Abstract
Background:
The aim of this study was to determine the regional heterogeneity and clinicopathological significance of microRNA-21 (miR-21) in advanced colorectal cancer (CRC) patients with distant metastasis.
Methods:
miR-21 expression was investigated by using locked nucleic acid– fluorescence in situ hybridization in the center and periphery of the primary cancer and in distant metastasis from 170 patients with advanced CRC. In addition, α-smooth muscle actin and desmin were evaluated to identify cancer-associated fibroblasts (CAFs) by using immunohistochemistry.
Results:
The miR-21 signal was observed in the cancer stroma. The expression of miR-21 (a score of 1–4) in the center and periphery of the primary cancer and in distant metastasis was observed in specimens from 133 (78.2%), 105 (61.8%), and 91 (53.5%) patients, respectively. miR-21 expression was heterogeneous in advanced CRC. Discordance between miR-21 expression in the center of the primary cancer and either the periphery of the primary cancer or distant metastasis was 31.7% or 44.7%, respectively. miR-21 stromal expression in the periphery of the primary cancer was significantly associated with a better prognosis (p=.004). miR-21 expression was significantly associated with CAFs in the center of the primary cancer (p=.001) and distant metastases (p=.041).
Conclusions:
miR-21 expression is observed in cancer stroma related to the CAF quantity and frequently presents regional heterogeneity in CRC. Our findings indicate that the role of miR-21 in predicting prognosis may be controversial but provide a new perspective of miR-21 level measurement in cancer specimens.
Keywords: Colorectal neoplasms, MicroRNA-21, Neoplasm metastasis, Genetic heterogeneity
MicroRNAs (miRNAs) are small (18–25 nucleotides), endogenous, non-coding RNAs that act as post-transcriptional modulators of all cellular processes, including proliferation, differentiation, and apoptosis [1]. Alterations in miRNA expression are associated with the deregulation of oncogenes and tumor suppressor genes [2]. The discovery that miRNA expression is deregulated in cancer suggests the potential of miRNA as biomarkers for cancer [3]. Therefore, targeting miRNAs is regarded as a potential therapeutic strategy and may be a promising diagnostic tool.
MiRNA-21 (miR-21) is consistently upregulated in various cancers, including the colon, stomach, lung, and breast cancer [4-7]. Previous studies proved that miR-21 expression correlates with carcinogenesis [8-11]. Some studies indicated that miR-21 expression is closely associated with poor prognosis in various cancers [4,12,13]. In colorectal cancer (CRC), miR-21 functions as an onco-miRNA due to its key roles in proliferation, invasion, and metastasis [10,14]. Although the mechanisms underlying the regulation of miR-21 in CRC remain to be defined, miR-21 can promote tumorigenesis in CRC. Some studies reported that plasma miR-21 is a potential noninvasive biomarker for early detection and prognosis of CRC [15-17]. Additionally, miR-21 expression is greatly increased in chemotherapy-resistant CRC cells [18,19]. Recent reports suggest that increased expression of miR-21 predicts poor prognosis in patients with CRC [20-22]. However, these results were restricted to patients with stage II CRC [21-24]. Thus, the prognostic value of miR-21 alterations in patients with advanced CRC presenting metastases is controversial.
We previously evaluated cancer-associated fibroblasts (CAFs) in advanced CRC with synchronous or metachronous distant metastases [25]. CAFs facilitate the communication between tumor cells and the tumor microenvironment. In addition, they regulate tumor invasion and metastasis [26,27]. Interestingly, most studies reported that miR-21 was predominantly observed in CAFs, not in cancer cells [21,22,24]. These findings point to a dynamic malignant role of CAFs through miR-21 expression in CRC. Nevertheless, the correlation between miR-21 and CAF status is yet to be demonstrated in CRC.
Recently, systemic chemotherapy and targeted therapy have been used in patients with advanced CRC, increasing patient survival [28]. Despite advances in medicine, some patients with CRC respond poorly [29]. Although the reasons for drug resistance are not fully understood, they may be related to the presence of tumor heterogeneity [30-32]. Previous reports indicated that regional heterogeneity of miRNA expression is observed in various cancer types [33-35]. Therefore, variation of miR-21 expression between the primary tumor and metastatic sites needs to be elucidated in patients with advanced CRC.
The aim of this study was to evaluate the clinical significance of miR-21 and analyze its heterogeneity in patients with advanced CRC presenting metastases. Additionally, we analyzed the correlation between CAFs and miR-21 status.
