ABSTRACT
Genes of the major histocompatibility complex (MHC; also called HLA in human) are polymorphic elements in the genomes of sharks to humans. Class-I and class-II MHC loci appear responsible for much of the genetic linkage to myriad disease states via the capacity to bind short (~8-15 a.a.) peptides of a given pathogen's proteome, or in some cases, the altered proteomes of cancerous cells, and even (in autoimmunity) certain nominal ‘self’ peptides (Janeway, 2004).1 Unfortunately, little is known about how the canonical structure of the MHC-I/-II peptide-presenting gene evolved, particularly since beyond ~500 Mya (sharks) no paralogs exist.2,3 We previously reported that HLA-A isotype alleles with the α1-helix, R65 motif, are wide-spread in phylogeny, but that the α 2-helix, H151R motif, has apparently segregated out of most species. Surprisingly, an uncharacterized orf in T. syrichta (Loc-103275158) encoded R151, but within a truncated A-23 like gene containing 5′- and 3′- footprints of the transposon (TE), tigger-1; the extant tarsier A-23 allele is totally missing exon-3 and part-of exon-4; together, suggesting TE-mediated inactivation of an intact/ancestral A-23 allele (Murray, 2015a).4 The unique Loc-103275158 orf encodes a putative 15-exon transcript with no apparent paralogs throughout phylogeny. However, an HLA-A11 like gene in M. leucophaeus with a shortened C-terminal domain, and an HLA-A like orf in C. atys with two linked α1/α2/α3 domains, both contain a second transmembrane segment, which is conserved in Loc-103275158. Thus, we could model the putative protein with its Nef-like tail domain docked to its MHC-I like α3 domain (i.e., on the same side of a membrane). This modeled tertiary structure is strikingly similar to the solved structure of the Nef:MHC-I CD:AP1mu transporter (Jia, 2012).5 Nef:AP1mu binds the CD of MHC-I in trafficking MHC-I away from the trans-golgi and into the endocytic pathway in HIV-1 infected cells. The CD loop of the Loc-103275158 provisional protein conserved the nominal MHC-I CD tyrosine phosphorylation site, and it has an N-terminal SH3 domain that we docked in one conformation to its internal Nef-like domain. Here, we suggest that phosphorylation of the protein's CD-loop signals an exchange between the internal Nef-like domain and a lentiviral-Nef for binding the N-terminal SH3 domain - freeing the Nef-like domain to bind MHC-I CD. Since the 5′-tigger sequence encodes part of the pseudo α1/α2 MHC-I domain, and the 3′-tigger part of the Nef-like domain, we speculate that transposition proceeded phylogenetically disparate horizontal transfers, involving adjacent 5′- and 3′- parasitic footprints, which we also found in the Loc-103275158 orf.
KEYWORDS: HIV, intracellular trafficking, kinase, MHC, Nef, SH3 signaling, TCR, transposon
The adaptive immune system of all gnathostomes is thought to center (most fundamentally) on a tripartite cellular mechanism involving cells such as dendritic cells (DC), T lymphocytes (T cells) and B lymphocytes (B cells).1 Moreover, it is thought that an ancient retroviral transposition event initiated the split gene arrangement (and the DNA rearrangement mechanism) of the highly variable receptors used by T-cells (the TCR) and B-cells (the BCR, also known as the surface immunoglobulin receptor) in the immune recognition of pathogens and altered/self proteins (Teng and Shatz, 2015).6 The role of the DC [generically referred to as “antigen-presenting cells” (APC)] in this interaction is to “present” the said protein(s) to T cells, and this involves the MHC class-I and –II molecules, several non-classical MHC proteins, and at least two dedicated pathways that first “process” proteins into appropriate sized peptides.1 Once processed and transported into the endoplasmic reticulum, these peptides bind to MHC-I, or in the case of MHC-II, are processed in endosomal pathways and bind MHC-II in vesicles beyond the Golgi; in both cases, leading to the MHC-I/II molecules with their bound peptide on DC surfaces. Here, T cells of the CD4 type bind via their TCR to MHC-II:peptide complexes and CD8 T cells to MHC-I:peptide complexes (abbreviated as pMHC-I, or pMHC-II; or in humans, pHLA-A, -B, -DR, etc.), and it is only later that this impacts the activation of B cells. This is because the production of immunoglobulin (antibody) by the B cell against a given pathogen depends on the aforementioned recognition by CD4 T cells of the pathogen. Once CD4 T cells are activated, they interact via the same TCR with pMHC-II on B cells presenting the same (or highly similar) pathogen peptide(s). This activates the B cell to differentiate into plasma cells that secrete antibody against the pathogen, e.g., antibody against HIV gp60.1,7 On the other hand, CD8 T cells do not characteristically provide this “help” to B cells, but rather have the capacity to move from the DC after initial pMHC-I recognition to pathogen infected cells of all types (even B cells); and crucially, CD8 T cells have the programmed ability to directly kill these infected cells. In the case of viral immunity to pathogens like HIV, this mechanism is the most effective means of overcoming the pathogen, i.e., to destroy the production of new pathogens by infected cells (see Janeway, 20041 for a thorough review).
