ABSTRACT
Within the mammalian prion field, the existence of recombinant prion protein (PrP) conformers with self-replicating (ie. autocatalytic) activity in vitro but little to no infectious activity in vivo challenges a key prediction of the protein-only hypothesis of prion replication – that autocatalytic PrP conformers should be infectious. To understand this dissociation of autocatalysis from infectivity, we recently performed a structural and functional comparison between a highly infectious and non-infectious pair of autocatalytic recombinant PrP conformers derived from the same initial prion strain.1 We identified restricted, C-terminal structural differences between these 2 conformers and provided evidence that these relatively subtle differences prevent the non-infectious conformer from templating the conversion of native PrPC substrates containing a glycosylphosphatidylinositol (GPI) anchor.1 In this article we discuss a model, consistent with these findings, in which recombinant PrP, lacking post-translational modifications and associated folding constraints, is capable of adopting a wide variety of autocatalytic conformations. Only a subset of these recombinant conformers can be adopted by post-translationally modified native PrPC, and this subset represents the recombinant conformers with high specific infectivity. We examine this model's implications for the generation of highly infectious recombinant prions and the protein-only hypothesis of prion replication.
KEYWORDS: mammalian prions, scrapie prion protein, recombinant prions, cofactors, amyloid
A misfolded conformer of the prion protein (PrP) termed PrPSc is an essential component of infectious mammalian prions.2 Through an as yet unknown mechanism, PrPSc interacts with cellular PrP (PrPC) and templates the conversion of PrPC into additional PrPSc. This converting activity of PrPSc is called autocatalysis and is presumed to underlie the infectious nature of the prion diseases.
It is widely believed that autocatalytic PrPSc is the sole component of infectious prions; however, attempts to rigorously demonstrate a protein-only mechanism of prion replication have so far been unsuccessful. Interestingly, in the process of testing the protein-only hypothesis, researchers have generated a variety of autocatalytic recombinant PrP conformers in vitro, but only a select few of these conformers possess levels of specific infectivity in vivo approaching that of native prions (reviewed in3). This dissociation between the autocatalytic and infectious activities of recombinant PrP conformers has challenged a key prediction of the protein-only hypothesis – that autocatalytic PrP conformers should be infectious – and remained as yet without a satisfactory explanation.
To better understand the mechanism for the dissociation between PrP autocatalysis and infectivity we recently performed a structural and functional comparison between 2 autocatalytic recombinant PrP conformers generated from the same initial seed but differing >105-fold in specific infectivity for wild-type mice.1 By comparing this matched pair of autocatalytic recombinant PrP conformers, one fully infectious and the other without detectable infectivity, we sought to identify structural and functional features that correlate with PrPSc infectious activity.
Infectious and non-infectious recombinant PrP conformers differ structurally in restricted C-terminal domains
Using hydrogen/deuterium exchange mass spectrometry (DXMS) we found that our infectious and non-infectious autocatalytic PrP conformers were remarkably similar, as measured by solvent accessibility, throughout their C-terminal protease-resistant cores.1 Both conformers displayed a high degree of protection against solvent exchange, consistent with previous DXMS studies of mammalian prions and with those PrPSc models that propose a complete refolding of the PrP C-terminus to β-sheet secondary structure.1,4-8
Despite the striking structural similarities between the infectious and non-infectious PrP conformers, structural differences were detected in 2 restricted C-terminal domains, including residues 91-115 and 144-163 (mouse PrP numbering).1 Within these domains, the non-infectious conformer appears to have relatively increased solvent accessibility. Using conformation-specific immunoprecipitation and Raman spectroscopy we were able to confirm that structural differences between the infectious and non-infectious conformers reside within these 2 restricted domains.1
