Abstract
Recent advances in the field of mitochondrial DNA (mtDNA) replication highlight the diversity of both the mechanisms utilized and the structural and functional organization of the proteins at mtDNA replication fork, despite the simplicity of the animal mtDNA genome. DNA polymerase γ, mtDNA helicase and mitochondrial single-stranded DNA-binding protein- the key replisome proteins, have evolved distinct structural features and biochemical properties. These appear to be correlated with mtDNA genomic features in different metazoan taxa and with their modes of DNA replication, although a substantial integrative research is warranted to establish firmly these links. To date, several modes of mtDNA replication have been described for animals: rolling circle, theta, strand-displacement, and RITOLS/bootlace. Resolution of a continuing controversy relevant to mtDNA replication in mammals/vertebrates will have a direct impact on the mechanistic interpretation of mtDNA-related human diseases. Here we review these subjects, integrating earlier and recent data to provide a perspective on the major challenges for future research.
Keywords: mitochondria, mitochondrial DNA, replication, replisome, DNA polymerase, DNA helicase, single-stranded DNA-binding protein, animal phylogeny
1. Overview
The discovery of mitochondrial DNA (mtDNA) established the unique character of mitochondria as the only organelles in the animal cell with an “extrachromosomal” genome [1]. Its compact structure and organization engendered much interest in the study of its replication, expression, inheritance and evolution, and with the identification of pathogenic and heritable mutations that result in human disease, the field of mitochondrial medicine emerged [2–4].
We focus our attention here on the proteins and mechanisms involved in animal mtDNA replication, keeping in mind the essential and dynamic nuclear-mitochondrial interactions that drive its evolution. Indeed, mtDNA per se evolves at a relatively high rate as compared to the nuclear genome, and despite its similar general structure and organization, it varies even among closely related animal species, such as higher primates [5, 6]. Unveiling the mechanisms by which mtDNA sequence variation is introduced, inherited and fixed, and their relationship to mtDNA replication and repair provide a major challenge for future research.
2. Structure-function relationships in mtDNA replication proteins
Three nuclear-encoded proteins play key roles at the mtDNA replication fork (Figure 1): DNA polymerase γ (Pol γ), the replicative mtDNA helicase Twinkle, and the mitochondrial single-stranded DNA-binding protein (mtSSB). Together, these proteins are sufficient for effective DNA synthesis in vitro, and their knockdown or ablation in vivo results in replication defects and mtDNA depletion (reviewed in [7–9]). The replicative mtDNA helicase catalyzes dsDNA unwinding at the fork that is driven by NTP hydrolysis, releasing single-stranded DNA that is stabilized and protected by mtSSB, and is used subsequently as template by Pol γ for DNA synthesis. In this section, we review the biochemical properties of the proteins and their implications in human health and disease, with a final sub-section on features relevant to various mtDNA replication systems.
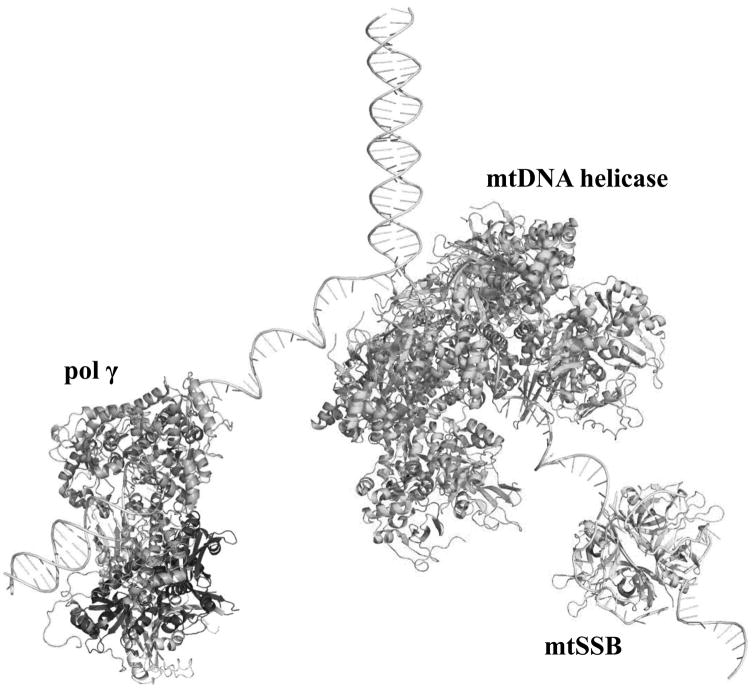
Figure 1.
Proteins at the mitochondrial DNA replication fork. The crystal structure of the heterotrimeric human Pol γ bound to primer-template DNA (PDB: 4ZTZ; [16]), the model of the homotetrameric human mtSSB wrapped around ssDNA [47], and the model of the ring-shaped heptameric human mtDNA helicase [27] were used to create a representation of the nuclear-encoded proteins that function at the mtDNA replication fork. Pol γ-α, the proximal Pol γ-β and the distal Pol γ-β are shown in pink, and light and dark gray, respectively; mtDNA helicase and mtSSB are shown in green and cyan, respectively. The diagram is to scale, but not meant to depict specific protein-protein interactions. Structures and models were analyzed and the figure was produced using Pymol (www.pymol.org).
2.1 DNA polymerase γ, the mitochondrial replicase
Three catalytic activities have been ascribed to Pol γ: 5′-3′ DNA polymerase, 3′-5′ exonuclease and 5′-dRP lyase (reviewed in [7, 10]). All are contained in its catalytic subunit, Pol γ-α (or POLGA, encoded by the POLG gene), although the residues required for its lyase activity have not yet been identified. These activities are regulated by its accessory subunit Pol γ-β (or POLGB, encoded by the POLG2 gene) (Figure 2), which itself has no catalytic activity. However, its effects on DNA synthesis are substantial: it enhances DNA and nucleotide binding, stimulating DNA synthesis by Pol γ-α, and increasing the processivity of the holoenzyme ~100 fold. Pol γ is highly accurate in nucleotide polymerization, with an in vitro error rate of only ~1 misincorporated nucleotide per ~half-million bases polymerized. However, replicative bypass of abasic sites and sites of oxidative damage by Pol γ is error prone. The high base-substitution fidelity of Pol γ is also compromised by nucleotide imbalances, a situation that may be physiologically relevant to the normal fluctuation of metabolites known to occur in the mitochondrial matrix, as well as pathogenic defects in the nuclear genes involved in nucleotide synthesis and transport to mitochondria. The high base-substitution fidelity in human Pol γ derives both from the high nucleotide selectivity of its polymerase (pol) domain, and from exonucleolytic proofreading catalyzed by its exonuclease (exo) domain. Unlike numerous nuclear animal DNA polymerases, Pol γ displays a remarkable ability to utilize diverse primer-template substrates, most likely because it is involved in all processes of DNA metabolism in mitochondria (reviewed in [7]).
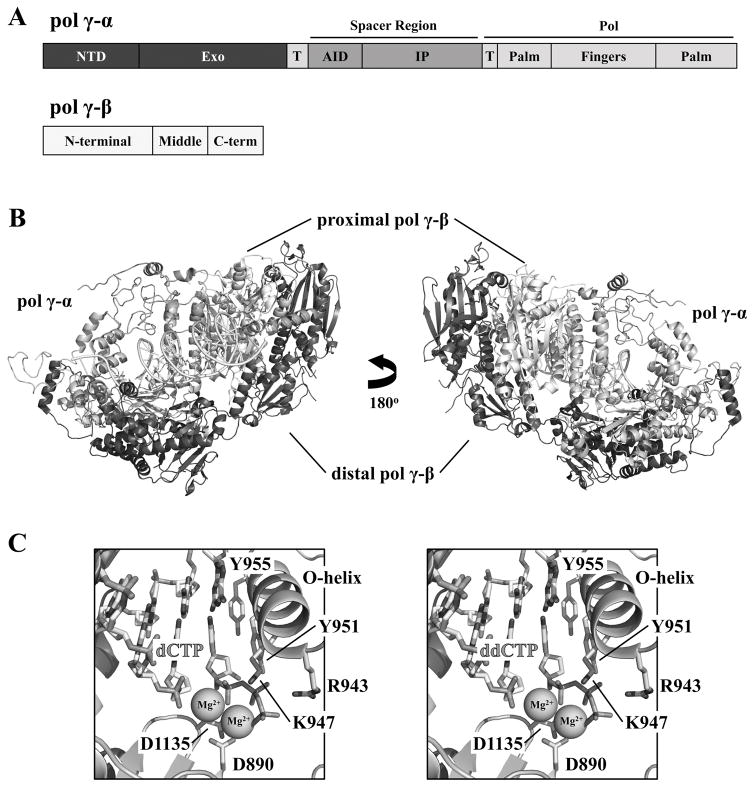
Figure 2.
Structure of human Pol γ bound to primer-template DNA and nucleotide. A, Representation (to scale) of domains and sub-domains in the Pol γ-α and β polypeptide sequences. NTD, N-terminal domain; Exo, exonuclease domain; AID, accessory-interacting determinant sub-domain; IP, intrinsic processivity sub-domain; Pol, DNA polymerase domain; T, the bipartite thumb sub-domain. B, Cartoon representation of the crystal structure of the heterotrimeric human Pol γ holoenzyme bound to primer-template DNA and nucleotide (PDB: 4ZTS; [16]). The Pol γ-α domains are colored as shown in A; the proximal and distal Pol γ-βs are shown in light and dark gray, respectively. C, Enlargement of the pol active site, showing conserved residues for Mg2+- and incoming nucleotide-binding, and the identical positioning of dCTP (left panel, PDB: 4ZTZ) and the inhibitor ddCTP (right panel, PDB: 4ZTU) [16].
Pol γ is the most extensively studied protein of the mtDNA replisome, and its biomedical relevance is well documented: pathogenic mutations in the genes encoding both the catalytic and accessory subunits have been identified in association with numerous human diseases (reviewed in [10]); impaired proofreading has been shown to cause premature aging in mammalian models [11, 12]; and the sensitivity of Pol γ to specific nucleoside analog reverse transcriptase inhibitors (NRTIs) used to treat HIV infection can result in mitochondrial toxicity (reviewed in [13, 14]). Structure-function relationships in Pol γ have been forged by a combination of many years of biochemical study and recent advances in the determination of the 3D structure of the human apo-holoenzyme [15] and of the holoenzyme bound in a ternary complex with primer-template DNA together with either normal or derivatized nucleotides [16, 17].
The heterotrimeric organization of the human replicase, comprising a single Pol γ-α and two Pol γ-β polypeptides, appears to be common to all vertebrate mtDNA polymerases [18]. Pol γ-α consists of three domains, arranged spatially to interact with the Pol γ-β dimer, and to facilitate the transition between its 5′-3′ polymerase and 3′-5′ exonuclease activities (Figure 2). The pol domain carries the canonical “right-hand” fold common to DNA polymerases, formed by palm, fingers and thumb sub-domains. Notably, the thumb subdomain is bipartite, a feature that is apparently unique to Pol γ-α as compared to other Family A DNA polymerases, and makes extensive interactions with the proximal Pol γ-β protomer [15]. However, the major binding site for the accessory subunit is in the accessory-interacting determinant (AID) sub-domain within the spacer domain (Figure 2). The spacer domain also contains an intrinsic processivity (IP) sub-domain not found in the homologous T7 gp5 DNA polymerase, and is implicated in the relatively high processivity of the catalytic subunit alone (reviewed in [7]). Interestingly, in the absence of DNA, Pol γ-α contacts the distal Pol γ-β protomer only by interaction of the Pol γ-α R232 and Q540 residues with Pol γ-β E394 and R122, respectively [15]. The contact region expands greatly upon primer-template DNA-binding due to conformational changes that rotate the dimeric Pol γ-β by 22° towards the polymerase domain of the catalytic subunit [16, 17]. The 3D structure of the DNA-bound Pol γ provides a mechanistic explanation for human disease-causing substitutions in both the catalytic and accessory subunits. For example, the deleterious effects of the Pol γ-β G451E mutation on subunit association and holoenzyme processivity [19, 20] are apparent only in the ternary complex: upon DNA-binding, the G451 residues in the proximal and distal Pol γ-βs interact with the hydrophobic L-helix and the R232 region of Pol γ-α, respectively [16]. Furthermore, the 3D structure enabled us to establish genotype-phenotype correlations that in turn have led to the development of a powerful pathogenicity prediction tool to evaluate the likely effects of chromosomal variants in the compound heterozygous form in which they are most often manifest in patients with POLG syndromes [21, 22].
The 3D structures of Pol γ have also shed light on how the holoenzyme is affected by NRTIs, and provide a strong basis for rational drug design. Figure 2C shows that ddCTP, the antiviral agent known as zalcitabine, is stabilized in the pol active site by R943, K947 and Y951, and a bent conformation of the template imparted by Y955. This configuration is highly similar to that observed upon dCTP binding, providing a molecular basis for the susceptibility of Pol γ to this HIV reverse transcriptase inhibitor [16]. On the other hand, new classes of NRTIs, such as (−)-FTC [(−)-2,3′-dideoxy-5-fluoro-3′-thiacytidine, emtricitabine] causes much reduced mitochondrial toxicity because Pol γ discriminates against it more efficiently than does the viral reverse transcriptase [23]; the nucleotide-binding site in the pol domain of the mitochondrial replicase can distinguish the ribose and base modifications in (−)-FTC as a result of the potential steric clashes between the rigid Y951 residue and the modified ribose. Moreover, the hydrophobic nature of the I948 residue precludes interactions with the 5-fluorine, forcing both the nucleoside and the α-phosphate of the inhibitor to be misaligned [17].
