Abstract
Endothelial cells (EC) represent an important target for pharmacologic intervention, given their central role in a wide variety of human pathophysiologic processes. Studies in lab animal species have established that conjugation of drugs and carriers with antibodies directed to surface targets like the Platelet Endothelial Cell Adhesion Molecule-1 (PECAM-1, a highly expressed endothelial transmembrane protein) help to achieve specific therapeutic interventions in ECs. To translate such “vascular immunotargeting” to clinical practice, it is necessary to replace antibodies by advanced ligands that are more amenable to use in humans. We report the molecular design of a single chain variable antibody fragment (scFv) that binds with high affinity to human PECAM-1 and cross-reacts with its counterpart in rats and other animal species, allowing parallel testing in vivo and in human endothelial cells in microfluidic model. Site-specific modification of the scFv allows conjugation of protein cargo and liposomes, enabling their endothelial targeting in these models. This study provides a template for molecular engineering of ligands, enabling studies of drug targeting in animal species and subsequent use in humans.
Graphical abstract
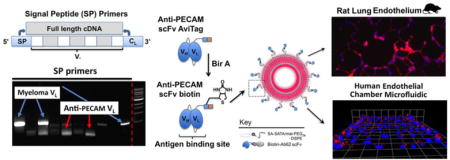
Introduction
The vascular endothelium has a variety of important physiologic roles, helping to control vascular tone and permeability, limiting activation of the coagulation system, and regulating the adhesion and infiltration of immune cells into tissue parenchyma[1–3]. Failure of these anti-thrombotic, anti-inflammatory, and barrier-stabilizing mechanisms is a key part of the pathophysiology of a number of human diseases, making endothelial cells (ECs) an important potential target for pharmacologic intervention[4–6]. While only a minute fraction of most drugs are taken up by ECs, several groups have developed strategies of “vascular targeting”, in which affinity ligands are attached to drugs or drug carriers to enable effective endothelial delivery[7–12].
Amongst the various classes of affinity ligands utilized to achieve vascular targeting, monoclonal antibodies (mAbs) are by far the most popular and best studied. Antibodies are robust molecules with high binding affinity, which is typically preserved even after conjugation to nanoparticles or therapeutic cargo[13,14]. They have proven themselves in the marketplace, accounting for billions of dollars in worldwide pharmaceutical sales[15]. Moreover, there is a large, pre-existing repertoire of hybridomas, including many that produce antibodies with well-characterized and favorable functional activities (e.g., receptor blockade, transcytosis, etc).
Despite their many advantages, antibodies have several features that complicate their use for endothelial drug delivery. In addition to the antigen targeting domain (Fab), mAbs have an effector domain (the fragment crystallizable, or FC), which can bind complement and/or various immune cells, inducing immune reactions or rapid clearance from the circulation[16–18]. Furthermore, mAbs are typically produced in mammalian cell lines, making protein expression relatively costly and complicating genetic manipulation for affinity maturation or the introduction of site-specific modifications. The latter often complicates attachment to cargo or drug carriers, necessitating the use of non-specific chemical conjugation, which can compromise antigen binding or result in crosslinking and the formation of large complexes[19].
Single-chain variable fragments (scFv) are antibody derivatives that represent an attractive alternative to intact mAb[20]. Typically, scFv retain most of the positive characteristics of their parental immunoglobulin while eliminating Fc-mediated immune responses and clearance mechanisms. Moreover, the recombinant construction of scFv and their expression in microbial systems allows efficient scale up of production, straightforward introduction of sequence modifications, direct fusion to therapeutic cargo, and the potential for affinity maturation via display techniques[8,21–24].
From the standpoint of translational medicine, however, scFv and mAb share an important limitation: species cross-reactivity is uncommon. In particular, the need to study therapeutics in rodent models necessitates the creation of high affinity mouse or rat-specific antibodies, which rarely recognize their human cognates. In the case of endothelial drug delivery, even subtle differences in epitope may have significant effects on the pharmacokinetics, cellular uptake, and functional properties of the targeting antibody[25–27]. As such, substitution of a human-specific antibody can dramatically alter the characteristics of the drug delivery system in ways that may be difficult prior to clinical testing.
We report here the development of an scFv derived from a rare antibody with both high affinity and wide species cross-reactivity[28]. The novel scFv, like its parental mAb, is specific for the Platelet Endothelial Cell Adhesion Molecule-1 (CD31/PECAM-1), a transmembrane glycoprotein concentrated at endothelial cell-cell borders[29,30]. The high level of PECAM-1 expression on endothelial cells (ECs) and its distribution across nearly all vascular beds make it a prime target for endothelial drug delivery, especially in the setting of acute inflammation, where its blockade has been shown to reduce neutrophil transmigration[31]. To demonstrate the potential utility of the Ab62 scFv in the development of translational nanomedicines, we introduce a site-specific modification, allowing oriented attachment to protein cargo and liposomes, and show delivery to the pulmonary endothelium in rats and binding to human endothelial cells in a microfluidic device perfused with whole blood.
Materials & Methods
Ethics Statement
Animal studies were conducted following the Guide for the Care and Use of Laboratory Animals as adopted by the NIH, under protocols 804858 AND 805013 approved by the IACUC of the University of Pennsylvania.
Cell lines
The Ab62 hybridoma was a generous gift of Marian Nakada[28]. Human umbilical vein endothelial cells (HUVECs) were purchased from Lonza (Walkersville, MD) and maintained using EGM™-2 BulletKit™ (Lonza, Walkersville, MD). REN-hPECAM cells, which stably express human PECAM-1[32] were maintained in RPMI-1640 (Life Technologies, Grand Island, NY) supplemented with 10% (v/v) fetal bovine serum (FBS), 1X antibiotic-antimycotic (Life Technologies, Grand Island, NY), and 200 μg/mL of Geneticin (Life Technologies, Grand Island, NY).
Antibodies and other reagents
Ab62 mAb was purified from hybridoma supernatant using Protein G Sepharose 4 Fast flow (GE Healthcare Life Sciences, Pittsburgh, PA) per manufacturer protocol. FITC-conjugated anti-FLAG (M2) and M2-agarose were purchased from Sigma Aldrich (St. Louis, MO). Streptavidin (SA) was purchased from Calbiochem (Billerica, MA). Biotin ligase, Bir A, was obtained from Avidity (Aurora, CO). Lipids including DPPC (1,2-dipalmitoyl-sn-glycero-3-phosphocholine), PC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, PG (1,2-di-O-tetradecyl-sn-glycero-3-phospho-(1′-rac-glycerol)), Liss Rhod PE (L-α-Phosphatidylethanolamine-N-(lissamine rhodamine B sulfonyl), and malemide PEG2000-DSPE (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] were purchased from Avanti Polar Lipids (Alabaster, AL). SATA (N-succinimidyl-S-acetylthioacetate) was from Pierce Biotechnology (Rockford, IL).
Cloning of scFv VH and VL cDNAs
Total cellular RNA was isolated from hybridoma cells using RNeasy kit (Qiagen, Valencia, CA). Reverse transcription was performed using High Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Foster City, CA). VH and VL cDNAs were generated using the Expand High Fidelity PCR system (Roche, Indianapolis, IN). Two set of 5′ primers were used for cloning of the light chain variable sequence: 1. FR1 primers previously reported in the literature[33], and 2. a novel set of degenerate signal peptide (SP) region primers.
Sequence Analysis
The abYsis database was queried using available online search parameters, resulting in a text file with 2676 light chain sequences[34]. A Python script was used to analyze this data, eliminating incomplete and duplicate sequences and comparing the amino acid sequences at the start of the SP and FR1 regions (Supplemental Document 1).
