Abstract
Treatment with abiraterone acetate and prednisone (AA/P) prolongs survival in metastatic castration-resistant prostate cancer (mCRPC) patients. We evaluated the genetic variation in CYP17A1 as predictive of response to AA/P. A prospective collection of germline DNA prior to AA/P initiation and follow-up of a mCRPC cohort was performed. Five common single-nucleotide polymorphisms (SNPs) in CYP17A1 identified using a haplotype-based tagging algorithm were genotyped. Clinical outcomes included biochemical response and time to biochemical progression on AA/P. Logistic regression was used to assess the association between tag SNPs and biochemical response. Proportional hazards regression was used to assess the association between tag SNPs and time to biochemical progression. Odds or hazard ratio per minor allele were estimated and p-values below 0.05 were considered statistically significant. Germline DNA was successfully genotyped for four tag SNPs in 87 patients. The median age was 73 years (54–90); the median prostate-specific antigen was 66 ng/dL (0.1–99.9). A single SNP, rs2486758, was associated with lower odds of experiencing a biochemical response (Odds ratio 0.22, 95% confidence interval 0.07–0.63, p = 0.005) and a shorter time to biochemical progression (Hazard ratio 2.23, 95% confidence interval 1.39–3.56, p < 0.001). This tag SNP located in the promoter region of CYP17A1 will need further validation as a predictive biomarker for AA/P therapy.
Keywords: metastatic castration-resistant prostate cancer, abiraterone acetate, single nucleotide polymorphism, CYP17A1, predictive biomarker
1. Introduction
Prostate cancer is the second leading cause of cancer-related mortality in patients in the United States, with an estimated 26,120 deaths in 2016 [1]. Almost all prostate cancer–related deaths occur in patients with advanced, metastatic disease. The initial treatment for this stage of the disease is androgen deprivation therapy, which provides effective control of the disease for variable time periods in patients with advanced metastatic hormone-sensitive prostate cancer by depleting testosterone [2,3,4]. Inevitably, disease progression occurs to a castration-resistant state (mCRPC) [5,6,7,8], although there is considerable variability in time to progression reflecting an underlying genetic heterogeneity. Once a patient has progressed to mCRPC, the median overall survival (OS) is limited to 15–35 months, despite the availability of several therapeutic options [9,10,11,12,13]. Recent therapeutic advances including the successful targeting of the testosterone-androgen receptor (AR) axis with several new drugs have resulted in slowing castration-resistant disease progression and improved survival. One of the novel drugs contributing to the increase in survival in the castration-resistant stage is the cytochrome P45017 (CYP17) inhibitor abiraterone acetate (AA) which has efficacy in combination with prednisone (P) in both the pre- and post-chemotherapy setting, although the response to treatment remains variable [14,15,16,17]. CYP17A1 is encoded by a single gene on chromosome 10q24.3 (CYP17A1), expressed in the adrenals and gonads, and a membrane-bound dual-function monooxygenase with a critical role in the synthesis of many human steroid hormones [18,19]. Defects in CYP17A1 were found to cause 17α-hydroxylase deficiency and 17,20-lyase deficiency [18]. CYP17A1 has also been implicated in the development of cardiovascular diseases through effects on blood pressure [20,21,22]. CYP17A1 is an important target for the treatment of prostate cancer that proliferates in response to androgens [19,23] Genetic variation of CPY17A1 was found to be associated with increased risk of prostate cancer [24,25,26,27,28]. Furthermore, alterations of CYP17A1 activity have been described as possible mechanisms of resistance to hormonal treatments in experimental models of prostate cancer [29].
We investigated the genetic variation in a key sex steroid pathway gene, CYP17A1, which results in the enzyme activity for metabolizing intracellular testosterone levels as a potential predictive factor for response to AA/P at the castration-resistant stage.
2. Results
The characteristics of the 87 patients identified to meet the inclusion criteria are shown in Table 1. The median time from the initiation of androgen deprivation therapy to the development of mCRPC was two years (0–17). The time from DNA collection to the initiation of AA/P was two years (0–6). Four of the five (80%) tag single nucleotide polymorphisms (SNPs) in CYP17A1 were successfully genotyped in all patients with an adequate call rate of 0.958. No markers were out of Hardy-Weinberg equilibrium (p > 0.001 for all comparisons), and the mean imputation R2 was 0.974. The four successfully genotyped SNPs (rs4919685, rs743572, rs17115100, and rs2486758) provided 100% coverage of the common genetic variations (minor allele frequency ≥ 0.05) in CYP17A1 (chromosome 10, base pairs 102,830,531–102,837,533).
Table 1.
