SUMMARY
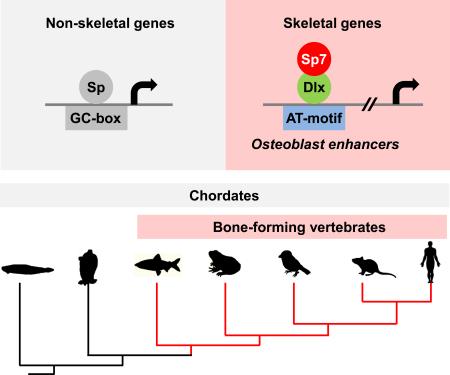
Bone formation in extant species is restricted to vertebrate species. Sp7/Osterix is a key transcriptional determinant of bone-secreting osteoblasts. We performed Sp7 ChIP-seq analysis identifying a large set of predicted osteoblast enhancers and validated a subset of these in cell culture and transgenic mouse assays. Sp-family members bind GC-rich target sequences through their zinc finger domain. Several lines of evidence suggest Sp7 acts differently, engaging osteoblast targets in Dlx-containing regulatory complexes bound to AT-rich motifs. Amino acid differences in the Sp7 zinc finger domain reduce Sp7's affinity for the Sp-family consensus GC-box target; Dlx5 binding maps to this domain of Sp7. The data support a model in which Dlx recruitment of Sp7 to osteoblast enhancers underlies Sp7-directed osteoblast specification. As an Sp7-like zinc finger variant is restricted to vertebrates, the emergence of an Sp7 member within the Sp family was likely closely coupled to evolution of the bone forming vertebrates.
Keywords: osteoblast specification, bone, Sp7/Osterix, Dlx, cis-regulation, evolution
Graphical Abstract
INTRODUCTION
Mammalian bones form from three distinct cellular populations through two different developmental mechanisms (Olsen et al., 2000). Cranially, neural crest cells form facial bones and much of the skull. Paraxial mesoderm also contributes to the skull and generates the axial skeleton of the trunk. Finally, lateral plate mesoderm generates the appendicular limb skeleton. In the face and skull, intramembranous ossification is the predominant mechanism of osteogenesis. Here, mesenchymal osteoblasts condense and give rise directly to bone. The remainder of the boney skeleton forms through the secondary ossification of a cartilage template: the process of endochondral ossification. Regardless of cellular origin or mode of ossification, a common bone matrix-secreting cell type, the osteoblast, underlies bone development. Genetic studies demonstrate Runt-related transcription factor 2 (Runx2) (Komori et al., 1997) and Sp7/Osterix (Nakashima et al., 2002) are essential regulators of the osteoblast program: both Runx2- and Sp7-null mutants lack all bones in mice and mutations in these genes underlie bone diseases in man (Lapunzina et al., 2010; Mundlos et al., 1997).
In the osteoblast program, mesenchymal progenitors are initially committed to Runx2-positive osteoblast precursors that transition to Runx2- and Sp7-double positive osteoblast progenitors before adopting a mature osteoblast phenotype. Runx2-positive osteoblast precursors are present but fail to differentiate into osteoblasts in Sp7-deficient mice whereas no Sp7 expression is observed in Runx2 deficient skeletal elements (Nakashima et al., 2002). Thus, Sp7 acts genetically downstream of Runx2 in the regulatory hierarchy of specifying an osteoblast cell fate.
The molecular targets of Sp7 action in the osteoblast program are not well understood. A limited number of non-coding regions flanking Col1a1 (Koga et al., 2005; Nakashima et al., 2002), Col1a2 (Yano et al., 2014), Col5a1(Wu et al., 2010), Col5a3 (Yun-Feng et al., 2010), Dkk1 (Zhang et al., 2012a), Mmp13 (Meyer et al., 2015; Zhang et al., 2012b), Sost (Zhou et al., 2010), VEGF (Tang et al., 2012), and Runx2 (Kawane et al., 2014) have been identified as potential Sp7 associated cis-regulatory osteoblast enhancers. However, there has been no genomic scale identification of Sp7-directed osteogenesis.