Transposon (TE) insertions are characterized by defined sequences which may or may not include terminal repeats, short target duplications and donor-gene excisions, as well as some portion of the transposase itself.8-11 Over the course of evolutionary time, a given TE insertion can become ‘domesticated’ (i.e., certain sequences excised/replaced/mutated) within its location, contributing functions associated with expression of affected host genes and even altering the structure of functional proteins.12-15
During the study of the HLA-A23 lineage in early primates, we encountered a surprising example of domestication involving the DNA transposon, tigger-1.4,15 In the tarsier, Tarsius syrichta, an apparent deletion of an ancestral/intact A23 allele led to exon-3 (encodes the α2 domain) being completely absent, and exon-4 almost absent (only the last two β-strands of the α3 domain remains encoded).4 By contrast, a highly unusual, uncharacterized orf in T. syrichta (Loc-103275158) displays a complete deletion of exon-2 (encodes α1) and contains only part of exon-3 (~ from the structurally crucial “kink” in the α2 helix at R15116 to the end of the α2 domain; this is encoded by exon-5 in the orf). Moreover, the full α3 domain, and the C-terminus of a nominal MHC-I, are encoded in the 3′-sequence of Loc-103275158.4,16 Indeed, there are several unique features to this uncharacterized orf (Fig. 1).
Figure 1.
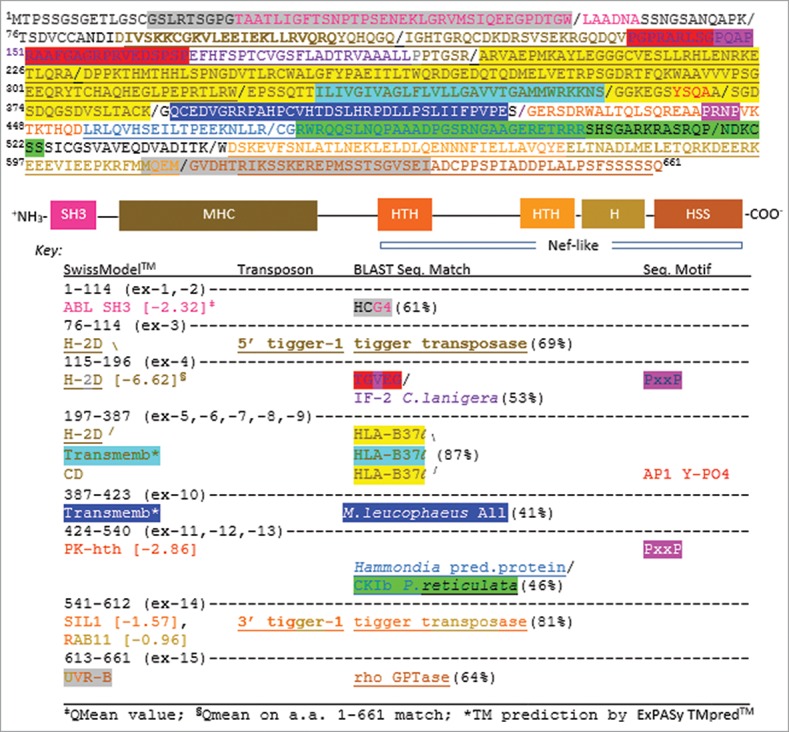
The predicted protein of Loc-103275158 annotated by sequence homology and structural homology. (Top) Amino acid (a.a.) sequence of the orf (exon separation indicated by “/ ” symbols). Color coding matches the (Middle) diagram of the full protein by structural homology to solved domains as determined by SwissModel™ searches on the amino acid sequence shown. (Bottom) The Key shows the locations within the a.a. sequence of the indicated structural domains, the TE footprints of tigger-1, the best sequence matches by BLAST, and motifs associated with known kinases and intracellular trafficking proteins. Gene abbreviations are GenBank annotations (www.ncbi.nlm.nih.gov). H (helix), HTH (helix-turn-helix), HSS (helix-strand-strand).
