Abstract
Aim
The aim of this work was to improve understanding about the mode, geography and tempo of diversification in deep‐sea organisms, using a time‐calibrated molecular phylogeny of the heterobranch gastropod genus Scaphander.
Location
Atlantic and Indo‐West Pacific (IWP) oceans.
Methods
Two mitochondrial gene markers (COI and 16S) and one nuclear ribosomal gene (28S) from six Atlantic species of Scaphander, and four IWP species were used to generate a multilocus phylogenetic hypothesis using uncorrelated relaxed‐clock Bayesian methods implemented in beast and calibrated with the first occurrence of Scaphander in the fossil record (58.7–55.8 Ma).
Results
Two main clades were supported: clade A, with sister relationships between species and subclades from the Atlantic and IWP; and clade B, with two western Atlantic sister species. Our estimates indicate that the two earliest divergences in clade A occurred between the middle Eocene and late Miocene and the most recent speciation occurred within the middle Miocene to Pleistocene. The divergence between the two western Atlantic species in clade B was estimated at late Oligocene–Pliocene.
Main conclusions
The prevailing mode of speciation in Scaphander was allopatric, but one possible case of sympatric speciation was detected between two western Atlantic species. Sister relationships between IWP and Atlantic lineages suggest the occurrence both of vicariance events caused by the closure of the Tethyan Seaway and of dispersal between the two ocean basins, probably around South Africa during episodic disruptions of the deep‐sea regional current system caused by glacial–interglacial cycles. Cladogenetic estimates do not support comparatively older diversification of deep‐sea faunas, but corroborate the hypothesis of a pulse of diversification centred in the Oligocene and Miocene epochs. Amphi‐Atlantic species were found to occur at deeper depths (bathyal–abyssal) and we hypothesize that trans‐Atlantic connectivity is maintained by dispersal between neighbouring reproductive populations inhabiting the abyssal sea floor and by dispersal across the shelf and slope of Arctic and sub‐Arctic regions.
Keywords: Allopatric speciation, amphi‐Atlantic, Cephalaspidea, dispersal, diversification pulse, Heterobranchia, marine biogeography, Mollusca, Tethyan vicariance
Introduction
Little is known about the biogeography and speciation mechanisms of deep‐sea (> 200 m deep) organisms (Thistle, 2003). This is in sharp contrast with shallow‐water faunas, where the advent of molecular phylogenetic methods during the last 20 years has led to a profusion of studies that improve our understanding of the origins of species and patterns of diversity (e.g. Williams & Reid, 2004; Williams & Duda, 2008; Malaquias & Reid, 2009; Bowen et al., 2013). Some factors commonly invoked to explain diversification in shallow water (e.g. sea‐level fluctuations, variation in water temperature) are unlikely to have the same impact in the deep sea, but a common period of diversification for both shallow and deep‐water faunas was recently suggested around the Oligocene and Miocene epochs, probably influenced by global warming, high levels of tectonic activity and changes in oceanic currents (Williams & Duda, 2008; Cabezas et al., 2012; Williams et al., 2013).
At a global scale, the effects of major tectonic events on the biogeography and diversification of shallow‐water marine faunas are well documented, and include the opening of the Drake Passage and establishment of the Antarctic Circumpolar Current (Beu et al., 1997), the closure of the Tethys Sea (Rögl, 1998), the opening of the Bering Strait (Marincovich & Gladenkov, 1999) and the uplift of the Isthmus of Panama (Lessios, 2008). Nevertheless, how these same events affected deep‐water fauna remains largely unknown.
The traditional view has been that deep‐sea species have large biogeographical ranges due to the apparent lack of barriers to dispersal and homogeneous environment (McClain & Hardy, 2010). The notion of the deep sea as a continuous and homogeneous ecosystem where diversity and biogeographical patterns are mostly controlled by biological interactions such as competition (the stability–time hypothesis; Sanders, 1968), has been challenged by the discoveries of chemosynthetic habitats [hydrothermal vents (Lonsdale, 1977), cold seeps (Paull et al., 1984) and whale‐falls (Smith et al., 1989)], seasonality (Billett et al., 1983) and deep‐sea storms (Hollister & McCave, 1984). Today, the deep sea is regarded as a patchy environment of great heterogeneity, created by disturbances at different scales, generating a mosaic of communities at different stages of succession (temporal mosaic hypothesis; Grassle & Sanders, 1973; Levin et al., 2001; Rex & Etter, 2010). These two hypotheses are mostly based on contemporaneous ecological factors, however, and do not take large‐scale temporal effects or an evolutionary perspective into account.
There are very few molecular phylogenies of benthic deep‐sea clades with complete (or even near‐complete) taxon sampling, and the few available works have a regional focus (e.g. Puillandre et al., 2010; Cabezas et al., 2012; Dueñas et al., 2014) or are concerned with the origins of major deep‐sea clades (genus‐level or higher; e.g. Strugnell et al., 2008; Lins et al., 2012; Williams et al., 2013; Corrigan et al., 2014). More attention has been paid to the genetic diversity of broadly distributed species, such as the bivalves Deminucula atacellana and Ledella ultima (Zardus et al., 2006; Etter et al., 2011) or the amphipod Eurythenes gryllus (Havermans et al., 2013). Differences in geographical ranges between organisms that inhabit the bathyal zone (c. 1000–3000 m) and those that inhabit the abyssal plains (c. 4000 m) have been recognized, and studies on deep‐sea bivalves and gastropods have shown that abyssal species have larger biogeographical ranges than bathyal species (Etter & Rex, 1990; Allen & Sanders, 1996; Etter et al., 2005, 2011; Zardus et al., 2006).
Despite the fact that evidence is slowly accumulating, biogeographical inferences on deep‐sea faunas are still hampered by three main factors.
Taxonomic impediment. Many deep‐sea species with broad geographical ranges have been identified based on similarities of their external morphology only (e.g. Allen & Sanders, 1996; Allen, 2008; Miljutin et al., 2010; Menzel et al., 2011), and there is growing recognition of cryptic speciation in the marine realm (Carmona et al., 2011; Claremont et al., 2011).
Sampling bias. It is acknowledged that very little of the deep sea has actually been sampled (less than 0.01%; Ramirez‐Llodra et al., 2010), and that sampling is uneven across ocean basins. For example, the South Atlantic is much less sampled than the North Atlantic (McClain & Hardy, 2010).
Sampling constraints. Deep‐sea sampling is expensive and the amount of material suitable for DNA analysis available in worldwide research institutions is very low. This is especially limiting for phylogenetic analyses where high levels of geographical and taxon coverage is desirable.
Scaphander is a globally distributed genus of predominantly deep‐sea cephalaspidean gastropods (Eilertsen & Malaquias, 2013a). Eight species of Scaphander are recognized in the Atlantic Ocean, and these are well supported based on shell morphology, internal anatomy (including characters of the male reproductive system) and reciprocal monophyly in a molecular phylogenetic tree (Eilertsen & Malaquias, 2013a). Because of its large, strong shell, Scaphander is relatively well documented in the fossil record. Palaeocene fossils are known from Europe, the USA and the east and west coasts of Argentina, with the oldest fossils that can confidently be identified as Scaphander dating from the late Palaeocene (Thanetian, 58.7–55.8 Ma) of Poland (Krach, 1963) and California (Weaver, 1949; Schoellhamer et al., 1981).
Our main goal is to contribute to a better understanding of the mode, causes and tempo of diversification of deep‐sea organisms, with an emphasis on the Atlantic Ocean and using Scaphander snails as our study group. We will produce a time‐calibrated molecular phylogeny to infer relationships and estimate the age of divergence between species. This phylogeny, together with knowledge about the geographical distributions of species, will be used to infer the prevalent geographical mode of speciation and test the hypothesis that the Oligocene and Miocene epochs correspond to a period of major diversification of deep‐sea fauna. We will test the prediction that, in deep‐sea groups, speciation between Atlantic and Indo‐Pacific lineages caused by the closure of the Tethyan Seaway is expected to pre‐date the diversification times of shallow‐water species. We will also assess whether species of Scaphander with deeper bathymetric ranges have broader geographical distributions, and then discuss the possible causes.