The mechanism of Sp7's mode of action is also unclear. Sp7 is a member of the Sp family of transcriptional regulators (Nakashima et al., 2002). Each Sp family member contains a zinc finger domain that comprises three zinc fingers. In Sp1, the best studied member, the zinc finger domain binds directly at a GC-rich specific DNA recognition sequence (GGGCGG; the GC-box) (Kadonaga et al., 1986). ChIP-seq analysis of Sp1 and Sp2 nuclear interactions (Wang et al., 2012) and high throughput screening of protein-DNA bindings for Sp1, Sp3, and Sp4 (Hume et al., 2015; Wingender et al., 2013) have highlighted Sp-factor binding to a GC-rich target site. However, data are less clear for Sp7. Several in vitro binding studies argued that Sp7 shares a GC-box preference with other family members (Nakashima et al., 2002; Yun-Feng et al., 2010; Zhang et al., 2012a; Zhang et al., 2012b; Yang et al.,2012), while other evidence failed to identify a GC-box preference (Hekmatnejad et al., 2013). Sp7 is reported to also complex with other transcriptional regulators (Kawane et al., 2014; Koga et al., 2005); however, the significance of these protein-protein interactions has not been rigorously addressed in vivo. Here, we use several approaches to analyze Sp7's molecular interactions in the osteoblast program. Collectively, these data support an alternative model for Sp7 action in osteoblast specification. In addition the studies highlight the association of an Sp7-variant of the Sp family with vertebrate restricted bone formation.
RESULTS
Genome-wide analysis for Sp7-DNA association in osteoblasts
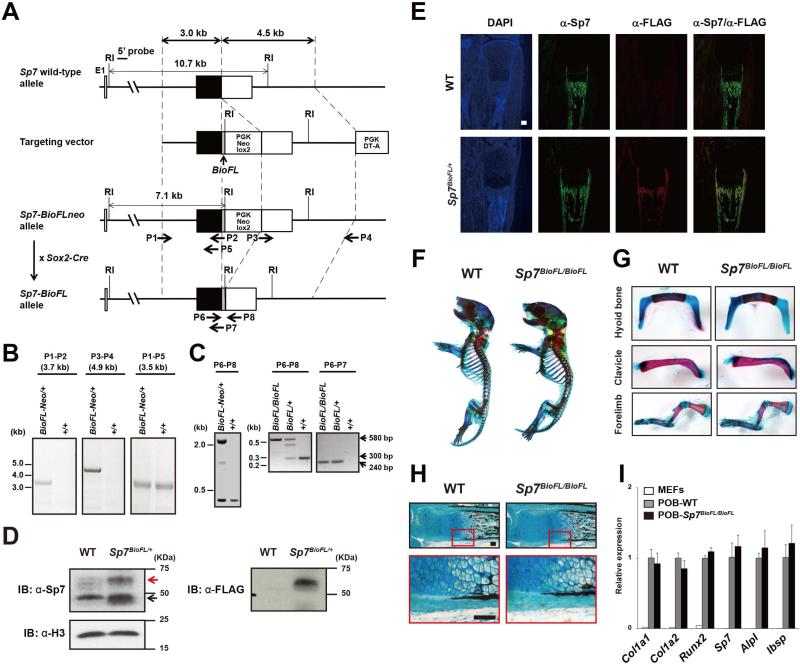
To identify Sp7's molecular interactions in vivo, we used gene-targeting strategies to generate an Sp7-Biotin-3xFLAG knock-in mouse (Sp7-BioFL mouse; Figure 1 A-C and Supplemental Experimental Procedure). This approach appends a biotin (Bio) recognition motif and 3 copies of the FLAG (FL) epitope, a motif that been broadly used in ChIP-seq studies (Yu et al., 2009), at the C-terminus of the Sp7 protein. Importantly, mice homozygous for the Sp7-BioFL allele were viable and displayed no obvious adult phenotype. Western blotting detected Sp7-BioFL at slightly reduced levels to wild-type Sp7 protein (Figure 1 D) and immunohistochemistry on tibial section showed an equivalent temporal and spatial distribution to the unmodified protein (Figure 1E). In addition, skeletal development appeared normal in mice homozygous for the Sp7-BioFl allele (Figure 1F-H), and calvarial osteoblasts from these mice displayed similar levels of expression of Sp7 itself, and a number of osteoblast-related genes to mice with unaltered Sp7 alleles (Figure 1I). These data, together with DNA binding studies (see later) suggest that the BioFL epitope does not obviously impact the normal regulation or action of Sp7.