Characteristics of 87 patients receiving abiraterone acetate and prednisone for metastatic castration-resistant prostate cancer.
| Demographics at the Time of Initial Prostate Cancer Diagnosis | |
|---|---|
| Median age (years) | 64 (43–89) |
| Median prostate-specific antigen (ng/dL) | 7 (0–3229) |
| Stage | |
| T1–T2b | 24 (28%) |
| T2c | 25 (29%) |
| T3+ | 38 (43%) |
| N+ | 23 (38%) |
| M+ | 18 (56%) |
| Gleason grade | |
| 5–6 | 11 (13%) |
| 7 | 27 (31%) |
| 8–10 | 49 (56%) |
| Radical prostatectomy | 51 (71%) |
| Patient Characteristics at the Time of Initiation of Abiraterone Acetate and Prednisone for Castration-Resistant Stage | |
| Median age (years) | 73 (54–90) |
| Median prostate-specific antigen (ng/dL) | 66 (0–100) |
| Median body mass index (kg/m2) | 30 (21–56) |
| Bone metastases | 73 (84%) |
| Soft tissue metastases | 43 (49%) |
| Previous androgen-deprivation therapy | 87 (100%) |
| Previous docetaxel | 56 (65%) |
| Previous enzalutamide | 5 (6%) |
| Previous cabazitaxel | 5 (6%) |
| Time from initial diagnosis (years) | 8 (1–24) |
| Patients alive at the time of data analysis | 38 (44%) |
Data are given as median (range) or count (percent of the entire cohort).
The associations between the four genotyped SNPs and the biochemical response to as well as the time to biochemical progression on AA/P are shown in Table 2. The minor allele frequency for rs2486758 was 0.33 in this cohort of patients with mCRPC.
Table 2.
Association between common genetic variations in CYP17A1 and biochemical response to abiraterone acetate and prednisone in 87 patients with metastatic castration-resistant prostate cancer.
| SNP | n | Biochemical Response to Treatment | p | |
|---|---|---|---|---|
| Response Rates 1 | Effect Estimate | |||
| rs2486758 | 87 | 50%/17% | OR 0.22 (0.07–0.63) | 0.005 |
| rs4919685 | 87 | 38%/39% | OR 1.24 (0.64–2.40) | 0.524 |
| rs17115100 | 87 | 40%/37% | OR 0.82 (0.31–2.19) | 0.689 |
| rs743572 | 87 | 33%/40% | OR 1.13 (0.58–2.20) | 0.718 |
| Time to Biochemical Progression | ||||
| Median Time to Progression 1 | Effect Estimate | |||
| rs2486758 | 87 | 7.0 months/2.9 months | HR 2.23 (1.39–3.56) | <0.001 |
| rs4919685 | 87 | 3.5 months/4.6 months | HR 0.82 (0.47–1.42) | 0.429 |
| rs17115100 | 87 | 4.5 months/5.3 months | HR 0.95 (0.67–1.35) | 0.693 |
| rs743572 | 87 | 3.5 months/4.6 months | HR 1.03 (0.73–1.46) | 0.889 |
Effect estimates are expressed as unadjusted odds ratio (OR) (95% confidence interval) or hazard ratio (HR) (95% confidence interval) per minor allele. 1 Biochemical (prostate-specific antigen) response rates and median time to biochemical progression are listed as major homozygotes versus hetero- and minor homozygotes; SNP, single nucleotide polymorphism.
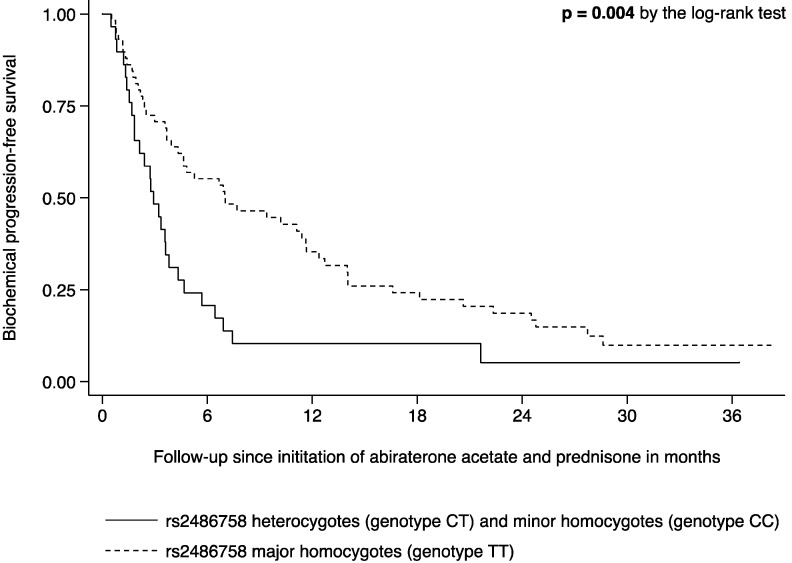
The genetic variant rs2486758 (T>C) in CYP17A1 was negatively associated with biochemical response to AA/P (Odds ratio 0.22, 95% confidence interval 0.07–0.63, p = 0.005), translating into a more-than-four-fold decrease in the odds of biochemical response to AA/P per minor allele. The biochemical response rate to AA/P was 50% in patients with wild-type rs2486758 and 17% in patients with at least one minor allele (p = 0.005). Unadjusted and multivariable-adjusted estimates for the effect of rs2486758 on biochemical response to AA/P are shown in Table 3. The genetic variant rs2486758 (T>C) in CYP17A1 was also associated with a shorter time to biochemical progression on AA/P (Hazard ratio 2.23, 95% confidence interval 1.39–3.56, p < 0.001), translating into a more-than-two-fold increase in the hazard of biochemical disease progression on AA/P per minor allele. After a median follow-up time of 4.6 months (3.4–6.9), 77 patients (89%) had progressed. The median time to biochemical progression was 7.0 (4.3–11.6) months in patients with wild-type rs2486758 and 2.9 months (1.8–3.8) in patients with at least one minor allele (p = 0.004, Figure 1). Unadjusted and multivariable-adjusted estimates for the effect of rs2486758 on time to biochemical progression on AA/P are shown in Table 3.