Assembly, Production, Purification, and Site-specific Biotinylation of Ab62 scFv
Ab62 VH and VL cDNAs were assembled into an scFv construct (Figure 2), with the two domains separated by a flexible (GGGGS)3 linker. A triple FLAG tag was included on the 3′ end for purification and detection purposes. Ab62 scFv was expressed in S2 insect cells and purified using anti-FLAG resin. To allow site specific biotinylation of the Ab62 scFv, a second construct was made, with an AviTag biotinylation cassette[35] inserted between an (SSSG)2AAA rigid linker and the triple FLAG tag (Figure 3). Biotinylation was performed in vitro using recombinant biotin ligase, BirA, and the company provided protocol. Biotinylated scFv was separated from free biotin by exhaustive dialysis.
Figure 2.
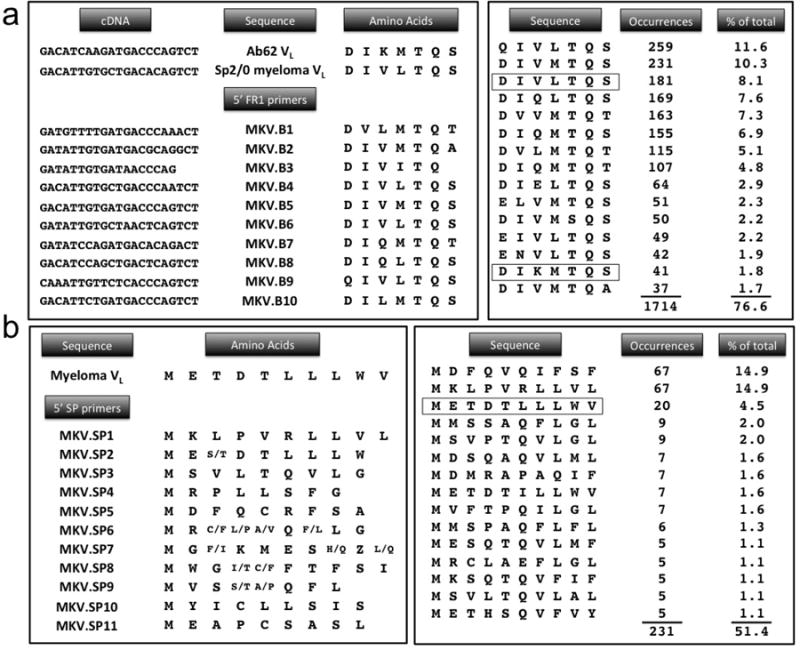
(a) Comparison of Ab62 and myeloma VL sequences in the region covered by the FR1 primers. Significant homology is seen, particularly at the 3′ end of the nucleic acid sequence, where the strength of primer annealing is largely determined. Right hand panel shows analysis of 2237 light chain sequences from the abYsis database[34]. The 15 most common 7-amino acid FR1 sequences are shown. Significant homology between these sequences explains why repeated amplification of the Sp2/0 myeloma transcript is frequently a problem. (b) Similar analysis of the SP region shows substantially greater variation in the amino acid sequences encoded by the 11 SP primers. While the SP sequence of the Ab62 VL is not known, its selective amplification by MKV.SP6 and SP9 suggests that it has greater homology to those primers. Degeneracy in the SP primers results in ambiguous amino acids at some positions, as shown. Analysis of 449 sequences from abYsis database also shows less homology in SP region, suggesting that SP primers will frequently be capable of selective amplification of antigen-specific cDNA.
Figure 3. Assembly of Ab62 scFv and validation of human PECAM-1 binding in vitro.
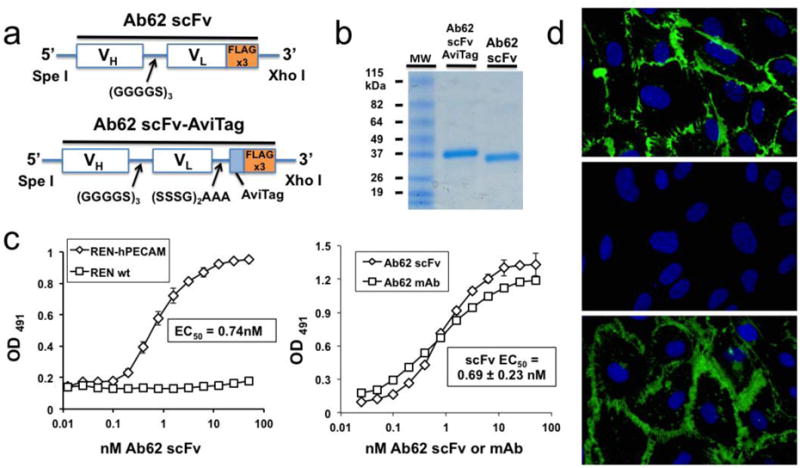
(a) Molecular assembly of VH and VL sequences into scFv construct, with and without AviTag biotinylation cassette. (b) SDS-PAGE gel electrophoresis of purified Ab62 scFv and scFv-AviTag. (c) Cell based ELISA demonstrates specific binding of Ab62 scFv to human PECAM expressing REN cells (REN-hPECAM), with no binding observed to wild type REN cells. (d) Ab62 scFv binds to primary human endothelial cells (HUVECs) with affinity similar to that of the parental antibody. Each ELISA experiment was done in triplicate (i.e., each point shown represents three wells), with SD shown. The EC50 of the Ab62 scFv ELISA was calculated based on three independent experiments – a representative binding curve is shown. (e) Immunofluorescence images demonstrate expected localization of Ab62 scFv to endothelial cell-cell borders, characteristic of PECAM-1 and analogous to the pattern of staining seen with parental Ab62 mAb. No staining is seen with anti-mouse PECAM-1 Mec13 scFv.
Cell binding experiments
Cell based Enzyme-linked Immunosorbent Assays (ELISAs) were performed as previously described[12]. Briefly, cells were grown to confluence in 96-well plates (BD Biosciences, Franklin Lakes, NJ). Wells were pre-coated with 1% gelatin in experiments involving endothelial cells. Cell monolayers were incubated for 2 hours with increasing concentration of scFv in assay buffer (cell culture media with 5% FBS) at 4°C, to prevent internalization. Cells were washed twice with assay buffer and then incubated with anti-FLAG (M2)-HRP for 1 hour at 4°C. After 3 additional washes, a phosphate buffered saline (PBS) solution containing the enzyme substrate, o-phenylenediamine (OPD; Sigma-Aldrich, St. Louis, MO), and hydrogen peroxide, was added. The reaction was quenched by the addition of 100 μL of 50% H2SO4. Absorbance readings at 490 nm (OD490) were performed on a Multiskan FC Microplate reader (Thermofisher, Waltham, MA) at room temperature (RT). The ELISA binding data were analyzed and binding parameters were determined using Prism 6.0 (GraphPad, San Diego, CA) software.
