Abstract
Oxidative phosphorylation (OXPHOS) is fundamental for life. OXPHOS complexes pose a unique challenge for the cell, because their subunits are encoded on two different genomes, the nuclear genome and the mitochondrial genome. Genomic approaches designed to study nuclear/cytosolic and bacterial gene expression have not been broadly applied to the mitochondrial system; thus the co-regulation of OXPHOS genes remains largely unexplored. Here we globally monitored mitochondrial and nuclear gene expression processes in Saccharomyces cerevisiae during mitochondrial biogenesis, when OXPHOS complexes are synthesized. Nuclear- and mitochondrial-encoded OXPHOS transcript levels do not increase concordantly. Instead, we observe that mitochondrial and cytosolic translation are rapidly and dynamically regulated in a strikingly synchronous fashion. Furthermore, the coordinated translation programs are controlled unidirectionally through the intricate and dynamic control of cytosolic translation. Thus the nuclear genome carefully directs the coordination of mitochondrial and cytosolic translation to orchestrate the timely synthesis of each OXPHOS complex, representing an unappreciated regulatory layer shaping the mitochondrial proteome. Our whole-cell genomic profiling approach establishes a foundation for global gene regulatory studies of mitochondrial biology.
The large majority of cellular energy is produced by oxidative phosphorylation (OXPHOS) complexes within the mitochondrial inner membrane, which consist of a mix of mitochondrial- and nuclear-encoded subunits. Their dual-origin nature requires the cell to coordinate completely orthogonal gene expression machineries to match expression with environmental demands for energy. The mitochondrial gene expression machinery is distinct from its nuclear/cytosolic counterparts, and has also diverged dramatically from its bacterial correlates. Transcription is carried out by a single-subunit phage-related RNA polymerase1 and translation by a dedicated ribosome (the mitoribosome) that is protein-rich compared to cytosolic and bacterial ribosomes2. Mitochondrial transcripts are polycistronic and mRNAs have neither 5’ caps nor Shine-Dalgarno sequences. In some species, including S. cerevisiae, poly(A) tails are also absent3. Mitochondria use modified genetic codes, deciphered by mitochondrial-encoded tRNAs4. Most notably, no gene-specific transcription factors have been characterized; instead there are mRNA-specific translational activators, generally present in limiting amounts, that have roles in initiation and\or elongation and in some cases feedback control of OXPHOS complex assembly on translation5-8. Thus the nuclear and mitochondrial genes are expressed by distinct machinery and controlled by disparate regulatory mechanisms. It remains unclear whether these radically different genomes coordinate their gene expression programs during any physiological response when OXPHOS synthesis is required, such as during mitochondrial biogenesis.
OXPHOS mRNAs are not coordinately induced
To comprehensively analyze OXPHOS expression we used a set of quantitative approaches to monitor the level and translation of mitochondrial- and nuclear-encoded RNA (Fig. 1a). To induce OXPHOS synthesis, we rapidly shifted S. cerevisiae cells from growth in the fermentable carbon source glucose to non-fermentable glycerol, requiring a reprogramming of gene expression to adapt for respiratory metabolism9,10 (Fig. 1b). As expected, steady-state protein levels of both mitochondrial- and nuclear-encoded OXPHOS subunits are induced as cells adapt to respiratory metabolism, and accumulate to high levels in cells undergoing log phase growth in glycerol (Extended Data Fig. 1). Mitochondrial transcripts accumulate in response to the shift11,12, as do nuclear-encoded OXPHOS mRNAs13,14, but whether the transcript abundances rise concordantly is not clear. To quantify levels of both nuclear- and mitochondrial-encoded mRNAs we used rRNA depletion, as poly(A) selection would not capture mitochondrial messages, and included spike-in standards to allow quantitation across samples. As is observed in most transcriptional programs, nuclear-encoded protein complex components are co-regulated at the RNA level15 (Extended Data Fig. 2a, full dataset provided in Supplementary Table 1). The mitochondrial genome encodes 8 major proteins that contribute to dual-origin complexes: the mitoribosome and the OXPHOS complexes III-V. At low levels, the genome also produces maturases required to process COB and COX1 mRNAs (Extended Data Fig. 2b). The nuclear- and mitochondrial- encoded RNAs of the mitoribosome are not significantly induced across the time series, and so by default display similar dynamics (Extended Data Fig. 2c). In contrast, nuclear- and mitochondrial-encoded RNA levels of the dual-origin OXPHOS complexes are induced and interestingly are not co-regulated (Fig. 1c). Whereas nuclear OXPHOS messages are induced rapidly in response to nutrient shift, mitochondrial OXPHOS messages are induced much more slowly. The difference in induction kinetics may reflect the absence of environment-responsive transcription factors from the mitochondria.
Figure 1. Synthesis of dual-origin OXPHOS complexes is induced upon adaptation to respiratory growth.
a, Whole-cell genomic profiling approach used to monitor gene expression during mitochondrial biogenesis; purple, cytoribosomes; orange, mitoribosomes. b, Experimental setup to rapidly induce respiratory adaptation. Solid line shows yeast culture grown to log phase in glucose media and shifted to glycerol media, where it is cultured for an additional 3 h. Dotted line shows parallel culture that is diluted and incubated ~16 h for log-phase respiratory growth. c, Cartoon highlighting the mitochondrial-encoded proteins of each OXPHOS complex (top panel), and line plots showing induction kinetics for mRNAs encoding each subunit of the OXPHOS complexes (bottom panels). Solid lines: nuclear-encoded mRNAs, dotted lines: mito-encoded mRNAs.