Materials and methods
Taxon sampling, geography and bathymetric distributions
Our set of specimens for molecular study was assembled from natural history museums and from fieldwork along the coast of Norway (Table 1). Geographical and bathymetric distributions of all Atlantic species of Scaphander were taken from Eilertsen & Malaquias (2013a). Only bathymetric records of live specimens were considered to be reliable, and therefore the bathymetric distribution of S. gracilis is not included here, because this species is only known from empty shells. Geographical and bathymetric distributions of Indo‐Pacific species were not plotted in detail because there is no comprehensive revision available for these species and they are not the focus of the present study.
Table 1.
List of specimens used for molecular phylogenetic analysis including sampling localities and voucher numbers. Sequences labelled with an asterisk (*) were generated for the present study, the remaining sequences were downloaded from GenBank. Abbreviations: MCZ, Museum of Comparative Zoology, Harvard University, Boston, MA, USA; MNHN, Muséum national d'Histoire naturelle, Paris, France; MZSP, Museu de Zoologia da Universidade de São Paulo, Brazil; NHMUK, Natural History Museum, London, UK; RMNH, National Museum of Natural History (Naturalis), Leiden, The Netherlands; USNM, National Museum of Natural History, Smithsonian Institution, Washington DC, USA; ZMBN, Natural History Collections, University Museum of Bergen, Norway
| Species | Specimen | Locality | Voucher number | COI | 16S | 28S |
|---|---|---|---|---|---|---|
| Scaphander lignarius | 1 | Cádiz, Spain | ZMBN 87998 | KC731431* | KC351524 | KC351543 |
| 2 | Bergen, Norway | ZMBN 87999 | KC351562 | KC351525 | KC351544 | |
| 19 | Barcelona, Spain | MCZ 371884 | KC351561 | KC351522 | ||
| 37 | Bergen, Norway | ZMBN 88000 | KC351563 | KC351526 | KC351545 | |
| 51 | Lofoten, Norway | ZMBN 88001 | KC351564 | KC351527 | KC731432* | |
| GB1 | Algarve, Portugal | NHMUK 20060325 | DQ974663 | DQ923454 | DQ927221 | |
| GB2 | Algarve, Portugal | NHMUK 20060114 | DQ974664 | DQ927212 | ||
| GB3 | Blanes, Spain | EED‐Phy‐442 | EF489372 | |||
| Scaphander punctostriatus | 3 | Bergen, Norway | ZMBN 88002 | KC351568 | KC351532 | KC351549 |
| 4 | Newfoundland, Canada | MNHN, Paris | KC351566 | KC351531 | KC351548 | |
| 34 | Lofoten, Norway | ZMBN 88006 | KC351571 | KC351536 | KC351553 | |
| 35 | Honningsvåg, Norway | ZMBN 88005 | KC351570 | KC351535 | KC351551 | |
| 36 | Skagerrak, Denmark | ZMBN 88004 | KC351569 | KC351534 | KC351552 | |
| 38 | Hauglandsosen, Norway | ZMBN 88003 | KC351567 | KC351533 | KC351550 | |
| Scaphander watsoni | 15 | Tampa, FL, USA | USNM 1151226 | KC351575 | KC351542 | KC351557 |
| 17 | New Orleans, LA, USA | USNM 1151240 | KC351576 | KC731433* | KC351558 | |
| Scaphander nobilis | 9 | Bay of Biscay, France | MNHN, Paris | KC351530 | ||
| Scaphander bathymophilus | 13 | Azores, Portugal | RMNH unnr. | KC351559 | KC351520 | |
| 52 | San Juan, Puerto Rico | MZSP 75708 | KC731430* | KC351519 | ||
| Scaphander darius | 21 | Guarapari, Brazil | MZSP 29016 | KC351560 | KC351521 | |
| Scaphander mundus | 29 | East of the Philippines | MNHN, IM‐2009‐4319 | KC351565 | KC351529 | KC351547 |
| 31 | East of the Philippines | MNHN, IM‐2009‐4318 | KC731429* | KC351528 | KC351546 | |
| Scaphander sp. A | 30 | Grand Passage, New Caledonia | MNHN, IM‐2009‐4317 | KC351572 | KC351537 | KC351554 |
| Scaphander sp. B | 32 | Grand Passage, New Caledonia | MNHN, IM‐2009‐4371 | KC351573 | KC351538 | KC351555 |
| Scaphander subglobosus | 33 | Bohol Sea, Philippines | MNHN, IM‐2009‐4339 | KC351574 | KC351539 | KC351556 |
| Sagaminopteron psychedelicum | Kalakajoro, Madagascar | Cas‐Cephas3 | DQ974667 | KJ022787 | DQ927225 |
Molecular phylogenetic analyses and estimation of divergence times
Sequences of the mitochondrial genes cytochrome oxidase c subunit I (COI) and 16S rRNA (16S) and the nuclear gene 28S rRNA (28S) of Scaphander were obtained from GenBank (previously generated by Eilertsen & Malaquias, 2013a) and some additional sequences were produced (Table 1). DNA was extracted, amplified and sequenced according to the protocol described by Eilertsen & Malaquias (2013a), except for some specimens where amplification of 28S failed. These were run again using LA Taq polymerase with GC buffer from TaKaRa (TaKaRa Bio, Otsu, Japan). The PCR reaction volume was 25 μL, comprising 5.35 μL ddH2O, 12.5 μL GC buffer, 4 μL dNTPs, 1 μL of each primer (10 μM concentration), 0.15 μL Taq and 1 μL DNA template. PCR thermal cycles were as follows: initial denaturation of 1 min at 94 °C, followed by 40 cycles with denaturation for 30 s at 94 °C, annealing for 30 s at 52 °C, and extension for 2 min at 72 °C. Final extension was 10 min at 72 °C. PCR products were cleaned and sequenced as described in Eilertsen & Malaquias (2013a).
sequencher 4.10.1 (Gene Codes, Ann Arbor, MI, USA) was used to assemble the forward and reverse strands and to assess the quality of the sequences, which were edited by careful examination of chromatograms. Sequences were checked for potential contamination using blast (Altschul et al., 1990) and have been deposited in GenBank (Table 1). The sequences were aligned using clustal X (Thompson et al., 1997) with a gap‐opening penalty of 60 and a gap‐extension penalty of 30. The single‐gene alignments were examined in bioedit (Hall, 1999), padded to equal the longest sequence and missing data at the ends were coded with question marks. Blocks of ambiguous data in the single‐gene alignments were identified and excluded using gblocks with relaxed settings (Talavera & Castresana, 2007; Kück et al., 2010; see Appendix S1 in Supporting Information). Pairwise uncorrected p‐distances were calculated using mega 5.2 (Tamura et al., 2011). The incongruence length difference test (ILD; Farris et al., 1995), implemented in paup* 4.0 b10 (Swofford, 2003) as the partition homogeneity test, was performed on the dataset with 100 replicates to test for incongruence between genetic markers. Saturation was tested for each gene and for the first, second and third codon positions of the COI gene, by plotting GTR pairwise distances against total substitutions (transitions + transversions). Substitution saturation analysis showed signs of saturation at the third codon position of COI, so two COI datasets were created: one including the third position (COI‐A) and one excluding it (COI‐B).
The best‐fitting models of evolution (see Appendix S2) were selected according to the Akaike information criterion (Akaike, 1974) implemented in mrmodeltest 2.3 (Nylander, 2008). For COI and 16S, the GTR+G+I model was the best model (Appendix S2), but because of statistical concerns regarding the coestimation of the gamma and invariant‐site parameters (discussed in the RaxML manual; Stamatakis, 2008) we chose to use GTR+G for these genes. Chronograms for each of the single‐gene datasets (COI, 16S and 28S) and for a concatenated dataset of all three genes with missing data coded as question marks were produced in beast 1.8.0 (Drummond et al., 2007). The analyses were set up in BEAUti 1.8.0 (Drummond et al., 2007) with the concatenated dataset partitioned by gene, using unlinked substitution models and clock models and linked tree priors. The ingroup (all species of Scaphander) was defined and set to be monophyletic. The sister lineage of Scaphander is not known and we therefore selected as an outgroup a representative of the family Gasteropteridae (Sagaminopteron psychedelicum), which has been shown to be closely related to Scaphandridae (Malaquias et al., 2009).