MATERIALS AND METHODS
Patients and samples
To evaluate the clinicopathological significance and heterogeneity of miR-21 status, 170 patients with advanced CRC presenting synchronous or metachronous metastases who had undergone surgical resection at Seoul National University Bundang Hospital between May 2003 and December 2009 were enrolled. The clinicopathological characteristics were obtained from the patients’ medical records and pathology reports. Follow-up information, including patient’s outcome and the interval between the date of surgical resection and death was collected. Data from patients lost to follow-up or those who had died from causes other than CRC were censored.
Ethical statement
All samples were obtained from surgically resected tumors examined pathologically at the Department of Pathology, Seoul National University Bundang Hospital. All samples and medical record data were anonymized before use in this study and the participants did not provide written informed consent. The use of medical record data and tissue samples for this study was approved by the Institutional Review Board of Seoul National University Bundang Hospital (reference No. B-1109/136-302).
Tissue array method
Surgically resected primary CRC specimens were formalin-fixed and paraffin-embedded (FFPE). For each case, representative areas of the donor blocks were obtained and rearranged into new recipient blocks (Superbiochips Laboratories, Seoul, Korea). A single core was 2 mm in diameter and those containing >20% tumor cells were considered valid cores.
miRNA in situ hybridization
Tissue microarray slides were processed by using locked nucleic acid (LNA)–fluorescence in situ hybridization (FISH) oligonucleotide probes for miR-21 and U6, both labeled with fluorescein at the 5'-end, according to the protocol described in the preparation protocol. After deparaffinization, the slides were incubated with proteinase-K and endogenous peroxidase was blocked with 3% H2O2. Next, the slides were incubated with hybridization mix containing 1 nM LNA U6 snRNA probe (Exiqon, Vedbaek, Denmark) and 20 nM doubled-DIG LNA miR-21 probe (Exiqon) in ThermoBrite (Abbott Laboratories, Abbott Park, IL, USA) for 1 hour at 50°C. Next, the slides were incubated in blocking solution and antifluorescein–horseradish peroxidase antibody (1:125, PerkinElmer, Shelton, CT, USA) for 1 hour. The signals were then amplified using Tyramide Signal Amplification (TSA-plus FITC, 1:50, PerkinElmer) for 10 minutes at room temperature. After incubation, the slides were mounted directly with SlowFade Gold antifade reagent with DAPI (Invitrogen, Carlsbad, CA, USA). All steps beginning with hybridization were performed in the dark.
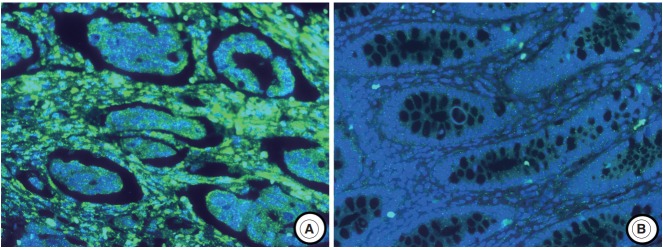
The experimental data were interpreted according to the instructions in the RNAscope FFPE Assay Kit (Advanced Cell Diagnostics, Hayward, CA, USA): no staining observed at 40× magnification (score of 0); difficult to see at 40× magnification (score of 1); difficult to see at 20× magnification, but detectable staining at 40× magnification (score of 2); difficult to see at 10× magnification, but detectable staining at 20× magnification (score of 3); and detectable staining at 10× magnification (score of 4). A score of 1–4 indicates miR-21 overexpression (Fig. 1).
Fig. 1.
MicroRNA-21 (miR-21) staining using locked nucleic acid–based in situ hybridization in colorectal cancer. (A) miR-21 expression is observed in cancer stromal tissue. (B) No miR-21 expression is observed in normal colonic mucosa.
Immunohistochemistry
Array slides were labeled by immunohistochemistry using antibodies for smooth muscle actin (SMA; 1:1,000, Neomarkers, Fremont, CA, USA) and desmin (1:300, Dako, Glostrup, Denmark) after a microwave antigen retrieval procedure, except for SMA. The staining procedures were carried out using the ultraView Universal DAB Kit (Ventana Medical Systems, Tucson, AZ, USA) and an automated stainer (BenchMark XT, Ventana Medical Systems), according to the manufacturer’s instructions.