Mitochondrial translation is dynamically regulated
Traditionally, mitochondrial translation has been monitored using metabolic labeling after inhibition of cytosolic translation by cycloheximide, but this method requires specific buffer conditions and has poor time resolution16. Thus, despite the existence of translational activators, it is not known whether translation of mitochondrial mRNAs is differentially regulated under normal physiological conditions, nor whether mitochondrial translation responds rapidly to environmental changes as does cytosolic translation17. To quantitatively monitor mitochondrial translation under any growth condition with high time resolution, we re-engineered the ribosome profiling approach originally developed for cytosolic ribosomes18 through three major modifications: (1) Affinity purification by FLAG-tagged mitoribosomal subunits replaced sucrose fractionation to separate 74S mitoribosomes from 80S cytosolic ribosomes (cytoribosomes) (Extended Data Fig. 3a-d). (2) Lysis and buffer conditions were optimized to solubilize the membrane-associated mitoribosomes while maintaining subunit association (Extended Data Fig. 3c,d). Although mitoribosome footprints have been captured previously19, mitoribosomes have strongly altered sensitivity to ionic composition compared to cytosolic ribosomes (cytoribosomes), and efficient purification of intact mitoribosomes requires optimized conditions20. (3) Size selection of footprints was modified as we found mitoribosome-protected fragments are ~38 nt (Fig. 2b,c) in contrast to the ~28 nt cytoribosome-protected fragments21. These adaptations enabled the quantitative capture of mitoribosome footprints (Fig 2a, Extended Data Fig. 4a).
Figure 2. Mitoribosome profiling provides genome-wide readout of mitochondrial translation.

a, Schematic of mitoribosome profiling protocol. Asterisks: steps in which major modifications are required to capture mitoribosome footprints in contrast to cytoribosome footprints. b, RNase I titration (0, 50, 125, 250, 500, 1000 U mL−1) followed by mitoribosome immunoprecipitation (IP) via MrpS17-F. For gel source data, see Supplementary Fig. 1. c, Length distribution for mitoribosome profiling reads that map to mitochondrial-encoded mRNA in comparison to contaminating reads that map to rRNA and tRNA. d, Genome-wide view of mitochondrial ORFs with mapped RNA-seq reads and mitoribosome profiling footprint reads (inferred A site). Lack of COX2-mapped reads in pet111Δ strain is highlighted. Major ORFs are colored. Gray annotations are maturase genes (note low level of translation) and tRNA genes. e, Zoom-in of the region encoding the polycistronic transcript ATP8 – ATP6.
Our approach resulted in reads mapping precisely to all known open reading frames of the 86-kb mitochondrial genome (Fig. 2d,e). Mapped reads show 3-nt periodicity (Extended Data Fig. 4b) and reproducibility is high between biological replicates at the nucleotide level (r = 0.96 for rpm values), and at the gene level (r= 0.9998 for RPKM values) (Extended Data Fig. 4c). We found mitochondrial translation inhibitors unnecessary when cells are rapidly collected and lysed while frozen (Extended Data Fig. 4d). Finally, mitoribosome profiling in a strain without the COX2 translational activator, Pet111, showed a lack of footprint reads mapping to COX2 (Fig. 2d), confirming genetic evidence that Pet111 affects translation initiation22.
We applied mitoribosome profiling to cells undergoing respiratory adaption, revealing that mitochondrial-encoded proteins are indeed differentially synthesized during OXPHOS biogenesis (Extended Data Fig. 5a, Supplementary Table 2). To isolate translation regulation from changes in RNA transcript levels, we calculated the translation efficiency (TE; footprint RPKM values / RNA-seq RPKM values) for each message. We found a rapid and reproducible redistribution of mitoribosomes within 15 min of shifting cells into glycerol (Fig. 3a, Extended Data Fig. 6, Supplementary Table 3), demonstrating that the differential synthesis is due to strong translation regulation. In the early response, mitoribosomes shift from ATP synthase complex mRNAs to Complex IV (cytochrome c oxidase complex) mRNAs. Furthermore, translation is dynamically regulated throughout the adaptation program with ATP synthase translation recovering over time, COB translation not fully induced until 1 h and COX1 translation not fully induced until 3 h. Thus, in contrast to the transcription of mitochondrial-encoded genes, mitochondrial translation is dynamically and differentially regulated.
Figure 3. Mitochondrial and cytosolic translation on OXPHOS mRNAs is rapidly and synchronously regulated.
a, b, -Fold change in translation efficiencies (TEs) compared to log-phase glucose growth for the OXPHOS subunits synthesized in (a) the mitochondria (values are averages of two experiments) and (b) the cytosol. Asterisks on heat maps indicate the subunits shown in the line plots.
Translation regulation is synchronized across compartments
To determine whether cytosolic translation is coordinated with the rapid shift in mitochondrial translation efficiencies, we determined relative synthesis and translation efficiencies for all cytosolic transcripts across our experimental conditions using cytoribosome profiling (Supplementary Tables 4 and 5, representative library characteristics shown in Extended Data Fig. 4a,b). Abrupt transfer from a fermentable to a non-fermentable carbon source results in a transient reduction in cytosolic translation17 (Extended Data Fig. 7), but select transcripts escape and are preferentially translated to produce proteins required for cell survival in the new condition23. As expected, synthesis of all OXPHOS subunits increases as cells adapt to respiratory growth (Extended Data Fig. 5b), which is likely driven by the large-scale increase in RNA transcript levels that occurs immediately after carbon source shift. Remarkably, after normalizing for these RNA changes, the pattern of translational regulation on nuclear-encoded OXPHOS subunits is the same as for their mitochondrial-encoded counterparts (Fig. 3b). Within 15 min after media shift, actively translating cytoribosomes dramatically redistribute off of ATP synthase mRNAs on to Complex III and Complex IV mRNAs. Despite the double membrane separating cyto- and mitoribosomes and a lack of any shared components, translation regulation of OXPHOS subunits is synchronized across cellular compartments.
Translation systems communicate unidirectionally
Is there communication between the two pools of ribosomes to ensure the synchronized regulation on OXPHOS mRNAs, or does each react independently to environmental signals? When isolated from cytosolic factors after treatment with an uncoupler, carbonyl cyanide m-chlorophenyl hydrazone (CCCP), which blocks mitochondrial import (Extended Data Fig. 8a), mitochondrial translation is inhibited across all transcripts, independent of cytosolic translation and carbon source (Extended Data Fig. 8b-e, data not shown). Consistently, in organello mitochondrial translation is repressed unless the purified mitochondria are charged with a mixture of amino acids and nucleotides24.