Notably, the footprint of a tigger-1 insertion domesticated between a sequence that models into an ABL-like SH3 domain (pink) (QMean -2.32/template PDB 1ju5) and the R151 position of the partially conserved A-23 α2 domain [see N-terminus of yellow highlighted sequence (exon-5), Fig. 1]. This 5′-tigger-1 footprint is shown in brown (underlined) and models into what would be the majority of the “missing” α-1 helix of an intact MHC-I like molecule (see also Fig. 2, below). Importantly, a proline-rich sequence from a.a. 138 – 167 interrupts this MHC-like β-sheet with a loop projected toward the α-3 domain [highlighted red sequence (PxxP in pink) in Fig. 1; see red β-loop in Fig. 2A]. Indeed, the consensus PxxP motif,5 at the tip of this loop informed our initial modeling of the CD peptide docked to the Loc-103215158 protein.
Figure 2.
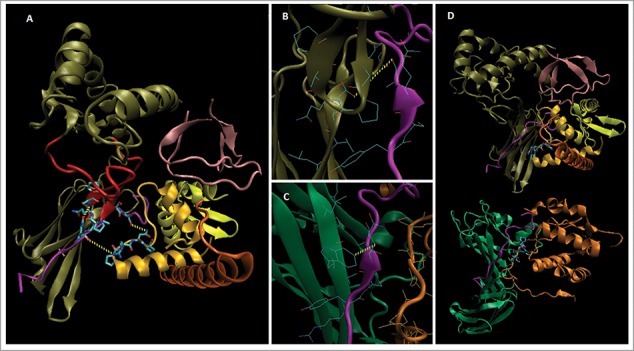
Model structure of Loc-103275158 predicted protein. SwissModel™ (www.expasy.org) was used to generate a homology-based model for 71% of the protein (A). The HLA-A CD from PDB 4EMZ (magenta) was docked at R282 based on homology with the W408:peptide region of 4EMZ (B, C; see text). The Nef-like C-terminal domain was built from matched sub-domain structures in SwissModel™ (see Fig. 1), and docked to the CD peptide on the opposite face from the α3 domain; thus, forming a CD binding groove similar to that of Nef:AP1mu (A-D). As in 4EMZ, a PxxP motif [in the PK-HTH (lt. orange), a.a. P442-P445 (licorice side chains)] was docked to the CD peptide, and the Nef-like structure (A) was built from subdomain homology with Nef (dark orange, D (bottom) after this placement (see D, for overall structural comparison with Nef:MHC-I CD:AP1mu). MHC-I like structure [tan (β-loop in red), CD contacts in licorice]; CD peptide (magenta, Q322 licorice); SH3-like N-terminus (pink); Nef-like C-terminus (lt. orange, yellow, dk. orange, PxxP licorice); AP1mu (green); Nef (dk. orange, PxxP licorice); not shown a.a. in black, including the two transmembrane regions (highlighted lt. blue and blue), Figure. 1 sequence.