Less experimental focus has been placed on the proofreading exonuclease activity of Pol γ, although it is also implicated strongly in human disease and in aging. The exo domain of human Pol γ-α contains the highly conserved residue D274 (D257 in the mouse, D263 in Drosophila melanogaster). Its substitution with alanine results in mtDNA mutator mouse lines [11, 12], and more recently a mutator fly line [24]. Whereas the mtDNA mutator mice have been analyzed extensively to show the substantial impact of a proofreading-deficient Pol γ in causing premature aging phenotypes and in shortening life span, the homozygous flies carrying the equivalent mutant enzyme surprisingly die during development in the late third larval instar stage, indicating potentially major differences in the requirements for the exonuclease activity of Pol γ in mtDNA maintenance in mammals and insects. Indeed, other major differences exist between the vertebrate and insect Pol γs, such as the heterodimeric structure of the latter [25, 26].
2.2 The replicative mtDNA helicase
Recently, we reviewed comprehensively the literature on the structure, catalytic activity, and evolution of the animal mtDNA helicase, including an evaluation of human pathogenic variants and a discussion of current animal models [27]. Among the important findings, two features of the replicative mtDNA helicase are notable: its remarkable resemblance to the bacteriophage T7 gp4, a bifunctional primase-helicase (see Chapter 3 of this Volume), and the numerous mutations in the human gene associated with mitochondrial disorders. Its primary sequence suggests a modular architecture as in T7 gp4 [28], comprising of a zinc-binding-like domain (ZBD), an RNA polymerase-like domain (RPD), a Linker region and a C-terminal helicase domain (CTD) (Figure 3). Molecular modeling and mapping of the disease-related residues reported to date in the human enzyme identified two major structural regions that we explored in detail. First, the abundance of pathogenic residues found in the Linker region and CTD argues that their effects on subunit interactions are a major cause of mtDNA replication defects leading to human disorders [27]. Oligomerization in replicative helicases is key to the formation of the nucleotide-binding pocket at the protomer-protomer interface that is required for proper positioning of the substrate for hydrolysis, which is coordinated with translocation of the helicase on DNA [29]. The putative role of these disease-related residues in maintaining the stability of the oligomeric mtDNA helicase is apparent only when two protomers are analyzed together, revealing that the Linker region of one protomer likely interacts with the CTD of the adjacent protomer. At present, several studies support such a hypothesis [30–33].
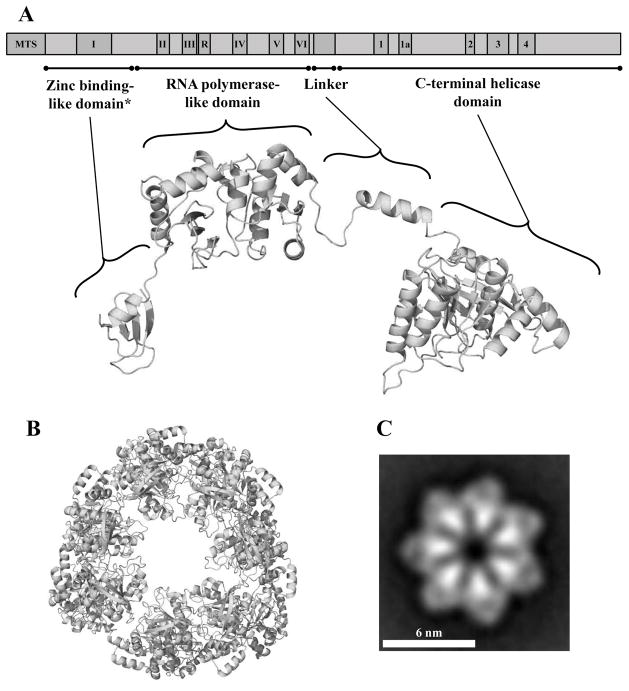
Figure 3.
Schematic representation and structural model of the human mtDNA helicase. A, Upper image, schematic representation of the conserved amino acid sequence motifs in human mtDNA helicase. MTS, mitochondrial target sequence; I–VI and R, conserved sequence motifs I–IV and RNAP basic motif of prokaryotic primases; 1–4 and 1a, conserved sequence motifs 1–4 and 1a of ring-shaped helicases [167]. The size and position of the conserved sequence motifs are represented to scale. Lower image, structural model of a protomer of the human mtDNA helicase highlighting its modular architecture organized in a zinc binding-like domain (ZBD), RNA polymerase-like domain (RPD), Linker region and a C-terminal helicase domain (CTD). * ZBD portion is represented as the polypeptide backbone of the bacteriophage T7 gp4 ZBD. B, Model of the heptameric human mtDNA helicase, CTD view. C, Electron microscopic image of the recombinant human mtDNA helicase at 100 mM NaCl in the presence of Mg2+ and ATPγS, showing its heptameric configuration. Reproduced with permission from “L.S. Kaguni, M.T. Oliveira: Structure, function and evolution of the animal mitochondrial replicative DNA helicase. Critical Reviews in Biochemistry and Molecular Biology (2016) 51, 53–64” and from “T.D. Ziebarth, R. Gonzalez-Soltero, M.M. Makowska-Grzyska, R. Núñez-Ramírez, J.M. Carazo, L.S. Kaguni: Dynamic effects of cofactors and DNA on the oligomeric state of human mitochondrial DNA helicase. Journal of Biological Chemistry (2010) 285, 14639–14647”.
Second, pathogenic residues that map onto a human RPD model cluster on a positively charged surface area, which might represent a new DNA-binding region. If so, the RPD in mtDNA helicase binds DNA in a configuration not described for T7 gp4 or other prokaryotic primases [27]. Evidence suggesting the importance of this putative DNA-binding region includes the findings: 1) expressing D. melanogaster mtDNA helicase bearing analogous mutations found in human patients causes severe mtDNA depletion in S2 cells [34], consistent with the fact that the recombinant N-terminal domain of the insect enzyme binds to both ss and dsDNA [35]; and 2) recombinant human mtDNA helicases lacking part or all of the RPD exhibited lower ssDNA-binding, ATPase and unwinding activities [36], in agreement with the observed decrease in mtDNA copy number in human cultured cells [37]. Interestingly, the RPD of the mtDNA helicase appears to have evolved these novel functions without losing either the polypeptide fold or some of the conserved amino acid residues of a prokaryotic primase, even though it does not synthesize RNA primers [27, 36].
Whereas oligomerization clearly involves residues in the Linker region and CTD, both the oligomeric form and the conformation of individual protomers in the human mtDNA helicase appear to be dynamic [33, 38, 39]. A high proportion of structurally-heterogeneous homohexamers with 3-fold symmetry was observed at high ionic strength with protein purified in the baculovirus expression system, which contrasts with a more homogeneous homoheptameric species with a clear 7-fold symmetry (Figure 3C) observed at low ionic strength and in the presence of Mg2+ and ATPγS [38]. Interestingly, the opposite balance between forms is found for T7 gp4, which is predominantly homohexameric upon nucleotide triphosphate binding [40]. Because the heptamers of T7 gp4 are unable to bind DNA efficiently, it is proposed that the loss of a protomer during the switch from heptamers to hexamers promotes ring-opening for loading of the enzyme on ssDNA. By analogy, and considering evidence that the mtDNA helicase can load on a circular ssDNA template in the absence of a helicase loader [41], it seems plausible that ring-opening in the hexameric mtDNA helicase occurs by acquiring an extra subunit upon ATP binding, suggesting a role for the heptamers form in DNA loading, nucleotide hydrolysis, and dsDNA unwinding in mtDNA replication. Clearly, substantial research is warranted to test this hypothesis in vitro and in vivo.
The most intriguing feature of the mtDNA helicase is perhaps the sequence and structural diversity of its ZBD among different species. Whereas the ZBD of T7 gp4 contains four cysteine residues that coordinate zinc that is essential for primase activity, it has been suggested that the absence of three of four cysteine residues in the ZBD of human (and other vertebrate) mtDNA helicases is the reason for their lack of primase activity [42]. Underscoring the structural differences among mtDNA helicases, we showed that a catalytic Mg2+-binding pocket in the RPD cannot be modeled in either the human or the D. melanogaster enzymes [27]. Although invertebrates do possess the conserved cysteine residues in the ZBD, physical characterization of the D. melanogaster ZBD showed that it binds an iron-sulfur cluster, coordinated by the homologous cysteine residues of T7 gp4 [35]. No evidence of zinc binding has been reported in human mtDNA helicase, and amino acid sequence analyses indicate that the ZBD in vertebrate mtDNA helicases has diverged significantly from their prokaryotic counterparts. Implying a potentially major shift in function, the evolutionary novelty of the iron-sulfur cluster makes the insect ZBD a candidate for sensing the redox state inside mitochondria, linking mtDNA replication to mitochondrial responses to excess reactive oxygen species. At present, it is not clear if the presence of an iron-sulfur cluster-binding domain is an ancestral condition in animal mtDNA helicases, or if it is a derived feature of the insect enzyme, but it is clearly absent in vertebrates, and could contribute to the diversification of mtDNA replication modes identified across animal taxa (see Section 3).
2.3. mtSSB
Single-stranded DNA-binding proteins (reviewed in [43]) including mtSSB are essential in DNA metabolism, serving to bind and protect ssDNA during replication, repair and recombination. At the mtDNA replication fork, mtSSB most likely coordinates interactions within the replisome, stimulating Pol γ and mtDNA helicase function. In vitro stimulation of the DNA polymerase and exonuclease activities of Pol γ by mtSSB has been documented in both the human and D. melanogaster systems [44–48]. Human mtSSB stimulates both the unwinding activity of the human mtDNA helicase up to 8-fold [46, 47, 49], and the concerted actions of the human Pol γ and mtDNA helicase in strand-displacement DNA synthesis [50]. The role of mtSSB in mtDNA replication has also been documented genetically in cultured cells and in whole animals. Disruption of the D. melanogaster gene (lopo) resulted in developmental lethality, accompanied by severe mtDNA depletion and loss of cellular respiratory capacity [51]. Interestingly, Sugimoto et al. showed that viability of the nematode Caenorhabditis elegans is not affected by systemic mtSSB knockdown, but mtDNA levels and animal fertility are reduced drastically [52]. mtDNA depletion is consistently observed when mtSSB protein is knocked down by RNAi either in Drosophila Schneider cells [47, 53, 54] or in human HeLa cells [55].
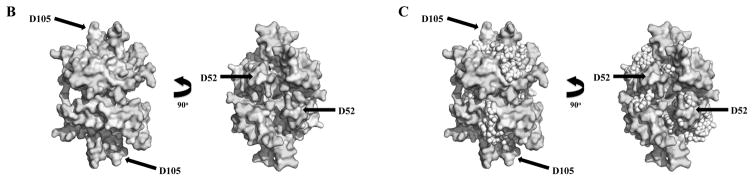
As a homologue of the well studied SSB from Escherichia coli (EcSSB), sequence analysis indicates that the mtSSB of all metazoans is likely homotetrameric and binds to ssDNA similarly to the bacterial protein (Figure 4). We retrieved 56 animal homologue sequences of the human and D. melanogaster proteins from diverse databanks, making structural predictions and drawing functional correlations, as recently reported for Pol γ [18] and the mtDNA helicase [27]. Because animal mtSSB sequences are highly similar, we also evaluated them using the Evolutionary Trace Server [56, 57] to identify important amino acid residues. These are highlighted in yellow in Figure 4A, B and C as those conserved only in metazoans. A comparison with the EcSSB residues that are known to be crucial for ssDNA binding and subunit interactions (red and light green arrows, respectively) reveals a remarkable overlap. This is consistent with the long-standing concept that bacterial and mitochondrial SSBs are structurally and functionally similar, especially in regard to ssDNA-binding activity [47, 58, 59]. The only residues identified by the Evolutionary Trace analysis that do not appear to be involved directly in DNA binding are D52 and D105 (in reference to the human sequence) (Figure 4B and C). Interestingly, we have shown that deletion of the loop containing D52 (loop 2,3), and alanine substitutions in the loop containing D105 (loop 4,5) of human mtSSB do not affect substantially its ssDNA-binding affinities, but instead compromise its ability to stimulate either human Pol γ or mtDNA helicase [47] (see Section 2.4).
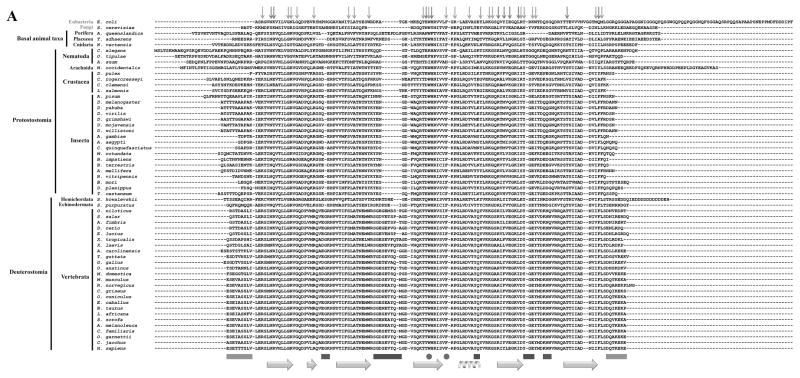
Figure 4.