A relaxed, uncorrelated lognormal clock model was selected, and substitution rates were left to be estimated (Drummond et al., 2006). The tree model was chosen using Bayes factor (BF) calculations comparing the Yule speciation model (Gernhard, 2008) and the birth–death model with incomplete sampling (Stadler, 2009) based on single‐run marginal likelihoods obtained by stepping‐stone (SS) sampling. The result [2 ln(BF) = 12.2] strongly favoured the birth–death model with incomplete sampling (Kass & Raftery, 1995). The prior for the proportion of taxa sampled was modelled as a normal distribution with a mean of 0.45 and a standard deviation of 0.05. The time to the most recent common ancestor (TMRCA) for Scaphander was given a lognormal prior with an offset of 55.8 Ma and, a mean of 1.5 Myr and a standard deviation of 1 Myr (Ho & Phillips, 2009). These settings were based on the oldest reliable fossils of Scaphander, which date from the late Palaeocene (55.8–58.7 Ma; Weaver, 1949; Krach, 1963; Schoellhamer et al., 1981). A vaguely informative prior (exponential distribution, mean 0.1) was set for the relaxed clock rates, and the remaining priors were left at the default settings (Drummond et al., 2007). Three independent runs were carried out and each analysis was run for 20 million generations for the single‐gene datasets, and 50 million generations for the combined dataset, with sampling every 1000 generations. The log files were examined in tracer 1.5 (Rambaut & Drummond, 2009) to ensure convergence was reached and to determine the burn‐in. The outputs were combined in logcombiner and maximum clade credibility trees created in treeannotator (Drummond & Rambaut, 2007) with a burn‐in of 10%. The resulting trees were converted to graphics in figtree 1.4.0 (Rambaut, 2012) and final adjustments were made in adobe illustrator CS6 (Adobe Systems, San Jose, CA, USA).
Results
DNA sequence analyses
The present dataset includes six of eight valid Atlantic species (75% of recognized diversity; Eilertsen & Malaquias, 2013a), and four Indo‐Pacific species (40% of recognized diversity; Valdés, 2008; Rosenberg et al., 2012; for the complete specimen list see Table 1). Seven sequences from S. lignarius were downloaded from GenBank and included in the dataset. The gblocks analysis excluded 10 positions from the COI alignment with third codon positions excluded (COI‐B), 27 positions from the 16S alignment and 25 positions from the 28S alignment (all positions excluded were at the ends of the alignments; see Appendix S1 for settings). Uncorrected p‐distances for COI (COI‐A, complete alignment) ranged from 0.1% to 5.9% within Scaphander species and 10.8–19.7% between species; however, two specimens of S. lignarius from Spain (specs 1 and 19) showed unusually high divergence from conspecifics (9.6–10.1%). For 16S, uncorrected p‐distances ranged between 0–2% within Scaphander species and 0.4–10.1% between species. It is interesting to note that the two S. lignarius specimens from Spain (specimens 1 and 19) also showed high intraspecific divergence in 16S, along with one specimen from Portugal (specimen GB1), with 1–2% differences from conspecifics. This may indicate some phylogeographical structure in S. lignarius, but it would require a much larger dataset with better geographical coverage to test this hypothesis.
The ILD test (Farris et al., 1995) performed on the concatenated dataset showed no incongruence between the genetic markers (P ≫ 0.05). Because substitution saturation analysis showed signs of saturation in the third codon position of COI, the alignment excluding this position (COI‐B) was used in the phylogenetic analyses.
Phylogenetic hypothesis
The individual gene trees were not in conflict (see Appendix S3), and the topology of the concatenated tree is consistent with the first phylogenetic analysis of Scaphander (Eilertsen & Malaquias, 2013a), with all morphological species forming monophyletic groups (Fig. 1). Scaphander lignarius is basal to the other species with high support, but the remaining deep divergences are not resolved [PP (posterior probabilities) < 0.9]. We focus on the species relationships within the genus that are well supported: clade A, with S. subglobosus (West Pacific) sister to a group containing S. punctostriatus (Atlantic) + S. mundus (WP) + S. nobilis (A), and clade B consisting of two West Atlantic sister species: S. darius and S. watsoni (Fig. 1).
Figure 1.
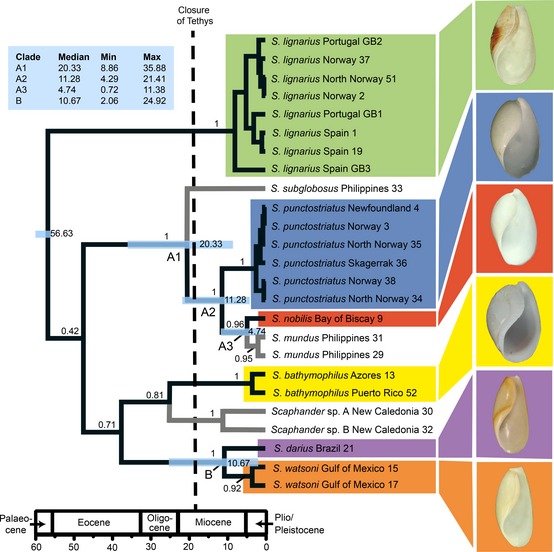
Chronogram produced by time‐calibrated Bayesian analysis of the concatenated three marker dataset (COI + 16S + 28S), using a relaxed molecular clock in BEAST). Branch labels show posterior probabilities, node labels show median ages of supported nodes and node bars represent 95% highest posterior density intervals (HPD). Median divergence times for nodes A1, A2, A3 and B in millions of years ago (Ma) are listed in the blue box with upper and lower limits of 95% HPD. The outgroup was pruned from the tree for clarity. The shells of the Atlantic species are illustrated (taken from Eilertsen & Malaquias, 2013a).
Divergence times and rates of evolution
The estimated age of divergence for node A1 representing the split between the West Pacific species S. subglobosus and clade A2, containing S. punctostriatus (amphi‐Atlantic), S. nobilis (amphi‐Atlantic), and S. mundus (IWP) is 20.33 Ma [highest posterior density (HPD) 35.88–8.86 Ma]. Node A2, the divergence between S. punctostriatus and the ancestral lineage of S. nobilis and S. mundus, is estimated at 11.28 Ma (HPD 21.41–4.29 Ma). The more recent split between S. nobilis and S. mundus (A3) is estimated at 4.74 Ma (HPD 11.38–0.72 Ma), and the divergence between the western Atlantic species S. darius and S. watsoni (clade B) is estimated to 13.16 Ma (HPD 27.02–2.4 Ma; see Fig. 1).
Average rates of substitution were highest for COI with 0.19% Myr−1, followed by 16S with 0.11% Myr−1 and 28S with 0.03% Myr−1. These rates are in line with those found for other gastropods under similar methodological approaches (Williams & Reid, 2004; Frey & Vermeij, 2008; Malaquias & Reid, 2009).
Geographical and bathymetric distributions
Two of the eight valid Atlantic species of Scaphander are amphi‐Atlantic (S. nobilis and S. punctostriatus), whereas one species, S. bathymophilus, is known from the western Atlantic and the mid‐Atlantic islands of the Azores (see Fig. 2). Scaphander punctostriatus and S. bathymophilus are represented in the molecular phylogeny by specimens from both east and west Atlantic (Table 1). For S. nobilis, only one specimen from the east Atlantic was suitable for molecular analysis, but specimens from the west Atlantic were studied and characters from the shell, digestive tract and reproductive system support that eastern and western populations are conspecific (Eilertsen & Malaquias, 2013a). Of the remaining Atlantic species, three are restricted to the western Atlantic (S. darius, S. watsoni and S. clavus), one is restricted to the eastern Atlantic (S. lignarius), and one is known only from the Azores (S. gracilis; Eilertsen & Malaquias, 2013a).
Figure 2.