Calculation of CAFs using digital pathology
CRC cells were considered as internal negative controls. Intestinal muscular layer or medium- to large-sized vessels were considered as internal positive controls for desmin and SMA. Samples showing inappropriate staining in internal negative or positive controls were considered non-informative and were excluded from the analysis. Slides were scanned using an Aperio ScanScopeH CS instrument (Aperio Technologies Inc., Vista, CA, USA) at 20 × magnification. Because desmin-positive muscularis mucosa and propria are positive for SMA staining, the area of CAFs (mm2) was calculated by subtracting the areas of desmin staining from that of SMA staining (SMA-desmin).
Microsatellite instability
Microsatellite instability (MSI) was assessed in 160 CRC cases with available tissue. MSI results were generated by comparing the allelic profiles of five microsatellite markers (BAT-26, BAT-25, D5S346, D17S250, and D2S123) in the tumors and corresponding normal samples. Polymerase chain reaction products from the FFPE tissues were analyzed using an automated DNA sequencer (ABI 3731 Genetic Analyzer, Applied Biosystems, Foster City, CA, USA) according to the protocol.
Statistical analyses
The association between the clinicopathological features and miR-21 status was analyzed by using the chi-square or Fisher exact test, as appropriate. The correlation between miR-21 expression and CAFs was examined by using the Mann-Whitney test. The patients’ survival was analyzed by using the Kaplan-Meier method and the log-rank test was used to determine if there were any significant differences between the survival curves. A p-value <.05 was considered statistically significant. All statistical analyses were performed using the SPSS statistics ver. 21 software (IBM Corp., Armonk, NY, USA).
RESULTS
miR-21 stromal expression and regional heterogeneity in advance CRC patients
The miR-21 signal was predominantly observed in the stromal compartment of the cancer and the normal mucosa was negative for miR-21 (Fig. 1). The snRNA U6 signal was observed in the nucleus of all cell types. To evaluate the regional heterogeneity of miR-21 expression, we examined tissues from three sites, including the center and periphery of primary cancer as well as distant metastases for each patient with advanced CRC. In the center of primary tumors, a miR-21 FISH score of 0 was observed in 37 (21.8%), a score of 1 in 16 (9.4%), a score of 2 in 56 (32.9%), a score of 3 in 46 (27.1%), and a score of 4 in 15 (8.8%) patients with CRC. In the periphery of primary tumors, a miR-21 FISH score of 0 was observed in 65 (38.2%), a score of 1 in 14 (8.2%), a score of 2 in 46 (27.1%), a score of 3 in 44 (25.9%), and a score of 4 in one (0.6%) patient with CRC. Additionally, in distant metastatic tumors, a miR-21 FISH score of 0 was observed in 79 (46.5%), a score of 1 in 13 (7.6%), a score of 2 in 43 (25.3%), a score of 3 in 32 (18.8%), and a score of 4 in three (1.8%) patients with CRC. miR-21 stromal expression (a score of 1–4) in the center and periphery of primary tumor as well as in distant metastasis was observed in 133 (78.2%), 105 (61.8%), and 91 (53.5%) patients with CRC, respectively.
The heterogeneity of miR-21 status according to tumor location is shown in Table 1. Of the 170 cases, discordance between miR-21 expression in the center and periphery was noted in 54 cases (31.7%). Discordance between the center and distant metastasis was detected in 76 cases (44.7%). Thus, regional heterogeneity of miR-21 stromal expression was very common in patients with advanced CRC.
Table 1.
Heterogeneity of miR-21 expression with respect to tumor location in advanced CRC
| miR-21 expression | Center |
Total | ||
|---|---|---|---|---|
| Negative | Positive | |||
| Periphery | Negative | 24 (14.1) | 41 (24.1) | 170 (100) |
| Positive | 13 (7.6) | 92 (54.1) | ||
| Distant metastasis | Negative | 20 (11.8) | 59 (34.7) | 170 (100) |
| Positive | 17 (10.0) | 74 (43.5) | ||
Values are presented as number (%). p-values are calculated by using chi-square test or Fisher exact test.
miR-21, microRNA-21; CRC, colorectal cancer.