Figure 1. Generation, characterization and validation of an Sp7 Biotin-3xFLAG knock-in mouse strain.
(A) Targeting strategy.
(B) Long-range PCR analysis of Sp7 knock in allele using primers indicated in (A). P1-P2 and P3-P4 are targeted allele specifc, whereas P1-P5 detects targeted and wild-type (WT) alleles.
(C) PCR genotyping of Sp7-BioFLneo and Sp7-BioFL mice obtained by crossing Sp7-BioFLneo mice to Sox2-Cre mice (see A). P6 and P8 primers yield amplification products of approximately 2 kb, 580 bp, and 300 bp for BioFL-neo, BioFL, and WT Sp7 alleles, respectively. P6 and P7 primers amplify a 240 bp BioFL dependent product.
(D) Western blot comparison of Sp7 and Sp7-BioFL in neonatal calvarial cells. Red arrow: Sp7-BioFL protein; black arrow: Sp7 protein.
(E) Sp7 and the FLAG-tag detection in tibial sections of P1 Sp7-BioFL pup. Scale bar, 300 μm
(F and G) Alcian blue (cartilage) and alizarin red (mineralized tissue) stained skeletal elements of P1 pups.
(H) Histological analysis of von Kossa stained calcified bone matrix (black) and alcian blue stained cartilage. Scale bar, 100 μm
(I) RT-qPCR of osteoblast markers in MEFs and primary osteoblasts isolated from neonatal calvaria of wild-type and Sp7BioFL/BioFL mice at P1.
See also Supplemental Experimental Procedures.
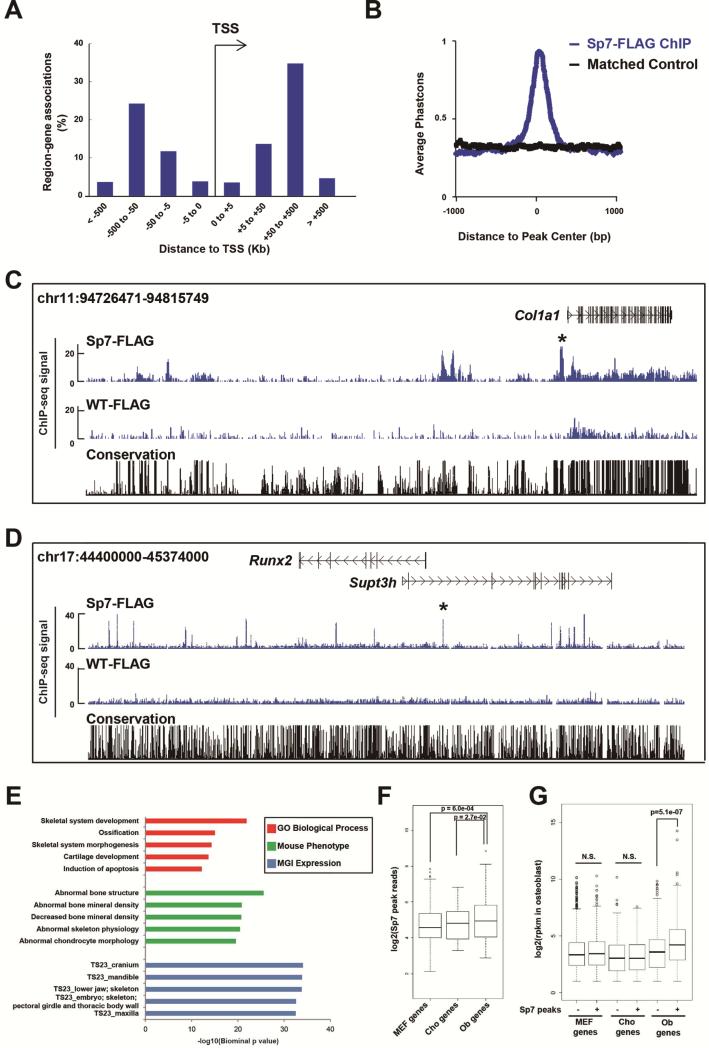
To obtain an Sp7-DNA association profile in an in vivo setting, we performed FLAG antibody-directed ChIP-seq analysis on Sp7-BioFL osteoblasts isolated from the mouse calvaria at postnatal day 1 (P1; Supplemental experimental procedures and Table. S1). Osteoblast isolation was optimized as shown in Figure S1A-C. The intersection of biological replicates identified a stringent set of 2,112 Sp7-BioFL specific ChIP-seq peaks with a false discovery rate [FDR] cutoff of 0.01. Peak distribution analysis revealed that less than 7% of the Sp7 associated regions occurred within 5 kilobases (kb) of a transcription start sites (TSSs), suggesting longer-range interactions are the predominant feature of the Sp7 regulatory program (Figure 2A). Sp7 associated regions were well conserved among different vertebrate species (Figure 2B). Figure 2C and 2D show screenshots of Sp7 engagement around Col1a1 (Figure 2C), an essential osteoblast matrix protein encoding gene, and Runx2 (Figure 2D). In addition to strong interactions at previously characterized osteoblast specific enhancers for these key osteoblast genes (Bedalov et al., 1995; Kawane et al., 2014), Sp7 engaged at multiple additional sites around both genes suggesting a complex regulatory control of these targets. A GREAT-GO analysis was performed to annotate the peak set (McLean et al., 2010). Sp7 peaks showed a strong association around genes related to biological processes in skeletal system development, skeletal tissue-specific phenotypes and expressions in bone tissues consistent with Sp7's expected role in osteoblast regulatory programs (Figure 2E).
