Abstract
Human proteins can exist as multiple proteoforms with potential diagnostic or prognostic significance. MS top-down approaches are ideally suited for proteoforms identification because there is no prerequisite for a priori knowledge of the specific proteoform. One such top-down approach, termed mass spectrometric immunoassay utilizes antibody-derivatized microcolumns for rapid and contained proteoforms isolation and detection via MALDI-TOF MS. The mass spectrometric immunoassay can also provide quantitative measurement of the proteoforms through inclusion of an internal reference standard into the analytical sample, serving as normalizer for all sample processing and data acquisition steps. Reviewed here are recent developments and results from the application of mass spectrometric immunoassays for discovery of clinical correlations of specific proteoforms for the protein biomarkers RANTES, retinol binding protein, serum amyloid A and apolipoprotein C-III.
Keywords: : immunoaffinity, MALDI, MS, plasma, proteoform
Background
MS in the form of ESI or MALDI TOF has played an important role in the past 30+ years in studying protein structure and function. A whole new field of ‘proteomics’ was coined to describe advances in technology and applications that were enabled by MS. That was followed-up by a significant undertaking to discover new protein biomarkers, producing hundreds of biomarker candidates. However, the pace of protein biomarker discovery has slowed down in recent years, partly attributed to the lack of robust MS methods and platforms for downstream biomarker validation. In another sign of weakness, MS has yet to make a significant inroad into clinical laboratories and find use in protein diagnostics applications.
While the MS detection itself in principle is quite simple, sample preparation and processing prior to MS detection is quite complex and time consuming. Various approaches and techniques have been used as front-end separations of proteins in preparation for MS analysis, including 2D gel electrophoresis, HPLC, capillary electrophoresis, immunoaffinity chromatography, etc. However, none of these were designed with MS detection in mind – they were simply existing technologies bolted in front of mass spectrometers, with the hope of creating multiplexed all-in-one approaches. The resulting complexity of these multiplexed approaches is what is hindering their widespread application in biomarker validation studies and preventing their adoption and translation into clinical laboratories.
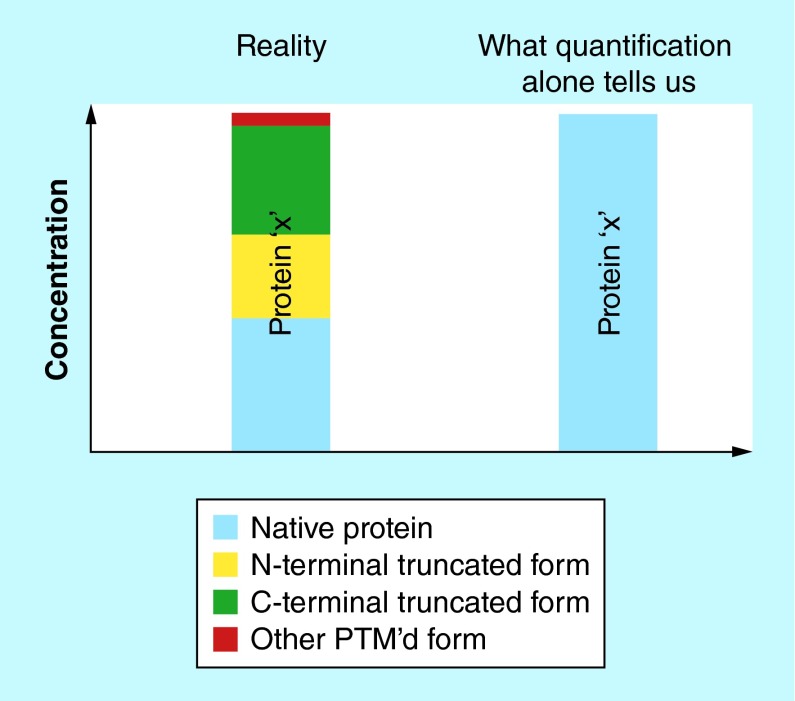
On the other end of the clinical assays spectrum are protein enzymatic immunoassays (EIAs), which have been a mainstay in clinical laboratories for the past 30+ years. These cost-effective and high-throughput quantitative protein tests have a simple modus operandi: a protein from human sample is bound to an immobilized antibody and then detected with another antibody labeled with a fluorophore, radioisotope, or reporter enzyme through generation of a measurable signal. While it is expected that EIAs will continue to dominate the clinical assays arena for the foreseeable future, they do have a certain limitation: EIAs are blind to the existence of protein variants (or proteoforms [1]). Proteoforms may exist in vivo for many proteins, as a result of alternative splicing [2], nonsynonymous single nucleotide polymorphisms (SNPs) [3] and post-translational modifications (PTMs) [4]. (Note: the term proteoform includes both SNP-derived forms of the proteins traditionally referred to as ‘isoforms’, and other PTM-derived protein species normally referred to as ‘variants’). In EIAs, the secondary (labeled) reporter antibody cannot discriminate between structural protein modifications, so the resulting quantitative signal is the sum of signals from all (or few) proteoforms for a given protein captured by the primary antibody (Figure 1). Because proteoforms may have potential diagnostic or prognostic significance, it is important to develop analytical methods for their detection and quantification.
Figure 1. . Limitation of enzymatic immunoassays for proteoforms quantification.
A protein may exist as several proteoforms, but enzymatic immunoassays cannot detect and quantify individual proteoforms.
Proteoforms were initially exposed in the early eighties with one-dimensional gel electrophoresis, starting with the identification of hemoglobin, α-antitrypsin, amylase and transthyretin proteoforms. With protein MS advances, proteoforms were detected via 2D gel electrophoresis coupled with mass spectrometric protein identification from excised and digested protein gel spots [5]. 2D difference gel electrophoresis (DIGE) was also used to examine proteoforms distributions across populations [6–8]. More recently, several bottom-up and top-down MS approaches have been used for decoding protein modifications [9–11]. The most popular bottom-up approach involves initial protein digestion with trypsin, followed by selected reaction monitoring (SRM) or multiple reaction monitoring (MRM) MS detection of the resulting peptides that contain the protein modifications (straight from the human plasma for higher-abundance proteins, or immunoenriched for lower abundance proteins). This approach has been used for identification of osteopontin splice proteoforms [12], novel proteoforms of prostate specific antigen [13] and single amino acid polymorphism of complement component C7, complement factor H and complement component C5 [14].
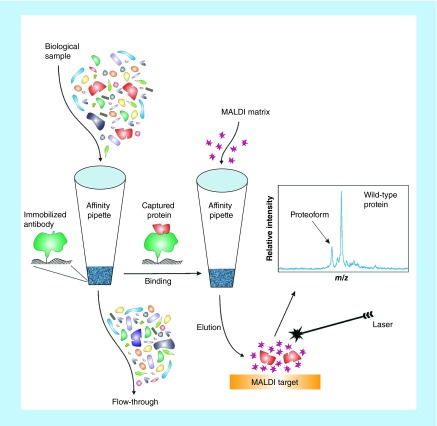
Top-down MS approaches are better suited for proteoforms identification because they do not require a priori knowledge of the protein modification in order to select the appropriate modification-specific peptide (as in MRM-MS). MS of intact proteins and their proteoforms can be challenging [15], but our group has developed a relatively straightforward top-down mass spectrometric immunoassay approach for proteoforms detection and quantification. The approach, termed mass spectrometric immunoassay [16,17] employs affinity microcolumns fitted at the entrance of pipettor tips that are derivatized with antibodies for fast and contained retrieval of proteins from complex biological samples, followed by MALDI-TOF MS detection of the eluted proteoforms (Figure 2). This two-step approach is similar to that of enzymatic immunoassays, but instead of secondary reporter antibody, MS is utilized to read out the masses of the captured proteins, and thus detect all proteoforms based on their distinct molecular mass. Most SNPs and post-translational modifications result in relatively small shifts in molecular mass of the proteoforms, producing signals in the mass spectra that are near the wild-type protein signal. These signals can be assigned to specific proteoforms by accurately measuring the observed mass shifts and matching them with the known protein sequence, and can be further verified via proteolytic digestion and detection of the modification-bearing peptides.