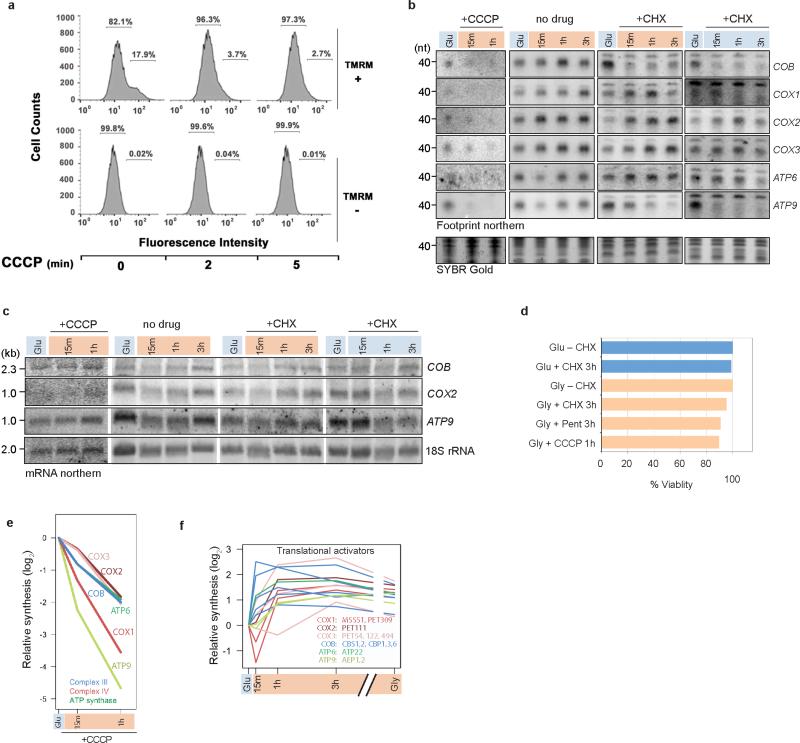
To directly determine whether there is communication between the two translation systems, we specifically inhibited cytosolic translation with cycloheximide (CHX)25 and observed the effect on mitochondrial translation (Fig. 4a-c, Extended Data Fig. 8b). CHX treatment does not affect mitochondrial mRNA levels (Extended Data Fig. 8c), and cells remain viable through the treatment course (Extended Data Fig. 8d). Upon complete inhibition of cytosolic translation, translation of mitochondrial messages is differentially affected: some messages gain translational capacity and others lose it (Fig. 4b). Further, there is a decrease in the dynamics, with only minor changes after the initial response, suggesting ongoing cytosolic translation is important for mitochondrial translation changes over the course of adaptation. Aside from COB and ATP6, CHX treatment does not significantly impact the rapid mitochondrial translational response (15 min). Surprisingly, CHX treatment similarly influences mitochondrial translation without a change in media carbon source (Fig. 4c). Thus, the early mitochondrial translation response is not directly responding to environmental inputs, but rather to the transient inhibition of cytosolic translation.
Figure 4. Communication between translation systems is unidirectional.
a, Schematic depicting action of drugs. b, c, Mitochondrial translation response, measured by northern blotting for footprints (Extended Data Fig. 8b), to cytosolic translation inhibition by CHX with (b) or without (c) carbon source shift. Note relative synthesis is a good proxy for translation efficiency (compare –CHX to Fig. 3a) because levels of mitochondrial mRNAs do not significantly change relative to each other during this time period (see Fig. 1c). Values in (b) are averages of two experiments. See Source Data for range values. d, e, -Fold change in TEs of nuclear-encoded OXPHOS subunits measured by cytoribosome profiling (d) without (–Pent) or with (+Pent) inhibition of mitochondrial translation, and (e) in ρ0 cells. The subset of OXPHOS subunits shown is the same as that shown in line plots in Fig. 3b.
The loss of COB translational capacity with CHX indicates that its translation requires one or more newly synthesized cytosolic products that escape the global stress-induced translation inhibition. Indeed, relative synthesis of two COB translational activators, Cbp6 and Cbs2, is nearly 4- and 6-fold increased, respectively, 15 min after the shift (Extended Data Fig. 8f). Compatible with the gain of ATP6 translational capacity with CHX, the ATP8/ATP6 transcript is the only one associated with a translational repressor26. These results suggest that the dynamic cytosolic translation control of translational activators contribute to the orchestration of mitochondrial OXPHOS protein synthesis.
Having established that the synchronization of translational programs is actively controlled by cytosolic translation, we next asked whether communication across compartments is unidirectional or bidirectional. We focused on the early response at 15 min when translational changes are maximal (Fig. 3b) and any secondary responses should be minimized. After specific inhibition of mitochondrial translation with the drug pentamidine27,28 (Fig. 4a, Extended Data Fig. 9a, left panel) ribosome profiling revealed that cytoribosomes respond independently of mitochondrial translation (Fig. 4d, Extended Data Fig. 9b). To test the possibility that the cytosolic translation response on OXPHOS mRNAs results from feedback from alterations in membrane potential or the OXPHOS complexes themselves rather than mitochondrial translation, we created a ρ0 yeast strain (Extended Data Fig. 10a,b). In this strain, which completely lacks mitochondrial DNA, mitochondrial translation, and functional OXPHOS complexes (Extended Data Fig. 9a, right panel), the cytosolic translation response on OXPHOS mRNAs is maintained (Fig. 4e, Extended Data Fig. 9c). Thus the synchronized translation programs are facilitated through unidirectional communication from cytosolic to mitochondrial ribosomes.
Discussion
Translational reprogramming allows rapid change in protein synthesis and conservation of cellular resources by focusing the expensive process of translation to where it is needed most29-31. The preference for Complex III and IV synthesis during adaptation may reflect their lower levels in fermenting cells compared to ATP synthase, which functions in reverse in the absence of the electron transport chain to maintain the mitochondrial inner membrane potential2. Interestingly, we observe that core subunits of Complex III and IV, Cob and Cox1 respectively, are translationally upregulated at later time points, which are likely due to feedback mechanisms that couple their translation to their respective complex assemblies32,33. Thus, the synchronized translation could serve in part to maximize assembly efficiency of the OXPHOS complexes and limit nonproductive or harmful off-target interactions. Important goals for future studies include unraveling the roles of translation activators and other factors in mediating this synchronized response, as well as analysis of metazoan systems where some mitochondrial gene regulatory mechanisms have diverged from those in yeast3. We expect that the whole-cell genomic profiling approach described here will usher in an era of global gene expression analyses that will shed light on many aspects of mitochondrial biology in health and disease.