The overall homology of the second β strand of the Loc-103215158 model's 3 domain to the a.a. 405-414 β-strand of AP1mu (in contact with MHC-I CD in PDB 4EMZ5); included the contact of the CD peptide with residue W408, which we modeled by docking the CD peptide to R282 of the Loc-103275158 model (Fig. 2Cand 2B). As shown in Fig. 2A, the CD peptide is docked in substantively similar orientation to the bound peptide in 4EMZ, and contacts the α3 β-strand face via the PxxP motif of the aforementioned a.a. 138-167 loop (loop, A149:N to CD, Q322:OE @ 3.63 Å; top dotted yellow line). In 4EMZ, Q322 contacts W408 @4.67 Å (see Fig. 2C). Also, in 4EMZ, the CD peptide contacts Nef from the face opposite the AP1mu β-strand, thus we docked the putative Nef-like (NefL) domain of Loc-103275158 to the face opposite the α3 β-strand, and formed a strikingly similar CD-peptide binding groove (Fig. 2A-D). Interestingly, this would require that the protein contain a second transmembrane segment, i.e., the NefL domain is encoded 3′ to the nominal MHC-I like transmembrane exon (exon-7, lt. blue highlighted seq. in Fig. 1). While such a second transmembrane segment is clearly not found in conventional MHC-I heavy chains, we used BLAST to find two early primate MHC-I like molecules with a possible second transmembrane segment. As shown in Figure. 3A, drills (Madrillus leucophaeus) have a slightly unusual A-11 like (A-11L) allele, which has a truncated MHC-I C domain and its C-terminus contains a non-conserved (in human HLA-A) sequence that is a predicted second transmembrane domain (ExPASy TMpred™ score: 1406io, a.a. 282-300; Figure. 3B; the nominal TM sequence @ 2638oi, a.a. 187-213). The inside (i) to outside (o) sequence is an 85% ID match with a segment from the highly unusual “dual” MHC-I like molecule of Cercocebus atys, Loc-105576907. The sooty mangabey (C. atys) predicted protein contains two α1/α2/α3 like domains that BLAST to HLA-A11 (Fig. 3A), and these are separated by two transmembrane regions; the first in the conventional location of an MHC-I (a.a. 305-329, ExPASy TMpred™ score: 2342oi), and a second one, which is included in the segment homologous to the M. leucophaeus A-11L protein (ExPASy TMpred™ score: 702io, a.a. 395-414).
Figure 3.
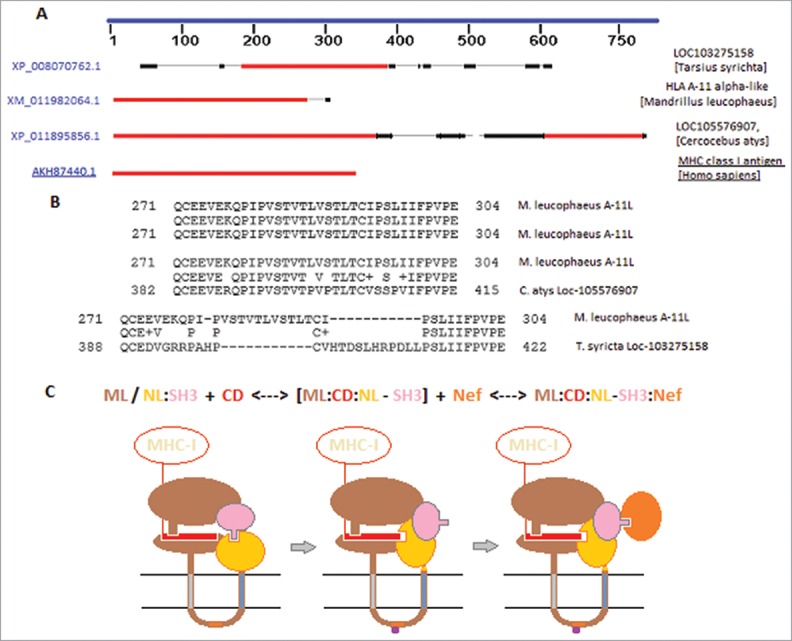
Suitable mechanism for Loc-103275158 predicted protein based on tandem transmembrane segments. BLAST of HLA-A11 versus M. leucophaeus, C. atys, and T. syrichta unusual MHC-I like molecules (A; regions of homology shown as blocks, red > > black). Sequence alignment of M. leucophaeus A-11L second transmembrane segment against putative dual MHC-I like molecule of C. atys and the T. syrichta, Loc-103275158 (B). Proposed mechanism for conformational change in Loc-103275158 provisional protein in the inhibition of Nef-mediated re-trafficking of MHC-I [C (domain rotations not scalar); see text]. ML (MHC-I like), NL (Nef-like), Y-motif (orange rectangle), phosphoryl (purple pentagon).