Endothelial cell staining and immunofluorescence
For static cell staining experiments, HUVEC cell monolayers were grown in 8 well μ-slides (Ibidi, Verona, WI) coated with 1% gelatin. Cells were treated for 1 hour with EGM-2 media supplemented with 3% BSA, to minimize nonspecific binding, and then stained with Ab62 scFv or mAb or Mec13 scFv (1 μg/mL) for 30 minutes at 37°C. Cells were washed with EGM-2 media and then fixed for 10 minutes with a freshly prepared 1:1 mixture of methanol and acetone. Fixed cells were washed 4 times with TBS and incubated with anti-FLAG (M2)-FITC (Sigma-Aldrich, St. Louis, MO) at 10 μg/mL or Alexa Fluor 488-labeled anti-mouse (Life Technologies, Grand Island, NY) at 1 μg/mL for 1 hour at RT. For fluorescent nanocarrier experiments, cells were treated with scFv-decorated liposomes for 30 minutes at 37°C and then washed three times with EGM-2 medium, followed by fixation with 2% paraformaldehyde for 10 minutes. In all experiments, fixed cells were washed three times with PBS prior to addition of ProLong Gold Antifade reagent with DAPI (Life Technologies, Grand Island, NY).
Radioiodination of proteins
Streptavidin (SA) was directly radioiodinated with [125I]NaI (Perkin Elmer, Waltham, MA) and purified using Zeba desalting spin columns (ThermoScientific). Radiolabeling efficiency was > 90% and free iodine was < 5% following purification.
Production and characterization of scFv-biotin-SA protein complexes
Site specifically biotinylated Ab62 and Mec13 scFv were combined with 125I-SA at a 10:1 molar excess, to drive the formation of multivalent complexes. Endothelial binding of scFv-SA complexes was tested using confluent monolayers of HUVEC blocked with EGM-2 media supplemented with 3% BSA to minimize non-specific binding. Cells were incubated with scFv-SA complexes for 30 minutes at 37°C. Following incubation, the cells were washed three times and lysed using 1N NaOH with 0.5% Triton X-100. Radioactivity was measured in cell lysate (“bound”) vs. supernatant and washes (“unbound”) using a WIZARD Automatic Gamma Counter (Perkin Elmer, Waltham, MA). In all cases, > 95% of the input radioactivity was recovered.
SA-Fluoroliposomes
Fluoroliposomes were prepared as previously described[11], modified to incorporate 1.25 mol% maleimide PEG2000-DSPE and 1 mol% fluorescent phospholipid. Liposomes were extruded through 200 μm polycarbonate filters (Avanti Polar Lipids, Alabaster, AL) and measured for hydrodynamic diameter and particle distribution by dynamic light scattering (Brookhaven Instruments, Holtsville, NY).
Streptavidin was attached to maleimide-functionalized fluoroliposomes by introduction of sulfhydryls using SATA. A 6-fold excess of SATA was added for 30 minutes at RT to achieve ~ 1 reactive group per SA molecule. SA-SATA was deprotected using hydroxylamine (50 mM concentration) and added to fluoroliposomes at a concentration calculated to conjugate approximately 250 SA molecules/liposome, assuming an efficiency of 75–80%[11]. Unreacted components were removed at each step using Zeba desalting columns (Thermo Scientific, Rockford, IL).
In vivo targeting of scFv-SA complexes and scFv-liposomes
Sprague Dawley rats (Charles River, Wilmington, MA) weighing 250g to 400g were anesthetized with 2–2.5% isoflurane. For biodistribution experiments, 125I-labeled Ab62 or Mec13 scFv-biotin-125I-SA complexes were injected via jugular vein. One hour post injection, blood was withdrawn from the inferior vena cava and animals were euthanized. Organs, including lung, liver, kidney, spleen, heart, brain, and thyroid were harvested, weighed, and gamma counted. In experiments involving scFv-decorated fluoroliposomes, rats were euthanized 30 minutes after intravenous injection of nanoparticles. The pulmonary vasculature was flushed with PBS to remove non-specifically bound particles and the lungs were inflated with a 1:1 mixture of PBS and Tissue Tek O.C.T. compound (Sakura, Alphen aan den Rijn, The Netherlands) prior to freezing and sectioning.
Microfluidic Experiments
“Endothelialized” microfluidic chambers were prepared as previously described[36]. Briefly, HUVECs were seeded in a polydimethylsiloxane flow chamber coated with fibronectin and grown until confluent. In some experiments, scFv-decorated SA-fluoroliposomes were mixed with cell culture media. In other experiments, liposomes were mixed with whole blood freshly collected from a volunteer in vacutainer tubes containing citrate and corn trypsin inhibitor (Haematologic Technologies, Inc., Essex Junction, VT). Both cell culture and whole blood mixtures were perfused into the microfluidic chamber at fluid shear stress of 5 dyne/cm2. Following cessation of flow, cells were fixed with 1% paraformaldehyde and washed prior to confocal imaging.
Results
Amplification of Ab62 variable region cDNAs
Construction of scFv requires the identification of variable heavy chain (VH) and light chain (VL) sequences from the parental hybridoma. The most commonly described method involves PCR amplification of VH and VL cDNAs using a single 3′ constant region primer and a set of 5′ primers which anneal to the most commonly occurring sequences in the first framework (FR1) region[33,37]. Several groups have reported difficulty using this method to clone VL, due to the repeated amplification of myeloma-derived light chain sequences[38,39]. The FR1 method also results in the incorporation of primer sequence at the 5′ end of each variable domain, introducing amino acid substitutions which can compromise stability and/or antigen binding[40,41]. In an effort to circumvent both of these issues, we utilized a novel set of degenerate primers homologous to the signal peptide region of VH and VL, and compared their performance to the traditional FR1 primers. Figure 1a illustrates both strategies in schematic form, whereas Figure 1b shows DNA gels with the PCR reactions done using each set of primers. For the FR1 approach, seven out of ten reactions generated an appropriately sized amplicon, but sequencing revealed only the previously reported, aberrantly rearranged light chain derived myeloma-derived and antigen-specific cDNA, each band on the gel was purified and subcloned, with subsequent sequencing of 10–20 clones. Despite this significant effort, no antigen-specific sequence was identified. Instead, the vast majority of clones contained the Sp2/0 myeloma VL, whereas a small minority contained truncated VL sequences or sequences with multiple in-frame stop codons. In contrast, the eleven SP primer reactions produced six appropriately sized amplicons, and sequencing of two of them yielded a novel, full-length cDNA that was ultimately found to be the Ab62 VL. Indeed, the SP primers appeared to selectively amplify either the Sp2/0 myeloma VL (MKV.SP1, SP2, SP4, and SP11) or the Ab62 VL (MKV.SP6 and SP9).
Figure 1.
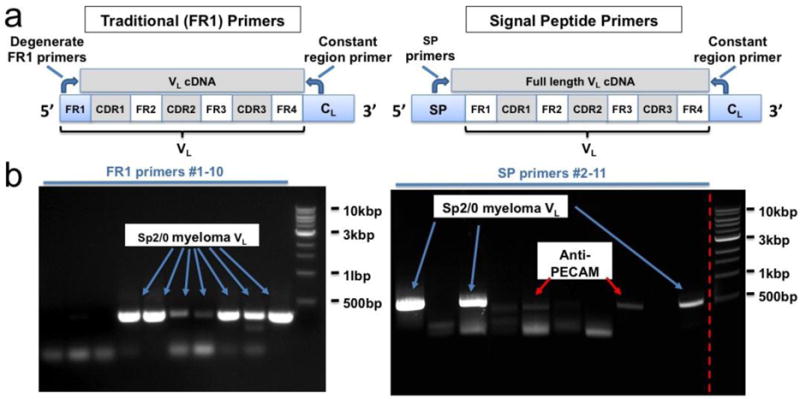
PCR amplication of Ab62 VH and VL cDNAs. (a) Schematic comparing use of traditional (FR1) vs. signal peptide (SP) primer sets. SP primers amplify full-length cDNA, without incorporation of primer-derived sequence, which can introduce amino acid substitutions at the N terminus of the FR1 region. (b) DNA gels comparing VL cDNAs amplified using FR1 vs. SP primer sets. Sequencing of each appropriately sized amplicon revealed an additional advantage of the SP primers – selective amplification of the Ab62 anti-PECAM VL by primers MKV.SP6 and SP9.