Methods
Strain construction, growth conditions, and petite frequency analysis
To ensure a robust and physiological response, we modified the S288c background typically used for genomic studies to make it amenable to mitochondrial studies by (1) correcting the HAP1 mutation that leads to lower mitochondrial biomass production34,35, and (2) creating a point change to restore a conserved amino acid important for the fidelity of the mitochondrial DNA polymerase36,37. These modifications significantly reduced the frequency of mtDNA loss (Extended Data Fig. 3b). S288c derivative DBY12045 (MATa, HAP1+, GAL2+, ura3Δ0, MIP1[S]) was a gift from M. Hickman and D. Botstein. It was further modified by generating a single point change using a loopout strategy to create MIP1[S]A661T, repairing a strictly conserved threonine that is mutated in S288c and responsible for the increased rate of mitochondrial mutations in this strain36. Epitope-tagged proteins were expressed from their endogenous loci and generated using a “scarless” loopout strategy38,39. Deletion of PET111 was performed using the delitto perfetto approach40. For ρ0 strain generation, mtDNA loss was induced by overnight growth in 10 μg/mL ethidium bromide at 25 °C.
Petite frequency was assayed essentially as described41. Briefly, fresh colonies from YPD (2% glucose) plates were resuspended and plated at low density on YPDG (0.1% glucose, 3% glycerol). ‘Petite’ and ‘grande’ colonies were counted after 5 days of growth at 30 °C.
Yeast strains were grown in YP (1% yeast extract, 2% peptone), pH 5.0, supplemented with 2% glucose (Glu) or 3% glycerol (Gly) as indicated. The pet111Δ strain is respiratory-deficient thus grown in YPGal (2% galactose) instead of YPGly. Controlling the pH of the media is essential for consistent growth on glycerol. Overnight liquid YPD cultures were grown to saturation and used to inoculate fresh media to OD600 = ≤0.06. Cultures were grown at 30 °C until OD600 reached 0.6–0.8. For rapid media transfers cultures were harvested by filtration, rinsed once in YP (containing drug as indicated), and scraped off filter into fresh media. All media and flasks were pre-warmed. Where indicated, CHX (Sigma) was used in cultures at a final concentration of 100 μg/mL, pentamidine (Sigma) at 10 μM, and CCCP (Sigma) at 40 μM.
FACS analysis
Yeast cultures were grown until they reached OD600 = 0.5 in YP (1% yeast extract, 2% peptone), pH 5.0 supplemented with 2% glucose. Where indicated, cells were treated with 40 μM CCCP for 2 or 5 min. After treatment, the drug was washed off with 1X PBS and cells were diluted to 106 cells mL−1 in 1X PBS. Where indicated, diluted cells were treated with tetramethylrhodamine (TMRM) at 1 μM for 30 min. Cells were washed 2x, resuspended 1X PBS to final concentration of 106 mL−1 and loaded into 96-well culture plates (CellTreat). FACS analysis was performed using a Stratedigm 1000 instrument. Emission wavelengths were recorded at 586 nm. Histograms were generated using FlowJo software.
General nucleic acid and protein methods
For footprint detection by northern blotting, RNA was isolated from purified mitoribosomes (FLAG eluate) and loaded on a 15% polyacrylamide TBE-urea gel. After transfer to nylon (Hybond N+), blots were probed at room temperature with internally labeled random hexamer-primed DNA fragments synthesized from 200–500 bp PCR-generated templates. For mRNA detection, RNA was separated on 1.2% formaldehyde agarose and blots were probed at 42 °C. Quantitative PCR (qPCR) was performed using a CFX BioRad Connection qPCR thermocycler with EvaGreen (BioRad) fluorescent dye.
Proteins were resolved before silver staining or western blotting on NuPAGE Novex Bis-Tris gels (Thermo Fisher Scientific). Staining was done with the SilverQuest Silver Staining Kit (Invitrogen) according to the manufacturer's instructions. Antibodies against OXPHOS proteins were purchased from Santa Cruz Biotechnology (anti-Cob, sc-11436) and Abcam Mitosciences (anti-Cox1, ab110270; anti-Cox2, ab110271; anti-Cox4, ab110272). Fluorescently-labeled secondary antibodies (IRDye, LI-COR) were detected using the LI-COR Odyssey.
Metabolic labeling was performed essentially as described42. Briefly, cultures were grown in YPGal (2% galactose) to log phase, washed and resuspended in potassium phosphate buffer at 30 °C, shaking, for 2 h. Pentamidine was added to 10 μM where indicated and incubation continued for 15 min. To assay mitochondrial translation, CHX was added to 500 μg mL−1 for 3 min prior to addition of 35S-labeled methionine and cysteine mix (Perkin Elmer). Labeling was continued for 20 min at 30 °C. Proteins were TCA precipitated, resolved on 17.5% Tris-glycine polyacrylamide, pH 8.3, and transferred to nitrocellulose. For visualizing cytosolic translation CHX was omitted and protein samples were diluted 1:15 before loading the gel.
Mitoribosome profiling
Cell culture (400 OD600 equivalents) was rapidly harvested by filtration onto 0.45 μm nitrocellulose (Whatman). Cell pellet was flash frozen in liquid nitrogen and combined with 4 mL of frozen lysis buffer (10 mM Tris, pH 8.0, 50 mM NH4Cl, 10 mM MgCl2, 0.5% lauryl maltoside, 0.25 mM DTT, 1.5x protease inhibitor cocktail (Complete, EDTA-free, Roche)). In optimizing buffer conditions to maintain mitoribosome subunit association we found the ratio of monovalent to divalent cations to be of vital importance20. The frozen cell mixture was pulverized in 50-mL canisters prechilled in liquid nitrogen for six cycles of 3 min each at 15 Hz, on a Retsch MM301 mixer mill. Upon thaw, fresh lysis buffer was added to bring lysate concentration to 25 OD600 equivalents mL–1. Lysate was digested for 30 min at 25 °C with 500 U mL–1 of recombinant RNase I (RNase If, NEB). The reaction was stopped with 100 U mL–1 SUPERase-In (Thermo Fisher Scientific) and clarified by centrifugation at 4 °C at 20,000g for 15 min. The supernatant was reserved for immunoprecipitation.