While the extant a.a. 388-422 sequence of Loc-103275158 is only a 41% match with the second TM segment of M. leucophaeus A-11L, there are two obvious changes (Fig. 3B): (i) an 11 a.a. deletion of a VSTLV-rich motif, and (ii) an insertion of an 11 a.a. segment including a VHTDSLHRP kinase phosphorylation site (NetPhos™ 2.0 prediction). Thus, it follows that either the Loc-103275158 protein evolved phospho-regulation in place of the second transmembrane span, or that some portion of the 388-422 segment retains this capacity. In favor of the latter, a.a. LLPSLIIFPVPE (411-422) has a high TM-tendency (ProtScale™) and is quite hydrophobic (GRAVY @ 1.525, ProtPram™, ExPASy); thus, it might be capable of associating with the inner membrane in the correct (i-o) orientation, and could be capable of spanning a bilayer in an extended conformation.17,18 With this potential caveat in mind, we have proposed a conformational mechanism (Fig. 3C) for how the Loc-103275158 protein might combat Nef down-regulation of MHC-I from the surface of tarsier cells infected by an appropriate primate lentivirus, i.e., after the mechanism of HIV-1 Nef in humans.5,7,19-21
Theoretically, a phosphorylation/dephosphorylation event within the CD loop would occur when the protein is in a “closed” conformation; based upon our modeled contacts, we prefer the notion that the MHC-I CD initially associates with some affinity to this closed conformation (left cartoon, Fig. 3C). The exact indel of 11 a.a. in the Loc-103275158 exon-10 suggests that this length is crucial to overall function, perhaps linked to disruption of the internal SH3:NefL interface as shown in the transition state of the mechanism (middle cartoon, Fig. 3C). In our homology model (Fig. 2A), an SH3 rotation of ~20˚ would both allow the NefL domain to bind the CD peptide opposite the β-loop/β-strand in a fashion highly similar to the Nef:CD:AP1mu complex, and would free the SH3 domain to bind exogenous Nef, i.e., encoded by an appropriate lentivirus (right cartoon, Fig. 3C). This novel Nef:Loc-103275158:CD complex would retain MHC-I in the trans-golgi pathway, ostensibly because it excludes AP1mu (i.e., the subunit subverted by Nef in virus-infected cells) and would provide a “sink” for Nef (i.e., subverting it away from AP1mu).5 Indeed, this might involve the previously indicated serine phosphorylation site in the 11 a.a. insertion (Fig. 3B).
Interestingly, there is a greater than 80% match of the tigger-1 transposase footprints with tigger-1 sequences in several species (not shown), but the near reciprocal deletion in the A-23 allele and the extant MHC-I sequence in Loc-103275158 suggest that gene conversion subsequent to excision of the parent insertion was involved; perhaps similar to the mechanism favored for new allele formation in the MHC.22-25 It is possible, of course, that the two footprints were separated in evolutionary time, with the 3′-tigger footprint resulting from a gene conversion involving the nascent SH3-MHC gene, e.g., via an ABL kinase exchange with a GTPase-like gene harboring the nascent NefL structures.26-28
Finally, the inherent resistance of C. atys to SIV disease,29-31 suggests that some biology linked to the Loc-105576907 orf might afford novel explanations, and quite possibly novel approaches to HIV-1 therapeutics. The T. syrichta Loc-103275158 orf is (as we have discussed) one such piece of biology. While the identity match is relatively modest compared to other components in the sequence (Fig. 1), the symmetrical Toxoplasma gondii (TGVEG) match, 5′; and the related parasite, Hammondia, sequence, 3′; suggest their possible involvement. Importantly, we can speculate that this may have involved recombination with the nearly identical IF-2 and rho GTPase genomic sequences of intermediate hosts (Fig. 4). Perhaps, the protozoan “vectors” T. gondii and Hammondia emanated from these distant intermediate species; rodents and fish, respectively, in what is a remarkably complex horizontal transfer mechanism (Fig. 4).32-34 Given the role of GTPases in intracellular immunity to T. gondii,34 it is an intriguing hypothesis that protozoans may have fortuitously enhanced immunity against retroviral pathogens. That such complexity is seemingly recapitulated within an MHC-I structural gene4 is profound, and might suggest a previously unrecognized level of evolutionary interplay between these two types of pathogens and the genesis of the human adaptive immune system.