The first of these 2 domains, residues 91-115, is located within the unstructured N-terminus of normally folded α-PrP.9 Not much is known about the role of this domain in PrP misfolding, although it contains 3 residues at which amino acid substitutions lead to familial prion disease,10 as well as the so-called 'fifth site' for copper binding, which was recently implicated in the PrP conversion process.11
The second domain within which our infectious and non-infectious PrP conformers structurally differ (residues 144-163) corresponds to the first α-helix and second β-strand of α-PrP.9 There are many existing lines of evidence to suggest that this α1- β2 domain plays a key role in the formation of infectious prions. Several PrPSc-specific antibodies have epitopes that reside within the α1- β2 domain.12,13 Small deletions toward the C-terminus of this domain inhibit PrP conversion and, in fact, yield PrP molecules with dominant negative activity against PrPSc formation.14 Furthermore, the complete absence of the α1- β2 domain in a redacted 'miniprion' does not prevent PrPSc formation, but such miniprions are non-infectious when inoculated into animals expressing full-length PrP.15 Finally, the α1- β2 domain also lies immediately adjacent to the β2-α2 loop, a region with notable influence on prion formation and interspecies prion transmission.16
Infectious and non-infectious autocatalytic recombinant PrP conformers differ in their ability to convert post-translationally modified PrP substrates
To understand the mechanistic basis for the dissociation of autocatalytic and infectious activities in recombinant PrP conformers, we undertook a series of in vitro conversion experiments utilizing our matched pair of infectious and non-infectious autocatalytic conformers to seed differentially modified PrP substrates. Native PrPC is post-translationally modified by the addition of 2 N-linked glycans and a C-terminal glycosylphosphatidylinositol (GPI) anchor.2 We reasoned that perhaps recombinant PrP, free of these bulky modifications, has relatively few conformational constraints and is capable of accessing a greater variety of autocatalytic states than native PrPC. By this logic, only those recombinant PrP conformations that are compatible with the presence of PrPC post-translational modifications would be capable of replication in vivo, and this group of recombinant PrP conformers would represent the potentially infectious subset (Fig. 1). \raster="rgFigKPRN_A_1123843_F0001_B"
FIGURE 1.
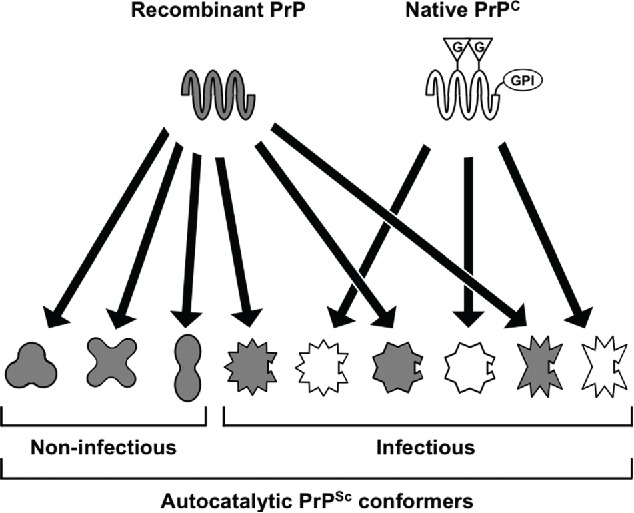
Model to account for the dissociation of autocatalytic activity from infectivity in some recombinant PrPSc conformers. We propose that recombinant PrP, lacking post-translational modifications and associated folding constraints, is capable of adopting a wide variety of autocatalytic conformations. Native PrPC, which is modified by a GPI anchor and up to 2 N-linked glycans, can only adopt a subset of these conformers. Those recombinant PrP conformers that are compatible with a post-translationally modified PrPC substrate are highly infectious. G = N-linked glycan; GPI = GPI anchor. Adapted from Noble et al.1
To test this model, we used our infectious and non-infectious recombinant PrP conformers to seed in vitro conversion reactions containing modified native PrPC substrates (summarized in Table 1). We found that the non-infectious recombinant PrP conformer was unable to template the conversion of native PrPC or deglycosylated PrPC,1 while the infectious conformer was a functional seed in both settings. Unfortunately, the complementary experiment, in which delipidated native PrPC is utilized as a conversion substrate is not possible for technical reasons.17,18 Nonetheless, the results of these conversion experiments are consistent with our proposed model (Fig. 1) and specifically suggest that our non-infectious recombinant PrP conformer fails to function as a seed in vivo because it is unable to template the conversion of GPI anchored PrP substrates.
TABLE 1.