Anti-FLAG M2 affinity gel (Sigma) was washed 3x with wash buffer (10 mM Tris, pH 8.0, 50 mM NH4Cl, 10 mM MgCl2, 0.1% Triton X-100), and added to clarified lysate at 12 μl 50% gel slurry mL–1. The mixture was rotated end-over-end at 4 °C for 3 h. The affinity gel was washed three times for 10 min in 10 mL of wash buffer at room temperature, then FLAG-tagged protein was eluted by incubation with 200 μg mL–1 3xFLAG peptide (Sigma) in 6x volumes affinity gel slurry for 40 min at room temperature. RNA was isolated using phenol/chloroform extraction.
Mitoribosome protected fragments were isolated by excision of ~36-42 nt fragments from 12% or 15% TBE-urea polyacrylamide gels. Sequencing library preparation was performed through a circular intermediate as described43, omitting the rRNA depletion step. Sequencing was performed on an Illumina MiSeq system using Reagent Kit v2 or v3. All experiments were performed in biological duplicate and averages reported.
Cytoribosome profiling
Cytoribosome profiling for data presented in Fig. 3b and Extended Data Fig. 8f was performed using sucrose density gradients as described43, except that CHX was omitted from cultures, and frozen cells were pulverized with lysis buffer containing CHX. Cytoribosome profiling for data presented in Fig. 4d,e and Extended Data Fig. 9b,c was performed using 1 M sucrose cushions in place of gradients. Sequencing libraries were prepared identically to those for mitoribosome profiling (above). Sequencing was performed on an Illumina NextSeq 500 system. Each experiment presented was performed once as cytoribosome profiling is highly reproducible44,45. Additionally, trends are reproducible between experiments performed with density gradients and cushions.
mRNA sequencing
Total RNA was isolated prior to RNase I digestion from a portion of the thawed lysates described above. ERCC RNA Spike-In Mix (Thermo Fisher Scientific), a mixture of 92 in vitro synthesized transcripts, was added in equal volume across samples that were prepared from equal cell numbers. 50 μg of the total RNA with Spike-in mix was digested with 3 U RQ1 RNase-free DNase (Promega) for 30 min at 37 °C. 5 μg of purified RNA was then subjected to rRNA depletion using the Ribo-Zero Magnetic Gold Kit for yeast (Epicentre) according to the manufacturer's instructions.
Sequencing libraries were prepared as above following fragmentation by alkaline hydrolysis in 5 mM Na2CO3, 45 mM NaHCO3, 1 mM EDTA, pH 9.3 for 25 min at 95 °C and gel isolation of 30–70 nt fragments unless otherwise noted (see Extended Data Fig. 4b). Sequencing was performed on an Illumina NextSeq 500 system. Each experiment was performed once.
Data analysis
Raw sequences were processed by first removing 3’ adaptor sequence using Cutadapt46 and removing the first nucleotide from the 5’ end of all reads (except where otherwise noted) because we observed, as has been previously reported43, that it frequently represents untemplated addition by reverse transcriptase Supercript III (Thermo Fisher). Next, reads mapping to non-coding RNAs were removed by aligning using Bowtie147 to a collection of RNA genes downloaded from Saccharomyces Genome Database. Notably, we allowed a 3-nt 3’ mismatch when mapping to tRNAs to account for non-templated CCA addition on mature tRNAs. Remaining reads were then aligned allowing two mismatches to the S. cerevisiae genome assembly R64 (UCSC: sacCer3) using Tophat248. We also aligned separately to the nuclear and mitochondrial genomes to determine proportions of each library mapping to each.
To determine the A-site position in mitoribosome footprints we observed the first reads mapping to each ORF, which overlap the start codons. Consistently for 36-40 nt reads, the 3’ ends of these reads were 19 nt downstream of the start codon (mitoribosome P site). The 5’ ends were more heterogeneous depending on length of the read. Thus we assigned A sites for each read as 16 nt from the 3’ end.
RNA-seq normalization to spike-ins was performed by dividing the raw read count at each position by the number of spike-in reads in the library (rps, reads per spike-in). The number of spike-in reads is a proxy for cell number (see above: mRNA sequencing). RNA-seq reads were also normalized to total nuclear or mitochondrial mRNA mappers, as were cytoribosome profiling and mitoribosome profiling reads, respectively (rpm, reads per million).
To determine expression values for each gene, rps or rpm values were summed across ORFs and normalized to ORF length (RPKS or RPKM, normalized reads per kb). For footprint reads, the first and last five codons were excluded to remove effects of translation initiation and termination49. Translation efficiency (TE) was calculated by dividing cytoribosome footprint RPKM values by nuclear-mapping RNA-seq RPKM values and mitoribosome footprint RPKM values by mito-mapping RNA-seq RPKM values.
Scripts for A-site assignment, normalization, TE calculation, and other text file manipulations were written for Python 2.7.5. Plots and genome browser visualization were generated using R version 3.2.2 and Bioconductor. Heat maps were generated with Matrix2png (http://www.chibi.ubc.ca/matrix2png/).
For analysis of mitoribosome footprints by northern blotting, phosphoimager signals were quantified using ImageJ (http://imagej.nih.gov/) and normalized to the amount of mitoribosome recovered as measured by MrpS17-FLAG in elution. Comparisons are made only between samples on the same membrane probed at the same time with the same probe.
Extended Data
Extended Data Figure 1. OXPHOS proteins are induced during mitochondrial biogenesis.