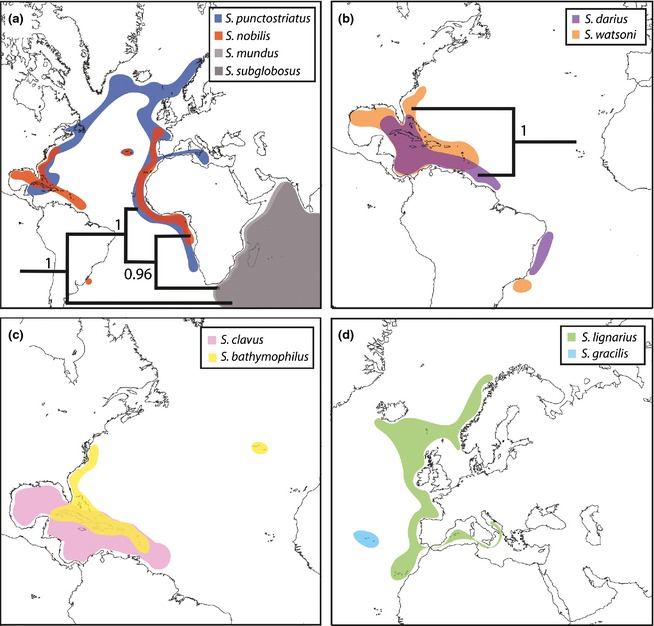
Geographical distributions and phylogenetic relationships of Atlantic Scaphander species. (a) clade A with species S. subglobosus, S. punctostriatus, S. mundus and S. nobilis; (b) clade B with species S. watsoni and S. darius; (c) S. clavus and S. bathymophilus; (d) S. lignarius and S. gracilis. Detailed maps and references to the literature surveyed for each species can be found in Eilertsen & Malaquias (2013a).
The bathymetric distributions of the Atlantic species of Scaphander are depicted in Fig. 3. The depth range of S. gracilis is not considered here because this species is only known from empty shells, which may be transported by currents or other animals. Three species are found on the continental shelf: S. darius has only been found between 16 m and 97 m, whereas the other two, S. watsoni and S. lignarius, occur at 110–476 m and 70–630 m, respectively. Two species have a mainly upper‐bathyal distribution (200–2000 m), namely S. clavus and S. punctostriatus found at 595–1056 m and 264–2730 m, respectively; but the latter also extends into the lower bathyal zone (2000–4000 m). Scaphander bathymophilus and S. nobilis have a wide bathyal to abyssal distribution with a bathymetric range of 805–5130 m and 1493–4255 m, respectively.
Figure 3.
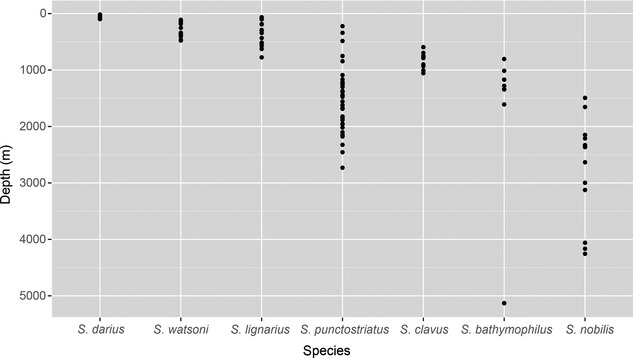
Depth distribution of Atlantic species of Scaphander, except S. gracilis, which is only known from shells. References to the literature surveyed for each species can be found in Eilertsen & Malaquias (2013a).
Discussion
Phylogeny, speciation and patterns of diversity in Scaphander
The monophyly of Scaphander has previously been confirmed by Eilertsen & Malaquias (2013a). That analysis showed that the Atlantic species‐group is not monophyletic, with several sister relationships between Atlantic and Indo‐Pacific lineages. Additionally, the systematics of the genus in the Atlantic was revised and species were delimited based on a combination of morpho‐anatomical characters and reciprocal monophyly of lineages rendered in a multilocus molecular phylogenetic analysis, which we have expanded here (Fig. 1). Eilertsen & Malaquias (2013a) found pronounced character displacement between the Atlantic species of Scaphander, particularly on reproductive structures, which, combined with shell shape, makes species identification reliable.
Only two Atlantic species of Scaphander were unavailable for molecular analysis: S. gracilis, known only from shells collected in the Azores; and S. clavus, from the west Atlantic (Eilertsen & Malaquias, 2013a). We acknowledge that incomplete taxon sampling can hamper inference of sister relationships and estimates of divergence times (Heath et al., 2008), but the present dataset remains one of the most complete datasets of a clade of deep‐sea invertebrates to have been assembled and analysed in a molecular phylogenetic context (but see Puillandre et al., 2010; Cabezas et al., 2012).
The combination of phylogenetic relationships and geographical distributions of species revealed little geographical overlap between sister taxa or clades (Fig. 2), which supports the prevailing view that allopatric speciation is dominant not only in shallow marine taxa (Meyer, 2003; Williams & Reid, 2004; Malaquias & Reid, 2009; Frey, 2010), but also in the deep sea. However, the overlap between the distributions of the western Atlantic sister species S. darius and S. watsoni, estimated to have diverged in the late Oligocene–Pliocene (median 10.67 Ma, HPD 24.92–2.06 Ma), hints at a possible case of sympatric speciation in Scaphander. Sympatric sister species are often characterized by ecological differentiation (e.g. Bolnick & Fitzpatrick, 2007; Frey, 2010), but no differences in bathymetric distribution, trophic ecology or habitat between these species were recognized, suggesting a similar ecological niche. We cannot, however, rule out the hypothesis of allopatric speciation followed by later geographical range shifts and secondary contact (Collin, 2003; Frey, 2010). Even though these two species are located on the same side of the Isthmus of Panama, speciation could be related to the uplift of the isthmus, which led to changes in current flow, salinity, temperature and primary production in the Atlantic and eastern Pacific, generating opportunities for transient allopatry and speciation (Williams & Reid, 2004; Lessios, 2008; Miura et al., 2010).
A pulse of diversification centred in the Oligocene and Miocene epochs has been suggested for shallow‐water organisms (e.g. Williams & Duda, 2008; Malaquias & Reid, 2009), and this has been also hypothesized more recently for deep‐sea fauna (Cabezas et al., 2012; Williams et al., 2013). An Oligocene/Miocene pulse of diversification is corroborated by the present results, in which median divergence time estimates of speciation events ranged between 26.64 and 7.38 Ma (Fig. 1).
There is a cline in diversity between the bathyal and abyssal zones with all but one Scaphander species (S. darius) present in the former bathymetric zone and only two (S. nobilis and S. bathymophilus) extending their range into the abyssal plains (> 4000 m; Fig. 3).
Tethyan vicariance and deep‐sea dispersal across oceans
The presence of sister relationships between species and clades from the Atlantic and IWP dating from the Eocene–Miocene (nodes A1 and A2, with two Atlantic and two IWP species; Fig. 1) may suggest vicariance associated with the closure of the Tethyan Seaway in the early Miocene (c. 18–19 Ma; Rögl, 1998). Diversification events that resulted from Tethyan vicariance have been widely documented for shallow‐water organisms, and both molecular estimates and the fossil record indicate that differentiation between the biogeographical regions of the proto‐Mediterranean and the proto‐IWP region was already present in the Oligocene (e.g. Williams & Reid, 2004; Harzhauser et al., 2007; Frey & Vermeij, 2008; Malaquias & Reid, 2009; Cowman & Bellwood, 2013).
It is to be expected that deep‐sea organisms may have been affected earlier by the closure of the Tethyan Seaway than organisms associated with shallow habitats, but it is not clear if this is the case in Scaphander. The age estimates for the cladogenetic events spanning the Tethys closure are equivocal (nodes A1 and A2 in Fig. 1; A1, median 20.33 Ma, HPD 35.88–8.86 Ma; A2, median 11.28 Ma, HPD 21.41–4.29 Ma) and speciation could therefore have been driven either by Tethyan vicariance or by post‐Tethyan dispersal between the Atlantic and the Indo‐Pacific. On the other hand, the split between the Indo‐West Pacific S. mundus and the Atlantic S. nobilis (median 4.74 Ma, HPD 11.38–0.72 Ma), seems too recent to be associated with Tethyan vicariance, suggesting dispersal between the two ocean basins followed by subsequent isolation and allopatric speciation.