Analysis of Ab62 and myeloma light chain sequences
To understand why the two approaches produced such disparate results, we first compared the sequences of the Ab62 and Sp2/0 myeloma light chains in the region recognized by the FR1 primers. Figure 2 shows marked similarity, with 16 out of 21 nucleic acids and 5 out of 7 amino acids shared between the two sequences. Both show homology to primers MKV.B4 through B10 – i.e., the seven primers that generated appropriately sized PCR products (Figure 1). This suggests that the myeloma and Ab62 light chains are similar enough in this region that none of FR1 primers are capable of selectively amplifying the latter. Interestingly, we had encountered similar issues amplifying VL cDNA from two other hybridomas from which we recently cloned scFv’s. In each of those cases, protein sequencing was used to identify a small portion of the antigen specific VL, with subsequent design of oligonucleotide primers based on this amino acid sequence[12,27].
Our recurrent difficulties with amplification of myeloma sequences and reports of similar problems in the literature prompted us to investigate the overall sequence diversity of light chains in the FR1 region. To address this question, we queried the Abysis database, an online resource which integrates immunoglobulin sequence data from the Kabat database, the International Immunogenetics Information System (IMGT), and the Protein Databank (PDB)[34]. Searching the database for all mouse kappa light chains with full length FR1 sequence information resulted in 2676 VL sequences (Supplemental Fig 1). After excluding duplicates, we compared the initial 7 amino acids of FR1 (the region encoded by PCR primers). The results shows that the sequence diversity is fairly limited – just 190 unique heptapeptides account for all 2237 sequences, with 15 of these accounting for 1714, or just over 76% of the light chains. Moreover, Figure 2 shows that the majority of these 15 sequences share significant homology with the Sp2/0 myeloma, the third most commonly occurring sequence.
A similar analysis was performed on the SP region, although fewer light chains in the database have complete signal peptide information. The first 10 amino acids (the area covered by the SP primers) of the SP region were analyzed for each of the 449 unique light chains identified, resulting in 141 unique decapeptide sequences. In contrast to the FR1 region, the top 15 sequences account for only 51% of the light chains. Interestingly, the Sp2/0 myeloma SP sequence again ranks as the third most common, although in this case, it accounts for < 5% of the total sequences. More importantly, significantly less sequence homology is present in this region, as compared to the start of the FR1 region. This is particularly notable towards the end of the sequence, corresponding to the 3′ end of DNA critical for annealing of PCR primers. Overall, this analysis supports our observations and suggests that individual SP primers are more likely to selectively amplify either myeloma or antigen-specific cDNAs than their FR1 counterparts.
Production and characterization of Ab62 scFv protein
Having successfully amplified VH and VL, the cDNAs were assembled into an scFv construct (Figure 3a) with a C-terminal triple FLAG tag for purification of the protein following production in S2 cells. A second construct was made, incorporating a 15-amino acid AviTag cassette for site-specific, enzymatic biotinylation (see below). SDS-PAGE gel electrophoresis of each protein demonstrates a single major band at the expected size (Figure 3b). The functional activity of the Ab62 scFv was first tested on REN-hPECAM cells, which stably express human PECAM-1[32]. REN wild type cells were used as a control. As shown in Figure 3c, high affinity binding was seen on REN-hPECAM cells, with little non-specific binding on REN wild type cells. A similar cell-based ELISA was performed using primary human endothelial cells (HUVECs), to confirm binding to endogenously expressed PECAM-1. In these experiments, Ab62 scFv demonstrated sub-nanomolar affinity (EC50 = 0.69±0.23 nM based on three independent ELISAs), with a binding curve similar to its parental mAb. Finally, we assessed the distribution of Ab62 scFv following binding to HUVECs. As shown in Figure 3d, the pattern of staining matches the known distribution of PECAM-1, which is concentrated at cell-cell borders, and is identical to that of the parental antibody. Essentially no staining is seen with the control Mec13 scFv, which binds mouse PECAM-1 but has no species cross-reactivity.
Site-specific modification and in vivo biodistribution in rats
Having established binding of the Ab62 scFv to human endothelial PECAM-1, our next goal was to evaluate its species cross-reactivity – specifically, the ability to target the pulmonary endothelium in rats. Prior experience in our laboratory suggested that monovalent binding to PECAM-1, combined with the rapid clearance of isolated scFv, might severely limit biodistribution to the lung. We hypothesized that increasing the size and avidity of the Ab62 scFv targeting moiety would enhance pulmonary endothelial delivery in vivo. To test this hypothesis, we constructed scFv-streptavidin (SA) complexes, making use of the AviTag biotinylation cassette. In addition to its high efficiency, enzymatic biotinylation is site-specific, preserving the scFv antigen binding site, and allows addition of only one biotin per molecule (Figure 4a)[35]. The latter prevents crosslinking of streptavidin (SA) and formation of large complexes. To ensure tracing of the cargo protein, streptavidin was radiolabeled first and then complexed with either Ab62 or Mec13 scFv-biotin. Binding of the scFv-SA complexes was tested in vitro using HUVECs (Figure 4b), followed by intravenous injection in rats. As shown in Figure 4c, Ab62 scFv-biotin-SA complexes selectively accumulate in the lung, as compared to Mec13 controls (3.70±0.39% vs. 0.46±0.05 ID/g). Both the lung biodistribution and the immunospecificity index were significantly higher for the scFv-biotin-SA complexes than the radiolabeled scFv alone. In addition to confirming the species cross-reactivity of Ab62 scFv, this result provided a first assessment of its potential for targeting therapeutic proteins to the lung vasculature.
Figure 4. Validation of Ab62 scFv species cross-reactivity and binding in vivo.
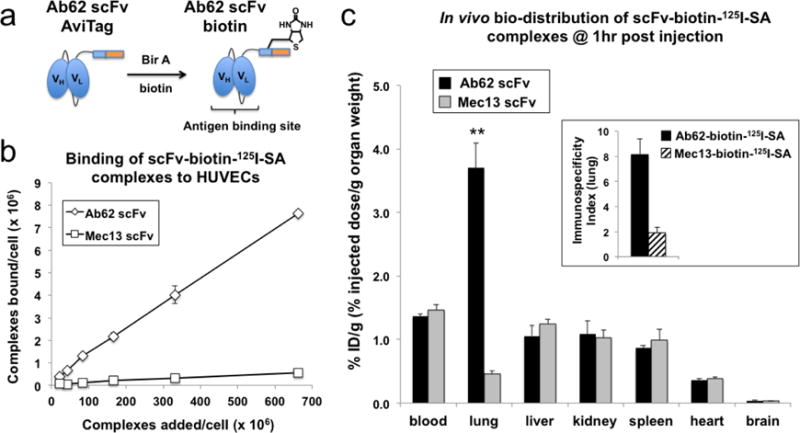
(a) Enzymatic biotinylation of Ab62 scFv modifies the protein site-specifically, preserving the antigen binding site. (b) Ab62-biotin-125I-SA (i.e., complexes of Ab62 scFv-biotin with 125I-labeled streptavidin) bound human ECs in vitro and (c) to rat lung endothelium in vivo. Mec13-biotin-125I-SA complexes were used as controls. N = 4 rats for each group, with data shown as mean ± SD. ** indicates statistical significance (p < 0.001). Inset figure shows the lung immunospecificity index, calculated as the ratio of %ID/g of targeted (Ab62) vs. nontargeted (Mec13) SA-complexes, normalized to blood levels.
