Abstract
The current decrease of new drugs brought to the market has fostered renewed interest in plant-based drug discovery. Given the alarming rate of biodiversity loss, systematic methodologies in finding new plant-derived drugs are urgently needed. Medicinal uses of plants were proposed as proxy for bioactivity, and phylogenetic patterns in medicinal plant uses have suggested that phylogeny can be used as predictive tool. However, the common practice of grouping medicinal plant uses into standardised categories may restrict the relevance of phylogenetic predictions. Standardised categories are mostly associated to systems of the human body and only poorly reflect biological responses to the treatment. Here we show that medicinal plant uses interpreted from a perspective of a biological response can reveal different phylogenetic patterns of presumed underlying bioactivity compared to standardised methods of medicinal plant use classification. In the cosmopolitan and pharmaceutically highly relevant genus Euphorbia L., identifying plant uses modulating the inflammatory response highlighted a greater phylogenetic diversity and number of potentially promising species than standardised categories. Our interpretation of medicinal plant uses may therefore allow for a more targeted approach for future phylogeny-guided drug discovery at an early screening stage, which will likely result in higher discovery rates of novel chemistry with functional biological activity.
Plants have played a central role in human health-care since ancient times1,2. Although the plant domain is widely regarded as natural capital with potential to yield new drugs, this potential is under threat due to the alarming biodiversity loss, with recent estimates indicating that every fifth plant species on earth is threatened with extinction3. Unlocking the potential of plants in health-care therefore urges for a time-efficient and systematic approach.
Following the assumption that plant-derived chemicals are constrained to evolutionary plant lineages4,5,6, phylogeny-guided approaches have been seen as one of the time-efficient and informed approaches to plant-based drug discovery7,8,9,10,11,12,13,14. Some of these approaches utilise information from ethnomedicine: the use of plants by humans as medicines15. Reports of medicinal plant uses are employed as a proxy for bioactivity and are superimposed on phylogenetic trees e.g.8,11,14. Evolutionary methodologies then predict potential bioactivity of different plant lineages, based on the distribution of medicinal plant use across the phylogeny.
One potential limitation of the phylogenetic approach lies in the classification of medicinal plant use. Prior to being analysed in a phylogenetic context, documented plant medicinal uses are collected and classified according to the diseases they are used to treat as typically done in ethnomedicinal research16. For this purpose, both internationally recognised medical standards such as the International Classification of Diseases (ICD; http://www.who.int/classifications/icd/en/) of the WHO, as well as a classification system developed in the field of Economic Botany (Economic Botany Data Collection Standard, EBDCS17) are widely used16. Although these classification systems are useful to guarantee consistency, data exchange and comparability among different studies16,18, they have two major drawbacks. First, they do not fully capture the complexity and idiosyncrasy of local plant-based healthcare6,19,20. Second, and most important, they are based on categories reflecting systems of the human body (e.g. digestive system) or symptoms. Not only are classifications based on systems of the body affected or common symptoms little informative for disease etiology21, but they also allow very little insight into the potential underlying biological activity of the medicinal plants. In recent years cellular and molecular mechanisms underlying diseases have been extensively studied22 and aided the discovery of disease etiology. We postulate that in a phylogenetic context a classification based on a biological response provoked by the treatment may reflect more accurately the cellular or molecular mechanisms underlying the disease’s treatment and thus provides a more accurate proxy of the biological activities of the plant-derived compounds and therefore enables a more accurate phylogenetic prediction of distinct biological activities triggered by the plants (Fig. 1).
Figure 1. Hypothetical distribution of medicinal plant uses across a phylogeny.
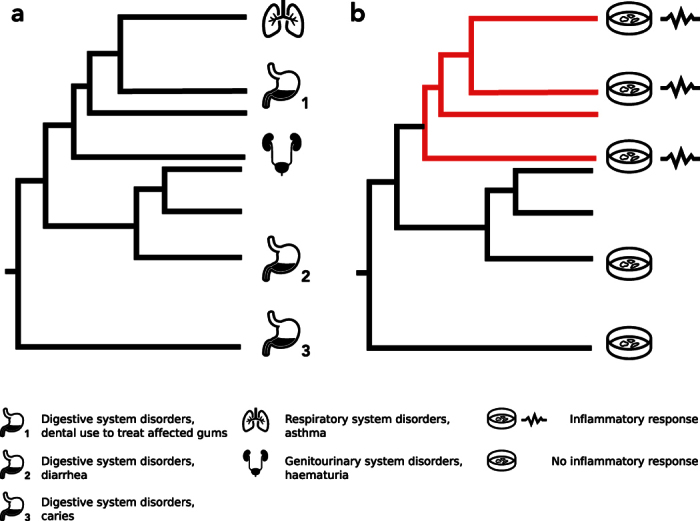
(a) Plant medicinal uses as classified by the Economic Botany Data Collection Standard based on systems of the body (b) Same plant medicinal uses classified based on a biological response. When seeking lineages with potential agents modulating an inflammatory response, the classification in (a) is not informative. Instead, the classification in (b) allows us to identify clades (marked in red in (b)) that are overrepresented in species potentially modulating an inflammatory response. Icons: thenounproject.com.
Here, we investigate the influence of medicinal plant use classification in a phylogenetic context, using the genus Euphorbia L. (Euphorbiaceae) as an example. With about 2,000 recognized species23,24, Euphorbia is among the three largest genera of angiosperms, with a near-cosmopolitan distribution and remarkable morphological diversity, including annual herbs, succulents and large trees, united by a unique, flower-like inflorescence and often poisonous, milky latex24,25. The molecular phylogenetics of the genus has been extensively studied24,26,27,28,29,30 and a large compendium of data on the medicinal uses is available31. The diterpenoid ingenol mebutate, a chemical compound isolated from Euphorbia peplus L. is marketed as a drug for the treatment of actinic keratosis, a precancerous skin condition32. Nevertheless ingenol mebutate is only obtained in extremely low quantities from the plant, making its production inefficient33. Alternative sources are therefore desired and investigated33. Euphorbia exemplifies the need for a systematic approach to plant-based drug discovery: with approximately 5% of species in the genus chemically investigated34, how do we go about prioritizing which of the remaining 1,900 species to investigate?