The shortest route of dispersal between the present‐day distributions of these two species is around the Cape of Good Hope (Fig. 2a). The southern tip of South Africa is a well‐known biogeographical barrier for shallow‐water tropical/temperate taxa because of the Benguela cold‐water system, which became established during the late Miocene (Siesser, 1980; Marlow et al., 2000; Teske et al., 2011). Nevertheless, the fossil record and molecular phylogenetics studies have documented successful dispersal and speciation events from the IWP to the Atlantic around South Africa in shallow‐water taxa after the establishment of the Benguela current system during the Plio‐Pleistocene (Vermeij & Rosenberg, 1993; Vermeij & Snyder, 2003; Rocha et al., 2005; Levy et al., 2013). These dispersal events have been attributed to changes in climate associated with the Plio‐Pleistocene glacial–interglacial cycles, which prompted modifications in ocean currents and water temperature (Vermeij & Rosenberg, 1993; Rocha et al., 2005).
The impact of these processes on the diversification of deep‐sea faunas is poorly understood, because of a general lack of knowledge about the distribution of deep‐sea organisms and the paucity of available phylogenetic hypotheses. Nevertheless, the lack of Scaphander species shared between the Atlantic and Indo‐Pacific realms suggests that barriers to dispersal are also present in the deep sea in this region. Evidence from deep‐sea foraminiferan assemblages and geochemistry suggest that the influx of fresh water caused by melting ice‐caps during the Plio‐Pleistocene strongly disrupted the deep‐sea current system in the area (e.g. Schnitker, 1974; Hodell et al., 1985; Gupta & Srinivasan, 1990), and this could have created opportunities for deep‐sea organisms to disperse.
Alternative dispersal routes between the Atlantic and the Indo‐Pacific are the Drake Passage in the southern Atlantic (opened in the late Oligocene; Beu et al., 1997), the Central American corridor before the final closure of the Isthmus of Panama (by the middle Miocene, there was a corridor about 2000 m deep; Coates & Obando, 1996; Lessios, 2008), and the trans‐Arctic route during the Pliocene (Marincovich & Gladenkov, 1999). However, these would have implied dramatic range shifts (see Eilertsen & Malaquias, 2013a; Fig. 2) or regional extinctions (but Palaeocene fossils are known from California; Weaver, 1949; Schoellhamer et al., 1981). Dispersal across the Eastern Pacific Barrier is known to be insurmountable for the majority of marine invertebrates (Williams & Reid, 2004; Frey, 2010) and further evidence, including a better representation of the diversity of Indo‐Pacific species, is necessary to further test these alternative hypotheses.
Trans‐Atlantic dispersal via abyssal plains and along Arctic slopes
Two species of Scaphander are amphi‐Atlantic (S. nobilis and S. punctostriatus) and a third is known from the western Atlantic and the mid‐Atlantic islands of the Azores (S. bathymophilus; see Fig. 2). The remaining five Atlantic Scaphander species (S. lignarius, S. darius, S. watsoni, S. clavus and S. gracilis) have more limited distributions, restricted to one side of the Atlantic or to the Azores (Fig. 2). This raises the question of how connectivity is maintained across these long distances. Nothing is known about the reproduction and larval development of Scaphander, but the life‐span of heterobranch planktotrophic larvae is usually 15–42 days (Schaefer, 1996). The journey across the Atlantic by larval drift, following one of the main trans‐Atlantic surface currents, is estimated to take 60–400 days (Scheltema, 1971). Deep‐sea currents are slow (2–20 cm s−1; Heezen et al., 1966) compared to surface currents (trans‐Atlantic surface currents 14–90 cm s−1; Scheltema, 1971), and deep‐sea currents do not usually reach higher velocities than surface currents even during benthic storms (15–40 cm s−1; Hollister & McCave, 1984; Hollister, 1993). This indicates that unless Scaphander has much greater than average dispersal potential, trans‐Atlantic dispersal is unlikely to maintain connectivity between the eastern and western populations.
Comparing geographical and bathymetric ranges, it becomes evident that the three broadly distributed species are those with deeper bathymetric distributions (Fig. 3), which supports the traditional view that deeper species have wider geographical ranges (McClain & Hardy, 2010). Two of these species (S. bathymophilus and S. nobilis) extend their bathymetric range into abyssal depths below 4000 m (Fig. 3). We speculate that if these species have reproducing populations scattered over the abyssal plains, gene flow could then be maintained without requiring long‐distance dispersal of larvae, but instead by dispersal between populations. There is unfortunately a scarcity of data from these regions, because deep‐sea sampling has been mostly focused on coastal areas, areas around islands, hydrothermal vents or other chemosynthetic habitats, whereas the open‐ocean abyssal plains have hardly been sampled (Ramirez‐Llodra et al., 2010).
The ‘source–sink’ hypothesis of Rex et al. (2005) suggests that abyssal populations are non‐reproducing and require larval supply from bathyal populations. The main argument for this hypothesis is the decline in nutrient input with increasing distance from the continental slope, but a patchy distribution of areas with sufficient food supply, as predicted by the ‘temporal mosaic’ hypothesis (Grassle & Sanders, 1973; Rex & Etter, 2010), could enable a dynamic mosaic of reproductive populations of Scaphander. In fact, food availability is unlikely to be a limiting factor in Scaphander, because these snails feed almost exclusively upon foraminiferans (Eilertsen & Malaquias, 2013b), which are among the most abundant inhabitants of the abyssal sea floor (Gooday et al., 1992).
An interesting case is the distribution of S. bathymophilus, which occurs at depths of 805–5130 m in the Caribbean Sea, along the coast of the US north to Cape Hatteras (North Carolina), and also in the Azores (Eilertsen & Malaquias, 2013a; Figs 2 & 3). The broad longitudinal distribution of this species, spanning the width of the western Atlantic basin can be explained analogously to S. nobilis, but its isolation in the western Atlantic could be a consequence of a barrier effect caused by the Mid‐Atlantic Ridge hindering eastward dispersal in this species. The role of the Mid‐Atlantic Ridge in the dispersal of bathyal and abyssal organisms across the Atlantic is, however, poorly understood (Mullineaux et al., 2002; Zardus et al., 2006; Etter et al., 2011).
Scaphander punctostriatus, the most widely distributed Scaphander species, has only been recorded down to 2730 m (Fig. 3), albeit at Arctic and sub‐Arctic latitudes, where the distance between the eastern and western margins is shorter, and the coasts of Iceland and Greenland could provide ‘staging‐posts’ for dispersal. In fact the percentage of amphi‐Atlantic species at Arctic and boreal latitudes has been documented to be higher than at temperate and tropical latitudes (Vermeij, 2005; García & Bertsch, 2009).
Conclusions
The sister relationships between Atlantic and IWP lineages dating from the middle Eocene to late Miocene suggest vicariance events caused by the closure of the Tethyan Seaway, but they do not support a comparatively older diversification of deep‐sea faunas. However, our results corroborate the hypothesis of a pulse of diversification centred in the Oligocene and Miocene epochs. A post‐Tethyan divergence between Atlantic and IWP species is hypothesized to result from dispersal around South Africa during episodic disruptions of the deep‐sea regional current system caused by glacial–interglacial cycles. Allopatric speciation was prevalent, but one potential case of sympatric speciation was detected between two western Atlantic species. Amphi‐Atlantic species have comparatively deeper distributions (inhabiting bathyal‐abyssal depths), and we hypothesize that trans‐Atlantic dispersal is attained by connectivity between reproductive populations inhabiting the abyssal sea floor and by dispersal across the shelf and slope of Artic and sub‐Arctic regions.
Biosketches
Mari H. Eilertsen is a PhD student at the Marine Biodiversity Research Group at the University of Bergen, Norway. Her main research interests are the biogeography and speciation of marine invertebrates in the deep sea. In addition to gastropods, her study groups include ampharetid polychaetes and calcareous sponges.
Manuel António E. Malaquias is an associate professor in invertebrate systematics at the Department of Natural History, University Museum of Bergen, University of Bergen, Norway. His main research interests are the diversity, systematics and phylogeny of cephalaspidean gastropods and the study of speciation and biogeography in the marine realm.
