking intronic sequences based on the reference sequence of the bovine FASN referring to Bos_taurus_UMD_3.1 assembly (NCBI Reference Sequence: AC_000176.1) using Primer3 web program (v.0.4.0) [32]. Following with the same method, a pair of specific primers was designed for selective amplification based on the exon 9 and partial intron 9 sequence of PPARGC1A (NCBI Reference Sequence: AC_000163.1): forward 5′- GCC GGT TTA TGT TAA GAC AG-3′ and reverse 5′- GGT ATT CTT CCC TCT TGA GC-3′. Primers were also designed from exon 7 and partial flanking intronic sequences of the ABCG2 gene (NCBI Reference Sequence: AC_000163.1): forward 5′- TAA AGG CAG GAG TAA TAA AG-3′ and reverse 5′- TAA CAC CAA ACT AAC CGA AG-3′, and the 5′-flanking region of the IGF1 gene (NCBI Reference Sequence: AC_000162.1): forward 5′- ATT ACA AAG CTG CCT GCC CC-3′ and reverse 5′- CAC ATC TGC TAA TAC ACC TTA CCC G-3′.
Polymerase chain reaction (PCR) amplifications for the pooled DNA from the 13 sires were performed in a final reaction volume of 25 μL comprising of 50 ng of genomic DNA, 0.5 μL of each primer (10 mM), 2.5 μL of 10 × PCR buffer, 2.5 mM each of dNTPs, and 1 U of Taq DNA polymerase (Takara, Dalian, China). The PCR protocol was 5 min at 94 °C for initial denaturation followed by 34 cycles at 94 °C for 30 s; 56 ~ 60 °C for 30 s; 72 °C for 30 s; and a final extension at 72 °C for 7 min for all primers. The PCR products were purified to remove residual primers, dNTPs and reagents from the amplification reaction. A gel purification kit (DNA Gel Extraction Kit, TransGen Biotech, China) was used to extract the target DNA band. Then, 15 μL of each purified PCR product with 1 μL of each forward and reverse primer, was bi-directionally sequenced using an ABI3730XL sequencer (Applied Biosystems, Foster City, CA, USA).
Matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS, Sequenom MassARRAY, Bioyong Technologies Inc. HK) was used for subsequent genotyping of the 346 Chinese Holstein cows.
Linkage disequilibrium (LD) analysis and haplotype construction
Pair-wise LD was measured between the genotyped SNPs of each gene and the corresponding adjacent SNPs that were significantly associated with target traits identified in our previous GWAS based on the criterion of D’ using the software Haploview [33]. Accordingly, haplotype blocks where SNPs are in high LD (D’ > 0.90) were also determined based on confidence interval methods [34]. A haplotype with a frequency >5 % was treated as a distinguishable haplotype, and those haplotypes each with relative frequency <5 % were pooled into a single group.
Association analyses
Hardy-Weinberg equilibrium tests were performed on each identified SNP. A goodness-of-fit test (Chi-square) was used to compare the number of expected and observed genotypes, using 0.05 as significant threshold value.
The mixed procedure of SAS 9.3 software (SAS Institute Inc., Cary, NC) with the following animal model was performed to estimate the genetic effects of each candidate SNP or haplotype on the milk fatty acid traits.
where, yijklmn was the phenotypic value of each trait of the cows; μ was the overall mean; Fi was the fixed effect of the farm; Pj was the fixed effect of parity; Lk was the fixed effect of the stage of lactation; Gl was the fixed effect corresponding to the genotype of polymorphisms or haplotype; αm was the random polygenic effect, distributed as N (0, Aσa2), with the additive genetic relationship matrix A and the additive genetic variance σ2a; and eijklmn was the random residual, distributed as N (0, Iσe2), with identity matrix I and residual error variance σ2e. Bonferroni correction was adopted to correct for multiple testing. The significance level of the multiple tests was equal to the raw P value divided by number of tests. In the present study, three genotypes were compared for each trait mean that three multiple comparisons needed to be performed, therefore, Bonferroni corrected significance levels of 0.05/3 = 0.0167 and 0.01/3 = 0.0033 were used. For the haplotype, the Bonferroni corrected significance levels were presented as 0.05/N, where N refers to the number of formed haplotypes. The additive (a), dominance (d) and allele substitution (α) effects were estimated according to the equation proposed by Falconer & Mackay [35], i.e. , and α = a + d(q − p), where AA and BB represent the two homozygous genotypes, AB is the heterozygous genotype, and p and q are the allele frequencies of the corresponding alleles.
Results
SNPs identification
After sequencing the PCR products directly using the pooled genomic DNA, a total of nine SNPs were identified for the FASN gene. Of these, three were located in the intronic region and six were in exons. The SNP in exon 39 (rs41919985) was predicted to result in an amino acid replacement (A2266T) from threonine (ACC) to alanine (GCC) in the FASN protein, and the other five SNPs in the coding region (rs136947640, rs134340637, rs41919992, rs41919984 and rs41919986) were synonymous mutations. Regarding PPARGC1A, ABCG2 and IGF1, only one SNP was detected in each gene (rs109579682, rs137757790 and rs109763947, respectively), of which rs109763947 is located in the 5′-untranslated region (UTR) and the other two SNPs are in intronic regions. The detailed SNP information is shown in Table 1, and the five significant SNPs for milk fatty acids that are close to FASN, PPARGC1A, ABCG2 and IGF1 identified in our previous GWAS [26] are listed as well. All the identified SNPs in this study were found to be in Hardy-Weinberg equilibrium (P > 0.01, Tables 2 and 3).
Table 1.
SNPs information identified in this study and in a previous GWA study
| CHR | RefSNP | Locus | Allele | Gene region | Positiona | Amino acid substitution | Gene | Origin |
|---|---|---|---|---|---|---|---|---|
| 5 | rs109763947 | g.1407C > T | C/T | 5'-UTR | 66605011 | IGF1 | This study | |
| 5 | rs41643203 | Hapmap50366-BTA-46960 | C/T | intron-2 | 68610818 | Close to IGF1 | [23] | |
| 6 | rs109579682 | g.85330C > T | C/T | Intron-9 | 44875251 | PPARGC1A | This study | |
| 6 | rs110131167 | Hapmap26001-BTC-038813 | A/G | intron-2 | 44926243 | PPARGC1A | [23] | |
| 6 | rs108967640 | Hapmap31284-BTC-039204 | C/T | - | 45096462 | PPARGC1A | [23] | |
| 6 | rs137757790 | g.45599A > C | A/C | Intron-7 | 38005668 | ABCG2 | This study | |
| 6 | rs43450879 | BTB-00246150 | A/G | Intron-1 | 20993424 | Close to ABCG2 | [23] | |
| 19 | rs136947640 | g.7709 T > C | T/C | Exon-10 | 51391830 | FASN | This study | |
| 19 | rs41919999 | g.8948C > T | C/T | Intron-12 | 51393068 | FASN | This study | |
| 19 | rs132865003 | g.10568 T > C | T/C | Intron-18 | 51394689 | FASN | This study | |
| 19 | rs134340637 | g.11280G > A | G/A | Exon-21 | 51395400 | FASN | This study | |
| 19 | rs41919992 | g.13965C > T | C/T | Exon-27 | 51398083 | FASN | This study | |
| 19 | rs133498277 | g.14439 T > C | T/C | Intron-28 | 51398557 | FASN | This study | |
| 19 | rs41919984 | g.16907 T > C | T/C | Exon-37 | 51401022 | FASN | This study | |
| 19 | rs41919985 | g.17924A > G | A/G | Exon-39 | 51402032 | A2266T | FASN | This study |
| 19 | rs41919986 | g.18663 T > C | T/C | Exon-42 | 51402774 | FASN | This study | |
| 19 | rs41921177 | ARS-BFGL-NGS-39328 | A/G | Intron-11 | 51326750 | Close to FASN | [23] |
Note: aAll SNP nucleotide positions were derived from the Bos_taurus_UMD_3.1 assembly (GenBank accession number: AC_000171.1)
Table 2.
Genotypic and allelic frequencies and Hardy-Weinberg equilibrium test of nine SNPs of the FASN gene in Chinese Holstein cattle
| Position | Locus | Genotypes | N | Frequency | Allele | Frequency | Hardy-Weinberg equilibrium χ2 test |
|---|---|---|---|---|---|---|---|
| Exon-10 | rs136947640 | CC | 248 | 0.790 | C | 0.892 | P > 0.05 |
| TT | 2 | 0.006 | T | 0.108 | |||
| CT | 64 | 0.204 | |||||
| Intron-12 | rs41919999 | CC | 64 | 0.204 | C | 0.462 | P > 0.05 |
| TT | 88 | 0.280 | T | 0.538 | |||
| CT | 162 | 0.516 | |||||
| Intron-18 | rs132865003 | CC | 220 | 0.698 | C | 0.833 | P > 0.05 |
| TT | 10 | 0.032 | T | 0.167 | |||
| CT | 85 | 0.270 | |||||
| Exon-21 | rs134340637 | AA | 10 | 0.032 | A | 0.167 | P > 0.05 |
| GG | 220 | 0.698 | G | 0.833 | |||
| AG | 85 | 0.270 | |||||
| Exon-27 | rs41919992 | CC | 157 | 0.500 | C | 0.712 | P > 0.05 |
| TT | 24 | 0.076 | T | 0.288 | |||
| CT | 133 | 0.424 | |||||
| Intron-28 | rs133498277 | CC | 157 | 0.500 | C | 0.713 | P > 0.05 |
| TT | 23 | 0.073 | T | 0.287 | |||
| CT | 134 | 0.427 | |||||
| Exon-37 | rs41919984 | CC | 157 | 0.498 | C | 0.711 | P > 0.05 |
| TT | 24 | 0.076 | T | 0.289 | |||
| CT | 134 | 0.425 | |||||
| Exon-39 | rs41919985 | AA | 25 | 0.079 | A | 0.290 | P > 0.05 |
| GG | 157 | 0.498 | G | 0.710 | |||
| AG | 133 | 0.422 | |||||
| Exon-42 | rs41919986 | CC | 155 | 0.497 | C | 0.708 | P > 0.05 |
| TT | 25 | 0.080 | T | 0.292 | |||
| CT | 132 | 0.423 |
Table 3.
