Abstract
The study of metabolism has had a long history. Metabolomics, a systems biology discipline representing analysis of known and unknown pathways of metabolism, has grown tremendously over the past 20 years. Because of its comprehensive nature, metabolomics requires careful consideration of the question(s) being asked, the scale needed to answer the question(s), collection and storage of the sample specimens, methods for extraction of the metabolites from biological matrices, the analytical method(s) to be employed and the quality control of the analyses, how collected data are correlated, the statistical methods to determine metabolites undergoing significant change, putative identification of metabolites and the use of stable isotopes to aid in verifying metabolite identity and establishing pathway connections and fluxes. The National Institutes of Health Common Fund Metabolomics Program was established in 2012 to stimulate interest in the approaches and technologies of metabolomics. To deliver one of the program’s goals, the University of Alabama at Birmingham has hosted an annual 4-day short course in metabolomics for faculty, postdoctoral fellows and graduate students from national and international institutions. This paper is the first part of a summary of the training materials presented in the course to be used as a resource for all those embarking on metabolomics research.
Keywords: metabolomics, GC-MS, LC-MS, CE-MS, NMR, study design, sample extraction
Introduction
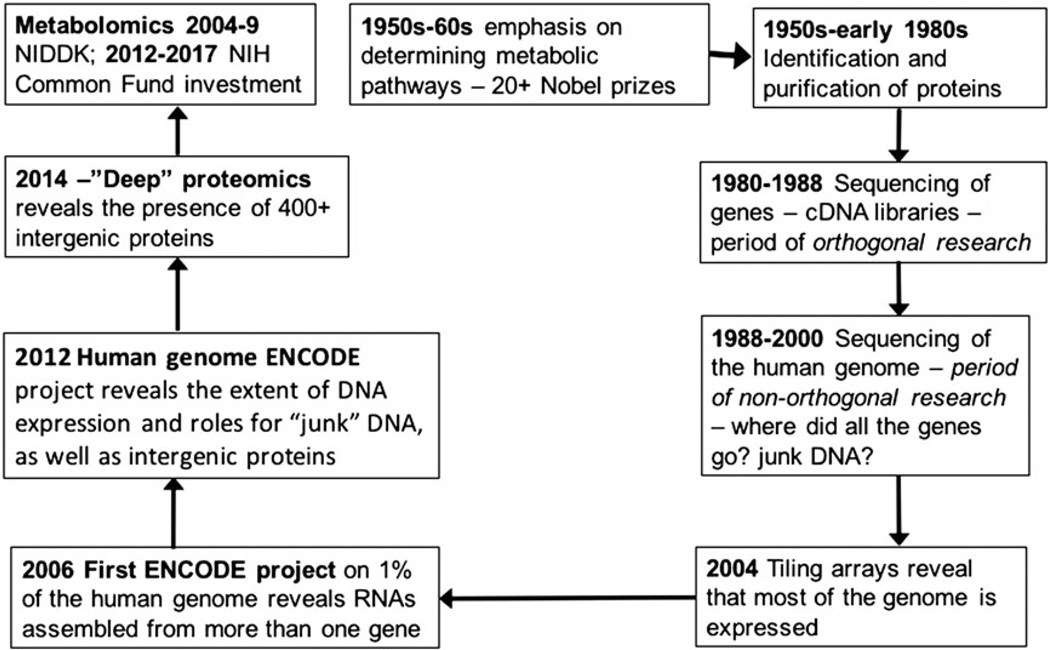
Studies of metabolism have been a major feature from the earliest days of NIH research (Fig. 1). As the metabolic pathways were defined, this led to the isolation and characterization of the enzymes catalyzing the formation of metabolites. In turn, the genes encoding the enzymes were recognized and sequenced. The arrival of cDNA libraries containing copies of all the mRNAs expressed in tissues or cells facilitated cloning and identification of the open reading frames of genes encoding the enzymes, as well as expression of recombinant forms of the enzymes. The excitement generated by this approach enabled large-scale funding of an engineering approach to the sequencing of the entire genome (the Human Genome Project). Subsequently, microarrays where unique oligonucleotides representing each expressed open reading frame were attached to the surface of glass chips were generated.[1] Now, using the microarrays, all the genes being transcribed could be viewed at one time and transcriptomics began. It was an exciting time although reproducibility and statistical analysis quickly revealed that the high-dimensional nature of transcriptomics nonetheless had to obey the rules of science.[2] Following the introduction of electrospray ionization (ESI)[3] and matrix-assisted laser desorption ionization[4] for vaporizing peptides and proteins, methods based on mass spectrometry for studying entire proteomes (proteomics) quickly emerged. However, it, too, has been fraught with statistical problems in experimental design, data acquisition and data analysis.
Figure 1.
The cyclic nature of NIH-funded biomedical research. The initial interest and investigation of the metabolic basis of heart disease, endocrine disorders and cancer was replaced by focused studies on the enzymes and transporters responsible for the formation and distribution of metabolites. In turn, the genes responsible for these proteins were isolated and sequenced to identify their open reading frames. Then, as cDNA and other high-throughput methods became available, the task of sequencing the human and other genomes was undertaken. Using that information, oligonucleotides specific for each gene were attached to microarray chips. Similar technologies allowed the whole genome to be attached to microarrays. This revealed that most of the genome was transcribed to RNAs, with only small portion being messenger RNA. More careful re-sequencing of the human genome in the ENCODE project showed that some of the expressed RNAs were intergenic. In 2012, confirmation of an intergenic protein by mass spectrometer was reported. Now, over 400 intergenic proteins have been described. In 2004, NIH began its investment in metabolomics and returned to its early roots.
Nonetheless, the concept of -omics has continued to spread throughout the biological research endeavor, and therefore, the development of a global approach to the measurement of all the metabolites (metabolomics) was not surprising. Following funding support by the NIH (by National Institute of Diabetes and Digestive and Kidney Diseases from 2004 to 2009[5] and by the NIH Common Fund Program from 2012 to 2017[6]), there has been a substantial increase in the application of metabolomics spanning basic science to clinical translational research, clinical trials and epidemiology through Regional Comprehensive Metabolomics Research Centers,[7] as well as Technology Development R01s, Mentored Research Awards, Administrative supplements to existing NIH grants, pilot grants from Regional Comprehensive Metabolomics Research Centers and to Training. One of these investments was sponsorship of a Metabolomics Short Course via the R25 mechanism held at the University of Alabama at Birmingham (UAB) to introduce investigators to the steps needed to design and perform a metabolomics experiment and to analyze the collected data.[8] There have also been several excellent practical descriptions of metabolomics.[9–14] In this paper, we have outlined the training provided in the NIH-sponsored Metabolomics Short Course at UAB. In the second part to be published in the following issue of the journal, the focus is on the statistical methods applied to metabolomics data, identification of metabolites and the pathways of metabolism, new areas of application and the future of metabolomics. It concludes by providing guidance to investigators wishing to pursue various and deeper aspects of metabolomics.
What is the metabolome?
The metabolome is more than the metabolites that are part of known metabolic pathways. Instead, it is all the chemicals with molecular weights less than 1500 Da1 that can be detected by mass spectrometry (MS) or nuclear magnetic resonance (NMR) analysis of biofluids or tissues. These chemicals include not only those that were originally in the biological sample but also chemicals coming from the tubing/vials used in collecting and storing the samples, the reagents used to process and extract the samples and the components in HPLC mobile phase or NMR solvent. If metabolites were extracted from cultured cells, then the culture medium may contain undocumented components, some of which come from other organisms/species (e.g. fetal calf serum). For serum and urine from humans and animal models, it is much more complex. Compounds may come not only from mammalian cells but also from the food that is eaten plus any chemical additives used in its production. Other compounds may result from the metabolic and metabolizing capability of the gut microbiome. For example, the complement of gut microorganisms may vary from individual to individual, and their populations are influenced by what is eaten. They are most prevalent in the large bowel where the oxygen tension is the lowest. This frequently leads to metabolites recovered from the bowel that are reduced. An example is S(−)-equol, a metabolite of the food isoflavone daidzein.[15] Additional compounds come from the anesthetics used to anesthetize experimental animals as well as analgesics to reduce pain.[16] In humans, careful attention has to be made regarding any drugs being taken by a patient (and their metabolites) because these may represent large peaks in a sample. Thus, the vast complexity of the microbiome and variance introduced by diet and exogenous compounds such as drugs requires substantial consideration in investigations.
Planning a metabolomics experiment
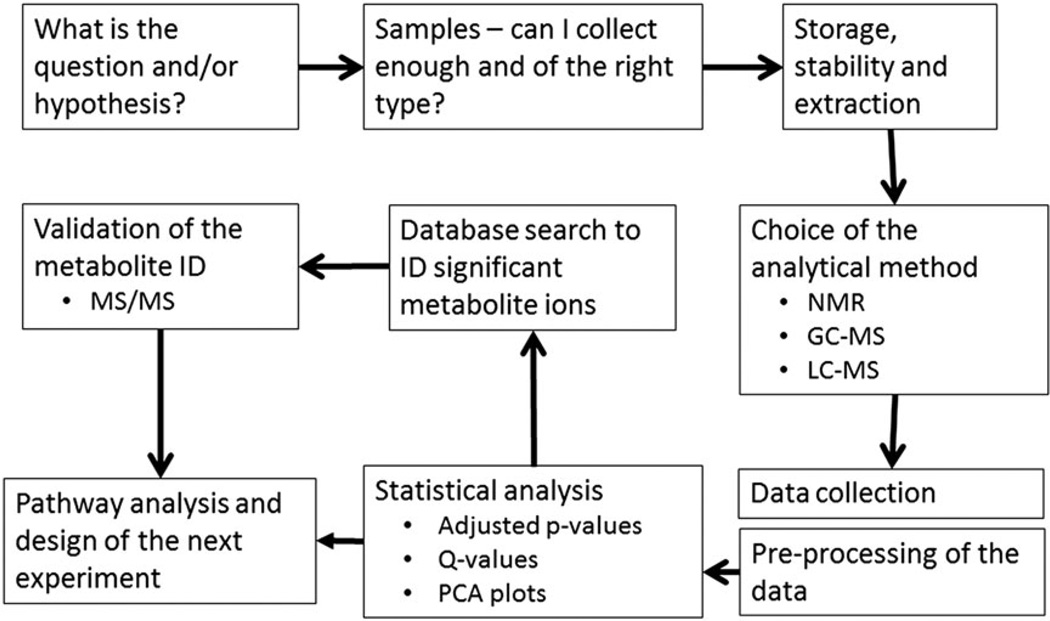
Developing a planned approach is the most critical part of a metabolomics experiment. A general overview of the steps comprising a metabolomics experiment is illustrated in Fig. 2. Sample integrity may alter the capacity of experimental design. If the samples have already been collected (as per an epidemiological or clinical study), then it is important to know (1) how have they been collected and stored, (2) how can control and treatment samples be matched, and (3) is there a clear phenotype between the control and experimental groups?
Figure 2.