We follow the core hypothesis that phylogenetic patterns in medicinal properties are underlied by similarities in phytochemical properties. We apply two interpretative approaches to the medicinal uses of Euphorbia – one following a standard classification system used in ethnomedicinal studies, the other aiming to identify plant uses modulating the inflammatory response. Inflammatory processes are highly relevant in the treatment of actinic keratosis by ingenol mebutate32. We scrutinize phylogenetic patterns in species recovered by both approaches and describe the influence of classification of medicinal plant use on the Euphorbia species identified for potential early drug discovery by selected phylogenetic measures. We refer to medicinal properties as properties described from ethnomedicinal research31. Data presented in this study are exclusively based on ethnomedicinal use reports of species of Euphorbia31 and do not include new chemical data on the genus Euphorbia. Instead, we refer to two extensive reviews on the chemical and pharmacological properties of species of the genus Euphorbia34,35. The context for the data presented in this study is therefore an assessment of an in vitro early stage screening in drug discovery, before parameters such as drugability, safety, formulation or comparative effectiveness of isolated compounds are considered.
Results
We classified ethnomedicinal data using two different approaches: one that is used commonly in ethnomedicinal studies and one based on the biological response to the treatment. In specific, we identified plant uses modulating the inflammatory response. Inflammation is a prominent biological response to ingenol mebutate, which plays an important role in the treatment of actinic keratosis32,36. The first approach (EBDCS) is widely used in ethnomedicinal studies18 and classifies uses of plants into standardized descriptors and terms in hierarchical level states17. Level 1 states are subdivided into 13 categories (e.g. food, materials, fuels, medicines, vertebrate poisons etc.), whereas level 2 states break down level 1 states in more detail17. The level 2 states of the use category medicines described by the EBDCS are mostly linked to systems of the body or symptoms of a disease, allowing only little insight into the disease’s cause or assumptions on a distinct biological activity or chemical nature of the medicinal plants. On contrary, our interpretation of plant medicinal uses gives a better insight into how the plant’s chemicals might interfere in the disease process (pharmacological effect37) and thus is a better proxy for the plant’s biological activity. Plant uses identified as modulating the inflammatory response were collected within a category we refer to as inflammatory response. We compared the category inflammatory response with the EBDCS categories investigating the effect of data organisation on evolutionary patterns relevant to early drug discovery.
First, using publicly available data, we produced the most comprehensive phylogenetic hypothesis of Euphorbia to date, including 560 Euphorbia species (>25% of the genus) representing all known subgeneric clades. In agreement with previous studies24,30 our topology (Supplementary Figure 1) confirms the presence of four subgeneric clades with posterior probability (PP) branch support values > 0.99 (except for subgenus Esula sensu lato PP = 0.90; subgenus Esula excluding E. lathyris, E. lagascae and E. phymatosperma PP = 0.99). Euphorbia subg. Esula (mainly herbaceous species with centre of diversity in temperate Europe29) is sister to the three remaining subgenera. Euphorbia subg. Athymalus (highly diverse succulent species with predominantly African distribution26,28) is sister to subgenera Euphorbia (most diverse species of the genus distributed across tropics and subtropics27) and Chamaesyce (diverse growth forms including many New World species26) (Supplementary Figure 1). With regard to relationships below the subgenus level, despite relatively low resolution as would be expected due to the use of only one marker (ndhF), no well supported (PP > 0.95) incongruences with previous studies were found26,27,28,29.
We then investigated the phylogenetic distribution of medicinal plants on the phylogeny of Euphorbia. We found that plants used medicinally in the genus are significantly phylogenetically clustered (median = 0.76, p(D < 1): ***, Table 1). The EBDCS level 2 state categories of medicines that showed weak to moderate phylogenetic signal included genitourinary system disorders and unspecified medicinal disorders (Table 2).
Table 1. Phylogenetic signal per EBDCS level 1 state categories.
| N | Prevalence | D-statistic |
Phylogenetic signal |
|||
|---|---|---|---|---|---|---|
| Median | Range | p(D < 1)a | strengthb | |||
| Animal food | 12 | 0.02 | 0.98 | 0.55–1.47 | ns | weak |
| Environmental uses | 18 | 0.03 | 0.90 | 0.56–1.27 | ns | weak |
| Materials | 15 | 0.03 | 0.88 | 0.61–1.21 | ns | weak |
| Medicines | 65 | 0.12 | 0.76 | 0.65–0.88 | *** | weak |
| Non-Vertebrate poisons | 9 | 0.02 | 0.85 | 0.39–1.57 | ns | weak |
| Social uses | 8 | 0.01 | 0.82 | 0.15–1.51 | ns | weak |
| Vertebrate poisons | 46 | 0.08 | 1.06 | 0.90–1.26 | ns | weak |
Phylogenetic signal (D-statistic) on a randomly selected subset of 1,000 Bayesian trees within the 95% credible set of uses of Euphorbia31 classified into level 1 state categories according to the Economic Botany Data Collection Standard (EBDCS). Out of all level 1 state categories only medicines shows phylogenetic signal (in bold). N: Number of species.
a* 95% p < 0.05; *** all p < 0.005.
bweak: <90% p(D > 0) > 0.05; moderate: 90% p(D > 0) > 0.05; strong: 95% p(D > 0) > 0.05; very strong: all p(D > 0) > 0.05.
