ABSTRACT
Prion diseases involve the conversion of the endogenous cellular prion protein, PrPC, into a misfolded infectious isoform, PrPSc. Several functions have been attributed to PrPC, and its role has also been investigated in the olfactory system. PrPC is expressed in both the olfactory bulb (OB) and olfactory epithelium (OE) and the nasal cavity is an important route of transmission of diseases caused by prions. Moreover, Prnp−/− mice showed impaired behavior in olfactory tests. Given the high PrPC expression in OE and its putative role in olfaction, we screened a mouse OE cDNA library to identify novel PrPC-binding partners. Ten different putative PrPC ligands were identified, which were involved in functions such as cellular proliferation and apoptosis, cytoskeleton and vesicle transport, ubiquitination of proteins, stress response, and other physiological processes. In vitro binding assays confirmed the interaction of PrPC with STIP1 homology and U-Box containing protein 1 (Stub1) and are reported here for the first time. Stub1 is a co-chaperone with ubiquitin E3-ligase activity, which is associated with neurodegenerative diseases characterized by protein misfolding and aggregation. Physiological and pathological implications of PrPC-Stub1 interaction are under investigation. The PrPC-binding proteins identified here are not exclusive to the OE, suggesting that these interactions may occur in other tissues and play general biological roles. These data corroborate the proposal that PrPC is part of a multiprotein complex that modulates several cellular functions and provide a platform for further studies on the physiological and pathological roles of prion protein.
Keywords: prion, yeast two-hybrid, olfactory epithelium, Stub1, protein interaction, CHIP
Abbreviations
- PrP
prion protein
- CHIP
C-terminus of Hsc70-interacting protein
- OB
olfactory bulb
- OE
olfactory epithelium
- OSN
olfactory sensory neurons
- Stub1
STIP1 homology and U-Box containing protein 1
- TSE
transmissible spongiform encephalopathies
- STI1
stress-inducible protein 1
Introduction
Transmissible spongiform encephalopathies (TSEs), also known as prion diseases, are fatal neurodegenerative disorders that include Creutzfeldt-Jakob disease (CJD) in humans, scrapie in sheep, and bovine spongiform encephalopathy (BSE) in cattle. These diseases involve the conversion of the endogenous cellular prion protein, PrPC, into a misfolded and infectious isoform named PrPSc.1,2
PrPC is predominantly an extracellular glicosylphosphatidilinositol (GPI)-anchored glycoprotein that is expressed in most tissues and is enriched in the central nervous system.3 Despite intensive research and the fact that PrPC is conserved across different species, a consensus about its physiological function has not yet been reached.2,4 Several functions have been attributed to PrPC, including stress and behavior, sleep-wake cycle, memory, neuritogenesis, neuroprotection, and cellular adhesion.2 Many of these roles were elucidated with the discovery of PrP-binding partners using different biophysical assays such as conventional yeast two-hybrid screening, co-immunoprecipitation, and other methods. These findings suggest that PrPC may serve as a scaffolding protein for the assembly of signaling modules and therefore it must be related to a variety of cellular processes, rather than having a specific function.2,5
The physiological role of PrPC has also been investigated in the olfactory system. PrPC is expressed in different areas of the olfactory system including the olfactory epithelium (OE) and the olfactory bulb (OB), and is localized in the axons of both olfactory sensory neurons (OSNs) and mitral cells.6 Interestingly, Prnp−/− mice showed impaired behavior in olfactory tests and have altered electrophysiological activity in the dendrodendritic synapse in the olfactory bulb. Both the behavioral and electrophysiological deficits observed in Prnp−/− mice were rescued by transgenic neuronal-specific expression of PrPC.7 It was reported that the OE from Prnp−/− mice showed no alterations in the odorant-evoked electro-olfactogram, suggesting that the PrPC plays an olfactory function in the OB, rather than in the OE.7 However, PrPC expression in olfactory sensory neurons, both in dendrites and cell bodies (in a lower abundance) and in axons (more abundantly) is evident6,8 and suggests that this protein should play a physiological role in these sensory cells. Moreover, the OE has been previously demonstrated to be a site of prion infection in humans9 and animals.10
We performed a yeast two-hybrid screen to address the physiological roles of PrPC using PrPC as bait. Given the high PrPC expression in OE and its putative role in olfaction, we screened a mouse OE cDNA library, in attempt to identify novel PrPC-binding partners. Ten different putative PrPC-binding proteins were identified to provide insights into the physiological and pathological functions of PrP in OE and other tissues. The PrPC interaction with STIP1 homology and U-Box containing protein 1 (Stub1) was also confirmed by in vitro binding assays and are reported here for the first time. Stub1 functions both as a molecular co-chaperone, and as an ubiquitin E3 ligase protein.11 Moreover, Stub1 is associated with several neurodegenerative diseases characterized by protein misfolding and aggregation.12 Hence, the interaction between PrPC and Stub1, identified in our study, may occur in vivo and modulate PrPC stability, which is implicated in the PrPSc conversion.