Workflow of a metabolomics experiment. The first step is to determine the question that is being addressed. It is important that a clear phenotype is to be studied. The next issue concerns obtaining the biological samples and in sufficient numbers to provide statistical power. Fresh samples from a clinical, animal model or cell culture study provide the most control over sample collection and storage. Samples from epidemiological studies may have varied isolation steps and an unknown storage history. Most metabolomics analyses require a sample extraction procedure, and the one chosen must be appropriate for the question being asked. Next comes the choice of analytical platform followed by the collection and storage of data. Most data require initial pre-processing to align the data in order to make statistical comparisons across samples. Once aligned, data are normalized prior to statistical analysis. Putative metabolite ions are searched for in metabolite databases and are confirmed with standards (if available), making use of MS/MS data of both. Metabolites, once identified, are then mapped to known metabolic pathways. In a new development, Mummichog analysis searches for metabolite networks and then associates these with existing pathways.
The number of samples and/or size of the groups needed for a metabolomics experiment depend on the biological variability associated with the system being studied compared with the analytical variability of the analytical system. Because the growth cells in culture can be carefully controlled, a sample size of three to five per group may give useful preliminary data that can be used to design more elaborate and/or targeted metabolomics studies. Laboratory animals on controlled diets are more complex than cells in culture, but depending on the degree of control, a sample size of 6–12 may be adequate, although if female animals undergoing estrus cycling are used, they should be studied during the same point in the estrus cycle. Controlled clinical trials where subjects provide multiple samples or where the subjects are carefully matched may be able to be carried out with as few as 10–20 patients, but this will depend on the variance of the disease traits, drug response or that which is introduced by an interventional procedure. These sample numbers (n = 3–20) in a project are suitable for generating preliminary and/or pilot data for which the analysis can be carried out in a single batch, thereby simplifying comparisons. For epidemiological studies, where the samples were collected from a general population, often over long periods of time, variance is a substantial issue and may require patient numbers in the thousands. In this case, the metabolomics assays have to be carried out in multiple batches over many months or even years. How to relate data from one batch to another remains highly challenging,[13] and methods are emerging to address this.[17–19]
Avoiding unintended bias in any sort of -omics analysis is a critical issue, and metabolomics is no exception. For instance, not controlling for the effect of diet or the time of day of sample collection can lead to excessive variation and/or differences between groups that masquerade as biologically relevant changes in metabolite levels.[19] As noted by Scalbert et al.,[20] metabolites arising from gut microbiome and the edible plant metabolome far outnumber metabolites produced by human cells. Rat chow diets are an uncontrolled admixture of soy, wheat and fishmeal.[21] A metabolomics experiment in rodents therefore should consider using a defined rodent diet, for example, a diet based on casein as the protein source such as AIN-93.[22] It is also important to utilize a single batch of the diet to avoid variability in the composition of the diet masquerading as a biological effect. In addition, diet manufacturers are now including exposing diets to gamma radiation to reduce microbiological contaminants. It is important to note that such diets may be depleted in critical micronutrients[23] as well as radiation-induced oxidative modifications to dietary components.
The constant flux of the metabolome involves a vast number of dynamic variables that are essential to consider for obtaining high-quality results. This includes the timing of sample collection. Blood samples are best taken in the fasting state or at least at the same time of day to minimize the effects of selective variations from the absorption of dietary components related to when rodents or other experimental animals have been fed.[24] In addition, diurnal variation in enzymatically driven metabolic pathways can obscure other biological events.[25] The metabolome of spot urine is the summation of excreted metabolites over the previous (undefined) several hours. However, by collecting the total urine output over a 24-h period (humans) or longer (experimental animals), this variation can be averaged out. If a 24-h collection is used, then provisions for storing the urine at 4 °C or lower or adding a preservative such as sodium azide to prevent bacterial growth during the collection period are essential. In the case of rats, using suspended metabolic cages, urine can be collected into beakers in Styrofoam containers packed with dry ice.
Another factor that may induce non-biological variation is the containers in which the samples are collected or stored. It is very important that the same batch of containers is used across all groups that are going to be compared. Similarly, samples should be stored in such a manner as to avoid creating group effects. When collecting plasma samples, the same type of coagulant (EDTA, heparin or citrate) must be used. Using barcodes for the samples allows samples to be randomly placed in a freezer without leading to confusion of their identity. At this time, storage of samples at −80 °C is preferred. Furthermore, samples with the same freeze–thaw cycles should be used for comparison purposes.
With the tremendous risk of confounding, it behooves investigators to consult with a biostatistician before conducting a metabolomics study to avoid the potential experimental design errors described in the preceding texts. The statistician can create a protocol for the order of processing and analyzing samples (refer in the succeeding texts). It is also essential to record all the steps taken in the design of the experiments, collection of samples, their storage and processing, the analytical platform and its performance, the pre-processing of collected data and the statistical and pathway tools used to interpret the data. This information is called the metadata and is critical for careful evaluation of the data. It is also an ethically correct approach to doing metabolomics research. Advice on organizing storage of metabolomics data as well as recording the versions of the computational procedures and metabolite databases is well described by Buffalo.[26]
Extracting and processing samples
The question being asked is essential to determine the extraction and processing technique to be used. Because of the extremely diverse chemical nature of the metabolome, there is no method that captures all of the metabolites. The simplest matrix is urine because the metabolites are water-soluble. It is important to remove any precipitates in urine using centrifugation at 12 000 × g at 4° C for 10 min. Additionally, the sample can be passed through a spin filter with a 3 or 10 kDa cutoff although there is a risk that metabolites could be selectively retained by the filter. In the case of rodent urines, it is important to remove the precipitable protein and large peptide components by the addition of four volumes of methanol, cooling the sample to −20 °C for 20 min and then centrifuging the mixture. The supernatant containing the metabolites is aspirated and evaporated to dryness under N2. For NMR analysis, the methanol supernatant is usually freeze-dried once the methanol has evaporated. The dried materials are dispersed in a solvent for NMR (D2O, CD3OD or CDCl3). For liquid chromatography-mass spectrometry (LC-MS), dried materials are dissolved in water or 0.1% formic acid and centrifuged, typically at 12 000×g to remove any particles prior to analysis.
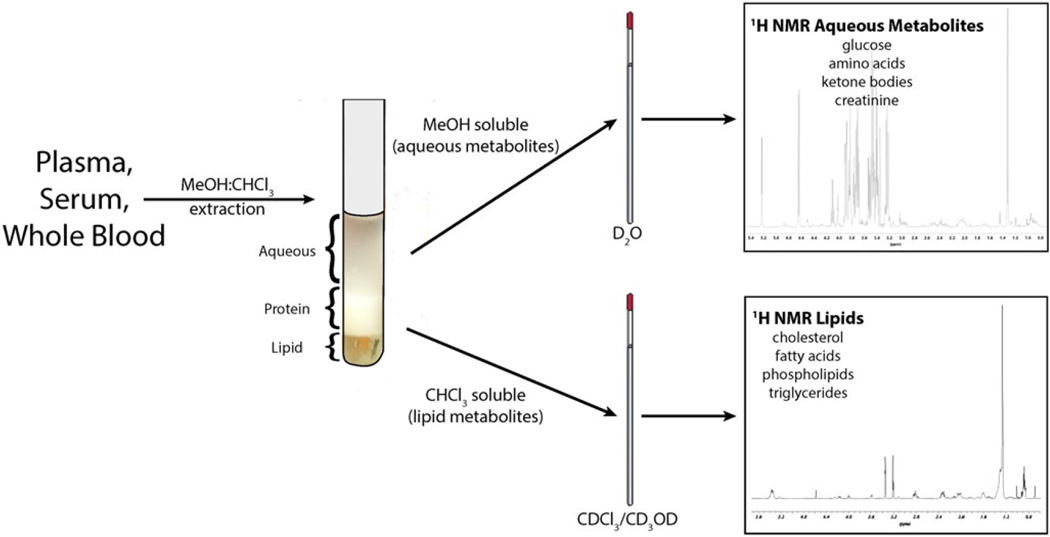
Blood and other biological fluids typically contain large amounts of proteins and phospholipids. To separate and recover the water-soluble and lipid metabolite fractions, these fluids are extracted by the Bligh–Dyer procedure[27] in the presence of butylated hydroxytoluene to prevent lipid oxidation. Most often though, deuterated chloroform/methanol (1 : 1, v/v) is added to blood (whole blood, serum or plasma) samples that are then subjected to a series of centrifugation steps.[28] This extraction yields aqueous (MeOH) and lipid (CHCl3) fractions that can be assayed by either 1H-NMR or LC-MS (Fig. 3). Alternatively, serum and plasma samples can be filtered. This results in the loss of the hydrophobic metabolites but permits more expeditious sample processing, and the resulting spectra can be quantified.[29] Tissue samples can be extracted using similar extraction protocols.[30] Precious blood samples can also be directly mixed with a solution containing saline in D2O and an internal chemical shift reference standard[30] for NMR metabolomics analysis so that the sample can be re-used for other experiments or assays.
Figure 3.
Fractionation of blood or plasma into water-soluble and lipid components. In this example, the biological fluid (blood, plasma, etc.) is partitioned between deuterated chloroform (CDCl3)/deuterated methanol (CD3OD) and deuterated water (D2O). Using deuterated solvents is ideal for NMR metabolomics. If the sample is analyzed by LC-MS, GC-MS or CE-MS, non-deuterated solvents can be used. The organic solvent phase contains fatty acids, triglycerides, cholesterol and other sterols, phospholipids and other fat-soluble compounds such as vitamins A, D, E and K. The aqueous phase contains water-soluble organic acids, amino acids, sugars, creatine, bile acids, β-glucuronides and sulfate esters of steroids, food phytochemicals and therapeutics.
It is also possible to further fractionate the protein-precipitated, water-soluble metabolome into sub-fractions based on extraction into ethyl acetate. We have found that this is very effective for decreasing the complexity of fecal water.2 The recovered metabolome in this case is highly dependent on the pH of the aqueous phase during extraction (Fig. 4). For example, fatty acids and other organic acids will pass into the ethyl acetate phase under acidic conditions, whereas amines will be extracted under basic conditions.
Figure 4.
Effect of pH on the extraction of the metabolome into ethyl acetate from fecal water. Aliquots (100 mg) of fecal material were mixed with 0.1% formic acid (purple trace), water (green trace) and 0.1 M NaOH (blue trace) and were centrifuged at 150 000×g at 4 °C for 30 min to yield fecal water. Fecal water was extracted with two volumes of ethyl acetate. The ethyl acetate upper layer was carefully removed and taken to dryness under N2. It was reconstituted in 100 µl of 5% acetonitrile/0.1% formic acid. Aliquots (5 µl) were analyzed by nanoLC-ESI-MS (SCIEX 5600 TripleTOF) on a 10 cm × 200 µm ID C18 reverse-phase Chip column, using a 20-min linear gradient (5–80%) of acetonitrile in 0.1% formic acid at a flow rate of 1 µl/min. The red trace is the background ions detected substituting double-distilled water for fecal water and extracting with ethyl acetate. The traces represent the total ion chromatogram for each extracted sample.