Table 2. Phylogenetic signal per EBDCS level 2 state categories medicines and the category inflammatory response.
| N | Prevalence | D-statistic |
Phylogenetic signal |
|||
|---|---|---|---|---|---|---|
| Median | Range | p(D < 1)a | strengthb | |||
| Abnormalities | 12 | 0.02 | 0.83 | 0.48–1.20 | ns | weak |
| Digestive system disorders | 26 | 0.05 | 0.78 | 0.51–1.04 | ns | weak |
| Genitourinary system disorders | 9 | 0.02 | 0.35 | −0.06–0.87 | * | moderate |
| Infections/infestations | 21 | 0.04 | 0.82 | 0.50–1.15 | ns | weak |
| Inflammation | 11 | 0.02 | 0.54 | 0.09–1.27 | ns | weak |
| Injuries | 16 | 0.03 | 0.88 | 0.52–1.25 | ns | weak |
| Pain | 15 | 0.03 | 0.78 | 0.43–1.19 | ns | weak |
| Respiratory system disorders | 10 | 0.02 | 0.89 | 0.39–1.38 | ns | weak |
| Skin-/subcutaneous cellular tissue disorders | 27 | 0.05 | 0.73 | 0.50–0.95 | ns | weak |
| Unspecified medicinal disorders | 27 | 0.05 | 0.65 | 0.43–0.88 | * | weak |
| Inflammatory response | 44 | 0.08 | 0.88 | 0.68–1.03 | ns | weak |
| No inflammatory response | 12 | 0.02 | 0.85 | 0.49–1.29 | ns | weak |
| Unknown | 23 | 0.04 | 1.05 | 0.77–1.37 | ns | weak |
Phylogenetic signal (D-statistic) on a randomly selected subset of 1,000 Bayesian trees within the 95% credible set of uses of Euphorbia31 classified into level 2 state categories of medicines according to the Economic Botany Data Collection Standard (EBDCS) and the category inflammatory response. EBDCS categories genitourinary system disorders and unspecified medicinal disorders show phylogenetic signal (in bold), while the EBDCS category inflammation and the category inflammatory response don’t. N: Number of species.
a* 95% p < 0.05; *** all p < 0.005.
bweak: <90% p(D > 0) > 0.05; moderate: 90% p(D > 0) > 0.05; strong: 95% p(D > 0) > 0.05; very strong: all p(D > 0) > 0.05.
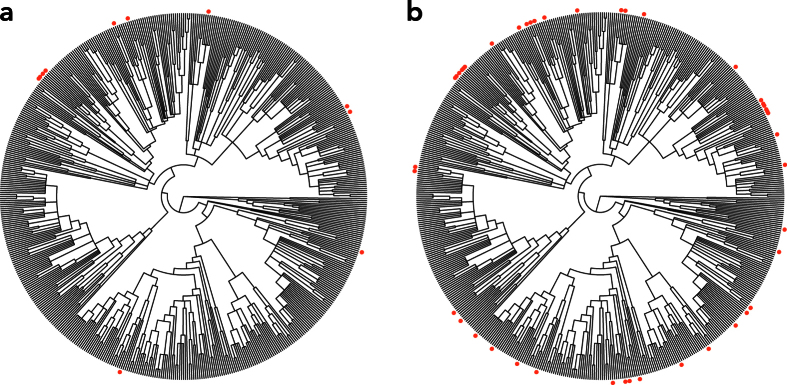
Of all EBDCS categories, the category inflammation is the most comparable to our interpretation of plant uses modulating the inflammatory response. However, in contrast to the EBDCS category inflammation, our category inflammatory response comprises not only plant species potentially causing an anti- but also a pro-inflammatory effect. In the interest of investigating differences resulting from the different classifications of medicinal plant uses, we subsequently compared the two inflammation-related categories. The category inflammatory response included 44 species, four times as many species as the EBDCS category inflammation (11 species; Fig. 2). There were also differences in the phylogenetic distribution of plants used for these two categories: A more than two-fold increase of the phylogenetic diversity index (PD) from the EBDCS category inflammation (median = 5.70, 7.40%; range = 4.71–6.79, 6.32–8.60%) to the category inflammatory response (median = 14.08, 18.36%; range = 11.96–16.58, 16.78–20.03%) was observed. Given the differences in the groups of plant species identified by the different inflammation-related categories, we further investigated whether plant species from these two categories come from the same lineages. We estimated the phylogenetic similarity between these two plant groups using the MNTD metric. The category inflammatory response showed no significant similarity to the EBDCS category inflammation (median = 0.40, p-value: ns, Table 3). It also showed no significant phylogenetic overlap with any of the other EBDCS level 2 state medicinal categories (Table 3). However, neither the EBDCS inflammation category, nor the inflammatory response category showed significant phylogenetic clustering (median = 0.54 and 0.88, p(D < 1): ns, Table 2). As a tentative approach to narrow down the number of species selected for bioactivity screening within the category inflammatory response, we identified nodes that are significantly overrepresented by species in this category (hot nodes8; Fig. 3). Hot nodes were mainly found within subgenera Chamaesyce and Euphorbia.
Figure 2.
Phylogenetic distribution of species for (a) the Economic Botany Data Collection Standard (EBDCS) category inflammation and (b) the category inflammatory response. Red dots indicate species with documented use described in the category.
Table 3. Phylogenetic similarity between the category inflammatory response and the EBDCS level 2 state categories medicines.
| N | Prevalence | MNTD |
|||
|---|---|---|---|---|---|
| Median | Range | p-value | |||
| Abnormalities | 12 | 0.02 | 0.35 | 0.29–0.42 | ns |
| Digestive system disorders | 26 | 0.05 | 0.19 | 0.16–0.22 | ns |
| Genitourinary system disorders | 9 | 0.02 | 0.59 | 0.46–0.71 | ns |
| Infections/infestations | 21 | 0.04 | 0.14 | 0.12–0.18 | ns |
| Inflammation | 11 | 0.02 | 0.40 | 0.33–0.49 | ns |
| Injuries | 16 | 0.03 | 0.25 | 0.21–0.30 | ns |
| Pain | 15 | 0.03 | 0.29 | 0.24–0.34 | ns |
| Respiratory system disorders | 10 | 0.02 | 0.40 | 0.34–0.47 | ns |
| Skin-/subcutaneous cellular tissue disorders | 27 | 0.05 | 0.15 | 0.13–0.19 | ns |
| Unspecified medicinal disorders | 27 | 0.05 | 0.18 | 0.16–0.21 | ns |
Mean nearest taxon distance (MNTD) between species in the Economic Botany Data Collection Standard (EBDCS) level 2 state medicines categories and the category inflammatory response. The category inflammatory response does not show significant phylogenetic similarity with any of the EBDCS categories and thus is sufficiently sensitive to highlight a different group of species, eventually reflecting unexplored medicinal potential not recovered by the EBDCS.