Genotypic and allelic frequencies and Hardy-Weinberg equilibrium test of SNPs of the PPARGC1A, ABCG2 and IGF1 genes in Chinese Holstein cattle
| Gene | Position | Locus | Genotypes | N | Frequency | Allele | Frequency | Hardy-Weinberg equilibrium χ2 test |
|---|---|---|---|---|---|---|---|---|
| PPARGC1A | Intron-9 | rs109579682 | CC | 27 | 0.078 | C | 0.292 | P > 0.05 |
| TT | 170 | 0.494 | T | 0.708 | ||||
| CT | 147 | 0.427 | ||||||
| ABCG2 | Intron-7 | rs137757790 | AA | 115 | 0.333 | A | 0.543 | P > 0.01 |
| CC | 85 | 0.246 | C | 0.457 | ||||
| AC | 145 | 0.420 | ||||||
| IGF1 | 5’-UTR | rs109763947 | CC | 58 | 0.168 | C | 0.439 | P > 0.05 |
| TT | 100 | 0.290 | T | 0.561 | ||||
| CT | 187 | 0.542 |
Associations between the four candidate genes and milk fatty acid traits
Associations between the nine SNPs of FASN and 16 milk fatty acid composition traits are presented in Table 4. We found that all nine SNPs showed significant associations with at least one milk fatty acid trait. Of these, three SNPs (rs136947640, rs132865003 and rs134340637) were only significantly associated with C18:2n6c (P < 0.0001, P = 0.0128, P = 0.0128), two SNPs (rs41919992 and rs133498277) showed strong associations with seven traits of C10:0, C12:0, C14:0, C18:1n9c, C16 index, SFA and UFA (P = 0.0190 to < 0.0001), three SNPs (rs41919984, rs41919985 and rs41919986) were strongly associated with the above seven traits plus SFA/UFA (P = 0.045 to P <0.0001), and one SNP (rs41919999) showed significant association with C10:0 (P = 0.0012), C12:0 (P = 0.0041) and C14:0 (P = 0.0071). Meanwhile, for C14:1, C16:0, C16:1, C18:0, CLA, C14 index and C18 index, no significant SNPs in FASN were detected. Furthermore, the results showed that heterozygous genotypes of these SNPs were the dominant type for saturated fatty acids (C10:0, C12:0, C14:0, SFA and SFA/UFA), and the homozygotic genotypes of these SNPs were dominant for unsaturated fatty acids (C18:1n9c, C16 index and UFA).
Table 4.
Associations of nine SNPs of the FASN gene with milk medium-chain fatty acids (MCFAs) in Chinese Holstein cattle (LSM ± SE)
| Locus | Genotypes | C10:0 | C12:0 | C14:0 | C14:1 | ||
|---|---|---|---|---|---|---|---|
| rs136947640 | CC(248) | 2.13 ± 0.06 | 2.63 ± 0.08 | 9.55 ± 0.13 | 0.79 ± 0.03 | ||
| TT(2) | 2.23 ± 0.24 | 2.66 ± 0.32 | 8.92 ± 0.54 | 0.65 ± 0.16 | |||
| CT(64) | 2.09 ± 0.07 | 2.56 ± 0.09 | 9.42 ± 0.15 | 0.78 ± 0.04 | |||
| P-value | 0.6139 | 0.5290 | 0.2611 | 0.6836 | |||
| rs41919999 | CC(64) | 2.13 ± 0.07AB | 2.68 ± 0.09AB | 9.64 ± 0.15A | 0.76 ± 0.04 | ||
| TT(88) | 1.99 ± 0.07B | 2.53 ± 0.09B | 9.25 ± 0.14B | 0.79 ± 0.04 | |||
| CT(162) | 2.15 ± 0.06A | 2.73 ± 0.08A | 9.52 ± 0.13A | 0.80 ± 0.03 | |||
| P-value | 0.0012 | 0.0041 | 0.0071 | 0.6264 | |||
| rs132865003 | CC(220) | 2.11 ± 0.06 | 2.68 ± 0.08 | 9.52 ± 0.13 | 0.80 ± 0.03 | ||
| TT(10) | 2.17 ± 0.12 | 2.72 ± 0.16 | 9.45 ± 0.26 | 0.75 ± 0.08 | |||
| CT(85) | 2.11 ± 0.06 | 2.68 ± 0.08 | 9.49 ± 0.14 | 0.78 ± 0.04 | |||
| P-value | 0.8601 | 0.9610 | 0.9217 | 0.7157 | |||
| rs134340637 | AA(10) | 2.17 ± 0.12 | 2.72 ± 0.16 | 9.45 ± 0.26 | 0.75 ± 0.08 | ||
| GG(220) | 2.11 ± 0.06 | 2.68 ± 0.08 | 9.52 ± 0.13 | 0.80 ± 0.03 | |||
| AG(85) | 2.11 ± 0.06 | 2.68 ± 0.08 | 9.49 ± 0.14 | 0.78 ± 0.04 | |||
| P-value | 0.8601 | 0.9610 | 0.9217 | 0.7157 | |||
| rs41919992 | CC(157) | 2.05 ± 0.06A | 2.53 ± 0.08A | 9.31 ± 0.13A | 0.77 ± 0.04 | ||
| TT(24) | 2.06 ± 0.09AB | 2.45 ± 0.12A | 9.35 ± 0.20A | 0.76 ± 0.06 | |||
| CT(133) | 2.20 ± 0.06B | 2.74 ± 0.08B | 9.75 ± 0.13B | 0.79 ± 0.04 | |||
| P-value | 0.0013 | <.0001 | <.0001 | 0.6169 | |||
| rs133498277 | CC(157) | 2.05 ± 0.06A | 2.53 ± 0.08A | 9.32 ± 0.13A | 0.77 ± 0.04 | ||
| TT(23) | 2.07 ± 0.09AB | 2.47 ± 0.12A | 9.42 ± 0.20AB | 0.75 ± 0.06 | |||
| CT(134) | 2.18 ± 0.06B | 2.73 ± 0.08B | 9.75 ± 0.13B | 0.79 ± 0.04 | |||
| P-value | 0.0043 | 0.0003 | <.0001 | 0.6826 | |||
| rs41919984 | CC(157) | 2.06 ± 0.09AB | 2.51 ± 0.11A | 9.38 ± 0.19AB | 0.76 ± 0.06 | ||
| TT(24) | 2.04 ± 0.06B | 2.58 ± 0.08A | 9.29 ± 0.13B | 0.78 ± 0.04 | |||
| TC(134) | 2.19 ± 0.06A | 2.81 ± 0.08B | 9.74 ± 0.13A | 0.80 ± 0.04 | |||
| P-value | 0.0010 | <.0001 | <.0001 | 0.6958 | |||
| rs41919985 | AA(25) | 2.08 ± 0.09AB | 2.54 ± 0.11A | 9.46 ± 0.19AB | 0.76 ± 0.06 | ||
| GG(157) | 2.04 ± 0.06B | 2.58 ± 0.08A | 9.28 ± 0.13B | 0.78 ± 0.04 | |||
| GA(133) | 2.18 ± 0.06A | 2.80 ± 0.08B | 9.73 ± 0.13A | 0.80 ± 0.04 | |||
| P-value | 0.0017 | <.0001 | <.0001 | 0.7268 | |||
| rs41919986 | CC(155) | 2.03 ± 0.06A | 2.54 ± 0.08A | 9.36 ± 0.13A | 0.77 ± 0.04 | ||
| TT(25) | 2.08 ± 0.09AB | 2.50 ± 0.11A | 9.50 ± 0.19AB | 0.76 ± 0.06 | |||
| CT(132) | 2.17 ± 0.06B | 2.74 ± 0.08B | 9.78 ± 0.13B | 0.79 ± 0.04 | |||
| P-value | 0.0015 | 0.0002 | <.0001 | 0.7225 | |||
| Locus | Genotypes | C16:0 | C16:1 | C18:0 | C18:1n9c | C18:2n6c | CLA |
| rs136947640 | CC(248) | 32.30 ± 0.33 | 1.75 ± 0.05 | 12.59 ± 0.17 | 29.36 ± 0.22 | 4.03 ± 0.03A | 0.38 ± 0.01 |
| TT(2) | 32.38 ± 1.52 | 1.86 ± 0.21 | 12.25 ± 0.85 | 30.18 ± 1.13 | 3.73 ± 0.13A | 0.38 ± 0.05 | |
| CT(64) | 32.17 ± 0.40 | 1.81 ± 0.06 | 12.54 ± 0.21 | 29.36 ± 0.28 | 4.12 ± 0.03B | 0.40 ± 0.01 | |
| P-value | 0.9155 | 0.3007 | 0.9028 | 0.7714 | <.0001 | 0.3536 | |
| rs41919999 | CC(64) | 32.22 ± 0.40 | 1.79 ± 0.06 | 12.31 ± 0.21 | 29.43 ± 0.28 | 4.09 ± 0.03 | 0.40 ± 0.01 |
| TT(88) | 32.23 ± 0.38 | 1.77 ± 0.05 | 12.68 ± 0.20 | 29.82 ± 0.27 | 4.08 ± 0.03 | 0.38 ± 0.01 | |
| CT(162) | 32.11 ± 0.34 | 1.76 ± 0.05 | 12.50 ± 0.17 | 29.36 ± 0.23 | 4.07 ± 0.03 | 0.39 ± 0.01 | |
| P-value | 0.8921 | 0.7478 | 0.2182 | 0.0982 | 0.5542 | 0.4618 | |
| rs132865003 | CC(220) | 32.24 ± 0.33 | 1.75 ± 0.05 | 12.54 ± 0.17 | 29.44 ± 0.22 | 4.06 ± 0.03a | 0.38 ± 0.01 |
| TT(10) | 32.65 ± 0.73 | 1.78 ± 0.10 | 12.43 ± 0.40 | 28.95 ± 0.54 | 4.00 ± 0.06ab | 0.41 ± 0.02 | |
| CT(85) | 32.19 ± 0.36 | 1.80 ± 0.05 | 12.46 ± 0.19 | 29.35 ± 0.25 | 4.12 ± 0.03b | 0.40 ± 0.01 | |
| P-value | 0.8073 | 0.4185 | 0.8504 | 0.6087 | 0.0128 | 0.1185 | |
| rs134340637 | AA(10) | 32.65 ± 0.73 | 1.78 ± 0.10 | 12.43 ± 0.40 | 28.95 ± 0.54 | 4.00 ± 0.06ab | 0.41 ± 0.02 |
| GG(220) | 32.24 ± 0.33 | 1.75 ± 0.05 | 12.54 ± 0.17 | 29.44 ± 0.22 | 4.06 ± 0.03b | 0.38 ± 0.01 | |
| AG(85) | 32.19 ± 0.36 | 1.80 ± 0.05 | 12.46 ± 0.19 | 29.35 ± 0.25 | 4.12 ± 0.03a | 0.40 ± 0.01 | |
| P-value | 0.8073 | 0.4185 | 0.8504 | 0.6087 | 0.0128 | 0.1185 | |