Figure 2. Genome-wide analysis of Sp7 association profiles in osteoblasts.
(A) Genome-wide distribution of Sp7 associated regions relative to transcriptional start sites (TSSs).
(B) Phastcon score highlighting conservation of aligned regions (Y-axis) relative to peak centers (X-axis).
(C and D) CisGenome browser screenshots showing Sp7 engagement around Col1a1 and Runx2. Asterisk (*) highlights a previously verified cis-regulatory region (Bedalov et al., 1995; Kawane et al., 2014). WT-FLAG shows FLAG immunoprecipitation in wild-type osteoblasts as a negative control.
(E) GREAT Gene Ontology (GO) and MGI expression annotations of Sp7 peaks showing top five enriched terms.
(F) Relationship of Sp7 ChIP-seq peak signals to expression level of nearest neighboring gene (10 Mb window) comparing MEFs, chondrocytes and osteoblasts.
(G) Comparison of MEF-, chondrocyte- and osteoblast-enriched gene sets (rpkm >2) to osteoblast-associated Sp7 engagement.
See also Figures S1, Table S2 and S3
To ensure that this in vivo ChIP-seq data represented an appropriate target gene set, we performed a number of control experiments to examine the effects of FLAG epitope tagging. First, we compared ChIP-seq results in the MC3T3-E1 osteoblast cell line introducing Sp7 forms epitope-tagged at either the N- (N-terminal FLAG tag) or C-terminus (C-terminal Biotin-FLAG tag; as in the in vivo targeted allele) (Figure S1D). An extensive overlap is observed in these data suggesting that FLAG tagging at different positions gives comparable outcomes. Second, we examined ChIP on wild-type calvarial osteoblasts using an anti-Sp7 antibody. Though we obtained a weaker peak set in this approach, the majority of Sp7-Ab ChIP-seq peaks (95%) overlapped with the larger set identified by Sp7-BioFL ChIP-seq (Figure S1E). Further, the Sp7-Ab recovered peaks enriched for highly ranked peaks in the Sp7-BioFL dataset, and a strong positive correlation of peak signal was observed between Sp7-Ab-peaks and Sp7-BioFL-peaks in the overlapping regions (Figure S1F and S1G). As in other systems, epitope-tagging introducing a high affinity epitope for a well-characterized antibody likely optimizes for target recovery capturing physiologically relevant Sp7-DNA associations with a higher sensitivity than Sp7-Ab ChIP-seq. Hence, we used the 2,112 Sp7-BioFL peaks for additional analysis.