The complexity of the metabolome warrants consideration of other fractionation methods. Based on previous methods of analysis, metabolites can be fractionated by ion exchange chromatography. Anion exchange resins are activated by washing with dilute sodium hydroxide to convert them to the hydroxide form and then washed with double-distilled water until neutral. The aqueous extracts of biofluids or tissues are slowly passed over a small column of the anion exchanger (Fig. 5). At pH 7, negatively charged metabolites bind to the resin. Amines and neutral sugars pass through the column unimpeded. After washing with water, the bound organic acids are eluted with sodium formate. It is necessary to remove the sodium ions, and this is accomplished by passing the eluate over a cation exchange column in the [H+] form. This converts sodium formate into formic acid that has a boiling point of 101 °C and therefore is readily removable by evaporation or freeze drying. A similar method can be used to isolate positively charged metabolites. The metabolite extracts are passed over a cation exchange column in the [H+] form (Fig. 5). Neutral and negatively charged ions pass through the column, and positively charged ions bind to the resin. They are eluted by 1 M ammonium hydroxide that is easily removed by evaporation.
Figure 5.
Separation of metabolites using ion exchange chromatography. The aqueous metabolite extract is either passed over an anion exchange column or a cation exchange column. The anion exchange column is pre-washed with NaOH and then with freshly prepared, double-distilled water until neutral. The extract is passed over the column, and the materials that do not bind (neutral and positively charged ions) eluted with further washing. Negatively charged metabolites are eluted with 1 M sodium formate, and the eluate passed over freshly prepared cation exchange resin in the H+ form (pre-washed with 4 M HCl and then with freshly prepared, double-distilled water until neutral) to convert the sodium formate to formic acid. The eluate is freeze-dried prior to LC-MS analysis. The cation exchange resin in the H+ form is also used to recover the positively charged metabolites from the metabolite extract. In this case, the neutral and negatively charged metabolites do not bind to the resin and pass through. After further washing, the positively charged metabolites are eluted with 2 M ammonium hydroxide. Excess ammonia is removed by evaporation under N2.
Another method is to exploit the chemistry of the metabolite classes. As used in proteomics analysis, metabolites containing aldehyde or keto groups can react with hydrazine derivatives such as biotin hydrazide.[31] This reagent attaches a tag that enables the metabolites to be recovered from a complex matrix by passage over an avidin affinity column. As occurred in the use of this type of reagent in proteomics,[32] it may be necessary to include an acid-cleavable group between the reactive group and the biotin group because elution of biotinylated derivatives from the avidin affinity phase is difficult and non-quantitative. Other forms of these reagents contain quaternary nitrogen atoms that make aldehyde, ketone and cis-diene-containing metabolites readily detectable in LC-MS analysis (in the positive mode).
When using whole tissues, samples should be flash frozen in liquid N2 at the time of collection to prevent further metabolism and stored at −80 °C. In some circumstances, it is convenient to flush tissues before harvesting with ice-cold physiological saline to minimize the contribution of blood metabolites to the tissue metabolome. In order to extract metabolites, stored, frozen tissue must be ground to a powder in a pestle and mortar cooled with liquid N2. Alternative methods include shock pulverization in liquid N2 and use of a French press (suited to plant tissues). The frozen powdered tissue is extracted with either ice-cold methanol (4 volumes/g) or 0.1% perchloric acid. These methods both precipitate and inactivate tissue enzymes.[33,34] The precipitated proteins are removed by centrifugation. The methanol extract can be processed to remove lipids by addition of chloroform and water as described in the preceding texts for the Bligh–Dyer extraction. Perchloric acid is removed from the extract by neutralization with potassium hydroxide. Potassium perchlorate has very low solubility in water[35] and is removed by centrifugation.
For adherent cells grown on culture plates, an effective procedure is to rapidly remove the culture medium with a Pasteur pipet attached to a vacuum line. The cells are immediately washed with ice-cold, isotonic physiological saline which after swirling for a few seconds is also aspirated.[36] The washed cells can either be instantly frozen with liquid N2 to be extracted at a later time[37] or immediately extracted with methanol pre-cooled to dry ice temperature (−43 °C). These steps serve to ‘freeze’ metabolism. The extraction temperature is allowed to rise to 0°°C, and the cells are released from the culture plates with a cell scraper. After 30 min, the mixtures are centrifuged (4000×g) and the supernatants aspirated to clean glass tubes. The methanol is removed by evaporation under N2. Depending on the type of cell being studied, some modifications of the previous generalized procedure may be necessary and investigators are advised to search the literature to determine the experiences of others.
Analysis of the metabolome
Historical methods
Before modern metabolomics came into being, several forms of metabolomics already existed, although not given that moniker. In the early 1960s, investigators at Medical Research Council Metabolic Reactions Research Unit at Imperial College in London led by Professor Sir Ernst Chain used an automated 2D paper chromatography method for the separation and detection of metabolites of 14C-glucose developed in the Instituto di Sanita Superiore in Rome, Italy in the 1950s.[38] Geiger counters mounted on a typewriter carriage moved incrementally across the dried paper chromatogram and recorded the measured radioactivity in each zone. Engineers built 10 of the radioactivity scanners that were placed in a dedicated room. Data from the Geiger counters were stored on data tape, and after computer processing with Digital PDP-9 computers, images of the location of the radioactivity on the dried sheet and its amounts were generated.[39] A similar approach connected gas chromatographs to radioactivity detectors. Suitably derivatized 14C-labeled metabolites were either oxidized to CO2 by passage over finely divided Fe/CuO and into an in-line Geiger counter[40] or were condensed out of the gas phase for measurement in liquid scintillation counters (offline or in-line).[41]
In the early 1980s, the introduction of superconducting magnets for NMR experiments allowed substantial improvements in resolution of the resonance peaks of metabolites, as did the introduction of pulse sequences that permitted further discrimination of otherwise overlapping resonance peaks.[42] Using iron-based magnets operating at 90 MHz, the 1H-NMR spectrum of the bile acid, cholic acid, consisted of strong signals for its C18, C19 and C21 methyl group protons and weaker signals from the protons epimeric to the C3α, C7α and C12α hydroxyl groups. The remainder of the methane and methylene protons from the steroid ring system and the side chain was grouped together into the ‘steroid hump’.[43] At 400 MHz using a superconducting magnet, these extensively overlapping resonances largely resolved with the use of proton decoupling and nuclear Overhauser effect experiments.[44] The application of a homonuclear-decoupled heteronuclear-correlated 2D NMR experiment introduced by Bax[45] allowed a much more efficient method for assigning proton resonances in bile acids.[46]
In addition, wide bore instruments became available allowing small animals to be placed in the analytical zone of the NMR instrument. This allowed for direct in situ measurement in the living animal of 31P-metabolites (ATP, ADP, AMP, phosphocreatine)[47] and metabolites of 13C-labeled precursors.[48]
Targeted and untargeted analysis
The study of metabolites associated with genetic disorders of metabolism has had a long history. Currently, this is routinely carried out using gas liquid chromatography-mass spectrometry (GC-MS) or by LC-MS. These analyses are on specific compounds already known to the (clinical) investigator. This type of analysis is termed targeted metabolomics. With modern mass spectrometers, 100 compounds or more can be measured in a single analysis.
A feature of the current applications of metabolomics is that the problem being investigated does not necessarily have a predetermined genetic/metabolic defect associated with it. Furthermore, even when there is information on the gene defect, the impact of the defect is hard to predict. For that reason, an open-minded, global approach to the analysis is taken. This is also often termed untargeted metabolomics. This approach allows investigators to gather information to propose novel hypotheses that could not be envisaged using targeted analysis.
Modern metabolomics
Two main analytical platforms in the global analysis of metabolites have emerged in the past 30 years – the use of high-resolution, 1D NMR (also referred to as metabonomics)[49] and MS with and without prior chromatographic separation.
1H nuclear magnetic resonance
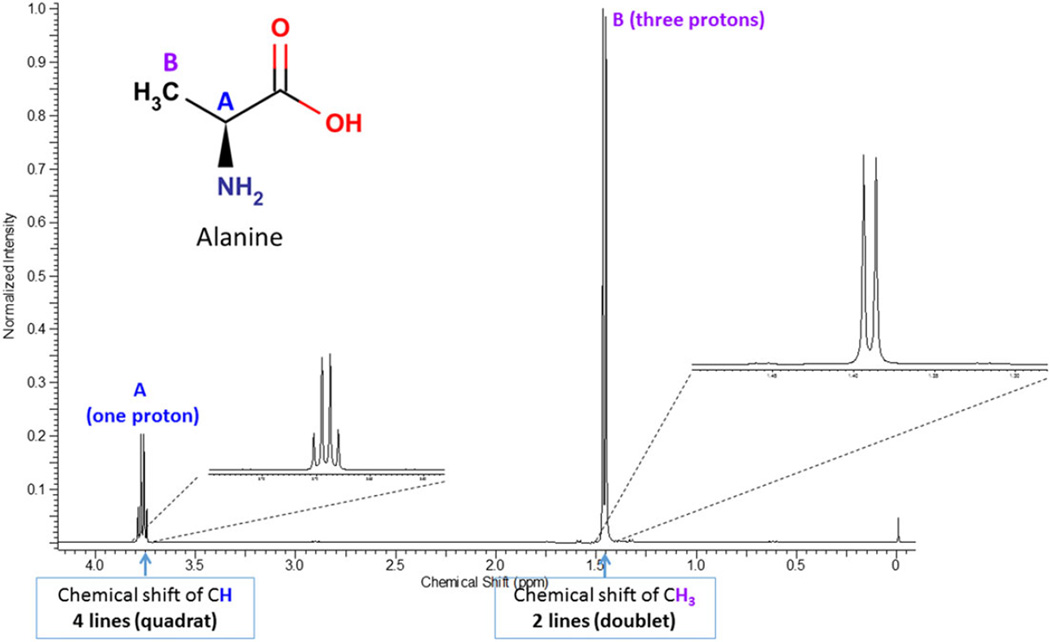
The use of proton NMR for metabolomics is based on the principle that protons resonate between energy states in a high magnetic field. A high-power short duration radio frequency pulse causes the absorption and subsequent release of electromagnetic radiation that varies for a compound based on the location (e.g. energy state) of its associated protons. This generates a free induction decay signal as a function of time (time domain), which is then Fourier transformed to convert it as a function of frequency (frequency domain), generating the more commonly recognized chemical shift plot spectrum that has units of parts per million (ppm) or δ versus peak height plot where each resonant frequency in the sample shows up as one peak. The positions (chemical shifts) of those peaks are affected by the proximity of electronegative groups such as nitrogen, oxygen, carbonyls, double bonds, halogens, etc., which will influence the place of each type of proton on the spectrum. Every metabolite has its own unique NMR spectrum that represents the environment of each proton in the metabolite. These resonances are further split by interaction with protons on neighboring carbon atoms. For example, if a methyl group is attached to a carbon atom that is not connected to a proton, it will produce a singlet resonance peak. If the carbon atom is attached to a single proton, then the resonance is split into two (doublet) (Fig. 6). More complex splitting is observed if the adjacent carbon atoms contain protons in different environments. As a result, the mixture of different metabolites in a metabolomics sample produces a complex NMR spectrum (Fig. 7).
Figure 6.