Supporting information
Appendix S1 gblocks settings.
Appendix S2 Best‐fit model and estimated parameters for phylogenetic analysis.
Appendix S3 Single gene chronograms.
Acknowledgements
We are very grateful to A. Baldinger, G. Giribet and P. Benson (Museum of Comparative Zoology, Harvard University, Cambridge, MA), E. Strong, R. Hershler and T. Nickens (National Museum of Natural History, Smithsonian Institution, Washington, DC), A. Salvador, H. Taylor, K. Way and J. Ablett (Natural History Museum, London), P. Bouchet (Muséum national d'Histoire naturelle, Paris), C. Mangenta and L. Simone (Museu de Zoologia, University of São Paulo, Brazil), J. Pohle (Atlantic Reference Centre, Canada), J. Goud (National Museum of Natural History Naturalis, Leiden, The Netherlands), J. Slapcinsky (Florida Museum of Natural History, Gainesville, FL), and A. Warén (Naturhistoriska Riksmuseet, Stockholm, Sweden) for their help with specimens and images of type material.
At the University of Bergen, we thank E. Willassen for discussions on phylogenetic analyses, K. Kongshavn for help with image editing and production of distribution maps, J. Kongsrud and L. Ohnheiser for facilitating access to specimens, E. Erichsen for help with SEM, and L. Lindblom and K. Meland for help with molecular work, which was conducted in the Biodiversity Laboratories, University of Bergen. We also want to thank three anonymous referees for their constructive feedback.
This work was funded by in part by Artsdatabanken (project number: 56–10, prosjekt 70184219) and a student grant from the Meltzer Foundation awarded to the first author, and benefited from material collected by the Norwegian National programme MAREANO. Additionally this research received support from the SYNTHESYS Project http://www.synthesys.info/, which is financed by European Community Research Infrastructure Action under the FP7 “Capacities” Program.
References
- Akaike, H. (1974) A new look at the statistical model identifications. IEEE Transactions on Automatic Control, 19, 716–723. [Google Scholar]
- Allen, J.A. (2008) Bivalvia of the deep Atlantic. Malacologia, 50, 57–173. [Google Scholar]
- Allen, J.A. & Sanders, H.L. (1996) The zoogeography, diversity and origin of the deep‐sea protobranch bivalves of the Atlantic: the epilogue. Progress in Oceanography, 38, 95–153. [Google Scholar]
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. & Lipman, D.J. (1990) Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Beu, A.G. , Griffin, M. & Maxwell, P.A. (1997) Opening of Drake Passage gateway and Late Miocene to Pleistocene cooling reflected in Southern Ocean molluscan dispersal: evidence from New Zealand and Argentina. Tectonophysics, 281, 83–97. [Google Scholar]
- Billett, D.S.M. , Lampitt, R.S. , Rice, A.L. & Mantoura, R.F.C. (1983) Seasonal sedimentation of phytoplankton to the deep‐sea benthos. Nature, 302, 520–522. [Google Scholar]
- Bolnick, D.I. & Fitzpatrick, B.M. (2007) Sympatric speciation: models and empirical evidence. Annual Review of Ecology, Evolution, and Systematics, 38, 459–487. [Google Scholar]
- Bowen, B.W. , Rocha, L.A. , Toonen, R.J. & Karl, S.A. (2013) The origins of tropical marine biodiversity. Trends in Ecology and Evolution, 28, 359–366. [DOI] [PubMed] [Google Scholar]
- Cabezas, P. , Sanmartín, I. , Paulay, G. , Macpherson, E. & Machordom, A. (2012) Deep under the sea: unraveling the evolutionary history of the deep‐sea squat lobster Paramunida (Decapoda, Munididae). Evolution, 66, 1878–1896. [DOI] [PubMed] [Google Scholar]
- Carmona, L. , Malaquias, M.A.E. , Gosliner, T.M. , Pola, M. & Cervera, J.L. (2011) Amphi‐Atlantic distributions and cryptic species in Sacoglossan sea slugs. Journal of Molluscan Studies, 77, 401–412. [Google Scholar]
- Claremont, M. , Williams, S.T. , Barraclough, T.G. & Reid, D.G. (2011) The geographic scale of speciation in a marine snail with high dispersal potential. Journal of Biogeography, 38, 1016–1032. [Google Scholar]
- Coates, A.G. & Obando, J.A. (1996) The geologic evolution of the Central American Isthmus Evolution and environment in tropical America (ed. by Jackson J.B.C., Budd A.F. and Coates A.G.), pp. 21–56. University of Chicago Press, Chicago. [Google Scholar]
- Collin, R. (2003) Phylogenetic relationships among calyptraeid gastropods and their implications for the biogeography of marine speciation. Systematic Biology, 52, 618–640. [DOI] [PubMed] [Google Scholar]
- Corrigan, L.J. , Horton, T. , Fotherby, H. , White, T.A. & Hoelzel, A.R. (2014) Adaptive evolution of deep‐sea amphipods from the superfamily Lysiassanoidea in the North Atlantic. Evolutionary Biology, 41, 154–165. [Google Scholar]
- Cowman, P.F. & Bellwood, R. (2013) Vicariance across major marine biogeographic barriers: temporal concordance and the relative intensity of hard versus soft barriers. Proceedings of the Royal Society B: Biological Sciences, 280, 2013–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond, A.J. & Rambaut, A. (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond, A.J. , Ho, S.Y.W. , Phillips, M.J. & Rambaut, A. (2006) Relaxed phylogenetics and dating with confidence. PLoS Biology, 4, 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond, A.J. , Ho, S.Y.W. , Rawlence, N. & Rambaut, A. (2007) A rough guide to BEAST 1.4. University of Auckland, New Zealand. [Google Scholar]
- Dueñas, L.F. , Alderslade, P. & Sánchez, J.A. (2014) Molecular systematics of the deep‐sea bamboo corals (Octocorallia: Isididae: Keratoisidinae) from New Zealand with descriptions of two new species of Keratoisis . Molecular Phylogenetics and Evolution, 74, 15–28. [DOI] [PubMed] [Google Scholar]
- Eilertsen, M.H. & Malaquias, M.A.E. (2013a) Systematic revision of the genus Scaphander (Gastropoda, Cephalaspidea) in the Atlantic Ocean with a molecular phylogenetic hypothesis. Zoological Journal of the Linnean Society, 167, 389–429. [Google Scholar]
- Eilertsen, M.H. & Malaquias, M.A.E. (2013b) Unique digestive system, trophic specialization, and diversification in the deep‐sea gastropod genus Scaphander . Biological Journal of the Linnean Society, 109, 512–525. [Google Scholar]
- Etter, R.J. & Rex, M.A. (1990) Population differentiation decreases with depth in deep‐sea gastropods. Deep Sea Research Part A: Oceanographic Research Papers, 37, 1251–1261. [Google Scholar]
- Etter, R.J. , Rex, M.A. , Chase, M.R. & Quattro, J.M. (2005) Population differentiation decreases with depth in deep‐sea bivalves. Evolution, 59, 1479–1491. [PubMed] [Google Scholar]
- Etter, R.J. , Boyle, E.E. , Glazier, A. , Jennings, R.M. , Dutra, E. & Chase, M.R. (2011) Phylogeography of a pan‐Atlantic abyssal protobranch bivalve: implications for evolution in the deep Atlantic. Molecular Ecology, 20, 829–843. [DOI] [PubMed] [Google Scholar]
- Farris, J.S. , Källesjö, M. , Kluge, A.G. & Bult, C. (1995) Testing significance of incongruence. Cladistics, 10, 315–319. [Google Scholar]
- Frey, M.A. (2010) The relative importance of geography and ecology in species diversification: evidence from a tropical marine intertidal snail (Nerita). Journal of Biogeography, 37, 1515–1528. [Google Scholar]