Results
Identification of PrPC-Interacting Proteins by Yeast Two-Hybrid Screen
We conducted a yeast two-hybrid screening of an OE cDNA library to identify new interactors for the cellular prion protein using PrPC as bait. Fifty-three clones were isolated for both LEU2 and Lac-z reporter genes. These clones were sequenced and their identities are listed in Table 1. Eighteen out of the total clones had their insert sequence in the correct frame. Most of the putative ligands were found in one single positive clone, except for a few that were found in 2 (Srpk2, Pcsk5, Stub1) or 4 (Dynlt1b) different clones.
TABLE 1.
Gene identities obtained from yeast two-hybrid screening
| Gene description | Symbol | Accession# | Gene ID | # of clones obtained |
|---|---|---|---|---|
| Dynein light chain Tctex type 1 D | Dynlt1b | NM_009342.2 | 42476289 | 4 |
| NEDD4 binding protein1 | N4bp1 | NM_030563.2 | 242117940 | 1 |
| Catenin (cadherin associated protein) beta 1 | Ctnnb1 | NM_001165902.1 | 260166641 | 1 |
| Collagen type3 alpha 1 | Col3a1 | NM_009930.2 | 226423932 | 1 |
| Collagen type1 alpha 2 | Col1a2 | NM_007743.2 | 111120328 | 1 |
| Ribosomal protein AS | Rpsa | NM_011029.4 | 224994259 | 1 |
| Charged multivesicular body protein 2A | Chmp2a | NM_026885.3 | 254826731 | 1 |
| Vimentin | Vim | NM_011701.4 | 227430362 | 1 |
| Heterogeneous nuclear ribonucleoprotein K | Hnrnpk | NM_025279.2 | 142350515 | 1 |
| Serine/Arginine-rich protein specific kinase 2 | Srpk2 | NM_009274.2 | 47059479 | 2 |
| DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3 x-linked | Ddx3x | NM_010028.3 | 164607180 | 1 |
| Proprotein convertase subtilisin/kexin type 5 | Pcsk5 | NM_001163144.1 | 253314508 | 2 |
| Transforming growth factor beta regulated gene 1 | Tgfb1 | NM_025289.3 | 224967131 | 1 |
| Glutathione S transferase | Gstm1 | NM_010358.5 | 239937552 | 1 |
| Pyruvate kinase | Pkm2 | NM_001253883.1 | 359807366 | 1 |
| Basic transcription factor 3 | Btf3 | NM_001170540.1 | 281485610 | 1 |
| STIP1 homology and U-Box containing protein 1 | Stub1 | NM_019719.3 | 118130581 | 2 |
| S100 calcium binding protein A5 | S100a5 | NM_011312.2 | 113930759 | 1 |
| Not in frame* | — | — | — | 35 |
Inserts sequences were not in frame with the B42 transcription activation domain in the library vector.
In order to confirm these interactions, we performed cross-mating assays. As shown in Figure 1, 10 of the interactions were validated, i.e., the reporter gene (β-galactosidase) was only expressed when diploid yeasts contained both the target and bait vectors.
Figure 1.
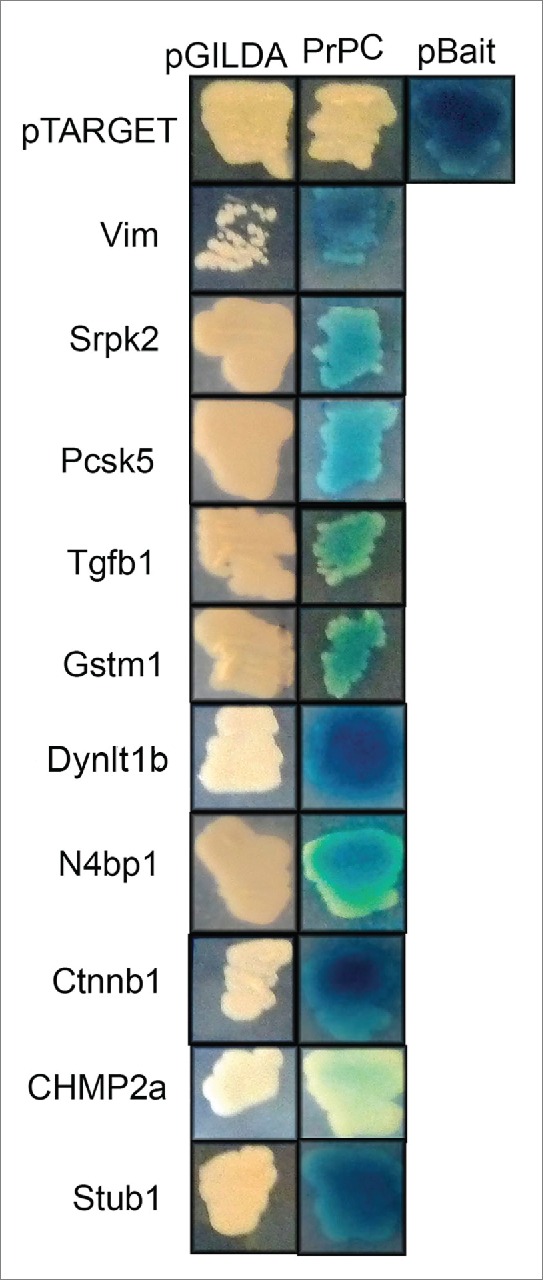
PrPC interacts with 10 different ligands in cross-mating assay. Bait strain expressing PrPC and the Lac-Z reporter gene were mated with target strains expressing the putative PrPC interactors. X-gal was used to score positive interactions (blue color development). pBait and pTarget were used as a positive control for interaction, since they express known interaction partners. The empty bait vector (pGilda), which expresses only Lex A domain, was used as negative control.