A 950 MHz proton [1H]-NMR spectrum of alanine. The spectrum shows the chemical shifts of the CH and CH3 protons. The CH signal is split into a quartet (four lines) and the CH3 signal is split into a doublet (two lines) because of the J-coupling with the neighboring protons. The upper traces show an expansion of the CH and CH3 NMR signals.
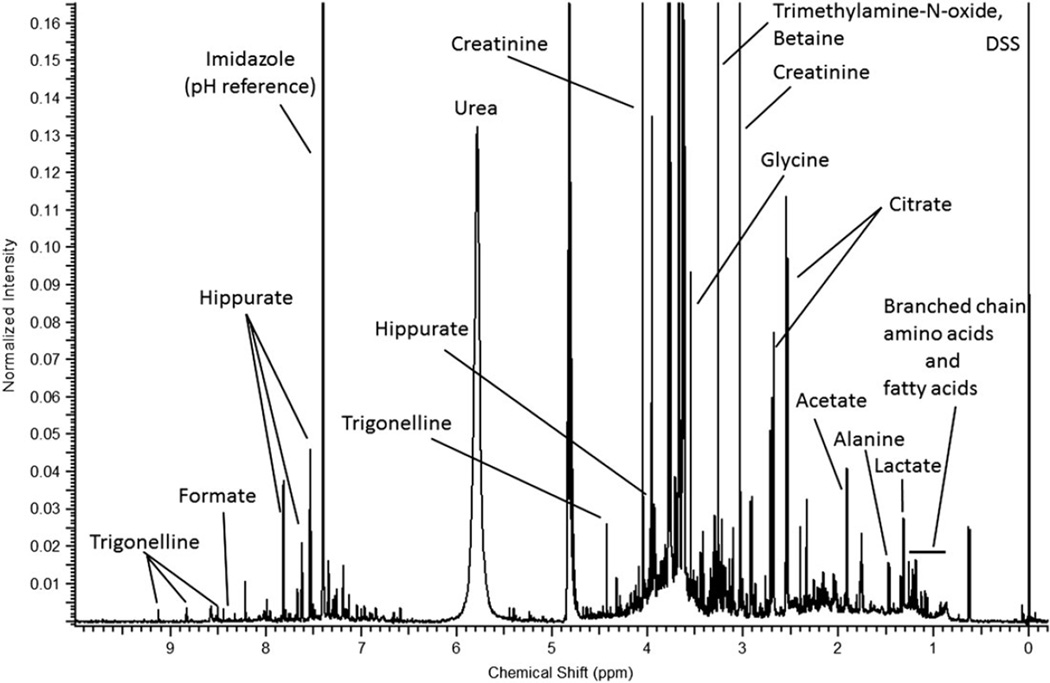
Figure 7.
A 950 MHz 1H-NMR spectrum of a human urine sample. It consists of thousands of features that can be deconvoluted into metabolites belonging to a wide range of compound classes. Urine sample consists of both endogenous and exogenous metabolites including many unknowns. 1H-NMR spectrum is referenced with respect to the 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) resonance at δ 0 ppm. The pH of the solution is among the factors that influence the chemical shift, and imidazole can be used as a pH reference, which helps assigning the NMR signals into metabolites using pH-dependent NMR libraries such Chenomx NMR Suite.
Since their initial introduction in the late 1970s, the increased field strength of the superconducting magnets has evolved to better resolve the resonances of compounds in a biological sample. Currently, 600 MHz NMR instruments are considered state of the art in metabolomics, although larger magnets are in use for other biochemical applications, and the higher the magnetic field strength, the greater the resolution. Compared with their early predecessors, they have the useful engineering feature of being able to automatically load/unload metabolite extracts. Some also have cryoprobes and microprobes to increase their sensitivity. NMR analysis of metabolites takes advantage of pulse sequences[42,45] and suppression of the intense 1H water resonance.[50,51]
There are several features of NMR that have led to its early and continued use in metabolomics – (1) the observed resonances are quantitative once calibrated with a known amount of a single internal standard such as 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS), which has nine identical methyl protons that results in a single high-intensity proton peak that can be set to 0 ppm[29]; (2) it is a non-destructive technique, so the sample can subsequently be analyzed by GC- or LC-MS; (3) it is highly reproducible and is less susceptible to instrument variability like LC-MS that makes it ideal for inter- and cross-center studies[50]; and (4) if observable (particularly if an unknown metabolite is first isolated), the fine structure of the resonances of a metabolite can provide information about its chemical nature. In addition, magic angle NMR allows for the observation of high-resolution metabolite signals directly in tissues without preliminary extraction.[52]
From a practical standpoint, the sample volumes needed for NMR analysis are higher than for MS. Typically, at least 0.5 ml of plasma, serum or urine or 0.5 g of feces or tissue are required for optimal analysis. For in vitro cell culture experiments, NMR metabolomics requires 10 million cells. Smaller amounts can be used but at the expense of losing detection of lower abundant metabolites. Use of smaller diameter NMR tubes will be helpful for samples with smaller amounts. For NMR analysis of plasma, heparin is the preferred preservative over EDTA or citrate because the latter two preservatives interfere with the 1H-NMR spectrum. Untargeted and targeted NMR metabolomics methods are used and a number of library matching tools are available for identifying metabolites.[53–55] Complementary 2D NMR methods (correlation spectroscopy, total correlation spectroscopy, heteronuclear single quantum correlation, heteronuclear multiple-bond correlation spectroscopy) are used in confirming the metabolite assignments. New pulse programs have been developed for measuring lipids by NMR.[56]
Mass spectrometry
Mass spectrometry is based on the metabolite forming ions (either inherently charged ions or ones that are formed under the conditions of analysis). There are several separation methods that are used prior to MS analysis. These include GC, LC and capillary electrophoresis (CE).
Gas chromatography-mass spectrometry
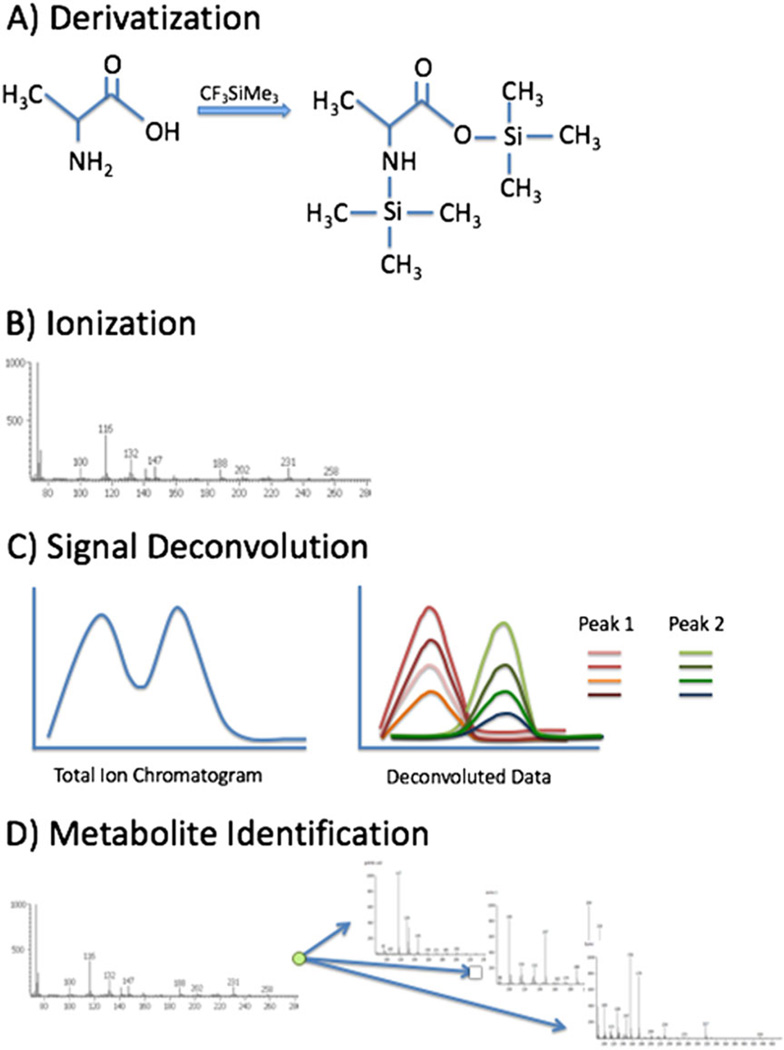
Gas chromatography is commonly used for analysis of volatile complex mixtures. GC-MS analysis of plant volatiles were among the earliest metabolomics publications.[57,58] Other common biological samples measured by GC-MS include human serum or plasma, urine and cultured bacterial or mammalian cells. Volatile compounds can be introduced into a column using headspace injection that measures only the gas-phase molecules trapped in the headspace (air trapped in a chromatography vial above the sample). However, most metabolites common to biological samples must be derivatized after extraction in order to enter the gas phase. In a typical GC-MS analysis, after extraction of metabolites from these samples, the metabolites are evaporated under vacuum and derivatized. The most common derivatization is trimethylsilylation with reagents such as N-methyl and N-trimethylsilyl-trifluoroacetamide (Fig. 8(A)). Trimethylsilylation is often combined withmethoximation to stabilize ketones and prevents cyclization in abundant metabolites like glucose, ultimately reducing the number of peaks measured for an individual metabolite (reviewed in[59]). After derivatization, the samples can be applied to the column using several methods. The simplest is direct injection that simply adds the sample to the column where it enters gas phase in the inlet, heated to a temperature of >250 °C. These derivatives can be chromatographically resolved using a variety of commercially available fused silica capillary columns. The stationary phases of these columns are diverse but most metabolomics applications take advantage of a long non-polar column chemistry that separates the complex mixtures so that individual components arrive at the mass spectrometer at different times.
Figure 8.
Elements of GC-MS analysis. GC-MS has several elements: (A) the formation of trimethylsilyl derivatives to cover polar (non-volatile) groups on a metabolite in order to carry out gas-phase chromatography; (B) post-column, electron impact ionization to cause dissociation of the metabolite derivatives into a spectrum of product ions; (C) signal deconvolution of the product ions to recognize individual peaks; and (D) from spectra of the aligned product, ions make assignments using metabolite databases.
Following separation by chromatography, molecules are measured by MS; however, because of the derivatization, compounds exiting the gas chromatograph do not carry a charge. To create ions, derivatized molecules are bombarded with energetic (70 eV) electrons. These electrons not only form molecular ions of the metabolite derivatives but also cause dissociation of the molecular ions into smaller fragment ions. Typically, the parent ion is lost, but the resulting pattern of fragment ions is used as a fingerprint in combination with indexed retention times to provide confident identification for many metabolites (Fig. 8(B)). Extensive reference libraries are available, for example, the National Institute of Standards and Technology database consists of over 50 000 standards[60,61] and smaller, more biologically focused libraries are also available.[62,63]
A significant advantage of GC-MS is the highly reproducible chromatography. Including internal standards in each sample allows a large number of samples to be compared with one another and also allows comparison of samples between laboratories using indexed retention times.[64] This includes using indexed retention times in reference libraries to improve metabolite identification.