To further understand the biological relevance of the Sp7 peaks to potential target gene expression in the osteoblast, we performed RNA-seq on FACS sorted GFP+ cranial osteoblasts isolated at P1 from Sp7-GFP transgenic mice (Rodda and McMahon, 2006; Figure S1H and S1I). Hierarchical clustering of osteoblast transcriptional profiles with profiles for chondrocytes and mouse embryonic fibroblasts (MEFs, GSM1173355 and GSM1173356 (Hou et al., 2013)) identified cell-type enriched gene expression signatures for osteoblasts, chondrocytes and MEFs (Figure S1J and Table S2; hereafter we refer to these gene sets as Ob-, Cho- and MEFs-genes, respectively). To examine engagement of Sp7 around these gene sets, we first compared enrichment of Sp7 associated regions flanking each cell-type enriched group of genes. The analysis showed that Sp7 ChIP-seq signals were most enriched around Ob-genes (Figure 2F). Next, we divided each cell type-enriched gene set into two groups, those with Sp7 associated peaks and those without, comparing gene expressions in osteoblasts between each group. Ob-genes with associated Sp7 peaks were more highly expressed than those without Sp7 peaks; no up-regulation of gene expression was observed in other comparisons (Figure 2G). These data suggest that Sp7 engagement with the osteoblast genome results in the positive regulation of Ob-enriched genes. Together these analyses indicate that the Sp7-DNA association profiles identify known regulatory elements and the broad features of the dataset suggest that Sp7 peaks provide a biologically relevant, genome-wide assessment of enhancer signatures underlying the Sp7 osteoblast program.
Enhancer activity of Sp7 peak regions
Integrating the conservation score around a ChIP-seq peak, osteoblast expression of the nearest gene to an Sp7 associated region, and the strength of the Sp7 association approximated by ChIP-seq peak reads, we predicted 194 high-value, putative enhancer regions regulated by an Sp7 interaction (Table S3). To test selected regions, we took a stepwise strategy. We first verified cell-type specific activity of selected enhancer regions identified in the literature, around Col1a1, Runx2 and MMP13 (Bedalov et al., 1995; Kawane et al., 2014; Meyer et al., 2015) and a subset of additional Sp7-associated putative enhancer modules around each of these genes. Enhancer activity on a luciferase reporter was assayed on transfection into either NIH3T3 fibroblasts (3T3), or the MC3T3E1 pre-osteoblast cell line, in the absence or presence of osteoblast-inducing osteogenic medium. Osteoblast marker genes were up-regulated upon osteoblast induction (Figure S2A). The assay confirmed osteoblast specific reporter activity for all known osteoblast enhancers and all newly identified putative cis-regulatory modules (Figure S2B-D; known enhancer were highlighted by asterisk). Thus, multiple enhancers likely mediate osteoblast activity of Col1a1 and Runx2 genes, although the regulatory strength of distinct cis-regulatory modules may vary considerably.
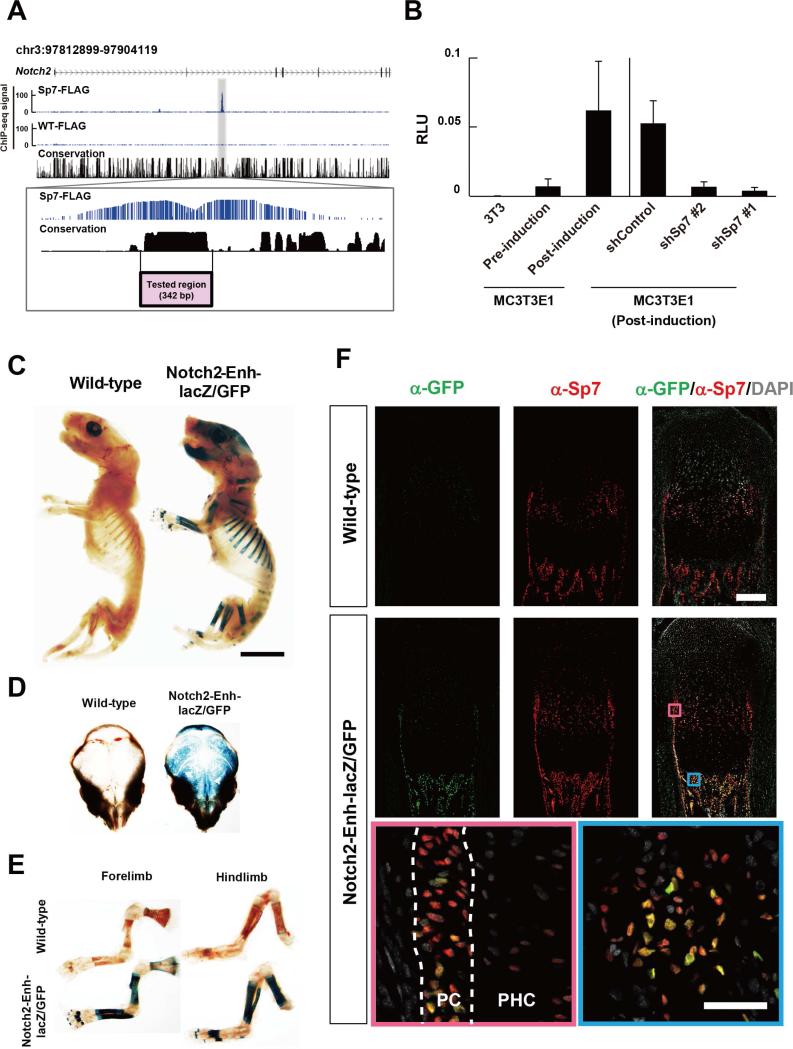