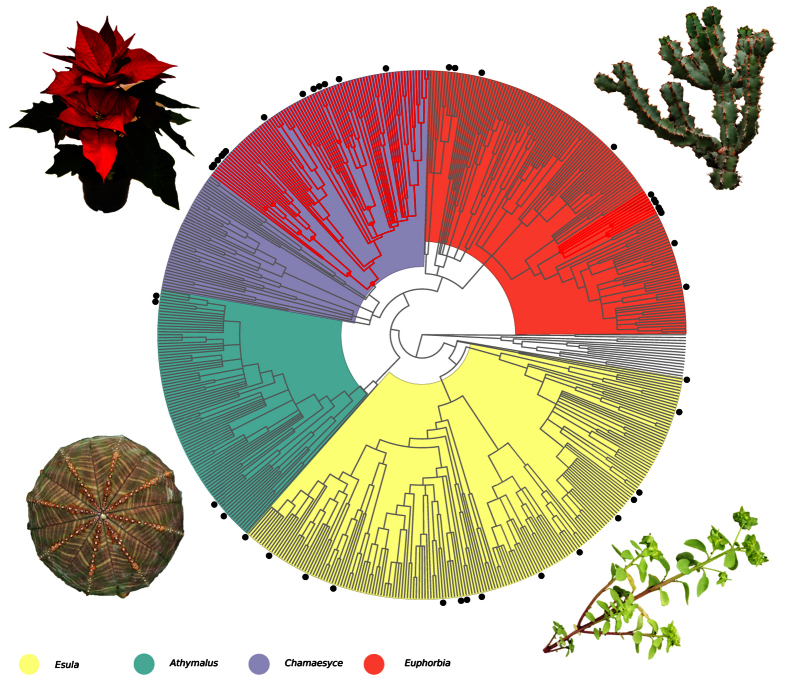
Figure 3. Hot nodes and corresponding clades of the category inflammatory response.
Hot nodes (red dots) were identified by the nodesigl command in PHYLOCOM v4.2 on the majority consensus tree. Hot nodes indicate that the observed number of species in the category in that node is higher than expected by chance. Black dots indicate species with documented uses in the category inflammatory response. Photo: Mogens Trolle and Madeleine Ernst (Euphorbia pulcherrima).
Discussion
Ethnomedicinal uses have inspired the discovery of many drugs in the past2,15,38. In recent decades, following the assumption that plant-derived chemicals are constrained to certain evolutionary plant lineages4,5,6, it was shown that plants used in ethnomedicine are not randomly distributed across taxonomic groups e.g.5,6,39. Taxonomic relationships were seen, therefore, as a way of predicting the occurrence and nature of useful chemicals in plants5,6. More recent studies have built upon this idea and with increasingly robust molecular phylogenetic methods at hand have added to the taxonomy-oriented approaches to informed drug discovery7,8,9,10,11,12,13,14.
In this study, we explored evolutionary patterns of medicinal properties in Euphorbia. Based on the mechanism of action of ingenol mebutate, we focused the investigation on described plant uses that possibly modulate an inflammatory response.
First, we looked at the phylogenetic distribution of medicinal plants on the phylogeny of Euphorbia. In agreement with previous studies on medicinal properties in other angiosperm lineages8,9,11,12,14, we found that Euphorbia species used medicinally are significantly phylogenetically clustered (Table 1). The EBDCS level 2 state categories of medicines that showed weak to moderate phylogenetic signal included genitourinary system disorders and unspecified medicinal disorders (Table 2). Biological activities specific to systems of the body (such as the genitourinary system) can be of highly diverse nature, therefore assumptions on distinct biological actions or chemical underpinnings of the signal cannot be made. In contrast, neither the EBDCS inflammation category, nor the inflammatory response category showed significant phylogenetic clustering. Our rationale in this study was that classification of medicinal plant use based on the biological response to the treatment can uncover phylogenetic patterns that reflect more accurately the presence of certain chemical compounds. Our findings suggest that inflammatory modulators are found in several Euphorbia lineages. This was recovered based on the EBDCS classification, but was even more pronounced by interpreting medicinal plant uses from a perspective of a biological response. There are several potential explanations for this pattern. First, plant chemicals can be homoplasious, occurring across distantly related lineages40. This may be due to convergent evolution, recycling of chemicals from the environment by the plant e.g.41 or production of chemicals by endophytic fungi, as well as environmental and ecological effects on plant chemistry10,40,42. Although some structural types of Euphorbia diterpenoids, to which also ingenol mebutate belongs, and which most likely exhibit pro-inflammatory properties36, have been described as taxonomically significant for the genus43, their distribution across the phylogeny of Euphorbia may still be random, since their production may be more strongly influenced by some of the above named factors, or is not significantly related to the evolutionary history of the genus. Conservation of gene clusters associated with diterpenoid biosynthesis was reported at a high taxonomic level of the plant family Euphorbiaceae44. This suggests that the ability of diterpenoid production is present throughout the genus Euphorbia44. However, especially some members of Euphorbia subg. Chamaesyce have been reported as not containing Euphorbia specific diterpenoids34, potentially having the responsible genes silenced according to ecological needs40. Second, bioactivity might not be related to any phylogenetic patterns, since different chemical compounds from different biosynthetic routes may show the same or similar bioactivities45. For example, structurally different chemicals derived from unrelated natural sources have shown inhibition of the tumour necrosis factor-α, a pro-inflammatory cytokine, which regulates inflammation and related disorders45. Inflammatory modulators targeted by the inflammatory response category can be of different chemical structural classes and might therefore not follow a distribution associated with the phylogeny of the genus. It is therefore plausible that the category inflammatory response comprises a broad spectrum of different chemical structures, which are not associated to the phylogenetic relationships of the plants producing them. Third, there is inherent bias in the collection of data on medicinal use, as it is a subjective process with several restrictions, such as underrepresentation of certain languages and access to literature not available online. Only a large-scale chemical exploration of Euphorbia species would be able to tease apart the underlying causes of the phylogenetic pattern observed here.