Figure 2. . Schematics of the mass spectrometric immunoassay.
The mass spectrometric immunoassay approach can also provide quantitative measurement of the proteoforms. An internal reference standard (IRS), introduced into the analytical sample, is affinity co-purified along with the protein and its proteoforms either by a polyclonal antibody with affinities toward the multiple forms of the protein and the IRS (if the IRS is a modified form of the protein), or with another antibody co-immobilized alongside the targeted protein antibody in the microcolumn. Mass spectrometric analysis produces signals from both the targeted intact protein and the IRS; the signal for the targeted protein is normalized to the signal of the IRS, and, with the use of a standard curve, the protein concentration is then determined [18–24]. It is important to note that the internal reference standard (IRS) is added to the sample at the beginning of the assay so that it goes through the same processing as does the protein target. This way, the IRS serves as a normalizer for all sample processing and data acquisition steps – from protein extraction and processing, to elution and MS response. The ideal IRS is a protein analog differing slightly in mass from the wild-type protein, for example, proteins from other species that differ very little in the amino acid sequence but are still recognized by the antibody [22,23].
We have developed assays for several proteins that exhibit proteoforms in vivo. Some notable examples include: a quantitative mass spectrometric immunoassay for detection of endogenous BNP with LOD and LOQ of <10 pg/ml, used for simultaneous detection of six BNP proteoforms (three newly discovered) in patients with heart failure at concentrations of 7–228 pg/ml [25]; and semiquantitative full-length PTH assay used for monitoring ten different proteoforms (five newly discovered) in renal failure patients [26]. In this review we summarize the latest results from the application of mass spectrometric immunoassays for discovery of clinical correlations of specific proteoforms for the protein biomarkers RANTES, retinol binding protein, serum amyloid A, and apolipoprotein C-III.
RANTES (CCL5) proteoforms
RANTES is a member of the CC chemokine subfamily (it is also called CCL5). RANTES is released from activated macrophages and T-lymphocytes, endothelial and epithelia cells, dermal fibroblast and renal tubular epithelium [27,28]. RANTES has been indicated in many diseases such as autoimmune [29], diabetes [30], obesity and metabolic syndrome [31], and cardiovascular disease [32–35]. RANTES is composed of 68 amino acids, with two intact disulfide bonds (MW = 7847) [36]. Its biological activity is modulated by two proteolysis pathways: the first pathway is mediated by DPP IV – a regulatory enzyme present on surface of many cell types, including activated T cells – which removes the two N-terminal amino acids, producing cleaved RANTES (Pro3–68) proteoform [37]; the second pathway is mediated by cathepsin G, producing an additional truncated proteoform (Tyr4–68) [38]. Because proteolysis modulates the RANTES activity, the study of the RANTES structure-function relationship is very important.
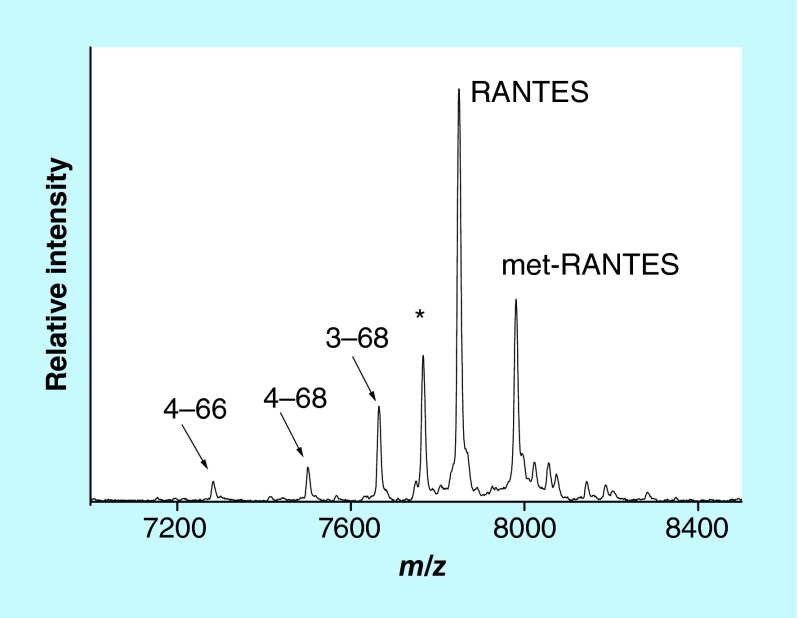
We have recently developed a mass spectrometric immunoassay for analysis of the RANTES proteoforms [39]. Representative mass spectrum resulting from the application of the assay to a human plasma sample is shown in Figure 3. Signals form several RANTES proteoforms are identified: full-length (wild-type) RANTES (residues 1–68, MW = 7847.9), RANTES missing a Ser-Pro N-terminal dipeptide as a result of DPP IV enzymatic cleavage (labeled 3–68, MW = 7663.7), RANTES missing three N-terminal residues (4–68, MW = 7500.6), and RANTES missing three N-terminal and two C-terminal residues (4–66, MW = 7282.3). In order to determine the individual concentration of each proteoform, the assay was made quantitative by inclusion of an internal reference standard (IRS). Consistent with our prior work using protein analogs [22,23], methionine-RANTES was chosen as an IRS. The met-RANTES differs from native RANTES in one additional N-terminus methionine, and it has a mass shift of + 131.2 Da from human RANTES in the mass spectrum (Figure 3). As required, met-RANTES is recognized (affinity-captured) by the antihuman RANTES antibody that binds the wild-type RANTES and its proteoforms. Most importantly, met-RANTES is not subject to cleavage and/or interactions with the enzymes present in human plasma (as previously determined [38], which was also confirmed in our laboratory through spike and recovery experiments of met-RANTES in human plasma).
Figure 3. . Mass spectrum of RANTES from human plasma sample obtained with the mass spectrometric immunoassay.
Signals are labeled according to the sequence of RANTES. Met-RANTES is the internal reference standard spiked into the sample.
*A nonspecifically bound protein.
The quantitative RANTES mass spectrometric immunoassay was applied to analysis of plasma samples obtained from 297 individuals with Type 2 diabetes (not treated with DPP IV inhibitor medication) [39]. The assays were performed on an automated high-throughput platform, 96 samples at a time. From the resulting mass spectra, individual proteoform/IRS peak heights ratios were determined, ratios were summed up for all proteoforms, total RANTES concentration was determined using the corresponding standard curve, and then the concentration of each proteoform was calculated based on its percentage of the total protein concentration. This approach worked especially well for the proteoform/IRS peak ratios that were below those of the standard curve. The mean concentration of all RANTES proteoforms in the cohort was 44.9 ng/ml (2.15–163 ng/ml range). The mean and the range were slightly higher than the reference levels for RANTES in normal plasma samples [40], which could be attributed to changes in inflammatory responses that are common in patients with abnormal glucose metabolism [41]. In majority of samples, the most abundant proteoforms were the full-length (wild-type) RANTES (mean = 37.4 ng/ml) and the DPP IV-cleaved RANTES 3–68 (6.64 ng/ml). These two proteoforms were detected in every plasma sample. The truncated proteoforms 4–68 and 4–66 were present less frequently (in 206 and 87 samples, respectively), and at much lower mean concentrations (0.94 ng/ml and 0.95 ng/ml, respectively). They, nevertheless, deserve full attention because extensive proteolytic cleavage can be indicator of a specific enzymatic activity associated with disease states. Studies will continue in this direction, with closer examination of the relationship of some of these proteoforms and the cohort's clinical and biochemical characteristics.