Another important aspect of GC-MS data analysis is signal deconvolution. Separation of a complex mixture, as in metabolomics, results in many peaks that co-elute or partially overlap. The basic principle of signal deconvolution is to consider all ions measured over time and identify groups of ions that within a region of the chromatogram show a highly correlated signal. Among those correlated ions, one or several unique ions will be chosen to represent that peak for the purposes of relative quantification (Fig. 8(C)). The Automated Mass Spectral Deconvolution and Identification System software is a common tool for signal deconvolution.[65] This algorithm assigns each measured ion to a peak and calculates similarity scores between the resulting fingerprints and any available library spectra to identify metabolites.
GC-MS is a useful tool for high-throughput metabolomics analysis. It allows relative quantification (or true quantification using standard curves) of a large number of metabolites in a single assay. GC-MS does have limitations: It is most successful in measuring small metabolites (<400 Da) and is not reliable for measurement of molecules that are unstable under the heating conditions used for derivatization and chromatography. Nonetheless, significant improvements in both chromatography and MS have been made to GC-MS for metabolomics analysis. 2D GC has been explored that improves chromatographic resolution allowing detection of more peaks of similar biochemical properties. High-resolution MS has not been used previously because metabolite identification was based on comparison with spectra of known standards, but recently, GC has been coupled with high-resolution instruments such as an Orbitrap that improves identification of unknown metabolites. This method continues to improve and offers a necessary complementary method for metabolomics by LC-MS and NMR.
Liquid chromatography-mass spectrometry
The key to the success of LC-MS has been the introduction of the ESI[66] and atmospheric pressure ionization interfaces.[67] Both allow the transfer of charged ions from the liquid phase to the gas phase without the need for derivatization. Indeed, ESI has been widely used in proteomics to transfer peptides and whole proteins into the gas phase. Many metabolites readily form ions, e.g. organic acids form negatively charged carboxylates at neutral pH and amines and nitrogen-containing compounds form positively charged ions under acidic conditions. Other metabolites, including neutral compounds, also form positively charged sodium, potassium and ammonium adducts or chloride and formate adducts. For neutral molecules, atmospheric pressure chemical ionization can be used to enhance the formation of [M+H]+ or [M−H]− or adduct molecular ions.
Liquid chromatography of metabolites
Because metabolites have such a wide range of hydrophobicities/hydrophilicities, it is impossible to separate them all on a single LC column. Typically, an in-depth analysis of a metabolomics sample will involve both reverse-phase (C4–C18) and some form of normal-phase column. Also, as noted in the preceding texts, metabolites form both negative and positive ions. At this time, a sample is typically run twice: first, to capture the negative ions and then, in a separate LC-MS experiment, to capture the positively charged ions (however, there are instruments in which fast switching between negative and positive ion modes is possible).
The size of the particles in the column is an important variable. Typically, HPLC phases contain particles that are 2.5–3 µm in diameter. To take advantage of the increased peak resolution that occurs with decreases in particle size, ultra-high performance LC uses 1.7–1.8 µmparticles. However, a penalty for this is a substantial increase in backpressure. Nonetheless, pumps that can reliably deliver accurate flow rates against such backpressure are commercially available. A newer development is in the area of solid core particles (1.3 µm) with the liquid phase coated on the outside – this gives rise to very high column efficiencies with a lower backpressure than existing LC columns. The future may lie in using, as occurred in GC 36 years ago, columns where the stationary phase only coats the wall of a quartz tube, thereby substantially lower the backpressure and allowing for long columns and hence much higher chromatographic resolution.[68] Such approach has already been introduced for LC-MS in proteomics.[69]
An important property of ESI is that its sensitivity goes up inversely with the flow rate. This has been exploited in proteomics by using columns with smaller internal diameters. Instead of the 4.6-mm ID columns used in LC-MS analysis of drugs and other compounds where sensitivity is not limiting, in proteomics, the column ID is typically reduced to 75 µm resulting in flow rates of 200–300 nl/min. Assuming that the same amount of sample was analyzed on both columns and the linear flow rate through the column is the same, the difference in sensitivity is the square of the ratio of the two diameters, i.e. (4.6/0.075)2 = 3762. Increasing the column diameter to 200 µm (and a flow rate of 1 µl/min) still gives a 529-fold sensitivity increase. The disadvantage of columns with such low diameters is that they are very sensitive to changes in temperature in a laboratory. Contemporary columns are prepared by machine etching two blocks of silica so that once joined, they create a channel for the column (column on a chip), vastly improving reproducibility. Improvements in the connections of the ChipLC column to the nanoLC pump and the electrospray tip have led to the use of 200-µmID ChipLC columns where the overall delay from sample loading to the electrospray tip is 5 min. Operation of these ChipLC columns at a fixed temperature (for example 45 °C) leads to excellent retention time reproducibility (typically 0.3%, which translates to ±2–3 s). These approaches have led to development of nanoLC-MSMS analysis of polyphenols.[70]
Untargeted liquid chromatography-mass spectrometry metabolomics
When the investigator does not have a priori information about the possible nature of the metabolites and wants to examine all possible compounds in a metabolomics sample, an untargeted approach is used. The selection of the analytical platform is a trade-off between the length of the analytical run, the peak width and mass resolution and mass accuracy. Within the constraints of an analysis time of 30 min or less per sample, the instrument of choice for untargeted metabolomics is the hybrid quadrupole orthogonal time-of-flight (QqTOF) mass spectrometer. The modern TOF analyzer provides a mass resolution of 30 000–60 000 and mass accuracy of 2–3 ppm (using external mass standards). Importantly, this performance can be achieved with data collection times of 100 ms or less. In this scenario, instruments with detectors whose mass resolution and mass accuracy is time-dependent (Orbitraps and Fourier transform-ion cyclotron resonance instruments) have much reduced performances compared with their use under optimal conditions. A typical LC-MS metabolomics experiment uses a 1.1-s cycle time composed of an initial 100 ms high-resolution TOF-MS spectrum, followed by 50 ms high-resolution MSMS spectra of 20 selected molecular ions observed in the TOF-MS spectrum. This allows accurate representation of the areas under 10 s wide chromatographic peaks. Ultra performance liquid chromatography that generates even narrower peaks would require correspondingly shorter data acquisition times for MSMS spectra.
Targeted metabolomics
When the metabolites suspected of being associated with a phenotype are known, then LC-multiple reaction ion monitoring mass spectrometry is used to quantify them. This method is heavily used for analysis of pharmaceuticals and known biochemical molecules and is typically carried out on a triple quadrupole or Qtrap mass spectrometer. Precursor molecular ions are filtered by the first quadrupole (through a 0.7 m/z mass window) and accelerated and collided with N2 gas. The resulting product ions are filtered by the third quadrupole or by a linear trap. The advantage of the linear trap is that further ion fragmentation can occur, and while this may result in lowered signal intensity, the signal to noise can nonetheless be improved. Because known compounds are to be measured in targeted analysis, standard curves representing differing concentrations of each metabolite can be prepared and therefore the concentrations of these metabolites in the biological samples can be determined. If an isotopically labeled form of the metabolite is available, then the assay has the greatest accuracy and precision.
Isotope dilution mass spectrometry
In the case of targeted metabolomics, where a suite of metabolites is pre-selected for analysis, it is possible to include an isotopically labeled form of each metabolite (incorporating 2H, 13C or 15N stable isotopes).[71] The labeled forms are added in a constant amount to the biological samples and to known, but varying amounts of unlabeled forms of each metabolite. The assumption is that the stable isotopically labeled form of the metabolite will be subjected to the same processing losses of its non-labeled partner and that both will be eluted from the LC column into the mass spectrometer and hence reach the detector at the same time. Of course, labeled and unlabeled forms of a metabolite do behave differently – indeed, there is a whole science of MS applied to paleontology that relies on such differences.[72] However, for the purposes of isotope dilution MS in metabolomics, such differences are small and the method represents a marked improvement in reproducibility over an analysis where they are not used. Nevertheless, there are examples where selective interference from the matrix of the sample extract can interfere with quantification using isotope dilution MS.[73]
13C-labeled standards versus 2H-labeled standards
An issue that an investigator will encounter is that incorporation of deuterium causes the metabolite to be less hydrophobic and will therefore elute slightly earlier than its unlabeled analog from a reverse-phase column. This effect is well known. In the early days of gas chromatography, it was found that 2H6-cyclohexane could be separated from 1H6-cyclohexane.[74] Similarly, 2H9-labeled anabolic steroids separated from unlabeled steroids[75] – the cure was to use 13C-labeled steroids that do not exhibit this chromatographic separation. The phenomenon was also seen in the use of the isotope-coded affinity tag reagent in proteomics.[76] The initial form of the reagent was labeled with deuterium (2H8) but was replaced by the 13C8 form.[77] The best way to avoid problems created by the chromatographic differences isotopically labeled and unlabeled metabolites is to compare the areas under each peak rather than to use the ratio of their intensities. There is also a risk with deuterated standards in that many are introduced by exchange reactions. Therefore, they may undergo back exchange in solvent during storage.[78]
Sequential window acquisition of all theoretical (SWATH)-MS
A new development in LC-MS/MS proteomics has been the use of complete collection of MS/MS spectra on ALL precursor ions, termed SWATH-MS.[79,80] This could be adapted to metabolomics analysis. In this method, the mass window for the selection of ions is expanded from 1 Da, as is usually done in qualitative analysis, to 25 Da. All the precursor ions within the window are fragmented, and the product ions are separated into groups based on retention time. If a mass range from 100 to 900 Da is to be examined, there needs to be 32 collection windows with an isolation width of 25 Da. If 50-ms acquisitions are used for each 25 Da isolation window, a full duty cycle takes 100 + 32 × 25 ms = 1.7 s. To capture enough data points in the MS/MS domain to estimate the area under a peak, the minimum peak width should be 15 s. If the peaks are narrower, then the MS/MS data acquisition times should be reduced in proportion. While on a Q-TOF instrument this will not reduce mass resolution or mass accuracy, it will lower sensitivity. Arnhard et al.[81] applied SWATH-MS to the analysis of samples typically used in forensic casework in order to detect toxic or harmful compounds. While they had a high rate of success and low false discovery (<0.06%), their study revealed the need for fully automated, deconvolution software to extract adequate ion maps from the SWATH-MS data.
Lipidomics
There are many, many lipids in biological samples, perhaps in excess of 180 000. Lipids can be separated and analyzed by LC-MS using methods described in the preceding texts for water-soluble metabolites. It may be necessary to use a more non-polar, but water-miscible, solvent such as isopropanol to elute the lipids. Alternatively, there are two infusion-based methods for lipid analyses. In the first, MS/MSALL[82,83] lipid extracts are diluted into methanol : chloroform (2 : 1 v/v) containing 5 mM ammonium acetate and infused at 7 µl/min into the ESI source. Data are recorded in the positive ion and negative ion modes over the mass range from 200 to 1200 m/z. A 250 ms high-resolution MS scan is initially acquired. It is then followed by a series of 500 ms MS/MS scans of the product ions of lipid molecular ion precursors. The latter are selected by the quadrupole filter increasing by 1 m/z at a time starting at 200 m/z and continuing to 1200 m/z. Each mass window is 1.2 m/z. The collision energy is adjusted to give an energy spread during product ion data acquisition. The data are processed with suitable software (LipidView™, SCIEX). While this approach allows the identification of thousands of lipid species, it nonetheless is only semi-quantitative. Lipid internal standards do not work because they will be at different masses and hence be measured at different times during the infusion. Also, the overlapping ion masses because of lipids having very similar masses and to natural abundance isotopic forms make data interpretation difficult. This can be offset by using high mass resolution Fourier transform-ion cyclotron resonance or Orbitrap instruments to record the MS and MSMS data.