Clinical implications of miR-21 stromal expression in advanced CRC patients
Table 2 shows the relationships between miR-21 status and the clinicopathological parameters of patients with advanced CRC. In the periphery of primary tumor, miR-21 expression was correlated with less aggressive features, including histologic low grade differentiation (p=.002). In addition, lymphatic invasion and lymph node metastasis was frequently observed in patients with CRC negative for miR-21 (p=.042 and p=.017, respectively). There was no statistically significant correlation between the clinicopathological factors and miR-21 expression in the center of the primary tumors and distant metastatic tumors (p>.05). The expression of miR-21 in distant metastasis was different according to metastatic site (Table 3). miR-21 stromal expression was more frequently observed in lung metastasis and peritoneal seeding (p = .046).
Table 2.
The association between clinicopathological parameters and expression of miR-21 in 170 advanced CRC patients with metastasis
| Variable | Total | Center |
p-value | Periphery |
p-value | Metastasis |
p-value | |||
|---|---|---|---|---|---|---|---|---|---|---|
| miR-21 |
miR-21 |
miR-21 |
||||||||
| Negative | Positive | Negative | Positive | Negative | Positive | |||||
| Age (yr) | ||||||||||
| Mean | 60.0 | 60.7 | 59.7 | .668 | 60.4 | 59.6 | .661 | 59.8 | 60.1 | .875 |
| Sex | ||||||||||
| Male | 90 | 21 (12.4) | 69 (40.6) | .599 | 31 (18.2) | 59 (34.7) | .281 | 36 (21.2) | 54 (31.8) | .073 |
| Female | 80 | 16 (9.4) | 64 (37.6) | 34 (20.0) | 46 (27.1) | 43 (25.3) | 37 (21.8) | |||
| pT stage | ||||||||||
| 0-2 | 103 | 23 (13.5) | 80 (47.1) | .825 | 36 (21.2) | 67 (39.4) | .275 | 45 (26.5) | 58 (34.1) | .367 |
| 3-4 | 67 | 14 (8.2) | 53 (31.2) | 29 (17.1) | 38 (22.4) | 34 (20.0) | 33 (19.4) | |||
| Differentiation | ||||||||||
| LG | 148 | 33 (19.4) | 115 (67.6) | .662 | 50 (29.4) | 98 (57.6) | .002 | 70 (41.2) | 78 (49.5) | .575 |
| HG | 22 | 4 (2.4) | 18 (10.6) | 15 (8.8) | 7 (4.1) | 9 (5.3) | 13 (7.6) | |||
| Location of primary tumor | .299 | .260 | .925 | |||||||
| Right colon | 41 | 8 (4.7) | 33 (19.4) | 20 (11.8) | 21 (12.4) | 20 (11.8) | 21 (12.4) | |||
| Left colon | 69 | 12 (7.1) | 57 (33.5) | 23 (13.5) | 46 (27.1) | 31 (18.2) | 38 (22.4) | |||
| Rectum | 60 | 17 (10.0) | 43 (25.3) | 22 (12.9) | 38 (22.4) | 28 (16.5) | 32 (18.8) | |||
| LN metastasis | ||||||||||
| Absent | 31 | 7 (4.1) | 24 (14.1) | .903 | 6 (3.5) | 25 (14.7) | .017 | 15 (8.8) | 16 (9.4) | .813 |
| Present | 139 | 30 (17.6) | 109 (64.1) | 59 (34.7) | 80 (47.1) | 64 (37.6) | 75 (44.1) | |||
| Lymphatic invasion | ||||||||||
| Absent | 55 | 9 (5.3) | 46 (27.1) | .238 | 15 (8.8) | 40 (23.5) | .042 | 28 (16.5) | 27 (15.9) | .422 |
| Present | 115 | 28 (16.5) | 87 (51.2) | 50 (29.4) | 65 (38.2) | 51 (30.0) | 64 (37.6) | |||
| Perineural invasion | ||||||||||
| Absent | 83 | 19 (11.2) | 64 (37.6) | .728 | 29 (17.1) | 54 (31.8) | .388 | 41 (24.1) | 42 (24.7) | .455 |