- Frey, M.A. & Vermeij, G.J. (2008) Molecular phylogenies and historical biogeography of a circumtropical group of gastropods (Genus: Nerita): implications for regional diversity patterns in the marine tropics. Molecular Phylogenetics and Evolution, 48, 1067–1086. [DOI] [PubMed] [Google Scholar]
- García, F.J. & Bertsch, H. (2009) Diversity and distribution of the Gastropoda Opisthobranchia from the Atlantic Ocean: a global biogeographic approach. Scientia Marina, 73, 153–160. [Google Scholar]
- Gernhard, T. (2008) The conditioned reconstructed process. Journal of Theoretical Biology, 253, 769–778. [DOI] [PubMed] [Google Scholar]
- Gooday, A.J. , Levin, L.A. , Linke, P. & Heeger, T. (1992) The role of benthic foraminifera in deep‐sea food webs and carbon cycling Deep‐sea food chains and the global carbon cycle (ed. by Rowe G.T. and Pariente V.), pp. 63–91. Kluwer Academic Publishers, Dordrecht. [Google Scholar]
- Grassle, J.F. & Sanders, H.L. (1973) Life histories and the role of disturbance. Deep Sea Research and Oceanographic Abstracts, 20, 643–659. [Google Scholar]
- Gupta, A.K. & Srinivasan, M.S. (1990) Response of Northern Indian Ocean deep‐sea benthic foraminifera to global climates during Pliocene‐Pleistocene. Marine Micropaleontology, 16, 77–91. [Google Scholar]
- Hall, T.A. (1999) BioEdit: a user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98. [Google Scholar]
- Harzhauser, M. , Kroh, A. , Mandic, O. , Piller, W.E. , Göhlich, U. , Reuter, M. & Berning, B. (2007) Biogeographic responses to geodynamics: a key study all around the Oligo‐Miocene Tethyan seaway. Zoologischer Anzeiger, 246, 241–256. [Google Scholar]
- Havermans, C. , Sonet, G. , d'Udekem d'Acoz, C. , Nagy, Z.T. , Martin, P. , Brix, S. , Riehl, T. , Agrawal, S. & Held, C. (2013) Genetic and morphological divergences in the cosmopolitan deep‐sea amphipod Eurythenes gryllus reveal a diverse abyss and a bipolar species. PLoS ONE, 8, e74218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath, T.A. , Zwickl, D.J. , Kim, J. & Hillis, D.M. (2008) Taxon sampling affects inferences of macroevolutionary processes from phylogenetic trees. Systematics Biology, 57, 160–166. [DOI] [PubMed] [Google Scholar]
- Heezen, B.C. , Hollister, C.D. & Ruddiman, W.F. (1966) Shaping of the continental rise by deep geostrophic contour currents. Science, 152, 502–508. [DOI] [PubMed] [Google Scholar]
- Ho, S.Y.W. & Phillips, M.J. (2009) Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Systematic Biology, 58, 367–380. [DOI] [PubMed] [Google Scholar]
- Hodell, D.A. , Williams, D.F. & Kennett, J.P. (1985) Late Pliocene reorganization of deep vertical water‐mass structure in the western south Atlantic: faunal and isotopic evidence. Geological Society of America Bulletin, 96, 495–503. [Google Scholar]
- Hollister, C.D. (1993) The concept of deep‐sea contourites. Sedimentary Geology, 82, 5–11. [Google Scholar]
- Hollister, C.D. & McCave, I.N. (1984) Sedimentations under deep‐sea storms. Nature, 309, 220–225. [Google Scholar]
- Kass, R.E. & Raftery, A.E. (1995) Bayes factors. Journal of the American Statistical Association, 430, 773–795. [Google Scholar]
- Krach, W. (1963) Mollusca of the Babica Clays (Paleocene) of the Middle Carpathians. Studia Geologica Polonica, 14, 1–151. [Google Scholar]
- Kück, P. , Meusemann, K. , Dambach, J. , Thormann, B. , von Reumont, B.M. , Wägele, J.W. & Misof, B. (2010) Parametric and non‐parametric masking of randomness in sequence alignments can be improved and leads to better resolved trees. Frontiers in Zoology, 7, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessios, H.A. (2008) The Great American Schism: divergence of marine organisms after the rise of the Central American Isthmus. Annual Review of Ecology, Evolution, and Systematics, 39, 63–91. [Google Scholar]
- Levin, L.A. , Etter, R.J. , Rex, M.A. , Gooday, A.J. , Smith, C.R. , Pineda, J. , Stuart, C.T. , Hessler, R.R. & Pawson, D. (2001) Environmental influences on regional deep‐sea species diversity. Annual Review of Ecology and Systematics, 32, 51–93. [Google Scholar]
- Levy, A. , von der Heyden, S. , Floeter, S.R. , Bernardi, G. & Almada, V.C. (2013) Phylogeny of Parablennius Miranda Ribeiro, 1915 reveals a paraphyletic genus and recent Indo‐Pacific diversification from an Atlantic ancestor. Molecular Phylogenetics and Evolution, 67, 1–8. [DOI] [PubMed] [Google Scholar]
- Lins, L.S.F. , Ho, S.Y.W. , Wilson, G.D.F. & Lo, N. (2012) Evidence for Permo‐Triassic colonization of the deep sea by isopods. Biology Letters, 8, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale, P. (1977) Clustering of suspension‐feeding macrobenthos near abyssal hydrothermal vents at oceanic spreading centers. Deep‐Sea Research, 24, 857–863. [Google Scholar]
- Malaquias, M.A.E. & Reid, D.G. (2009) Tethyan vicariance, relictualism and speciation: evidence from a global molecular phylogeny of the opisthobranch genus Bulla . Journal of Biogeography, 36, 1760–1777. [Google Scholar]
- Malaquias, M.A.E. , Mackenzie‐Dodds, J. , Bouchet, P. , Gosliner, T. & Reid, D.G. (2009) A molecular phylogeny of the Cephalaspidea sensu lato (Gastropoda: Euthyneura): Architectibranchia redefined and Runcinacea reinstated. Zoologica Scripta, 38, 23–41. [Google Scholar]
- Marincovich, L. & Gladenkov, A.Y. (1999) Evidence for an early opening of the Bering Strait. Nature, 397, 149–151. [Google Scholar]
- Marlow, J.R. , Lange, C.B. , Wefer, G. & Rosell‐Melé, A. (2000) Upwelling intensification as part of the Pliocene‐Pleistocene climate transition. Science, 290, 2288–2291. [DOI] [PubMed] [Google Scholar]
- McClain, C.R. & Hardy, S.M. (2010) The dynamics of biogeographic ranges in the deep sea. Proceedings of the Royal Society B: Biological Sciences, 277, 3533–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel, L. , George, K.H. & Martínez Arbizu, P. (2011) Submarine ridges do not prevent large‐scale dispersal of abyssal fauna: a case study of Mesocletodes (Crustacea, Copepoda, Harpacticoida). Deep‐Sea Research Part I: Oceanographic Research Papers, 58, 839–864. [Google Scholar]
- Meyer, C.P. (2003) Molecular systematics of cowries (Gastropoda: Cypraeidae) and diversification patterns in the tropics. Biological Journal of the Linnean Society, 79, 401–459. [Google Scholar]
- Miljutin, D.M. , Gad, G. , Miljutina, M.M. , Mokievsky, V.O. , Fonseca‐Genevois, V. & Esteves, A.M. (2010) The state of knowledge on deep‐sea nematode taxonomy: how many valid species are known down there? Marine Biodiversity, 40, 143–159. [Google Scholar]
- Miura, O. , Torchin, M.E. & Bermingham, E. (2010) Molecular phylogenetics reveals differential divergence of coastal snails separated by the Isthmus of Panama. Molecular Phylogenetics and Evolution, 56, 40–48. [DOI] [PubMed] [Google Scholar]
- Mullineaux, L.S. , Speer, K.G. , Thurnherr, A.M. , Maltrud, M.E. & Vangriesheim, A. (2002) Implications of cross‐axis flow for larval dispersal along mid‐ocean ridges. Cahiers de Biologie Marine, 43, 281–284. [Google Scholar]
- Nylander, J.A.A . (2008) MrModeltest v2.3. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- Paull, C.K. , Hecker, B. , Commeau, R. , Freeman‐Lynde, R.P. , Neumann, C. , Corso, W.P. , Golubic, S. , Hook, J.E. , Sikes, E. & Curray, J. (1984) Biological communities at the Florida escarpment resemble hydrothermal vent taxa. Science, 226, 965–967. [DOI] [PubMed] [Google Scholar]