Western blot analysis of mitochondrial (Cob, Cox1, Cox2) and nuclear (Cox4) OXPHOS proteins compared to FLAG-tagged mitoribosome small subunit protein MrpS17 and GAPDH. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 2. Dynamics of non-OXPHOS RNAs through mitochondrial biogenesis.
a, b, c, RNA levels (reads per kb) normalized to spike-in controls and plotted as -fold change compared to levels in log phase glucose growth for (a) all nuclear-encoded structural components of the complexes shown, (b) intron-encoded maturases, and (c) nuclear and mitochondrial-encoded mitoribosome subunits. To calculate values for maturase transcripts, only reads not overlapping the main ORF (COX1 or COB) were considered. Group II intron splicing intermediates stably accumulate and may not represent translation-competent transcripts.
Extended Data Figure 3. Optimization of affinity purification for intact mitoribosomes.
a, Spot tests verifying tagged mitoribosome subunits are functional as they support respiratory growth on glycerol (YPG). ρ0 is a strain without mitochondrial DNA. b, Frequency of petite colonies in our corrected S288c strain (see Methods) after growth for 5 days on 0.1% glucose + 3% glycerol. BY4742 is S288c background with designer auxotrophies. Σ1278b is a strain with wild-type HAP1, and a high-fidelity allele of MIP1, MIP1[Σ], along with other differences compared to S288c. Error bars show variation due to counting, with 175–750 colonies counted for each sample. c, Lysis and IP buffer conditions affect mitoribosome subunit association and thus footprint retention. Left panel: silver staining after IP of the large subunit (LSU) with Mrp20-F and of the small subunit (SSU) with MrpS17-F in Condition 1 (20 mM Tris, pH 8.0, 200 mM KCl, 5 mM MgCl2, 0.5% lauryl maltoside), and in Condition 2 (10 mM Tris, pH 8.0, 50 mM NH4Cl, 10 mM MgCl2, 0.5% lauryl maltoside). Arrowheads indicate bands that appear in both IPs in Condition 2 that can be assigned to the LSU or SSU by comparison to Condition 1. Asterisks mark the expected mobility of the tagged proteins. Right panel: Northern blotting of the co-purifying RNA in each condition. For gel source data, see Supplementary Fig. 1. d, Western blot showing fractions from IP using optimized buffer conditions. FLAG IP targeting the mitoribosome SSU co-purifies an HA-tagged LSU protein. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 4. Mitoribosome profiling is robust, reproducible, and does not require translation inhibitors.
a, Mapping statistics for representative mitoribosome and cytoribosome profiling libraries from log phase glycerol-grown cells. b, Fraction of reads mapping to each frame of mitochondrial ORFs (left panel) and nuclear ORFs (right panel) in mitoribosome profiling and cytoribosome profiling data, respectively. RNA-seq reads in the left panel were treated identically to footprint reads, including size selection for library generation. c, Reproducibility between biological replicates. Each dot corresponds to the number of reads mapped to a particular position on mRNA (rpm, left panel), or summed number of reads mapped across each mRNA then normalized to length (RPKM, right panel). d, Reproducibility with and without translation inhibitors thiamphenicol (50 μg/mL) and GMPPNP (1 mM).
Extended Data Figure 5. Mitochondrial and cytosolic protein synthesis on OXPHOS mRNAs is rapidly regulated.
a, b, -Fold change in relative protein synthesis (footprint RPKM values) compared to log phase glucose growth for the OXPHOS subunits synthesized in (a) the mitochondria (values are averages of two experiments) and (b) the cytosol. Asterisks on heat maps indicate the subunits shown in the line plots.
Extended Data Figure 6. Mitoribosome TE -fold changes are reproducible.
–Fold change data identical to that shown in Fig. 3a, but including range bars for two experiments performed from independent cultures on different days (left panel), and -fold change TE data plotted as a scatter with the Pearson correlation coefficient (right panel). Dotted lines mark 2-fold difference. RNA-seq data used in calculating TE is from a single experiment.
Extended Data Figure 7. Global translation is transiently inhibited upon shift to glycerol.
Polysome profiles from samples used for cytoribosome profiling, but without RNase I treatment. Gradients were loaded with lysate from equal cell numbers, allowing overall ribosome abundances to be compared between samples. Doubling time during log phase in glucose is ~1.2 h, and in glycerol is ~3.7 h.
Extended Data Figure 8. Cytosolic translation controls mitochondrial translation response.
a, FACS analysis of yeast cultures treated with CCCP. Wild-type cultures were grown in glucose to mid-log phase and treated with 40 μM of CCCP for the indicated times. Mitochondrial membrane potential (ΔΨm) was assessed using 1 μM tetramethylrhodamine (TMRM) that only a fraction of the cell population takes up (17.9% in this experiment). TMRM accumulates inside negatively charged mitochondria producing increased fluorescence intensity (102). Loss of membrane potential dissipates probe, measured as loss of high-intensity fluorescence. b, Representative northern blots for data in Fig. 4b,c and Extended Data Fig. 8e. For quantification, northern signals were normalized by relative mitoribosome recovery measured by MrpS17-F signal in western blots. For gel source data, see Supplementary Fig. 1. c, Northern blotting of total RNA for the indicated transcripts. For gel source data, see Supplementary Fig. 1. d, Quantification of viability assay. Cells were grown in YPD (Glu) or YPG (Gly) with or without drug for the time indicated. Cells were washed out of drug and plated on YPD for calculation of colony-forming units. CHX (100 μg mL−1). Pent: pentamidine (10 μM). CCCP (40 μM). e, Mitochondrial translation response, measured by northern blotting for footprints (see b), to inhibition of mitochondrial import with CCCP. f, -Fold change in synthesis measured by cytoribosome profiling of the nuclear-encoded mitochondrial mRNA-specific translational activators (color-coded by mRNA target). For each mitochondrial mRNA, the names of the known translation activators is listed.
