Abstract
Although p53 is the most frequently mutated gene in cancer, half of human tumors retain wild-type p53, whereby it is unknown whether normal p53 function is compromised by other cancer-associated alterations. One example is Ewing’s sarcoma family tumors (ESFT), where 90% express wild-type p53. ESFT are characterized by EWS-FLI1 oncogene fusions. Studying 6 ESFT cell lines, silencing of EWS-FLI1 in a wild-type p53 context resulted in increased p53 and p21WAF1/CIP1 levels, causing cell cycle arrest. Using a candidate gene approach, HEY1 was linked to p53 induction. HEY1 was rarely expressed in 59 primary tumors, but consistently induced upon EWS-FLI1 knockdown in ESFT cell lines. The NOTCH signaling pathway targets HEY1, and we show NOTCH2 and NOTCH3 to be expressed in ESFT primary tumors and cell lines. Upon EWS-FLI1 silencing, NOTCH3 processing accompanied by nuclear translocation of the activated intracellular domain was observed in all but one p53-mutant cell line. In cell lines with the highest HEY1 induction, NOTCH3 activation was the consequence of JAG1 transcriptional induction. JAG1 modulation by specific siRNA, NOTCH-processing inhibition by either GSI or ectopic NUMB1, and siRNA-mediated HEY1 knockdown all inhibited p53 and p21WAF1/CIP1 induction. Conversely, forced expression of JAG1, activated NOTCH3, or HEY1 induced p53 and p21WAF1/CIP1. These results indicate that suppression of EWS-FLI1 reactivates NOTCH signaling in ESFT cells, resulting in p53-dependent cell cycle arrest. Our data link EWS-FLI1 to the NOTCH and p53 pathways and provide a plausible basis both for NOTCH tumor suppressor effects and oncogenesis of cancers that retain wild-type p53.
Introduction
Ewing’s sarcoma and related tumors (Ewing’s sarcoma family of tumors, ESFT) affect bone and soft tissue in children and young adults, and arise from a presumably mesenchymal precursor. The only recurrent aberration in this disease is a chromosomal translocation fusing the EWS gene to an ETS oncogene, most frequently FLI1. As a result, a chimeric transcription factor is generated. Sustained expression of the EWS-FLI1 fusion protein is required for ESFT cell proliferation and tumor growth. However, ectopically expressed EWS-FLI1 transcriptionally induces p53 in most primary cell types tested, resulting in cell cycle arrest or apoptosis. In these cases, loss of INK4A or p53 gene function can rescue cells from EWS-FLI1–induced toxicity and facilitate tumorigenesis ( for review, see ref. 1). Paradoxically, p53 mutations and INK4A deletions are rare in ESFT (<10% and ~20%, respectively), although associated with poor outcome (2–5). Similarly, bone marrow–derived mesenchymal progenitor cells, currently the only primary cell type tolerant to the oncogenic stress imposed by ectopic EWS-FLI1 expression, retain wild-type p53 (wt-p53) upon transformation with the chimeric gene (6).
p53 serves an important function as a checkpoint control protein regulating cellular fate in response to different stresses (7). Consistent with the absence of p53-pathway alterations, we have previously shown that wt-p53–expressing ESFT cell lines show largely normal DNA damage signal integration, p53-induced cell cycle arrest, and apoptosis in response to ionizing irradiation (8). However, p53 also plays a surveillance role in protection against activated oncogenes and little is known about mechanisms by which wt-p53–expressing tumors escape oncogenic stress.
We performed EWS-FLI1 gene–silencing experiments in a series of ESFT cell lines to identify pathways that may play a role in the mechanisms of escape from EWS-FLI1–induced oncogenic stress and to better understand deregulated tumor cell growth in the presence of wt-p53 (9). Here, we report on the identification of EWS-FLI1–mediated NOTCH pathway suppression as a mechanism of keeping p53 in check in ESFT.
Materials and Methods
Cell lines
All ESFT cell lines used in this study have previously been described (2, 4). Their EWS-FLI1 fusion type, p53, and INK4A gene status are indicated in Fig. 1.
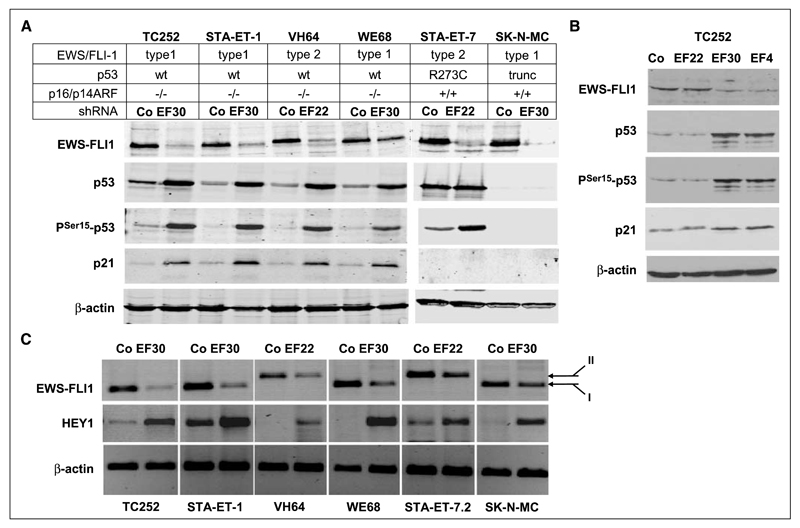
Figure 1.
EWS-FLI1 silencing induces expression of activated p53, p21WAF1/CIP1, and HEY1 expression in wild-type p53 ESFT cell lines. A, p53, PSer15-p53, and p21WAF1/CIP1 accumulation after EWS-FLI1 silencing in four wt-p53 and two mt-p53 ESFT cell lines. Immunoblot analysis 4 d after transfection of the indicated shRNA expression vectors, demonstrating induction of Ser 15-phosphorylated p53 and p21WAF1/CIP1 restricted to the wt-p53 cell lines upon introduction of shRNA specific for the respective EWS-FLI1 fusion types (EF30 for type 1 EWS-FLI1 in TC252, STA-ET-1, WE68, and SK-N-MC cells; EF22 for type 2 EWS-FLI1 in VH64 and STA-ET-7.2 cells) compared with control transfections with a nontargeting shRNA (Co). β-Actin is shown as a loading control. B, p53, PSer15-p53, and p21WAF1/CIP1 accumulation are specifically associated with EWS-FLI1–modulating shRNAs. TC252 cells were transfected with either nontargeting shRNA, shRNAs targeting the EWS-FLI1 type 1 fusion region (EF30) or the FLI1 3′ portion (EF4), or with a shRNA to the type 2 fusion region as a mismatched control. EWS-FLI1 and p53 protein expression was monitored by Western blotting. C, demonstration of HEY1 induction upon EWS-FLI1 silencing by RT-PCR 96 h after EWS-FLI1 silencing. For control, a nontargeting shRNA was transfected. β-actin PCR was used as a control for equal input. EWS-FLI1 fusion types I and II are indicated.