Ab62 scFv targeting of liposomes to human and rat endothelium
To further assess the translational potential of Ab62 scFv, we evaluated its capacity as a targeting ligand on PEGylated liposomes – well characterized, uniform, and biocompatible nanocarriers successfully used to deliver antioxidant enzymes, antithrombotic agents, and a wide variety of other cargoes to therapeutic sites in animal models of cardiovascular disease[42–44]. Fluorescent SA-liposomes were decorated with Ab62 or Mec13 scFv-biotin and bound to HUVECs under static conditions (Supplemental Fig 2). The pattern of staining in Ab62 scFv liposome treated cells suggests endosomal uptake, consistent with previous reports that nanoparticles bearing multiple anti-PECAM antibodies trigger a non-canonical vesicular uptake pathway[25,45]. The present results suggest that multivalent binding by Ab62 scFv-decorated nanocarriers also induces endocytosis.
We next studied binding of scFv-decorated liposomes to ECs under flow. HUVECs were grown in a continuous 360° monolayer in a fibronectin-coated microfluidic chamber. Liposomes dispersed in cell culture medium were flowed into the chamber at a rate calculated to produce a constant shear stress of 5 dynes/cm2, roughly equivalent to the conditions in pulmonary capillaries and post-capillary venules. Video 1 shows a typical experiment in time-lapse format (each frame represents 15 seconds), with two side-by-side chambers to allow direct comparison of Ab62 and Mec13 scFv-liposomes. The former accumulate rapidly on the endothelial monolayer (top chamber), whereas the latter show minimal binding (bottom panel). To better simulate physiological conditions, we conducted a series of experiments in which liposomes were added to anti-coagulated whole blood, which was immediately perfused into the microfluidic chamber. Again, the flow rate was adjusted to maintain a constant shear stress of 5 dynes/cm2. In this case, binding of nanoparticles was difficult to visualize in real time due to signal attenuation by the blood cells (Video 2 and 3). After flushing the channel with saline, however, adherent and internalized liposomes were apparent (Figure 5). Projected z-stacks of the bottom half of the channel (Figure 5a) show a clear difference in the overall binding of Ab62 scFv-targeted and Mec13 scFv-control liposomes. Figure 5b shows 3-D reconstructions of confocal images from Ab62 scFv-liposome treated channels and demonstrates the same perinuclear staining pattern seen in Supplemental Figure 2, again suggesting endosomal uptake of the multivalent PECAM-targeted nanocarriers.
Figure 5. Microfluidic testing of anti-PECAM scFv liposomes.
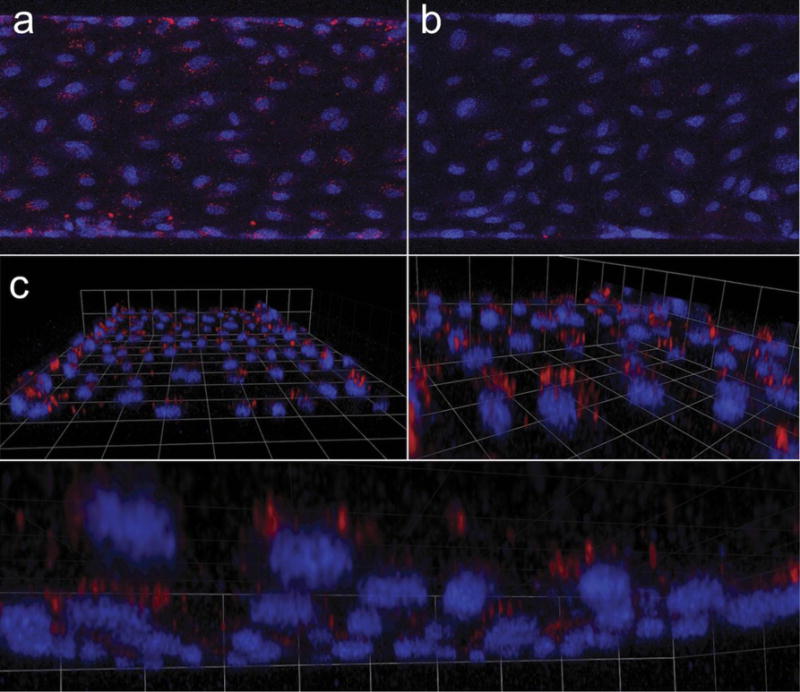
scFv-biotin decorated SA-fluoroliposomes were mixed with human whole blood and infused in a fully endothelialized microfluidic device at a flow rate calculated to produce shear stress (5 dynes/cm2) typical of capillaries and post-capillary venules. (a) Projected confocal stacks show that Ab62 scFv, but not Mec13 scFv, liposomes bound to HUVEC. (b) 3D reconstruction reveals punctate, perinuclear localization, suggesting endosomal uptake of the fluoroliposomes.
Finally, we tested targeting of scFv-decorated liposomes to the lung endothelium in vivo. Figure 6 shows staining of the pulmonary capillaries in rats injected with Ab62 scFv-liposomes, whereas no fluorescence is seen in animals receiving control liposomes. The diffuse pattern of staining is distinct from what has been observed with other types of fluorescent nanoparticles, e.g., antibody-coated polystyrene beads[45]. It is possible that this reflects the diffusion of fluorescent lipid throughout the EC membrane, but this interesting observation clearly merits further investigation.
Figure 6. Localization of Ab62 scFv-fluoroliposomes in rat lung.
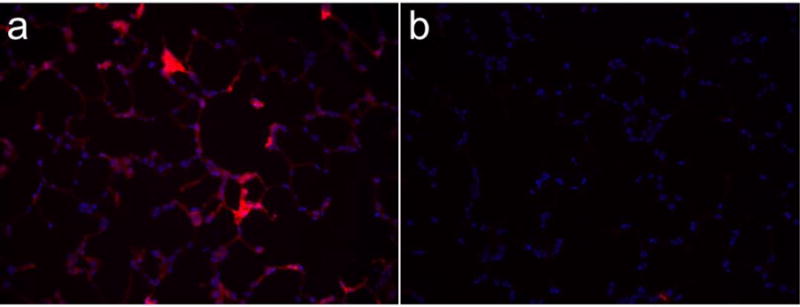
Rats were injected with (a) Ab62 scFv-decorated liposomes, or (b) control liposomes. 30 minutes later, the pulmonary vasculature was flushed with saline and lungs were inflated and frozen. Fluorescence imaging was performed on 5μm frozen sections.
Discussion
The endothelium, the cellular interface regulating interactions between blood and tissues, is a critically important target for therapeutic interventions in a wide variety of acute cardiovascular and inflammatory diseases. Vascular immunotargeting is a pharmacologic strategy to direct delivery of drugs to the endothelium and, in some cases, to further control their therapeutic effect via precise localization within or around these cells. Several promising endothelial targets have been explored, including PECAM-1 and other cell adhesion molecules, angiotensin converting enzyme, and caveolar proteins such as aminopeptidase P and the plasmalemma vesicle associated protein (PLVAP or PV-1)[7,9,26,46,47]. Drug delivery systems targeted to these and other determinants have enabled significant improvement of therapeutic interventions in numerous animal models of human diseases, but translation to the clinical domain has been lacking. Amongst other challenges, efforts have been hampered by a lack of affinity ligands that react across multiple species. Emerging evidence that antibody epitope is an important determinant of drug carrier accessibility, cellular uptake, and recycling, has further complicated this issue[25,26,36]. In this light, simple substitution of an anti-human antibody specific for the equivalent target protein may not be a viable strategy for translation.