A second method of performing infusion-based lipidomics is to carry out differential ion mobility prior to the entry of molecular ions into the mass spectrometer.[84–86] This method separates lipid classes before they are subjected to quadrupole selection of molecular ions and then collision-induced formation of product ions, thereby substantially deconvoluting complex overlapping ions. It has been applied to the separation of leukotriene and protectins.[86] This approach has been combined with the use of an internal standard containing >1000 deuterated lipid species (commercially available from Metabolon). Extracts are analyzed on a SCIEX 5500 Qtrap.[87] This method results in much improved quantification because the standards are added prior to the Bligh–Dyer extraction step and thereby take into account differences in extractability when partitioning lipids across a chloroform-methanol-water two-phase system. It therefore generates absolute quantitative data. However, the high expense of the software needed to process these data may preclude its wide application until open source software becomes available.
Capillary electrophoresis-mass spectrometry
Capillary electrophoresis has had a long history in the analysis of individual metabolites.[88,89] It is excellent for the separation of hydrophilic compounds and is comparable with, but different from, hydrophilic interaction liquid chromatography and normal-phase chromatography. Importantly, it can be scaled down to very low sample volumes (1–20 nl). Its limitation for many years with regard to CE-MS applications has been the nature of the CE-ESI interface. The sheath-liquid interface design, whereby fluid eluting from the capillary is mixed with a makeup fluid, caused dilution of the sample eluting from the capillary and thereby considerable loss of sensitivity. This restricted its applicability to metabolomics. However, the introduction of the sheathless porous tip interface[90] has considerably improved the sensitivity possible with CE-MS, and a commercial form suitable for attachment to mass spectrometers of several leading mass spectrometer manufacturers has been developed by Beckman Coulter-SCIEX. While CE-MS is very effective for hydrophilic metabolites, it is also possible to perform the electrophoresis using capillaries packed with a reverse-phase packing.[91]
Direct use of cerebrospinal fluid[92] and serum[93] in CE-MS led to protein precipitation within the capillary. It was necessary to use ultrafiltration of these fluids to avoid this issue. On the other hand, using sheathless fluid CE-MS, simple dilution of cerebrospinal fluid, plasma and urine 1 : 1 with water gave excellent results[94] with sensitivities in the low nanometer. For urine, ~3500 features were detected using an injection volume of 9 nl. CE-MS with sheathless fluid interface has a great future and certainly complements existing GC-MS and LC-MS methods. However, it has yet to be rigorously studied from the standpoint of reproducibility and robustness. So far, it has been limited to the study of cationic metabolites although a non-aqueous CE-MS method has been described for drug β-glucuronides.[95]
Quality control
Monitoring recovery
The response of a MS detector is not absolute, nor do the methods of processing a biological sample in preparation for analysis yield 100% recovery of all the metabolites. These two effects present a considerable challenge in obtaining an accurate assessment of the concentrations of the metabolites comprising the metabolome. Unless the investigator makes serious efforts to control these variables, it is not only difficult to uncover the real differences between sample groups but it also encourages ‘discovery’ of false effects.
Although there are efforts to provide investigators with kits containing a wide range of isotopically labeled metabolites, for many investigators, either the labeled forms of the metabolites do not exist or they are unable to afford the considerable expense involved. This is the case for untargeted (or global) metabolomics. However, even in this approach, it is advisable to use two to three isotopically labeled internal standards that represent the range of metabolites that would be separated by the LC-MS method chosen for the analysis. Examples of those that have been used for reverse-phase LC-MS are 13C11-tryptophan, 13C4-succinate and 13C16-palmitate. The advantage of using these particular compounds is that they are chemically stable. The total cost of purchasing at least 100 mg of each is ~$3000. If 1 µg of each is added to samples to be tested, the standards could be used for over 10 000 assays. Therefore, this cost could be spread across multiple investigators. It is probable that commercial companies will provide products such as this that would enable comparison of studies from different institutions.
Sample pooling
A second important quality control procedure is to generate a pooled sample to represent all the samples to be analyzed.[96] For example, if the lab receives ~0.5 ml of a biofluid (serum/plasma/urine), 50 µl should be taken from each and combined to make a pooled sample that is well mixed and then divided into aliquot volumes that are the same as those to be used for analysis of the unknown samples. These aliquots are frozen along with the unknown samples and thawed once when needed to be included for analysis. This pooled quality control sample should be run with each group of unknown samples. For a large sample set, the quality control sample should be run every 10 unknown samples to determine within-assay variability.
A laboratory should establish the variation that occurs for repeated analysis of the same sample extract, for multiple extractions of the same sample on 1 day and for multiple extractions of the same sample over a period of time.
Distinguishing metabolites from artifacts
A typical untargeted LC-MS analysis may result in the detection of 2000–10 000 ions – this number of metabolite ions depends on the amount of the sample loaded and its complexity, the chromatographic resolution of the LC system and the speed and mass resolution of the mass spectrometer. Some of the ions have nothing to do with the biological samples. A way to distinguish some of the contaminants is to run several dilutions of a pooled sample. Ions that do not change with respect the amount of the sample can be eliminated from further analysis. This is most easily done after pre-processing software (XCMS – refer to paper to be published in the next issue of the Journal of Mass Spectrometry) has identified reproducible peaks across all samples.
Because of the soft ionization aspect of ESI, a metabolite may appear in several precursor ion forms. These include 13C isotope ions ([M+H+ 13Cn]+ or [M−H+ 13Cn]−), with and without adduct ions ([M+Na]+, [M+K]+, [M+NH4]+, [M+H+CH3OH]+, [M+H+CH3CN]+; [M−H+ Cl]−, [M−H+ HCOO]−, [M−H+CH3COO]−). Metabolites present in the highest concentrations may also form ions such as [2 M+H]+ and [2 M−H]+. Although this may at first seem complex, the mass differences between these forms are easy to recognize in a high mass resolution mass spectrometer. An important issue is that all of these precursor ions of a metabolite will elute with the same retention time, thereby allowing the investigator or the software they are using to distinguish between the various forms of the molecular ion and other metabolite(s) having similar m/z values but different retention times.
Acknowledgments
Funds for the National Workshop in Metabolomics came from NIH grant R25 GM103798-03. The workshop also received support from the UAB Office of the Vice-President for Research and Economic Development, the UAB School of Medicine and College of Arts and Science, the UAB Comprehensive Cancer Center (P30 CA13148), the UAB Comprehensive Diabetes Center, the UAB Diabetes Research Center (P60 DK079626), the UAB Center for Free Radical Biology and the UAB-UCSD O′Brien Center for Acute Kidney Disease (P30 DK079337), and the Departments of Chemistry and Pharmacology and Toxicology. We are also grateful for unrestricted support from Metabolon, Sciex and Waters. Acknowledgements are also given to Dr N. Rama Krishna for the use of the Central Alabama NMR facility and to staff who assisted in the running of the workshop (UAB: Jennifer Spears, Lynn Waddell, Mikako Kawai, D. Ray Moore II, Landon Wilson, Ali Arabshahi and Ronald Shin; RTI International, Rodney Snyder) and student trainees, Haley Albright, Kelly Walters and Sean Wilkinson. We are indebted to faculty who have also previously contributed to the development of the workshop: Dr Kathleen Stringer (University of Michigan), Dr Dean P. Jones (Emory University), Dr Grier P. Page (RTI International), Dr Olga Ilkayeva (Duke University), Dr Nikolaos Psychogios (Shire Pharmaceuticals) and Dr Natalie Serkova (University of Colorado-Denver) and to faculty in receipt of NIH Administrative Supplements (Dr Lalita Shevde-Sumant, R01 CA138850 and Dr Adam Wende R00 HL111322). Finally, the workshop has been enhanced by several plenary speakers: Dr Stanley Hazen (Cleveland Clinic), Dr Keith Baggerly (MD-Anderson), Dr Richard Caprioli (Vanderbilt University), Dr Arthur Edison (University of Florida) and Dr David Wishart (University of Alberta), as well as speakers from several companies: Brigitte Simons and Jeremiah Tipton (SCIEX), Tom Beaty and John Shockcor (Waters) and Edward Karoly and Rob Mohney (Metabolon).
Footnotes
The definition of the metabolome being compounds up to 1500 Da is somewhat arbitrary. Compounds defined this way may not be part of metabolic pathways, such as most, but not all, short peptides. It also omits compounds formed by metabolic reactions, e.g. cardiolipins, that have molecular weights greater than 1500 Da.
Stoll M, Wilson LS, Arabshahi A and Barnes S, unpublished observations
The complete set of training materials including slide sets and videos can be viewed at http://www.uab.edu/proteomics/metabolomics/workshop/workshop_june_2015.php.
References
- 1.Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. U. S. A. 1996;93:10614. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mehta T, Tanik M, Allison DB. Towards sound epistemological foundations of statistical methods for high-dimensional biology. Nat. Genet. 2004;36:943. doi: 10.1038/ng1422. [DOI] [PubMed] [Google Scholar]
- 3.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 4.Hillenkamp F, Karas M. Mass spectrometry of peptides and proteins by matrix-assisted ultraviolet laser desorption/ionization. Methods Enzymol. 1990;193:280. doi: 10.1016/0076-6879(90)93420-p. [DOI] [PubMed] [Google Scholar]
- 5. http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-04-002.html.
- 6. https://commonfund.nih.gov/metabolomics/index.