RNA interference, cDNA expression, and inhibitor treatment
Cells were transfected with Lipofectamine Plus reagent (Invitrogen) and subjected to puromycin selection (1 μg/mL) the next day. On day 4 after transfection, puromycin-selected cells were processed for RNA and protein extraction.
Sequences targeted by previously validated siRNA sequences used in this study were GCAGAACCCTTCTTATGAC for EWS-FLI1–specific EF30 (9, 10), GCTACGGGCAGCAGAGTTC for EWS-FLI1 type 2–specific EF22 (9), CACCCACGTGCCTTCACAC for EF4 targeting the FLI1 3′ portion (9), GACTCCAGTGGTAATCTAC for the shRNA against p53 (11), GCTAGAAAAAGCTGAGATC for the HEY1-specific siRNA (12), and CGCCAAATCCTGTAAGAAT for the siRNA against JAG1 (13). For negative control in siRNA transfections, the ON-TARGETplus nontargeting siRNA pool was used (Dharmacon).
SiRNAs against EWS-FLI1 and p53 were expressed as small hairpin (sh) RNAs from pSUPER-based retroviral expression constructs as previously described (9, 11). For negative control, pSTNeg (Ambion; Applied Biosystems) encoding a scrambled shRNA with no significant similarity to human sequences was used.
Expression vectors for the human NOTCH3 intracellular domain (NICD3) and NUMB1 cDNA were kindly provided by M. Bonafé (Dipartimento di Patologica Sperimentale, Bologna, Italy) and K. Brennan (Welcome Trust Centre for Cell Matrix Research, Manchester, UK; ref. 14). Human JAG1 and DLL1 cDNAs were obtained from Dr. I. Alcobia (Institute of Molecular Medicine, Lisbon, Portugal; ref. 15). The pSPORT-based expression construct for human HEY1 was a gift from M.T. Chin (Brigham and Woman’s Hospital, Cambridge, MA; ref. 16).
γ Secretase inhibitor (GSI; ZLeuLeuNleCHO; Calbiochem) was used at a concentration of 1 to 10 μmol/L.
Gene expression analysis by microarray technology
Changes in gene expression profiles upon knockdown of EWSFLI1 were followed on Affymetrix HGU133A arrays (Affymetrix, Inc.). cRNA target synthesis and GeneChip processing were performed according to standard protocols (Affymetrix, Inc.). All further analyses were performed in R statistical environment using Bioconductor packages (17).
CEL files for (a) 5 Ewing Sarcoma cell lines (TC252, SKNMC, STAET7, STAET1, WE68, knockdown and control), (b) expression experiments from 3 different data sets of primary Ewing Sarcoma tumors (n = 5, ref. 18; n = 27, ref. 19; n = 27, ref. 20), and (c) the ‘‘Novartis gene expression atlas,’’ which comprises 79 tissues (21) were normalized together using gcrma (22). For each gene, only one probeset was selected for further analysis by the criterion of maximizing the expression variation across arrays.
Cell line data were further filtered by (a) excluding probesets with very low expression by comparison to the probability density distribution of ‘‘negative’’ probesets with no significant EST hits (Bioconductor package panp) and (b) excluding probsets with very small variation (interquartile range <0.3) across the 10 arrays. Then, for each cell line and each gene, ratios of knockdown versus control were calculated, and these five ratios were used as replicas for a moderated one-sample t test, and P values were corrected for multiple testing by the Benjamini-Hochberg method (Bioconductor package limma).
Real-time and quantitative reverse transcription-PCR
RNA samples (1 μg each) were reverse-transcribed using the Moloney murine leukemia virus reverse transcriptase and random hexamers. Ten nanograms of cDNA were used in PCR reactions. β-actin was amplified for 22 cycles, and EWS-FLI1 and HEY1 were amplified for 35 cycles.
For quantitative analysis, cDNA samples were analyzed by Taqman quantitative reverse-transcription PCR (qRT-PCR). Ten nanograms of cDNA were used per reaction, and expression of ACTA2, JAG1, HES1, and HEY1 was calculated as a percentage of β2-microglobulin content. Fold changes upon EWS-FLI1 knockdown for these genes were calculated by dividing their normalized expression levels in knockdown versus control conditions. Primer sequences and PCR reaction conditions are available upon request.
Protein extracts, Western blot, and immunofluorescence analyses
For immunoblot analysis, total proteins were resolved by SDS-PAGE and processed for Western blotting according to standard procedures. For subcellular fractionation experiments, cells were swollen in hypotonic buffer [10 mmol/L Tris (pH 7.5) and 10 mmol/L KCl] including complete protease inhibitor cocktail (Roche) for 10 min on ice before lysis with 0.25% NP40 for 15 min. The cytosolic fraction was separated from the crude nuclear cell pellet by centrifugation. After a second 15-min incubation of the pellet in the hypotonic lysis buffer, nuclei were resuspended in high salt buffer [20 mmol/L Tris (pH 7.5), 400 mmol/L NaCl, 0.5% NP40, 0.3% Triton X100, and protease inhibitors] for 15 min, and the supernatant containing nuclear proteins was collected after centrifugation at 10.000 g for 10 min at 4°C.
The following reagents were used for immunoblot and immunofluorescence studies: The FLI1 monoclonal antibody (mAb) 7.3 was kindly provided by O. Delattre (Institute Curie, Paris, France). P53-specific mAb DO-1 was a gift from B. Vojtesek (Masaryk Memorial Cancer Institute, Brno, Czech Republic). Human p21WAF1/CIP1 mAb F-5, polyclonal antibodies to Jagged1 (H-114), NOTCH 2 (25-255), and NOTCH 3 (M-134) were from Santa Cruz Biotechnology. MAb Ab-1 to smooth muscle α actin protein (ACTA2) was from NeoMarkers (LabVision), to Ser 15-phospho-p53 from Cell Signaling Technologies, to α-tubulin (DM1A) from Calbiochem, to β-actin from Abcam, to lamin A from Santa Cruz Biotechnology, and the polyclonal antibody to PARP was from Boehringer Mannheim.
For subcellular localization studies of p53 protein, cells were washed twice with PBS, fixed for 2 min in acetone/methanol (1:1) solution at −20°C, rehydrated in PBS, and incubated with DO1 antibody for 1 h at room temperature, followed by FITC-labeled second step antibody incubation.
Cell proliferation assay
Cell proliferation was quantified by measuring DNA synthesis using the Click-iT EdU CellCycle 633-red flow cytometry assay kit (Molecular Probes). Cells were incubated with the thymidine analogue 5-ethynyl-2′-deoxyuridine (EdU; 10 μmol/L) for 45 min and processed for flow cytometry according to the manufacturer’s instructions. DNA counter staining was achieved with CellCycle 405-blue (Molecular Probes, Invitrogen). Flow cytometry was performed on a LSR2 flow cytometer (Becton Dickinson) with excitation at 633 and 405 nm. The proportion of S-phase cells was calculated relative to the total number of vital cells.