| Present | 87 | 18 (10.6) | 69 (40.6) | 36 (21.2) | 51 (30.0) | 38 (22.4) | 49 (28.8) | |||
| Venous invasion | ||||||||||
| Absent | 118 | 24 (14.1) | 94 (55.3) | .497 | 42 (24.7) | 76 (44.7) | .286 | 58 (34.1) | 60 (35.3) | .291 |
| Present | 52 | 13 (7.6) | 39 (22.9) | 23 (13.5) | 29 (17.1) | 21 (12.4) | 31 (18.2) | |||
| Tumor border | ||||||||||
| Expanding | 12 | 4 (2.4) | 8 (4.7) | .314 | 4 (2.4) | 8 (4.7) | .717 | 6 (3.5) | 6 (3.5) | .799 |
| Infiltrative | 158 | 33 (19.4) | 125 (73.5) | 61 (35.9) | 97 (57.1) | 73 (42.9) | 85 (50.0) | |||
| Distant metastasis | ||||||||||
| Synchronous | 110 | 23 (13.5) | 87 (51.2) | .714 | 45 (26.5) | 65 (38.2) | .331 | 52 (30.6) | 58 (34.1) | .776 |
| Metachronous | 60 | 14 (8.2) | 46 (21.7) | 20 (11.8) | 40 (23.5) | 27 (15.9) | 33 (19.4) | |||
| pTNM stage at initial diagnosis | ||||||||||
| I, II | 20 | 5 (2.9) | 15 (8.8) | .709 | 4 (2.4) | 16 (9.4) | .074 | 9 (5.3) | 11 (6.5) | .888 |
| III, IV | 150 | 32 (18.8) | 118 (69.4) | 61 (35.9) | 89 (52.4) | 70 (41.2) | 80 (47.1) | |||
| MSI status | ||||||||||
| MSS/MSI-L | 157 | 35 (21.9) | 122 (76.3) | .650 | 59 (36.9) | 98 (61.3) | .304 | 74 (46.3) | 83 (51.9) | .069 |
| MSI-H | 3 | 1 (0.6) | 2 (1.3) | 2 (1.3) | 1 (0.6) | 3 (1.9) | 0 | |||
Values are presented as number (%). p-values are calculated by using chi-square test or Fisher exact test.
miR-21, microRNA-21; CRC, colorectal cancer; T, tumor; LG, low grade; HG, high grade; LN, lymph node; MSI, microsatellite instability; MSS, microsatellite stable; MSI-L, microsatellite instability–low; MSI-H, microsatellite instability–high.
Table 3.
Expression of miR-21 in distant metastasis according to metastatic site in 170 advanced CRC patients
| Site of metastasis | Total | miR-21 in metastasis |
p-value | |
|---|---|---|---|---|
| Negative | Positive | |||
| Liver | 76 | 42 (24.7) | 34 (20.0) | .046 |
| Lung | 37 | 13 (7.6) | 24 (14.1) | |
| Seeding | 37 | 13 (7.6) | 24 (14.1) | |
| Distant nodes | 2 | 0 | 2 (1.2) | |
| Ovary | 18 | 11 (6.5) | 7 (4.1) | |
| Total | 170 | 79 | 91 | |
Values are presented as number (%).
miR-21, microRNA-21; CRC, colorectal cancer.
Prognostic significance of miR-21 stromal expression in advanced CRC patients
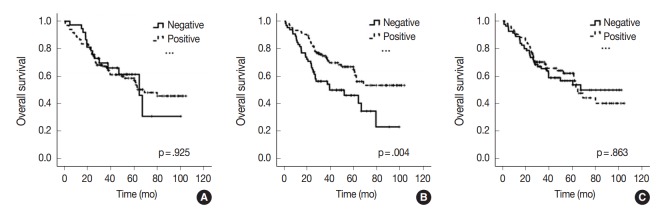
All 170 patients with advanced CRC were successfully followed up for inclusion in the survival analysis (Fig. 2). The mean follow-up time was 42 months (range, 1 to 105 months) and 73 patients (42.9%) died from cancer during the follow-up period. Kaplan-Meier analysis showed that miR-21 stromal expression in the periphery of primary tumors was significantly associated with a better prognosis (p=.004). There was no significant correlation between the patients’ prognosis and miR-21 expression in the center of primary tumors and distant metastases (p=.925 and p=.863, respectively).
Fig. 2.
Kaplan-Meier curves illustrating the overall survival of patients with advanced colorectal cancer in relation to microRNA-21 expression. (A) Center. (B) Periphery. (C) Distant metastasis.