Summary of functional data comparing our infectious and non-infectious recombinant PrP conformers derived from the same initial prion strain.1,29 Delipidated PrPC PMCA was not performed due to previously described technical challenges.17,18 In previous papers, our infectious conformer is referred to as cofactor PrPSc and the non-infectious conformer as protein-only PrPSc.1,29
| Functional Experiment |
|||||
|---|---|---|---|---|---|
| Recombinant PrP PMCA | Delipidated PrPC PMCA | Deglycosylated PrPC PMCA | Native PrPC PMCA | Bioassay | |
| Infectious Conformer | + | n/a | + | + | + |
| Non-infectious Conformer | + | n/a | − | − | − |
Influence of post-translational modifications on protein folding and misfolding
It is well known that post-translational modifications can influence protein folding pathways. Indeed, N-linked glycans are known to have a chaperone-like activity during protein folding and to enhance overall protein conformational stability.19 In addition, covalently linked PrP glycans have been shown to alter the rate of amyloid fibril formation,20 limit the prion strain susceptibility of host animals,21 and even influence interspecies transmission barriers.22
The role of the GPI anchor modification in protein folding is less well studied. In the case of PrP, in vitro removal of the PrP GPI anchor interferes with PrPSc formation.17 In vivo, loss of the PrP GPI anchor appears to have only a modest effect on PrPSc structure,23 but significantly alters its biochemical properties, leading to the production of large, fibrillar aggregates.24 Interestingly, the experimental addition of a GPI anchor to the amyloidogenic yeast protein Sup35p appears to have an analogous biochemical influence, promoting the formation of non-fibrillar aggregates with ultrastructural similarity to PrPSc.25 Recent work has shown that anchorless PrPSc more readily crosses a species barrier.26 Finally, the co-expression of anchorless and wild-type PrPC molecules in recipient animals appears to allow for the detection of infectious activity in PrP amyloid fibril preparations.27
Although our recent experiments and the model outlined above suggest that a key determinant of in vivo activity for recombinant PrP conformers is the ability to accommodate a native PrPC GPI anchor, it should be emphasized that the influence of substrate post-translational modifications may be context dependent. For alternative autocatalytic recombinant PrP conformers, one or both host PrPC glycans may serve as an obstacle to in vivo activity. Furthermore, compatibility between recombinant seed and post-translationally modified substrate may be relative rather than absolute. It is known that non-infectious recombinant PrP amyloids can evolve into infectious prions after repeated in vivo passages, possibly by a deformed templating mechanism (reviewed in28). Our recent results1 would suggest that perhaps an early step during the deformed templating process involves serial alterations to the input recombinant PrP amyloid structure that more and more efficiently accommodate PrPC post-translational modifications.
Implications for the production of fully infectious recombinant prions and the protein-only hypothesis
The production of fully infectious recombinant prions has been a long-standing goal of the prion field, as such material is required for the definitive determination of PrPSc structure by techniques like solid state NMR. To date, highly infectious recombinant PrP preparations, such as the one utilized in our recent study, have only been obtained in conversion systems that include accessory cofactor molecules, specifically phospholipids.29,30 These phospholipids appear to maintain the infectious conformation of PrPSc,29 but the mechanism by which this occurs is not yet known. One possible explanation, based on the model proposed in our recent study (Fig. 1), is that cofactor molecules favor the formation of autocatalytic recombinant PrP conformers with structural features that accommodate native PrPC post-translational modifications. Such a model is consistent with the existence of highly infectious recombinant PrP conformers that do not require a cofactor, so long as native PrPC is able to fold into that particular structure. The identification of such a cofactor-free conformer would represent a definitive proof of the protein-only hypothesis of prion replication.
The existence of autocatalytic recombinant PrP conformers lacking significant levels of specific infectivity has been a persistent obstacle to advancement within many fields of mammalian prion research. Based on our recent work, we propose that the difficulty in producing highly infectious recombinant PrP conformers may be related to the relatively broad conformational space available to recombinant PrP as compared to post-translationally modified native PrPC. We conclude that matching between a recombinant PrP conformer's structure and the ability of natively modified PrPC to adopt that structure governs recombinant PrPSc infectivity.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
Funding
This work was supported by NIH grant RO1 NS046478 to SS.