Gene reporter assays
Sequences that span from −1700 to −1 and from −574 to −1 relative of JAG1 gene start site were amplified by PCR from human genomic DNA and cloned into firefly luciferase–containing pGL4.10 vector (Promega). Cells were transfected with either −1700/-1-JAG1.Luc, -574/-1-JAG1.Luc, or thymidine kinase promoter–driven firefly luciferase (TKp.Luc), along with thymidine kinase promoter–driven renilla luciferase (TKp.rhL) for transfection efficiency normalization purposes. Cells were harvested 96 h posttransfection, and gene reporter assays were carried out with the Dual Glo Luciferase assay kit (Promega).
Results
P53 is activated and induces p21WAF1/CIP1 upon silencing of EWS-FLI1 in wild-type p53 ESFT cells
We performed EWS-FLI1– silencing experiments in four wt-p53 and two mutant p53 (mt-p53) ESFT cell lines using previously validated vector-expressed shRNAs to the chimeric EWS-FLI1 transcripts (9). ShRNA EF30 was used to modulate type 1 EWS-FLI1 (exon 7/6 fusion) in the cell lines STA-ET-1, SK-N-MC, TC252, and WE68, whereas type 2 EWS-FLI1 (exon 7/5 fusion) was specifically targeted with EF22 in the cell lines STA-ET-7.2 and VH64. To exclude off-target effects, and because these cells do not express FLI1, additional experiments were performed using EF4, an shRNA directed to the FLI1-encoding portion common to all EWS-FLI1 fusion types (9). Negative control experiments were generally performed by transfection of pSTNeg coding for shRNA with no significant similarity to human gene sequences.
Gene expression profiling performed in five of the cell lines identified a p53 signature upon silencing of EWS-FLI1 in the wt-p53 but not in the mt-p53 ESFT cell lines, with strong transcriptional expression of p21WAF1/CIP1 (data not shown). This result suggested activation of a p53 response when EWS-FLI1 was down-modulated. Because p53 RNA levels were not affected by silencing of EWS-FLI1 (data not shown), we tested for p53 protein expression. ShRNA-mediated down-modulation of EWS-FLI1 resulted in a marked increase in p53 and Ser 15–phosphorylated p53 levels associated with p21WAF1/CIP1 induction in all four wt-p53 ESFT cell lines. In contrast, in the cell line STA-ET-7.2 expressing high levels of mutant p53, and in SK-N-MC cells lacking p53 due to a truncation mutation, p21WAF1/CIP1 protein levels remained undetectable (Fig. 1A). This result suggested that p21WAF1/CIP1 induction in response to EWS-FLI1 silencing was the consequence of activated p53. As shown in TC252 cells, the increase in Ser 15–phosphorylated p53 and in p21WAF1/CIP1 proteins was only seen with EWS-FLI1–silencing shRNAs (EF30 and EF4) but not with mismatched shRNA (EF22) or negative control shRNA excluding a nonspecific induction mechanism by shRNA treatment (Fig. 1B).
p53 and p21WAF1/CIP1 induction by EWS-FLI1 silencing in wild-type p53 ESFT cells is driven by HEY1 gene expression
Affymetrix gene expression profiling in wt-p53 ESFT cell lines suggested that among genes previously reported to encode positive modulators of p53 (23), HEY1, encoding a member of the hairy enhancer of split-related transcription factor family, was strongly induced after RNAi-mediated silencing of EWS-FLI1 (data not shown). Because none of three tested commercially available HEY1 antibodies was sufficiently sensitive and specific to detect the protein on Western blots, HEY1 induction was confirmed by conventional (Fig. 1C) and qRT-PCR (see below).
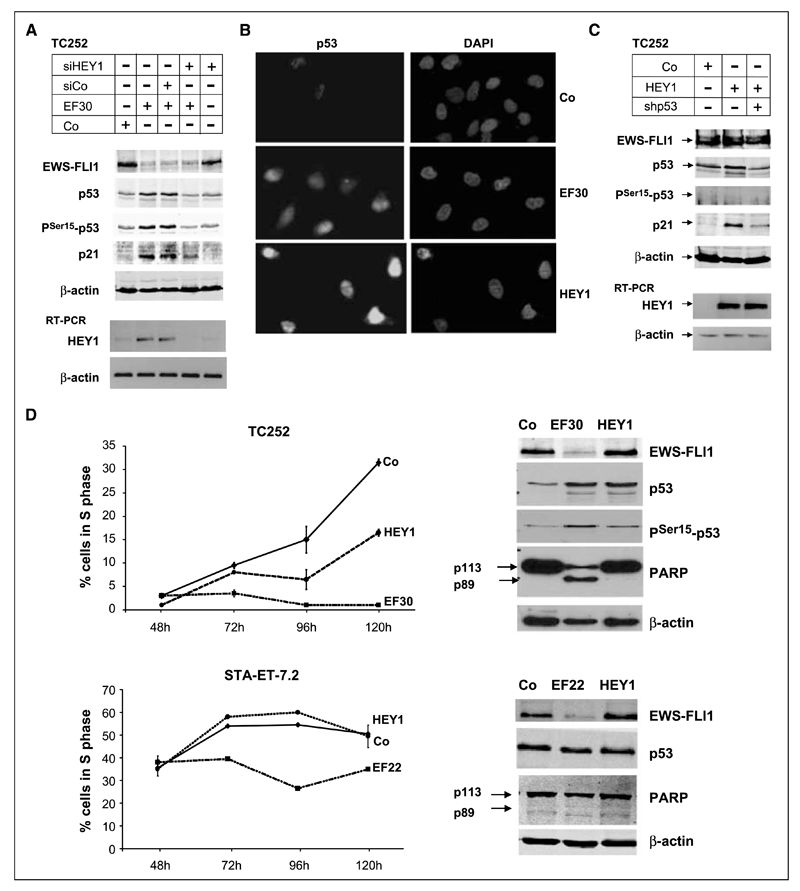
Next, we tested if HEY1 induction was involved in p53 and subsequent p21WAF1/CIP1 induction in EWS-FLI1–silenced ESFT cells. Cotransfection of a previously validated siRNA against HEY1 (12), but not of control siRNA, with EF30 into TC252 cells reduced p53 and p21WAF1/CIP1 protein to basal levels (Fig. 2A). Conversely, ectopic expression of a HEY1 cDNA in TC252 cells was sufficient to induce nuclear p53 accumulation similar to EWS-FLI1 silencing (Fig. 2B). P53 accumulation was accompanied by p21WAF1/CIP1 induction, which was reduced by coexpression of shRNA to p53 (Fig. 2C). Together, these results implicate HEY1 in p53 accumulation and p21WAF1/CIP1 expression in response to EWS-FLI1 silencing. In contrast to EWS-FLI1 silencing; however, HEY1 did not result in Ser 15 phosphorylation of p53 (Fig. 2C), suggesting additional mechanisms involved into p53 modification in response to EWS-FLI1 silencing.
Figure 2.