Correlation between miR-21 stromal expression and CAF value
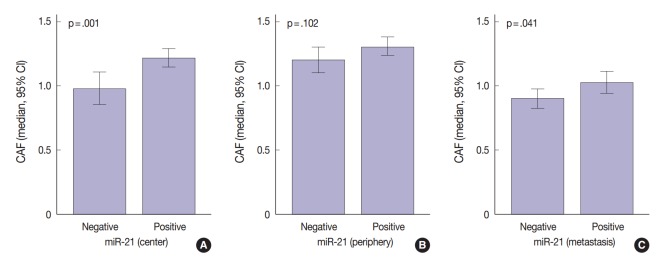
The regional heterogeneous values for CAF in CRC are shown in our previous study [25]. The area occupied by CAFs was the lowest in distant metastases (median, 0.91; interquartile range [IQR], 0.68 to 1.18) than any other sites (median, 1.12; IQR, 0.88 to 1.41 in the center of primary tumors and median, 1.22; IQR, 0.96 to 1.54 in the periphery of primary tumors). Mann-Whitney test showed that miR-21 expression was significantly associated with CAFs in the center of primary tumors (p=.001) and distant metastases (p=.041). In the periphery of primary tumors, miR-21 overexpression was not correlated with CAFs (p=.102) (Fig. 3).
Fig. 3.
Correlation between microRNA-21 (miR-21) expression and cancer associated fibroblasts (CAFs) by Mann-Whitney analysis. (A) Center. (B) Periphery. (C) Distant metastasis. CI, confidence interval.
DISCUSSION
Many predictive and prognostic molecular markers have been suggested for CRC. However, they are not reliably accepted due to a lack of reproducibility and validation. Previous studies proposed that high expression of miR-21 might predict poor survival in patients with CRC [20-24]. However, elevated miR-21 level and poor prognosis correlated only in a subgroup of patients with stage II CRC [21-24]. Interestingly, our results contradict those of previous miR-21 expression studies. In our study, miR-21 expression in the periphery of primary tumors was significantly associated with a better prognosis in patients with advanced stage CRC (p=.004) (Fig. 2). Otherwise, there was no significant correlation between the prognosis and miR-21 expression in the center of primary tumors and in distant metastases (p=.925 and p=.863, respectively) (Fig. 2). This discrepancy may be explained by the fact that our cohort was largely comprised of patients with stage IV CRC (98 cases, 57.6%). They received various personalized treatments and these might reflect the statistical difference.
Bullock et al. [36] recently demonstrated that stromal miR-21 expression induced a pro-metastatic mechanism of CRC via activation of matrix metalloproteinase-2. These data highlight the importance of miR-21 deregulation in CRC metastasis. Because all patients in our study presented with advanced CRC with metastasis, miR-21 may be expressed at higher levels than that observed in previous reports. In our study, miR-21 expression in the center and periphery of primary tumors and in distant metastasis was observed in 78.2%, 61.8%, and 53.5% of the patients, respectively, whereas it was observed in 27.4% of the patients in another study [22]. Our data suggest that the lack of miR-21 expression in patients presenting with CRC with metastasis is associated with a rather poor prognosis and more frequent lymphatic invasion and lymph node metastasis (Table 2, Fig. 1), presumably because, in these patients, the metastatic mechanism is controlled by other regulatory pathways. Further studies are necessary to prove that other mechanisms induce CRC metastasis, independently of miR-21. The evaluation of the prognostic value of miR-21 expression in patients with advanced CRC presenting distant metastasis might be of little importance.
Previous studies in various cancers indicated that miR-21 localizes mainly in the cancer stroma and more particularly in the stromal fibroblast-like cells [21,22,24]. This localization may be due to molecules secreted by cancer cells, which influence the microenvironment [37-39]. Meanwhile, the mechanisms of regulation of miR-21 in CAFs remain unknown. Only few studies examined the correlation between the value of CAFs and miR-21 expression [40]. Our study revealed that miR-21 expression was associated with CAF value in the center of primary tumors and distant metastases (p = .001 and p = .041, respectively) (Fig. 3). As expected, the value of CAF was greater in stroma of CRC specimens presenting miR-21 expression. Therefore, we should be careful when evaluating stromal miR-21 expression. In most previous studies, miR-21 expression was evaluated quantitatively by image analysis [21,24,36]. This method may mislead us when distinguishing miR-21 high expression from a simple increase in the number of CAFs. Thus, accurate observation should be performed to distinguish miR-21 expression from CAF increase.