This study reports several relevant and critical results to the field of vascular immunotargeting. First, we reintroduce a method for cloning VH and VL cDNAs from hybridoma cells, highlighting a previously unrecognized advantage of this approach in the creation of scFv and other recombinant antibody derivatives. SP primers have been reported previously, amongst the earliest methods described for PCR-based cloning of variable regions[48,49]. In these early reports, however, the signal peptide, or “leader sequence”, was singled out not because of its ability to selectively amplify antigen specific cDNAs, but because of its relatively conserved sequence, which led to speculation that it might allow creation of a universal degenerate PCR primer capable of amplifying any variable region cDNA[50]. Potential interference by myeloma sequences, however, was not mentioned and, in fact, the primers were reportedly mixed together and run in a single PCR reaction, such that the product presumably contained a mixture of a number of different sequences.
Since the publication of the original reports of SP primers, FR1 primer sets have become the mainstay of variable region cloning and the existence of multiple distinct heavy and light chain transcripts within a single hybridoma clone has been identified as a significant challenge[33,39]. Of note, these transcripts exist even in hybridomas where the tumor cell fusion partner (e.g., the Sp2/0 myeloma) has been selected to make no immunoglobulin[51]. One review cites the existence of as many as nine different VL sequences in a single hybridoma and notes that pseudogenes containing internal stop codons, such as the Sp2/0 transcript, “may represent the most abundant PCR product in some cases”[33]. Data from the current study certainly supports this notion, as the majority of our PCR reactions yielded the Sp2/0-derived variable light chain upon sequencing (Figure 1). While it is likely that at least some of these PCR products were mixtures of Sp2/0 and Ab62 cDNAs, our results suggest a significant excess of myeloma-derived sequence, leaving us with the unappetizing option of “trial-and-error” modulation of PCR parameters in an effort to increase the relative amplification of antigen-specific cDNA, or the equally unpalatable brute force sequencing of hundreds of clones.
A number of different techniques have been developed to deal with this situation. Several of these involve destruction of myeloma mRNA, e.g. via RNase H digestion or ribozyme cleavage, whereas others involve “clamping” of the undesired cDNA to prevent its amplification[39,52,53]. Other techniques use high-throughput screening methods such as PCR or cell free expression systems to rapidly separate clones incorporating myeloma sequence from those conferring antigen specificity[38,52]. Still other methods circumvent the entire process of annealing primers to the V-region, using techniques such as inverted PCR or the so-called “Rapid Amplification of cDNA ends”, or RACE technique[54,55]. Finally, our group and others have made use of protein sequencing of the parental mAb, with subsequent design of selective oligonucleotide primers based on this amino acid sequence[12,27]. While each of these techniques has been successfully reported as a means of identifying VH and VL cDNAs, they are all potentially time consuming, labor intensive, and fraught with technical difficulties.
In contrast, we report here that SP primers may offer a straightforward and simple means for selectively amplifying antigen-specific cDNA, at least for mouse-derived mAbs. The sequence analysis in Figure 2 suggests that the majority of mouse kappa monoclonal antibodies will share little homology in the SP region with the Sp2/0 myeloma transcript, found not only in hybridomas generated using this fusion partner, but also MOPC-21 and P3-X63-Ag8.653-derived cell lines. In this case, creation of an scFv becomes a relatively straightforward and rapid process, achievable not only in specialized antibody engineering laboratories, but also those primarily focused on the design and testing of drug delivery systems. Beyond the speed and ease of this process, the SP primers have another significant advantage, that being the identification of full-length cDNAs, without the incorporation of primer sequence. While this is infrequently discussed in the literature, many reported scFv sequences obtained with FR1 primers have amino acid substitutions at the start of both heavy and light chain variable regions. Reports exist in the literature of both decreased stability and complete loss of antigen binding due to FR1 substitutions[40,41].
Apart from the techniques used in cloning, the current study is significant in demonstrating the potential of Ab62 scFv as a translational affinity ligand for endothelial drug delivery. Our results indicate that the scFv functions similarly to its parental immunoglobulin, with nearly equivalent binding affinity and distribution on human endothelial cells. Likewise, when the scFv is assembled on a liposomal drug carrier, it demonstrates behavior characteristic of anti-PECAM-1 antibody decorated nanoparticles: namely, targeted delivery to the pulmonary vasculature in animals and endosomal uptake by endothelial cells under flow. One caveat with regards to our current approach is the use of biotin-streptavidin as a means of attaching scFv to protein cargo and drug carrier. To be fully translational, future systems will require covalent attachment of scFv, ideally without sacrificing the site-specific and directionally oriented approach utilized here. This limitation aside, our results support Ab62 scFv as a valuable affinity ligand, which will allow study of pharmacokinetics, therapeutic efficacy, and toxicity in rat models of lung disease, with the potential for straightforward translation to large animals and the clinical domain. The identification of similar species cross-reactive endothelial targeting ligands should be a priority.
Conclusion
In this study we report the engineering of a high affinity scFv that binds human PECAM-1 and cross reacts with the analogous endothelial cell adhesion molecule in rats. We modify this recombinant affinity ligand site-specifically, allowing oriented attachment to protein cargo and liposomal drug carriers. We demonstrate endothelial targeting in vivo in rodents and in a microfluidic model of a perfused human capillary. Our approach provides a template for the generation of translational affinity ligands with potential application to a wide variety of advanced drug delivery systems.
Supplementary Material
Acknowledgments
This work was supported by a Center for Targeted Therapeutics and Translational Nanomedicine (CT3N) Pilot Grant Award (ITMAT) at the University of Pennsylvania (CFG), as well as an American Heart Association Fellow to Faculty grant 10FTF4150053 (CFG), and National Institutes of Health Grant R01-HL091950 (VRM). The authors would like to thank Dr. Marian Nakada for the generous gift of Ab62 hybridoma.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authorship
C.F.G., V.R.M., and C.G. conceived the study. C.G. designed the SP primers. C.F.G. conducted the experiments. E.D.H. assisted in radiolabeling proteins and produced SA-fluoroliposomes. J.S.B. assisted in preparing and imaging rat lungs. A.Y. and M.K. assisted with scFv cloning, protein production, and purification. I.H.J. and M.P. assisted with the microfluidic experiments. C.F.G., C.G., E.D.H., and V.R.M wrote and edited the manuscript.