Evolutionary Trace analysis of animal mtSSBs. A, Multiple amino acid sequence alignment using selected animal mtSSBs retrieved from public databases (NCBI, Ensembl Metazoa and 959 Nematode Genomes). TBLASTN [168] and HMMR3 BLAST [169] searches were performed using the translated mRNA reference sequences for the Homo sapiens (AK313033.1) and Drosophila melanogaster (BT016028.1) mtSSB as queries. The alignment was performed using the MUSCLE algorithm built into the MEGA6 software [170]. The outgroup sequences used here were the Saccharomyces cerevisiae mtSSB (S43128.1) and the Escherichia coli SSB (J01704.1). Red and light green arrows above the alignment indicate the residues that are crucial for ssDNA binding and subunit interactions in the E. coli SSB protein, respectively [171, 172]. Gray, purple and dark green bars below the alignment indicate, respectively, three mtSSB regions grouped by their functional properties: N- and C- termini (which have negative effects on Pol γ stimulation [46]); loop 2,3, alpha-1 and loop 4,5-1 (which have positive effects on Pol γ stimulation [47]); and loop 1,2 and loop 4,5-2 (which have positive effects on mtDNA helicase stimulation [47]). The cyan arrows and helix below the alignment indicate the residues that form the β sheets and the only α-helix in the mtSSB polypeptide, according to the crystal structure (PDB: 3ULL, [59]). The residues highlighted in yellow were identified by the Evolutionary Trace Server [56, 57] as conserved in metazoans. These have been mapped onto the crystal structure of the human mtSSB (B) and onto a model of ssDNA-bound mtSSB (C). The model is as reported by Oliveira and Kaguni [47]. D52 and D105 are the only residues identified by the Evolutionary Trace analysis that are most likely not involved in ssDNA binding (see text for details).
2.4. Functional interactions among replisome proteins and evolutionary implications
The protein components of the minimal mtDNA replisome have distinct evolutionary origins, prompting the question of how they have co-evolved to function coordinately to promote mtDNA replication. The mtDNA helicase and the catalytic subunit of Pol γ share ancestry with the gp4 primase-helicase and the gp5 DNA polymerase of bacteriophage T7, respectively [60]. mtSSB and Pol γ-β are eubacterial-like proteins with strong structural similarity to the homotetrameric E. coli SSB [61] and a class II aminoacyl-tRNA synthetase [62–64], respectively. However, unlike mtSSB, which most likely originated from the endosymbiotic α-proteobacterium that became the eukaryotic mitochondrion, sequence alignments indicate that Pol γ-β evolved as the accessory subunit of the mitochondrial replicase by lateral gene transfer involving a eubacterial species and early metazoans [18, 64].
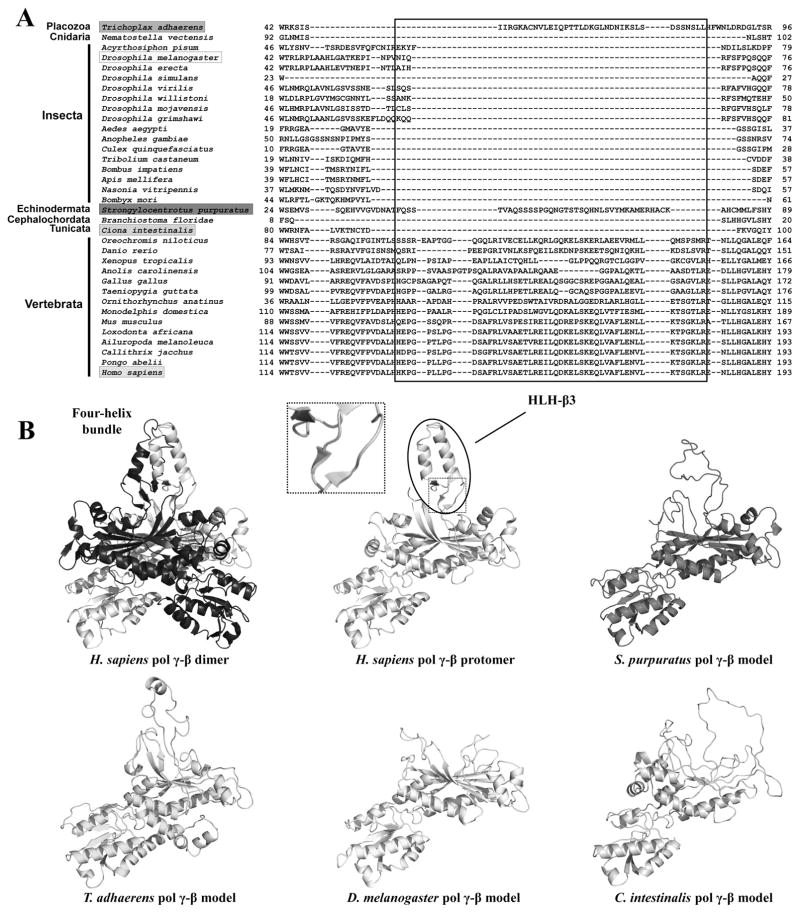
We recently analyzed the largest number of Pol γ-α and β sequences currently available for animal species, taking advantage of the ever-increasing volume of genomic and transcriptomic data in public databases [18]. We showed that the mitochondrial replicase presents distinct patterns of molecular evolution throughout the animal phylogenetic distribution, which might reflect distinct mechanisms for replicating mtDNA (see Section 3). The most striking finding relates to the oligomeric composition of Pol γ, which extends beyond a simplistic view of one Pol γ-α and one or two Pol γ-β protomers forming the holoenzyme. The presence of the helix-loop-helix (HLH)-3β domain in Pol γ-β (Figure 5) that enables its homodimerization of the human and mouse replicases [15, 65] argues that the heterotrimeric nature is conserved in all vertebrate species. On the other hand, a holoenzyme comprising one Pol γ-α and one Pol γ-β, represented by the Drosophila melanogaster enzyme [25, 26], is most likely the ancestral form for all animals. However, our analysis revealed exceptions to this ancestral heterodimeric form. Specifically, we were unable to find the Pol γ-β gene in the genome and/or transcriptome of any nematode species, a finding that correlates with the absence of the AID domain in the Pol γ-α polypeptide, indicating that the mitochondrial replicase in this animal group does not have a β accessory subunit, and may be a single-subunit enzyme [18]. The nematode mtDNA polymerase resembles the enzyme of other eukaryotic taxa such as Saccharomyces cerevisiae. Notably, mtDNA replication in the nematode C. elegans proceeds via a rolling circle mechanism [66], which is apparently the main mode of mtDNA replication in S. cerevisiae [67], and represents a mechanism distinct from that shown for other animals (see Section 3).
Figure 5.
Dimerization of vertebrate Pol γ-β via the formation of the four-helix bundle structure. A, Amino acid sequence alignment indicates the presence of the HLH-3β domain (boxed) in all species of Vertebrata and possibly in a few other animal groups. B, Comparison of the crystal structure of the human Pol γ-β dimer and structural models for Pol γ-β of Trichoplax adhaerens, Strongylocentrotus purpuratus, Drosophila melanogaster and Ciona intestinalis, showing that only vertebrate Pol γ-β can fold into a HLH-3β structure and therefore, form the four-helix bundle dimerization interface. The inset shows the three short β-sheets at the base of the HLH-3β structure. Reproduced with permission from “M.T. Oliveira, J. Haukka, L.S. Kaguni: Evolution of the metazoan mitochondrial replicase. Genome Biology and Evolution (2015) 7, 943–959”.
Clearly, the evolution of the oligomeric form of Pol γ has taken several routes that may correlate with variations in the mechanisms of mtDNA replication in vivo [18]. We reported recently a direct comparison of recombinant Pol γ holoenzymes from a vertebrate (human) and an insect (D. melanogaster) species, which showed that the extent of DNA synthesis on a singly-primed, circular ssDNA template is ~5-fold higher with the human enzyme [48] (Figure 6C). In comparison, when variants of the human and mouse Pol γ-β that are unable to homodimerize were examined in similar in vitro assays in a heterodimeric state with Pol γ-α, thus resembling insect Pol γ, the decrease in DNA polymerase activity observed as compared to that of their native heterotrimeric forms ranged from ~4- to 7-fold [65, 68, 69]. The heterotrimeric holoenzyme form might provide a functional advantage to vertebrates during mtDNA replication to increase the rate of nucleotide incorporation. In comparing the crystal structure of the human Pol γ apo-holoenzyme with that of the replicase in a ternary complex with a primer-template DNA and nucleotide, Yin and collaborators noted that the Pol γ-β dimer is rotated 22° towards the pol domain of Pol γ-α upon DNA binding, allowing the catalytic subunit to interact extensively with the distal Pol γ-β protomer, and enabling the dimeric accessory subunit to regulate DNA synthesis allosterically [16, 17]. In the ternary complex, human Pol γ-α undergoes conformational changes in several subdomains including the fingers, thumb, L-helix and IP subdomain. These changes are perhaps more limited in the single β-containing animal Pol γs, including that of D. melanogaster, such that its intrinsic DNA synthesis capacity is lower in the absence of other replisome proteins.
Figure 6.
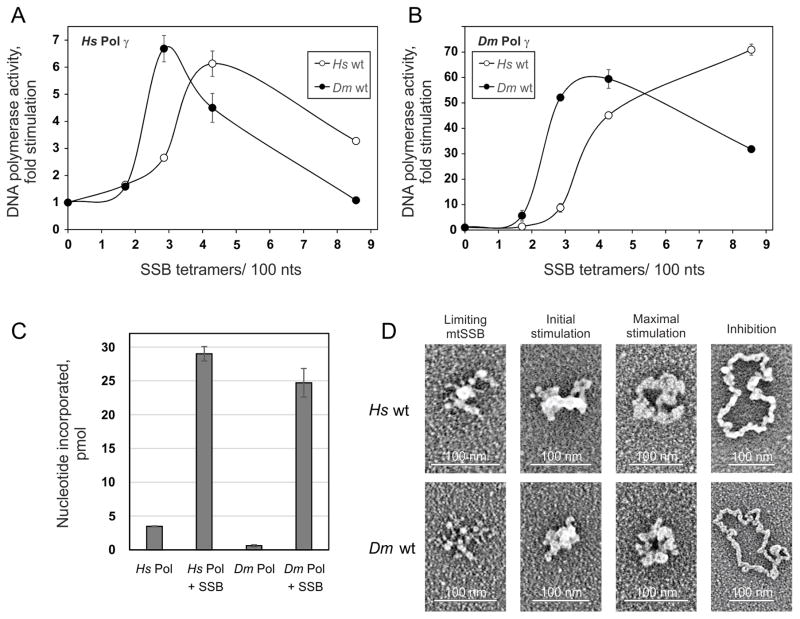
Stimulation of DNA synthesis catalyzed by Pol γ correlates with specific ssDNA template organization by mtSSB. A and B, DNA polymerase assays were performed using 58.5 fmol of singly primed M13 DNA, 35 fmol of human Pol γ-α, 220 fmol of human Pol γ-β (A) or 40 fmol of D. melanogaster Pol γ (as Pol γ-α) (B), and 0, 6.4, 10.7, 16, or 32 pmol of either human (open circles) or D. melanogaster (closed circles) wild-type mtSSB. Assays were performed at 30 mM KCl and 4 mM MgCl2. The data were normalized to the amount of nucleotide incorporated by human Pol γ in the absence of mtSSB (arbitrarily set to 1 in each case). C, Comparison of nucleotide incorporation by human and D. melanogaster Pol γ in the absence or presence of their cognate mtSSBs at the concentrations resulting in maximal stimulation. D, electron microscopy of human (top) and D. melanogaster (bottom) wild-type mtSSB proteins bound to M13 DNA. The binding reaction was performed at 30 mM KCl and 4 mM MgCl2. The images are representative of template species formed at the following ratios of mtSSB tetramers per 100 nucleotides, which correspond to the indicated individual phases of the stimulation of human Pol γ activity: limiting mtSSB, 1.6 human and 1.2 D. melanogaster mtSSB; initial stimulation, 3.2 human and 1.8 D. melanogaster mtSSB; maximal stimulation, 3.8 human and 2.5 D. melanogaster mtSSB; inhibition, 6.4 human and 7 D. melanogaster mtSSB. Reproduced with permission from “G.L. Ciesielski, O. Bermek, F.A. Rosado-Ruiz, S.L. Hovde, O.J. Neitzke, J.D. Griffith, L.S. Kaguni: Mitochondrial single-stranded DNA-binding proteins stimulate the activity of DNA polymerase γ by organization of the template DNA. Journal of Biological Chemistry (2015) 290, 28697–28707”.