When comparing the EBDCS category inflammation to the inflammatory response category we observed a more than two-fold increase of the phylogenetic diversity index (PD). Not only were more species included in the category inflammatory response, but also their distribution over the phylogeny was wider. Furthermore, the category inflammatory response showed no significant similarity to the EBDCS category inflammation, according to the MNTD metric. It also showed no significant phylogenetic overlap with any of the other EBDCS level 2 state medicinal categories. Our findings thus indicate that the category inflammatory response highlighted a different group of species, and may reflect unexplored medicinal potential not recovered by a standard method of classification such as the EBDCS. As a tentative approach to narrow down the number of species to be screened at an early stage of drug discovery within the category inflammatory response, we identified nodes that are significantly overrepresented by species in this category (hot nodes; Fig. 3). Hot nodes were mainly found within subgenera Chamaesyce and Euphorbia, which together comprise over half of the diversity in Euphorbia (about 1,200 species24). These two subgenera have not been as extensively studied chemically as subgenus Esula (the subject of 70% chemical studies to date34,35). Despite mentioned limitations, we were able to highlight specific lineages with a potential overrepresentation of chemical compounds modulating an inflammatory response in humans, and these highlighted lineages are poorly studied for their chemistry. Therefore, our approach shows potential for novel discoveries of pharmacologically relevant compounds from understudied plant lineages.
Although plant-derived chemicals play a relatively minor role in drug discovery in the pharmaceutical industry nowadays, due to reasons related to the relatively labor-, time- and cost-intensive work with naturally derived chemicals, fear of duplication, intellectual property concerns or biodiversity conservation issues46,47,48 plant-derived drugs continue to be part of the list of essential medicines for priority diseases published by the WHO (WHO Model List of Essential Medicines, April 2015). Given the current decrease of new drugs brought to the market46, and the huge potential of plant-derived chemicals in providing medicinally relevant bioactivity, there is undoubtedly scope for innovation in identifying drug candidates. Our approach highlights clusters of species in Euphorbia subgenera Chamaesyce and Euphorbia, which compared to the European subgenus Esula, have largely been chemically under-investigated. Out of a total of 91 Euphorbia species, which have been investigated for their chemistry and pharmacology34,35, there are only five species of Euphorbia subgenus Chamaesyce and 24 species of Euphorbia subgenus Euphorbia (based on recent taxonomic and molecular phylogenetic studies24,26,27,28,29,30,49). The remaining 62 species form part of the European Euphorbia subgenus Esula.
Future large-scale chemical and pharmacological investigations of previously untested species will be able to show if selection based on a classification system reflecting the biological response to the treatment efficiently results in improved hit rates. The classification of plant medicinal uses proposed here was associated to the inflammatory response. The inflammatory response plays an important role in the efficacy of the treatment of actinic keratosis by ingenol mebutate, a diterpenoid isolated from Euphorbia peplus32,36. Despite its release to the market, ingenol mebutate is not sourced efficiently from Euphorbia peplus and alternative sources such as synthetic or biosynthetic approaches are being investigated33,44. Little is known about the biosynthetic pathway of Euphorbia diterpenoids and a synthetic route for ingenol mebutate production is not feasible yet33,44. Given the low percentage and uneven subgeneric distribution of species of the genus Euphorbia investigated chemically, it is likely to find species with higher production of ingenol mebutate or compounds with similar or other medicinally relevant bioactivity profiles. Based on our findings, chemical characterization and investigation of biological activities related to the inflammatory response of selected Euphorbia species will follow, using the evolutionary approach and methodologies of classification proposed in this study.
Methods
Phylogenetic hypothesis
In this study, we produced a more densely sampled phylogenetic hypothesis of Euphorbia compiling DNA sequence data from the plastid marker ndhF from a series of studies focusing on subgenera of the genus26,27,28,29. Our matrix included sequences (1789 base pairs) of 560 Euphorbia species (>25% of the genus) representing all known subgeneric clades (Supplementary Table 1). To root the tree and allow comparison with previous studies24,26,27,28,29,30, exemplars of 15 related genera representing the remainder of the Euphorbiaceae family were included as outgroup. Species names and corresponding GenBank accessions are listed in Supplementary Table 2. We produced a Bayesian phylogenetic hypothesis using the GTR + I + G model. All comparative phylogenetic analyses described below were performed on a randomly selected subset of 1,000 trees within the 95% credible set.
Medicinal uses of species of the genus Euphorbia
Information on uses of species of the genus Euphorbia was drawn from an extensive database31, including plants used medicinally as well as for other purposes; such as animal food, environmental uses, materials, (non-) vertebrate poisons and social uses. The database contains over 1,000 use records referring to 156 Euphorbia species, of which 92 (63%) were included in our phylogenetic tree. The use records from the database were coded according to the Economic Botany Data Collection Standard (EBDCS)17, recommended by the Biodiversity Information Standards TDWG (http://www.tdwg.org).
Identifying Euphorbia uses modulating an inflammatory response
In the present study we argue that standard approaches to medicinal plant use classification used in previous studies are potentially misleading for predictive purposes in early drug discovery. We propose that, when studying medicinal plant uses in a phylogenetic context, a classification system that reflects the biological response to the treatment may reflect more accurately the cellular or molecular mechanism of the condition that a plant is used to treat and thus can reveal more successfully underlying biological activities and chemical properties. To illustrate this we used the EBDCS as an example. The level 2 states of the use category medicines described by the EBDCS17 are mostly linked to systems of the human body or symptoms of a disease, allowing only little insight into the disease’s cause or assumptions on a distinct biological activity or chemical nature of the medicinal plants. Here, we explore an alternative way of classifying diseases treated by Euphorbia species. We suggest that the proposed classification gives a better insight into how the plant’s chemicals might interfere in the disease process (pharmacological effect37) and thus is a better proxy for the plant’s biological activity than a classification based on systems of the body.