- Puillandre, N. , Sysoev, A.V. , Olivera, B.M. , Couloux, A. & Bouchet, P. (2010) Loss of planktotrophy and speciation: geographical fragmentation in the deep‐water gastropod genus Bathytoma (Gastropoda, Conoidea) in the western Pacific. Systematics and Biodiversity, 8, 371–394. [Google Scholar]
- Rambaut, A. (2012) FigTree. Version 1.4.0. University of Edinburgh, Edinburgh, UK: Available at: http://tree.bio.ed.ac.uk/software/figtree (last accessed May 2014). [Google Scholar]
- Rambaut, A. & Drummond, A.J. (2009) Tracer. MCMC trace analysis tool. Version 1.5. University of Edinburgh, Edinburgh, UK: Available at: http://beast.bio.ed.ac.uk/Tracer (last accessed May 2014). [Google Scholar]
- Ramirez‐Llodra, E. , Brandt, A. , Danovaro, R. , De Mol, B. , Escobar, E. , German, C.R. , Levin, L.A. , Martinez Arbizu, P. , Menot, L. , Buhl‐Mortensen, P. , Narayanaswamy, B.E. , Smith, C.R. , Tittensor, D.P. , Tyler, P.A. , Vanreusel, A. & Vecchione, M. (2010) Deep, diverse and definitely different: unique attributes of the world's largest ecosystem. Biogeosciences, 7, 2851–2899. [Google Scholar]
- Rex, M.A. & Etter, R.J. (2010) Deep‐sea biodiversity – pattern and scale. Harvard University Press, Cambridge, MA. [Google Scholar]
- Rex, M.A. , McClain, C.R. , Johnson, N.A. , Etter, R.J. , Allen, J.A. , Bouchet, P. & Warén, A. (2005) A source‐sink hypothesis for abyssal biodiversity. The American Naturalist, 165, 163–178. [DOI] [PubMed] [Google Scholar]
- Rocha, L.A. , Robertson, D.R. , Rocha, C.R. , Van Tassell, J.L. , Craig, M. & Bowen, B.W. (2005) Recent invasion of the tropical Atlantic by an Indo‐Pacific coral reef fish. Molecular Ecology, 14, 3921–3928. [DOI] [PubMed] [Google Scholar]
- Rögl, F. (1998) Palaeogeographic considerations for Mediterranean and Paratethys seaways (Oligocene to Miocene). Annalen des Naturhistorischen Museums in Wien, 99A, 279–310. [Google Scholar]
- Rosenberg, G. , Bouchet, P. & Gofas, S. (2012) Scaphander Montfort, 1810. World Register of Marine Species; Available at: http://www.marinespecies.org/aphia.php?p=taxdetails&id=137871 (last accessed September 2012). [Google Scholar]
- Sanders, H.L. (1968) Marine benthic diversity: a comparative study. The American Naturalist, 102, 243–282. [Google Scholar]
- Schaefer, K. (1996) Review of data on cephalaspid reproduction, with special reference to the genus Haminaea (Gastropoda, Opisthobranchia). Ophelia, 45, 17–37. [Google Scholar]
- Scheltema, R.S. (1971) Larval dispersal as a means of genetic exchange between geographically separated populations of shallow‐water benthic marine gastropods. Biological Bulletin, 140, 284–322. [Google Scholar]
- Schnitker, D. (1974) West Atlantic abyssal circulation during the past 120,000 years. Nature, 248, 385–387. [Google Scholar]
- Schoellhamer, J.E. , Vedder, J.G. , Yerkes, R.F. & Kinney, D.M. (1981) Geology of the northern Santa Ana mountains, California. Professional Paper 420‐D. United States Geological Survey, Washington, DC. [Google Scholar]
- Siesser, W.G. (1980) Late Miocene origin of the Benguela upwelling system off northern Namibia. Science, 208, 283–285. [DOI] [PubMed] [Google Scholar]
- Smith, C.R. , Kukert, H. , Wheatcroft, R.A. , Jumars, P.A. & Deming, J.W. (1989) Vent fauna on whale remains. Nature, 341, 27–28. [Google Scholar]
- Stadler, T. (2009) On incomplete sampling under birth–death models and connections to the sampling‐based coalescent. Journal of Theoretical Biology, 261, 58–66. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2008) The RAxML 7.0.3 manual. Exelixis Lab, Heidelberg Institute for Theoretical Studies, Heidelberg. http://www.trex.uqam.ca/documents/RAxML-Manual.7.0.3.pdf. [Google Scholar]
- Strugnell, J.M. , Rogers, A.D. , Prodöhl, P.A. , Collins, M.A. & Allcock, A.L. (2008) The thermohaline expressway: the Southern Ocean as a centre of origin for deep‐sea octopuses. Cladistics, 24, 853–860. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (2003) PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Talavera, G. & Castresana, J. (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology, 56, 564–577. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. & Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske, P.R. , von der Heyden, S. , McQuaid, C.D. & Barker, N.P. (2011) A review of marine phylogeography in southern Africa. South African Journal of Science, 107(5/6), 43–53. [Google Scholar]
- Thistle, D. (2003) The deep‐sea floor: an overview Ecosystems of the world, Vol. 28, Ecosystems of the deep ocean (ed. by Tyler P.A.). Elsevier, Amsterdam. [Google Scholar]
- Thompson, J.D. , Gibson, T.J. , Plewniak, F. , Jeanmougin, F. & Higgins, D.G. (1997) The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés, A. (2008) Deep‐sea “cephalaspidean” heterobranchs (Gastropoda) from the tropical southwest Pacific Tropical deep‐sea benthos, Vol. 25 (ed. by Héros V., Cowie R.H. and Bouchet P.). émoires du Muséum national d?Histoire Naturelle, 196, 587–792. [Google Scholar]
- Vermeij, G.J. (2005) From Europe to America: Pliocene to Recent trans‐Atlantic expansion of cold‐water North Atlantic molluscs. Proceedings of the Royal Society B: Biological Sciences, 272, 2545–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeij, G.J. & Rosenberg, G. (1993) Giving and receiving: the tropical Atlantic as donor and recipient region for invading species. American Malacological Bulletin, 10, 181–194. [Google Scholar]
- Vermeij, G. & Snyder, M.A. (2003) The fasciolariid gastropod genus Benimakia: new species and a discussion of Indo‐Pacific genera in Brazil. Proceedings of the Academy of Natural Sciences of Philadelphia, 153, 15–22. [Google Scholar]
- Weaver, C.E. (1949) Geology of the coast ranges immediately north of the San Francisco bay region. Geological Society of America, Boulder, CO. [Google Scholar]
- Williams, S.T. & Duda, T.F. Jr. (2008) Did tectonic activity stimulate Oligo‐Miocene speciation in the Indo‐West Pacific? Evolution, 62, 1618–1634. [DOI] [PubMed] [Google Scholar]
- Williams, S.T. & Reid, D.G. (2004) Speciation and diversity on tropical rocky shores: a global phylogeny of snails of the genus Echinolittorina . Evolution, 58, 227–2251. [DOI] [PubMed] [Google Scholar]
- Williams, S.T. , Smith, L.M. , Herbert, D.G. , Marshall, B.A. , Warén, A. , Kiel, S. , Dyal, P. , Linse, K. , Vilvens, C. & Kano, Y. (2013) Cenozoic climate change and diversification on the continental shelf and slope: evolution of gastropod diversity in the family Solariellidae (Trochoidea). Ecology and Evolution, 3, 887–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zardus, J.D. , Etter, R.J. , Chase, M.R. , Rex, M.A. & Boyle, E.E. (2006) Bathymetric and geographic population structure in the pan‐Atlantic deep‐sea bivalve Deminucula atacellana (Schenck, 1939). Molecular Ecology, 15, 639–651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 gblocks settings.
Appendix S2 Best‐fit model and estimated parameters for phylogenetic analysis.
Appendix S3 Single gene chronograms.