| rs41919992 | CC(157) | 32.21 ± 0.35 | 1.79 ± 0.05 | 12.67 ± 0.18 | 29.68 ± 0.24A | 4.05 ± 0.03 | 0.39 ± 0.01 |
| TT(24) | 31.62 ± 0.54 | 1.80 ± 0.08 | 12.69 ± 0.29 | 30.64 ± 0.39C | 4.02 ± 0.04 | 0.38 ± 0.02 | |
| CT(133) | 32.42 ± 0.35 | 1.73 ± 0.05 | 12.45 ± 0.18 | 28.89 ± 0.24B | 4.05 ± 0.03 | 0.38 ± 0.01 | |
| P-value | 0.2037 | 0.2115 | 0.2499 | <.0001 | 0.8456 | 0.7406 | |
| rs133498277 | CC(157) | 32.22 ± 0.35 | 1.80 ± 0.05 | 12.67 ± 0.18 | 29.71 ± 0.24A | 4.05 ± 0.03 | 0.39 ± 0.01 |
| TT(23) | 31.57 ± 0.54 | 1.82 ± 0.08 | 12.59 ± 0.29 | 30.62 ± 0.39C | 4.06 ± 0.04 | 0.38 ± 0.02 | |
| CT(134) | 32.44 ± 0.35 | 1.74 ± 0.05 | 12.47 ± 0.18 | 28.92 ± 0.24B | 4.05 ± 0.03 | 0.38 ± 0.01 | |
| P-value | 0.1847 | 0.2131 | 0.3740 | <.0001 | 0.9503 | 0.8353 | |
| rs41919984 | CC(157) | 31.54 ± 0.53 | 1.79 ± 0.07 | 12.64 ± 0.29 | 30.68 ± 0.38A | 4.06 ± 0.04 | 0.38 ± 0.02 |
| TT(24) | 32.16 ± 0.34 | 1.80 ± 0.05 | 12.62 ± 0.18 | 29.75 ± 0.24C | 4.08 ± 0.03 | 0.39 ± 0.01 | |
| TC(134) | 32.41 ± 0.34 | 1.73 ± 0.05 | 12.39 ± 0.17 | 28.88 ± 0.23B | 4.08 ± 0.03 | 0.39 ± 0.01 | |
| P-value | 0.1507 | 0.2088 | 0.2673 | <.0001 | 0.8309 | 0.6460 | |
| rs41919985 | AA(25) | 31.55 ± 0.53 | 1.8 ± 0.07 | 12.52 ± 0.29 | 30.59 ± 0.38A | 4.07 ± 0.04 | 0.38 ± 0.02 |
| GG(157) | 32.16 ± 0.34 | 1.80 ± 0.05 | 12.61 ± 0.18 | 29.75 ± 0.24A | 4.08 ± 0.03 | 0.39 ± 0.01 | |
| GA(133) | 32.41 ± 0.34 | 1.73 ± 0.05 | 12.41 ± 0.17 | 28.88 ± 0.23B | 4.08 ± 0.03 | 0.39 ± 0.01 | |
| P-value | 0.1446 | 0.1888 | 0.3716 | <.0001 | 0.9818 | 0.6229 | |
| rs41919986 | CC(155) | 32.11 ± 0.34 | 1.80 ± 0.05 | 12.67 ± 0.18 | 29.77 ± 0.24A | 4.05 ± 0.03 | 0.39 ± 0.01 |
| TT(25) | 31.53 ± 0.53 | 1.81 ± 0.07 | 12.54 ± 0.29 | 30.60 ± 0.38A | 4.05 ± 0.04 | 0.38 ± 0.02 | |
| CT(132) | 32.38 ± 0.35 | 1.74 ± 0.05 | 12.50 ± 0.18 | 28.88 ± 0.24B | 4.03 ± 0.03 | 0.38 ± 0.01 | |
| P-value | 0.1502 | 0.1425 | 0.4945 | <.0001 | 0.7292 | 0.6757 | |
| Locus | Genotypes | C14INDEX | C16INDEX | C18INDEX | SFA | UFA | SFA/UFA |
| rs136947640 | CC(248) | 7.62 ± 0.26 | 5.15 ± 0.12 | 69.96 ± 0.52 | 61.45 ± 0.31 | 36.89 ± 0.28 | 1.70 ± 0.04 |
| TT(2) | 6.69 ± 1.19 | 5.44 ± 0.54 | 71.10 ± 2.50 | 60.67 ± 1.52 | 37.49 ± 1.39 | 1.62 ± 0.20 | |
| CT(64) | 7.62 ± 0.31 | 5.35 ± 0.14 | 70.06 ± 0.63 | 61.09 ± 0.38 | 37.08 ± 0.34 | 1.68 ± 0.05 | |
| P-value | 0.7344 | 0.1582 | 0.8863 | 0.4488 | 0.7360 | 0.8360 | |
| rs41919999 | CC(64) | 7.39 ± 0.31 | 5.27 ± 0.14 | 70.57 ± 0.63 | 61.22 ± 0.38 | 37.11 ± 0.35 | 1.67 ± 0.05 |
| TT(88) | 7.84 ± 0.30 | 5.22 ± 0.14 | 70.18 ± 0.60 | 60.92 ± 0.36 | 37.43 ± 0.33 | 1.66 ± 0.05 | |
| CT(162) | 7.73 ± 0.26 | 5.20 ± 0.12 | 70.13 ± 0.53 | 61.29 ± 0.31 | 36.96 ± 0.29 | 1.69 ± 0.04 | |
| P-value | 0.2796 | 0.8450 | 0.7036 | 0.4242 | 0.2017 | 0.7246 | |
| rs132865003 | CC(220) | 7.74 ± 0.26 | 5.16 ± 0.12 | 70.10 ± 0.51 | 61.31 ± 0.31 | 37.02 ± 0.28 | 1.69 ± 0.04 |
| TT(10) | 7.36 ± 0.57 | 5.17 ± 0.26 | 70.04 ± 1.19 | 61.77 ± 0.72 | 36.54 ± 0.66 | 1.71 ± 0.10 | |
| CT(85) | 7.60 ± 0.28 | 5.30 ± 0.13 | 70.22 ± 0.57 | 61.17 ± 0.34 | 37.06 ± 0.31 | 1.68 ± 0.05 | |
| P-value | 0.6364 | 0.2951 | 0.9567 | 0.6591 | 0.7296 | 0.9378 | |
| rs134340637 | AA(10) | 7.36 ± 0.57 | 5.17 ± 0.26 | 70.04 ± 1.19 | 61.77 ± 0.72 | 36.54 ± 0.66 | 1.71 ± 0.10 |
| GG(220) | 7.74 ± 0.26 | 5.16 ± 0.12 | 70.10 ± 0.51 | 61.31 ± 0.31 | 37.02 ± 0.28 | 1.69 ± 0.04 | |
| AG(85) | 7.60 ± 0.28 | 5.30 ± 0.13 | 70.22 ± 0.57 | 61.17 ± 0.34 | 37.06 ± 0.31 | 1.68 ± 0.05 | |
| P-value | 0.6364 | 0.2951 | 0.9567 | 0.6591 | 0.7296 | 0.9378 | |
| rs41919992 | CC(157) | 7.66 ± 0.27 | 5.30 ± 0.12a | 70.06 ± 0.55 | 61.03 ± 0.33A | 37.27 ± 0.30A | 1.67 ± 0.04 |
| TT(24) | 7.50 ± 0.42 | 5.40 ± 0.19ab | 70.71 ± 0.87 | 60.33 ± 0.52A | 38.22 ± 0.48A | 1.60 ± 0.07 | |
| CT(133) | 7.56 ± 0.28 | 5.08 ± 0.13b | 69.86 ± 0.55 | 61.82 ± 0.33B | 36.44 ± 0.30B | 1.72 ± 0.04 | |
| P-value | 0.8491 | 0.0190 | 0.5297 | 0.0004 | <.0001 | 0.0612 | |
| rs133498277 | CC(157) | 7.58 ± 0.27 | 5.31 ± 0.12a | 70.07 ± 0.54 | 61.05 ± 0.32A | 37.31 ± 0.29A | 1.67 ± 0.04 |
| TT(23) | 7.42 ± 0.42 | 5.45 ± 0.19a | 70.84 ± 0.87 | 60.29 ± 0.53A | 38.25 ± 0.48A | 1.60 ± 0.07 | |
| CT(134) | 7.48 ± 0.27 | 5.10 ± 0.12b | 69.81 ± 0.54 | 61.84 ± 0.32B | 36.46 ± 0.30B | 1.73 ± 0.04 | |
| P-value | 0.8410 | 0.0178 | 0.4115 | 0.0004 | <.0001 | 0.0617 | |
| rs41919984 | CC(157) | 7.53 ± 0.42 | 5.37 ± 0.19ab | 70.82 ± 0.86 | 60.27 ± 0.52A | 38.29 ± 0.47A | 1.60 ± 0.07a |
| TT(24) | 7.77 ± 0.27 | 5.32 ± 0.12b | 70.24 ± 0.54 | 60.91 ± 0.32A | 37.40 ± 0.29A | 1.66 ± 0.04ab | |
| TC(134) | 7.61 ± 0.27 | 5.08 ± 0.12a | 69.95 ± 0.53 | 61.77 ± 0.32B | 36.48 ± 0.29B | 1.72 ± 0.04b | |
| P-value | 0.6745 | 0.0154 | 0.4795 | 0.0002 | <.0001 | 0.0450 | |
| rs41919985 | AA(25) | 7.51 ± 0.41 | 5.38 ± 0.19ab | 70.97 ± 0.85 | 60.31 ± 0.51A | 38.23 ± 0.47A | 1.60 ± 0.07a |
| GG(157) | 7.77 ± 0.27 | 5.32 ± 0.12b | 70.24 ± 0.54 | 60.91 ± 0.32A | 37.4 ± 0.29A | 1.66 ± 0.04ab | |
| GA(133) | 7.61 ± 0.27 | 5.07 ± 0.12a | 69.93 ± 0.53 | 61.77 ± 0.32B | 36.47 ± 0.29B | 1.72 ± 0.04b | |
| P-value | 0.6642 | 0.0122 | 0.3517 | 0.0002 | <.0001 | 0.0472 | |
| rs41919986 | CC(155) | 7.61 ± 0.27 | 5.34 ± 0.12A | 70.09 ± 0.54 | 60.97 ± 0.32A | 37.37 ± 0.29A | 1.66 ± 0.04ab |
| TT(25) | 7.43 ± 0.41 | 5.45 ± 0.19A | 70.92 ± 0.85 | 60.33 ± 0.51A | 38.22 ± 0.47A | 1.60 ± 0.07b | |
| CT(132) | 7.49 ± 0.27 | 5.09 ± 0.12B | 69.71 ± 0.54 | 61.86 ± 0.32B | 36.41 ± 0.29B | 1.73 ± 0.04a | |
| P-value | 0.7937 | 0.0074 | 0.2439 | 0.0001 | <.0001 | 0.0393 |
Notes: P-value refers to the results of the association analysis between each SNP and milk fatty acid traits. Different letter (small letters: P < 0.05; capital letters: P < 0.01) superscripts (adjusted value after correction for multiple testing) indicate significant differences among the genotypes
The effects of the three genotyped polymorphisms in PPARGC1A, ABCG2 and IGF1 on 16 milk fatty acid compositions are shown in Table 5. SNP rs109579682 in PPARGC1A was significantly associated with eight milk fatty acid traits, such as C10:0 (P = 0.0251), C12:0 (P = 0.0340), C14:0 (P = 0.0188), C16:1 (P = 0.0401), C18:1n9c (P = 0.0015), C16 index (P = 0.0010), SFA (P = 0.0065) and UFA (P = 0.0038). Correspondingly, the CC genotype was the dominant type for saturated fatty acids (C10:0, C12:0, C14:0 and SFA), and the TT genotype was dominant for unsaturated fatty acids (C16:1, C18:1n9c, C16 index and UFA).
Table 5.