Extended Data Figure 9. Cytosolic OXPHOS translation response is independent of mitochondrial gene expression.
a, Metabolic labeling to measure mitochondrial translation (Mito Tln), detectable only in the presence of CHX, and cytosolic translation (Cyto Tln). Samples generated in the absence of CHX were diluted 15-fold prior to loading the gel compared to samples generated with CHX. Mitochondrial translation products are labeled. b,c, Full dataset for experiment presented in Fig. 4d,e, showing -fold change in translation efficiencies (TEs) of all nuclear-encoded Complex III, Complex IV, and ATP synthase subunits measured by cytoribosome profiling (b) without (–Pent) or with (+Pent) inhibition of mitochondrial translation, and (c) in ρ0 cells, which have neither mitochondrial translation nor functional OXPHOS complexes.
Extended Data Figure 10. Verification of mtDNA loss in ρ0 strain.
a, Spot tests verifying that the ρ0 strain generated by overnight growth in ethidium bromide (see Methods) cannot respire (no growth on YPG). b, PCR (left panel) and qPCR (right panel) verifying loss of mitochondrial-encoded genes COX1, COX3, and 21S mitochondrial rRNA gene. MRPS17 is nuclear-encoded. Bars show s.e.m for technical triplicates.
Supplementary Material
Acknowledgements
We thank F. Winston, T. Fox, G. Brar and members of the Churchman lab for advice and discussions. We thank members of the O'Shea, Novina, and Springer labs for use of equipment and advice. We thank M. Hickman and D. Botstein for sharing the HAP1+ strain. Research supported by a Damon Runyon Cancer Research Foundation Frey Award (to L.S.C.), a Burroughs Wellcome Fund Career Award at the Scientific Interface (to L.S.C.), and an Ellison Medical Foundation New Scholar in Aging Award (to L.S.C.), the National Institutes of Health F32 (to M.T.C.), and a Boehringer Ingelheim Fonds PhD Fellowship (to G.S.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions M.T.C. and L.S.C. designed the research and wrote the manuscript. M.T.C. conducted the experiments with help from I.C.S. who performed mitoribosome profiling with drug treatments and G.S. who created and performed mitoribosome profiling on the pet111Δ strain. M.T.C. analyzed the data. All authors discussed the results and commented on the manuscript.
Author Information All data are deposited in Gene Expression Omnibus (accession number GSE74454). The authors declare no competing financial interests.
References
- 1.Masters BS, Stohl LL, Clayton DA. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 2.Faye G, Sor F. Analysis of mitochondrial ribosomal proteins of Saccharomyces cerevisiae by two dimensional polyacrylamide gel electrophoresis. Mol. Gen. Genet. 1977;155:27–34. doi: 10.1007/BF00268557. [DOI] [PubMed] [Google Scholar]
- 3.Kehrein K, Bonnefoy N, Ott M. Mitochondrial protein synthesis: efficiency and accuracy. Antioxidants & redox signaling. 2013;19:1928–1939. doi: 10.1089/ars.2012.4896. doi:10.1089/ars.2012.4896. [DOI] [PubMed] [Google Scholar]
- 4.Bonitz SG, et al. Codon recognition rules in yeast mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 1980;77:3167–3170. doi: 10.1073/pnas.77.6.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costanzo MC, Fox TD. Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annual review of genetics. 1990;24:91–113. doi: 10.1146/annurev.ge.24.120190.000515. doi:10.1146/annurev.ge.24.120190.000515. [DOI] [PubMed] [Google Scholar]
- 6.Green-Willms NS, Butler CA, Dunstan HM, Fox TD. Pet111p, an inner membrane-bound translational activator that limits expression of the Saccharomyces cerevisiae mitochondrial gene COX2. The Journal of biological chemistry. 2001;276:6392–6397. doi: 10.1074/jbc.M009856200. doi:10.1074/jbc.M009856200. [DOI] [PubMed] [Google Scholar]
- 7.Herrmann JM, Woellhaf MW, Bonnefoy N. Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochimica et biophysica acta. 2013;1833:286–294. doi: 10.1016/j.bbamcr.2012.03.007. doi:10.1016/j.bbamcr.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Muller PP, et al. A nuclear mutation that post-transcriptionally blocks accumulation of a yeast mitochondrial gene product can be suppressed by a mitochondrial gene rearrangement. Journal of molecular biology. 1984;175:431–452. doi: 10.1016/0022-2836(84)90178-5. [DOI] [PubMed] [Google Scholar]
- 9.Fraenkel DG. Yeast Intermediary Metabolism. Cold Spring Harbor Press; 2011. [Google Scholar]
- 10.Kuhn KM, DeRisi JL, Brown PO, Sarnow P. Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Molecular and cellular biology. 2001;21:916–927. doi: 10.1128/MCB.21.3.916-927.2001. doi:10.1128/MCB.21.3.916-927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amiott EA, Jaehning JA. Mitochondrial transcription is regulated via an ATP “sensing” mechanism that couples RNA abundance to respiration. Molecular cell. 2006;22:329–338. doi: 10.1016/j.molcel.2006.03.031. doi:10.1016/j.molcel.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 12.Mueller DM, Getz GS. Steady state analysis of mitochondrial RNA after growth of yeast Saccharomyces cerevisiae under catabolite repression and derepression. The Journal of biological chemistry. 1986;261:11816–11822. [PubMed] [Google Scholar]
- 13.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 14.Roberts GG, Hudson AP. Transcriptome profiling of Saccharomyces cerevisiae during a transition from fermentative to glycerol-based respiratory growth reveals extensive metabolic and structural remodeling. Molecular genetics and genomics : MGG. 2006;276:170–186. doi: 10.1007/s00438-006-0133-9. doi:10.1007/s00438-006-0133-9. [DOI] [PubMed] [Google Scholar]
- 15.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fox TD, et al. Analysis and manipulation of yeast mitochondrial genes. Methods in enzymology. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- 17.Ashe MP, De Long SK, Sachs AB. Glucose depletion rapidly inhibits translation initiation in yeast. Molecular biology of the cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. doi:10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rooijers K, Loayza-Puch F, Nijtmans LG, Agami R. Ribosome profiling reveals features of normal and disease-associated mitochondrial translation. Nature communications. 2013;4:2886. doi: 10.1038/ncomms3886. doi:10.1038/ncomms3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vignais PV, Stevens BJ, Huet J, Andre J. Mitoribosomes from Candida utilis. Morphological, physical, and chemical characterization of the monomer form and of its subunits. The Journal of cell biology. 1972;54:468–492. doi: 10.1083/jcb.54.3.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolin SL, Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. The EMBO journal. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poutre CG, Fox TD. PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics. 1987;115:637–647. doi: 10.1093/genetics/115.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simpson CE, Ashe MP. Adaptation to stress in yeast: to translate or not? Biochemical Society transactions. 2012;40:794–799. doi: 10.1042/BST20120078. doi:10.1042/BST20120078. [DOI] [PubMed] [Google Scholar]