References
- 1.Cines DB, Pollak ES, Buck CA, Loscalzo J, Zimmerman GA, McEver RP, et al. Endothelial Cells in Physiology and in the Pathophysiology of Vascular Disorders. Blood. 1998;91:3527–3561. [PubMed] [Google Scholar]
- 2.Komarova Y, Malik AB. Regulation of Endothelial Permeability via Paracellular and Transcellular Transport Pathways. Annu Rev Physiol. 2010;72:463–493. doi: 10.1146/annurev-physiol-021909-135833. [DOI] [PubMed] [Google Scholar]
- 3.Kazmi R, Boyce S, Lwaleed B. Homeostasis of Hemostasis: The Role of Endothelium. Seminars in Thrombosis and Hemostasis. 2015;41:549–555. doi: 10.1055/s-0035-1556586. [DOI] [PubMed] [Google Scholar]
- 4.Aird WC. Endothelium as a therapeutic target in sepsis. Curr Drug Targets. 2007;8:501–507. doi: 10.2174/138945007780362782. [DOI] [PubMed] [Google Scholar]
- 5.Mather KJ. The vascular endothelium in diabetes–a therapeutic target? Rev Endocr Metab Disord. 2013;14:87–99. doi: 10.1007/s11154-013-9237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budhiraja R, Tuder RM, Hassoun PM. Endothelial Dysfunction in Pulmonary Hypertension. Circulation. 2004;109:159–165. doi: 10.1161/01.CIR.0000102381.57477.50. [DOI] [PubMed] [Google Scholar]
- 7.Danilov SM, Gavrilyuk VD, Franke FE, Pauls K, Harshaw DW, McDonald TD, et al. Lung uptake of antibodies to endothelial antigens: key determinants of vascular immunotargeting. Am J Physiol Lung Cell Mol Physiol. 2001;280:L1335–1347. doi: 10.1152/ajplung.2001.280.6.L1335. [DOI] [PubMed] [Google Scholar]
- 8.Gottstein C, Wels W, Ober B, Thorpe PE. Generation and characterization of recombinant vascular targeting agents from hybridoma cell lines. BioTechniques. 2001;30:190–194. 196. doi: 10.2144/01301dd03. 198 passim. [DOI] [PubMed] [Google Scholar]
- 9.Chrastina A, Valadon P, Massey KA, Schnitzer JE. Lung vascular targeting using antibody to aminopeptidase P: CT-SPECT imaging, biodistribution and pharmacokinetic analysis. J Vasc Res. 2010;47:531–543. doi: 10.1159/000313880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kowalski PS, Lintermans LL, Morselt HWM, Leus NGJ, Ruiters MHJ, Molema G, et al. Anti-VCAM-1 and anti-E-selectin SAINT-O-Somes for selective delivery of siRNA into inflammation-activated primary endothelial cells. Mol Pharm. 2013;10:3033–3044. doi: 10.1021/mp4001124. [DOI] [PubMed] [Google Scholar]
- 11.Hood ED, Greineder CF, Dodia C, Han J, Mesaros C, Shuvaev VV, et al. Antioxidant protection by PECAM-targeted delivery of a novel NADPH-oxidase inhibitor to the endothelium in vitro and in vivo. J Control Release. 2012;163:161–169. doi: 10.1016/j.jconrel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greineder CF, Chacko AM, Zaytsev S, Zern BJ, Carnemolla R, Hood ED, et al. Vascular Immunotargeting to Endothelial Determinant ICAM-1 Enables Optimal Partnering of Recombinant scFv-Thrombomodulin Fusion with Endogenous Cofactor. PLoS ONE. 2013;8:e80110. doi: 10.1371/journal.pone.0080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shuvaev VV, Christofidou-Solomidou M, Scherpereel A, Simone E, Arguiri E, Tliba S, et al. Factors modulating the delivery and effect of enzymatic cargo conjugated with antibodies targeted to the pulmonary endothelium. J Control Release. 2007;118:235–244. doi: 10.1016/j.jconrel.2006.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alley SC, Okeley NM, Senter PD. Antibody-drug conjugates: targeted drug delivery for cancer. Curr Opin Chem Biol. 2010;14:529–537. doi: 10.1016/j.cbpa.2010.06.170. [DOI] [PubMed] [Google Scholar]
- 15.Reichert JM. Marketed therapeutic antibodies compendium. mAbs. 2012;4:413–415. doi: 10.4161/mabs.19931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurlander RJ, Ellison DM, Hall J. The blockade of Fc receptor-mediated clearance of immune complexes in vivo by a monoclonal antibody (2.4G2) directed against Fc receptors on murine leukocytes. J Immunol. 1984;133:855–862. [PubMed] [Google Scholar]
- 17.Aragnol D, Leserman LD. Immune clearance of liposomes inhibited by an anti-Fc receptor antibody in vivo. Proc Natl Acad Sci USA. 1986;83:2699–2703. doi: 10.1073/pnas.83.8.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, Shuvaev VV, Muzykantov VR. Catalase and superoxide dismutase conjugated with platelet-endothelial cell adhesion molecule antibody distinctly alleviate abnormal endothelial permeability caused by exogenous reactive oxygen species and vascular endothelial growth factor. J Pharmacol Exp Ther. 2011;338:82–91. doi: 10.1124/jpet.111.180620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson AL. Antibody fragments. MAbs. 2010;2:77–83. doi: 10.4161/mabs.2.1.10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanes J, Schaffitzel C, Knappik A, Plückthun A. Picomolar affinity antibodies from a fully synthetic naive library selected and evolved by ribosome display. Nature Biotechnology. 2000;18:1287–1292. doi: 10.1038/82407. [DOI] [PubMed] [Google Scholar]
- 22.Ding BS, Hong N, Christofidou-Solomidou M, Gottstein C, Albelda SM, Cines DB, et al. Anchoring fusion thrombomodulin to the endothelial lumen protects against injury-induced lung thrombosis and inflammation. Am J Respir Crit Care Med. 2009;180:247–256. doi: 10.1164/rccm.200809-1433OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Damasceno LM, Lee F, Ritter G, Old L, Batt C. High-level expression of a phage display-derived scFv in Pichia pastoris. Methods Mol Biol. 2009;562:225–236. doi: 10.1007/978-1-60327-302-2_18. [DOI] [PubMed] [Google Scholar]
- 24.Cherf GM, Cochran JR. Applications of Yeast Surface Display for Protein Engineering. Methods Mol Biol. 2015;1319:155–175. doi: 10.1007/978-1-4939-2748-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garnacho C, Albelda SM, Muzykantov VR, Muro S. Differential intra-endothelial delivery of polymer nanocarriers targeted to distinct PECAM-1 epitopes. J Control Release. 2008;130:226–233. doi: 10.1016/j.jconrel.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muzykantov VR. Targeted Drug Delivery to Endothelial Adhesion Molecules. ISRN Vascular Medicine. 2013;2013:1–27. doi: 10.1155/2013/916254. [DOI] [Google Scholar]
- 27.Greineder CF, Brenza JB, Carnemolla R, Zaitsev S, Hood ED, Pan DC, et al. Dual targeting of therapeutics to endothelial cells: collaborative enhancement of delivery and effect. FASEB J. 2015;29:3483–3492. doi: 10.1096/fj.15-271213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakada MT, Amin K, Christofidou-Solomidou M, O’Brien CD, Sun J, Gurubhagavatula I, et al. Antibodies Against the First Ig-Like Domain of Human Platelet Endothelial Cell Adhesion Molecule-1 (PECAM-1) That Inhibit PECAM-1-Dependent Homophilic Adhesion Block In Vivo Neutrophil Recruitment. J Immunol. 2000;164:452–462. doi: 10.4049/jimmunol.164.1.452. [DOI] [PubMed] [Google Scholar]