Retinol binding protein proteoforms in insulin resistance & coronary heart disease risk
RBP is a member of the lipocalin family synthesized in liver and adipose tissues [42]. RBP is secreted into circulation where it serves as a major transport protein for retinol (vitamin A). RBP also forms a complex with transthyretin, avoiding the glomerular filtration process and excretion through the kidneys. Circulating RBP concentrations are influenced by several factors, most notably renal function [43,44]. However, elevated RBP plasma levels have also been positively associated with insulin resistance and Type 2 diabetes [45]. Moreover, RBP has been implicated as an emerging cardiometabolic risk factor [46,47].
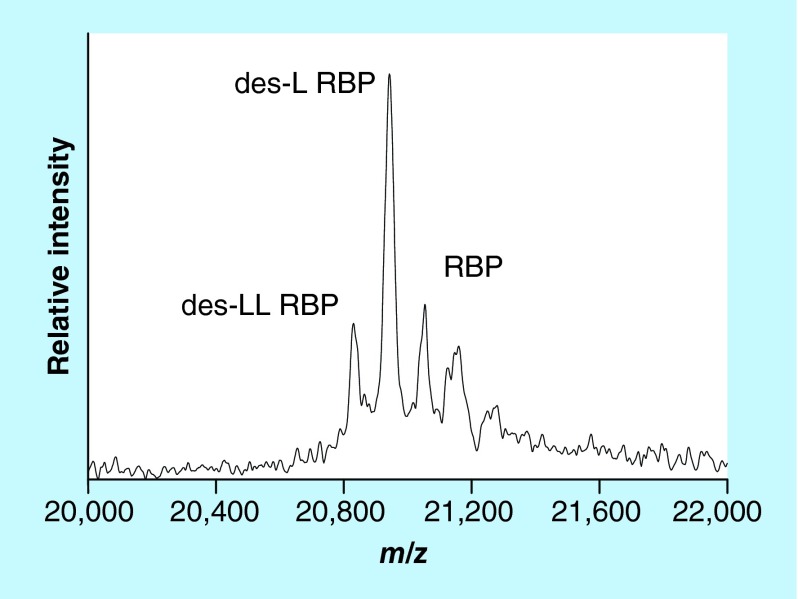
Wild-type (full-length) RBP is comprised of 123 amino acids (MW = 21,065). Using SDS-PAGE separation along with MS, Jaconi et al. were the first to identify two post-translationally modified proteoforms of RBP – one missing a single C-terminal leucine residue (des-L RBP, MW = 20,952), and the other missing two C-terminal leucines (des-LL RBP, MW = 20,839) [48]. The levels of these truncated forms were found to be increased in patients with chronic renal failure, suggesting impaired clearance of these proteoforms by the kidneys in those patients. Our group developed targeted RBP mass spectrometric immunoassay that also detected these two proteoforms [49]. Representative mass spectrum resulting from the application of the assay to a human plasma sample is shown in Figure 4. When the RBP mass spectrometric immunoassay was applied to two large cohorts of healthy subjects, additional truncated proteoforms were detected, including des-NLL, des-RNLL and des-SERNLL RBP proteoforms [50,51]. A fully quantitative RBP mass spectrometric immunoassay was subsequently developed [18] and first applied to determine the concentration of all RBP proteoforms in a population of 500 healthy individuals [52]. Wild-type (full-length) RBP was present at the highest concentration; des-L was less abundant but still present in all 500 samples, whereas des-LL RBP was detected and quantified in half of the cohort. The des-RNLL and des-RNLL proteoforms were present in only few of the samples.
Figure 4. . Mass spectrum of RBP from human plasma sample obtained with the mass spectrometric immunoassay.
des-L RBP: Missing one C-terminal leucine residue (MW = 20,952); des-LL RBP: Missing two C-terminal leucines (MW = 20,839); RBP: Wild-type protein (MW = 21,065).
The most recent applications of the quantitative RBP mass spectrometric immunoassay revealed some very interesting clinical implications for the truncated RBP proteoforms in specific medical conditions. In the first study, full-length, des-L and des-LL RBP proteoforms were measured in serum samples obtained from individuals with normal glucose tolerance (NGT), insulin-resistant with impaired glucose tolerance (IGT) and individuals that had Type 2 diabetes (T2D) [53]. It was found that RBP des-L correlated highly with insulin resistance – its levels were as much as fourfold higher in IGT and T2D patients than in NGT patients. In a second study, the RBP proteoforms were evaluated in relation to the development of a coronary heart disease among women in the Nurses’ Health Study [54]. The nested case–control study design consisted of 468 women who developed coronary heart disease and 472 matched controls, during 16 years of follow-up. The examination of the relationship between the RBP proteoforms levels and other cardiovascular disease factors in the control group revealed a positive correlation between the des-L RBP proteoform and total cholesterol and fasting triacylglycerol, and negative correlation between the des-L RBP proteoform and the estimated glomerular filtration rate (eGFR). The key finding of the study was that only full-length RBP levels (and not the des-L and des-LL RBP proteoforms) were associated with higher coronary heart disease risk, suggesting that the full-length RBP may be the most biologically active proteoform. It is important to note that the C-terminus of RBP is nestled in a hydrophobic patch at the RBP-transthyretin protein complex interface [55], so it is quite possible that truncation of RBP diminishes its binding to transthyretin. These results illustrate the importance of measuring individual proteoforms, and more importantly – the biologically active proteoforms. Such measurement cannot be done with other approaches, such as quantitative Western blotting or ELISAs in the case of RBP. It should be mentioned that in order to mimic the experimental conditions used for the existing quantitative Western blotting RBP assay, we have modified the RBP mass spectrometric immunoassay protocol to include a denaturing agent (SDS), thus eliminating any remote possibility of RBP epitope masking by the RBP-TTR complex. When compared, the total serum levels of RBP measured with the mass spectrometric immunoassay (sum of all proteoforms concentrations) correlated highly with those measured by quantitative Western blotting [53]; however, quantitative western blotting cannot distinguish among the individual RBP proteoforms and is thus unfit for studies where their clinical correlations and significance are evaluated.