Figure 4.
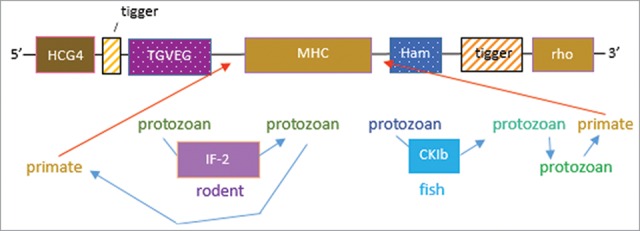
Hypothesis on horizontal transfer in evolution of Loc-103275158. BLAST match of the orf segments as shown in Figure. 1, suggest intermediate species as hosts for protozoan vectors of the tigger TE. Thus, the 5′-block matches the T.gondii TGVEG to IF-2 in C. lanigera, which could have passed the HCG4-tigger block into T. syrichta via T. gondii. The 3′-block is suggested to involve P. reticulata, wherein the CK1b footprint matches an uncharacterized orf of Hammondia (a close relative of T. gondii). Ostensibly, this would require protozoan:protozoan transfer from a fish protozoan to Hammondia, which could have then infected T. syrichta. The red arrows indicate possible gene conversions in T. syrichta leading to the symmetrical, MHC central location in the orf.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Janeway CA., Jr Immunobiology, 6th ed 2004. Garland Science; New York, NY: ISBN: 9780815341017 [Google Scholar]
- [2].Shen T, Lei M, Wang J, He X, Li X, Li J. Molecular cloning, organization, expression and 3D structural analysis of the MHC class Ia gene in the whitespotted bamboo shark (Chiloscyllium plagiosum). Vet Immunol Immunopathol 2014; 157(1-2):111-8; PMID:24315118; http://dx.doi.org/ 10.1016/j.vetimm.2013.10.012 [DOI] [PubMed] [Google Scholar]
- [3].Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet 2010; 11(1):47-59; PMID:19997068; http://dx.doi.org/ 10.1038/nrg2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Murray JS. Transposon-mediated death of an ancestral A-23-like allele: Evolution of TCR-positioning motifs in the HLA-A lineage. Immuno Genetics 2015; 67(8):473-6; PMID:26063599; http://dx.doi.org/ 10.1007/s00251-015-0852-3 [DOI] [PubMed] [Google Scholar]
- [5].Jia X, Singh R, Homann S, Yang H, Guatelli J, Xiong Y. Structural basis of evasion of cellular adaptive immunity by HIV-1 Nef. Nat Struct Mol Biol 2012; 19(7):701-6; PMID:22705789; http://dx.doi.org/ 10.1038/nsmb.2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Teng G, Schatz DG. Regulation and Evolution of the RAG Recombinase. Adv Immunol 2015; 128:1-39; PMID:26477364; http://dx.doi.org/ 10.1016/bs.ai.2015.07.002 [DOI] [PubMed] [Google Scholar]
- [7].Mann JK, Chopera D, Omarjee S, Kuang XT, Le AQ, Anmole G, Danroth R, Mwimanzi P, Reddy T, Carlson J, et al.. Nef-mediated downregulation of CD4 and HLA class I in HIV-1 subtype C infection: association with disease progression and influence of immune pressure. Virol 2014; 468-470:214-25; PMID:25193656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mateo L, González J. Pogo-like transposases have been repeatedly domesticated into CENP-B-related proteins. Genome Biol Evol 2014; 6(8):2008-16; PMID:25062917; http://dx.doi.org/ 10.1093/gbe/evu153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hayward A, Ghazal A, Andersson G, Andersson L, Jern P. ZBED evolution: repeated utilization of DNA transposons as regulators of diverse host functions. PLoS One 2013; 8(3):e59940; PMID:23533661; http://dx.doi.org/ 10.1371/journal.pone.0059940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Abrusán G, Zhang Y, Szilágyi A. Structure prediction and analysis of DNA transposon and LINE retrotransposon proteins. J Biol Chem 2013; 288(22):16127-38; http://dx.doi.org/ 10.1074/jbc.M113.451500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Riordan JD, Keng VW, Tschida BR, Scheetz TE, Bell JB, Podetz-Pedersen KM, Moser