- 29.Albelda SM, Muller WA, Buck CA, Newman PJ. Molecular and cellular properties of PECAM-1 (endoCAM/CD31): a novel vascular cell-cell adhesion molecule. J Cell Biol. 1991;114:1059–1068. doi: 10.1083/jcb.114.5.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman PJ. The biology of PECAM-1. J Clin Invest. 1997;99:3–8. doi: 10.1172/JCI119129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaporciyan AA, DeLisser HM, Yan HC, Mendiguren, Thom, Jones ML, et al. Involvement of platelet-endothelial cell adhesion molecule-1 in neutrophil recruitment in vivo. Science. 1993;262:1580–1582. doi: 10.1126/science.8248808. [DOI] [PubMed] [Google Scholar]
- 32.Gurubhagavatula I, Amrani Y, Pratico D, Ruberg FL, Albelda SM, Panettieri RA. Engagement of human PECAM-1 (CD31) on human endothelial cells increases intracellular calcium ion concentration and stimulates prostacyclin release. J Clin Invest. 1998;101:212–222. doi: 10.1172/JCI269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toleikis L, Broders O, Dübel S. Cloning single-chain antibody fragments (scFv) from hybridoma cells. Methods Mol Med. 2004;94:447–458. doi: 10.1385/1-59259-679-7:447. [DOI] [PubMed] [Google Scholar]
- 34.Martin ACR, Allen J. Bioinformatics Tools for Antibody Engineering. In: Dübel S, Reichert JM, editors. Wiley: Handbook of Therapeutic Antibodies. 2nd. Wiley-VCH Verlag GmbH; 2014. [Google Scholar]
- 35.Cull MG, Schatz PJ. Biotinylation of proteins in vivo and in vitro using small peptide tags. Meth Enzymol. 2000;326:430–440. doi: 10.1016/s0076-6879(00)26068-0. [DOI] [PubMed] [Google Scholar]
- 36.Chacko AM, Han J, Greineder CF, Zern BJ, Mikitsh JL, Nayak M, et al. Collaborative Enhancement of Endothelial Targeting of Nanocarriers by Modulating Platelet-Endothelial Cell Adhesion Molecule-1/CD31 Epitope Engagement. ACS Nano. 2015;9:6785–6793. doi: 10.1021/nn505672x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dübel S, Breitling F, Fuchs P, Zewe M, Gotter S, Welschof M, et al. Isolation of IgG antibody Fv-DNA from various mouse and rat hybridoma cell lines using the polymerase chain reaction with a simple set of primers. Journal of Immunological Methods. 1994;175:89–95. doi: 10.1016/0022-1759(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 38.Nicholls PJ, Johnson VG, Blanford MD, Andrew SM. An improved method for generating single-chain antibodies from hybridomas. J Immunol Methods. 1993;165:81–91. doi: 10.1016/0022-1759(93)90109-k. [DOI] [PubMed] [Google Scholar]
- 39.Cochet O, Martin E, Fridman WH, Teillaud JL. Selective PCR amplification of functional immunoglobulin light chain from hybridoma containing the aberrant MOPC 21-derived V kappa by PNA-mediated PCR clamping. BioTechniques. 1999;26:818–820. 822. doi: 10.2144/99265bm04. [DOI] [PubMed] [Google Scholar]
- 40.Li J, Wang Y, Wang Z, Dong Z. Influences of amino acid sequences in FR1 region on binding activity of the scFv and Fab of an antibody to human gastric cancer cells. Immunol Lett. 2000;71:157–165. doi: 10.1016/s0165-2478(00)00152-8. [DOI] [PubMed] [Google Scholar]
- 41.de Haard HJ, Kazemier B, van der Bent A, Oudshoorn P, Boender P, van Gemen B, et al. Absolute conservation of residue 6 of immunoglobulin heavy chain variable regions of class IIA is required for correct folding. Protein Eng. 1998;11:1267–1276. doi: 10.1093/protein/11.12.1267. [DOI] [PubMed] [Google Scholar]
- 42.Yun X, Maximov VD, Yu J, Zhu H, Vertegel AA, Kindy MS. Nanoparticles for targeted delivery of antioxidant enzymes to the brain after cerebral ischemia and reperfusion injury. J Cereb Blood Flow Metab. 2013;33:583–592. doi: 10.1038/jcbfm.2012.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Absar S, Nahar K, Kwon YM, Ahsan F. Thrombus-targeted nanocarrier attenuates bleeding complications associated with conventional thrombolytic therapy. Pharm Res. 2013;30:1663–1676. doi: 10.1007/s11095-013-1011-x. [DOI] [PubMed] [Google Scholar]
- 44.Howard MD, Greineder CF, Hood ED, Muzykantov VR. Endothelial targeting of liposomes encapsulating SOD/catalase mimetic EUK-134 alleviates acute pulmonary inflammation. J Control Release. 2014;177:34–41. doi: 10.1016/j.jconrel.2013.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han J, Zern BJ, Shuvaev VV, Davies PF, Muro S, Muzykantov V. Acute and chronic shear stress differently regulate endothelial internalization of nanocarriers targeted to platelet-endothelial cell adhesion molecule-1. ACS Nano. 2012;6:8824–8836. doi: 10.1021/nn302687n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greineder CF, Howard MD, Carnemolla R, Cines DB, Muzykantov VR. Advanced drug delivery systems for antithrombotic agents. Blood. 2013;122:1565–1575. doi: 10.1182/blood-2013-03-453498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simone EA, Zern BJ, Chacko AM, Mikitsh JL, Blankemeyer ER, Muro S, et al. Endothelial targeting of polymeric nanoparticles stably labeled with the PET imaging radioisotope iodine-124. Biomaterials. 2012;33:5406–5413. doi: 10.1016/j.biomaterials.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larrick JW, Danielsson L, Brenner CA, Abrahamson M, Fry KE, Borrebaeck CA. Rapid cloning of rearranged immunoglobulin genes from human hybridoma cells using mixed primers and the polymerase chain reaction. Biochem Biophys Res Commun. 1989;160:1250–1256. doi: 10.1016/s0006-291x(89)80138-x. [DOI] [PubMed] [Google Scholar]
- 49.Gavilondo-Cowley JV, Coloma MJ, Vazquez J, Ayala M, Macías A, Fry KE, et al. Specific amplification of rearranged immunoglobulin variable region genes from mouse hybridoma cells. Hybridoma. 1990;9:407–417. doi: 10.1089/hyb.1990.9.407. [DOI] [PubMed] [Google Scholar]
- 50.Larrick JW, Danielsson L, Brenner CA, Wallace EF, Abrahamson M, Fry KE, et al. Polymerase Chain Reaction Using Mixed Primers: Cloning of Human Monoclonal Antibody Variable Region Genes from Single Hybridoma Cells. Nat Biotech. 1989;7:934–938. doi: 10.1038/nbt0989-934. [DOI] [Google Scholar]
- 51.Shulman M, Wilde CD, Köhler G. A better cell line for making hybridomas secreting specific antibodies. Nature. 1978;276:269–270. doi: 10.1038/276269a0. [DOI] [PubMed] [Google Scholar]
- 52.Duan L, Pomerantz RJ. Elimination of endogenous aberrant kappa chain transcripts from sp2/0-derived hybridoma cells by specific ribozyme cleavage: utility in genetic therapy of HIV-1 infections. Nucleic Acids Res. 1994;22:5433–5438. doi: 10.1093/nar/22.24.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ostermeier C, Michel H. Improved cloning of antibody variable regions from hybridomas by an antisense-directed RNase H digestion of the P3-X63-Ag8.653 derived pseudogene mRNA. Nucleic Acids Res. 1996;24:1979–1980. doi: 10.1093/nar/24.10.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zwickl M, Zaninetta D, McMaster GK, Hardman N. Selective cloning of B cell hybridoma-specific rearranged immunoglobulin gene loci using the polymerase chain reaction. J Immunol Methods. 1990;130:49–55. doi: 10.1016/0022-1759(90)90298-a. [DOI] [PubMed] [Google Scholar]
- 55.Ruberti F, Cattaneo A, Bradbury A. The use of the RACE method to clone hybridoma cDNA when V region primers fail. J Immunol Methods. 1994;173:33–39. doi: 10.1016/0022-1759(94)90280-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