Additional information about these 10 PrPC-binding proteins is given in Table 2. Several inserts contain most of the coding region sequence, but only the clones corresponding to Dynlt1b have the entire coding sequence.
TABLE 2.
Ten putative PrPC ligands
| Name/Symbol | Protein region contained in the target clonea | Function |
|---|---|---|
| Vimentin (Vim) |  |
Class-III intermediate filaments found in various non-epithelial cells |
| Serine/arginine-rich protein specific kinase 2 (Srpk2) |  |
Protein involved in the phosphorylation of SR-splicing factors and the regulation of splicing. Promotes neuronal apoptosis by upregulating cyclin-D1 (CCND1) expression |
| Proprotein convertase subtilisin/kexin type 5 (Pcsk5) |  |
Likely to represent a widespread endoprotease activity within the constitutive and regulated secretory pathway |
| Transforming growth factor beta-1 (Tgfb1) |  |
Multifunctional protein that controls proliferation, differentiation, and other functions in many cell types. Many cells synthesize TGFB1 and have specific receptors for it |
| Glutathione S-Transferase Mu 1 (Gstm1) |  |
Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles |
| Dynein light chain Tctex type 1 D 1 (Dynlt1b) |  |
Acts as one of several noncatalytic accessory components of the cytoplasmic dynein 1 complex that are thought to be involved in linking dynein to cargos and to adapter proteins that regulate dynein function. Cytoplasmic dynein 1 acts as a motor for the intracellular retrograde motility of vesicles and organelles along microtubules |
| NEDD4 binding protein 1 (N4bp1) |  |
Inhibitor of the E3 ubiquitin-protein ligase ITCH |
| Catenin (cadherin-associated protein) beta 1 (Ctnnb1) |  |
Downstream component of the canonical Wnt signaling pathway. Component of an E-cadherin/catenin adhesion complex located to adherens junctions |
| Charged multivesicular body protein 2a (Chmp2a) |  |
Core component of the endosomal sorting required for transport complex III (ESCRT-III) which is involved in multivesicular bodies (MVBs) formation and sorting of endosomal cargo proteins into MVBs |
| STIP1 homology and U-Box containing protein 1 (Stub1) |  |
E3 ubiquitin-protein ligase which targets misfolded chaperone substrates toward proteasomal degradation |
Gray color indicates the region found in the target clone.
The identified PrPC ligands are involved in many functions such as cellular proliferation and apoptosis, cytoskeleton and vesicle transport, ubiquitination of proteins, stress response, and other physiological processes (Table 2). Interestingly, these putative PrPC-binding proteins are not exclusive to the OE, suggesting that these interactions may also occur in other tissues and play general biological functions.
Validation of PrPC Interaction with Stub1 Using In Vitro Binding Assays
Out of the 10 ligands, the following 4 showed strong interaction (based on the intensity of blue color) with PrPC: vimentin (Vim), dynein light chain Tctex-type 1B (Dynlt1b), catenin beta 1 (Ctnnb1), and STIP1 homology and U-Box containing protein 1 (Stub1) (Fig. 1). Since Stub1 is homologous to stress-inducible protein 1 (STI1), a well-known PrPC ligand,13,14 we decided to further investigate the putative interaction between Stub1 and PrPC. Western blotting assays were performed using protein extracts prepared from these mouse tissues to examine protein expression of Stub1 in the OE and the brain. As shown in Figure 2, Stub1 is expressed both in the OE and the brain with similar expression levels.
Figure 2.
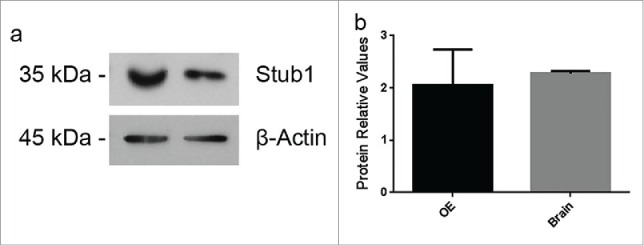
Stub1 is expressed in both olfactory epithelium (OE) and brain. (a) Protein extracts prepared from mouse brain and OE were analyzed by western blotting with anti-Stub1 (upper panel) antibodies. Anti-β-actin was used as loading control (lower panel). (b) Protein relative values were quantified using ImageJ Software® and normalized to β-actin. The results are representative of 4 independent experiments. Statistical analysis was conducted with GraphPad Prism (values were expressed as mean ±SE. Student's t test was used for statistical analysis and P<0.05 was considered significant).
We conducted pull-down assays to further analyze the interactions of PrPC with Stub1. Recombinant His6-PrPC was incubated with protein extract prepared from mouse brain, as a source of endogenous Stub1. Protein complexes were then pulled down using Ni-NTA-agarose beads. Western blotting analysis showed that recombinant PrPC was recovered from the resin, attested by the detection of a 25 kDa band when probed with anti-His tag (Fig. 3a). Stub1 were pulled down together with recombinant PrPC. No reaction was observed when the extract was incubated with Ni-NTA-agarose beads alone.