- 7.Regional Comprehensive Metabolomics Research Centers: Michigan Regional Comprehensive Metabolomics Resource Core. ( http://mrc2.umich.edu/index.php); NIH West Coast Metabolomics Center at UC Davis ( http://metabolomics.ucdavis.edu); NIH Eastern Regional Comprehensive Metabolomics Resource Core at RTI International ( http://www.rti.org/page.cfm?obj=3BC41B11-068E-1405-9A6F79D91D8D69EC); Southeast Center for Integrated Metabolomics (SECIM) ( http://secim.ufl.edu); Resource Center for Stable Isotope-Resolved Metabolomics (RC-SIRM) ( http://bioinformatics.cesb.uky.edu/bin/view/RCSIRM/); Metabolomics Core at Mayo Clinic ( http://www.mayo.edu/research/core-resources/metabolomics-resource-core/overview) [Google Scholar]
- 8. http://www.uab.edu/proteomics/metabolomics/workshop.php. [Google Scholar]
- 9.Want EJ, Wilson ID, Gika H, Theodoridis G, Plumb RS, Shockcor J, Holmes E, Nicholson JK. Global metabolic profiling procedures for urine using UPLC-MS. Nat. Protoc. 2010;5:1005–1018. doi: 10.1038/nprot.2010.50. [DOI] [PubMed] [Google Scholar]
- 10.Beckonert O, Coen M, Keun HC, Wang Y, Ebbels TM, Holmes E, Lindon JC, Nicholson JK. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat. Protoc. 2010;5:1019. doi: 10.1038/nprot.2010.45. [DOI] [PubMed] [Google Scholar]
- 11.Smart KF, Aggio RB, Van Houtte JR, Villas-Bôas SG. Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography-mass spectrometry. Nat. Protoc. 2010;5:1709. doi: 10.1038/nprot.2010.108. [DOI] [PubMed] [Google Scholar]
- 12.Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 2011;6:743. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 13.Dunn WB, Broadhurst D, Begle P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN, Nicholls AW, Wilson ID, Kell DB, Goodacre R Human Serum Metabolome (HUSERMET) Consortium. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011;6:1060. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 14.Zhu ZJ, Schultz AW, Wang J, Johnson CH, Yannone SM, Patti GJ, Siuzdak G. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat. Protoc. 2013;8:451. doi: 10.1038/nprot.2013.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Axelson M, Kirk DN, Farrant RD, Cooley G, Lawson AM, Setchell KD. The identification of the weak oestrogen equol [7-hydroxy-3-(4’-hydroxyphenyl)chroman] in human urine. Biochem. J. 1982;201:353. doi: 10.1042/bj2010353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Overmyer KA, Thonusin C, Qi NR, Burant CF, Evans CR. Impact of anesthesia and euthanasia on metabolomics of mammalian tissues: studies in a C57BL/6J mouse model. PLoS One. 2015;10:e0117232. doi: 10.1371/journal.pone.0117232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dane AD, Hendriks MMWB, Reijmers TH, Harms AC, Troost J, Vreeken RJ, Boomsma DI, van Duijn CM, Slagboom EP, Hankemeier T. Integrating metabolomics profiling measurements across multiple biobanks. Anal. Chem. 2014;86:4110. doi: 10.1021/ac404191a. [DOI] [PubMed] [Google Scholar]
- 18.Smilde AK. Analysis of high-dimensional data from designed metabolomics studies. Metab. Profiling: Disease and Xenobiotics. 2014;21:117. [Google Scholar]
- 19.Park Y, Kim SB, Wang B, Blanco RA, Le NA, Wu S, Accardi CJ, Alexander RW, Ziegler TR, Jones DP. Individual variation in macronutrient regulation measured by proton magnetic resonance spectroscopy of human plasma. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009;297:R202. doi: 10.1152/ajpregu.90757.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scalbert A, Brennan L, Manach C, Andres-Lacueva C, Dragsted LO, Draper J, Rappaport SM, van der Hooft JJ, Wishart DS. The food metabolome: a window over dietary exposure. Am. J. Clin. Nutr. 2014;99:1286. doi: 10.3945/ajcn.113.076133. [DOI] [PubMed] [Google Scholar]
- 21.Brown NM, Setchell KD. Animal models impacted by phytoestrogens in commercial chow: implications for pathways influenced by hormones. Lab. Invest. 2001;81:735. doi: 10.1038/labinvest.3780282. [DOI] [PubMed] [Google Scholar]
- 22.Reeves PG, Nielsen FH, Fahey GC., Jr AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 1993;123:1939. doi: 10.1093/jn/123.11.1939. [DOI] [PubMed] [Google Scholar]
- 23.Lewis M, Ullrey DE, Barnard DE, Napka JJ. Nutrition. In: Suckow MA, Weisbroth SH, Franklin CL, editors. The Laboratory rat. 2nd. Chapter 9. Boston: Academic Press; 2005. pp. 219–302. [Google Scholar]
- 24.Robertson DG, Ruepp SU, Stryker SA, Hnatyshyn SY, Shipkova PA, Aranibar N, Mcnaney CA, Fiehn O, Reily MD. Metabolomic and transcriptomic changes induced by overnight (16 h) fasting in male and female Sprague-Dawley rats. Chem. Res. Toxicol. 2011;24:481. doi: 10.1021/tx200074f. [DOI] [PubMed] [Google Scholar]
- 25.Minami Y, Kasukawa T, Kakazu Y, Iigo M, Sugimoto M, Ikeda S, Yasui A, van der Horst GT, Soga T, Ueda HR. Measurement of internal body time by blood metabolomics. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9890. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buffalo V. Bioinformatics data skills. Sebastopol, CA: O’Reilly Media Inc.; 2015. [Google Scholar]
- 27.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 28.Serkova NJ, Brown MS. Quantitative analysis in magnetic resonance spectroscopy: from metabolic profiling to in vivo biomarkers. Bioanalysis. 2012;4:321. doi: 10.4155/bio.11.320. [DOI] [PubMed] [Google Scholar]
- 29.Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B, Young N, Xia J, Knox C, Dong E, Huang P, Hollander Z, Pedersen TL, Smith SR, Bamforth F, Greiner R, McManus B, Newman JW, Goodfriend T, Wishart DS. The human serum metabolome. PLoS One. 2011;6:e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beckonert O, Keun HC, Ebbels TMD, Bundy J, Holmes E, Lindon JC, Nicholson JK. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007;2:2692. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 31.Hensley K. Detection of protein carbonyls by means of biotin hydrazide-streptavidin affinity methods. Methods Mol. Biol. 2015;1314:95. doi: 10.1007/978-1-4939-2718-0_11. [DOI] [PubMed] [Google Scholar]
- 32.Hansen KC, Schmitt-Ulms G, Chalkley RJ, Hirsch J, Baldwin MA, Burlingame AL. Mass spectrometric analysis of protein mixtures at low levels using cleavable 13C-isotope-coded affinity tag and multidimensional chromatography. Mol. Cell. Proteomics. 2003;2:299. doi: 10.1074/mcp.M300021-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Viant MR. Revealing the metabolome of animal tissues using 1H nuclear magnetic resonance spectroscopy. Methods Mol. Biol. 2007;358:229. doi: 10.1007/978-1-59745-244-1_13. [DOI] [PubMed] [Google Scholar]
- 34.Martineau E, Tea I, Loaëc G, Giraudeau P, Akoka S. Strategy for choosing extraction procedures for NMR-based metabolomic analysis of mammalian cells. Anal. Bioanal. Chem. 2011;401:2133. doi: 10.1007/s00216-011-5310-y. [DOI] [PubMed] [Google Scholar]
- 35.Dulski TR. A Manual for the Chemical Analysis of Metals. West Conshohocken: ASTM; 1942. p. 34. [Google Scholar]
- 36.Martano G, Delmotte N, Kiefer P, Christen P, Kentner D, Bumann D, Vorholt JA. Fast sampling method for mammalian cell metabolic analyses using liquid chromatography-mass spectrometry. Nat. Protoc. 2015;10:1. doi: 10.1038/nprot.2014.198. [DOI] [PubMed] [Google Scholar]
- 37.Lorenz MA, Burant CF, Kennedy RT. Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Anal. Chem. 2011;83:3406. doi: 10.1021/ac103313x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chain EB, Frank M, Pocchiari F, Rossi C, Ugolini F, Ugolini G. Quantitative bidimensional radiochromatography. Instituto Superiore di Sanita. 1956;1:241. [Google Scholar]
- 39.Chain EB, Lowe AE, Mansford KRL. A computerised scanner for bidimensional radiochromatograms. J. Chromatogr. 1970;53:293. [Google Scholar]
- 40.Hitchcock C, James AT. Oxidation of unsaturated fatty acids by leaf tissue. J. Lipid Res. 1964;5:593. [PubMed] [Google Scholar]
- 41.Popjak G, Lowe AE, Moore D, Brown L, Smith FA. Scintillation counter for the measurement of radioactivity of vapors in conjunction with gas-liquid chromatography. J. Lipid Res. 1959;1:29. [Google Scholar]
- 42.Bax A, Svererenyi NM, Maciel GE. Two-dimensional NMR spectroscopy. In: Petrakis L, Fraissard JP, editors. Magnetic Resonance, Advanced topics and Applications to Fossil Energy. Dordrecht: Reidel Publishing Co; 1984. pp. 137–145. [Google Scholar]
- 43.Barnes S, Kirk DN. Nuclear magnetic resonance spectroscopy of bile acids. In: Setchell KDR, Kritchevsky D, Nair PP, editors. “The Bile Acids-Chemistry, Physiology, and Metabolism” (Vol. 4: Methods and Applications) New York: Plenum Press; 1988. pp. 65–136. [Google Scholar]
- 44.Barnes S, Geckle JM. High resolution nuclear magnetic resonance spectroscopy of bile salts: individual proton assignments for sodium cholate in aqueous solution at 400 MHz. J. Lipid Res. 1982;23:161. [PubMed] [Google Scholar]
- 45.Bax A. Broadband homonuclear decoupling in heteronuclear shift correlation NMR spectroscopy. J. Magn. Reson. 1983;53:517. [Google Scholar]
- 46.Waterhous DV, Barnes S, Muccio DD. Nuclear magnetic resonance spectroscopy of bile acids. Development of two-dimensional NMR methods for the elucidation of proton resonance assignments for five common hydroxylated bile acids, and their parent bile acid, 5 beta-cholanoic acid. J. Lipid Res. 1985;26:1068. [PubMed] [Google Scholar]
- 47.Evanochko WT, Ng TC, Lilly MB, Lawson AJ, Corbett TH, Durant JR, Glickson JD. In vivo 31P NMR study of the metabolism of murine mammary 16/C adenocarcinoma and its response to chemotherapy, x-radiation, and hyperthermia. Proc. Natl. Acad. Sci. U. S. A. 1983;80:334. doi: 10.1073/pnas.80.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neurohr KJ, Barrett EJ, Shulman RG. In vivo carbon-13 nuclear magnetic resonance studies of heart metabolism. Proc. Natl. Acad. Sci. U. S. A. 1983;80:1603. doi: 10.1073/pnas.80.6.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nicholson JK, Lindon JC, Holmes E. ’Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 50.Lacy P, McKay RT, Finkel M, Karnovsky A, Woehler S, Lewis MJ, Chang D, Stringer KA. Signal intensities derived from different NMR probes and parameters contribute to variations in quantification of metabolites. PLoS One. 2014;9:e85732. doi: 10.1371/journal.pone.0085732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giraudeau P, Silvestre V, Akoka S. Optimizing water suppression for quantitative NMR in metabolomics: a tutorial review. Metabolomics. 2015;11:1041. [Google Scholar]
- 52.Cheng LL, Ma MJ, Becerra L, Ptak T, Tracey I, Lackner A, Gonzalez RG. Quantitative neuropathology by high-resolution magic angle spinning proton magnetic resonance spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6408. doi: 10.1073/pnas.94.12.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal. Chem. 2006;78:4430. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 54.Jie H, Astle W, De Iorio M, Ebbels TMD. BATMAN—an R package for the automated quantification of metabolites from nuclear magnetic resonance spectra using a Bayesian model. Bioinformatics. 2012;28:2088. doi: 10.1093/bioinformatics/bts308. [DOI] [PubMed] [Google Scholar]
- 55.Siamak R, Liu P, Mandal R, Grant JR, Wilson M, Eisner R, Sinelnikov I, Hu X, Luchinat C, Greiner R, Wishart DS. Accurate, fully-automated NMR spectral profiling for metabolomics. PLoS One. 2015;10:e0132873. doi: 10.1371/journal.pone.0124219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beckwith-Hall BM, Thompson NA, Nicholson JK, Lindon JC, Holmes E. A metabonomic investigation of hepatotoxicity using diffusion-edited 1H NMR spectroscopy of blood serum. Analyst. 2003;128:814. doi: 10.1039/b302360p. [DOI] [PubMed] [Google Scholar]
- 57.Sumner LW, Mendes P, Dixon RA. Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry. 2003;62:817. doi: 10.1016/s0031-9422(02)00708-2. [DOI] [PubMed] [Google Scholar]
- 58.Tikunov Y, Lommen A, de Vos CHR, Verhoeven HA, Bino RJ, Hall RD, Bovy AG. A novel approach for nontargeted data analysis for metabolomics. Large-scale profiling of tomato fruit volatiles. Plant Physiol. 2005;139:1125. doi: 10.1104/pp.105.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Halket JM, Waterman D, Przyborowska AM, Patel RKP, Fraser PD, Bramley PM. Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. J. Exp. Bot. 2005;56:219. doi: 10.1093/jxb/eri069. [DOI] [PubMed] [Google Scholar]
- 60. http://chemdata.nist.gov/mass-spc/amdis/docs/amdis.pdf.