REFERENCES
- 1.Noble GP, Wang DW, Walsh DJ, Barone JR, Miller MB, Nishina KA, Li S, Supattapone S. A Structural and Functional Comparison Between Infectious and Non-Infectious Autocatalytic Recombinant PrP Conformers. PLoS Pathog 2015; 11:e1005017; PMID:26125623; http://dx.doi.org/ 10.1371/journal.ppat.1005017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prusiner SB. Prions. Proc Natl Acad Sci U S A 1998; 95:13363–83; PMID:9811807; http://dx.doi.org/ 10.1073/pnas.95.23.13363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Supattapone S. Synthesis of High Titer Infectious Prions with Cofactor Molecules. J Biol Chem 2014; 289:19850–4; PMID:24860097; http://dx.doi.org/ 10.1074/jbc.R113.511329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cobb NJ, Sonnichsen FD, McHaourab H, Surewicz WK. Molecular architecture of human prion protein amyloid: a parallel, in-register β-structure. Proc Natl Acad Sci U S A 2007; 104:18946–51; PMID:18025469; http://dx.doi.org/ 10.1073/pnas.0706522104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groveman BR, Dolan MA, Taubner LM, Kraus A, Wickner RB, Caughey B. Parallel In-register Intermolecular β-Sheet Architectures for Prion-seeded Prion Protein (PrP) Amyloids. J Biol Chem 2014; 289:24129–42; PMID:25028516; http://dx.doi.org/ 10.1074/jbc.M114.578344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller MB, Wang DW, Wang F, Noble GP, Ma J, Woods VL Jr., Li S, Supattapone S. Cofactor Molecules Induce Structural Transformation during Infectious Prion Formation. Structure 2013; 21(11):2061–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smirnovas V, Baron GS, Offerdahl DK, Raymond GJ, Caughey B, Surewicz WK. Structural organization of brain-derived mammalian prions examined by hydrogen-deuterium exchange. Nat Struct Mol Biol 2011; 18:504–6; PMID:21441913; http://dx.doi.org/ 10.1038/nsmb.2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Safar JG, Xiao X, Kabir ME, Chen S, Kim C, Haldiman T, Cohen Y, Chen W, Cohen ML, Surewicz WK. Structural determinants of phenotypic diversity and replication rate of human prions. PLoS Pathog 2015; 11:e1004832; PMID:25875953; http://dx.doi.org/ 10.1371/journal.ppat.1004832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wuthrich K. NMR structure of the mouse prion protein domain PrP(121-321). Nature 1996; 382:180–2; PMID:8700211; http://dx.doi.org/ 10.1038/382180a0 [DOI] [PubMed] [Google Scholar]
- 10.Lloyd SE, Mead S, Collinge J. Genetics of prion diseases. Curr Opin Genet Dev 2013; 23:345–51; PMID:23518043; http://dx.doi.org/ 10.1016/j.gde.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giachin G, Mai PT, Tran TH, Salzano G, Benetti F, Migliorati V, Arcovito A, Longa SD, Mancini G, D'Angelo P, et al. The non-octarepeat copper binding site of the prion protein is a key regulator of prion conversion. Sci Rep 2015; 5:15253; PMID:26482532; http://dx.doi.org/ 10.1038/srep15253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korth C, Stierli B, Streit P, Moser M, Schaller O, Fischer R, Schulz-Schaeffer W, Kretzschmar H, Raeber A, Braun U, et al. Prion (PrPSc)-specific epitope defined by a monoclonal antibody. Nature 1997; 390:74–7; PMID:9363892; http://dx.doi.org/ 10.1038/36337 [DOI] [PubMed] [Google Scholar]
- 13.Paramithiotis E, Pinard M, Lawton T, LaBoissiere S, Leathers VL, Zou WQ, Estey LA, Lamontagne J, Lehto MT, Kondejewski LH, et al. A prion protein epitope selective for the pathologically misfolded conformation. Nat Med 2003; 9:893–9; PMID:12778138; http://dx.doi.org/ 10.1038/nm883 [DOI] [PubMed] [Google Scholar]
- 14.Taguchi Y, Mistica AM, Kitamoto T, Schatzl HM. Critical significance of the region between Helix 1 and 2 for efficient dominant-negative inhibition by conversion-incompetent prion protein. PLoS Pathog 2013; 9:e1003466; PMID:23825952; http://dx.doi.org/ 10.1371/journal.ppat.1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Supattapone S, Bosque P, Muramoto T, Wille H, Aagaard C, Peretz D, Nguyen HO, Heinrich C, Torchia M, Safar J, et al. Prion protein of 106 residues creates an artifical transmission barrier for prion replication in transgenic mice. Cell 1999; 96:869–78; PMID:10102274; http://dx.doi.org/ 10.1016/S0092-8674(00)80596-6 [DOI] [PubMed] [Google Scholar]