Induction of p53 and p21WAF1/CIP1 is dependent on HEY1 expression and associated with cell cycle arrest. A, TC252 cells were transfected with either a nontargeting shRNA or EF30, in the absence or presence of a control nontargeting siRNA pool (siCo) or a siRNA to HEY1 (siHEY1), as indicated. Protein expression of EWS-FLI1, p53, PSer15-p53, p21, and β-actin for loading control was monitored on the Western blot; HEY1 expression was followed on the RNA level by RT-PCR. B, nuclear accumulation of p53 in TC252 cells upon EWS-FLI1 silencing (EF30) and ectopic HEY1 expression (HEY1), monitored by immunostaining with the p53-specific mAb DO1. For control, cells were tansfected with a nontargeting shRNA. Left, p53 staining; right, 4′,6-diamidino-2-phenylindole staining of nuclei. C, HEY1-mediated induction of p21WAF1/CIP1 expression is dependent on the presence of p53. TC252 cells were transfected with either 4-μg pSPORT empty vector (Co) or pSPORT-based HEY1 expression vector in the absence or presence of 1.5 μg of shp53. p53, PSer15-p53, p21, and β-actin protein expression were followed on the Western blot 96 h after transfection. Expression of the HEY1 transgene was monitored by RT-PCR. Note that in contrast to EWS-FLI1 silencing (Fig. 1), HEY1 does not induce p53 phosphorylation. D, cell fate determination after EWS-FLI1 silencing or ectopic HEY1 expression compared with control transfection. TC252 cells and STA-ET-7.2 cells were transfected with the indicated EWS-FLI1 silencing vectors (EF30 or EF22) or with a nontargeting shRNA in duplicates, subjected to puromycin selection 24 h after transfection, and the fraction of surviving cells incorporating the nucleoside analogue EdU within a 45-min incubation period was monitored in 24 h intervals over 4 d starting 48 h after transfection. EdU incorporation was measured by flow cytometry, exclusively gating on living cells. At 96 h posttransfection, protein extracts were prepared from all puromycin-resistant cells and tested for EWS-FLI1, p53, and PARP expression. The reduction of the p113 full length PARP band and the occurrence of a p89 PARP cleavage band were taken as an indication for apoptosis.
HEY1 expression results in growth arrest of p53 wild-type ESFT cells
To compare the physiologic consequences of EWS-FLI1 silencing and of HEY1 expression in ESFT cells, we performed cell cycle analyses of control, EF30, and HEY1-transfected TC252 (wt-p53) and STA-ET-7.2 (mt-p53) cells in a time course experiment (Fig. 2D). The size of the S-phase fraction among living cells was assessed by flow-cytometric measurement of cells incorporating the thymidine analogue EdU. Consistent with the p53-dependent induction of p21WAF1/CIP1, EWS-FLI1 silencing resulted in a marked decrease in S-phases in TC252 cells. Note that a similar growth arrest was observed in the STA-ET-7.2 cell line. This might be due to an induction of the cell cycle inhibitor p57KIP2, which we observed in all mt-p53 ESFT cell lines upon EWS-FLI1 silencing (data not shown). In contrast, HEY1 slowed down the cell cycle only in TC252 but not in STA-ET-7.2, suggesting a strict p53 dependence of HEY1-mediated cell cycle arrest. In addition, silencing of EWS-FLI1 induced apoptosis in TC252 but not in STA-ET-7.2 cells, as indicated by the occurrence of the apoptotic PARP cleavage product p89 on the Western blot of TC252 cells (Fig. 2D). In contrast, ectopic HEY1 expression did not elicit an apoptotic response in any of the two cell lines. Consistent with earlier studies, the induction of apoptosis by EWS-FLI1 silencing seems to be p53 independent and may be related to the induction of IGFBP3 (24), which is activated in TC252 cells but not in STA-ET-7.2 cells (data not shown).
EWS-FLI1 silencing results in NOTCH pathway activation
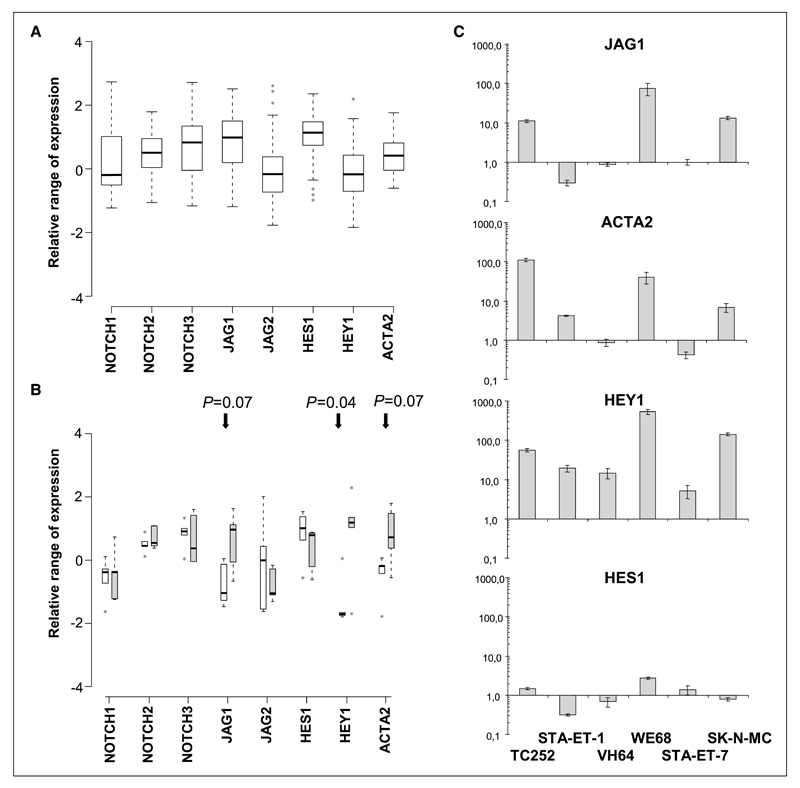
HEY1 is a well-known effector of NOTCH signaling (25). Upon ligand binding, NOTCH receptors are sequentially processed by the ADAM metallopeptidase TACE and γ secretase, releasing the NOTCH intracellular domain (NICD) into the cytoplasm. NICD translocates to the nucleus where it activates the transcription factor CBF1/CSL, which in turn activates NOTCH target genes, including HEY1. We analyzed the expression of NOTCH pathway components on Affymetrix arrays of 59 primary ESFT (Fig. 3A) and in the cell lines (Fig. 3B). To be able to compare expression levels from different genes and different data sets, we used the mean expression of each gene in the Novartis gene expression atlas as a baseline (21). Among NOTCH receptors, NOTCH2 and NOTCH3 were mostly expressed above the baseline in both primary tumors and cell lines. Among the ligands, JAG1 was variably expressed but mostly above the baseline in the primary tumors, whereas it was present at only low levels in the cell lines. JAG2 showed baseline expression compared with the gene expression atlas. Interestingly, among the transcriptional effectors, HES1 was predominantly highly expressed in the tumors and the cell lines, whereas HEY1 expression was low in the primary tumors and absent in most cell lines. As a surrogate marker for activated NOTCH signaling, we included the smooth muscle actin gene ACTA2, an established target of CBF1 (26), into our analysis. Although its expression was slightly elevated in the primary tumors, ACTA2 basal levels were low in the cell lines. Upon EWS-FLI1 silencing, however, a statistically significant increase in HEY1 expression (P = 0.04) associated with a marginally significant increase in JAG1 and ACTA2 expression (P = 0.07 each) was observed in the cell lines as a group (Fig. 3B).