Table 3.
Multivariable-adjusted effect estimates for rs2486758 and biochemical response to abiraterone acetate and prednisone as well as time to biochemical progression in 87 patients with metastatic castration-resistant prostate cancer.
| Model | n | Response to Treatment | p |
|---|---|---|---|
| Unadjusted | 87 | OR 0.22 (0.07–0.63) | 0.005 |
| Age-adjusted (I) | 87 | OR 0.21 (0.07–0.62) | 0.005 |
| Multivariable-adjusted (II) | 87 | OR 0.22 (0.07–0.63) | 0.005 |
| Multivariable-adjusted (III) | 87 | OR 0.21 (0.07–0.62) | 0.005 |
| Multivariable-adjusted (IV) | 87 | OR 0.18 (0.06–0.57) | 0.003 |
| Time to Progression | |||
| Unadjusted | 87 | HR 2.23 (1.39–3.56) | <0.001 |
| Age-adjusted (I) | 87 | HR 2.22 (1.40–3.55) | 0.001 |
| Multivariable-adjusted (II) | 87 | HR 2.26 (1.41–3.62) | 0.001 |
| Multivariable-adjusted (III) | 87 | HR 2.29 (1.43–3.67) | 0.001 |
| Multivariable-adjusted (IV) | 87 | HR 2.67 (1.64–4.36) | <0.001 |
Effect estimates are expressed as unadjusted and multivariable-adjusted odds ratio (OR) (95% confidence interval) or hazard ratio (95% confidence interval) per minor allele. Model I was additionally adjusted for age at the time of abiraterone acetate initiation. Model II was additionally adjusted for Gleason score at the time of initial diagnosis. Model III was additionally adjusted for prostate-specific antigen at the time of abiraterone acetate initiation. Model IV was additionally adjusted for prior administration of docetaxel.
Figure 1.
Kaplan-Meier estimates of time to biochemical progression on abiraterone acetate and prednisone stratified by rs2486758 genotype (hetero- and minor homozygotes versus major homozygotes).
3. Discussion
AA/P was studied in patients with metastatic castration-resistant prostate cancer both prior to and after administration of cytotoxic chemotherapy [14,15]. The biochemical response rates in these studies were lower in patients who received AA/P after cytotoxic chemotherapy (38%) compared to patients treated with pre-chemotherapy AA/P (62%). The observed biochemical response rate in our cohort (39%) is very similar to the expected biochemical response rate (46%), taking into account the large proportion of our patients who had been treated with prior cytotoxic chemotherapy. No predictive factors have been identified previously for response to AA/P, including the Gleason score [30] or type and duration of prior endocrine therapies [31]. Another smaller study showed a poorer response to AA/P in patients with shorter responses to previous prostate cancer therapies [32]. In our study cohort, 29 of the 87 patients (33%) receiving AA/P for mCRPC harbored the rs2486758 variant of CYP17A1 which was associated with a risk for lower biochemical response and shorter time to biochemical progression when taking AA/P. The rs2486758 minor allele which is located in the intergene region near the 5’ of the CYP17 gene remained statistically significant for the association after adjusting for multiple testing as well as potential confounding clinical variables including the Gleason score at the time of initial diagnosis, prostate-specific antigen at the time of initiating treatment, and whether the treatment was administered pre- or post-chemotherapy. Our data suggests a more-than-four-fold decrease in the odds of biochemical response to AA/P in patients with mCRPC per rs2486758 minor allele (C versus T). Furthermore, each minor allele was associated with a more-than-two-fold increase in the hazard ratio of biochemical disease progression. In previous studies, the rs2486758 polymorphism of CPY17A1 was associated with a significant increased risk of prostate cancer [24,25,26,27]. A meta-analysis of these studies confirmed this association (C versus T, odds ratio 1.07, 95% confidence interval 1.03–1.12, p = 0.002) [28]. Mechanistically, this 7% increase in the risk of prostate cancer was attributed to the location of rs2486758 in the promoter region of CYP17A1. Alterations of CYP17A1 activity including up-regulation have been described as a mechanism of resistance to AA/P in human prostate cancer xenografts, making the enzyme a potential candidate as a predictive biomarker [29]. With the up-regulation of CYP17A1 being described as a mechanism of resistance to AA/P and functional studies confirming significantly increased transcriptional activity of CYP17A1 with the minor allele (C versus T), up-regulation is a likely explanation for the observed effects [33]. Likewise, the shorter duration of the response may be explained by the up-regulation of CYP17A1 overcoming the effects of AA/P treatment. The rs2486758 minor allele has been reported to be associated with higher serum 17β-estradiol levels in premenopausal women [34], but it is unknown if sex steroids in men who have undergone castration are also altered in the presence of the polymorphism and if that will affect the response to treatment with AA/P.