Figure 3.
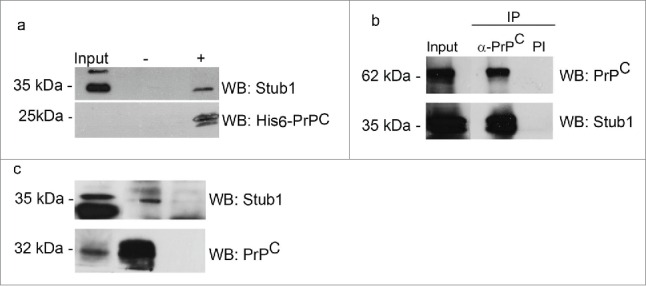
PrPC interacts with Stub1. (a) A pull-down assay was performed using mouse brain extract incubated with His6-PrPC bound to Ni-NTA-agarose beads (+) or Ni-NTA-agarose alone (−). Pulled-down proteins were analyzed by western blotting (WB) employing anti-Stub1 antibody (upper panel). The same membranes subjected to pull-down assay were re-probed with anti-his-tag antibody to attest that recombinant PrPC was recovered from the beads (lower panel). HEK293T cells protein extracts overexpressing GFP-PrPC and Myc-Stub1 (b) or olfactory epithelium (OE) extracts (c) were immunoprecipitated with anti-PrPC or pre-immune serum (PI). Co-precipitated proteins were analyzed using an anti-Stub1 antibody (upper panels). The same membranes were re-probed with anti-PrPC (lower panels) to confirm that PrPC was precipitated during IP-reaction.
Co-immunoprecipitation assays were also performed to confirm the pull-down results. Protein extracts of HEK293T cells were co-transfected with pEGFP-PrPC and pcDNA3-Myc-Stub1 expression vectors were incubated with anti-PrPC antibody, followed by immunoprecipitation (IP). The blotting reaction of the immunoprecipitated material with anti-Stub1 antibody revealed 2 bands of 35–40 kDa, corresponding to the endogenous Stub1 (lower band) and the myc-tagged Stub1 (higher band) (Fig. 3b). No bands were observed when pre-immune serum was employed in the immunoprecipitation reaction (Fig. 3b, PI). The same membrane was re-probed with anti-PrPC antibody to confirm that GFP-PrPC was precipitated during the IP-reaction.
To determine whether endogenous PrPC could bind endogenous Stub1, similar co-immunoprecipitation experiments were carried out using mouse OE extract. When PrPC was immunoprecipitated from OE lysates and subjected to western blot analysis, a specific band corresponding to Stub1 was detected (Fig. 3c). This band is slightly higher than in the input and this finding was verified in other co-immunoprecipitation assays, with different source of tissue samples. A weak signal of Stub1 was also obtained when beads coupled with pre-immune serum were used (negative control), suggesting residual nonspecific binding. Co-immunoprecipitation assays with anti-Stub1 were also done and produced similar results as shown in Figure 3c, confirming the specific interaction between endogenous PrPC and Stub1 (data not shown).
Discussion
This study aimed to identify novel PrPC binding proteins using two-hybrid screening of an OE cDNA library. A group of 10 proteins that potentially interact with PrPC was identified, which provided clues about the enigmatic function of PrPC. As PrPC is a GPI-anchored protein, localized predominantly on the extracellular side of the plasma membrane, the relevance of yeast two-hybrid system to analyze interactions of PrPC could be questioned. However, it is known that a subset of PrPC is present in the cytosol, designated as cytosolic PrPC (cytPrPC).15 There are also 2 PrPC transmembrane forms that are partially exposed to the cytosol: CtmPrP with the N-terminus in the cytosol and NtmPrP with the C-terminus in the cytosol.16 Therefore, interaction of PrPC with cytosolic proteins is possible and the identification of these ligands should help in understanding its physiological role(s).
Indeed, most of the putative PrPC ligands identified in the present study are cytoplasmic proteins, which belong to different protein classes and are associated to distinct cellular mechanisms. These potential interactions are consistent with the proposal that PrPC is part of a multiprotein complex that modulates several cellular functions.2 A number of other proteins have also been identified that could interact with PrP in the cytosol. Some of them mediate neuroprotection, including the anti-apoptotic protein Bcl-217 and the neurotrophin receptor-interacting MAGE homologue, NRAGE.18 Restelli and colleagues found that PrP molecules that escaped translocation into the ER were associated with an increase in the resistance of cortical and hippocampal cells to apoptosis, suggesting that this form of cytPrP has neuroprotective function.19,20 The putative interactions obtained that could modulate the neuroprotective role of cytPrP should be investigated.
Several studies indicated that cytPrP can acquire neurotoxic potential. Cytotoxic effects of cytPrP were observed in several mammalian cell lines and animal models.16 Thus, the interactions found in the present study may have some physiological role in the cytosol or they may participate in the induction of PrPSc and are potential targets for the treatment of TSEs.