To clarify miR-21 expression in CRC, being aware of the heterogeneity in miR-21 expression level is crucial because regional heterogeneity can lead to sampling bias. In recent studies, intra-tumor heterogeneity of various miRNAs was detected in colorectal, pancreatic, and breast cancer [34,35,41]. Our study indicates miR-21 regional heterogeneity, which constitutes approximately 40% of the total cohort (Table 1). Thus, a reliable assessment of CRC miR-21 expression may include sampling of the primary tumor in several locations and metastasis.
In conclusion, we demonstrated that miR-21 is expressed in advanced CRC and that this upregulation is mainly confined to the cancer stroma. Our FISH data indicated that miR-21 expression is related to CAF value in the center of primary tumors and distant metastasis. We determined that miR-21 expression frequently presents regional heterogeneity in CRC. Such data could lead to the new perspective of miR-21 level measurement. The evaluation of miR-21 expression for the prediction of survival in patient with advanced CRC is a matter of debate.
Acknowledgments
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI14C1813, https://www.khidi.or.kr/eps).
Footnotes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–8. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 2.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 3.Kwak PB, Iwasaki S, Tomari Y. The microRNA pathway and cancer. Cancer Sci. 2010;101:2309–15. doi: 10.1111/j.1349-7006.2010.01683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan SH, Wu CW, Li AF, Chi CW, Lin WC. miR-21 microRNA expression in human gastric carcinomas and its clinical association. Anticancer Res. 2008;28:907–11. [PubMed] [Google Scholar]
- 5.Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–80. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qian B, Katsaros D, Lu L, et al. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res Treat. 2009;117:131–40. doi: 10.1007/s10549-008-0219-7. [DOI] [PubMed] [Google Scholar]
- 7.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696–704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 8.Zhang A, Liu Y, Shen Y, Xu Y, Li X. miR-21 modulates cell apoptosis by targeting multiple genes in renal cell carcinoma. Urology. 2011;78:474. doi: 10.1016/j.urology.2011.03.030. [DOI] [PubMed] [Google Scholar]
- 9.Hiyoshi Y, Kamohara H, Karashima R, et al. MicroRNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clin Cancer Res. 2009;15:1915–22. doi: 10.1158/1078-0432.CCR-08-2545. [DOI] [PubMed] [Google Scholar]
- 10.Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 11.Lu Z, Liu M, Stribinskis V, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–9. doi: 10.1038/onc.2008.72. [DOI] [PubMed] [Google Scholar]
- 12.Mathé EA, Nguyen GH, Bowman ED, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15:6192–200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan LX, Huang XF, Shao Q, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–60. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Y, Nangia-Makker P, Farhana L, Rajendra SG, Levi E, Majumdar AP. miR-21 and miR-145 cooperation in regulation of colon cancer stem cells. Mol Cancer. 2015;14:98. doi: 10.1186/s12943-015-0372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanaan Z, Rai SN, Eichenberger MR, et al. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256:544–51. doi: 10.1097/SLA.0b013e318265bd6f. [DOI] [PubMed] [Google Scholar]
- 16.Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–81. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 17.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–26. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 18.Yu Y, Sarkar FH, Majumdar AP. Down-regulation of miR-21 induces differentiation of chemoresistant colon cancer cells and enhances susceptibility to therapeutic regimens. Transl Oncol. 2013;6:180–6. doi: 10.1593/tlo.12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valeri N, Gasparini P, Braconi C, et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc Natl Acad Sci U S A. 2010;107:21098–103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia X, Yang B, Zhai X, et al. Prognostic role of microRNA-21 in colorectal cancer: a meta-analysis. PLoS One. 2013;8: doi: 10.1371/journal.pone.0080426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kjaer-Frifeldt S, Hansen TF, Nielsen BS, et al. The prognostic importance of miR-21 in stage II colon cancer: a population-based study. Br J Cancer. 2012;107:1169–74. doi: 10.1038/bjc.2012.