Other genome-based predictive factors for AA/P that have been investigated include copy number variations of CYP17A1 in the serum of patients with mCRPC [35]. These may have contributed to the observed effects but were not evaluated in our study, which investigated germline factors for determining response to treatment. Furthermore, in a small study, germline variation in other signaling pathways such as the SULT1E1 variants described by Agarwal et al. [36] has been shown to have effects on duration of response and warrant validation. The aforementioned factors will need to be evaluated in concert with our findings to gain a better understanding of the different factors affecting response to treatment. Mostaghel et al. [29] also demonstrated the induction of ligand-independent androgen receptor (AR) variants in response to AA/P treatment, which is another plausible explanation for disease progression in this setting. Furthermore, differential expression of AR-V7 is another plausible explanation for the difference in observed response rates [37]. The mechanism described by Antonarakis et al. is biologically plausible, although the results have not been validated [38]. The technology to detect AR-V7 expression detection in circulating tumor cells is becoming available for clinical use, but is yet to be adopted in clinical practice.
A strength of our study was that it included prospectively followed patients after enrollment in a study devised to identify biomarkers in advanced prostate cancer in a uniform manner and therefore did not involve samples of convenience. The limitations include small sample size, lack of a validation cohort, and a candidate approach which restricts information of other germline or somatic genetic aberrations such as AR copy number variations and AR mutations on response to treatment. These will need to be evaluated in larger cohorts treated with AA/P in order to develop predictive models for individualizing treatment with AA/P in advanced prostate cancer.
4. Materials and Methods
4.1. Cohort and Clinical Data
This study included patients with mCRPC who were enrolled in a prospective and uniform collection of bio-specimens at Mayo Clinic (Rochester, MN, USA); the study was approved by the institutional review board (IRB# 09-007355). Written consent was obtained from all patients prior to enrollment. The prostate cancer cohort in this hospital-based registry includes patients with advanced-stage prostate cancer who provide blood and urine specimens at the time of enrollment for the purpose of biomarker identification to treatments and are followed for outcomes including overall survival as has been previously reported in detail [39,40,41]. All patients were enrolled between September 2009 and December 2012 and followed for outcomes after enrollment. The study involves collection of blood and urine specimens including the collection of peripheral blood mononuclear cells, which were used to extract germline DNA. Only those patients receiving the combination of AA/P for mCRPC stage were included in this analysis. Patients with missing clinical or unavailable genetic information were excluded from the analysis if they had received this drug combination. All patients had follow-up data abstracted from medical records for treatment and disease outcomes. Patient characteristics collected included demographic information, age at the time of DNA specimen collection, initial cancer diagnosis date, Gleason score at the time of initial diagnosis, time between initial prostate treatment and initiation of castration for mCRPC stage after failure of androgen deprivation therapy, date of castration, date of progression while receiving AA/P for mCRPC, prostate-specific antigen at the time of disease progression while receiving AA/P. In addition, we also collected relevant clinical information before androgen deprivation therapy, including stage at the time of initial prostate cancer diagnosis and primary prostate treatments received previously. Follow-up and care for all patients was performed by physicians specializing in prostate cancer as per the standard of care.