Previous studies also used the yeast two-hybrid methodology to screen for PrPC ligands; however the interactions found in these studies do not completely overlap with our findings.17,22-24 A number of possible explanations could account for the different results. First, most of the screens were not exhaustive and may not have reached saturation. Second, it is well known that a significant number of false-positive interactions can be obtained in the yeast two-hybrid method. Moreover, variations in the details of the protocol, such as the vectors used, the nature of the reconstituted transcription factor, and the nature of the libraries screened, have a great impact on the interactions that can be retrieved.25 This is the first screening for PrPC-binding proteins that used a cDNA library from OE in the yeast two-hybrid assay. This approach provided the identification of new PrPC interactions, although they were not limited to the OE.
In addition to yeast two-hybrid experiments, the interaction between PrPC and Stub1 was also confirmed by pull-down and co-immunoprecipitation assays.
Stub1, also known as CHIP (C-terminus of Hsc70-interacting protein), is a 35 kDa cytoplasmatic protein that functions both as a molecular co-chaperone and as an ubiquitin E3 ligase.11 The N-terminal region of Stub1/CHIP has 3 tetratricopeptide repeat (TPR) domains responsible for protein-protein interactions with Hsps 70/90 and other molecular chaperones. The C-terminus contains a U-box, which is the site of its ubiquitin E3 ligase activity.26 In cooperation with heat-shock chaperone proteins, including Hsc70, Hsp70, and Hsp90, Stub1/CHIP plays a crucial role in recognizing and modulating the degradation of numerous chaperone-client proteins.27
Stub1/CHIP is associated with several neurodegenerative disorders characterized by protein misfolding and aggregation, such as Alzheimer and Parkinson diseases.12 Since prion diseases are also characterized by protein misfolding and aggregations, it would be logical to expect that PrPC is a substrate for Stub1/CHIP. It is possible that the interaction identified between PrPC and Stub1/CHIP can occur in vivo and modulate PrPC stability, which is implicated in the PrPSc conversion.
Interestingly, Stub1/CHIP is homologous to stress-inducible protein 1 (STI1) or Hsp70/Hsp90-organizing protein (Hop), a well-known PrPC ligand.13,14 Like Stub1/CHIP, STI1/Hop is also a co-chaperone and contains TPR domains that interact with the C-terminal sequence of Hsp70 and Hsp90. Its interaction with PrPC at the cell surface is implicated in neuritogenesis and neuroprotection.2,13,14 Some works have demonstrated that both STI1/Hop and Stub1/CHIP are involved together, modulating the balance between folding and degradation for Hsp70/90 client proteins.28,29 It is possible that a similar mechanism is involved in PrPC quality control, since PrPC binds both STI1/Hop and Stub1/CHIP. This regulation of PrPC by STI1/Hop and Stub1/CHIP is currently under investigation in our group.
To date, there is no evidence in the literature showing that PrPC would be a substrate for Stub1/CHIP. This possibility was suggested in a study by Zhang and colleagues30, who reported a specific interaction between Hsp70 and cytosolic PrP and that mutant PrPs are the targets of Hsp70 in the cytosol. They also found that overexpression or activation of Hsp70 in cultured cells selectively mediated the degradation of cytosolic PrPs, contributing to the protective effect against cytotoxicity. They also suggested that the conversion of Hsp70's function in protein folding to its function as a degradation factor could be mediated by the co-chaperone Stub1/CHIP.30
Since Stub1 is a cytoplasmic protein and PrPC is located in the extracellular side of cell membrane, the sub-localization of the identified interactions awaits further investigation. The presence of PrPC in the cytosol might be a normal feature of PrPC metabolism, either by retro-translocation or by direct transfer from the ribosomes without entering the endoplasmic reticulum (ER).31 As mentioned above, some studies indicated that the cytoplasmic PrPC has neurotoxic effects, but other findings suggested a neuroprotective role for these PrP molecules. Hence, interaction of PrPC with Stub1 may occur in the cytosol and may be involved in neuroprotective or neurotoxic effects.
The characterization of novel PrPC ligands should contribute to the understanding of both the physiological and pathological roles played by PrPC. The functional implications of PrPC interaction with Stub1 in olfaction and other biological processes should be further investigated.
Materials and Methods
Antibodies and Plasmids
The following antibodies were used in this study: rabbit monoclonal anti-Stub1 (ABCAM, #134064) and mouse monoclonal anti- β-actin (Sigma Aldrich, #A5441). Polyclonal anti-PrPC antibodies were raised in PrPC-null mice (a kind gift from Dr. Marilene H. Lopes, University of Sao Paulo); the specificity of this antiserum has already been described.13,32
Plasmids used for the cells transfections were as follows: pCDNA 3.1-Myc-Stub1, which harbored a full-length mouse Stub1 cDNA insert; pEGFP-PrPC, which has been previously described33, was kindly provided by Dr. Vilma Regina Martins (International Research Center, A.C. Camargo Hospital, São Paulo, Brazil). The yeast two-hybrid vectors are described below.
Production of the Yeast Bait Strain
The sequence encoding the mature mouse PrPC (residues 23–231) was amplified by PCR using the recombinant plasmid pEGFP-PrPC as template and subcloned into the EcoRI/BamHI restrictions sites of the yeast bait expression vector pGILDA (Clontech, USA). The construct was then transformed into the yeast strain RFY206 together with pSH18-34, a Lac-Z reporter vector.