To address additional Sp7 targets, we focused on osteoblast-expressed genes with a strong association of Sp7: these included Notch2, Fgfr2, Col1a2, Gli2, and Kremmen1 (Figure 3A and Figure S2E-H). Of these, Notch2 is uniformly expressed in each cell type and broadly throughout the developing mouse skeleton where Notch2 is reported to maintain skeletal progenitors and regulate osteoblast differentiation (Long, 2012). Sp7 shows a very strong association with a conserved block of DNA in intron 2 of the Notch2 gene suggesting this region may direct Sp7-dependent, osteoblast specific regulation of Notch2 (Figure 3A).
Figure 3. Enhancer analysis for Sp7 targets.
(A) CisGenome browser view of Sp7 associated peak within Notch2 (zoom view below).
(B) Luciferase reporter comparing the activity of the putative Notch2 enhancer region in (A) in 3T3 cells, pre-osteogenic MC3T3E1 cells (pre-induction) and MC3T3E1 cells post osteoblast induction. Sp7 dependence was demonstrated by lentiviral infection of MC3T3E1 cells expressing short hairpin (sh)-Control or two independent sh-Sp7 (#1 and #2) RNAs. Data show means ± SDs from triplicate experiments.
(C, D, E) β-galacotsidase staining to visualize Notch2 enhancer-driven lacZ reporter activity in P1 transgenic mice. Two of four independent transgenic lines showed similar reporter activities. Scale bar, 5 mm.
(F) Immuno-analysis of Sp7 and Notch2 enhancer-driven GFP reporter activity in P1 tibia of transgenic mice. Zoom in view highlights boxed regions. Pink box: perichondrium (PC) and prehypertrophic chondrocytes (PHC); blue box: bone forming primary spongiosa. Nuclei are highlighted by DAPI. Upper panel scale bar: 200 μm; bottom panel: 50 μm. See also Figure S2 and Table S3
To examine Sp7's role in driving reporter activity through the Notch2 enhancer, we examined enhancer driven reporter expression following osteoblast induction and lentiviral shRNA knock down of Sp7 in MC3T3E1 cells. Effective knockdown of Sp7 mRNA and Sp7 protein levels was accomplished by two independent shSp7 target RNAs (shSp7#1 and shRNA#2; Figure S2I and S2J). The Notch2 intronic enhancer driven reporter activity was elevated in pre-osteoblastic MC3T3E1 cells compared to 3T3 cells and underwent a markedly enhanced activation upon osteogenic induction, and was suppressed by Sp7 knockdown, consistent with Sp7-mediated osteoblast specific regulation through the identified Notch2 cis-regulatory region (Figure 3B).
To further examine enhancer activity, we generated a transgenic reporter mouse line expressing a lacZ-GFP fusion cassette (Peterson et al., 2012) under control of the identified Notch2 enhancer region. Whole mount β-galactosidase staining showed reporter activity specific to developing skeletal tissues (Figure 3C-E). Further, immunohistochemistry on tissue sections revealed reporter activity restricted to a subset of Sp7 positive osteoblasts in both calvaria and long bones (Figure 3F and Figure S2K). Importantly, although endogenous Sp7 is expressed in both osteoblasts and pre-hypertrophic chondrocytes in the long bone, enhancer driven reporter activity was only detected in osteoblasts (Figure 3F).