Associations of SNPs of PPARGC1A, ABCG2 and IGF1 genes with milk medium-chain fatty acids (MCFAs) in Chinese Holstein cattle (LSM ± SE)
| Gene | Locus | Genotypes | C10:0 | C12:0 | C14:0 | C14:1 | ||
| PPARGC1A | rs109579682 | CC(27) | 2.10 ± 0.06ab | 2.66 ± 0.07a | 9.50 ± 0.13a | 0.77 ± 0.03 | ||
| TT(170) | 1.94 ± 0.08b | 2.42 ± 0.11b | 9.19 ± 0.19ab | 0.79 ± 0.05 | ||||
| CT(147) | 2.13 ± 0.06a | 2.62 ± 0.08ab | 9.30 ± 0.13b | 0.78 ± 0.03 | ||||
| P-value | 0.0251 | 0.034 | 0.0188 | 0.8281 | ||||
| ABCG2 | rs137757790 | AA(115) | 2.13 ± 0.06 | 2.67 ± 0.08 | 9.58 ± 0.13A | 0.78 ± 0.04 | ||
| CC(85) | 2.06 ± 0.06 | 2.58 ± 0.08 | 9.21 ± 0.14B | 0.76 ± 0.04 | ||||
| CA(145) | 2.12 ± 0.06 | 2.64 ± 0.08 | 9.50 ± 0.13A | 0.78 ± 0.03 | ||||
| P-value | 0.2206 | 0.3385 | 0.0026 | 0.7772 | ||||
| IGF1 | rs109763947 | CC(58) | 2.06 ± 0.07a | 2.64 ± 0.09 | 9.47 ± 0.15 | 0.77 ± 0.04 | ||
| TT(100) | 2.19 ± 0.06b | 2.72 ± 0.08 | 9.57 ± 0.14 | 0.77 ± 0.04 | ||||
| CT(187) | 2.10 ± 0.06ab | 2.60 ± 0.07 | 9.42 ± 0.13 | 0.78 ± 0.03 | ||||
| P-value | 0.0342 | 0.0764 | 0.2805 | 0.9454 | ||||
| Gene | Locus | Genotypes | C16:0 | C16:1 | C18:0 | C18:1n9c | C18:2n6c | CLA |
| PPARGC1A | rs109579682 | CC(27) | 32.44 ± 0.33 | 1.71 ± 0.05a | 12.61 ± 0.17 | 29.19 ± 0.22A | 4.07 ± 0.03 | 0.39 ± 0.01 |
| TT(170) | 32.40 ± 0.51 | 1.82 ± 0.07ab | 12.59 ± 0.27 | 30.08 ± 0.36B | 4.08 ± 0.04 | 0.37 ± 0.02 | ||
| CT(147) | 31.99 ± 0.34 | 1.79 ± 0.05b | 12.59 ± 0.17 | 29.74 ± 0.23B | 4.07 ± 0.03 | 0.38 ± 0.01 | ||
| P-value | 0.1541 | 0.0401 | 0.9845 | 0.0015 | 0.9515 | 0.6788 | ||
| ABCG2 | rs137757790 | AA(115) | 32.45 ± 0.35 | 1.72 ± 0.05 | 12.52 ± 0.18 | 29.12 ± 0.24A | 4.05 ± 0.03 | 0.37 ± 0.01 |
| CC(85) | 31.99 ± 0.37 | 1.71 ± 0.05 | 12.79 ± 0.19 | 29.91 ± 0.26B | 4.08 ± 0.03 | 0.38 ± 0.01 | ||
| CA(145) | 32.33 ± 0.33 | 1.76 ± 0.05 | 12.48 ± 0.17 | 29.50 ± 0.22AB | 4.07 ± 0.03 | 0.39 ± 0.01 | ||
| P-value | 0.3251 | 0.3431 | 0.1475 | 0.0048 | 0.6085 | 0.2071 | ||
| IGF1 | rs109763947 | CC(58) | 32.29 ± 0.39 | 1.81 ± 0.05 | 12.44 ± 0.20 | 29.42 ± 0.27AB | 4.08 ± 0.03A | 0.39 ± 0.01 |
| TT(100) | 32.52 ± 0.36 | 1.7 ± 0.05 | 12.62 ± 0.18 | 29.02 ± 0.25B | 3.99 ± 0.03B | 0.38 ± 0.01 | ||
| CT(187) | 32.19 ± 0.33 | 1.73 ± 0.05 | 12.57 ± 0.16 | 29.70 ± 0.22A | 4.10 ± 0.03A | 0.38 ± 0.01 | ||
| P-value | 0.4406 | 0.0797 | 0.6613 | 0.0024 | <.0001 | 0.5835 | ||
| Gene | Locus | Genotypes | C14INDEX | C16INDEX | C18INDEX | SFA | UFA | SFA/UFA |
| PPARGC1A | rs109579682 | CC(27) | 7.50 ± 0.26 | 5.03 ± 0.12A | 69.81 ± 0.51 | 61.62 ± 0.30A | 36.75 ± 0.28A | 1.70 ± 0.04 |
| TT(170) | 7.89 ± 0.40 | 5.33 ± 0.18B | 70.52 ± 0.82 | 60.75 ± 0.49B | 37.74 ± 0.45B | 1.64 ± 0.07 | ||
| CT(147) | 7.75 ± 0.27 | 5.33 ± 0.12B | 70.24 ± 0.53 | 60.92 ± 0.32B | 37.38 ± 0.29B | 1.66 ± 0.04 | ||
| P-value | 0.3099 | 0.0010 | 0.4283 | 0.0065 | 0.0038 | 0.2931 | ||
| ABCG2 | rs137757790 | AA(115) | 7.51 ± 0.28 | 5.05 ± 0.13 | 69.96 ± 0.55 | 61.64 ± 0.33A | 36.67 ± 0.30a | 1.71 ± 0.04 |
| CC(85) | 7.62 ± 0.29 | 5.10 ± 0.13 | 70.00 ± 0.59 | 60.81 ± 0.35B | 37.45 ± 0.32b | 1.65 ± 0.05 | ||
| CA(145) | 7.57 ± 0.26 | 5.19 ± 0.12 | 70.29 ± 0.52 | 61.33 ± 0.31AB | 37.11 ± 0.28ab | 1.68 ± 0.04 | ||
| P-value | 0.8925 | 0.3350 | 0.7065 | 0.0343 | 0.0266 | 0.4267 | ||
| IGF1 | rs109763947 | CC(58) | 7.50 ± 0.31 | 5.32 ± 0.14a | 70.26 ± 0.62 | 61.14 ± 0.37A | 37.10 ± 0.34AB | 1.67 ± 0.05 |
| TT(100) | 7.5 ± 0.28 | 4.98 ± 0.13b | 69.68 ± 0.56 | 61.88 ± 0.33B | 36.47 ± 0.30B | 1.73 ± 0.04 | ||
| CT(187) | 7.62 ± 0.26 | 5.13 ± 0.12ab | 70.31 ± 0.51 | 61.10 ± 0.30A | 37.31 ± 0.28A | 1.66 ± 0.04 | ||
| P-value | 0.8036 | 0.0239 | 0.3301 | 0.009 | 0.0023 | 0.1970 |
Notes: P-value refers to the results of the association analysis between each SNP and milk fatty acid traits. Different letter (small letters: P < 0.05; capital letters: P < 0.01) superscripts (adjusted value after correction for multiple testing) indicate significant differences among the genotypes
For ABCG2, SNP rs137757790 was significantly associated with C14:0 (P = 0.0026), C18:1n9c (P = 0.0048), SFA (P = 0.0343) and UFA (P = 0.0266). The AA genotype was dominant for saturated fatty acids (C14:0 and SFA), and the CC genotype was dominant for unsaturated fatty acids (C18:1n9c and UFA).
For IGF1, SNP rs109763947 was significantly associated with C10:0 (P = 0.0342), C18:1n9c (P = 0.0024), C18:2n6c (P < 0.0001), C16 index (P = 0.0239), SFA (P = 0.0090) and UFA (P = 0.0023). The homozygous genotype of TT was the dominant type for saturated fatty acids (C10:0 and SFA), and the heterozygous genotype of CT was the dominant type for unsaturated fatty acids (C18:1n9c, C16 index, C18:2n6c and UFA).
Additionally, the significant dominant, additive and allele substitution effects of the significant SNPs on the target milk fatty acid traits were observed (Tables 6 and 7).
Table 6.
Additive, dominant and allele substitution effects of the nine SNPs on milk fatty acids traits of FASN in Chinese Holstein cattle
| Locus | Genetic effect | C10:0 | C12:0 | C14:0 | C14:1 | C16:0 | C16:1 | C18:0 | C18:1n9c | C18:2n6c | CLA | C14 INDEX | C16 INDEX | C18 INDEX | SFA | UFA | SFA/ UFA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs136947640 | a | −0.052 | −0.011 | 0.315 | 0.070 | −0.039 | −0.056 | 0.169 | −0.409 | 0.147* | 0.000 | 0.462 | −0.143 | −0.570 | 0.390 | −0.301 | 0.036 |
| d | −0.093 | −0.082 | 0.186 | 0.063 | −0.164 | 0.007 | 0.127 | −0.409 | 0.244** | 0.015 | 0.470 | 0.057 | −0.465 | 0.029 | −0.113 | 0.016 | |
| α | −0.125 | −0.075 | 0.461 | 0.120 | −0.167 | −0.050 | 0.269 | −0.729 | 0.339** | 0.012 | 0.831 | −0.099 | −0.934 | 0.412 | −0.390 | 0.049 | |
| rs41919999 | a | 0.072* | 0.074 | 0.192** | −0.011 | −0.008 | 0.013 | −0.186 | −0.193 | 0.007 | 0.008 | −0.226 | 0.024 | 0.194 | 0.150 | −0.160 | 0.008 |
| d | 0.090* | 0.123* | 0.080 | 0.022 | −0.111 | −0.021 | 0.012 | −0.267 | −0.021 | −0.003 | 0.116 | −0.041 | −0.245 | 0.224 | −0.308 | 0.022 | |
| α | 0.079** | 0.084* | 0.198** | −0.009 | −0.017 | 0.012 | −0.185 | −0.214 | 0.005 | 0.007 | −0.217 | 0.020 | 0.175 | 0.167 | −0.183 | 0.010 | |
| rs132865003 | a | −0.029 | −0.020 | 0.034 | 0.024 | −0.206 | −0.014 | 0.058 | 0.246 | 0.027 | −0.012 | 0.195 | −0.009 | 0.030 | −0.230 | 0.238 | −0.011 |
| d | −0.026 | −0.018 | 0.004 | 0.006 | −0.256 | 0.035 | −0.022 | 0.162 | 0.089* | 0.005 | 0.046 | 0.139 | 0.157 | −0.373 | 0.274 | −0.019 | |
| α | −0.047 | −0.032 | 0.037 | 0.028 | −0.376 | 0.009 | 0.043 | 0.354 | 0.087 | −0.009 | 0.225 | 0.083 | 0.135 | −0.479 | 0.421 | −0.024 | |
| rs134340637 | a | 0.029 | 0.020 | −0.034 | −0.024 | 0.206 | 0.014 | −0.058 | −0.246 | −0.027 | 0.012 | −0.195 | 0.009 | −0.030 | 0.230 | −0.238 | 0.011 |
| d | −0.026 | −0.018 | 0.004 | 0.006 | −0.256 | 0.035 | −0.022 | 0.162 | 0.089* | 0.005 | 0.046 | 0.139 | 0.157 | −0.373 | 0.274 | −0.019 | |