Comparison of the heterodimeric and heterotrimeric Pol γs in in vitro DNA polymerase assays in the absence of other components of the mtDNA replisome provides an incomplete picture. Indeed, mtSSB has been shown to stimulate both the DNA polymerase and exonuclease activities of Pol γ ~20-fold, although this value varies substantially between the human and D. melanogaster systems [44–47]. To explore this, we pursued a comparative analysis of the human and D. melanogaster Pol γ holoenzymes with their cognate and heterologous mtSSBs using a combination of biochemical assays and transmission electron microscopy [48]. Under optimal conditions for each, both holoenzymes showed comparable DNA synthesis in the presence of their cognate SSBs. D. melanogaster Pol γ alone is ~5-fold less efficient than the human replicase. Notwithstanding their different oligomeric forms, the human and fly Pol γs behave similarly at maximum stimulation by mtSSB. In addition, D. melanogaster mtSSB stimulates human Pol γ as efficiently as does human mtSSB, and vice versa (Figure 6A and B). However, the stimulation profile is mtSSB specific: both Pol γs reach their maximal activity at a concentration of the insect mtSSB lower than that required in the presence of human mtSSB. This corresponds to a similar shift in the concentrations required for generation of specific mtSSB-template complexes (Figure 6D) that correlate with individual phases of the stimulatory effect [48]. That the ssDNA binding affinity of both mtSSBs are similar suggests the more efficient DNA template organization by D. melanogaster mtSSB compensates for the lower DNA synthetic capacity of D. melanogaster Pol γ. In composite, our results suggest that structural differences between the human and D. melanogaster mtSSBs give rise to their differential stimulatory effects on Pol γ. We identified by site-directed mutagenesis regions of human mtSSB that are important for maximal stimulation of human Pol γ in vitro, including loop 2,3 (S51-L59), the α-helix 1 (Y83-Q84), and loop 4,5 (Y100-E102) [47]. In fact, loop 2,3 and α-helix 1 show little sequence similarity between vertebrate and invertebrate species, and an extension of 6–7 amino acid residues in loop 2,3, which is disordered in the crystal structure of the human mtSSB, appears predominantly in vertebrate species (Figure 4A). Our recent electron microscopic study shows that loop 2,3 may be important for human mtSSB to organize the template in a competent configuration for Pol γ [48]. The extended loop 2,3 of vertebrate mtSSB, together with the presence of the additional Pol γ-β protomer in the vertebrate mitochondrial replicase, and other putative structural evolutionary novelties described for the catalytic subunit [18] may then explain the biochemical differences in the vertebrate and insect systems. This may in turn correlate with different modes of mtDNA replication in vivo (see Section 3).
Animal mtSSBs lack the long acidic C-terminal tail present in bacterial SSBs (Figure 4A), which protrudes into the ssDNA-binding channel in the absence of DNA, and is then rendered available upon ssDNA binding for protein-protein interactions with other components of the replication machinery (reviewed in [43]). By contrast, animal mtSSBs contain an N-terminal extension (notwithstanding the mitochondrial pre-sequence) that is absent in the eubacterial homologues (Figure 4A, gray bars). Removal of both termini of the human mtSSB, independently or in combination, revealed that they regulate negatively the stimulation of Pol γ while exerting no effects on ssDNA-binding affinity [46]. It remains to be determined what effects the termini of mtSSB might exhibit within the mtDNA replisome, and because the terminal regions are extremely variable among metazoans, molecular evolutionary analysis fails to shed light on their specific roles in other taxa. At present, site-directed mutagenesis of the human protein suggests that amino acid residues in mtSSB that are important for the functional interactions with Pol γ and the mtDNA helicase are distinct (Figure 4A, purple and dark green bars, respectively) [47]. Maximal stimulation of the human mtDNA helicase requires residues E33-K35 in loop 1,2 and K106-N108 in loop 4,5 of the human mtSSB. dsDNA unwinding produces ssDNA that is bound by mtSSB (consistent with recent data in vivo [70]). In contrast, in mtSSB-Pol γ interactions, mtSSB first binds to the template DNA strand, organizes it in a competent conformation for Pol γ, and then is displaced by the replicase as it synthesizes the complementary strand. This is consistent with the fact that excess mtSSB inhibits DNA synthesis by Pol γ, but not dsDNA unwinding by the mtDNA helicase [47, 48].
Falkenberg and coworkers first reconstituted a minimal mtDNA replisome, combining human Pol γ, mtDNA helicase and mtSSB in in vitro DNA synthesis using a 70-nt single-stranded circular DNA template annealed to a primer with a 5′-extension [50]. In this system, DNA synthesis occurs by a rolling circle-like mechanism, producing linear fragments much longer than the template (~2000 nt). The substantial stimulation by mtSSB may reflect functional interactions with Pol γ and/or its binding to the leading strand and lagging strands in a competent conformation at the fork. Strand-displacement DNA synthesis by the minimal mtDNA replisome has been employed to elucidate the replication defects in a number of disease-related enzyme variants of both human Pol γ-α and mtDNA helicase in the context of a functional mtDNA replication fork [30, 71, 72]. Furthermore, addition of the mitochondrial RNA polymerase engenders priming and DNA synthesis on the lagging DNA strand, to generate nicked dsDNA products [73–75]. In subsequent studies, a double-stranded circular DNA template containing a displacement loop (bubble template) was used to show that the mtDNA helicase can load in the absence of a helicase loader such as those required in prokaryotic and nuclear systems, leading to DNA synthesis by the concerted actions of Pol γ, mtDNA helicase and mtSSB [41]. Notably, the reaction efficiency on this template was low as compared with the earlier substrates. To date, only the elongation phase of mtDNA replication has been reconstituted. Thus, there remains much to learn about the initiation and termination of mtDNA replication, and the proteins that function in those processes.
In sum, biochemical studies have demonstrated important functional interactions at the mtDNA replication fork, and we have attempted to provide an overview with evolutionary perspective. At present, we know little about how these functional interactions rely on physical contacts, and if and how they may vary in different animals. We also acknowledge the importance of other factors in mtDNA maintenance: TFAM in organization of mtDNA nucleoids, RNase H1, FEN1, DNA2 and MGME1 in primer processing, Top1mt, Top2β and Top3α in altering mtDNA topology, DNA ligase III, mitochondrial RNA polymerase and PrimPol (possible primase activity), and a diverse array of transcription and repair proteins (reviewed in [8]). With the diversity in the protein requirements for mtDNA synthesis in replication, repair and recombination, it is likely that other factors also participate and as a result, the approach of large-scale screening for new proteins with mtDNA maintenance functions, such as that recently reported by Fukuoh et al. [54], is especially compelling.
3. Mechanisms of mitochondrial DNA replication in vivo
The recent application of diverse techniques to evaluate mtDNA replication in various physiological systems has prompted new models for animal mtDNA replication, highlighting the mechanistic diversity found in vivo to ensure appropriate mtDNA copy number, mitochondrial gene expression, and ATP production via oxidative phosphorylation. Here, we provide an overview of the current models of mtDNA replication in nematodes, insects, echinoderms, and vertebrates, though these may not represent all throughout the animal phylogenetic distribution.
3.1 The rolling circle model in Caenorhabditis elegans
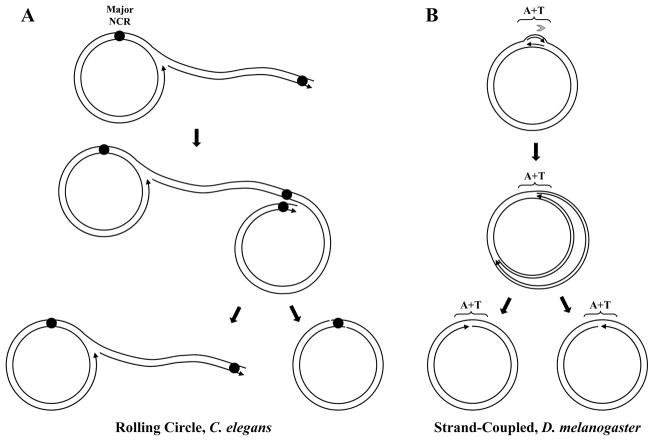
Although the round worm Caenorhabditis elegans has been used as a model organism in many studies, including those on mitochondrial diseases [76–78], replication of its mtDNA has only recently been investigated. Two dimensional agarose gel electrophoresis (2DAGE) of mitochondrial nucleic acids (mtNA) isolated from sucrose gradient-purified mitochondria revealed prominent signals for Y and X arcs. These are representative of fragments containing elongating replication forks and cruciform structures, respectively [79–82]. No structure corresponding to a replication initiation bubble (bubble arc) was detected [66]. Direct observation of the isolated mtDNA by transmission electron microscopy (TEM) identified branched-circular lariat molecules with concatemeric tails, which are characteristic of intermediates during rolling circle replication (Figure 7). The X arcs observed by 2DAGE, which traditionally correspond to various cruciform structures that may result from recombination or replication termination [81, 83–85], were abundant primarily when the fragment harboring the major non-coding region (NCR) of the nematode mtDNA was probed. Treatment of the mtNA with a combination of S1 nuclease and E.coli resolvase RusA, highly specific for resolving Holiday junctions, reduced the X arc signal substantially, suggesting that a sub-fraction of the X-shaped DNA molecules are possibly hemicatenanes, a DNA species resulting from the convergence of two Holiday junctions, or replication fork stalling [86]. The rolling circle mechanism is a robust mechanism to assure efficient production of genomes as is exploited by various bacteriophages, such as Phi29, T4, λ and M13 [87–90]. Interestingly, mitochondria of plants [91, 92] and fungi [67, 93] employ this mechanism of DNA replication, which suggests it’s ancestral origin. Nematodes represented by C. elegans appear to have maintained it [66], although all other animals analyzed to date seem to have evolved distinct mechanisms to replicate their mtDNA (see below).
Figure 7.
Models of invertebrate mtDNA replication. A, The rolling circle mechanism of nematode mtDNA replication proposed by Lewis et al. [66]. Circular progeny mtDNA molecules are proposed to be formed by a recombination-based resolution that involves the major non-coding region (NCR). B, The strand-coupled theta-like model of insect mtDNA replication. In the predominant mode described by Joers and Jacobs [96], replication initiates within the non-coding A+T region (A+T) and proceeds unidirectionally. The partially- or completely- strand-uncoupled models for Drosophila mtDNA replication [94, 96] (see text for details) are not represented. Arrows associated with replicating mtDNA indicate the 5′- to 3-′ direction of DNA synthesis. The gray arrowhead indicates the number and directionality of replication forks generated at the origin.
3.2 The theta model in Drosophila melanogaster
EM studies as early as the 1970s identified replication bubble-like structures, indicative of the initiation site within the repetitive non-coding A+T region in replicating mtDNA isolated from ovaries and embryos of various Drosophila species, including D. melanogaster [94, 95]. Recent analysis of D. melanogaster mtDNA from S2 cells in culture and whole animals using the 2DAGE technique corroborated these findings [96], and both the early and current reports concluded that replication starts at and proceeds unidirectionally from the A+T region in a theta-like fashion (Figure 7). The relative contribution of replication intermediates (RIs) that are products of uncoupled- or coupled- leading and lagging-strand synthesis is a point of disagreement; whereas the EM studies reported that an asynchronous mode of DNA synthesis predominates (with up to 99% of leading DNA strand synthesis completed before the initiation of the lagging strand synthesis), the 2DAGE analyses suggest the presence of only a subset of replicating molecules with stretches of ssDNA, arguing that D. melanogaster mtDNA replicates via a mechanism of coupled leading and lagging strand synthesis. Because duplex DNA replication intermediates observed by EM in an earlier study [97] are consistent with the 2DAGE data, the differences reported do not appear to be related to the techniques used; rather differences may lie in the preparation of mtDNA, and remain to be reconciled.
The 2DAGE analysis also revealed strong signals within the D. melanogaster mtDNA for two distinct replication pause sites or slow replicating zones, which were mapped to the binding sites of the transcription termination factor DmTTF [96, 98]. This implies that as with mammalian mtDNA [99] the transcription apparatus in Drosophila mitochondria also regulates mtDNA replication rate. Similar to the analysis of C. elegans mtDNA RIs, cruciform species (X arcs) are prominent intermediates in the Drosophila 2DAGE analysis, and map to the middle of the A+T region [96, 98]. In 1977, Rubenstein et al. reported that a significant fraction of D. melanogaster mtDNA was interlinked physically in two predominant forms of either supercoiled or relaxed catenated molecules. Interestingly, the frequency of catenanes increased during development from ~6.2% of the closed circular mtDNA molecules in 1–6 hr-old embryos to ~9.5% in 20 hr-old embryos, and ~12.5% in tissue culture cells in which mtDNA is replicated actively [97]. Thus, it appears that catenanes are related to the mtDNA replication process, suggesting that the cruciform intermediates observed by 2DAGE are derived by their digestion with the restriction endonucleases used in mtNA processing. Prominent RusA-sensitive X arcs and the presence of catenanes among the D. melanogaster mtRIs might be explained by a template switching-like mechanism, which has been proposed recently for replication of the 2μ plasmid-based high copy-number minichromosome of the yeast Saccharomyces cerevisiae [85]. Template-switching replication mediates damage bypass via a recombination-based mechanism in which a dynamic range of cruciform structures ultimately result in formation of hemicatenanes or similar forms. Further studies that engage both 2DAGE and EM are needed to explain the presence of cruciform structures and/or catenanes in D. melanogaster mtDNA.
3.3 The theta model in sea urchin
Sea urchins, such as Strongylocentrotus purpuratus, have been used as invertebrate models of the Deuterostome superphylum for many decades [100]. Their mtDNA contains a structure called the D-loop (alternately, R-loop) that is formed by the stable association of transcripts of ~60 bp with the major non-coding region (NCR), to generate a short triplex structure [101]. The D-loop RNA segments are found covalently linked to an ~15 bp long DNA, implying that the D-loop is the site of transition from priming to nascent leading DNA strand synthesis [101]. As with vertebrates (see below), D-loop strand synthesis events terminate frequently at the replication termination sequence downstream of the D-loop, but the mechanism allowing read-through and subsequent processive DNA synthesis remains unknown [102]. Using EM and 2DAGE, replication was shown to initiate in the D-loop region and proceed unidirectionally, implying that echinoderm mtDNA replication advances by D-loop expansion in a theta-like fashion [102, 103]. Characterization of RIs showed a high frequency of multiple duplex DNA segments on the lagging strand, and a less abundant fraction of ssDNA-containing species. Replication pause sites were found at widely scattered positions in the genome. The most prominent are at a distance of ~1/3 from the leading strand origin that coincides with the proposed origin of lagging strand synthesis. Interestingly, lagging strand synthesis appears to pause at the leading strand origin, suggesting that synthesis from, and pausing events in both sites could produce species that would appear as Cairns’ forms in EM analysis [102]. By contrast, Matsumoto et al. [103] observed RIs with expanded D-loops that were exclusively single-stranded.