We focus on described plant uses that suggest to modulate an inflammatory response. The inflammatory response is a protective response, which eliminates offending agents of cell injury (e.g., microbes, toxins) and its consequences (e.g., necrotic cells and tissues). On the other hand, inflammatory reactions also underlie many pathologic conditions and they are thought to contribute to a variety of diseases such as metabolic, degenerative, or genetic disorders (e.g. type 2 diabetes, Alzheimer, cancer)50,51. Inflammatory modulators can thus interact at many different levels of the inflammatory cascade, including many different mechanisms of action. By targeting a biological response with many possible molecular mechanisms of action of the drug candidates we increase the possibility of discovering new chemical compounds in future early stage drug discovery screening approaches without being restricted to chemical compounds with very similar structures, which would most likely be targeted by aiming for more specific mechanisms of action. Within the genus Euphorbia, almost 70% of all chemical compounds described to date are new34. The chance of finding previously undiscovered compounds within Euphorbia is therefore considerably high. Also, an inflammatory response can be deducted relatively easily from ethnomedicinal descriptions, in comparison to more specific mode of actions. We were particularly interested in medicinal plant uses modulating the inflammatory response because inflammatory processes have shown relevance in the treatment of actinic keratosis by ingenol mebutate32. In vitro and in vivo studies of the molecular and cellular mode of action of the treatment showed that initial cell death is followed by a complex inflammatory response crucial for preventing tumor relapse and responsible for the high efficacy of the treatment32,36. Given that the inflammatory response, in particular, plays an important role in the efficacy of the treatment, we aimed to identify species of Euphorbia, suggesting the ability to modulate an inflammatory response in humans.
With special focus on whether the described treatment by the medicinal plant triggers an inflammatory response in humans, we investigated all records in the database of uses of Euphorbia. We looked for indications of the presence of a potential anti- or pro-inflammatory agent. We used the definitions of diseases given on the PubMed Health Diseases and Conditions Database52 and Dorland’s Illustrated Medical Dictionary53, which include either descriptions of diseases or their treatment. Use reports were thereafter classified into three categories: inflammatory response (treatment that can be related to an inflammatory response; 44 species), no inflammatory response (treatment that can not be related to an inflammatory response; 12 species), and unknown (description of medicinal use that contains insufficient information for classification; 23 species). Besides records describing medicinal uses of Euphorbia, we also included records of toxicity found in the EBDCS level 1 state category vertebrate poisons. Toxicity is separated from medicinality only by dosage and may therefore be a convincing indicator of bioactivity54. An overview of the use data interpreted with the EBDCS categories and the inflammatory response categories is shown in Supplementary Tables 3 and 4.
Evolutionary patterns of medicinal properties in Euphorbia
Here, we explored whether our interpretation of medicinal plant use can reveal different phylogenetic patterns, compared to a standard approach of classification. We performed three different comparative phylogenetic analyses.
First, we investigated the strength in phylogenetic signal of the EBDCS categories and the inflammatory response category using the D statistic55, a measure of phylogenetic signal, implemented by the function phylo.d in the R package caper56. Two p-values are calculated for the D statistic, p(D < 1) indicating whether the D metric is significantly smaller than 1, meaning that the trait (species’ medicinal properties) is not randomly distributed over the phylogeny. The second p-value, p(D > 0) indicates whether the D metric is significantly greater than 0, meaning that the trait (species’ medicinal properties) has a significantly different distribution on the phylogeny from the standard Brownian model of evolution. The phylogenetic signal is considered strong if p(D < 1) < 0.05 and p(D > 0) > 0.0555. In our study, phylogenetic signal was considered significant if >95% of the 1,000 trees showed a p(D < 1) value < 0.05 and the signal was considered strong if >95% of the 1,000 trees showed a p(D > 0) value > 0.05.
Second, we compared the phylogenetic diversity captured by plant species identified by the two different classification methods. To evaluate the phylogenetic diversity (PD) of species identified by the inflammatory response category and the EBDCS categories, we calculated the PD index (measuring the total branch length spanned by the tree of species in a given category) proposed by Faith57, and implemented in the function pd in the R package picante v.1.6-258. The PD index was expressed as absolute value as well as a percentage of the PD of the total phylogenetic tree. High PD percentage means that species included in this category are spread across the whole tree, while low percentage means that the species are found in only few, clustered parts of the tree.
Third, we investigated the overlap in the plant lineages identified by the two classification methods. To do that, we compared the phylogenetic similarity of the inflammatory response category and the EBDCS categories, by calculating the mean nearest taxon distance (MNTD)59 – a measure showing the phylogenetic proximity of species between two categories on the tree - using the comdistnt function in the R package picante v.1.6-258. P-values for the MNTD were calculated by comparing the MNTD value between the inflammatory response category and an EBDCS category to 1,000 randomly generated categories of the same size within the species pool of the inflammatory response category and the EBDCS category medicines. The two categories were considered phylogenetically significantly similar if at least 95% of the 1,000 trees showed a p-value < 0.05.
Further, as tentative approach to narrow down the number of species chosen for an early stage drug discovery screening, we identified the position (nodes in phylogeny) of phylogenetic clustering for the inflammatory response category. We highlighted so-called “hot nodes” on the phylogeny, i.e. nodes that are significantly overrepresented by species in a given category8,11, using the nodesigl command in PHYLOCOM v4.259.
All analyses we describe were performed with in-house scripts in the R environment Version 3.0.3 (http://www.R- project.org/), which are available in the Supplementary Information. Additional information on all method subsections is presented in Supplementary Methods. The alignment file as well as the set of 1,000 Bayesian trees used for analysis was deposited on dryad (http://dx.doi.org/10.5061/dryad.s2df3).
Additional Information
How to cite this article: Ernst, M. et al. Evolutionary prediction of medicinal properties in the genus Euphorbia L. Sci. Rep. 6, 30531; doi: 10.1038/srep30531 (2016).
Supplementary Material
Acknowledgments
This work was supported by the Marie Curie Actions of the 7th European Community Framework Programme: FP7/2007-2013/, REA grant agreement n° 606895-MedPlant to NR and PIEF-GA-2012-328637-BiodiversityAltitude to CHSL and NR. No financial support was received from LEO Pharma A/S. The authors thank Mogens Trolle for photos of Euphorbia spp. for Figure 3.