| α | 0.047 | 0.032 | −0.037 | −0.028 | 0.376 | −0.009 | −0.043 | −0.354 | −0.087 | 0.009 | −0.225 | −0.083 | −0.135 | 0.479 | −0.421 | 0.024 | |
| rs41919992 | a | −0.001 | 0.042 | −0.022 | 0.008 | 0.297 | −0.006 | −0.007 | −0.482** | 0.012 | 0.004 | 0.080 | −0.049 | −0.326 | 0.350 | −0.475* | 0.033 |
| d | 0.139** | 0.250** | 0.418** | 0.030 | 0.510 | −0.064 | −0.235 | −1.266** | 0.010 | −0.002 | −0.022 | −0.271** | −0.524 | 1.142** | −1.309** | 0.091 | |
| α | 0.058 | 0.148* | 0.155 | 0.021 | 0.513 | −0.033 | −0.107 | −1.018** | 0.016 | 0.003 | 0.071 | −0.163 | −0.547 | 0.834* | −1.029** | 0.071 | |
| rs133498277 | a | −0.011 | 0.032 | −0.048 | 0.006 | 0.324 | 1.796 | 0.041 | −0.453* | −0.003 | 0.003 | 0.081 | −0.068 | −0.384 | 0.376 | −0.471* | 0.033 |
| d | 0.120** | 0.229** | 0.373** | 0.027 | 0.537 | 1.740 | −0.164 | −1.249** | −0.008 | −0.002 | −0.026 | −0.284** | −0.639 | 1.167** | −1.318** | 0.092 | |
| α | −0.062 | −0.066 | −0.208** | −0.005 | 0.094 | 1.819 | 0.111 | 0.080 | 0.000 | 0.004 | 0.092 | 0.053 | −0.111 | −0.122 | 0.092 | −0.006 | |
| rs41919984 | a | 0.009 | −0.036 | 0.048 | −0.010 | −0.308 | −0.003 | 0.011 | 0.463* | −0.012 | −0.005 | −0.121 | 0.027 | 0.292 | −0.323 | 0.445* | −0.030 |
| d | 0.138** | 0.258** | 0.409** | 0.027 | 0.559 | −0.059 | −0.233 | −1.333** | 0.012 | −0.003 | −0.042 | −0.268** | −0.577 | 1.183** | −1.372** | 0.095* | |
| α | 0.067* | 0.073 | 0.220** | 0.001 | −0.072 | −0.028 | −0.087 | −0.100 | −0.006 | −0.006 | −0.138 | −0.086 | 0.049 | 0.176 | −0.135 | 0.010 | |
| rs41919985 | a | 0.020 | −0.020 | 0.087 | −0.009 | −0.305 | 0.000 | −0.045 | 0.421* | −0.004 | −0.006 | −0.130 | 0.033 | 0.366 | −0.302 | 0.415 | −0.029 |
| d | 0.125** | 0.239** | 0.361** | 0.025 | 0.560 | −0.063 | −0.162 | −1.290** | 0.003 | −0.001 | −0.030 | −0.278** | −0.677 | 1.164** | −1.344** | 0.094* | |
| α | −0.032 | −0.120 | −0.064 | −0.019 | −0.539 | 0.026 | 0.022 | 0.962** | −0.005 | −0.005 | −0.117 | 0.150 | 0.650 | −0.790* | 0.978** | −0.068 | |
| rs41919986 | a | −0.025 | 0.020 | −0.073 | 0.006 | 0.293 | −0.006 | 0.068 | −0.415* | −0.001 | 0.004 | 0.092 | −0.050 | −0.414 | 0.318 | −0.425 | 0.029 |
| d | 0.122** | 0.224** | 0.350** | 0.024 | 0.563 | −0.072 | −0.106 | −1.301** | −0.017 | −0.004 | −0.032 | −0.301** | −0.798 | 1.210** | −1.390** | 0.097* | |
| α | 0.026 | 0.113 | 0.072 | 0.016 | 0.528 | −0.036 | 0.024 | −0.957** | −0.008 | 0.002 | 0.079 | −0.176 | −0.746 | 0.822* | −1.004** | 0.070 |
Note: a means additive effect; d means dominant effect; α means allele substitution effect. The asterisk (*) means the additive, dominant or allele substitution effect of the locus indicated differ at P < 0.05 and the asterisk (**) means the additive, dominant or allele substitution effect of the locus indicated differ at P < 0.01
Table 7.
Additive, dominant and allele substitution effects of the SNPs on milk fatty acids traits of PPARGC1A, ABCG2 and IGF1 in Chinese Holstein cattle
| Gene | Locus | Genetic effect | C10:0 | C12:0 | C14:0 | C14:1 | C16:0 | C16:1 | C18:0 | C18:1n9c | C18:2n6c | CLA | C14 INDEX | C16 INDEX | C18 INDEX | SFA | UFA | SFA/UFA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PPARGC1A | rs109579682 | a | 0.081* | 0.120** | 0.159* | −0.012 | 0.019 | −0.052 | 0.008 | −0.445** | −0.005** | 0.006 | −0.193 | −0.153* | −0.356 | 0.436* | −0.496* | 0.034 |
| d | 0.107* | 0.080 | −0.049 | −0.000 | −0.435 | 0.025 | −0.016 | 0.108 | −0.001** | 0.003 | 0.052 | 0.150 | 0.075 | −0.270 | 0.134 | −0.009 | ||
| α | 0.036 | 0.086* | 0.179** | −0.012 | 0.200 | −0.062* | 0.015 | −0.490** | −0.005* | 0.005 | −0.215 | −0.216** | −0.387 | 0.549** | −0.552** | 0.037 | ||
| ABCG2 | rs137757790 | a | 0.039 | 0.047 | 0.185** | 0.011 | 0.232 | 0.004 | −0.136 | −0.392** | −0.013 | −0.003 | −0.059 | −0.025 | −0.021 | 0.413** | −0.390** | 0.028 |
| d | 0.031 | 0.022 | 0.106 | 0.008 | 0.106 | 0.047 | −0.173 | −0.020 | 0.002 | 0.012 | 0.005 | 0.112 | 0.312 | 0.104 | 0.055 | 0.000 | ||
| α | 0.041 | 0.049 | 0.195** | 0.012 | 0.241 | 0.008 | −0.151 | −0.393** | −0.013 | −0.002 | −0.058 | −0.015 | 0.007 | 0.422** | −0.385** | 0.028 | ||
| IGF-1 | rs109763947 | a | −0.063* | −0.040 | −0.050 | −0.004 | −0.115 | 0.055 | −0.088 | 0.201 | 0.045 | 0.005 | 0.003 | 0.168** | 0.290 | −0.369* | 0.317* | −0.028 |
| d | −0.026 | −0.083 | −0.097 | 0.007 | −0.220 | −0.023 | 0.037 | 0.482** | 0.064 | −0.006 | 0.117 | −0.023 | 0.334 | −0.414 | 0.523* | −0.033 | ||
| α | −0.060* | −0.030 | −0.038 | −0.005 | −0.088 | 0.057 | −0.093 | 0.142 | 0.037 | 0.006 | −0.012 | 0.171** | 0.250 | −0.319 | 0.253 | −0.024 |
Note: a means additive effect; d means dominant effect; α means allele substitution effect. The asterisk (*) means the additive, dominant or allele substitution effect of the locus indicated differ at P < 0.05 and the asterisk (**) means the additive, dominant or allele substitution effect of the locus indicated differ at P < 0.01
LD between the SNPs identified in the four candidate genes and our previous GWAS
Pair-wise D’ measures showed that all nine SNPs in FASN were highly linked (D’ > 0.9), and one haplotype block comprising eight SNPs was inferred (Fig. 1) in which three haplotypes were formed. The common haplotypes TCGCCTGC, CCGTTCAT and CTACCTGC occurred at a frequency of 54.2 %, 27.8 % and 17.2 %, respectively (Table 8). Most importantly, the significant SNP (rs41921177) identified in our previous GWAS [26] showed strong linkage with the three FASN SNPs (rs136947640, rs132865003 and rs134340637). Subsequently, haplotype-based analysis showed significant associations of the haplotypes encompassing the eight FASN SNPs (rs41919999, rs132865003, rs134340637, rs41919992, rs133498277, rs41919984, rs41919985 and rs41919986) with C10:0, C12:0, C14:0, C18:1n9c, SFA and UFA (P = 0.0204 to P < 0.0001; Table 9).
Fig. 1.
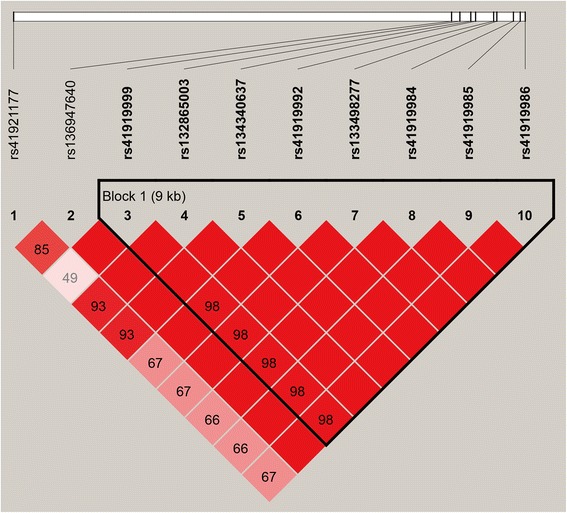
Linkage disequilibrium (LD) plot for 10 SNPs close to or within FASN. The values in boxes are pair-wise SNP correlations (D’), bright red boxes without numbers indicate complete LD (D’ = 1). The blocks indicate haplotype blocks and the texts above the horizontal numbers are the SNP names
Table 8.
Main haplotypes and their frequencies observed in the FASN gene
| FASN Haplotypes | SNP3 C > T | SNP4 T > C | SNP5 G > A | SNP6 C > T | SNP7 T > C | SNP8 T > C | SNP9 A > G | SNP10 T > C | Frequency (%) |
|---|---|---|---|---|---|---|---|---|---|
| TCGCCTGC | T | C | G | C | C | T | G | C | 54.2 |
| CCGTTCAT | C | C | G | T | T | C | A | T | 27.8 |
| CTACCTGC | C | T | A | C | C | T | G | C | 17.2 |
Note: The Ref number of each SNP can be found in the haplotype plot. Also, SNP3 = rs41919999, SNP4 = rs132865003, SNP5 = rs134340637, SNP6 = rs41919992, SNP7 = rs133498277, SNP8 = rs41919984, SNP9 = rs41919985, SNP10 = rs41919986
Table 9.