Apolipoprotein C-III proteoforms’ association with plasma triglycerides
ApoC-III is a component of the triglyceride-rich very low density lipoproteins (VLDL) and high density lipoproteins (HDL) in plasma [56]. ApoC-III plays an important role in triglyceride metabolism by inhibiting the lipolysis of triglyceride-rich lipoproteins. Recent studies revealed that a loss-of-function mutation in the apoC-III gene is associated with reduced triglyceride levels and cardiovascular risk [57–59]. This has spurred drug development programs for lowering the apoC-III levels, and consequently the levels of the triglycerides [60,61]. The sequence of apoC-III contains 79 amino acids, with a single glycosylation site at Threonine 74. The glycosylation produces three major proteoforms of apoC-III. All three share a core glycan comprised of an O-linked disaccharide galactose linked to N-acetylgalactosamine (Gal-GalNAc). This core is further modified with one or two sialic residues (Sia), giving rise to what's commonly known as apoC-III1 and apoC-III2, respectively (the proteoform containing just the core, without the sialic acid residues, is known as apoC-III0b; the wild-type protein (without the core glycan chain) is usually referred to as apoC-III0a). These three major proteoforms were first delineated in the early 80s, using isoelectric focusing techniques. When mass spectrometric methods of detection were applied in the late 90s, numerous other truncated apoC-III proteoforms were detected in dilapidated fractions obtained from very low density lipoprotein (VLDL), including C-terminally truncated forms missing one or two alanine residues [62]. Several other groups have applied MS-based methods for studying the apoCs proteoforms [63–65], discovering additional low-abundance proteoforms containing fucose in the glycan chain [66,67].
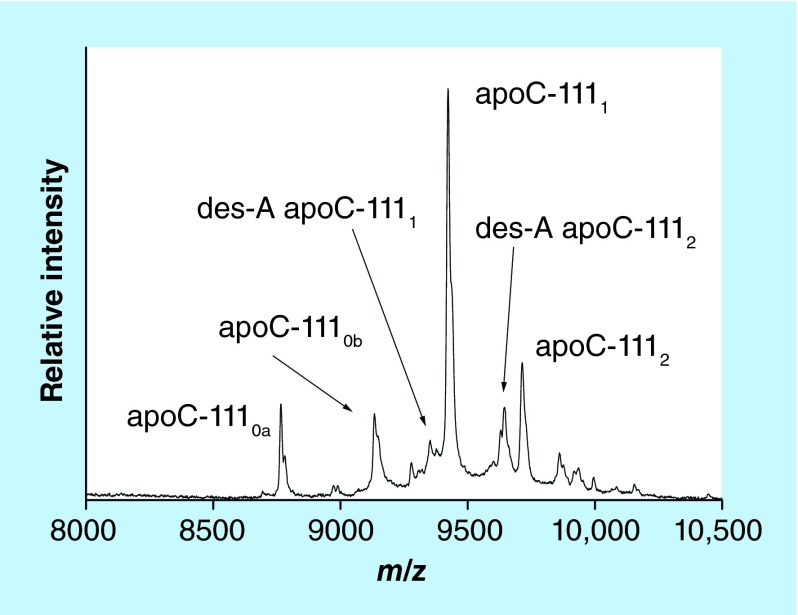
Our group developed an apoC-III mass spectrometric immunoassay for direct and high-throughput analysis of these proteoforms from plasma or serum samples. Representative mass spectrum resulting from the application of the assay to a human plasma sample is shown in Figure 5. The assay was initially applied to a cohort of healthy patients, detecting all three major apoC-III glycoproteoforms in all of the samples [50]. Recently our group has advanced the assay to a fully quantitative mode in order to determine the individual proteoforms concentrations, and applied it to a cohort of 82 individuals [24]. A total of 12 apoC-III proteoforms were quantified in the cohort. The main glycosylated proteoforms (apoC-III0b, C-III1 and C-III2) accounted for 90% of the total apoC-III concentration; alanine-cleaved proteoforms were also present in majority of the samples, at 1–5% of the total concentration. Least abundant were the fucosylated proteoforms, detected in approximately 20% of the samples; their different distribution among samples with similar total apoC-III concentrations implies different pathways for the fucosylation process.
Figure 5. . Mass spectrum of apo C-III from human plasma sample obtained with the mass spectrometric immunoassay.
apoC-III0a: wild-type apoC-III (no glycan); apoC-III0b: apoC-III-Gal-GalNac; apoC- III1: apoC-III-Gal-GalNac-Sia; des-A apoC- III1: C-terminally truncated alanine from apoC-III1; apoC- III2: apoC-III-Gal-GalNac-Sia-Sia; des-A apoC- III2: C-terminally truncated alanine from apoC-III2.
Because apoC-III regulates triglycerides, we decided to examine the role of the apoC-III sialylation in triglycerides metabolism using several different cohorts. In the first study, plasma samples from a cohort of 204 adolescent Hispanic nondiabetic children were analyzed with the apoC-III mass spectrometric immunoassay [68]. It was found that the ratios of apoC-III0a, apoC-III0b and apoC-III1 to apoC-III2 were significantly greater in overweight and obese participants versus the control group. Total apoC-III correlated strongly with plasma triglycerides concentrations, but this association was driven primarily by apoC-III0a, apoC-III0b and apoC-III1, and not by apoC-III2. ApoC-III2 was also not correlated to the other apoC-III proteoforms, suggesting different mechanism for the addition of the second sialic acid residue. These results indicate that apoC-III0 and C-III1 are under metabolic control.
In the second study, cross-sectional and longitudinal associations between apoC-III proteoforms and plasma lipids were examined in two randomized clinical trials in subjects with impaired glucose tolerance (n = 531) and Type 2 diabetes (n = 296). At baseline, higher relative apoC-III2 abundance was associated with lower triglycerides and total cholesterol, and higher HDL cholesterol concentrations in both cohorts [69]. Relative abundances of the other apoC-III proteoforms showed positive associations with the triglycerides. Furthermore, longitudinal changes in apoC-III2/apoC-III1 ratios were inversely associated with changes in triglycerides in both cohorts. The association between relative apoC-III2 abundance and triglycerides remained present after adjustments for demographic factors and lipid lowering medications, suggesting that apoC-III2 abundance may be an independent determinant of plasma triglyceride levels. These strong cross-sectional and longitudinal inverse relationships between the relative abundance of apoC-III2 proteoform and plasma triglyceride concentrations were very similar to the results from the adolescent cohort study described above, despite differences in the cohorts’ demographic and clinical characteristics. These results imply that different apoC-III proteoforms may have different effects on lipid metabolism.