4.2. Genetic Data
Germline DNA was purified from buffy coats isolated from whole blood obtained from the study patients. Briefly, germline DNA was extracted from buffy coats isolated from whole blood obtained from the identified (or selected) patients using a standardized organic (phenol-chloroform) purification procedure. Incubation of RNase A was also used to remove any contaminating RNA. Following extraction, DNA quality and quantity was assessed using spectrophotometry and the picogreen assay. All DNA samples utilized for genotyping required 260/280 spectrophotometric ratios of ≥1.8. Double stranded DNA concentrations obtained from the picogreen assay were all ≥50 ng/µL. DNA samples were diluted to a working concentration of 20 ng/µL prior to genotyping using the MassARRAY platform. Forty-eight SNPs in five candidate genes CYP17A1, HSD3B2, ESR1, ESR2, and AR were identified using a haplotype-based tagging algorithm [42]. Genotyping of these tag SNPs using the MassARRAY platform (Agena Bioscience, San Diego, CA, USA) was performed at the Mayo Clinic Genotyping Facility. The MaCH imputation program [43] was used to carry out multiple genotype imputation for missing genotypes using the HapMap Phase II release #22 CEU reference panel [44]. The a priori hypothesis for this study was an association between common genetic variation in CYP17A1 and biochemical response to AA/P. The four remaining candidate genes were genotyped for different outcomes in a larger cohort of men with advanced prostate cancer.
4.3. Outcomes
The clinical outcomes of interest included biochemical response to treatment as defined by ≥50% prostate-specific antigen decrease after initiating AA/P treatment. The second response outcome of interest for association with genotypes was time to biochemical progression as defined by >25% increase in prostate-specific antigen on two consecutive tests at least two weeks apart. Patients who had not progressed at the time of data analysis were censored.
4.4. Statistical Analysis
Fisher’s exact test was used to compare biochemical response to AA/P in subgroups. Progression-free survival estimates were calculated using the method described by Kaplan and Meier [45]. The log-rank test was used to compare time to biochemical progression in subgroups. Unadjusted and multivariable-adjusted logistic regression was used to assess the association between genetic variation and biochemical response. Unadjusted and multivariable-adjusted Cox proportional hazards regression was used to analyze the association between genetic variation and time to biochemical progression [46]. Multivariable-adjusted regression models included age at the time of AA/P initiation, Gleason Score at the time of initial diagnosis, prostate-specific antigen at the time of AA/P initiation, and whether AA/P was initiated pre- or post-chemotherapy in addition to the SNP of interest. Effect measures are expressed as odds or hazard ratio per minor allele. In order to assess associations at the SNP-level, the ProbABEL software was used to analyze the biochemical response and time to biochemical progression data using the imputed allele dosage [47]. In order to correct for multiple testing in the presence of linkage disequilibrium, a modified effective number of independent tests approach was employed [48,49,50]. The inferred Meff for the four SNPs was 3 (adjusted α-level = 0.017). Data management and statistical analyses were performed using the SAS software (version 9.3, SAS Institute, Cary, NC, USA) and the R environment (R Foundation for Statistical Computing, Vienna, Austria). Graphs were created using the Stata software (version 13.1, StataCorp, College Station, TX, USA).
5. Conclusions
We identified the common genetic variant rs2486758 in CYP17A1 that was negatively associated with a biochemical response to and time to biochemical progression in patients on AA/P with mCRPC. The identified variant is known to increase transcriptional activity, likely conferring resistance to AA/P through up-regulation of CYP17A1. These findings warrant external validation to verify the predictive power of this readily available biomarker.
Acknowledgments
We thank the Mayo Clinic Medical Genome Facility Genotyping Core (Rochester, MN, USA) for providing the DNA sequencing service. This study was supported by Joseph and Gail Gassner and John P. Vaile development funds for prostate cancer research (Manish Kohli) and Mayo CCaTS grant number UL1TR000135 (Manish Kohli). No funds were received to cover costs to publish in open access.