3.4 mtDNA replication in vertebrates
3.4.1 Common concepts
We begin our discussion of vertebrate mtDNA replication with a description of initiation of leading strand synthesis, its relation to transcriptional start sites, and the structure and function of the D-loop and 7S DNA. Our focus will be primarily on mammals because they are represented by the bulk of the reported results, and cite data available on other systems where relevant.
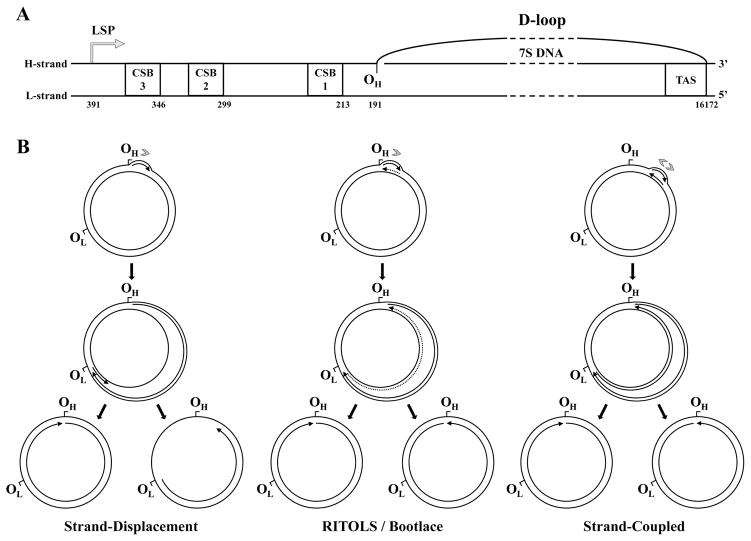
The complementary strands of vertebrate mtDNA are denoted as heavy (H) and light (L) due to their distinct base composition, which results in different sedimentation patterns in CsCl gradients [104]. The single NCR of the mammalian mitochondrial genome spans ~1 kb on average and contains three conserved sequence blocks (CSBs), assigned such that CSB1 is located near the tRNAPro gene and CSB3 lies towards the middle of the NCR [105–107]. In 1971, Kasamatsu and colleagues demonstrated that up to 50% of closed circular mtDNA molecules isolated from mouse L cells in culture contain a D-loop structure comprising a displaced heavy strand, and a light strand hybridized to a DNA segment with a sedimentation velocity of 7S [108] (Figure 8A). Further analysis demonstrated that the D-loop is located in the NCR [105–107]. The actual length of the D-loop depends on the length of the 7S DNA, which in humans is generally ~600 nt but may vary in size at its 5′-end that lies adjacent to CSB1 [109]. The 3′-end of the 7S DNA maps specifically between nucleotides 16104–16106, adjacent to the termination-associated sequence (TAS) (Figure 8A) on the other end of the NCR, near the tRNAPhe gene [110, 111]. The TAS is likely engaged in formation of a secondary structure, and represents a termination site for both RNA and DNA synthesis in vivo [110–112].
Figure 8.
Current models of vertebrate mtDNA replication. A, Structural organization of the D-loop and the adjacent cis-elements present within the non-coding region of vertebrate mtDNA. CSB1, 2 and 3, conserved sequence blocks 1, 2 and 3; TAS, termination associated sequence; LSP, light strand promoter; OH, origin of heavy strand DNA synthesis. The numbers below each element represent genomic positions in the human mtDNA reference sequence. The schematic is to scale, except for the region represented by the dashed lines. B, Strand-displacement, RITOLS/bootlace and strand-coupled models of vertebrate mtDNA replication (see text for details). The sites OH and OL (origin of light strand DNA synthesis) are represented as reference points on the genome map, although these sites are important primarily for the strand-displacement model. Arrows associated with replicating mtDNA indicate the 5′- to 3′-direction of DNA synthesis; continuous and dotted lines represent DNA and RNA, respectively. Only the long stretches of RNA described in the RITOLS model are represented; the putative short RNA primers of the other models are not shown. Gray arrowheads indicate the number and directionality of replication forks generated at the origin according to each model. Adapted from “E.A. McKinney, M.T. Oliveira: Replicating animal mitochondrial DNA. Genetics and Molecular Biology (2013) 36, 308–315”.
Several comparative studies of mitochondrial NCR region demonstrated that the presence of individual CSB sites or length of the D-loop region vary among mammals [113, 114]. For example, the mouse and bovine D-loop regions are ~200 nt shorter than in human mtDNA, and the bovine NCR appears to lack CSB1 and 3 [115]. These differences extend to other classes of Vertebrates. The D-loop region of Xenopus laevis mtDNA is ~1.6 kb long, and CSB1 appears to be absent [116]. The galliform species of birds contain a mitochondrial genome ~200 nt larger than that of most mammals, and a D-loop of ~780 nt accounts for much of the length difference [117, 118]. The chicken NCR has been shown to contain a single bidirectional promoter as compared to the two separate transcriptional promoters as in mammals [119].
It is commonly accepted that synthesis of the nascent heavy strand in mammals, which is the leading strand in mtDNA replication [120, 121], originates in the NCR with the majority of replication initiation events occurring downstream of CBS1 at OH (nt position 191) [82, 105–107]. The free 5′-end of the longest 7S DNA identified maps to OH [109, 122], implying that 7S DNA is a prematurely-terminated leading strand. Consistent with this, the 5′-end of nascent (elongated beyond NCR) and total (including 7S DNA) heavy strands have been mapped to OH (nucleotide position 190 +/−1) [122]. Synthesis of the leading strand from OH is primed from the light strand transcriptional promoter (LSP) located at nucleotide position 400 [123, 124]. Indeed, in addition to the long transcripts produced from LSP, transcripts of around 200 nt have also been documented as 7S RNA. The location of the free 5′-end of 7S RNA corresponds to the LSP, whereas its 3′-ends map to all three CSB sites with CSB1 predominating [107, 111, 125]). That mitochondrial transcripts are typically of genome length suggests a mechanism exists to inhibit processive synthesis, thereby enabling synthesis of short primers for replication initiation. It was shown recently that up to two-thirds of all transcription events initiated at LSP are terminated prematurely due to stacking interactions of guanine residues of nascent RNA and the non-template displaced heavy strand, forming a G-quadruplex at the CSB2 site [126, 127]. In vitro, the mitochondrial transcription elongation factor (TEFM) promotes processive synthesis by the mitochondrial RNA polymerase (POLRMT), abolishing premature termination at CSB2 and increasing dramatically the abundance of longer transcripts [128–130]. It has been proposed that TEFM binds the G-quadruplex region directly, serving as a molecular switch between primer synthesis and processive transcription [130]. In contrast, RNA primers remain in the NCR upon depletion of mitochondrial RNase H in mouse embryonic fibroblasts, resulting in generation of double-stranded breaks at the origin, thus highlighting the essential role of RNase H in primer processing at the OH site [131]. To date, the roles of the R-loop at CSB2, and the mechanism(s) for bypassing it by the priming apparatus are not fully understood, and warrant further investigation.
Because the half-life of the 7S DNA in rodent cells has been documented to be ~45 min [132], one can argue that the frequent replacement of the 7S DNA contradicts the notion of it serving to “prime” further synthesis. If so, a high rate of initiation events from OH rarely results in synthesis through the TAS site. The terminated DNA is likely removed by the mitochondrial ssDNA nuclease MGME1 [133, 134]; MGME1 knockdown in cultured cells or MGME1 deficiency in human patients results in a large accumulation of 7S DNA [133, 134]. Pol γ appears to bind preferentially at OH in vivo, and at the site corresponding to the 3′-end of the 7S DNA, which suggests a mechanism for replisome restart from the latter [111]. Upon ddCTP treatment, increased binding of the mtDNA helicase at the TAS site is also observed, suggesting that it also represents replication restart from the 3′-end of 7S DNA. However, this treatment leads to substantially reduced occupancy of Pol γ at this site, which would in any case be nonproductive due to its documented sensitivity to ddCTP incorporation. Perhaps the mtDNA helicase simply accumulates at the transition point from ssDNA to dsDNA when DNA synthesis stalls or is terminated. In support, data indicates that mtDNA helicase is enriched at the OH region and not at the 3′-end of the 7S DNA under normal conditions. The reason for the rapid turnover of 7S DNA remains unclear, but as suggested previously [122] may relate to functions of the D-loop in processes not associated with replication initiation. In that regard, a novel mitochondrial protein, ATAD3, binds the D-loop specifically and facilitates the interaction and segregation of mtDNA molecules in dividing mitochondria, and possibly anchors the mtDNA molecules to a portion of the mitochondrial inner membrane that is cholesterol-rich [135, 136].
A site for protein complex formation and plausibly for interactions of mtDNA with the mitochondrial inner membrane, the D-loop is also a natural substrate for mtSSB binding [137]. Given that the presence of mtSSB is a hallmark of replicating mtDNA molecules, the D-loop likely serves as the site for replisome assembly [70]. Moreover, Pol γ-β binds dsDNA [138] and has been demonstrated to bind with high affinity to the D-loop [139]. Furthermore, cells depleted of mtSSB and mtDNA helicase show loss of 7S DNA [55, 140], implying that the D-loop structure is maintained by the replisome, akin to the formation of the replication initiation bubble in other replication systems [141].
3.4.2 The strand-displacement, RITOLS/bootlace and strand-coupled models
On the basis of EM analyses of mtDNA isolated from mouse cells in culture [104], it was proposed over three decades ago that mammalian mtDNA is replicated via a strand displacement mechanism that is unidirectional and asynchronous mechanism [142] (Figure 8B). In this model, replication proceeds from OH continuously and unidirectionally, displacing the parental heavy strand, which remains single-stranded and bound by mtSSB [75, 111, 142, 143]. When the replication fork approaches approximately two-thirds of the genome length, the initiation site for lagging strand synthesis, OL, is exposed forming a stem-and-loop structure [142, 144, 145]. POLRMT has been shown to bind to the OL structure to initiate primer synthesis up to 25 nts, followed by binding by Pol γ to catalyze light strand DNA synthesis [74, 144–146]. The specific mechanism by which POLRMT recognizes OL and activates primer synthesis in vivo remains unknown, although a role for POLRMT in priming DNA synthesis from both strands has been shown in vitro [73, 74]. After initiation at OL, synthesis on both strands proceeds continuously, until two fully replicated daughter molecules are formed and segregated (Figure 8B). Notably, a stem-loop structure equivalent to OL appears to be absent in the mtDNA of galliform species, which is also the case in for species from many classes of Vertebrates, including for example, short-beaked dolphins and snake-neck turtles [118, 146]. Thus one might expect variations in the replication process among vertebrates, and in lagging DNA strand synthesis in particular.
In the year 2000, Holt and colleagues published the first results involving analyses of human and rodent RIs by 2DAGE [80]. They demonstrated that RIs contain a ssDNA-specific nuclease-resistant Y arc, suggesting that the long single-stranded stretches of the parental heavy strand predicted by the strand displacement model were absent. Instead, the authors proposed that the RIs observed in their analysis resulted from a strand-coupled replication mechanism. Holt and colleagues noted that a small subset of the Y arc intermediates were sensitive to ssDNA-specific nucleases. Thereafter, a refined protocol for isolation of mtNA to include a step of sucrose density-gradient purification led to the elimination of ssDNA-containing species in 2DAGE analysis, revealing a substantial presence of ribonucleotides in mtDNA replication intermediates [147]. Novel, unusually large DNA molecules forming slow-moving arcs in RIs were also identified. Whereas they were found to be largely resistant to restriction endonuclease digestion, they were sensitive to RNase H, suggesting that the incorporated RNA tracts were inhibiting the restriction enzymes. This led to a new proposed mechanism of mtDNA replication, similar in principle to the strand displacement model, but positing Ribonucleotides Incorporated Through Out the Lagging Strand (RITOLS) prior to initiation of the light strand synthesis [121, 148] (Figure 8B). Later, it was demonstrated that inhibition of transcription with cordycepin triphosphate did not affect replication, arguing that during leading strand synthesis preexisting RNA is incorporated on the lagging strand via a “bootlace” strategy, rather than being synthesized concomitant with leading strand synthesis [149]. Indeed, mature RNA is stored in mammalian mitochondria in RNA granules juxtaposed to the mitochondrial nucleoids [150, 151], and could feasibly serve as a source of the RITOLS. It remains unclear how the incorporated RNA is removed in maturation of progeny molecules [149, 152]. Although mitochondrial RNase H may be involved, recent data on the effects of its depletion in mouse embryonic fibroblasts show accumulation of RNA at OH and OL, suggesting a role in primer removal, though accumulation of the bootlace RNA was not apparent [131].