Serum amyloid A proteoforms in Type 2 diabetes
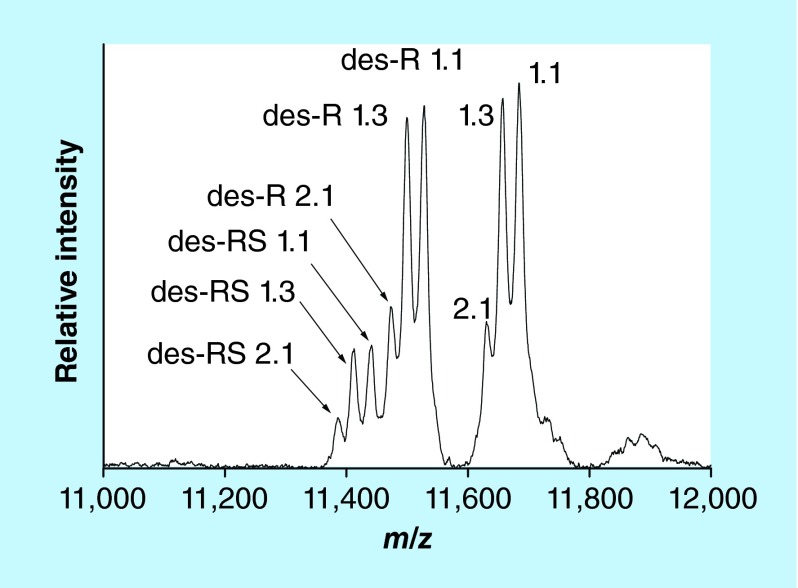
SAA is an acute phase protein with an important role in inflammatory processes [70,71]. SAA encompasses a group of closely related proteins produced by four genes, three of which are expressed in humans (SAA1, SAA2 and SAA4) [72]. The SAA1 and SAA2 genes express five (designated 1.1, 1.2, 1.3, 1.4 and 1.5) and two (2.1 and 2.2) acute phase allelic variants, resulting in multiple SAA proteoforms with highly homologous protein sequences [73]. The N-terminal amino-acids of SAA are subject to post-translational truncation [71]. Commercially available ELISAs cannot detect these truncations, and neither can genotyping and allele frequency analyses which are commonly used to investigate SAA polymorphism. Hence, MS is ideally suited for the analysis of these truncated SAA proteoforms. Several MS-based methods for SAA have been developed, including 2D-gel electrophoresis coupled with liquid chromatography/ESI MS [74], protein chip immunoaffinity capture with SELDI MS [75] and in-gel trypsin digestion followed by ESI-MS of SAA peptides [76]. Our group developed mass spectrometric immunoassay for analysis of the SAA proteoforms from plasma or serum samples [77]. Representative mass spectrum resulting from the application of the assay to a human plasma sample is shown in Figure 6. The assay was initially applied to a cohort of healthy patients (n = 96), revealing several allelic variants (1.1, 1.3, and 2.1) and N-terminally truncated (des-R and des-RS) proteoforms [50]. Recently, the abundance of the SAA truncations relative to the wild-type proteins was examined in plasma samples obtained from a cohort of 91 patients with Type 2 diabetes and chronic kidney disease and 69 healthy controls [78]. Four allelic variants were detected in the cohort – SAA 1.1, 1.3, 2.1, and 2.2 – expressed either as single or multiple proteoforms. The 1.1 proteoform was detected in all but one individual, with the other three being expressed less frequently (59% for SAA 2.1, 13% SAA 2.2 and 12% SAA 1.3). The des-R and des-RS N-terminal truncations were detected for all allelic variants. The relative abundances of each truncated proteoform (calculated as a percent of the wild-type) were compared between subjects with and without diabetes. After adjusting for age and body mass index, the des-R SAA 1.1 proteoform was found to be lower in subjects with Type 2 diabetes. Also, the relative abundance of the same proteoform was found to be lower in males compared with females in the entire cohort. Measuring these SAA proteoforms is important because the SAA truncations may modify SAA function and clearance. Furthermore, because the N-terminus of SAA determines SAA amyloid fibril formation [79], the truncations may alter the susceptibility to reactive amyloidosis in chronic conditions such as Type 2 diabetes.
Figure 6. . Mass spectrum of SAA from human plasma sample obtained with the mass spectrometric immunoassay.
The numbers designate allelic genes; des-R and des-RS designate N-terminally cleaved arginine and serine.
Conclusion
Presented here were several examples of mass spectrometric immunoassays for clinically relevant proteoforms. These studies were made possible by the relatively simple and high-throughput method of mass spectrometric immunoassay. The method relies on the use of antibodies for retrieval of multiple proteoforms, which is followed by their mass spectrometric detection. In its current iteration the method is ideally suited for high-throughput analysis of proteins and their proteoforms from human plasma and serum. When put together with well-characterized clinical cohorts, these methods can provide rapid assessment of proteoforms and their roles in specific diseases.
Future perspective
Proteoforms are formed in vivo and may have clinical significance. The two most prominent examples of clinically relevant proteoforms are carbohydrate-deficient transferrin which is monitored to detect congenital disorders of glycosylation and chronic alcohol abuse [80], and glycated hemoglobin A1c which is used to monitor long-term glycemic control [81]. Development of methods for proteoforms detection and quantification is thus of great significance. Certain proteoforms can be detected at the gene level (alternative splicing- and SNP-products), but for all others detection at the protein level is needed. MS-based methods are ideally suited for this task, especially top-down methods that provide direct readout of the intact molecular mass – an intrinsic property of each proteoform. The discovery of proteoforms with such methods will have to be followed by extensive validation studies, using large and well-defined clinical cohorts. Our group has recently made progress in associating certain proteoforms of serum amyloid A and apolipoprotein CIII with glycemic control and triglyceride metabolism [68,69,78]. More proteoforms discoveries and validations are certain to occur in the near term, but there is still a significant technological bottleneck facing these studies. Most proteomics work today is executed with LC–MS workflows that are geared toward detection of proteolytic peptides. Such peptides do not provide a whole view of the wild-type parent protein and its proteoforms. Furthermore, high cost and complexity hinders the application of these methods in proteoforms discovery and validation. On the other hand, MALDI-TOF MS is ideally suited for intact protein analysis, and with its robustness, ease-of-use, and low maintenance cost, could become the platform of choice for large-scale proteoforms detection [52] and future clinical protein tests [82]. MALDI platforms (FDA 510(k) approved) are already in microbiology hospital laboratories where they are used for microbial identification [83]. But regardless of the MS platform, the central bottleneck for protein (and proteoforms) MS analysis is the sample preparation – the fastest and cheapest way to bring a protein from solution to the inlet of the mass spectrometer. This is especially important in biomarker validation studies where the number of samples and assays is much higher than in the discovery phase, and where cost can play a significant role in go/no-go decisions. Until this sample preparation bottleneck is removed, mass spectrometric studies of proteoforms and their utility as the next generation biomarkers will remain a random pursuit
Executive summary.
Proteoforms may exist in vivo for many proteins, as a result of alternative splicing, nonsynonymous single nucleotide polymorphisms and post-translational modifications.
Important clinically relevant proteoforms include carbohydrate-deficient transferrin and glycated hemoglobin A1c.
Enzymatic immunoassay cannot detect and quantify individual proteoforms.
MS plays an important role in proteoforms detection as it provides direct readout of the molecular mass – an intrinsic property of each proteoform/modification.
Top-down mass spectrometric approaches are ideally suited for accurate and complete proteforms identification, without a priori knowledge of the protein modification.
One such top-down approach, termed mass spectrometric immunoassay, employs tip-fitted microcolumns derivatized with antibodies for fast and contained protein binding and elution for subsequent MALDI-TOF MS proteoforms detection.
The mass spectrometric immunoassay can also quantify individual proteoforms, through incorporation of an internal reference standard (IRS) introduced into the analytical sample, serving as a normalizer for all sample processing and data acquisition steps.
Screening of proteoforms can reveal their clinical association with specific disease states.
Footnotes
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Diabetes and Digestive and Kidney Diseases or the NIH.