Yeast Two-Hybrid Screening
The pre-transformed OE cDNA library has already been described by Von Dannecker and colleagues and Kerr and colleagues in their studies.34,35 Yeast two-hybrid screening was carried out using a DupLex-A Yeast Two-Hybrid System (OriGene Technologies, Rockville, MD) and was performed as previously described using a mating assay.34 Cross-mating test was conducted as described by Von Dannecker et al.34
Detection of Stub1 in Mouse Brain and OE Protein Homogenates
Mouse brain and OE tissues were removed and lysed in ice-cold lysis buffer (50 mM Tris-HCl pH 7.4, 1% NP-40, 0.2% sodium deoxycholate, 1 mM PMSF and 1 mM NEM). The extracts were centrifuged at 19,000 x g for 30 min at 4°C and the supernatants were saved. All animal procedures undertaken in this study were approved by the Animal Care and Use Committee (Federal University of Paraná) and are in accordance to the ethical guidelines established by CONCEA (National Council of Animal Experiment Control, Brazil).
Protein homogenates (100 μg) were separated in SDS-PAGE, and analyzed by western blotting using antibodies anti-Stub1 (1:2,000) and anti-β actin (1:3,000). The reaction was developed using a chemiluminescent substrate (WestPico, Pierce Co.).
Quantification of band intensity was performed using the Image J Software (NIH, Bethesda, MD, USA). The signal intensity of each protein was normalized with the corresponding β-actin signal. Experiments were conducted at least 3 times and values were expressed as mean ± standard error. Student's t test was used for statistical analysis and P < 0.05 was considered significant.
Pull-Down Assays
Mouse brain homogenate diluted in buffer A (10 mM Tris, pH 7.4; 100 mM NaH2PO4; 25 mM imidazole; 1% NP-40) was pre-cleared twice in 30 μl of Ni-NTA-agarose. The resulting supernatant (input; 250 μg) was incubated with 45 μg of mouse recombinant His6-PrPC (kindly provided by Dr. Vilma Regina Martins, International Research Center, A.C. Camargo Hospital, São Paulo, Brazil) for 2 h, at 4°C. The same amount of input (without recombinant protein) was used as a negative control. After that, 30 μl of Ni-NTA-agarose was added to each sample and incubated for 1 h, at room temperature. The resins were washed 7 times in buffer A (with 50 mM imidazole) and the last wash was in buffer A with 0.01% SDS. Pulled-down material was eluted with Laemmli sample buffer, resolved by a 12% SDS-PAGE, and analyzed by western blotting using the antibody anti-Stub1, as described above, and with monoclonal anti-penta His antibodies (Invitrogen, Carlsbad, CA).
Immunoprecipitation Assays
HEK 293T cells were co-transfected as previously described32 with pEGFP-PrP and pcDNA3.1-Myc-Stub1. After 48 h, cells were lysed in 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM CaCl2, 1 mM de MgCl2, 1% NP40, and 0.2% sodium deoxycholate plus complete protease inhibitor cocktail and centrifuged at 20,000 × g, for 15 min at 4°C. Protein extracts (800 μg of protein, 0.4 μg/μL) were pre-cleared using protein A/G-Sepharose (Sigma-Aldrich), for 3 h, at 4°C. The pre-cleared extract was aliquoted into 2 samples containing 400 μg of protein each. Both samples were incubated for 4 h at 4°C under gentle agitation; one of them was added with 30 μl of protein A/G-Sepharose cross-linked to anti-PrPC polyclonal antibodies, the other one (negative control) was added with an equal volume of protein A/G-Sepharose cross-linked to pre-immune mice serum. Beads were washed 5 times in lysis buffer, and bound proteins were eluted with 1x Laemmli buffer at 100°C. Recovered proteins were separated using 10% SDS-PAGE followed by western blot analysis using anti-PrPC (1:1,000) or anti-Stub1 (1:2,000).
The same assay described above was performed using protein lysates from mouse OE (500 μg, 0.5 μg/μL).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Evelyn Castillo Lima, Fernanda Miwa Yamasaka, and Bruno Nakayabu (UFPR) for technical assistance.
Funding
This investigation was supported by grants from the National Council for Scientific and Technological Development (CNPq, Brazil) (476849/2011-0) and Coordination for the Improvement of Higher Education Personnel (CAPES, Brazil) fellowships to APLG, LMLR, BCBB, and CFJ. MCA was supported by a CNPq-PiBIC fellowship.