Author Contributions
Moritz Binder, Ben Y. Zhang, Rhea Kohli, Tanvi Kohli, and Manish Kohli conceived and designed the study; Ben Y. Zhang, David W. Hillman and Manish Kohli performed the data collection; Adam Lee, Rhea Kohli and Manish Kohli were responsible for genetic data generation and Adam Lee and Manish Kohli were responsible for overseeing quality control of data; Moritz Binder analyzed the data; Moritz Binder and Manish Kohli wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Huggins C. Effect of orchiectomy and irradiation on cancer of the prostate. Ann. Surg. 1942;115:1192–1200. doi: 10.1097/00000658-194206000-00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huggins C. Prostatic cancer treated by orchiectomy; the five year results. J. Am. Med. Assoc. 1946;131:576–581. doi: 10.1001/jama.1946.02870240008003. [DOI] [PubMed] [Google Scholar]
- 4.Huggins C., Hodges C.V. Studies on prostatic cancer: I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. 1941. J. Urol. 2002;168:9–12. doi: 10.1016/S0022-5347(05)64820-3. [DOI] [PubMed] [Google Scholar]
- 5.Crawford E.D., Eisenberger M.A., McLeod D.G., Spaulding J.T., Benson R., Dorr F.A., Blumenstein B.A., Davis M.A., Goodman P.J. A controlled trial of leuprolide with and without flutamide in prostatic carcinoma. N. Engl. J. Med. 1989;321:419–424. doi: 10.1056/NEJM198908173210702. [DOI] [PubMed] [Google Scholar]
- 6.Eisenberger M.A., Blumenstein B.A., Crawford E.D., Miller G., McLeod D.G., Loehrer P.J., Wilding G., Sears K., Culkin D.J., Thompson I.M., et al. Bilateral orchiectomy with or without flutamide for metastatic prostate cancer. N. Engl. J. Med. 1998;339:1036–1042. doi: 10.1056/NEJM199810083391504. [DOI] [PubMed] [Google Scholar]
- 7.Denis L.J., Keuppens F., Smith P.H., Whelan P., de Moura J.L., Newling D., Bono A., Sylvester R. Maximal androgen blockade: Final analysis of eortc phase iii trial 30853. Eortc genito-urinary tract cancer cooperative group and the eortc data center. Eur. Urol. 1998;33:144–151. doi: 10.1159/000019546. [DOI] [PubMed] [Google Scholar]
- 8.Prostate Cancer Trialists’ Collaborative Group Maximum androgen blockade in advanced prostate cancer: An overview of the randomised trials. Lancet. 2000;355:1491–1498. [PubMed] [Google Scholar]
- 9.Kantoff P.W., Higano C.S., Shore N.D., Berger E.R., Small E.J., Penson D.F., Redfern C.H., Ferrari A.C., Dreicer R., Sims R.B., et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N. Engl. J. Med. 2010;363:411–422. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 10.Fizazi K., Scher H.I., Molina A., Logothetis C.J., Chi K.N., Jones R.J., Staffurth J.N., North S., Vogelzang N.J., Saad F., et al. Abiraterone acetate for treatment of metastatic castration-resistant prostate cancer: Final overall survival analysis of the COU-AA-301 randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 2012;13:983–992. doi: 10.1016/S1470-2045(12)70379-0. [DOI] [PubMed] [Google Scholar]
- 11.Scher H.I., Fizazi K., Saad F., Taplin M.-E., Sternberg C.N., Miller K., de Wit R., Mulders P., Chi K.N., Shore N.D., et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 12.Beer T.M., Armstrong A.J., Rathkopf D.E., Loriot Y., Sternberg C.N., Higano C.S., Iversen P., Bhattacharya S., Carles J., Chowdhury S., et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N. Engl. J. Med. 2014;371:424–433. doi: 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sartor O., Coleman R., Nilsson S., Heinrich D., Helle S.I., O’Sullivan J.M., Fosså S.D., Chodacki A., Wiechno P., Logue J., et al. Effect of radium-223 dichloride on symptomatic skeletal events in patients with castration-resistant prostate cancer and bone metastases: Results from a phase 3, double-blind, randomised trial. Lancet Oncol. 2014;15:738–746. doi: 10.1016/S1470-2045(14)70183-4. [DOI] [PubMed] [Google Scholar]
- 14.De Bono J.S., Logothetis C.J., Molina A., Fizazi K., North S., Chu L., Chi K.N., Jones R.J., Goodman O.B., Saad F., et al. Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan C.J., Smith M.R., de Bono J.S., Molina A., Logothetis C.J., de Souza P., Fizazi K., Mainwaring P., Piulats J.M., Ng S., et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N. Engl. J. Med. 2013;368:138–148. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rathkopf D.E., Smith M.R., de Bono J.S., Logothetis C.J., Shore N.D., de Souza P., Fizazi K., Mulders P.F.A., Mainwaring P., Hainsworth J.D., et al. Updated interim efficacy analysis and long-term safety of abiraterone acetate in metastatic castration-resistant prostate cancer patients without prior chemotherapy (COU-AA-302) Eur. Urol. 2014;66:815–825. doi: 10.1016/j.eururo.2014.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan C.J., Smith M.R., Fizazi K., Saad F., Mulders P.F.A., Sternberg C.N., Miller K., Logothetis C.J., Shore N.D., Small E.J., et al. Abiraterone acetate plus prednisone versus placebo plus prednisone in chemotherapy-naive men with metastatic castration-resistant prostate cancer (COU-AA-302): Final overall survival analysis of a randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 2015;16:152–160. doi: 10.1016/S1470-2045(14)71205-7. [DOI] [PubMed] [Google Scholar]