Haplotype associations of the eight SNPs in FASN with milk production traits in Chinese Holstein cattle (LSM ± SE)
| FASN haplotypes | C10:0 | C12:0 | C14:0 | C14:1 | C16:0 | C16:1 |
| H1H1(88) | 2.02 ± 0.05a | 2.54 ± 0.07A | 9.27 ± 0.12A | 0.78 ± 0.03 | 32.45 ± 0.33 | 1.79 ± 0.05 |
| H2H1(103) | 2.23 ± 0.05b | 2.81 ± 0.06B | 9.75 ± 0.11B | 0.80 ± 0.03 | 32.41 ± 0.29 | 1.72 ± 0.04 |
| H2H2(24) | 2.12 ± 0.09b | 2.54 ± 0.12B | 9.53 ± 0.20AB | 0.76 ± 0.06 | 31.54 ± 0.53 | 1.79 ± 0.07 |
| H2H3(28) | 2.16 ± 0.08b | 2.80 ± 0.10B | 9.95 ± 0.17B | 0.76 ± 0.05 | 32.90 ± 0.48 | 1.76 ± 0.07 |
| H3H1(57) | 2.09 ± 0.06b | 2.60 ± 0.08B | 9.22 ± 0.14A | 0.77 ± 0.04 | 31.84 ± 0.37 | 1.82 ± 0.05 |
| H3H3(10) | 2.16 ± 0.13b | 2.72 ± 0.17B | 9.43 ± 0.29AB | 0.74 ± 0.08 | 32.59 ± 0.77 | 1.80 ± 0.11 |
| P-value | 0.0204 | 0.0057 | 0.0001 | 0.9268 | 0.2257 | 0.4522 |
| FASN haplotypes | C18:0 | C18:1n9c | C18:2n6c | CLA | C14index | C16index |
| H1H1(88) | 12.69 ± 0.18 | 29.59 ± 0.24AC | 4.07 ± 0.03 | 0.38 ± 0.01 | 7.79 ± 0.26 | 5.26 ± 0.12 |
| H2H1(103) | 12.51 ± 0.16 | 28.89 ± 0.21BC | 4.04 ± 0.02 | 0.37 ± 0.01 | 7.61 ± 0.23 | 5.03 ± 0.10 |
| H2H2(24) | 12.62 ± 0.29 | 30.56 ± 0.38A | 4.05 ± 0.04 | 0.37 ± 0.02 | 7.46 ± 0.42 | 5.36 ± 0.19 |
| H2H3(28) | 12.22 ± 0.26 | 28.31 ± 0.34B | 4.14 ± 0.04 | 0.40 ± 0.02 | 7.16 ± 0.37 | 5.08 ± 0.17 |
| H3H1(57) | 12.65 ± 0.20 | 29.85 ± 0.27A | 4.11 ± 0.03 | 0.39 ± 0.01 | 7.68 ± 0.29 | 5.45 ± 0.13 |
| H3H3(10) | 12.38 ± 0.41 | 28.91 ± 0.55ABC | 4.03 ± 0.06 | 0.40 ± 0.03 | 7.23 ± 0.60 | 5.25 ± 0.27 |
| P-value | 0.6616 | <.0001 | 0.0792 | 0.4543 | 0.7264 | 0.0548 |
| FASN Haplotypes | C18index | SFA | UFA | SFA/UFA | ||
| H1H1(88) | 69.95 ± 0.53 | 61.15 ± 0.32AB | 37.20 ± 0.29AB | 1.68 ± 0.04 | ||
| H2H1(103) | 69.71 ± 0.47 | 61.87 ± 0.29A | 36.40 ± 0.26A | 1.73 ± 0.04 | ||
| H2H2(24) | 70.75 ± 0.86 | 60.39 ± 0.52AB | 38.15 ± 0.47B | 1.61 ± 0.07 | ||
| H2H3(28) | 69.85 ± 0.77 | 62.20 ± 0.46AB | 35.99 ± 0.42A | 1.75 ± 0.06 | ||
| H3H1(57) | 70.25 ± 0.60 | 60.69 ± 0.36B | 37.55 ± 0.33B | 1.64 ± 0.05 | ||
| H3H3(10) | 70.11 ± 1.24 | 61.75 ± 0.74AB | 36.54 ± 0.68AB | 1.71 ± 0.10 | ||
| P-value | 0.8619 | 0.0025 | <.0001 | 0.2846 | ||
Notes: P-value refers to the results of the association analysis between each haplotype and milk fatty acid traits. Different letter (small letters: P < 0.05; capital letters: P < 0.01) superscripts (adjusted value after correction for multiple testing) indicate significant differences among the haplotypes. H1 = TCGCCTGC, H2 = CCGTTCAT, H3 = CTACCTGC
Strong linkage among the two significant SNPs (rs110131167 and rs108967640) detected in our previous GWAS [26] and the SNP (rs109579682) in PPARGC1A was also observed (D’ > 0.9, Fig. 2). However, no LD was observed between the SNPs located in the ABCG2 and IGF1 genes.
Fig. 2.
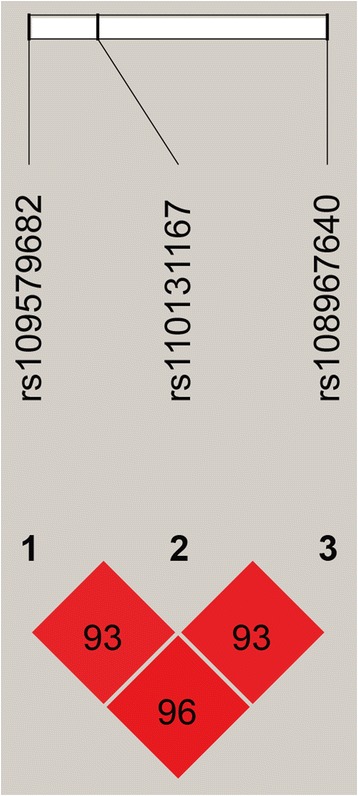
Linkage disequilibrium (LD) plot for three SNPs in PPARGC1A. The values in boxes are pair-wise SNP correlations (D’), the brighter shade of red indicates higher linkage disequilibrium
Discussion
Information on the effects of DNA polymorphisms on milk fatty acid composition is scarce, because milk fatty acid composition data, unlike those of milk fat percentage and fat yield, are not collected routinely in milk recording schemes. Therefore, we attempted to explore the genetic variants of candidate genes identified by our previous GWAS on milk fatty acid composition [26]. In this study, we first investigated the associations between the tested SNPs of FASN, PPARGC1A, ABCG2 and IGF1 and milk fatty acid traits in Chinese Holstein cows.
In our previous GWAS, the SNP rs41921177, at a distance of 58,172 bp away from FASN, showed significant association with C10:0 (P = 8.54E-06), C12:0 (P = 1.16E-07) and C14:0 (P = 6.01E-06) [26]. As expected, we found that this SNP was also strongly linked with the three SNPs in FASN (rs136947640, rs132865003 and rs134340637) that were significantly associated with C18:2n6c. Furthermore, if the haplotype block was defined based on the solid spine of the LD method, one haplotype block was constructed by the above three SNPs plus two SNPs, rs41921177 and rs41919999, that were associated with C10:0, C12:0 and C14:0. Similarly, strong linkages between the two significant SNPs (rs110131167 and rs108967640) for the C18 index, UFA and SFA/UFA identified in our previous GWAS and the SNP (rs109579682) in PPARGC1A for UFA and SFA identified in this study were observed. Probably as a result of the limited number of SNPs identified for ABCG2 and IGF1, and the farther distance between SNPs in the previous GWAS and their adjacent SNPs identified for ABCG2 and IGF1 in this study, no linkages with the significant SNPs identified in GWAS were observed.
Six out of nine SNPs in FASN (rs41919999, rs41919992, rs133498277, rs41919984, rs41919985 and rs41919986) were markedly associated with C10:0, C12:0 and C14:0, and five of these six SNPs (rs41919992, rs133498277, rs41919984, rs41919985 and rs41919986) also showed significant associations with SFA, which suggested that the FASN gene mainly affects the medium-long chain saturated fatty acid traits. FASN is a complex, multifunctional enzyme that catalyzes de novo biosynthesis of long-chain saturated fatty acids [36] and plays an essential role in the determination of fatty acid synthesis and release of newly synthesized SFAs [37, 38]. In addition, several previous linkage studies [8, 39, 40] and GWA studies [13–15] have reported that the FASN gene is located in a quite large region associated with the medium-chain saturated milk fatty acids on BTA19, which is in agreement with our results that the SNPs in FASN mainly showed significant associations with C10:0, C12:0 and C14:0. Moreover, the five SNPs (rs41919992, rs133498277, rs41919984, rs41919985 and rs41919986) also showed associations with the C18:1n9c, C16 index and UFA, and three SNPs (rs136947640, rs132865003 and rs134340637) showed associations with C18:2n6, revealing that the FASN gene affects the long-chain unsaturated fatty acid traits. The haplotype-based association analysis showed their significant associations with C10:0, C12:0, C14:0, C18:1n9c, SFA and UFA, also confirming the genetic effects of the FASN gene on the medium-chain saturated and long-chain unsaturated milk fatty acids. Kim & Ntambi [41] reported that FASN is a key gene involved in the pathway for MUFAs synthesis and incorporation into triacylglycerols and phospholipids, which is consistent with our results. However, the effect of FASN on PUFAs has not been reported elsewhere.
It was reported that the SNPs in different exons of the FASN gene were associated with milk-fat percentage [9] and with the medium- and long-chain fatty acid content of milk [8] and beef [42]. Morris et al. [8] identified five SNPs in FASN, including the non-synonymous SNP, rs41919985, observed in this study, which had been reported in different studies. The allele frequency of rs41919985 A (0.29) in our population is lower than that reported in Friesian and Jersey cattle (0.31 and 0.13, respectively) [8], 0.53 in Dutch Holstein–Friesian population [43] and 0.62 in Angus beef cattle [42]. Morris et al. [8] also reported that rs41919985 affected the C18:1cis9 and the total index, while other SNPs in FASN affected C14:0 and C18:2, which were consistent with our findings. Associations of the rs41919985 G allele with higher C14:0 and lower C18:1cis9 were also found in beef cattle [42]. Abe et al. [44] revealed that the FASN gene had a significant effect on the fatty acid composition of backfat, intramuscular and intermuscular fat in an F2 population from Japanese Black and Limousin cattle. For all nine significant SNPs in FASN, the heterozygous genotypes were associated with a higher proportion of milk SFAs, while the homozygous genotypes were associated with much higher levels of long-chain MUFAs and PUFAs. Thus, decreasing the number of individuals with heterozygous genotypes for these target SNPs in FASN will be beneficial to produce high-quality milk with a high proportion of unsaturated fatty acids (UFAs).
PPARGC1A is involved in mammary gland metabolism, and the expression of PPARGC1A correlates with milk fat content [45]. Moreover, it is a key factor in energy metabolism and plays a central role in thermogenesis, gluconeogenesis, glucose transport and β-oxidation of fatty acids [46]. The finding that PPAR agonists are able to increase stearoyl-CoA desaturase (SCD) mRNA levels in humans, mice and rats suggested that PPARs are able to regulate SCD [47]. As the SCD enzyme is involved in the desaturation of saturated fatty acids into cis9-unsaturated fatty acids, PPARs might have an effect on unsaturation indices via their regulation of SCD [43]. Our findings supported the above research that PPARGC1A was significantly associated with the C16 index. In our study, PPARGC1A mainly affected medium-chain saturated fatty acids and long-chain unsaturated fatty acids. Only a few studies have reported associations between PPARGC1A and milk fatty acid composition [13, 43]. Schennink et al. [43] found that one SNP in PPARGC1A, c.1790 + 514G > A, was associated with the C16:1 and C16 index, and Bouwman et al. [13] reported another significant SNP associated with C16:1, which are in agreement with the results in this study that rs109579682 in PPARGC1A is associated significantly with the C16:1 and C16 index. The significant associations between PPARGC1A c.1790 + 514G > A and the C14:1, C14 index, and C18 index [43] were not found in this study. The conflicting findings could be explained by the two different genetic backgrounds of the studied populations or by the different number of individuals included in each study. Phenotypic data were available from 1,905 cows in the study reported by Schennink et al. [43], while 346 cows were available in our study.