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang WK, Lee JK, Oh ST, Lee SH, Jung CK. Stromal expression of miR-21 in T3-4a colorectal cancer is an independent predictor of early tumor relapse. BMC Gastroenterol. 2015;15:2. doi: 10.1186/s12876-015-0227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang JX, Song W, Chen ZH, et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295–306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen BS, Jørgensen S, Fog JU, et al. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis. 2011;28:27–38. doi: 10.1007/s10585-010-9355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwak Y, Lee HE, Kim WH, Kim DW, Kang SB, Lee HS. The clinical implication of cancer-associated microvasculature and fibroblast in advanced colorectal cancer patients with synchronous or metachronous metastases. PLoS One. 2014;9: doi: 10.1371/journal.pone.0091811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsujino T, Seshimo I, Yamamoto H, et al. Stromal myofibroblasts predict disease recurrence for colorectal cancer. Clin Cancer Res. 2007;13:2082–90. doi: 10.1158/1078-0432.CCR-06-2191. [DOI] [PubMed] [Google Scholar]
- 27.Trimboli AJ, Cantemir-Stone CZ, Li F, et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature. 2009;461:1084–91. doi: 10.1038/nature08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knijn N, Tol J, Punt CJ. Current issues in the targeted therapy of advanced colorectal cancer. Discov Med. 2010;9:328–36. [PubMed] [Google Scholar]
- 29.O’Connell MJ, Campbell ME, Goldberg RM, et al. Survival following recurrence in stage II and III colon cancer: findings from the ACCENT data set. J Clin Oncol. 2008;26:2336–41. doi: 10.1200/JCO.2007.15.8261. [DOI] [PubMed] [Google Scholar]
- 30.Albanese I, Scibetta AG, Migliavacca M, et al. Heterogeneity within and between primary colorectal carcinomas and matched metastases as revealed by analysis of Ki-ras and p53 mutations. Biochem Biophys Res Commun. 2004;325:784–91. doi: 10.1016/j.bbrc.2004.10.111. [DOI] [PubMed] [Google Scholar]
- 31.Park JH, Han SW, Oh DY, et al. Analysis of KRAS, BRAF, PTEN, IGF1R, EGFR intron 1 CA status in both primary tumors and paired metastases in determining benefit from cetuximab therapy in colon cancer. Cancer Chemother Pharmacol. 2011;68:1045–55. doi: 10.1007/s00280-011-1586-z. [DOI] [PubMed] [Google Scholar]
- 32.Baldus SE, Schaefer KL, Engers R, Hartleb D, Stoecklein NH, Gabbert HE. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res. 2010;16:790–9. doi: 10.1158/1078-0432.CCR-09-2446. [DOI] [PubMed] [Google Scholar]
- 33.Villegas-Ruiz V, Juárez-Méndez S, Pérez-González OA, et al. Heterogeneity of microRNAs expression in cervical cancer cells: over-expression of miR-196a. Int J Clin Exp Pathol. 2014;7:1389–401. [PMC free article] [PubMed] [Google Scholar]
- 34.Jepsen RK, Novotny GW, Klarskov LL, Christensen IJ, Riis LB, Høgdall E. Intra-tumor heterogeneity of microRNA-92a, microRNA-375 and microRNA-424 in colorectal cancer. Exp Mol Pathol. 2016;100:125–31. doi: 10.1016/j.yexmp.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 35.Raychaudhuri M, Schuster T, Buchner T, et al. Intratumoral heterogeneity of microRNA expression in breast cancer. J Mol Diagn. 2012;14:376–84. doi: 10.1016/j.jmoldx.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 36.Bullock MD, Pickard KM, Nielsen BS, et al. Pleiotropic actions of miR-21 highlight the critical role of deregulated stromal microRNAs during colorectal cancer progression. Cell Death Dis. 2013;4: doi: 10.1038/cddis.2013.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamichi N, Shimomura R, Inada K, et al. Locked nucleic acid in situ hybridization analysis of miR-21 expression during colorectal cancer development. Clin Cancer Res. 2009;15:4009–16. doi: 10.1158/1078-0432.CCR-08-3257. [DOI] [PubMed] [Google Scholar]
- 38.Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–78. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 39.Fassan M, Pizzi M, Giacomelli L, et al. PDCD4 nuclear loss inversely correlates with miR-21 levels in colon carcinogenesis. Virchows Arch. 2011;458:413–9. doi: 10.1007/s00428-011-1046-5. [DOI] [PubMed] [Google Scholar]
- 40.Aprelikova O, Green JE. MicroRNA regulation in cancer-associated fibroblasts. Cancer Immunol Immunother. 2012;61:231–7. doi: 10.1007/s00262-011-1139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lou E, Subramanian S, Steer CJ. Pancreatic cancer: modulation of KRAS, microRNAs, and intercellular communication in the setting of tumor heterogeneity. Pancreas. 2013;42:1218–26. doi: 10.1097/MPA.0000000000000007. [DOI] [PubMed] [Google Scholar]