The 2DAGE technique has been used extensively in the analysis of various plasmid replication mechanisms [79, 81, 83, 153–157], but it differs from the traditional, well-established EM analyses using CsCl-EtBr gradient-purified mtDNA [104, 108]. Providing experimental support for the RITOLS model, Pohjoismäki and colleagues demonstrated that various methods of sample preparation affected substantially the composition of RIs. In particular, the use of CsCl gradient sedimentation and E. coli SSB binding (common in EM analysis) lead to systematic loss of RNA from the RNA/DNA hybrids. Moreover, EM analyses of the samples prepared as for 2DAGE showed that intact mtDNA replication intermediates are predominantly fully duplex. Rather, RNase H treatment resulted in accumulation of single-stranded intermediates consistent with those observed earlier by EM analysis [158]. In contrast, analysis by atomic force microscopy of rat liver mtNA that was isolated by a protocol similar to that used for the 2DAGE analysis, but involving CsCl-EtBr gradient sedimentation, lead to the conclusion that the majority of mtDNA RIs are formed by the strand-displacement mechanism [143]. Fusté et al. recently evaluated existing 2DAGE data in comparison with mtNAs treated with RNases or with DNase I; they observed that simple mixing of RNA- and DNA-free mtNA samples restores the bubble arc that had disappeared upon removal of the RNA, implying that the RIs giving rise to the RITOLS model could be artifacts of the isolation procedure. This evidence does not disprove the presence of the RNA/DNA hybrids in vivo [75], and their study did not reconstitute fully the slow-moving species that represent a hallmark of the RITOLS model [148]. Compelling evidence for the strand-displacement model derives from recent data generated using ChIP-Seq on the distribution of mtSSB bound to mtDNA [75]. mtSSB was found bound almost exclusively to the heavy strand with high concentrations found in the D-loop, and decreasing proportionally towards OL, which is consistent with it being single-stranded during nascent heavy strand synthesis. In contrast, Reyes et al. used psoralen/UV cross-linking and in organello labeling to provide strong evidence that RNA/DNA hybrids do indeed form in vivo during the process of mammalian mtDNA replication, and are later matured into fully duplex DNA [149]. Clearly, further experimentation is required to resolve these issues.
The findings published in the early 2000s suggest that a fraction of the mtRIs comprise fully-duplex dsDNA, the presumptive products of theta-like replication produced by coupled leading and lagging strand DNA synthesis [80, 159]. This mode, however, appears to occur primarily when cells are recovering from EtBr-induced mtDNA depletion, a condition that stimulates mtDNA replication. In this case, a broad zone containing the CYTB, ND5 and ND6 genes constitutes the initiation site from which replication proceeds bidirectionally until termination occurs in the the D-loop region [159] (Figure 8B). The model invokes the presence of Okazaki fragments, and though they have not yet been demonstrated directly, Okazaki fragment-processing enzymes including Pif1, FEN1, and Dna2 have been documented in mitochondria [160–162]. One might argue that the multiple sites of lagging strand initiation found by Brown et al. [143] are evidence for Okazaki fragment-like species in replicating mtDNA. Although the strand-coupled model of mammalian mtDNA replication is now considered to be a secondary mechanism, 2DAGE analysis of mtRIs from the chicken Gallus gallus suggested that a strand-coupled theta replication mode predominates, and similar to that in mammals, initiates over a broad zone, ori-Z [159, 163]. In contrast to mammals, in which the majority of the replication initiation events occur in the OH region, the prominent replication initiation site in chicken maps to the ND6 gene, and OH likely serves a role as a termination site. Interestingly, the ND6 and CYTB genes are transposed in bird mtDNA, such that ND6 maps adjacent to the NCR [117, 163].
Termination of mtDNA replication in vertebrates is a subject of current studies that suggest a possible involvement of four-way junctions. These are proposed to arise when replication forks arrest in the NCR [159]. Three proteins of the mTERF family may contribute to the termination of mtDNA replication in human cells in culture [99, 164]. Four-way structures that resemble Holliday junctions have been shown by EM to occur frequently in replicating mtDNA from human hearts, which also appears to be organized in multimeric catenated networks [165], suggestive of a recombination-based replication mechanism. The increased abundance of Holliday junctions and complex mtDNA forms obseved upon overexpression of TFAM or the mtDNA helicase in the mouse heart [165] suggests a similar recombinational DNA replication pathway. Similarly to that found in D. melanogaster, formation of more complex forms of mtDNA in the human heart correlates with high mtDNA copy number [166]. Interestingly, the mtDNA from mouse, rat, rabbit and infant human hearts has a less complex organization [165, 166].
4. Perspectives
Since the discovery of mitochondrial DNA 50 years ago and the identification of the mitochondrial replicase a decade later, a broad base of knowledge has been established on key elements of mtDNA replication. Yet, many fascinating questions remain relevant to the expanding repertoire of proteins involved in mtDNA metabolism and its regulation. Understanding the interplay of key proteins at the replication fork with new protein players will require development of new substrates for in vitro assays that are validated by novel approaches to probe the changing physiological environment in mitochondria in both normal and disease states. A major hurdle remains to elucidate the mechanism(s) of replication initiation across taxa. The processes of termination and segregation to ensure the integrity of mtDNA inheritance also warrant future study. Exploring the roles of recombinational intermediates and catenanes in replication may link the physiological processes of DNA replication and recombination, and perhaps also post-replicational repair.
Acknowledgments
We thank Dr. Jon Kaguni for critical reading of the manuscript. Research cited from the L.S.K. lab was supported by grant GM45295 from the National Institutes of Health. L.S.K. was supported partially by the Academy of Finland. G.L.C. was supported by the University of Tampere. M.T.O. acknowledges support from the Fundação de Amparo à Pesquisa do Estado de São Paulo (Grant 2014/02253-6).
References
- 1.Nass MM, Nass S. Intramitochondrial Fibers with DNA Characteristics. I. Fixation and Electron Staining Reactions. J Cell Biol. 1963;19:593–611. doi: 10.1083/jcb.19.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace DC, et al. Familial mitochondrial encephalomyopathy (MERRF): genetic, pathophysiological, and biochemical characterization of a mitochondrial DNA disease. Cell. 1988;55(4):601–10. doi: 10.1016/0092-8674(88)90218-8. [DOI] [PubMed] [Google Scholar]
- 3.Wallace DC, et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242(4884):1427–30. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 4.Holt IJ, Harding AE, Morgan-Hughes JA. Mitochondrial DNA polymorphism in mitochondrial myopathy. Hum Genet. 1988;79(1):53–7. doi: 10.1007/BF00291710. [DOI] [PubMed] [Google Scholar]
- 5.Brown WM, George M, Jr, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979;76(4):1967–71. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gissi C, Iannelli F, Pesole G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity (Edinb) 2008;101(4):301–20. doi: 10.1038/hdy.2008.62. [DOI] [PubMed] [Google Scholar]
- 7.Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu Rev Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- 8.Oliveira MT, Garesse R, Kaguni LS. Animal models of mitochondrial DNA transactions in disease and ageing. Exp Gerontol. 2010;45(7–8):489–502. doi: 10.1016/j.exger.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKinney EA, Oliveira MT. Replicating animal mitochondrial DNA. Genet Mol Biol. 2013;36(3):308–15. doi: 10.1590/S1415-47572013000300002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stumpf JD, Saneto RP, Copeland WC. Clinical and molecular features of POLG-related mitochondrial disease. Cold Spring Harb Perspect Biol. 2013;5(4):a011395. doi: 10.1101/cshperspect.a011395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–23. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 12.Kujoth GC, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309(5733):481–4. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 13.Brinkman K, Kakuda TN. Mitochondrial toxicity of nucleoside analogue reverse transcriptase inhibitors: a looming obstacle for long-term antiretroviral therapy? Curr Opin Infect Dis. 2000;13(1):5–11. doi: 10.1097/00001432-200002000-00002. [DOI] [PubMed] [Google Scholar]
- 14.Lewis W, Day BJ, Copeland WC. Mitochondrial toxicity of NRTI antiviral drugs: an integrated cellular perspective. Nat Rev Drug Discov. 2003;2(10):812–22. doi: 10.1038/nrd1201. [DOI] [PubMed] [Google Scholar]
- 15.Lee YS, Kennedy WD, Yin YW. Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations. Cell. 2009;139(2):312–24. doi: 10.1016/j.cell.2009.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szymanski MR, et al. Structural basis for processivity and antiviral drug toxicity in human mitochondrial DNA replicase. EMBO J. 2015;34(14):1959–70. doi: 10.15252/embj.201591520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sohl CD, et al. Probing the structural and molecular basis of nucleotide selectivity by human mitochondrial DNA polymerase gamma. Proc Natl Acad Sci U S A. 2015;112(28):8596–601. doi: 10.1073/pnas.1421733112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliveira MT, Haukka J, Kaguni LS. Evolution of the metazoan mitochondrial replicase. Genome Biol Evol. 2015;7(4):943–59. doi: 10.1093/gbe/evv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Longley MJ, et al. Mutant POLG2 disrupts DNA polymerase gamma subunits and causes progressive external ophthalmoplegia. Am J Hum Genet. 2006;78(6):1026–34. doi: 10.1086/504303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young MJ, et al. Biochemical analysis of human POLG2 variants associated with mitochondrial disease. Hum Mol Genet. 2011;20(15):3052–66. doi: 10.1093/hmg/ddr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farnum GA, Nurminen A, Kaguni LS. Mapping 136 pathogenic mutations into functional modules in human DNA polymerase gamma establishes predictive genotype-phenotype correlations for the complete spectrum of POLG syndromes. Biochim Biophys Acta. 2014;1837(7):1113–21. doi: 10.1016/j.bbabio.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Euro L, et al. Clustering of Alpers disease mutations and catalytic defects in biochemical variants reveal new features of molecular mechanism of the human mitochondrial replicase, Pol gamma. Nucleic Acids Res. 2011;39(21):9072–84. doi: 10.1093/nar/gkr618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bienstock RJ, Copeland WC. Molecular insights into NRTI inhibition and mitochondrial toxicity revealed from a structural model of the human mitochondrial DNA polymerase. Mitochondrion. 2004;4(2–3):203–13. doi: 10.1016/j.mito.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 24.Bratic A, et al. Complementation between polymerase- and exonuclease-deficient mitochondrial DNA polymerase mutants in genomically engineered flies. Nat Commun. 2015;6:8808. doi: 10.1038/ncomms9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olson MW, et al. Subunit structure of mitochondrial DNA polymerase from Drosophila embryos. Physical and immunological studies. J Biol Chem. 1995;270(48):28932–7. doi: 10.1074/jbc.270.48.28932. [DOI] [PubMed] [Google Scholar]
- 26.Wernette CM, Kaguni LS. A mitochondrial DNA polymerase from embryos of Drosophila melanogaster. Purification, subunit structure, and partial characterization. J Biol Chem. 1986;261(31):14764–70. [PubMed] [Google Scholar]
- 27.Kaguni LS, Oliveira MT. Structure, function and evolution of the animal mitochondrial replicative DNA helicase. Crit Rev Biochem Mol Biol. 2015:1–12. doi: 10.3109/10409238.2015.1117056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toth EA, et al. The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol Cell. 2003;12(5):1113–23. doi: 10.1016/s1097-2765(03)00442-8. [DOI] [PubMed] [Google Scholar]
- 29.Caruthers JM, McKay DB. Helicase structure and mechanism. Curr Opin Struct Biol. 2002;12(1):123–33. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 30.Korhonen JA, et al. Structure-function defects of the TWINKLE linker region in progressive external ophthalmoplegia. J Mol Biol. 2008;377(3):691–705. doi: 10.1016/j.jmb.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 31.Matsushima Y, Kaguni LS. Differential phenotypes of active site and human autosomal dominant progressive external ophthalmoplegia mutations in Drosophila mitochondrial DNA helicase expressed in Schneider cells. J Biol Chem. 2007;282(13):9436–44. doi: 10.1074/jbc.M610550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longley MJ, et al. Disease variants of the human mitochondrial DNA helicase encoded by C10orf2 differentially alter protein stability, nucleotide hydrolysis, and helicase activity. J Biol Chem. 2010;285(39):29690–702. doi: 10.1074/jbc.M110.151795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandez-Millan P, et al. The hexameric structure of the human mitochondrial replicative helicase Twinkle. Nucleic Acids Res. 2015;43(8):4284–95. doi: 10.1093/nar/gkv189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsushima Y, Kaguni LS. Functional importance of the conserved N-terminal domain of the mitochondrial replicative DNA helicase. Biochim Biophys Acta. 2009;1787(5):290–5. doi: 10.1016/j.bbabio.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stiban J, et al. The N-terminal domain of the Drosophila mitochondrial replicative DNA helicase contains an iron-sulfur cluster and binds DNA. J Biol Chem. 2014;289(35):24032–42. doi: 10.1074/jbc.M114.587774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farge G, et al. The N-terminal domain of TWINKLE contributes to single- stranded DNA binding and DNA helicase activities. Nucleic Acids Res. 2008;36(2):393–403. doi: 10.1093/nar/gkm1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wanrooij S, et al. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35(10):3238–51. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ziebarth TD, et al. Dynamic effects of cofactors and DNA on the oligomeric state of human mitochondrial DNA helicase. J Biol Chem. 2010;285(19):14639–47. doi: 10.1074/jbc.M109.099663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaguni LS, Oliveira MT. Structure, function and evolution of the animal mitochondrial replicative DNA helicase. Crit Rev Biochem Mol Biol. 2016;51(1):53–64. doi: 10.3109/10409238.2015.1117056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crampton DJ, et al. Oligomeric states of bacteriophage T7 gene 4 primase/helicase. J Mol Biol. 2006;360(3):667–77. doi: 10.1016/j.jmb.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 41.Jemt E, et al. The mitochondrial DNA helicase TWINKLE can assemble on a closed circular template and support initiation of DNA synthesis. Nucleic Acids Res. 2011;39(21):9238–49. doi: 10.1093/nar/gkr653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shutt TE, Gray MW. Twinkle, the mitochondrial replicative DNA helicase, is widespread in the eukaryotic radiation and may also be the mitochondrial DNA primase in most eukaryotes. J Mol Evol. 2006;62(5):588–99. doi: 10.1007/s00239-005-0162-8. [DOI] [PubMed] [Google Scholar]