CD, Copeland NG, Jenkins NA, Roberts LR, et al.. Identification of rtl1, a retrotransposon-derived imprinted gene, as a novel driver of hepatocarcinogenesis. PLoS Genet 2013; 9(4):e1003441; PMID:23593033; http://dx.doi.org/ 10.1371/journal.pgen.1003441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Campos-Sánchez R, Kapusta A, Feschotte C, Chiaromonte F, Makova KD. Genomic landscape of human, bat, and ex vivo DNA transposon integrations. Mol Biol Evol 2014; 31(7):1816-32; http://dx.doi.org/ 10.1093/molbev/msu138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Malik HS, Henikoff S. Positive selection of Iris, a retroviral envelope-derived host gene in Drosophila melanogaster. PLoS Genet 2005; 1(4):e44; PMID:16244705; http://dx.doi.org/ 10.1371/journal.pgen.0010044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sinzelle L, Izsvák Z, Ivics Z. Molecular domestication of transposable elements: from detrimental parasites to useful host genes. Cell Mol Life Sci 2009; 66(6):1073-93; PMID:19132291; http://dx.doi.org/ 10.1007/s00018-009-8376-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Smit AF, Riggs AD. Tiggers and DNA transposon fossils in the human genome. Proc Natl Acad Sci U S A 1996; 93(4):1443-8; PMID:8643651; http://dx.doi.org/ 10.1073/pnas.93.4.1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Murray JS. An old Twist in HLA-A: CDR3α Hook up at an R65-joint. Front Immunol 2015; 6:268; PMID:26074926; http://dx.doi.org/ 10.3389/fimmu.2015.00268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lewis MJ, Chang JA, Simoni RD. A topological analysis of subunit α from Escherichia coli F1F0-ATP synthase predicts eight transmembrane segments. J Biol Chem 1990; 265(18):10541-50; PMID:2162353 [PubMed] [Google Scholar]
- [18].Mateja A, Paduch M, Chang HY, Szydlowska A, Kossiakoff AA, Hegde RS, Keenan RJ. Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science 2015; 347(6226):1152-5; PMID:25745174; http://dx.doi.org/ 10.1126/science.1261671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lubben NB, Sahlender DA, Motley AM, Lehner PJ, Benaroch P, Robinson MS. HIV-1 Nef-induced down-regulation of MHC class I requires AP-1 and clathrin but not PACS-1 and is impeded by AP-2. Mol Biol Cell 2007; 18(9):3351-65; PMID:17581864; http://dx.doi.org/ 10.1091/mbc.E07-03-0218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pizzato N, Derrien M, Lenfant F. The short cytoplasmic tail of HLA-G determines its resistance to HIV-1 Nef-mediated cell surface downregulation. Hum Immunol 2004; 65(11):1389-96; PMID:15556689; http://dx.doi.org/ 10.1016/j.humimm.2004.07.239 [DOI] [PubMed] [Google Scholar]
- [21].Chang AH, O'Shaughnessy MV, Jirik FR. Hck SH3 domain-dependent abrogation of Nef-induced class 1 MHC down-regulation. Eur J Immunol 2001; 31(8):2382-7; PMID:11500821; http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- [22].Shum BP, Guethlein L, Flodin LR, Adkison MA, Hedrick RP, Nehring RB, Stet RJ, Secombes C, Parham P. Modes of salmonid MHC class I and II evolution differ from the primate paradigm. J Immunol 2001; 166(5):3297-308; PMID:11207285; http://dx.doi.org/ 10.4049/jimmunol.166.5.3297 [DOI] [PubMed] [Google Scholar]