- 18.Miller W.L., Auchus R.J. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr. Rev. 2011;32:81–151. doi: 10.1210/er.2010-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeVore N.M., Scott E.E. Structures of cytochrome p450 17a1 with prostate cancer drugs abiraterone and tok-001. Nature. 2012;482:116–119. doi: 10.1038/nature10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newton-Cheh C., Johnson T., Gateva V., Tobin M.D., Bochud M., Coin L., Najjar S.S., Zhao J.H., Heath S.C., Eyheramendy S., et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy D., Ehret G.B., Rice K., Verwoert G.C., Launer L.J., Dehghan A., Glazer N.L., Morrison A.C., Johnson A.D., Aspelund T., et al. Genome-wide association study of blood pressure and hypertension. Nat. Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ehret G.B., Munroe P.B., Rice K.M., Bochud M., Johnson A.D., Chasman D.I., Smith A.V., Tobin M.D., Verwoert G.C., Hwang S.-J., et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Attard G., Reid A.H.M., Olmos D., de Bono J.S. Antitumor activity with CYP17 blockade indicates that castration-resistant prostate cancer frequently remains hormone driven. Cancer Res. 2009;69:4937–4940. doi: 10.1158/0008-5472.CAN-08-4531. [DOI] [PubMed] [Google Scholar]
- 24.Beuten J., Gelfond J.A.L., Franke J.L., Weldon K.S., Crandall A.C., Johnson-Pais T.L., Thompson I.M., Leach R.J. Single and multigenic analysis of the association between variants in 12 steroid hormone metabolism genes and risk of prostate cancer. Cancer Epidemiol. Biomark. Prev. 2009;18:1869–1880. doi: 10.1158/1055-9965.EPI-09-0076. [DOI] [PubMed] [Google Scholar]
- 25.Setiawan V.W., Schumacher F.R., Haiman C.A., Stram D.O., Albanes D., Altshuler D., Berglund G., Buring J., Calle E.E., Clavel-Chapelon F., et al. CYP17 genetic variation and risk of breast and prostate cancer from the national cancer institute breast and prostate cancer cohort consortium (BPC3) Cancer Epidemiol. Biomark. Prev. 2007;16:2237–2246. doi: 10.1158/1055-9965.EPI-07-0589. [DOI] [PubMed] [Google Scholar]
- 26.Lindström S., Wiklund F., Adami H.-O., Bälter K.A., Adolfsson J., Grönberg H. Germ-line genetic variation in the key androgen-regulating genes androgen receptor, cytochrome p450, and steroid-5-α-reductase type 2 is important for prostate cancer development. Cancer Res. 2006;66:11077–11083. doi: 10.1158/0008-5472.CAN-06-3024. [DOI] [PubMed] [Google Scholar]
- 27.Lindström S., Adami H.-O., Bälter K.A., Xu J., Zheng S.L., Stattin P., Grönberg H., Wiklund F. Inherited variation in hormone-regulating genes and prostate cancer survival. Clin. Cancer Res. 2007;13:5156–5161. doi: 10.1158/1078-0432.CCR-07-0669. [DOI] [PubMed] [Google Scholar]
- 28.Wang F., Zou Y.-F., Feng X.-L., Su H., Huang F. CYP17 gene polymorphisms and prostate cancer risk: A meta-analysis based on 38 independent studies. Prostate. 2011;71:1167–1177. doi: 10.1002/pros.21332. [DOI] [PubMed] [Google Scholar]
- 29.Mostaghel E.A., Marck B.T., Plymate S.R., Vessella R.L., Balk S., Matsumoto A.M., Nelson P.S., Montgomery R.B. Resistance to CYP17A1 inhibition with abiraterone in castration-resistant prostate cancer: Induction of steroidogenesis and androgen receptor splice variants. Clin. Cancer Res. 2011;17:5913–5925. doi: 10.1158/1078-0432.CCR-11-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fizazi K., Flaig T.W., Stöckle M., Scher H.I., de Bono J.S., Rathkopf D.E., Ryan C.J., Kheoh T., Li J., Todd M.B., et al. Does gleason score at initial diagnosis predict efficacy of abiraterone acetate therapy in patients with metastatic castration-resistant prostate cancer? An analysis of abiraterone acetate phase iii trials. Ann. Oncol. 2016;27:699–705. doi: 10.1093/annonc/mdv545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bellmunt J., Kheoh T., Yu M.K., Smith M.R., Small E.J., Mulders P.F.A., Fizazi K., Rathkopf D.E., Saad F., Scher H.I., et al. Prior endocrine therapy impact on abiraterone acetate clinical efficacy in metastatic castration-resistant prostate cancer: Post-hoc analysis of randomised phase 3 studies. Eur. Urol. 2015;69:924–932. doi: 10.1016/j.eururo.2015.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies R.S., Smith C., Button M.R., Tanguay J., Barber J., Palaniappan N., Staffurth J., Lester J.F. What predicts minimal response to abiraterone in metastatic castrate-resistant prostate cancer? Anticancer Res. 2015;35:5615–5621. [PubMed] [Google Scholar]