- 43.Shereda RD, et al. SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol. 2008;43(5):289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thommes P, et al. Mitochondrial single-stranded DNA-binding protein from Drosophila embryos. Physical and biochemical characterization. J Biol Chem. 1995;270(36):21137–43. doi: 10.1074/jbc.270.36.21137. [DOI] [PubMed] [Google Scholar]
- 45.Farr CL, Wang Y, Kaguni LS. Functional interactions of mitochondrial DNA polymerase and single-stranded DNA-binding protein. Template-primer DNA binding and initiation and elongation of DNA strand synthesis. J Biol Chem. 1999;274(21):14779–85. doi: 10.1074/jbc.274.21.14779. [DOI] [PubMed] [Google Scholar]
- 46.Oliveira MT, Kaguni LS. Functional roles of the N- and C-terminal regions of the human mitochondrial single-stranded DNA-binding protein. PLoS One. 2010;5(10):e15379. doi: 10.1371/journal.pone.0015379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oliveira MT, Kaguni LS. Reduced stimulation of recombinant DNA polymerase gamma and mitochondrial DNA (mtDNA) helicase by variants of mitochondrial single-stranded DNA-binding protein (mtSSB) correlates with defects in mtDNA replication in animal cells. J Biol Chem. 2011;286(47):40649–58. doi: 10.1074/jbc.M111.289983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ciesielski GL, et al. Mitochondrial Single-stranded DNA-binding Proteins Stimulate the Activity of DNA Polymerase gamma by Organization of the Template DNA. J Biol Chem. 2015;290(48):28697–707. doi: 10.1074/jbc.M115.673707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korhonen JA, Gaspari M, Falkenberg M. TWINKLE Has 5′ -> 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J Biol Chem. 2003;278(49):48627–32. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 50.Korhonen JA, et al. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23(12):2423–9. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maier D, et al. Mitochondrial single-stranded DNA-binding protein is required for mitochondrial DNA replication and development in Drosophila melanogaster. Mol Biol Cell. 2001;12(4):821–30. doi: 10.1091/mbc.12.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugimoto T, et al. Caenorhabditis elegans par2.1/mtssb-1 is essential for mitochondrial DNA replication and its defect causes comprehensive transcriptional alterations including a hypoxia response. Exp Cell Res. 2008;314(1):103–14. doi: 10.1016/j.yexcr.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 53.Farr CL, et al. Physiological and biochemical defects in functional interactions of mitochondrial DNA polymerase and DNA-binding mutants of single-stranded DNA-binding protein. J Biol Chem. 2004;279(17):17047–53. doi: 10.1074/jbc.M400283200. [DOI] [PubMed] [Google Scholar]
- 54.Fukuoh A, et al. Screen for mitochondrial DNA copy number maintenance genes reveals essential role for ATP synthase. Mol Syst Biol. 2014;10:734. doi: 10.15252/msb.20145117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruhanen H, et al. Mitochondrial single-stranded DNA binding protein is required for maintenance of mitochondrial DNA and 7S DNA but is not required for mitochondrial nucleoid organisation. Biochim Biophys Acta. 2010;1803(8):931–9. doi: 10.1016/j.bbamcr.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 56.Innis CA, Shi J, Blundell TL. Evolutionary trace analysis of TGF-beta and related growth factors: implications for site-directed mutagenesis. Protein Eng. 2000;13(12):839–47. doi: 10.1093/protein/13.12.839. [DOI] [PubMed] [Google Scholar]
- 57.Lichtarge O, Bourne HR, Cohen FE. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol. 1996;257(2):342–58. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 58.Curth U, et al. Single-stranded-DNA-binding proteins from human mitochondria and Escherichia coli have analogous physicochemical properties. Eur J Biochem. 1994;221(1):435–43. doi: 10.1111/j.1432-1033.1994.tb18756.x. [DOI] [PubMed] [Google Scholar]
- 59.Yang C, et al. Crystal structure of human mitochondrial single-stranded DNA binding protein at 2.4 A resolution. Nat Struct Biol. 1997;4(2):153–7. doi: 10.1038/nsb0297-153. [DOI] [PubMed] [Google Scholar]
- 60.Shutt TE, Gray MW. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet. 2006;22(2):90–5. doi: 10.1016/j.tig.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 61.Mahoungou C, et al. The amino-terminal sequence of the Xenopus laevis mitochondrial SSB is homologous to that of the Escherichia coli protein. FEBS Lett. 1988;235(1–2):267–70. doi: 10.1016/0014-5793(88)81276-6. [DOI] [PubMed] [Google Scholar]
- 62.Fan L, et al. The accessory subunit of mtDNA polymerase shares structural homology with aminoacyl-tRNA synthetases: implications for a dual role as a primer recognition factor and processivity clamp. Proc Natl Acad Sci U S A. 1999;96(17):9527–32. doi: 10.1073/pnas.96.17.9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carrodeguas JA, et al. The accessory subunit of Xenopus laevis mitochondrial DNA polymerase gamma increases processivity of the catalytic subunit of human DNA polymerase gamma and is related to class II aminoacyl-tRNA synthetases. Mol Cell Biol. 1999;19(6):4039–46. doi: 10.1128/mcb.19.6.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolf YI, Koonin EV. Origin of an animal mitochondrial DNA polymerase subunit via lineage-specific acquisition of a glycyl-tRNA synthetase from bacteria of the Thermus-Deinococcus group. Trends Genet. 2001;17(8):431–3. doi: 10.1016/s0168-9525(01)02370-8. [DOI] [PubMed] [Google Scholar]
- 65.Yakubovskaya E, et al. Functional human mitochondrial DNA polymerase gamma forms a heterotrimer. J Biol Chem. 2006;281(1):374–82. doi: 10.1074/jbc.M509730200. [DOI] [PubMed] [Google Scholar]
- 66.Lewis SC, et al. A rolling circle replication mechanism produces multimeric lariats of mitochondrial DNA in Caenorhabditis elegans. PLoS Genet. 2015;11(2):e1004985. doi: 10.1371/journal.pgen.1004985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maleszka R, Skelly PJ, Clark-Walker GD. Rolling circle replication of DNA in yeast mitochondria. EMBO J. 1991;10(12):3923–9. doi: 10.1002/j.1460-2075.1991.tb04962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee YS, et al. Each monomer of the dimeric accessory protein for human mitochondrial DNA polymerase has a distinct role in conferring processivity. J Biol Chem. 2010;285(2):1490–9. doi: 10.1074/jbc.M109.062752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carrodeguas JA, et al. Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer. Mol Cell. 2001;7(1):43–54. doi: 10.1016/s1097-2765(01)00153-8. [DOI] [PubMed] [Google Scholar]
- 70.Rajala N, et al. Replication factors transiently associate with mtDNA at the mitochondrial inner membrane to facilitate replication. Nucleic Acids Res. 2014;42(2):952–67. doi: 10.1093/nar/gkt988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Atanassova N, et al. Sequence-specific stalling of DNA polymerase gamma and the effects of mutations causing progressive ophthalmoplegia. Hum Mol Genet. 2011;20(6):1212–23. doi: 10.1093/hmg/ddq565. [DOI] [PubMed] [Google Scholar]
- 72.Holmlund T, et al. Structure-function defects of the twinkle amino-terminal region in progressive external ophthalmoplegia. Biochim Biophys Acta. 2009;1792(2):132–9. doi: 10.1016/j.bbadis.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 73.Wanrooij S, et al. Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc Natl Acad Sci U S A. 2008;105(32):11122–7. doi: 10.1073/pnas.0805399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fuste JM, et al. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol Cell. 2010;37(1):67–78. doi: 10.1016/j.molcel.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 75.Miralles Fuste J, et al. In vivo occupancy of mitochondrial single-stranded DNA binding protein supports the strand displacement mode of DNA replication. PLoS Genet. 2014;10(12):e1004832. doi: 10.1371/journal.pgen.1004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ventura N, Rea SL. Caenorhabditis elegans mitochondrial mutants as an investigative tool to study human neurodegenerative diseases associated with mitochondrial dysfunction. Biotechnol J. 2007;2(5):584–95. doi: 10.1002/biot.200600248. [DOI] [PubMed] [Google Scholar]
- 77.Grad LI, Lemire BD. Mitochondrial complex I mutations in Caenorhabditis elegans produce cytochrome c oxidase deficiency, oxidative stress and vitamin-responsive lactic acidosis. Hum Mol Genet. 2004;13(3):303–14. doi: 10.1093/hmg/ddh027. [DOI] [PubMed] [Google Scholar]
- 78.Bratic I, Hench J, Trifunovic A. Caenorhabditis elegans as a model system for mtDNA replication defects. Methods. 2010;51(4):437–43. doi: 10.1016/j.ymeth.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 79.Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51(3):463–71. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 80.Holt IJ, Lorimer HE, Jacobs HT. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100(5):515–24. doi: 10.1016/s0092-8674(00)80688-1. [DOI] [PubMed] [Google Scholar]
- 81.Bell L, Byers B. Separation of branched from linear DNA by two-dimensional gel electrophoresis. Anal Biochem. 1983;130(2):527–35. doi: 10.1016/0003-2697(83)90628-0. [DOI] [PubMed] [Google Scholar]
- 82.Yasukawa T, et al. A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol Cell. 2005;18(6):651–62. doi: 10.1016/j.molcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 83.Brewer BJ, Fangman WL. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55(4):637–43. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 84.Kajander OA, et al. Prominent mitochondrial DNA recombination intermediates in human heart muscle. EMBO Rep. 2001;2(11):1007–12. doi: 10.1093/embo-reports/kve233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Giannattasio M, et al. Visualization of recombination-mediated damage bypass by template switching. Nat Struct Mol Biol. 2014;21(10):884–92. doi: 10.1038/nsmb.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee SH, et al. Synthesis and dissolution of hemicatenanes by type IA DNA topoisomerases. Proc Natl Acad Sci U S A. 2013;110(38):E3587–94. doi: 10.1073/pnas.1304103110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blanco L, et al. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. 1989;264(15):8935–40. [PubMed] [Google Scholar]
- 88.Morris CF, Sinha NK, Alberts BM. Reconstruction of bacteriophage T4 DNA replication apparatus from purified components: rolling circle replication following de novo chain initiation on a single-stranded circular DNA template. Proc Natl Acad Sci U S A. 1975;72(12):4800–4. doi: 10.1073/pnas.72.12.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Konopa G, et al. Bacteriophage and host mutants causing the rolling-circle lambda DNA replication early after infection. FEBS Lett. 2000;472(2–3):217–20. doi: 10.1016/s0014-5793(00)01451-4. [DOI] [PubMed] [Google Scholar]
- 90.Nomura N, Ray DS. Replication of bacteriophage M13. XV. Location of the specific nick in M13 replicative form II accumulated in Escherichia coli polAex1. J Virol. 1980;34(1):162–7. doi: 10.1128/jvi.34.1.162-167.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Backert S, et al. Rolling-circle replication of mitochondrial DNA in the higher plant Chenopodium album (L.) Mol Cell Biol. 1996;16(11):6285–94. doi: 10.1128/mcb.16.11.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cupp JD, Nielsen BL. Minireview: DNA replication in plant mitochondria. Mitochondrion. 2014;19(Pt B):231–7. doi: 10.1016/j.mito.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ling F, Shibata T. Mhr1p-dependent concatemeric mitochondrial DNA formation for generating yeast mitochondrial homoplasmic cells. Mol Biol Cell. 2004;15(1):310–22. doi: 10.1091/mbc.E03-07-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Goddard JM, Wolstenholme DR. Origin and direction of replication in mitochondrial DNA molecules from Drosophila melanogaster. Proc Natl Acad Sci U S A. 1978;75(8):3886–90. doi: 10.1073/pnas.75.8.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Goddard JM, Wolstenholme DR. Origin and direction of replication in mitochondrial DNA molecules from the genus Drosophila. Nucleic Acids Res. 1980;8(4):741–57. [PMC free article] [PubMed] [Google Scholar]
- 96.Joers P, Jacobs HT. Analysis of replication intermediates indicates that Drosophila melanogaster mitochondrial DNA replicates by a strand-coupled theta mechanism. PLoS One. 2013;8(1):e53249. doi: 10.1371/journal.pone.0053249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rubenstein JL, Brutlag D, Clayton DA. The mitochondrial DNA of Drosophila melanogaster exists in two distinct and stable superhelical forms. Cell. 1977;12(2):471–82. doi: 10.1016/0092-8674(77)90123-4. [DOI] [PubMed] [Google Scholar]