Footnotes
The authors declare no competing financial interests.
Author Contributions C.H.S.-L., N.R., O.M.G., N.N. and M.E. designed the study. M.E. collected data. M.E. and N.N. classified medicinal plant uses into the inflammatory response category. M.E. wrote the R scripts. M.E. and C.H.S.-L. analysed the data. M.E. wrote the manuscript together with C.H.S.-L., O.M.G. and N.R. All authors discussed the results, commented on the manuscript, and approved the final version.
References
- Newman D. J., Cragg G. M. & Snader K. M. The influence of natural products upon drug discovery. Nat. Prod. Rep. 17, 215–234 (2000). [DOI] [PubMed] [Google Scholar]
- Cragg G. M. & Newman D. J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta 1830, 3670–3695 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plants under pressure – a global assessment. IUCN Sampled Red List Index for Plants (Royal Botanic Gardens, Kew, UK, 2012).
- Dahlgren R. M. T. A revised system of classification of the angiosperms. Bot. J. Linn. Soc. 80, 91–124 (1980). [Google Scholar]
- Gottlieb O. R. Ethnopharmacology versus chemosytematics in the search for biologically active principles in plants. J. Ethnopharmacol. 6, 227–238 (1982). [DOI] [PubMed] [Google Scholar]
- Gottlieb O. R., Borin M. R. de M. B. & De Brito N. R. S. Integration of ethnobotany and phytochemistry: dream or reality? Phytochemistry 60, 145–152 (2002). [DOI] [PubMed] [Google Scholar]
- Rønsted N., Savolainen V., Mølgaard P. & Jäger A. K. Phylogenetic selection of Narcissus species for drug discovery. Biochem. Syst. Ecol. 36, 417–422 (2008). [Google Scholar]
- Saslis-Lagoudakis C. H., Klitgaard B. B., Forest F., Francis L., Savolainen V., Williamson E. M. & Hawkins J. A. The use of phylogeny to interpret cross-cultural patterns in plant use and guide medicinal plant discovery: an example from Pterocarpus (Leguminosae). PLoS One 6, e22275 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F. et al. Clustered patterns of species origins of nature-derived drugs and clues for future prospecting. Proc. Natl. Acad. Sci. USA 108, 12943–12948 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rønsted N. et al. Can phylogeny predict chemical diversity and potential medicinal activity of plants? A case study of Amaryllidaceae. BMC Evol. Biol. 12, 182 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saslis-Lagoudakis C. H. et al. Phylogenies reveal predictive power of traditional medicine in bioprospecting. Proc. Natl. Acad. Sci. USA 109, 15835–15840 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace O. M. et al. Evolutionary history and leaf succulence as explanations for medicinal use in aloes and the global popularity of Aloe vera. BMC Evol. Biol. 15, 29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L. et al. Clustered distribution of natural product leads of drugs in chemical space as influenced by the privileged target-sites. Sci. Rep. 5, 9325 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yessoufou K., Daru B. H. & Muasya A. M. Phylogenetic exploration of commonly used medicinal plants in South Africa. Mol. Ecol. Resour. 15, 405–413 (2015). [DOI] [PubMed] [Google Scholar]
- Fabricant D. S. & Farnsworth N. R. The value of plants used in traditional medicine for drug discovery. Environ. Health Perspect 109, 69–75 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich M., Edwards S., Moerman D. E. & Leonti M. Ethnopharmacological field studies: A critical assessment of their conceptual basis and methods. J. Ethnopharmacol. 124, 1–17 (2009). [DOI] [PubMed] [Google Scholar]
- Cook F. E. M. Economic Botany Data Collection Standard (Royal Botanic Gardens, Kew, UK, 1995). [Google Scholar]
- Gruca M., Cámara-Leret R., Macía M. J. & Balslev H. New categories for traditional medicine in the Economic Botany Data Collection Standard. J. Ethnopharmacol. 155, 1388–1392 (2014). [DOI] [PubMed] [Google Scholar]
- Oritz de Montellano B. Empirical Aztec medicine. Science 188, 215–220 (1975). [DOI] [PubMed] [Google Scholar]
- Staub O. P., Geck M. S., Weckerle C. S., Casu L. & Leonti M. Classifying diseases and remedies in ethnomedicine and ethnopharmacology. J. Ethnopharmacol. 174, 514–519 (2015). [DOI] [PubMed] [Google Scholar]
- Snider G. L. Nosology for our day: Its application to chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 167, 678–683 (2003). [DOI] [PubMed] [Google Scholar]
- Janssens A. C. J. W. & Van Duijn M. Genome-based prediction of common diseases: advances and prospects. Human Molecular Genetics 17, R166–R173 (2008). [DOI] [PubMed] [Google Scholar]
- Govaerts R., Frodin D. G. & Radcliffe-Smith A. World checklist and bibliography of Euphorbiaceae (with Pandaceae). Vol. 2 (Royal Botanic Gardens, Kew, UK, 2000). [Google Scholar]
- Horn J. W. et al. Phylogenetics and the evolution of major structural characters in the giant genus Euphorbia L. (Euphorbiaceae). Mol. Phylogenet. Evol. 63, 305–326 (2012). [DOI] [PubMed] [Google Scholar]
- Frodin D. G. History and concepts of big plant genera. TAXON 53, 753–776 (2004). [Google Scholar]
- Yang Y. et al. Molecular phylogenetics and classification of Euphorbia subgenus Chamaesyce (Euphorbiaceae). TAXON 61, 764–789 (2012). [Google Scholar]
- Dorsey B. L. et al. Phylogenetics, morphological evolution, and classification of Euphorbia subgenus Euphorbia (Euphorbiaceae). TAXON 62, 291–315 (2013). [Google Scholar]