- 33.Diver L.A., MacKenzie S.M., Fraser R., McManus F., Freel E.M., Alvarez-Madrazo S., McClure J.D., Friel E.C., Hanley N.A., Dominiczak A.F., et al. Common polymorphisms at the CYP17A1 locus associate with steroid phenotype: Support for blood pressure genome-wide association study signals at this locus. Hypertension. 2016;67:724–732. doi: 10.1161/HYPERTENSIONAHA.115.06925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iversen A., Thune I., McTiernan A., Makar K.W., Wilsgaard T., Ellison P.T., Jasienska G., Flote V., Poole E.M., Furberg A.-S. Genetic polymorphism CYP17 rs2486758 and metabolic risk factors predict daily salivary 17β-estradiol concentration in healthy premenopausal norwegian women. The EBBA-I study. J. Clin. Endocrinol. Metab. 2012;97:E852–E857. doi: 10.1210/jc.2011-2577. [DOI] [PubMed] [Google Scholar]
- 35.Salvi S., Casadio V., Conteduca V., Burgio S.L., Menna C., Bianchi E., Rossi L., Carretta E., Masini C., Amadori D., et al. Circulating cell-free ar and CYP17A1 copy number variations may associate with outcome of metastatic castration-resistant prostate cancer patients treated with abiraterone. Br. J. Cancer. 2015;112:1717–1724. doi: 10.1038/bjc.2015.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agarwal N., Alex A.B., Farnham J.M., Patel S., Gill D., Buckley T.H., Stephenson R.A., Cannon-Albright L. Inherited variants in sult1e1 and response to abiraterone acetate in men with metastatic castration refractory prostate cancer (mCRPC) J. Urol. 2016 doi: 10.1016/j.juro.2016.04.079. [DOI] [PubMed] [Google Scholar]
- 37.Antonarakis E.S., Lu C., Wang H., Luber B., Nakazawa M., Roeser J.C., Chen Y., Mohammad T.A., Chen Y., Fedor H.L., et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeuchi T., Okuno Y., Hattori-Kato M., Zaitsu M., Mikami K. Detection of AR-V7 mrna in whole blood may not predict the effectiveness of novel endocrine drugs for castration-resistant prostate cancer. Res. Rep. Urol. 2016;8:21–25. doi: 10.2147/RRU.S98877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohli M., Riska S.M., Mahoney D.W., Chai H.S., Hillman D.W., Rider D.N., Costello B.A., Qin R., Lamba J., Sahasrabudhe D.M., et al. Germline predictors of androgen deprivation therapy response in advanced prostate cancer. Mayo Clin. Proc. 2012;87:240–246. doi: 10.1016/j.mayocp.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia S., Kohli M., Du M., Dittmar R.L., Lee A., Nandy D., Yuan T., Guo Y., Wang Y., Tschannen M.R., et al. Plasma genetic and genomic abnormalities predict treatment response and clinical outcome in advanced prostate cancer. Oncotarget. 2015;6:16411–16421. doi: 10.18632/oncotarget.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang X., Yuan T., Liang M., Du M., Xia S., Dittmar R., Wang D., See W., Costello B.A., Quevedo F., et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur. Urol. 2015;67:33–41. doi: 10.1016/j.eururo.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Bakker P.I.W., Yelensky R., Pe'er I., Gabriel S.B., Daly M.J., Altshuler D. Efficiency and power in genetic association studies. Nat. Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 43.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. Mach: Using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.International HapMap Consortium The international hapmap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 45.Kaplan E.L., Meier P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958;53:457–481. doi: 10.1080/01621459.1958.10501452. [DOI] [Google Scholar]
- 46.Cox D.R. Regression models and life-tables. J. R. Stat. Soc. B. 1972;34:187–220. [Google Scholar]
- 47.Aulchenko Y.S., Struchalin M.V., van Duijn C.M. Probabel package for genome-wide association analysis of imputed data. BMC Bioinform. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao X., Starmer J., Martin E.R. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet. Epidemiol. 2008;32:361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 49.Gao X., Becker L.C., Becker D.M., Starmer J.D., Province M.A. Avoiding the high bonferroni penalty in genome-wide association studies. Genet. Epidemiol. 2010;34:100–105. doi: 10.1002/gepi.20430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao X. Multiple testing corrections for imputed snps. Genet. Epidemiol. 2011;35:154–158. doi: 10.1002/gepi.20563. [DOI] [PMC free article] [PubMed] [Google Scholar]