- 98.Joers P, et al. Mitochondrial transcription terminator family members mTTF and mTerf5 have opposing roles in coordination of mtDNA synthesis. PLoS Genet. 2013;9(9):e1003800. doi: 10.1371/journal.pgen.1003800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hyvarinen AK, et al. The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA. Nucleic Acids Res. 2007;35(19):6458–74. doi: 10.1093/nar/gkm676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rast JP, et al. Genomic insights into the immune system of the sea urchin. Science. 2006;314(5801):952–6. doi: 10.1126/science.1134301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jacobs HT, Herbert ER, Rankine J. Sea urchin egg mitochondrial DNA contains a short displacement loop (D-loop) in the replication origin region. Nucleic Acids Res. 1989;17(22):8949–65. doi: 10.1093/nar/17.22.8949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mayhook AG, Rinaldi AM, Jacobs HT. Replication origins and pause sites in sea urchin mitochondrial DNA. Proc Biol Sci. 1992;248(1321):85–94. doi: 10.1098/rspb.1992.0046. [DOI] [PubMed] [Google Scholar]
- 103.Matsumoto L, et al. Mitochondrial DNA replication in sea urchin oocytes. J Cell Biol. 1974;63(1):146–59. doi: 10.1083/jcb.63.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Robberson DL, Kasamatsu H, Vinograd J. Replication of mitochondrial DNA. Circular replicative intermediates in mouse L cells. Proc Natl Acad Sci U S A. 1972;69(3):737–41. doi: 10.1073/pnas.69.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Crews S, et al. Nucleotide sequence of a region of human mitochondrial DNA containing the precisely identified origin of replication. Nature. 1979;277(5693):192–8. doi: 10.1038/277192a0. [DOI] [PubMed] [Google Scholar]
- 106.Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–65. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 107.Nicholls TJ, Minczuk M. In D-loop: 40 years of mitochondrial 7S DNA. Exp Gerontol. 2014;56:175–81. doi: 10.1016/j.exger.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 108.Kasamatsu H, Robberson DL, Vinograd J. A novel closed-circular mitochondrial DNA with properties of a replicating intermediate. Proc Natl Acad Sci U S A. 1971;68(9):2252–7. doi: 10.1073/pnas.68.9.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brown WM, Shine J, Goodman HM. Human mitochondrial DNA: analysis of 7S DNA from the origin of replication. Proc Natl Acad Sci U S A. 1978;75(2):735–9. doi: 10.1073/pnas.75.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Doda JN, Wright CT, Clayton DA. Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proc Natl Acad Sci U S A. 1981;78(10):6116–20. doi: 10.1073/pnas.78.10.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jemt E, et al. Regulation of DNA replication at the end of the mitochondrial D-loop involves the helicase TWINKLE and a conserved sequence element. Nucleic Acids Res. 2015;43(19):9262–75. doi: 10.1093/nar/gkv804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Roberti M, et al. Multiple protein-binding sites in the TAS-region of human and rat mitochondrial DNA. Biochem Biophys Res Commun. 1998;243(1):36–40. doi: 10.1006/bbrc.1997.8052. [DOI] [PubMed] [Google Scholar]
- 113.Douzery E, Randi E. The mitochondrial control region of Cervidae: evolutionary patterns and phylogenetic content. Mol Biol Evol. 1997;14(11):1154–66. doi: 10.1093/oxfordjournals.molbev.a025725. [DOI] [PubMed] [Google Scholar]
- 114.Saccone C, Pesole G, Sbisa E. The main regulatory region of mammalian mitochondrial DNA: structure-function model and evolutionary pattern. J Mol Evol. 1991;33(1):83–91. doi: 10.1007/BF02100199. [DOI] [PubMed] [Google Scholar]
- 115.King TC, Low RL. Mapping of control elements in the displacement loop region of bovine mitochondrial DNA. J Biol Chem. 1987;262(13):6204–13. [PubMed] [Google Scholar]
- 116.Bogenhagen DF, Morvillo MV. Mapping light strand transcripts near the origin of replication of Xenopus laevis mitochondrial DNA. Nucleic Acids Res. 1990;18(21):6377–83. doi: 10.1093/nar/18.21.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nass MM. Precise sequence assignment of replication origin in the control region of chick mitochondrial DNA relative to 5′ and 3′ D-loop ends, secondary structure, DNA synthesis, and protein binding. Curr Genet. 1995;28(5):401–9. doi: 10.1007/BF00310807. [DOI] [PubMed] [Google Scholar]
- 118.Desjardins P, Morais R. Sequence and gene organization of the chicken mitochondrial genome. A novel gene order in higher vertebrates. J Mol Biol. 1990;212(4):599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- 119.L’Abbe D, et al. The transcription of DNA in chicken mitochondria initiates from one major bidirectional promoter. J Biol Chem. 1991;266(17):10844–50. [PubMed] [Google Scholar]
- 120.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–35. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 121.Holt IJ. Mitochondrial DNA replication and repair: all a flap. Trends Biochem Sci. 2009;34(7):358–65. doi: 10.1016/j.tibs.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 122.Kang D, et al. In vivo determination of replication origins of human mitochondrial DNA by ligation-mediated polymerase chain reaction. J Biol Chem. 1997;272(24):15275–9. doi: 10.1074/jbc.272.24.15275. [DOI] [PubMed] [Google Scholar]
- 123.Montoya J, et al. Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc Natl Acad Sci U S A. 1982;79(23):7195–9. doi: 10.1073/pnas.79.23.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chang DD, Hauswirth WW, Clayton DA. Replication priming and transcription initiate from precisely the same site in mouse mitochondrial DNA. EMBO J. 1985;4(6):1559–67. doi: 10.1002/j.1460-2075.1985.tb03817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chang DD, Clayton DA. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci U S A. 1985;82(2):351–5. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pham XH, et al. Conserved sequence box II directs transcription termination and primer formation in mitochondria. J Biol Chem. 2006;281(34):24647–52. doi: 10.1074/jbc.M602429200. [DOI] [PubMed] [Google Scholar]
- 127.Wanrooij PH, et al. A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop. Nucleic Acids Res. 2012;40(20):10334–44. doi: 10.1093/nar/gks802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Minczuk M, et al. TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res. 2011;39(10):4284–99. doi: 10.1093/nar/gkq1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Posse V, et al. TEFM is a potent stimulator of mitochondrial transcription elongation in vitro. Nucleic Acids Res. 2015;43(5):2615–24. doi: 10.1093/nar/gkv105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Agaronyan K, et al. Mitochondrial biology. Replication-transcription switch in human mitochondria. Science. 2015;347(6221):548–51. doi: 10.1126/science.aaa0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Holmes JB, et al. Primer retention owing to the absence of RNase H1 is catastrophic for mitochondrial DNA replication. Proc Natl Acad Sci U S A. 2015;112(30):9334–9. doi: 10.1073/pnas.1503653112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gensler S, et al. Mechanism of mammalian mitochondrial DNA replication: import of mitochondrial transcription factor A into isolated mitochondria stimulates 7S DNA synthesis. Nucleic Acids Res. 2001;29(17):3657–63. doi: 10.1093/nar/29.17.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kornblum C, et al. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat Genet. 2013;45(2):214–9. doi: 10.1038/ng.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Szczesny RJ, et al. Identification of a novel human mitochondrial endo-/exonuclease Ddk1/c20orf72 necessary for maintenance of proper 7S DNA levels. Nucleic Acids Res. 2013;41(5):3144–61. doi: 10.1093/nar/gkt029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.He J, et al. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J Cell Biol. 2007;176(2):141–6. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gerhold JM, et al. Human Mitochondrial DNA-Protein Complexes Attach to a Cholesterol-Rich Membrane Structure. Sci Rep. 2015;5:15292. doi: 10.1038/srep15292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Takamatsu C, et al. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002;3(5):451–6. doi: 10.1093/embo-reports/kvf099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Carrodeguas JA, Pinz KG, Bogenhagen DF. DNA binding properties of human pol gammaB. J Biol Chem. 2002;277(51):50008–14. doi: 10.1074/jbc.M207030200. [DOI] [PubMed] [Google Scholar]
- 139.Di Re M, et al. The accessory subunit of mitochondrial DNA polymerase gamma determines the DNA content of mitochondrial nucleoids in human cultured cells. Nucleic Acids Res. 2009;37(17):5701–13. doi: 10.1093/nar/gkp614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Milenkovic D, et al. TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum Mol Genet. 2013;22(10):1983–93. doi: 10.1093/hmg/ddt051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kaguni JM. Replication initiation at the Escherichia coli chromosomal origin. Curr Opin Chem Biol. 2011;15(5):606–13. doi: 10.1016/j.cbpa.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28(4):693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 143.Brown TA, et al. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev. 2005;19(20):2466–76. doi: 10.1101/gad.1352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hixson JE, Wong TW, Clayton DA. Both the conserved stem-loop and divergent 5′-flanking sequences are required for initiation at the human mitochondrial origin of light-strand DNA replication. J Biol Chem. 1986;261(5):2384–90. [PubMed] [Google Scholar]
- 145.Wong TW, Clayton DA. In vitro replication of human mitochondrial DNA: accurate initiation at the origin of light-strand synthesis. Cell. 1985;42(3):951–8. doi: 10.1016/0092-8674(85)90291-0. [DOI] [PubMed] [Google Scholar]
- 146.Wanrooij S, et al. In vivo mutagenesis reveals that OriL is essential for mitochondrial DNA replication. EMBO Rep. 2012;13(12):1130–7. doi: 10.1038/embor.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yang MY, et al. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell. 2002;111(4):495–505. doi: 10.1016/s0092-8674(02)01075-9. [DOI] [PubMed] [Google Scholar]
- 148.Holt IJ, Reyes A. Human mitochondrial DNA replication. Cold Spring Harb Perspect Biol. 2012;4(12) doi: 10.1101/cshperspect.a012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Reyes A, et al. Mitochondrial DNA replication proceeds via a ‘bootlace’ mechanism involving the incorporation of processed transcripts. Nucleic Acids Res. 2013;41(11):5837–50. doi: 10.1093/nar/gkt196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Iborra FJ, Kimura H, Cook PR. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004;2:9. doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Antonicka H, et al. The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab. 2013;17(3):386–98. doi: 10.1016/j.cmet.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 152.Yasukawa T, et al. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25(22):5358–71. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Brewer BJ, Fangman WL. Mapping replication origins in yeast chromosomes. Bioessays. 1991;13(7):317–22. doi: 10.1002/bies.950130702. [DOI] [PubMed] [Google Scholar]
- 154.Brewer BJ, Fangman WL. Initiation preference at a yeast origin of replication. Proc Natl Acad Sci U S A. 1994;91(8):3418–22. doi: 10.1073/pnas.91.8.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Martin-Parras L, et al. Unidirectional replication as visualized by two-dimensional agarose gel electrophoresis. J Mol Biol. 1991;220(4):843–53. doi: 10.1016/0022-2836(91)90357-c. [DOI] [PubMed] [Google Scholar]
- 156.Brun C, et al. SARs, ARSs, and A,T-rich regions evidenced by restriction mapping on an 835-kb DNA fragment from Drosophila. Exp Cell Res. 1995;220(2):338–47. doi: 10.1006/excr.1995.1324. [DOI] [PubMed] [Google Scholar]
- 157.Lunyak VV, et al. Developmental changes in the Sciara II/9A initiation zone for DNA replication. Mol Cell Biol. 2002;22(24):8426–37. doi: 10.1128/MCB.22.24.8426-8437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pohjoismaki JL, et al. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J Mol Biol. 2010;397(5):1144–55. doi: 10.1016/j.jmb.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bowmaker M, et al. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J Biol Chem. 2003;278(51):50961–9. doi: 10.1074/jbc.M308028200. [DOI] [PubMed] [Google Scholar]
- 160.Futami K, Shimamoto A, Furuichi Y. Mitochondrial and nuclear localization of human Pif1 helicase. Biol Pharm Bull. 2007;30(9):1685–92. doi: 10.1248/bpb.30.1685. [DOI] [PubMed] [Google Scholar]
- 161.Liu P, et al. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol. 2008;28(16):4975–87. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zheng L, et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell. 2008;32(3):325–36. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Reyes A, et al. Bidirectional replication initiates at sites throughout the mitochondrial genome of birds. J Biol Chem. 2005;280(5):3242–50. doi: 10.1074/jbc.M411916200. [DOI] [PubMed] [Google Scholar]
- 164.Hyvarinen AK, et al. Overexpression of MTERFD1 or MTERFD3 impairs the completion of mitochondrial DNA replication. Mol Biol Rep. 2011;38(2):1321–8. doi: 10.1007/s11033-010-0233-9. [DOI] [PubMed] [Google Scholar]
- 165.Pohjoismaki JL, et al. Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J Biol Chem. 2009;284(32):21446–57. doi: 10.1074/jbc.M109.016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pohjoismaki JL, et al. Developmental and pathological changes in the human cardiac muscle mitochondrial DNA organization, replication and copy number. PLoS One. 2010;5(5):e10426. doi: 10.1371/journal.pone.0010426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ilyina TV, Gorbalenya AE, Koonin EV. Organization and evolution of bacterial and bacteriophage primase-helicase systems. J Mol Evol. 1992;34(4):351–7. doi: 10.1007/BF00160243. [DOI] [PubMed] [Google Scholar]
- 168.Altschul SF, et al. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 169.Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011;7(10):e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tamura K, et al. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Raghunathan S, et al. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proc Natl Acad Sci U S A. 1997;94(13):6652–7. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Raghunathan S, et al. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Biol. 2000;7(8):648–52. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]