The bovine ABCG2 gene is located in the narrow region of chromosome 6 (BTA6), harboring a QTL with a large impact on milk production traits [48, 49]. The ABCG2 protein is responsible for the secretion of xenobiotics and some quantitatively minor nutrients, such as vitamin K3 or cholesterol, into milk [50, 51]. The insulin-like growth factor (IGF) signaling pathway plays a crucial role in the regulation of growth and development of mammals. Liang et al. [52] reported that IGF1 stimulates de novo fatty acid biosynthesis by Schwann cells during myelination. For ABCG2 and IGF1, most studies focused on investigating the association between the identified SNPs in these two genes and milk fat traits [4, 53–58], while limited studies on their association with milk fatty acid composition have been reported [13]. Bouwman et al. [13] reported that one QTL region underlying the ABCG2 gene showed significant effects on C12:1, C14:1 and C16:1. No association between IGF1 and milk fatty acids composition has been reported. Further studies will be necessary to confirm our results in different cattle population and to elucidate the mechanisms underlying the association found in this study.
Conclusions
In this study, we not only confirmed the deduction that the significant SNPs close to the FASN and PPARGC1A genes identified in our previous GWAS were strongly linked with the key mutations in these two candidate genes, but also presented a link of several variants of FASN, PPARGC1A, ABCG2 and IGF1 with milk fatty acid traits. In particular, FASN and PPARGC1A mainly affected medium-chain saturated fatty acids and long-chain unsaturated fatty acids. Our findings regarding genes and polymorphisms responsible for the variation of milk fatty acids composition provide useful information that can be combined with breeding programs to tailor the fatty acid content in cow’s milk.
Abbreviations
ABCG2, ATP-binding cassette, sub-family G, member 2; CAD, coronary artery disease; FASN, fatty acid synthase; GWAS, genome-wide association study; IACUC, institutional animal care and use committee; IGF1, insulin-like growth factor 1; LD, linkage disequilibrium; LDL, low density lipoprotein; MUFA, monounsaturated fatty acid; PPARGC1A, peroxisome proliferator-activated receptor gamma, coactivator 1 alpha; PUFA, polyunsaturated fatty acid; QTL, quantitative trait locus; SCD, stearoyl-CoA desaturase; SFA, saturated fatty acid; UFA, unsaturated fatty acids; UTR, untranslated region.
Acknowledgements
We appreciate the Dairy Data Center of China and Beijing Dairy Cattle Center for providing pedigree and milk samples for the Chinese Holstein cows.
Funding
This work was supported by the National Science and Technology Programs of China (2013AA102504, 2011BAD28B02, 2014ZX08009-053B), Beijing Natural Science Foundation (6152013), Beijing Dairy Industry Innovation Team, earmarked fund for Modern Agro-industry Technology Research System (CARS-37), and Program for Changjiang Scholar and Innovation Research Team in University (IRT1191).
Availability of data and materials
All relevant data are available within the manuscript and its Supporting Information files.
Authors’ contributions
CL conducted the association analysis and wrote the manuscript. DS and SZ designed the study and revised the manuscript. SY and MA prepared the DNA samples for SNP identification and genotyping. QZ participated in the data analysis and provided suggestions for the manuscript. YL and LL provided milk samples and participated in the result interpretation. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Animal handling and sample collection procedures were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) at China Agricultural University.
Additional file
Primers used to identify SNPs in the FASN gene. (PDF 104 kb)
Contributor Information
Cong Li, Email: li10020902@163.com.
Dongxiao Sun, Email: sundx@cau.edu.cn.
Shengli Zhang, Email: zhangslcau@cau.edu.cn.
Shaohua Yang, Email: yangshaohua309@126.com.
M. A. Alim, Email: alimmdcau@gmail.com
Qin Zhang, Email: qzhang@cau.edu.cn.
Yanhua Li, Email: yhli1976@163.com.
Lin Liu, Email: liulin@bdcc.com.cn.
References
- 1.Blott S, Kim JJ, Moisio S, Schmidt-Kuntzel A, Cornet A, Berzi P, Cambisano N, Ford C, Grisart B, Johnson D, et al. Molecular dissection of a quantitative trait locus: a phenylalanine-to-tyrosine substitution in the transmembrane domain of the bovine growth hormone receptor is associated with a major effect on milk yield and composition. Genetics. 2003;163(1):253–66. doi: 10.1093/genetics/163.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brym P, Kaminski S, Rusc A. New SSCP polymorphism within bovine STAT5A gene and its associations with milk performance traits in Black-and-White and Jersey cattle. J Appl Genet. 2004;45(4):445–52. [PubMed] [Google Scholar]
- 3.Brym P, Kaminski S, Wojcik E. Nucleotide sequence polymorphism within exon 4 of the bovine prolactin gene and its associations with milk performance traits. J Appl Genet. 2005;46(2):179–85. [PubMed] [Google Scholar]
- 4.Cohen-Zinder M, Seroussi E, Larkin DM, Loor JJ, Everts-van der Wind A, Lee JH, Drackley JK, Band MR, Hernandez AG, Shani M, et al. Identification of a missense mutation in the bovine ABCG2 gene with a major effect on the QTL on chromosome 6 affecting milk yield and composition in Holstein cattle. Genome Res. 2005;15(7):936–44. doi: 10.1101/gr.3806705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dybus A, Grzesiak W, Kamieniecki H, Szatkowska I, Sobek Z, Blaszczyk P, Czerniawska-Piatkowska E, Zych S, Muszynska M. Association of genetic variants of bovine prolactin with milk production traits of Black-and-White and Jersey cattle. Arch Fur Tierzucht-Arch Anim Breed. 2005;48(2):149–56. [Google Scholar]
- 6.Grisart B, Coppieters W, Farnir F, Karim L, Ford C, Berzi P, Cambisano N, Mni M, Reid S, Simon P, et al. Positional candidate cloning of a QTL in dairy cattle: Identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res. 2002;12(2):222–31. doi: 10.1101/gr.224202. [DOI] [PubMed] [Google Scholar]
- 7.Khatib H, Zaitoun I, Wiebelhaus-Finger J, Chang YM, Rosa GJM. The association of bovine PPARGC1A and OPN genes with milk composition in two independent holstein cattle populations. J Dairy Sci. 2007;90(6):2966–70. doi: 10.3168/jds.2006-812. [DOI] [PubMed] [Google Scholar]
- 8.Morris CA, Cullen NG, Glass BC, Hyndman DL, Manley TR, Hickey SM, McEwan JC, Pitchford WS, Bottema CDK, Lee MAH. Fatty acid synthase effects on bovine adipose fat and milk fat. Mamm Genome. 2007;18(1):64–74. doi: 10.1007/s00335-006-0102-y. [DOI] [PubMed] [Google Scholar]
- 9.Roy R, Ordovas L, Zaragoza P, Romero A, Moreno C, Altarriba J, Rodellar C. Association of polymorphisms in the bovine FASN gene with milk-fat content. Anim Genet. 2006;37(3):215–8. doi: 10.1111/j.1365-2052.2006.01434.x. [DOI] [PubMed] [Google Scholar]
- 10.Viitala S, Szyda J, Blott S, Schulman N, Lidauer M, Maki-Tanila A, George M, Vilkki J. The role of the bovine growth hormone receptor and prolactin receptor genes in milk, fat and protein production in Finnish Ayrshire dairy cattle. Genetics. 2006;173(4):2151–64. doi: 10.1534/genetics.105.046730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weikard R, Kuhn C, Goldammer T, Freyer G, Schwerin M. The bovine PPARGC1A gene: molecular characterization and association of an SNP with variation of milk fat synthesis. Physiol Genomics. 2005;21(1):1–13. doi: 10.1152/physiolgenomics.00103.2004. [DOI] [PubMed] [Google Scholar]
- 12.Winter A, Kramer W, Werner FAO, Kollers S, Kata S, Durstewitz G, Buitkamp J, Womack JE, Thaller G, Fries R. Association of a lysine-232/alanine polymorphism in a bovine gene encoding acyl-CoA : diacylglycerol acyltransferase (DGAT1) with variation at a quantitative trait locus for milk fat content. Proc Natl Acad Sci U S A. 2002;99(14):9300–5. doi: 10.1073/pnas.142293799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouwman AC, Bovenhuis H, Visker MHPW, van Arendonk JAM. Genome-wide association of milk fatty acids in Dutch dairy cattle. BMC Genet. 2011;12:43. doi: 10.1186/1471-2156-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouwman AC, Visker MHPW, van Arendonk JAM, Bovenhuis H. Genomic regions associated with bovine milk fatty acids in both summer and winter milk samples. BMC Genet. 2012;13:1–13. doi: 10.1186/1471-2156-13-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buitenhuis B, Janss LLG, Poulsen NA, Larsen LB, Larsen MK, Sorensen P. Genome-wide association and biological pathway analysis for milk-fat composition in Danish Holstein and Danish Jersey cattle. BMC Genomics. 2014;15:1112. doi: 10.1186/1471-2164-15-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conte G, Mele M, Chessa S, Castiglioni B, Serra A, Pagnacco G, Secchiari P. Diacylglycerol acyltransferase 1, stearoyl-CoA desaturase 1, and sterol regulatory element binding protein 1 gene polymorphisms and milk fatty acid composition in Italian Brown cattle. J Dairy Sci. 2010;93(2):753–63. doi: 10.3168/jds.2009-2581. [DOI] [PubMed] [Google Scholar]
- 17.Kgwatalala PM, Ibeagha-Awemu EM, Hayes JF, Zhao X. Stearoyl-CoA desaturase 1 3'UTR SNPs and their influence on milk fatty acid composition of Canadian Holstein cows. J Anim Breed Genet. 2009;126(5):394–403. doi: 10.1111/j.1439-0388.2008.00796.x. [DOI] [PubMed] [Google Scholar]
- 18.Macciotta NPP, Mele M, Conte G, Serra A, Cassandro M, Dal Zotto R, Borlino AC, Pagnacco G, Secchiari P. Association between a polymorphism at the stearoyl CoA desaturase locus and milk production traits in Italian Holsteins. J Dairy Sci. 2008;91(8):3184–9. doi: 10.3168/jds.2007-0947. [DOI] [PubMed] [Google Scholar]
- 19.Mele M, Conte G, Castiglioni B, Chessa S, Macciotta NPP, Serra A, Buccioni A, Pagnacco G, Secchiari P. Stearoyl-Coenzyme A desaturase gene polymorphism and milk fatty acid composition in Italian Holsteins. J Dairy Sci. 2007;90(9):4458–65. doi: 10.3168/jds.2006-617. [DOI] [PubMed] [Google Scholar]