- 24.Poyton RO, Bellus G, McKee EE, Sevarino KA, Goehring B. In organello mitochondrial protein and RNA synthesis systems from Saccharomyces cerevisiae. Methods in enzymology. 1996;264:36–42. doi: 10.1016/s0076-6879(96)64006-3. [DOI] [PubMed] [Google Scholar]
- 25.Lamb AJ, Clark-Walker GD, Linnane AW. The biogenesis of mitochondria. 4. The differentiation of mitochondrial and cytoplasmic protein synthesizing systems in vitro by antibiotics. Biochimica et biophysica acta. 1968;161:415–427. [PubMed] [Google Scholar]
- 26.Rak M, Su CH, Xu JT, Azpiroz R, Singh AM, Tzagoloff A. Regulation of mitochondrial translation of the ATP8/ATP6 mRNA by Smt1p. Mol. Biol. Cell. 2016 doi: 10.1091/mbc.E15-09-0642. doi:10.1091/mbc.E15-09-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun T, Zhang Y. Pentamidine binds to tRNA through non-specific hydrophobic interactions and inhibits aminoacylation and translation. Nucleic acids research. 2008;36:1654–1664. doi: 10.1093/nar/gkm1180. doi:10.1093/nar/gkm1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Bell A, Perlman PS, Leibowitz MJ. Pentamidine inhibits mitochondrial intron splicing and translation in Saccharomyces cerevisiae. Rna. 2000;6:937–951. doi: 10.1017/s1355838200991726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu B, Qian SB. Translational reprogramming in cellular stress response. Wiley interdisciplinary reviews. RNA. 2014;5:301–315. doi: 10.1002/wrna.1212. doi:10.1002/wrna.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pavlov MY, Ehrenberg M. Optimal control of gene expression for fast proteome adaptation to environmental change. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20527–20532. doi: 10.1073/pnas.1309356110. doi:10.1073/pnas.1309356110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. doi:10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gruschke S, et al. The Cbp3-Cbp6 complex coordinates cytochrome b synthesis with bc(1) complex assembly in yeast mitochondria. The Journal of cell biology. 2012;199:137–150. doi: 10.1083/jcb.201206040. doi:10.1083/jcb.201206040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez-Martinez X, Butler CA, Shingu-Vazquez M, Fox TD. Dual functions of Mss51 couple synthesis of Cox1 to assembly of cytochrome c oxidase in Saccharomyces cerevisiae mitochondria. Molecular biology of the cell. 2009;20:4371–4380. doi: 10.1091/mbc.E09-06-0522. doi:10.1091/mbc.E09-06-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehrenreich IM, et al. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature. 2010;464:1039–1042. doi: 10.1038/nature08923. doi:10.1038/nature08923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaisne M, Becam AM, Verdiere J, Herbert CJ. A ‘natural’ mutation in Saccharomyces cerevisiae strains derived from S288c affects the complex regulatory gene HAP1 (CYP1). Current genetics. 1999;36:195–200. doi: 10.1007/s002940050490. [DOI] [PubMed] [Google Scholar]
- 36.Baruffini E, Lodi T, Dallabona C, Foury F. A single nucleotide polymorphism in the DNA polymerase gamma gene of Saccharomyces cerevisiae laboratory strains is responsible for increased mitochondrial DNA mutability. Genetics. 2007;177:1227–1231. doi: 10.1534/genetics.107.079293. doi:10.1534/genetics.107.079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young MJ, Court DA. Effects of the S288c genetic background and common auxotrophic markers on mitochondrial DNA function in Saccharomyces cerevisiae. Yeast. 2008;25:903–912. doi: 10.1002/yea.1644. doi:10.1002/yea.1644. [DOI] [PubMed] [Google Scholar]
- 38.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moqtaderi Z, Struhl K. Expanding the repertoire of plasmids for PCR-mediated epitope tagging in yeast. Yeast. 2008;25:287–292. doi: 10.1002/yea.1581. doi:10.1002/yea.1581. [DOI] [PubMed] [Google Scholar]
- 40.Storici F, Resnick MA. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods in enzymology. 2006;409:329–345. doi: 10.1016/S0076-6879(05)09019-1. doi:10.1016/S0076-6879(05)09019-1. [DOI] [PubMed] [Google Scholar]
- 41.Dimitrov LN, Brem RB, Kruglyak L, Gottschling DE. Polymorphisms in multiple genes contribute to the spontaneous mitochondrial genome instability of Saccharomyces cerevisiae S288C strains. Genetics. 2009;183:365–383. doi: 10.1534/genetics.109.104497. doi:10.1534/genetics.109.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gouget K, Verde F, Barrientos A. In vivo labeling and analysis of mitochondrial translation products in budding and in fission yeasts. Methods in molecular biology. 2008;457:113–124. doi: 10.1007/978-1-59745-261-8_8. [DOI] [PubMed] [Google Scholar]
- 43.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosomeprotected mRNA fragments. Nature protocols. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. doi:10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brar GA, et al. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335:552–557. doi: 10.1126/science.1215110. doi:10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ingolia NT. Genome-wide translational profiling by ribosome footprinting. Methods in enzymology. 2010;470:119–142. doi: 10.1016/S0076-6879(10)70006-9. doi:10.1016/S0076-6879(10)70006-9. [DOI] [PubMed] [Google Scholar]
- 46.Martin M. Cutadapt removes adaptor sequences from high-thoughput sequencing reads. EMBnet.journal. 2011;17 doi:DOI:10.14806/ej.17.1.200. [Google Scholar]
- 47.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. doi:10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. doi:10.1186/gb-2013- 14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. doi:10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.