- Peirson J. A., Bruyns P. V., Riina R., Morawetz J. J. & Berry P. E. A molecular phylogeny and classification of the largely succulent and mainly African Euphorbia subg. Athymalus (Euphorbiaceae). TAXON 62, 1178–1199 (2013). [Google Scholar]
- Riina R. et al. A worldwide molecular phylogeny and classification of the leafy spurges, Euphorbia subgenus Esula (Euphorbiaceae). TAXON 62, 316–342 (2013). [Google Scholar]
- Horn J. W. et al. Evolutionary bursts in Euphorbia (Euphorbiaceae) are linked with photosynthetic pathway. Evolution 68, 3485–3504 (2014). [DOI] [PubMed] [Google Scholar]
- Ernst M. et al. Global medicinal uses of Euphorbia L. (Euphorbiaceae). J. Ethnopharmacol. 176, 90–101 (2015). [DOI] [PubMed] [Google Scholar]
- Berman B. New developments in the treatment of actinic keratosis: focus on ingenol mebutate gel. Clin. Cosmet. Investig. Dermatol. 20, 111–122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen L. et al. 14-step synthesis of (+)-ingenol from (+)-3-carene. Science 341, 878–882 (2013). [DOI] [PubMed] [Google Scholar]
- Vasas A. & Hohmann J. Euphorbia diterpenes: isolation, structure, biological activity, and synthesis (2008–2012). Chem. Rev. 114, 8579–8612 (2014). [DOI] [PubMed] [Google Scholar]
- Shi Q. W., Su X. H. & Kiyota H. Chemical and pharmacological research of the plants in genus Euphorbia. Chem. Rev. 108, 4295–4327 (2008). [DOI] [PubMed] [Google Scholar]
- Kedei N. et al. Characterization of the interaction of ingenol 3-angelate with Protein Kinase C. Cancer Res. 64, 3243–3255 (2004). [DOI] [PubMed] [Google Scholar]
- Vallance P. & Smart T. G. The future of pharmacology. Br. J. Pharmacol. 147, S304–S307 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balandrin M. F., Kinghorn A. D. & Farnsworth N. R. Plant-derived natural products in drug discovery and development. In Human Medicinal Agents from plant s, Vol. 534 (ed. Kinghorn D. & Balandrin M. F.) Ch. 1, 2–12 (American Chemical Society, 10.1021/bk-1993-0534.ch001, 1993). [DOI] [Google Scholar]
- Moerman D. E. The medicinal flora of native North America: An analysis. J. Ethnopharmacol. 31, 1–42 (1991). [DOI] [PubMed] [Google Scholar]
- Wink M., Botschen F., Gosmann C., Schäfer H. & Waterman P. G. Chemotaxonomy seen from a phylogenetic perspective and evolution of secondary metabolism. In Biochemistry of plant secondary metabolism Edn. 2 Annual Plant Reviews Vol. 40, 364–33 (ed. Wink M.) (Wiley-Blackwell, Oxford, UK, 2010). [Google Scholar]
- Kusari S. et al. Tramadol – A true natural product? Angew. Chem. Int. Ed. 53, 1–5 (2014). [DOI] [PubMed] [Google Scholar]
- Ernst M. et al. A metabolomics protocol for plant systematics by matrix-assisted laser-desorption/ionization time-of flight mass spectrometry. Anal. Chim. Acta 859, 46–58 (2015). [DOI] [PubMed] [Google Scholar]
- Evans F. J. & Kinghorn A. D. A comparative phytochemical study of the diterpenes of some species of the genera Euphorbia and Elaeophorbia (Euphorbiaceae). Bot. J. Linn. Soc. 74, 23–35 (1977). [Google Scholar]
- King A. J., Brown G. D., Gilday A. D., Larson T. R. & Graham I. A. Production of bioactive diterpenoids in the Euphorbiaceae depends on evolutionary conserved gene clusters. Plant Cell 26, 3286–3298 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A. T., Gohil V. M. & Bhutani K. K. Modulating TNF-α signalling with natural products. Drug Discov. Today 11, 725–732 (2006). [DOI] [PubMed] [Google Scholar]
- Li J. W. H. & Vederas J. C. Drug discovery and natural products: End of an era or an endless frontier? Science 325, 161–165 (2009). [DOI] [PubMed] [Google Scholar]
- Kingston D. G. I. Modern natural products drug discovery and its relevance to biodiversity conservation. J. Nat. Prod. 74, 496–511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell J. W., Blanckley A., Boldon H. & Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 11, 191–200 (2012). [DOI] [PubMed] [Google Scholar]
- Riina R. & Berry P. E. (coordinators), Euphorbia planetary biodiversity inventory database. (2012) Available at: http://app.tolkin.org/projects/72/taxa. (Accessed: 29th April 2015).
- Medzhitov R. Origin and physiological roles of inflammation. Nature 454, 428–435 (2008). [DOI] [PubMed] [Google Scholar]
- Kumar V., Abbas A. K. & Aster J. C. Robbins and Cotran pathologic basis of disease 9th edn, (ed. Kumar V., Abbas A. K. & Aster J. C.) Ch. 3, 69–112 (Elsevier, 2015). [Google Scholar]
- A.D.A.M. Medical Encyclopedia. (1997-2011) Available at: http://www.ncbi.nlm.nih.gov/pubmedhealth/s/diseases_and_conditions/. (Accessed: 29th April 2015).
- Dorland W. A. N. Dorland’s illustrated medical dictionary Edn. 32 (Elsevier, Philadelphia, USA, 2012). [Google Scholar]
- Stumpf W. E. The dose makes the medicine. Drug Discov. Today 11, 550–555 (2006). [DOI] [PubMed] [Google Scholar]
- Fritz S. A. & Purvis A. Selectivity in mammalian extinction risk and threat types: A new measure of phylogenetic signal strength in binary traits. Conserv. Biol. 24, 1042–1051 (2010). [DOI] [PubMed] [Google Scholar]
- Orme D. et al. caper: Comparative analyses of phylogenetics and evolution in R. R package version 0.5.2. (2013) Available at: http://CRAN.R-project.org/package=caper. (Accessed: 15th January 2016).
- Faith D. P. Conservation evaluation and phylogenetic diversity. Biol. Cons. 61, 1–10 (1992). [Google Scholar]
- Kembel S. W. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464 (2010). [DOI] [PubMed] [Google Scholar]
- Webb C. O., Ackerly D. D. & Kembel S. W. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24, 2098–2100 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.