Figure 3.
Range of relative expression values for selected NOTCH pathway genes shown as boxplots of (A) 3 pooled primary ESFT data sets (18–20; 59 tumors) and (B) 5 ESFT cell lines before (control-treated for 96 h, white bars) and after silencing of EWS-FLI1 (EF-treated for 96 h, gray bars). Expression values are standardized relative to the mean of 79 tissues in the Novartis Gene Expression Atlas, so that zero denotes the mean expression across all analyzed tissues. Therefore, for a given gene in the plot, a deviation from zero indicates that this gene is expressed at higher or lower levels in ESFT cell lines or primary tumors than in the tissue collection, and this deviation can be compared across different genes. Boxes, middle 50% of the values; black horizontal bars, median; the ‘‘whiskers’’ extend to the most extreme values not defined as outliers (1.5 interquartile range from the box); gray circles, outliers. In the cell lines, changes in gene expression upon EWS-FLI1 silencing were significant for HEY1 and marginally significant for JAG1 and ACTA2 with the indicated P values. C, qRT-PCR results for NOTCH pathway genes JAG1, ACTA2, HEY1, and HES1 in the six ESFT cell lines comparing EWS-FLI1 knockdown with control-treated (nontargeting shRNA) cells. Results are presented as fold change on a logarithmic scale; columns, mean relative values of triplicate experiments normalized for β2 microglobulin expression; bars, SD.
These results were confirmed by real-time quantitative PCR that revealed a strong induction (11- to 75-fold) of JAG1 mRNA in 3 of 6 EWS-FLI1–silenced ESFT cell lines (TC252, WE68, and SK-N-MC). Importantly, ACTA2 induction largely mirrored JAG1 and was paralleled by the highest levels of HEY1 induction in TC252 (56-fold), WE68 (532-fold), and SK-N-MC cells (141-fold; Fig. 3C). In cell lines lacking JAG1 induction, HEY1 activation upon EWS-FLI1 silencing was much lower (19-fold in STA-ET-1, 15-fold in VH64, and 5-fold in STA-ET-7). No correlation was observed between HES1 expression and JAG1 or ACTA2 expression.
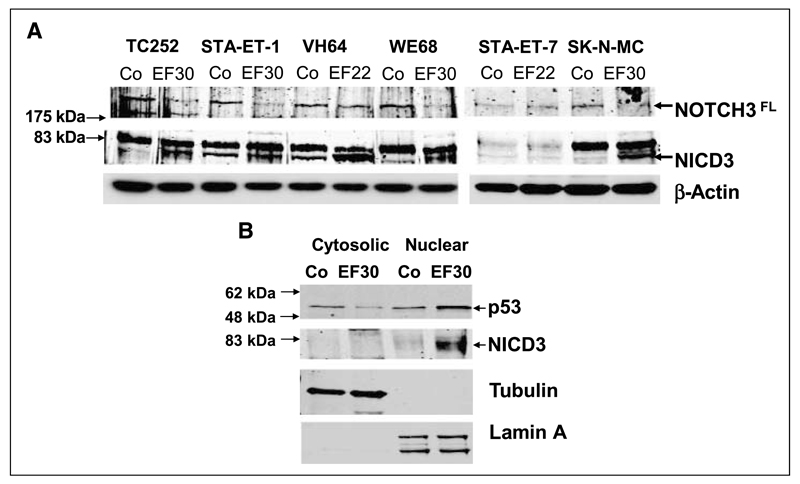
These data suggested that HEY1 activation after silencing of EWS-FLI1 might be the result of NOTCH pathway activation through ligand induction in at least three of the six ESFT cell lines. Because among NOTCH receptors, NOTCH3 was found to be most highly expressed in the ESFT cell lines, we tested for proteolytic processing of the NICD3 in all six ESFT cell lines. Upon EWS-FLI1 down-modulation, a remarkable increase in the ~80-kDa NICD3 band intensity was observed in all ESFT cell lines but STA-ET-7.2, which had shown the lowest increase in HEY1 expression (Fig. 4A). In STA-ET-1 cells, NICD3 was already detectable in the absence of EWS-FLI1 silencing, consistent with low constitutive expression of HEY1 in these cells (Fig. 1C). As shown for the cell line TC252 in a cell fractionation experiment, activated NOTCH3 efficiently translocated to the nucleus upon EWS-FLI1 silencing accompanied by nuclear p53 accumulation (Fig. 4B). Together, these results show activation of the NOTCH-signaling cascade by EWS-FLI1 silencing in at least 5 of 6 ESFT cell lines.
Figure 4.
Silencing of EWS-FLI1 induces NOTCH3 activation. A, protein extracts from the EWS-FLI1 knockdown experiment presented in Fig. 1 were probed for the expression of the 250-kDa full-length NOTCH3 (NOTCH3FL) and the presence of the ~80-kDa processed intracellular domain (NICD3). B, EWS-FLI1 silencing induces translocation of processed NICD3 to the nucleus. Control nontargeting shRNA and EF30-transfected TC252 were subjected to subcellular fractionation 96 h after transfection. Cytosolic and nuclear extracts were probed for the presence of processed NICD3 and p53. The purity of the extracts was tested using antibodies to cytoplasmic α-tubulin and nuclear lamin A.
NOTCH activation is responsible for HEY1, p53, and p21WAF1/CIP1 induction after EWS-FLI1 silencing
To study the effect of the NOTCH pathway on p53 and p21WAF1/CIP1 induction upon EWS-FLI1 silencing, we focused on the TC252 and WE68 cell lines. GSI readily blocked NOTCH3 receptor cleavage after EWS-FLI1 silencing in both cell lines (Fig. 5A). Concomitantly, it abrogated induction of p53 and p21WAF1/CIP1. This result suggested that the p53-inducing effect of EWS-FLI1 silencing was mediated by activation of the NOTCH signaling cascade. However, p53 Ser 15 phosphorylation upon EWS-FLI1 silencing was not affected by GSI treatment, consistent with our supposition that EWS-FLI1–suppressed mechanisms other than NOTCH pathway–induced HEY1 expression are responsible for p53 phosphorylation.
Figure 5.
Because, in TC252 cells, JAG1 expression was found to be induced by modulation of EWS-FLI1 (Fig. 3B and C), we tested the dependence of p53 activation on JAG1 expression (Fig. 5B): Cotransfection of EF30 with a validated siRNA to JAG1 (13) but not with control siRNA interfered with p53 accumulation and abrogated p21WAF1/CIP1 induction. Similarly, ectopic expression of the adaptor protein NUMB, which promotes rapid proteasomal degradation of the NICD, interfered with p53 and subsequent p21WAF1/CIP1 induction by EWS-FLI1 silencing. Conversely, ectopic expression of NICD3 induced p53 and p21WAF1/CIP1 to a level similar to that of EF30 shRNA–mediated silencing of EWS-FLI1. Consistent with these findings, ectopic expression of the NOTCH ligands JAG1 or DLL1 was sufficient to induce accumulation of p53 (Fig. 5B).