Financial & competing interests disclosure
This work was supported in part by Grants R01DK082542 and R24DK090958 from the National Institute of Diabetes and Digestive and Kidney Diseases. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Smith LM, Kelleher NL Proteomics CfTD. Proteoform: a single term describing protein complexity. Nat. Methods. 2013;10(3):186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roy B, Haupt LM, Griffiths LR. Review: alternative splicing (AS) of genes as an approach for generating protein complexity. Curr. Genomics. 2013;14(3):182–194. doi: 10.2174/1389202911314030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu JR, Zeng R. Molecular basis for population variation: from SNPs to SAPs. FEBS Lett. 2012;586(18):2841–2845. doi: 10.1016/j.febslet.2012.07.036. [DOI] [PubMed] [Google Scholar]
- 4.Farley AR, Link AJ. Identification and quantification of protein posttranslational modifications. Methods Enzymol. 2009;463:725–763. doi: 10.1016/S0076-6879(09)63040-8. [DOI] [PubMed] [Google Scholar]
- 5.Kim H, Eliuk S, Deshane J, et al. 2D gel proteomics: an approach to study age-related differences in protein abundance or isoform complexity in biological samples. Methods Mol. Biol. 2007;371:349–391. doi: 10.1007/978-1-59745-361-5_24. [DOI] [PubMed] [Google Scholar]
- 6.Corzett TH, Fodor IK, Choi MW, et al. Statistical analysis of variation in the human plasma proteome. J. Biomed. Biotechnol. 2010:258494. doi: 10.1155/2010/258494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson D, Herath A, Swinton J, et al. Considerations for powering a clinical proteomics study: Normal variability in the human plasma proteome. Proteomics Clin. Appl. 2009;3(3):394–407. doi: 10.1002/prca.200800066. [DOI] [PubMed] [Google Scholar]
- 8.Ignjatovic V, Lai C, Summerhayes R, et al. Age-related differences in plasma proteins: how plasma proteins change from neonates to adults. PLoS ONE. 2011;6(2):e17213. doi: 10.1371/journal.pone.0017213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siuti N, Kelleher NL. Decoding protein modifications using top-down mass spectrometry. Nat. Methods. 2007;4(10):817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Ge Y. Comprehensive analysis of protein modifications by top-down mass spectrometry. Circ. Cardiovasc. Genet. 2011;4(6):711. doi: 10.1161/CIRCGENETICS.110.957829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stastna M, Van Eyk JE. Analysis of protein isoforms: can we do it better? Proteomics. 2012;12(19–20):2937–2948. doi: 10.1002/pmic.201200161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J, Pungaliya P, Kraynov E, Bates B. Identification and quantification of osteopontin splice variants in the plasma of lung cancer patients using immunoaffinity capture and targeted mass spectrometry. Biomarkers. 2012;17(2):125–133. doi: 10.3109/1354750X.2011.643485. [DOI] [PubMed] [Google Scholar]
- 13.Végvári A, Sjödin K, Rezeli M, et al. Identification of a novel proteoform of prostate specific antigen (SNP-L132I) in clinical samples by multiple reaction monitoring. Mol. Cell Proteomics. 2013;12(10):2761–2773. doi: 10.1074/mcp.M113.028365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su ZD, Sun L, Yu DX, et al. Quantitative detection of single amino acid polymorphisms by targeted proteomics. J. Mol. Cell Biol. 2011;3(5):309–315. doi: 10.1093/jmcb/mjr024. [DOI] [PubMed] [Google Scholar]
- 15.Tipton JD, Tran JC, Catherman AD, Ahlf DR, Durbin KR, Kelleher NL. Analysis of intact protein isoforms by mass spectrometry. J. Biol. Chem. 2011;286(29):25451–25458. doi: 10.1074/jbc.R111.239442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson RW, Borges CR. Mass spectrometric immunoassay revisited. J. Am. Soc. Mass Spectrom. 2011;22(6):960–968. doi: 10.1007/s13361-011-0094-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nedelkov D. Mass spectrometry-based protein assays for in vitro diagnostic testing. Expert Rev. Mol. Diagn. 2012;12(3):235–239. doi: 10.1586/erm.12.15. [DOI] [PubMed] [Google Scholar]
- 18.Kiernan UA, Phillips DA, Trenchevska O, Nedelkov D. Quantitative mass spectrometry evaluation of human retinol binding protein 4 and related variants. PLoS ONE. 2011;6(3):e17282. doi: 10.1371/journal.pone.0017282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trenchevska O, Kamcheva E, Nedelkov D. Mass spectrometric immunoassay for quantitative determination of protein biomarker isoforms. J. Proteome Res. 2010;9(11):5969–5973. doi: 10.1021/pr1007587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trenchevska O, Kamcheva E, Nedelkov D. Mass spectrometric immunoassay for quantitative determination of transthyretin and its variants. Proteomics. 2011;11(18):3633–3641. doi: 10.1002/pmic.201100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trenchevska O, Nedelkov D. Targeted quantitative mass spectrometric immunoassay for human protein variants. Proteome Sci. 2011;9(1):19. doi: 10.1186/1477-5956-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oran PE, Trenchevska O, Nedelkov D, et al. Parallel workflow for high-throughput (>1,000 samples/day) quantitative analysis of human insulin-like growth factor 1 using mass spectrometric immunoassay. PLoS ONE. 2014;9(3):e92801. doi: 10.1371/journal.pone.0092801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherma ND, Borges CR, Trenchevska O, et al. Mass spectrometric immunoassay for the qualitative and quantitative analysis of the cytokine macrophage migration inhibitory factor (MIF) Proteome Sci. 2014;12(1):52. doi: 10.1186/s12953-014-0052-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trenchevska O, Schaab MR, Nelson RW, Nedelkov D. Development of multiplex mass spectrometric immunoassay for detection and quantification of apolipoproteins C-I, C-II, C-III and their proteoforms. Methods. 2015;81:86–92. doi: 10.1016/j.ymeth.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niederkofler EE, Kiernan UA, O'Rear J, et al. Detection of endogenous B-type natriuretic peptide at very low concentrations in patients with heart failure. Circ. Heart Fail. 2008;1(4):258–264. doi: 10.1161/CIRCHEARTFAILURE.108.790774. [DOI] [PubMed] [Google Scholar]
- 26.Lopez MF, Rezai T, Sarracino DA, et al. Selected reaction monitoring-mass spectrometric immunoassay responsive to parathyroid hormone and related variants. Clin. Chem. 2010;56(2):281–290. doi: 10.1373/clinchem.2009.137323. [DOI] [PubMed] [Google Scholar]
- 27.Blanpain C, Doranz BJ, Vakili J, et al. Multiple charged and aromatic residues in CCR5 amino-terminal domain are involved in high affinity binding of both chemokines and HIV-1 Env protein. J. Biol. Chem. 1999;274(49):34719–34727. doi: 10.1074/jbc.274.49.34719. [DOI] [PubMed] [Google Scholar]
- 28.Brandt SM, Mariani R, Holland AU, Hope TJ, Landau NR. Association of chemokine-mediated block to HIV entry with coreceptor internalization. J. Biol. Chem. 2002;277(19):17291–17299. doi: 10.1074/jbc.M108232200. [DOI] [PubMed] [Google Scholar]