- 61. http://www.nist.gov/srd/upload/NIST1a11Ver2-0Man.pdf.
- 62.Kind T, Wohlgemuth G, Lee DY, Lu Y, Palazoglu M, Shahbaz S, Fiehn O. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009;81:10038. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, Willmitzer L, Fernie AR, Steinhauser D. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics. 2005;21:1635. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- 64.Kovats E. Gas-chromatographische Charakterisierung organischer Verbindungen. Teil 1: Retentionsindices aliphatischer Halogenide, Alkohole, Aldehyde und Ketone. Helv. Chim. Acta. 1958;41:1915. [Google Scholar]
- 65. http://chemdata.nist.gov/dokuwiki/doku.php?id=chemdata:amdis.
- 66.Whitehouse CM, Dreyer RN, Yamashita M, Fenn JB. Electrospray interface for liquid chromatographs and mass spectrometers. Anal. Chem. 1985;57:675. doi: 10.1021/ac00280a023. [DOI] [PubMed] [Google Scholar]
- 67.Bruins AP. Liquid chromatography-mass spectrometry with ionspray and electrospray interfaces in pharmaceutical and biomedical research. J. Chromatogr. 1991;554:39. doi: 10.1016/s0021-9673(01)88435-1. [DOI] [PubMed] [Google Scholar]
- 68.Dandedau RD, Zenner EM. J. High Res. Chromatogr. 1979;2:351. [Google Scholar]
- 69.Luo Q, Gu Y, Wu SL, Rejtar T, Karger BL. Two-dimensional strong cation exchange/porous layer open tubular/mass spectrometry for ultratrace proteomic analysis using a 10 microm id poly(styrene-divinylbenzene) porous layer open tubular column with an on-line triphasic trapping column. Electrophoresis. 2008;29:1604. doi: 10.1002/elps.200700741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilson L, Arabshahi A, Simons B, Prasain JK, Barnes S. Improved high sensitivity analysis of polyphenols and their metabolites by nano-liquid chromatography-mass spectrometry. Arch. Biochem. Biophys. 2014;559:3. doi: 10.1016/j.abb.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thompson DK, Sloane R, Bain JR, Stevens RD, Newgard CB, Pieper CF, Kraus VB. Daily variation of serum acylcarnitines and amino acids. Metabolomics. 2012;8:556. doi: 10.1007/s11306-011-0345-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Layman CA, Araujo MS, Boucek R, Hammerschlag-Peyer CM, Harrison E, Jud ZR, Matich P, Rosenblatt AE, Vaudo J, Yeager LA, Post DM, Bearhop S. Applying stable isotopes to examine food web structure: an overview of analytical tools. Biol. Rev. 2012;87:545. doi: 10.1111/j.1469-185X.2011.00208.x. [DOI] [PubMed] [Google Scholar]
- 73.Wang S, Cyronak M, Yang E. Does a stable isotopically labeled internal standard always correct analyte response? A matrix effect study on a LC/MS/MS method for the determination of carvedilol enantiomers in human plasma. J. Pharm. Biomed. Anal. 2007;43:701. doi: 10.1016/j.jpba.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 74.Wilzbach KE, Riesz P. Isotope effects in gas-liquid chromatography. Science. 1957;126:748. doi: 10.1126/science.126.3277.748. [DOI] [PubMed] [Google Scholar]
- 75.Dehennin L, Reiffsteck A, Scholler R. Simple methods for the synthesis of twenty different, highly enriched deuterium labelled steroids, suitable as internal standards for isotope dilution mass spectrometry. Biol. Mass Spec. 1980;7:493. [Google Scholar]
- 76.Zhang R, Sioma CS, Thompson RA, Xiong L, Regnier FE. Controlling deuterium isotope effects in comparative proteomics. Anal. Chem. 2002;74:3662. doi: 10.1021/ac025614w. [DOI] [PubMed] [Google Scholar]
- 77.Bottari P, Aebersold R, Turecek F, Gelb MH. Design and synthesis of visible isotope-coded affinity tags for the absolute quantification of specific proteins in complex mixtures. Bioconjug. Chem. 2004;15:380. doi: 10.1021/bc034174s. [DOI] [PubMed] [Google Scholar]
- 78.Davison AS, Milan AM, Dutton JJ. Potential problems with using deuterated internal standards for liquid chromatography-tandem mass spectrometry. Ann. Clin. Biochem. 2013;50:274. doi: 10.1177/0004563213478938. [DOI] [PubMed] [Google Scholar]
- 79.Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.O111.016717. O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sajic T, Liu Y, Aebersold R. Using data-independent, high-resolution mass spectrometry in protein biomarker research: perspectives and clinical applications. Proteomics Clin. Appl. 2015;9:307. doi: 10.1002/prca.201400117. [DOI] [PubMed] [Google Scholar]
- 81.Arnhard K, Gottschall A, Pitterl F, Oberacher H. Applying ’Sequential Windowed Acquisition of All Theoretical Fragment Ion Mass Spectra’ (SWATH) for systematic toxicological analysis with liquid chromatography-high-resolution tandem mass spectrometry. Anal. Bioanal. Chem. 2015;407:405. doi: 10.1007/s00216-014-8262-1. [DOI] [PubMed] [Google Scholar]
- 82.Simons B, Kauhanen D, Sylvänne T, Tarasov K, Duchoslav E, Ekroos K. Shotgun lipidomics by sequential precursor ion fragmentation on a hybrid quadrupole time-of-flight mass spectrometer. Metabolites. 2012;2:195. doi: 10.3390/metabo2010195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Prasain JK, Wilson LS, Hoang HD, Moore DR, II, Miller MA. Comparative lipidomics of Caenorhabditis elegans metabolomics disease models by SWATH non-targeted tandem mass spectrometry. Metabolites. 2015;5:677. doi: 10.3390/metabo5040677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lintonen TP, Baker PR, Suoniemi M, Ubhi BK, Koistinen KM, Duchoslav E, Campbell JL, Ekroos K. Differential mobility spectrometry-driven shotgun lipidomics. Anal. Chem. 2014;86:9662. doi: 10.1021/ac5021744. [DOI] [PubMed] [Google Scholar]
- 85.Baker PR, Armando AM, Campbell JL, Quehenberger O, Dennis EA. Three-dimensional enhanced lipidomics analysis combining UPLC, differential ion mobility spectrometry, and mass spectrometric separation strategies. J. Lipid Res. 2014;55:2432. doi: 10.1194/jlr.D051581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jónasdóttir HS, Papan C, Fabritz S, Balas L, Durand T, Hardardottir I, Freysdottir J, Giera M. Differential mobility separation of leukotrienes and protectins. Anal. Chem. 2015;87:5036. doi: 10.1021/acs.analchem.5b00786. [DOI] [PubMed] [Google Scholar]
- 87.Conner A, Ubhi BK, Duchoslav E, Wang L, Baker PR, Watkins S. A novel lipid screening platform allowing a complete solution for lipidomics research. Poster abstract at 63rd American Society for Mass Spectrometry National Meeting; St. Louis, MO. 2015. [Google Scholar]
- 88.Wakayama M, Hirayama A, Soga T. Capillary electrophoresis-mass spectrometry. Methods Mol. Biol. 2015;1277:113. doi: 10.1007/978-1-4939-2377-9_9. [DOI] [PubMed] [Google Scholar]
- 89.Ramautar R, Somsen GW, de Jong GJ. CE-MS for metabolomics: developments and applications in the period 2012–2014. Electrophoresis. 2015;36:212. doi: 10.1002/elps.201400388. [DOI] [PubMed] [Google Scholar]
- 90.Moini M. Simplifying CE-MS operation. 2. Interfacing low-flow separation techniques to mass spectrometry using a porous tip. Anal. Chem. 2007;79:4241. doi: 10.1021/ac0704560. [DOI] [PubMed] [Google Scholar]
- 91.Wu Q, Yu X, Wang Y, Gu X, Ma X, Lv W, Chen Z, Yan C. Pressurized CEC coupled with QTOF-MS for urinary metabolomics. Electrophoresis. 2014;35:2470. doi: 10.1002/elps.201400117. [DOI] [PubMed] [Google Scholar]
- 92.Ibáñez C, Simó C, Martín-Álvarez PJ, Kivipelto M, Winblad B, Cedazo-Mínguez A, Cifuentes A. Toward a predictive model of Alzheimer’s disease progression using capillary electrophoresis-mass spectrometry metabolomics. Anal. Chem. 2012;84:8532. doi: 10.1021/ac301243k. [DOI] [PubMed] [Google Scholar]
- 93.Naz S, Garcia A, Rusak M, Barbas C. Method development and validation for rat serum fingerprinting with CE-MS: application to ventilator-induced-lung-injury study. Anal. Bioanal. Chem. 2013;405:4849. doi: 10.1007/s00216-013-6882-5. [DOI] [PubMed] [Google Scholar]
- 94.Ramautar RI, Shyti R, Schoenmaker B, de Groote L, Derks RJ, Ferrari MD, van den Maagdenberg AM, Deelder AM, Mayboroda OA. Metabolic profiling of mouse cerebrospinal fluid by sheathless CE-MS. Anal. Bioanal. Chem. 2012;404:2895. doi: 10.1007/s00216-012-6431-7. [DOI] [PubMed] [Google Scholar]
- 95.Bonvin G, Schappler J, Rudaz S. Non-aqueous capillary electrophoresis for the analysis of acidic compounds using negative electrospray ionization mass spectrometry. J. Chromatogr. A. 2014;1323:163. doi: 10.1016/j.chroma.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 96.Ejigu BA, Valkenborg D, Baggerman G, Vanaerschot M, Witters E, Dujardin JC, Burzykowski T, Berg M. Evaluation of normalization methods to pave the way towards large-scale LC-MS-based metabolomics profiling experiments. OMICS. 2013;17:473. doi: 10.1089/omi.2013.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]