Because JAG1 seemed to act upstream of EWS-FLI1 silencing– induced NOTCH pathway activation at least in the cell lines TC252, WE68, and SK-N-MC, we tested EWS-FLI1 dependency of JAG1 promoter activity in reporter gene assays in these cell lines. Two genomic fragments spanning the JAG1 promoter region from −1.7 or −0.57 kb to the transcriptional start site were cloned to drive luciferase reporter activity from the vector pGL4. In all three cell lines, silencing of EWS-FLI1 resulted in a 2- to >4-fold induction of normalized firefly luciferase activity with both the −1.7-kb (data not shown) and the −0.57-kb JAG1 promoter fragment (Fig. 5C). These results suggested that the JAG1 promoter elements responsible for EWS-FLI1–mediated regulation localize to the first 574 bp upstream of the JAG1 transcription unit. Future studies will attempt to uncover the cis-acting elements within this region that mediate EWS-FLI1 repressive activities.
Lastly, we asked whether the induction of p53 by JAG1 depended on HEY1 expression in TC252 cells. Coexpression of siRNA to HEY1 with JAG1 reduced p53 protein to basal levels comparable with those obtained by inhibition of NOTCH receptor cleavage with GSI (Fig. 5D).
Together, these data are consistent with a model in which EWS-FLI1 silencing in these cells results in the induction of the NOTCH ligand JAG1, which stimulates NOTCH3 cleavage by γ secretase, leading to the transcriptional induction of HEY1. The consequence is nuclear p53 accumulation and subsequent activation of CDKN1A (Fig. 6).
Figure 6.
Model for p53 and p21WAF1/CIP1 regulation by EWS-FLI1 in TC252 cells. Suppression of EWS-FLI1 results in JAG1 expression by a thus far unknown mechanism. Presumably intercellular interaction between JAG1 and constitutively expressed NOTCH3 (and possibly also NOTCH2) results in γ-secretase–mediated NOTCH cleavage. The NICD translocates to the nucleus and activates CBF/CSL-dependent HEY1 expression. This leads to posttranscriptional stabilization of p53 via a thus far undefined mechanism, and p53-dependent p21WAF1/CIP1 expression. There is evidence that HEY1 (9) and CDKN1A (46) are directly bound by EWS-FLI1. However, evidence presented in this paper indicates that activation of HEY1 and CDKN1A is dependent on stimulation of NOTCH signaling. The NOTCH pathway modulators and components ectopically expressed to establish the sequence of events in this study are indicated at the top of the figure.
Discussion
This is the first report that describes a mechanism of p53 suppression involving the NOTCH pathway by a tumor-derived chimeric oncogene, EWS-FLI1 in ESFT. Thus far, all but one published EWS-FLI1 knockdown study had used mt-p53 ESFT cell lines, and therefore, the tumor suppressive role of the NOTCH/p53 pathway had previously escaped attention (20, 24, 27–29). Here, we show that NOTCH-mediated tumor suppression requires the presence of wt-p53. This finding might explain why in the mt-p53 ESFT cell lines RDES and SK-ES-1, NOTCH activation was reported to moderately promote tumor cell growth (30). This same study reported frequent HES1 expression in ESFT and interpreted it as a sign for constitutively active NOTCH signaling in these highly proliferative tumors. In contrast, we find NOTCH signaling to be suppressed in ESFT and reactivated only upon EWS-FLI1 silencing or NOTCH ligand expression. Although we confirm frequently elevated HES1 expression in primary ESFT and cell lines (Fig. 3A and B), HES1 expression did not change after EWS-FLI1 knockdown and NOTCH activation in all six ESFT cell lines tested (Fig. 3B; data not shown). Thus, NOTCH-independent pathways, as previously described (31–33), are likely responsible for HES1 expression in ESFT.
After EWS-FLI1 knockdown, we found HEY1 to be strongly induced in a NOTCH3 activation–dependent fashion in three of six ESFT cell lines, which in turn was a consequence of JAG1 induction. Here, NOTCH pathway activation was also confirmed by ACTA2 induction. These results suggest that in these cell lines, EWS-FLI1 modulates NOTCH signaling in ESFT by JAG1 suppression. We delineated the regulatory sequences responsible for JAG1 modulation to a 574-bp promoter fragment, which is devoid of a canonical ETS binding motif (data not shown). Also, EWS-FLI1 was found to bind to the JAG1 upstream region in regular chromatin immunoprecipitation (ChIP) and ChIP-chip experiments,3 suggesting that EWS-FLI1 might directly regulate JAG1 transcription. Further studies are needed to elucidate the exact mechanism of JAG1 regulation by EWS-FLI1.
HEY1 was also induced, but to a much lesser extent, when no increase in JAG1 expression was observed. Here, two of three cell lines showed an increase in NOTCH3 processing suggesting alternative mechanisms of NOTCH activation in these cells. Whether the expression of other NOTCH ligands or modulators of NOTCH are affected by EWS-FLI1 silencing in these cases has not been investigated. In only one of six cell lines (STA-ET-7.2) did EWS-FLI1 silencing not result in detectable NOTCH3 processing. Interestingly, this p53 mutant cell line showed the lowest HEY1 induction levels (10–100 times less than in JAG1-induced ESFT cell lines). This result may indicate the existence of other less effective mechanisms of HEY1 activation. In two cell lines (STA-ET-1 and VH64), low-level constitutive NOTCH3 cleavage was observed even in the absence of EWS-FLI1 silencing, which in STA-ET-1 cells was accompanied by detectable HEY1 expression. However, these levels seemed to be below the necessary threshold for p53 activation, which was a posteriori reached only upon EWS-FLI1 knockdown, eliciting an at least 15-fold HEY1 induction in the wt-p53 cell lines.
The NOTCH signaling pathway is an evolutionary conserved developmental mechanism mediating cell fate selection via lateral inhibition (34). Forced NOTCH-signaling prevents progenitors from undergoing neurogenesis in the vertebrate nervous system, whereas blocking this pathway leads to excessive neurogenesis and depletion of the progenitor pool. In contrast, in mouse and human embryonal stem cells, NOTCH was shown to promote neural fates, underscoring the importance of the cellular context (35). Similarly in cancer, NOTCH signaling can either promote or suppress tumor formation. NOTCH1 is constitutively activated by mutation in >50% of T-cell acute lymphoblastic leukemias. Other cancers, in which NOTCH acts as an activated oncogene are carcinomas of the cervix, colon, lung, pancreas, breast, and brain tumors (34). As NOTCH signaling can maintain cells in the proliferative and undifferentiated state, it is thought that its role in these cancers is to prevent neoplastic cells from responding to differentiation cues in their immediate environment.