- 29.Lit LC, Wong CK, Tam LS, Li EK, Lam CW. Raised plasma concentration and ex vivo production of inflammatory chemokines in patients with systemic lupus erythematosus. Ann. Rheum. Dis. 2006;65(2):209–215. doi: 10.1136/ard.2005.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghanim H, Korzeniewski K, Sia CL, et al. Suppressive effect of insulin infusion on chemokines and chemokine receptors. Diabetes Care. 2010;33(5):1103–1108. doi: 10.2337/dc09-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matter CM, Handschin C. RANTES (regulated on activation, normal T cell expressed and secreted), inflammation, obesity, and the metabolic syndrome. Circulation. 2007;115(8):946–948. doi: 10.1161/CIRCULATIONAHA.106.685230. [DOI] [PubMed] [Google Scholar]
- 32.Winnik S, Klingenberg R, Matter CM. Plasma RANTES: a molecular fingerprint of the unstable carotid plaque? Eur. Heart J. 2011;32(4):393–395. doi: 10.1093/eurheartj/ehq376. [DOI] [PubMed] [Google Scholar]
- 33.von Hundelshausen P, Weber C. Platelets as immune cells: bridging inflammation and cardiovascular disease. Circ. Res. 2007;100(1):27–40. doi: 10.1161/01.RES.0000252802.25497.b7. [DOI] [PubMed] [Google Scholar]
- 34.Koenen RR, Weber C. Platelet-derived chemokines in vascular remodeling and atherosclerosis. Semin. Thromb. Hemost. 2010;36(2):163–169. doi: 10.1055/s-0030-1251500. [DOI] [PubMed] [Google Scholar]
- 35.Weber C. Chemokines in atherosclerosis, thrombosis, and vascular biology. Arterioscler. Thromb. Vasc. Biol. 2008;28(11):1896. doi: 10.1161/ATVBAHA.108.177311. [DOI] [PubMed] [Google Scholar]
- 36.Oran PE, Sherma ND, Borges CR, Jarvis JW, Nelson RW. Intrapersonal and populational heterogeneity of the chemokine RANTES. Clin. Chem. 2010;56(9):1432–1441. doi: 10.1373/clinchem.2010.147884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim JK, Burns JM, Lu W, DeVico AL. Multiple pathways of amino terminal processing produce two truncated variants of RANTES/CCL5. J. Leukoc. Biol. 2005;78(2):442–452. doi: 10.1189/jlb.0305161. [DOI] [PubMed] [Google Scholar]
- 38.Lim JK, Lu W, Hartley O, DeVico AL. N-terminal proteolytic processing by cathepsin G converts RANTES/CCL5 and related analogs into a truncated 4–68 variant. J. Leukoc. Biol. 2006;80(6):1395–1404. doi: 10.1189/jlb.0406290. [DOI] [PubMed] [Google Scholar]
- 39.Trenchevska O, Sherma ND, Oran PE, Reaven PD, Nelson RW, Nedelkov D. Quantitative mass spectrometric immunoassay for the chemokine RANTES and its variants. J. Proteomics. 2015;116:15–23. doi: 10.1016/j.jprot.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biancotto A, Wank A, Perl S, et al. Baseline levels and temporal stability of 27 multiplexed serum cytokine concentrations in healthy subjects. PLoS ONE. 2013;8(12):e76091. doi: 10.1371/journal.pone.0076091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herder C, Haastert B, Müller-Scholze S, et al. Association of systemic chemokine concentrations with impaired glucose tolerance and Type 2 diabetes: results from the Cooperative Health Research in the Region of Augsburg Survey S4 (KORA S4) Diabetes. 2005;54(Suppl. 2):S11–S17. doi: 10.2337/diabetes.54.suppl_2.s11. [DOI] [PubMed] [Google Scholar]
- 42.Kotnik P, Fischer-Posovszky P, Wabitsch M. RBP4: a controversial adipokine. Eur. J. Endocrinol. 2011;165(5):703–711. doi: 10.1530/EJE-11-0431. [DOI] [PubMed] [Google Scholar]
- 43.Ziegelmeier M, Bachmann A, Seeger J, et al. Serum levels of adipokine retinol-binding protein-4 in relation to renal function. Diabetes Care. 2007;30(10):2588–2592. doi: 10.2337/dc07-0275. [DOI] [PubMed] [Google Scholar]
- 44.Masaki T, Anan F, Tsubone T, et al. Retinol binding protein 4 concentrations are influenced by renal function in patients with Type 2 diabetes mellitus. Metabolism. 2008;57(10):1340–1344. doi: 10.1016/j.metabol.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 45.Yang Q, Graham TE, Mody N, et al. Serum retinol binding protein 4 contributes to insulin resistance in obesity and Type 2 diabetes. Nature. 2005;436(7049):356–362. doi: 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
- 46.Cabré A, Lázaro I, Girona J, et al. Retinol-binding protein 4 as a plasma biomarker of renal dysfunction and cardiovascular disease in Type 2 diabetes. J. Intern. Med. 2007;262(4):496–503. doi: 10.1111/j.1365-2796.2007.01849.x. [DOI] [PubMed] [Google Scholar]
- 47.Chavarria N, Kato TS, Khan R, et al. Increased levels of retinol binding protein 4 in patients with advanced heart failure correct after hemodynamic improvement through ventricular assist device placement. Circ. J. 2012;76(9):2148–2152. doi: 10.1253/circj.cj-12-0350. [DOI] [PubMed] [Google Scholar]
- 48.Jaconi S, Rose K, Hughes GJ, Saurat JH, Siegenthaler G. Characterization of two post-translationally processed forms of human serum retinol-binding protein: altered ratios in chronic renal failure. J. Lipid Res. 1995;36(6):1247–1253. [PubMed] [Google Scholar]
- 49.Kiernan UA, Tubbs KA, Nedelkov D, Niederkofler EE, Nelson RW. Comparative phenotypic analyses of human plasma and urinary retinol binding protein using mass spectrometric immunoassay. Biochem. Biophys. Res. Commun. 2002;297(2):401–405. doi: 10.1016/s0006-291x(02)02212-x. [DOI] [PubMed] [Google Scholar]
- 50.Nedelkov D, Kiernan UA, Niederkofler EE, Tubbs KA, Nelson RW. Investigating diversity in human plasma proteins. Proc. Natl Acad. Sci. USA. 2005;102(31):10852–10857. doi: 10.1073/pnas.0500426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nedelkov D, Phillips DA, Tubbs KA, Nelson RW. Investigation of human protein variants and their frequency in the general population. Mol. Cell Proteomics. 2007;6(7):1183–1187. doi: 10.1074/mcp.M700023-MCP200. [DOI] [PubMed] [Google Scholar]
- 52.Trenchevska O, Phillips DA, Nelson RW, Nedelkov D. Delineation of concentration ranges and longitudinal changes of human plasma protein variants. PLoS ONE. 2014;9(6):e100713. doi: 10.1371/journal.pone.0100713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Q, Eskurza I, Kiernan UA, et al. Quantitative measurement of full-length and C-terminal proteolyzed RBP4 in serum of normal and insulin-resistant humans using a novel mass spectrometry immunoassay. Endocrinology. 2012;153(3):1519–1527. doi: 10.1210/en.2011-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun Q, Kiernan UA, Shi L, et al. Plasma retinol-binding protein 4 (RBP4) levels and risk of coronary heart disease: a prospective analysis among women in the nurses’ health study. Circulation. 2013;127(19):1938–1947. doi: 10.1161/CIRCULATIONAHA.113.002073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naylor HM, Newcomer ME. The structure of human retinol-binding protein (RBP) with its carrier protein transthyretin reveals an interaction with the carboxy terminus of RBP. Biochemistry. 1999;38(9):2647–2653. doi: 10.1021/bi982291i. [DOI] [PubMed] [Google Scholar]