REFERENCES
- 1.Prusiner SB. Prions. Proc Natl Acad Sci U S A 1998; 95:13363-83; PMID:9811807; http://dx.doi.org/ 10.1073/pnas.95.23.13363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linden R, Martins VR, Prado MAM, Cammarota M, Izquierdo I, Brentani RR. Physiology of the prion protein. Physiol Rev 2008; 88:673-728; PMID:18391177; http://dx.doi.org/ 10.1152/physrev.00007.2007 [DOI] [PubMed] [Google Scholar]
- 3.Stahl N, Prusiner SB. Prions and prion proteins. FASEB J 1991; 5:2799-807; PMID:1916104 [DOI] [PubMed] [Google Scholar]
- 4.Martins VR, Beraldo FH, Hajj GN, Lopes MH, Lee KS, Prado MM, Linden R. Prion protein: orchestrating neurotrophic activities. Curr Issues Mol Biol 2010; 12:63-86; PMID:19767651 [PubMed] [Google Scholar]
- 5.Linden R, Mariante RM, Nóbrega A, Martins RA, Areal RB, Bellio M. Neuroimmunoendocrine regulation of the prion protein in neutrophils. J Biol Chem 2012; 287:35506-15; PMID:22910907; http://dx.doi.org/ 10.1074/jbc.M112.394924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Pichon CE, Firestein S. Expression and localization of the prion protein PrP(C) in the olfactory system of the mouse. J Comp Neurol 2008; 508:487-99; PMID:18338400; http://dx.doi.org/ 10.1002/cne.21698 [DOI] [PubMed] [Google Scholar]
- 7.Le Pichon CE, Valley MT, Polymenidou M, Chesler AT, Sagdullaev BT, Aguzzi A, Firestein S. Olfactory behavior and physiology are disrupted in prion protein knockout mice. Nat Neurosci 2009; 12:60-9; PMID:19098904; http://dx.doi.org/ 10.1038/nn.2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dibattista M, Massimino ML, Maurya DK, Menini A, Bertoli A, Sorgato MC. The cellular prion protein is expressed in olfactory sensory neurons of adult mice but does not affect the early events of the olfactory transduction pathway. Chem Senses 2011; 36:791-7; PMID:21680753; http://dx.doi.org/ 10.1093/chemse/bjr054 [DOI] [PubMed] [Google Scholar]
- 9.Orrú CD, Bongianni M, Tonoli G, Ferrari S, Hughson AG, Groveman BR, Fiorini M, Pocchiari M, Monaco S, Caughey B, et al.. A test for Creutzfeldt-Jakob disease using nasal brushings. N Engl J Med 2014; 371:519-29; PMID:25099576; http://dx.doi.org/ 10.1056/NEJMoa1315200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bessen RA, Shearin H, Martinka S, Boharski R, Lowe D, Wilham JM, Caughey B, Wiley JA. Prion shedding from olfactory neurons into nasal secretions. PLoS Pathog 2010; 6:e1000837; PMID:20419120; http://dx.doi.org/ 10.1371/journal.ppat.1000837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol 1999; 19:4535-45; PMID:10330192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickey CA, Patterson C, Dickson D, Petrucelli L. Brain CHIP: removing the culprits in neurodegenerative diseases. Trend Mol Med 2006; 13:32-8; PMID:17127096; http://dx.doi.org/ 10.1016/j.molmed.2006.11.003 [DOI] [PubMed] [Google Scholar]
- 13.Zanata SM, Lopes MH, Mercadante AF, Hajj GNM, Chiarini LB, Nomizo R, Freitas ARO, Cabral ALB, Lee KS, Juliano MA, et al.. Stress-inducible protein 1 is a cell surface ligand for cellular prion that triggers neuroprotection. EMBO J 2002; 21:3307-16; PMID:12093732; http://dx.doi.org/ 10.1093/emboj/cdf325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiarini LB, Freitas AR, Zanata SM, Brentani RR, Martins VR, Linden R. Cellular prion protein transduces neuroprotective signals. EMBO J 2002; 21:3317-26; PMID:12093733; http://dx.doi.org/ 10.1093/emboj/cdf324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mironov A Jr, Latawiec D, Wille H, Bouzamondo-Bernstein E, Legname G, Williamson RA, Burton D, DeArmond SJ, Prusiner SB, Peters PJ. Cytosolic prion protein in neurons. J Neurosci 2003; 23:7183-93; PMID:12904479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hegde RS, Mastrianni JA, Scott MR, DeFea KA, Tremblay P, Torchia M, DeArmond SJ, Prusiner SB, Lingappa VR. A transmembrane form of the prion protein in neurodegenerative disease. Science 1998; 279:827-34; PMID:9452375; http://dx.doi.org/ 10.1126/science.279.5352.827 [DOI] [PubMed] [Google Scholar]
- 17.Kurschner C, Morgan JI. The cellular prion protein (PrP) selectively binds to Bcl-2 in the yeast two-hybrid system. Brain Res Mol Brain Res 1995; 30:165-8; PMID:7609638; http://dx.doi.org/ 10.1016/0169-328X(95)00013-I [DOI] [PubMed] [Google Scholar]
- 18.Bragason BT, Palsdottir A. Interaction of PrP with NRAGE, a protein involved in neuronal apoptosis. Mol Cell Neurosci 2005; 29:232-44; PMID:15911347; http://dx.doi.org/ 10.1016/j.mcn.2005.02.013 [DOI] [PubMed] [Google Scholar]