- [23].Hughes AL, Hughes MK, Watkins DI. Contrasting roles of interallelic recombination at the HLA-A and HLA-B loci. Genetics 1993; 133(3):669-80; PMID:8454208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Satta Y, O'hUigin C, Takahata N, Klein J. Intensity of natural selection at the major histocompatibility complex loci. Proc Natl Acad Sci U S A 1994; 91(15):7184-8; PMID:8041766; http://dx.doi.org/ 10.1073/pnas.91.15.7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Widera G, Flavell RA. The nucleotide sequence of the murine I-E β b immune response gene: evidence for gene conversion events in class II genes of the major histocompatibility complex. EMBO J 1984; 3(6):1221-5; PMID:6086309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bianchi C, Torsello B, Angeloni V, Bombelli S, Soldi M, Invernizzi L, Brambilla P, Perego RA. Eight full-length abelson related gene (Arg) isoforms are constitutively expressed in caki-1 cell line and cell distribution of two isoforms has been analyzed after transfection. J Cell Bio Chem 2008; 105(5):1219-27; PMID:18810762; http://dx.doi.org/ 10.1002/jcb.21922 [DOI] [PubMed] [Google Scholar]
- [27].Chang X, Izumchenko E, Solis LM, Kim MS, Chatterjee A, Ling S, Monitto CL, Harari PM, Hidalgo M, Goodman SN, et al.. The relative expression of Mig6 and EGFR is associated with resistance to EGFR kinase inhibitors. PLoS One 2013; 8(7):e68966; PMID:23935914; http://dx.doi.org/ 10.1371/journal.pone.0068966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chen YC, Colvin ES, Griffin KE, Maier BF, Fueger PT. Mig6 haploinsufficiency protects mice against streptozotocin-induced diabetes. Diabetologia 2014; 57(10):2066-75; PMID:24989997; http://dx.doi.org/ 10.1007/s00125-014-3311-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang Z, Metcalf B, Kasheta M, Kasala-Hallinan C, Tran D, Johnson RP, Else JG, Karl J, O'Connor D, Apetrei C, et al.. Characterization of MHC class I alleles in sooty mangabeys as a tool for evaluating cellular immunity in natural hosts of SIV infection. Immuno Genetics 2015; 67(8):447-61; PMID:26129855; http://dx.doi.org/ 10.1007/s00251-015-0853-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jones AC, Herndon JG, Courtney CL, Collura L, Cohen JK. Clinicopathologic characteristics, prevalence, and risk factors of spontaneous diabetes in sooty mangabeys (Cercocebus atys). Comp Med 2014; 64(3):200-10; PMID:24956212 [PMC free article] [PubMed] [Google Scholar]
- [31].DeGottardi MQ, Specht A, Metcalf B, Kaur A, Kirchhoff F, Evans DT. Selective downregulation of rhesus macaque and sooty mangabey major histocompatibility complex class I molecules by Nef alleles of simian immunodeficiency virus and human immunodeficiency virus type 2. J Virol 2008; 82(6):3139-46; PMID:18199657; http://dx.doi.org/ 10.1128/JVI.02102-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in bdelloid rotifers. Science 2008; 320(5880):1210-3; PMID:18511688; http://dx.doi.org/ 10.1126/science.1156407 [DOI] [PubMed] [Google Scholar]
- [33].Pombert JF, Selman M, Burki F, Bardell FT, Farinelli L, Solter LF, Whitman DW, Weiss LM, Corradi N, Keeling PJ. Gain and loss of multiple functionally related, horizontally transferred genes in the reduced genomes of two microsporidian parasites. Proc Natl Acad Sci 2012; 109(31):12638-43; PMID:22802648; http://dx.doi.org/ 10.1073/pnas.1205020109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gazzinelli RT, Mendonça-Neto R, Lilue J, Howard J, Sher A. Innate resistance against Toxoplasma gondii: an evolutionary tale of mice, cats, and men. Cell Host Microbe 2014; 15(2):132-8; PMID:24528860; http://dx.doi.org/ 10.1016/j.chom.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]