- 56.Jong MC, Hofker MH, Havekes LM. Role of ApoCs in lipoprotein metabolism: functional differences between ApoC1, ApoC2, and ApoC3. Arterioscler. Thromb. Vasc. Biol. 1999;19(3):472–484. doi: 10.1161/01.atv.19.3.472. [DOI] [PubMed] [Google Scholar]
- 57.Hegele RA, Connelly PW, Hanley AJG, Sun F, Harris SB, Zinman B. Common genomic variation in the APOC3 promoter associated with variation in plasma lipoproteins. Arterioscler. Thromb. Vasc. Biol. 1997;17(11):2753–2758. doi: 10.1161/01.atv.17.11.2753. [DOI] [PubMed] [Google Scholar]
- 58.Jørgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjærg-Hansen A. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N. Engl. J. Med. 2014;371(1):32–41. doi: 10.1056/NEJMoa1308027. [DOI] [PubMed] [Google Scholar]
- 59.Crosby J, Peloso GM, Auer PL, et al. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N. Engl. J. Med. 2014;371(1):22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huff MW, Hegele RA. Apolipoprotein C-III: going back to the future for a lipid drug target. Circ. Res. 2013;112(11):1405–1408. doi: 10.1161/CIRCRESAHA.113.301464. [DOI] [PubMed] [Google Scholar]
- 61.Graham MJ, Lee RG, Bell TA, et al. Antisense oligonucleotide inhibition of apolipoprotein C-III reduces plasma triglycerides in rodents, nonhuman primates, and humans. Circ. Res. 2013;112(11):1479–1490. doi: 10.1161/CIRCRESAHA.111.300367. [DOI] [PubMed] [Google Scholar]
- 62.Bondarenko PV, Cockrill SL, Watkins LK, Cruzado ID, Macfarlane RD. Mass spectral study of polymorphism of the apolipoproteins of very low density lipoprotein. J. Lipid Res. 1999;40(3):543–555. [PubMed] [Google Scholar]
- 63.Nicolardi S, van der Burgt YE, Wuhrer M, Deelder AM. Mapping O-glycosylation of apolipoprotein C-III in MALDI-FT-ICR protein profiles. Proteomics. 2013;13(6):992–1001. doi: 10.1002/pmic.201200293. [DOI] [PubMed] [Google Scholar]
- 64.Wada Y, Kadoya M, Okamoto N. Mass spectrometry of apolipoprotein C-III, a simple analytical method for mucin-type O-glycosylation and its application to an autosomal recessive cutis laxa type-2 (ARCL2) patient. Glycobiology. 2012;22(8):1140–1144. doi: 10.1093/glycob/cws086. [DOI] [PubMed] [Google Scholar]
- 65.Jian W, Edom RW, Wang D, Weng N, Zhang SW. Relative quantitation of glycoisoforms of intact apolipoprotein C3 in human plasma by liquid chromatography-high-resolution mass spectrometry. Anal. Chem. 2013;85(5):2867–2874. doi: 10.1021/ac3034757. [DOI] [PubMed] [Google Scholar]
- 66.Balog CI, Mayboroda OA, Wuhrer M, Hokke CH, Deelder AM, Hensbergen PJ. Mass spectrometric identification of aberrantly glycosylated human apolipoprotein C-III peptides in urine from Schistosoma mansoni-infected individuals. Mol. Cell Proteomics. 2010;9(4):667–681. doi: 10.1074/mcp.M900537-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nicolardi S, van der Burgt YE, Dragan I, Hensbergen PJ, Deelder AM. Identification of new apolipoprotein-CIII glycoforms with ultrahigh resolution MALDI-FTICR mass spectrometry of human sera. J. Proteome Res. 2013;12(5):2260–2268. doi: 10.1021/pr400136p. [DOI] [PubMed] [Google Scholar]
- 68.Yassine HN, Trenchevska O, Ramrakhiani A, et al. The association of human apolipoprotein C-III sialylation proteoforms with plasma triglycerides. PLoS ONE. 2015;10(12):e0144138. doi: 10.1371/journal.pone.0144138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koska J, Yassine H, Trenchevska O, et al. Disialylated apolipoprotein C-III proteoform is associated with improved lipids in prediabetes and Type 2 diabetes. J. Lipid Res. 2016;57(5):894–905. doi: 10.1194/jlr.P064816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O'Brien KD, Chait A. Serum amyloid A: the ‘other’ inflammatory protein. Curr. Atheroscler. Rep. 2006;8(1):62–68. doi: 10.1007/s11883-006-0066-0. [DOI] [PubMed] [Google Scholar]
- 71.Uhlar CM, Whitehead AS. Serum amyloid A, the major vertebrate acute-phase reactant. Eur. J. Biochem. 1999;265(2):501–523. doi: 10.1046/j.1432-1327.1999.00657.x. [DOI] [PubMed] [Google Scholar]
- 72.Sellar GC, Oghene K, Boyle S, Bickmore WA, Whitehead AS. Organization of the region encompassing the human serum amyloid A (SAA) gene family on chromosome 11p15.1. Genomics. 1994;23(2):492–495. doi: 10.1006/geno.1994.1530. [DOI] [PubMed] [Google Scholar]
- 73.Sipe J. Revised nomenclature for serum amyloid A (SAA). Nomenclature Committee of the International Society of Amyloidosis. Part 2. Amyloid. 1999;6(1):67–70. doi: 10.3109/13506129908993291. [DOI] [PubMed] [Google Scholar]
- 74.Ducret A, Bruun CF, Bures EJ, Marhaug G, Husby G, Aebersold R. Characterization of human serum amyloid A protein isoforms separated by two-dimensional electrophoresis by liquid chromatography/electrospray ionization tandem mass spectrometry. Electrophoresis. 1996;17(5):866–876. doi: 10.1002/elps.1150170508. [DOI] [PubMed] [Google Scholar]
- 75.Tolson J, Bogumil R, Brunst E, et al. Serum protein profiling by SELDI mass spectrometry: detection of multiple variants of serum amyloid alpha in renal cancer patients. Lab. Invest. 2004;84(7):845–856. doi: 10.1038/labinvest.3700097. [DOI] [PubMed] [Google Scholar]
- 76.Loo D, Mollee PN, Renaut P, Hill MM. Proteomics in molecular diagnosis: typing of amyloidosis. J. Biomed. Biotechnol. 2011:754109. doi: 10.1155/2011/754109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kiernan UA, Tubbs KA, Nedelkov D, Niederkofler EE, Nelson RW. Detection of novel truncated forms of human serum amyloid A protein in human plasma. FEBS Lett. 2003;537(1–3):166–170. doi: 10.1016/s0014-5793(03)00097-8. [DOI] [PubMed] [Google Scholar]
- 78.Yassine HN, Trenchevska O, He H, et al. Serum amyloid a truncations in Type 2 diabetes mellitus. PLoS ONE. 2015;10(1):e0115320. doi: 10.1371/journal.pone.0115320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Egashira M, Takase H, Yamamoto I, Tanaka M, Saito H. Identification of regions responsible for heparin-induced amyloidogenesis of human serum amyloid A using its fragment peptides. Arch. Biochem. Biophys. 2011;511(1–2):101–106. doi: 10.1016/j.abb.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 80.Lacey JM, Bergen HR, Magera MJ, Naylor S, O'Brien JF. Rapid determination of transferrin isoforms by immunoaffinity liquid chromatography and electrospray mass spectrometry. Clin. Chem. 2001;47(3):513–518. [PubMed] [Google Scholar]
- 81.Lenters-Westra E, Schindhelm RK, Bilo HJ, Slingerland RJ. Haemoglobin A1c: historical overview and current concepts. Diabetes Res. Clin. Pract. 2013;99(2):75–84. doi: 10.1016/j.diabres.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 82.Duncan MW, Nedelkov D, Walsh R, Hattan SJ. Applications of MALDI mass spectrometry in clinical chemistry. Clin. Chem. 2016;62(1):134–143. doi: 10.1373/clinchem.2015.239491. [DOI] [PubMed] [Google Scholar]
- 83.Cheng K, Chui H, Domish L, Hernandez D, Wang G. Recent development of mass spectrometry and proteomics applications in identification and typing of bacteria. Proteomics Clin. Appl. 2016;10(4):346–357. doi: 10.1002/prca.201500086. [DOI] [PMC free article] [PubMed] [Google Scholar]