In contrast, we found that activation of the NOTCH signaling pathway in ESFT has tumor suppressive-like effects, leading to nuclear accumulation of p53, CDKN1A activation, and cell cycle arrest. Similarly, NOTCH signaling is tumor suppressive in a large range of B-cell malignancies (36), in small-cell lung carcinoma (SCLC; ref. 37) and, interestingly, in prostate cancer (38), which also carries ETS-gene rearrangements in a high proportion of cases. Consistent with our findings in ESFT, re-expression of NOTCH1 or NICD1 in SCLC cell lines resulted in p21WAF1/CIP1 expression (37). It remains to be seen whether the low levels of p53 and p21 in these cancers are caused by an oncogenic suppression of the NOTCH pathway. Similarly, in normal keratinocytes, JAG1-mediated NOTCH1 signaling was shown to induce p21WAF1/CIP1 expression, causing proliferating cells to withdraw from the cell cycle and helping to initiate terminal differentiation (39, 40). One might thus speculate that in the enigmatic ESFT precursor cell, NOTCH-activated p21WAF1/CIP1 expression is a step toward differentiation that is blocked by the consequences of the EWS-FLI1 gene rearrangement, thereby initiating tumorigenesis. This hypothesis is supported by the fact that in the vast majority of ESFT, HEY1 is suppressed. It will be of interest to characterize those rare cases of ESFT where HEY1 is activated. Based on our hypothesis, we expect enrichment in this subgroup of tumors showing either a slow growing phenotype in combination with a wt-p53 allele, or of tumors carrying p53 mutations.
The increase in p53 levels observed upon knockdown of EWS-FLI1 by RNAi was lower than was achieved by ionizing radiation or ectopic p53 expression (data not shown). Consistent with a model suggesting p53 levels as one determinant of cellular fate (41), we hypothesize that the levels and activities obtained by EWS-FLI1 silencing are only sufficient to activate high-affinity p53 targets such as the CDKN1A promoter, whereas low-affinity promoters such as those of the cell death genes BAX and PIG3 were not induced (data not shown).
Although this result is consistent with the observation that the EWS-FLI1 downstream target HEY1 causes cell cycle arrest but not cell death downstream of silenced EWS-FLI1, apoptosis was observed in the wt-p53 cell line TC252 but not in mt-p53 STA-ET-7.2 cells upon EWS-FLI1 knockdown. However, cell death induction in response to EWS-FLI1 modulation may not be p53 dependent because previous studies have shown apoptosis in the mt-p53 ESFT cell lines A673 and SK-ES (24, 42), and lack of apoptosis but induction of senescence in a wt-p53 cell line (29).
EWS-FLI1 knockdown induced p53 Ser 15 phosphorylation, whereas HEY1 ectopic expression did not. This result suggests that additional EWS-FLI1 regulated mechanisms effect on p53. Candidates are the transforming growth factor β pathway and loss of EWS-FLI1–mediated telomerase expression, potentially resulting in a DNA damage signal (43, 44).
Speculating ESFT cells resume characteristics of their tissue of origin upon silencing of EWS-FLI1 expression, our data suggest that the enigmatic ESFT precursor cell is a resting cell expressing p21WAF1/CIP1 due to elevated basal p53 levels, and which expresses components of the NOTCH signaling pathway as part of its normal differentiation regulatory program. The result of the chromosomal translocation t(11;22)(q24;q12) would enable an escape from normal developmental cues and allow for sustained proliferation due to NOTCH pathway suppression. Because we have shown that ESFT generally express NOTCH2 and NOTCH3 to variable extents and that re-expression of JAG1 or DLL1 was sufficient to elicit a p53 response via NOTCH pathway stimulation, ESFT cells likely retain their capability to respond to environmental cues that stimulate NOTCH. Such signals are provided by osteoblasts in the bone, stromal cells in the bone marrow, and by vascular endothelial cells (45). The lung, bone, and bone marrow are the primary sites of ESFT metastasis. It is tempting to speculate, that homing of ESFT cells to such NOTCH ligand–expressing niches may restrict tumor growth via a p53-dependent mechanism, providing a possible explanation for tumor dormancy and late relapse, two challenging problems in ESFT. If so, a better understanding of EWS-FLI1, NOTCH, and p53 cross-talk might provide us with tools to better manage progression of this disease.
Acknowledgments
Grant support: Austrian Science Fund FWF (#P18046-B12), the Austrian government (GEN-AU-Ch.I.L.D., contract 200.136/1-VI/1/2005), and the European Commission (PROTHETS, contract LSHC-CT-2004-503036; H. Kovar).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
We thank Fan Yang, Paul Meltzer, and Marc Ladanyi for giving permission to refer to their unpublished EWS-FLI1 ChIP-chip data.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Fan Yang and Paul Meltzer, National Cancer Institute, Bethesda, MD; Marc Ladanyi, MSKCC New York, personal communications.
References
- 1.Kovar H. Context matters: the hen or egg problem in Ewing’s sarcoma. Semin Cancer Biol. 2005;15:189–96. doi: 10.1016/j.semcancer.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Kovar H, Auinger A, Jug G, et al. Narrow spectrum of infrequent p53 mutations and absence of MDM2 amplification in Ewing tumours. Oncogene. 1993;8:2683–90. [PubMed] [Google Scholar]
- 3.De Alava E, Antonescu CR, Panizo A, et al. Prognostic impact of P53 status in Ewing sarcoma. Cancer. 2000;89:783–92. [PubMed] [Google Scholar]
- 4.Kovar H, Jug G, Aryee DNT, et al. Among genes involved in the RB dependent cell cycle regulatory cascade, the p16 tumor suppressor gene is frequently lost in the Ewing family of tumors. Oncogene. 1997;15:2225–32. doi: 10.1038/sj.onc.1201397. [DOI] [PubMed] [Google Scholar]
- 5.Huang HY, Illei PB, Zhao Z, et al. Ewing sarcomas with p53 mutation or p16/p14ARF homozygous deletion: a highly lethal subset associated with poor chemoresponse. J Clin Oncol. 2005;23:548–58. doi: 10.1200/JCO.2005.02.081. [DOI] [PubMed] [Google Scholar]
- 6.Riggi N, Cironi L, Provero P, et al. Development of Ewing’s sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Res. 2005;65:11459–68. doi: 10.1158/0008-5472.CAN-05-1696. [DOI] [PubMed] [Google Scholar]
- 7.Lu X. p53: a heavily dictated dictator of life and death. Curr Opin Genet Dev. 2005;15:27–33. doi: 10.1016/j.gde.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Kovar H, Pospisilova S, Jug G, Printz D, Gadner H. Response of Ewing tumor cells to forced and activated p53 expression. Oncogene. 2003;22:3193–204. doi: 10.1038/sj.onc.1206391. [DOI] [PubMed] [Google Scholar]
- 9.Siligan C, Ban J, Bachmaier R, et al. EWS-FLI1 target genes recovered from Ewing’s sarcoma chromatin. Oncogene. 2005;24:2512–24. doi: 10.1038/sj.onc.1208455. [DOI] [PubMed] [Google Scholar]
- 10.Dohjima T, Sook LN, Li H, Ohno T, Rossi JJ. Small interfering RNAs expressed from a Pol III promoter suppress the EWS/Fli-1 transcript in an Ewing sarcoma cell line. Mol Ther. 2003;7:811–6. doi: 10.1016/s1525-0016(03)00101-1. [DOI] [PubMed] [Google Scholar]