- 20.Moioli B, Contarini G, Avalli A, Catillo G, Orru L, De Matteis G, Masoero G, Napolitano F. Short communication: Effect of stearoyl-coenzyme A desaturase polymorphism on fatty acid composition of milk. J Dairy Sci. 2007;90(7):3553–8. doi: 10.3168/jds.2006-855. [DOI] [PubMed] [Google Scholar]
- 21.Rincon G, Islas-Trejo A, Castillo AR, Bauman DE, German BJ, Medrano JF. Polymorphisms in genes in the SREBP1 signalling pathway and SCD are associated with milk fatty acid composition in Holstein cattle. J Dairy Res. 2012;79(1):66–75. doi: 10.1017/S002202991100080X. [DOI] [PubMed] [Google Scholar]
- 22.Schennink A, Heck JML, Bovenhuis H, Visker MHPW, van Valenberg HJF, van Arendonk JAM. Milk fatty acid unsaturation: Genetic parameters and effects of stearoyl-CoA desaturase (SCD1) and acyl CoA : diacylglycerol acyltransferase 1 (DGAT1) J Dairy Sci. 2008;91(5):2135–43. doi: 10.3168/jds.2007-0825. [DOI] [PubMed] [Google Scholar]
- 23.Mensink RP, Zock PL, Kester AD, Katan MB. Effects of dietary fatty acids and carbohydrates on the ratio of serum total to HDL cholesterol and on serum lipids and apolipoproteins: a meta-analysis of 60 controlled trials. Am J Clin Nutr. 2003;77(5):1146–55. doi: 10.1093/ajcn/77.5.1146. [DOI] [PubMed] [Google Scholar]
- 24.Rasmussen BM, Vessby B, Uusitupa M, Berglund L, Pedersen E, Riccardi G, Rivellese AA, Tapsell L, Hermansen K, Grp KS. Effects of dietary saturated, monounsaturated, and n-3 fatty acids on blood pressure in healthy subjects. Am J Clin Nutr. 2006;83(2):221–6. doi: 10.1093/ajcn/83.2.221. [DOI] [PubMed] [Google Scholar]
- 25.Wijendran V, Hayes KC. Dietary n-6 and n-3 fatty acid balance and cardiovascular health. Annu Rev Nutr. 2004;24:597–615. doi: 10.1146/annurev.nutr.24.012003.132106. [DOI] [PubMed] [Google Scholar]
- 26.Li C, Sun DX, Zhang SL, Wang S, Wu XP, Zhang Q, Liu L, Li YH, Qiao L. Genome wide association study identifies 20 novel promising genes associated with milk fatty acid traits in chinese holstein. PLoS One. 2014;9(5):e96186. doi: 10.1371/journal.pone.0096186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alim MA, Wang P, Wu XP, Li C, Cui XG, Zhang SL, Zhang Q, Zhang Y, Sun DX. Effect of FASN gene on milk yield and milk composition in the Chinese Holstein dairy population. Anim Genet. 2014;45(1):111–3. doi: 10.1111/age.12089. [DOI] [PubMed] [Google Scholar]
- 28.Alim MA, Fan YP, Xie Y, Wu XP, Sun DX, Zhang Y, Zhang SL, Zhang Y, Zhang Q, Liu L. Single Nucleotide Polymorphism (SNP) in PPARGC1A Gene Associates Milk Production Traits in Chinese Holstein Cattle. Pakistan Vet J. 2012;32(4):609–12. [Google Scholar]
- 29.Alim MA, Xie Y, Fan YP, Wu XP, Sun DX, Zhang Y, Zhang SL, Zhang Y, Zhang Q, Liu L. Genetic effects of ABCG2 polymorphism on milk production traits in the Chinese Holstein cattle. J Appl Anim Res. 2013;41(3):333–8. doi: 10.1080/09712119.2013.782873. [DOI] [Google Scholar]
- 30.Alim MA, Xie Y, Fan YP, Wu XP, Zhang Y, Sun DX, Zhang SL, Zhang Y, Zhang Q, Liu L. Effects of polymorphism in the 5 '-flanking region of the IGF-I gene on milk-production traits in Chinese Holstein cattle. Anim Reprod Sci. 2012;52(9):795–8. doi: 10.1071/AN12018. [DOI] [Google Scholar]
- 31.Kelsey JA, Corl BA, Collier RJ, Bauman DE. The effect of breed, parity, and stage of lactation on conjugated linoleic acid (CLA) in milk fat from dairy cows. J Dairy Sci. 2003;86(8):2588–97. doi: 10.3168/jds.S0022-0302(03)73854-5. [DOI] [PubMed] [Google Scholar]
- 32.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23(10):1289–91. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 33.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 34.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 35.Falconer DS, Mackay TFC. Introduction to quantitative genetics. 4. New York: Longman Scientific and Technical; 1996. [Google Scholar]
- 36.Smith S. The animal fatty-acid synthase - one gene, one polypeptide, 7 enzymes. FASEB J. 1994;8(15):1248–59. [PubMed] [Google Scholar]
- 37.Chakravarty B, Gu ZW, Chirala SS, Wakil SJ, Quiocho FA. Human fatty acid synthase: structure and substrate selectivity of the thioesterase domain. Proc Natl Acad Sci U S A. 2004;101(44):15567–72. doi: 10.1073/pnas.0406901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin CY, Smith S. Properties of thioesterase component obtained by limited trypsinization of fatty-acid synthetase multi-enzyme complex. J Biol Chem. 1978;253(6):1954–62. [PubMed] [Google Scholar]
- 39.Schennink A, Stoop WM, Visker MHPW, van der Poel JJ, Bovenhuis H, van Arendonk JAM. Genome-wide scan for bovine milk-fat composition. II. Quantitative trait loci for long-chain fatty acids. J Dairy Sci. 2009;92(9):4676–82. doi: 10.3168/jds.2008-1965. [DOI] [PubMed] [Google Scholar]
- 40.Stoop WM, Schennink A, Visker MHPW, Mullaart E, van Arendonk JAM, Bovenhuis H. Genome-wide scan for bovine milk-fat composition. I. Quantitative trait loci for short- and medium-chain fatty acids. J Dairy Sci. 2009;92(9):4664–75. doi: 10.3168/jds.2008-1966. [DOI] [PubMed] [Google Scholar]
- 41.Kim YC, Ntambi JM. Regulation of stearoyl-CoA desaturase genes: role in cellular metabolism and preadipocyte differentiation. Biochem Biophys Res Commun. 1999;266(1):1–4. doi: 10.1006/bbrc.1999.1704. [DOI] [PubMed] [Google Scholar]
- 42.Zhang S, Knight TJ, Reecy JM, Beitz DC. DNA polymorphisms in bovine fatty acid synthase are associated with beef fatty acid composition. Anim Genet. 2008;39(1):62–70. doi: 10.1111/j.1365-2052.2007.01681.x. [DOI] [PubMed] [Google Scholar]
- 43.Schennink A, Bovenhuis H, Leon-Kloosterziel KM, van Arendonk JAM, Visker MHPW. Effect of polymorphisms in the FASN, OLR1, PPARGC1A, PRL and STAT5A genes on bovine milk-fat composition. Anim Genet. 2009;40(6):909–16. doi: 10.1111/j.1365-2052.2009.01940.x. [DOI] [PubMed] [Google Scholar]
- 44.Abe T, Saburi J, Hasebe H, Nakagawa T, Misumi S, Nade T, Nakajima H, Shoji N, Kobayashi M, Kobayashi E. Novel Mutations of the FASN gene and their effect on fatty acid composition in Japanese black beef. Biochem Genet. 2009;47(5–6):397–411. doi: 10.1007/s10528-009-9235-5. [DOI] [PubMed] [Google Scholar]
- 45.Bionaz M, Loor JJ. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genomics. 2008;9:366. doi: 10.1186/1471-2164-9-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benton CR, Nickerson JG, Lally J, Han XX, Holloway GP, Glatz JFC, Luiken JJFP, Graham TE, Heikkila JJ, Bonen A. Modest PGC-1 alpha overexpression in muscle in vivo is sufficient to increase insulin sensitivity and palmitate oxidation in subsarcolemmal, not intermyofibrillar, mitochondria. J Biol Chem. 2008;283(7):4228–40. doi: 10.1074/jbc.M704332200. [DOI] [PubMed] [Google Scholar]
- 47.Popeijus HE, Saris WHM, Mensink RP. Role of stearoyl-CoA desaturases in obesity and the metabolic syndrome. Int J Obes (Lond) 2008;32(7):1076–82. doi: 10.1038/ijo.2008.55. [DOI] [PubMed] [Google Scholar]
- 48.Olsen HG, Lien S, Gautier M, Nilsen H, Roseth A, Berg PR, Sundsaasen KK, Svendsen M, Meuwissen THE. Mapping of a milk production quantitative trait locus to a 420-kb region on bovine chromosome 6. Genetics. 2005;169(1):275–83. doi: 10.1534/genetics.104.031559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ron M, Kliger D, Feldmesser E, Seroussi E, Ezra E, Weller JI. Multiple quantitative trait locus analysis of bovine chromosome 6 in the Israeli Holstein population by a daughter design. Genetics. 2001;159(2):727–35. doi: 10.1093/genetics/159.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farke C, Meyer HHD, Bruckmaier RM, Albrecht C. Differential expression of ABC transporters and their regulatory genes during lactation and dry period in bovine mammary tissue. J Dairy Res. 2008;75(4):406–14. doi: 10.1017/S002202990800335X. [DOI] [PubMed] [Google Scholar]
- 51.van Herwaarden AE, Wagenaar E, Merino G, Jonker JW, Rosing H, Beijnen JH, Schinkel AH. Multidrug transporter ABCG2/breast cancer resistance protein secretes riboflavin (vitamin B-2) into milk. Mol Cell Biol. 2007;27(4):1247–53. doi: 10.1128/MCB.01621-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang GY, Cline GW, Macica CM. IGF-1 stimulates de novo fatty acid biosynthesis by Schwann cells during myelination. Glia. 2007;55(6):632–41. doi: 10.1002/glia.20496. [DOI] [PubMed] [Google Scholar]
- 53.Hinesh C, Ge W, Zhao Q, Davis ME. Association of genetic markers in growth hormone and insulin-like growth factor I loci with lactation traits in Holsteins. Anim Genet. 1998;29(Suppl.1):63. [Google Scholar]
- 54.Komisarek J, Dorynek Z. Effect of ABCG2, PPARGC1A, OLR1 and SCD1 gene polymorphism on estimated breeding values for functional and production traits in Polish Holstein-Friesian bulls. J Appl Genet. 2009;50(2):125–32. doi: 10.1007/BF03195663. [DOI] [PubMed] [Google Scholar]
- 55.Olsen HG, Nilsen H, Hayes B, Berg PR, Svendsen M, Lien S, Meuwissen T. Genetic support for a quantitative trait nucleotide in the ABCG2 gene affecting milk composition of dairy cattle. BMC Genet. 2007;8:32. doi: 10.1186/1471-2156-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ron M, Cohen-Zinder M, Peter C, Weller JI, Erhardt G. Short communication: a polymorphism in ABCG2 in Bos indicus and Bos taurus cattle breeds. J Dairy Sci. 2006;89(12):4921–3. doi: 10.3168/jds.S0022-0302(06)72542-5. [DOI] [PubMed] [Google Scholar]
- 57.Siadkowska E, Zwierzchowski L, Oprzadek J, Strzalkowska N, Bagnicka E, Krzyzewski J. Effect of polymorphism in IGF-1 gene on production traits in Polish Holstein-Friesian cattle. Anim Sci Pap Rep. 2006;24(3):225–37. [Google Scholar]
- 58.Szewczuk M, Zych S, Czerniawska-Piatkowska E, Wojcik J. Association between IGF1R/i16/TaqI and IGF1/SnaBI polymorphisms and milk production traits in Polish Holstein-Friesian cows. Anim Sci Pap Rep. 2012;30(1):13–24. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are available within the manuscript and its Supporting Information files.