- 16.Sigurdson CJ, Nilsson KP, Hornemann S, Manco G, Fernandez-Borges N, Schwarz P, Castilla J, Wüthrich K, Aguzzi A. A molecular switch controls interspecies prion disease transmission in mice. J Clin Invest 2010; 120:2590–9; PMID:20551516; http://dx.doi.org/ 10.1172/JCI42051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JI, Surewicz K, Gambetti P, Surewicz WK. The role of glycophosphatidylinositol anchor in the amplification of the scrapie isoform of prion protein in vitro. FEBS Lett 2009; 583:3671–5; PMID:19854187; http://dx.doi.org/ 10.1016/j.febslet.2009.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishina KA, Supattapone S. Immunodetection of glycophosphatidylinositol-anchored proteins following treatment with phospholipase C. Anal Biochem 2007; 363:318–20; PMID:17321480; http://dx.doi.org/ 10.1016/j.ab.2007.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitra N, Sinha S, Ramya TN, Surolia A. N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem Sci 2006; 31:156–63; PMID:16473013; http://dx.doi.org/ 10.1016/j.tibs.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 20.Chen PY, Lin CC, Chang YT, Lin SC, Chan SI. One O-linked sugar can affect the coil-to-β structural transition of the prion peptide. Proc Natl Acad Sci U S A 2002; 99:12633–8; PMID:12235358; http://dx.doi.org/ 10.1073/pnas.192137799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancellotti E, Mahal SP, Somerville R, Diack A, Brown D, Piccardo P, Weissmann C, Manson JC. Post-translational changes to PrP alter transmissible spongiform encephalopathy strain properties. EMBO J 2013; 32:756–69; PMID:23395905; http://dx.doi.org/ 10.1038/emboj.2013.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiseman FK, Cancellotti E, Piccardo P, Iremonger K, Boyle A, Brown D, Ironside JW, Manson JC, Diack AB. The glycosylation status of PrPC is a key factor in determining transmissible spongiform encephalopathy transmission between species. J Virol 2015; 89:4738–47; PMID:25673720; http://dx.doi.org/ 10.1128/JVI.02296-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baron GS, Hughson AG, Raymond GJ, Offerdahl DK, Barton KA, Raymond LD, Dorward DW, Caughey B. Effect of glycans and the glycophosphatidylinositol anchor on strain dependent conformations of scrapie prion protein: improved purifications and infrared spectra. Biochemistry 2011; 50:4479–90; PMID:21539311; http://dx.doi.org/ 10.1021/bi2003907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chesebro B, Race B, Meade-White K, Lacasse R, Race R, Klingeborn M, Striebel J, Dorward D, McGovern G, Jeffrey M. Fatal transmissible amyloid encephalopathy: a new type of prion disease associated with lack of prion protein membrane anchoring. PLoS Pathog 2010; 6:e1000800; PMID:20221436; http://dx.doi.org/ 10.1371/journal.ppat.1000800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall KE, Offerdahl DK, Speare JO, Dorward DW, Hasenkrug A, Carmody AB, Baron GS. Glycosylphosphatidylinositol anchoring directs the assembly of Sup35NM protein into non-fibrillar, membrane-bound aggregates. J Biol Chem 2014; 289:12245–63; PMID:24627481; http://dx.doi.org/ 10.1074/jbc.M114.556639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Race B, Phillips K, Meade-White K, Striebel J, Chesebro B. Increased infectivity of anchorless mouse scrapie prions in transgenic mice overexpressing human prion protein. J Virol 2015; 89:6022–32; PMID:25810548; http://dx.doi.org/ 10.1128/JVI.00362-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raymond GJ, Race B, Hollister JR, Offerdahl DK, Moore RA, Kodali R, Raymond LD, Hughson AG, Rosenke R, Long D, et al. Isolation of novel synthetic prion strains by amplification in transgenic mice coexpressing wild-type and anchorless prion proteins. J Virol 2012; 86:11763–78; PMID:22915801; http://dx.doi.org/ 10.1128/JVI.01353-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makarava N, Baskakov IV. The evolution of transmissible prions: the role of deformed templating. PLoS Pathog 2013; 9:e1003759; PMID:24339773; http://dx.doi.org/ 10.1371/journal.ppat.1003759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deleault NR, Walsh DJ, Piro JR, Wang F, Wang X, Ma J, Rees JR, Supattapone S. Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc Natl Acad Sci U S A 2012; 109:E1938–E46; PMID:22711839; http://dx.doi.org/ 10.1073/pnas.1206999109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F, Wang X, Yuan CG, Ma J. Generating a Prion with Bacterially Expressed Recombinant Prion Protein. Science 2010; 327:1132–5; PMID:20110469; http://dx.doi.org/ 10.1126/science.1183748 [DOI] [PMC free article] [PubMed] [Google Scholar]