- 11.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–3. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 12.Zamurovic N, Cappellen D, Rohner D, Susa M. Coordinated activation of notch, Wnt, and transforming growth factor-β signaling pathways in bone morphogenic protein 2-induced osteogenesis. Notch target gene Hey1 inhibits mineralization and Runx2 transcriptional activity. J Biol Chem. 2004;279:37704–15. doi: 10.1074/jbc.M403813200. [DOI] [PubMed] [Google Scholar]
- 13.Purow BW, Haque RM, Noel MW, et al. Expression of Notch-1 and its ligands, Δ-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 2005;65:2353–63. doi: 10.1158/0008-5472.CAN-04-1890. [DOI] [PubMed] [Google Scholar]
- 14.Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66:1517–25. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 15.Jaleco AC, Neves H, Hooijberg E, et al. Differential effects of Notch ligands Δ-1 and Jagged-1 in human lymphoid differentiation. J Exp Med. 2001;194:991–1002. doi: 10.1084/jem.194.7.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chin MT, Maemura K, Fukumoto S, et al. Cardiovascular basic helix loop helix factor 1, a novel transcriptional repressor expressed preferentially in the developing and adult cardiovascular system. J Biol Chem. 2000;275:6381–7. doi: 10.1074/jbc.275.9.6381. [DOI] [PubMed] [Google Scholar]
- 17.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson SR, Guiliano D, Presneau N, et al. A molecular map of mesenchymal tumors. Genome Biol. 2005;6:R76. doi: 10.1186/gb-2005-6-9-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaefer KL, Eisenacher M, Braun Y, et al. Microarray analysis of Ewing’s sarcoma family of tumours reveals characteristic gene expression signatures associated with metastasis and resistance to chemotherapy. Eur J Cancer. 2008;44:699–709. doi: 10.1016/j.ejca.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 20.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11:421–9. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 21.Su AI, Wiltshire T, Batalov S, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A Model Based Background Adjustment for Oligonucleotide Expression Arrays. J American Statistical Association. 2004;99:909–17. [Google Scholar]
- 23.Huang Q, Raya A, DeJesus P, et al. Identification of p53 regulators by genome-wide functional analysis. Proc Natl Acad Sci U S A. 2004;101:3456–61. doi: 10.1073/pnas.0308562100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prieur A, Tirode F, Cohen P, Delattre O. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol Cell Biol. 2004;24:7275–83. doi: 10.1128/MCB.24.16.7275-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier MM, Gessler M. Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem Biophys Res Commun. 2000;275:652–60. doi: 10.1006/bbrc.2000.3354. [DOI] [PubMed] [Google Scholar]
- 26.Noseda M, Fu Y, Niessen K, et al. Smooth Muscle α-actin is a direct target of Notch/CSL. Circ Res. 2006;98:1468–70. doi: 10.1161/01.RES.0000229683.81357.26. [DOI] [PubMed] [Google Scholar]
- 27.Carrillo J, Garcia-Aragoncillo E, Azorin D, et al. Cholecystokinin down-regulation by RNA interference impairs Ewing tumor growth. Clin Cancer Res. 2007;13:2429–40. doi: 10.1158/1078-0432.CCR-06-1762. [DOI] [PubMed] [Google Scholar]
- 28.Smith R, Owen LA, Trem DJ, et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing’s sarcoma. Cancer Cell. 2006;9:405–16. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 29.Matsunobu T, Tanaka K, Nakamura T, et al. The possible role of EWS-Fli1 in evasion of senescence in Ewing family tumors. Cancer Res. 2006;66:803–11. doi: 10.1158/0008-5472.CAN-05-1972. [DOI] [PubMed] [Google Scholar]
- 30.Baliko F, Bright T, Poon R, Cohen B, Egan SE, Alman BA. Inhibition of notch signaling induces neural differentiation in Ewing sarcoma. Am J Pathol. 2007;170:1686–94. doi: 10.2353/ajpath.2007.060971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curry CL, Reed LL, Nickoloff BJ, Miele L, Foreman KE. Notch-independent regulation of Hes-1 expression by c-Jun N-terminal kinase signaling in human endothelial cells. Lab Invest. 2006;86:842–52. doi: 10.1038/labinvest.3700442. [DOI] [PubMed] [Google Scholar]
- 32.Ingram WJ, McCue KI, Tran TH, Hallahan AR, Wainwright BJ. Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signaling. Oncogene. 2008;27:1489–500. doi: 10.1038/sj.onc.1210767. [DOI] [PubMed] [Google Scholar]
- 33.Katoh M, Katoh M. Integrative genomic analyses on HES/HEY family: Notch-independent HES1, HES3 transcription in undifferentiated ES cells, and Notch-dependent HES1, HES5, HEY1, HEY2, HEYL transcription in fetal tissues, adult tissues, or cancer. Int J Oncol. 2007;31:461–6. [PubMed] [Google Scholar]
- 34.Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer. 2003;3:756–67. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 35.Bray SJ. Notch signaling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–89. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 36.Zweidler-McKay PA, He Y, Xu L, et al. Notch signaling is a potent inducer of growth arrest and apoptosis in a wide range of B-cell malignancies. Blood. 2005;106:3898–906. doi: 10.1182/blood-2005-01-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sriuranpong V, Borges MW, Ravi RK, et al. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res. 2001;61:3200–5. [PubMed] [Google Scholar]
- 38.Wang XD, Leow CC, Zha J, et al. Notch signaling is required for normal prostatic epithelial cell proliferation and differentiation. Dev Biol. 2006;290:66–80. doi: 10.1016/j.ydbio.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 39.Rangarajan A, Talora C, Okuyama R, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20:3427–36. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nickoloff BJ, Qin JZ, Chaturvedi V, Denning MF, Bonish B, Miele L. Jagged-1 mediated activation of notch signaling induces complete maturation of human keratinocytes through NF-κB and PPARγ. Cell Death Differ. 2002;9:842–55. doi: 10.1038/sj.cdd.4401036. [DOI] [PubMed] [Google Scholar]
- 41.Chen X, Ko LJ, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;10:2438–51. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- 42.Chansky HA, Barahmand-Pour F, Mei Q, et al. Targeting of EWS/FLI-1 by RNA interference attenuates the tumor phenotype of Ewing’s sarcoma cells in vitro. J Orthop Res. 2004;22:910–7. doi: 10.1016/j.orthres.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 43.Cordenonsi M, Montagner M, Adorno M, et al. Integration of TGF-β and Ras/MAPK signaling through p53 phosphorylation. Science. 2007;315:840–3. doi: 10.1126/science.1135961. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi A, Higashino F, Aoyagi M, et al. EWS/ETS fusions activate telomerase in Ewing’s tumors. Cancer Res. 2003;63:8338–44. [PubMed] [Google Scholar]
- 45.Zhu J, Emerson SG. A new bone to pick: osteoblasts and the haematopoietic stem-cell niche. Bioessays. 2004;26:595–9. doi: 10.1002/bies.20052. [DOI] [PubMed] [Google Scholar]
- 46.Nakatani F, Tanaka K, Sakimura R, et al. Identification of p21WAF1/CIP1 as a direct target of EWS-Fli1 oncogenic fusion protein. JBiolChem. 2003;278:15105–15. doi: 10.1074/jbc.M211470200. [DOI] [PubMed] [Google Scholar]