- 19.Restelli E, Fioriti L, Mantovani S, Airaghi S, Forloni G, Chiesa R. Cell type-specific neuroprotective activity of untranslocated prion protein. PLoS One 2010; 5:e13725; PMID:21060848; http://dx.doi.org/ 10.1371/journal.pone.0013725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roucou X, Guo Q, Zhang Y, Goodyer CG, LeBlanc AC. Cytosolic prion protein is not toxic and protects against Bax-mediated cell death in human primary neurons. J Biol Chem 2003; 278:40877-81; PMID:12917444; http://dx.doi.org/ 10.1074/jbc.M306177200 [DOI] [PubMed] [Google Scholar]
- 21.Miesbauer M, Rambold AS, Winklhofer KF, Tatzelt J. Targeting of the prion protein to the cytosol: mechanisms and consequences. Curr Issues Mol Biol 2010; 12:109-18; PMID:19767654 [PubMed] [Google Scholar]
- 22.Spielhaupter C, Schätzl HM. PrPC directly interacts with proteins involved in signaling pathways. J Biol Chem 2001; 276:44604-12; PMID:11571277; http://dx.doi.org/ 10.1074/jbc.M103289200 [DOI] [PubMed] [Google Scholar]
- 23.Xu F, Karnaukhova E, Vostal JG. Human cellular prion protein interacts directly with clusterin protein. Biochim Biophys Acta 2008; 1782:615-20; PMID:18786636; http://dx.doi.org/ 10.1016/j.bbadis.2008.08.004 [DOI] [PubMed] [Google Scholar]
- 24.Huang T, Xu J, Xiang J, Lu Y, Chen R, Huang L, Xiao G, Sun G. PrPC interacts with potassium channel tetramerization domain containing 1 (KCTD1) protein through the PrP (51-136) region containing octapeptide repeats. Biochem Biophys Res Commun 2012; 417:182-6; PMID:22138399; http://dx.doi.org/ 10.1016/j.bbrc.2011.11.081 [DOI] [PubMed] [Google Scholar]
- 25.Koegl M, Uetz P. Improving yeast two-hybrid screening systems. Brief Funct Genomic Proteomic 2007; 6:302-12; PMID:18218650; http://dx.doi.org/ 10.1093/bfgp/elm035 [DOI] [PubMed] [Google Scholar]
- 26.Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem 2001; 276:33111-20; PMID:11435423; http://dx.doi.org/ 10.1074/jbc.M102755200 [DOI] [PubMed] [Google Scholar]
- 27.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Höhfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol 2001; 3:93-6; PMID:11146632; http://dx.doi.org/ 10.1038/35070170 [DOI] [PubMed] [Google Scholar]
- 28.Muller P, Ruckova E, Halada P, Coates PJ, Hrstka R, Lane DP, Vojtesek B. C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances. Oncogene 2013; 32:3101-10; PMID:22824801; http://dx.doi.org/ 10.1038/onc.2012.314 [DOI] [PubMed] [Google Scholar]
- 29.Ruckova E, Muller P, Nenutil R, Vojtesek B. Alterations of the Hsp70/Hsp90 chaperone and the HOP/CHIP co-chaperone system in cancer. Cell Mol Biol Lett 2012; 17:446-58; PMID:22669480; http://dx.doi.org/ 10.2478/s11658-012-0021-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Wang K, Guo Y, Shi Q, Tian C, Chen C, Gao C, Zhang BY, Dong XP. Heat shock protein 70 selectively mediates the degradation of cytosolic PrPs and restores the cytosolic PrP-induced cytotoxicity via a molecular interaction. Virol J 2012; 9:303; PMID:23216755; http://dx.doi.org/ 10.1186/1743-422X-9-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakrabarti O, Ashok A, Hegde RS. Prion protein biosynthesis and its emerging role in neurodegeneration. Trends Biochem Sci 2009; 34:287-95; PMID:19447626; http://dx.doi.org/ 10.1016/j.tibs.2009.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costa MDM, Paludo KS, Klassen G, Lopes MH, Mercadante AF, Martins VR, Camargo AA, Nakao LS, Zanata SM. Characterization of a specific interaction between ADAM23 and cellular prion protein. Neurosci Lett 2009; 461:16-20; PMID:19477226; http://dx.doi.org/ 10.1016/j.neulet.2009.05.049 [DOI] [PubMed] [Google Scholar]
- 33.Lee KS, Magalhaes AC, Zanata SM, Brentani RR, Martins VR, Prado MA. Internalization of mammalian fluorescent cellular prion protein and N-terminal deletion mutants in living cells. J Neurochem 2001; 79:79-87; PMID:11595760; http://dx.doi.org/ 10.1046/j.1471-4159.2001.00529.x [DOI] [PubMed] [Google Scholar]
- 34.Von Dannecker LE, Mercadante AF, Malnic B. Ric-8B, an olfactory putative GTP exchange factor, amplifies signal transduction through the olfactory-specific G-protein Galphaolf. J Neurosci 2005; 25:3793-800; PMID:15829631; http://dx.doi.org/ 10.1523/JNEUROSCI.4595-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerr DS, Von Dannecker LE, Davalos M, Michaloski JS, Malnic B. Ric-8B interacts with G alpha olf and G gamma 13 and co-localizes with G alpha olf, G beta 1 and G gamma 13 in the cilia of olfactory sensory neurons. Mol Cell Neurosci 2008; 38:341-8; PMID:18462949; http://dx.doi.org/ 10.1016/j.mcn.2008.03.006 [DOI] [PubMed] [Google Scholar]