In summary, the collective data indicate that the conserved Sp7-associated region within the intron of Notch2 encompasses a bona fide Sp7-dependent osteoblast-specific enhancer.
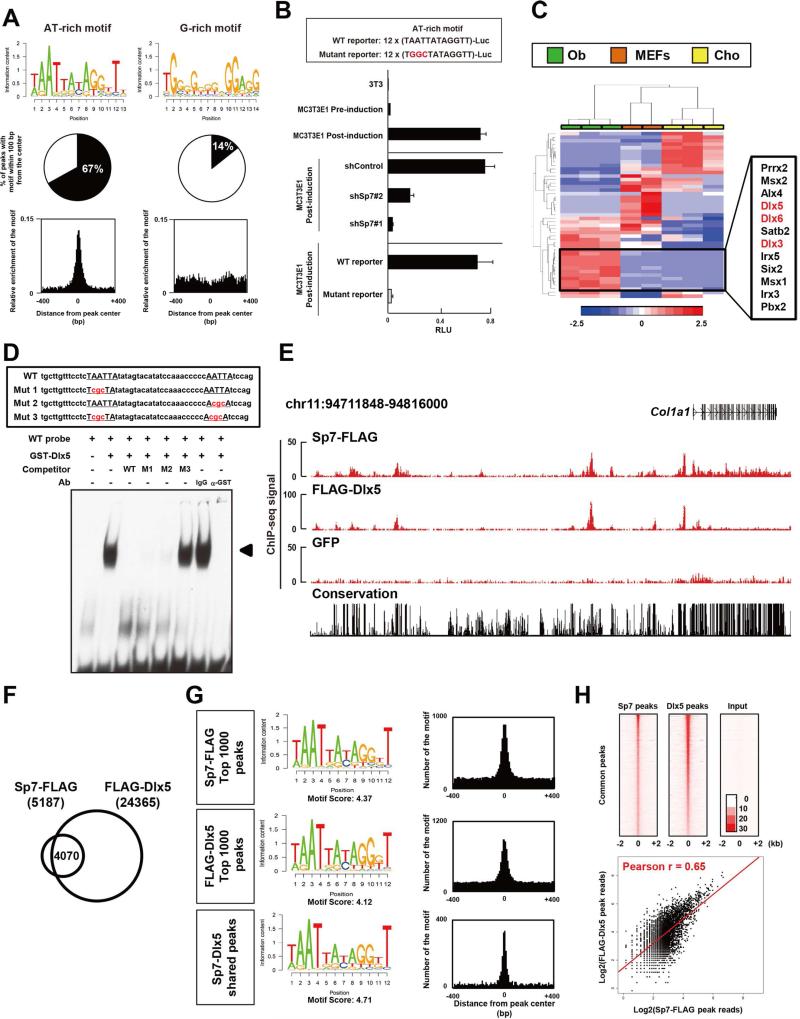
De novo motif analysis predicts an AT-rich motif as the primary Sp7 genomic interaction site
Sp-family members share a conserved DNA binding zinc finger domain that binds a GC-box sequence (Suske, 1999). Given reports from in vitro studies that Sp7 interacts with a GC-box sequence (Nakashima et al., 2002), we expected this to be the primary enriched motif in the Sp7 ChIP-seq datasets. In contrast, both CisGenome (Ji et al., 2008) and DREME (Bailey, 2011) analyses identified an AT-rich motif containing a putative homeodomain-response element as the most highly over-represented motif in de novo motif analysis mapping to 67% of all Sp7 peaks (Figure 4A). In addition to this sequence, multiple other AT-rich motifs containing the homeodomain-response element showed a more modest enrichment in Sp7 peaks (Figure S3A). Importantly, all of these AT-rich motifs are positioned at the peak centers consistent with the motif regulating the primary association of Sp7 at its target site. A G-rich sequence, distinct from the GC-box, was weakly enriched but showed no centering within the Sp7 peaks (Figure 4A and Figure S3A). These data indicate Sp7 is unlikely to interact with a GC-box within target osteoblast enhancers but most likely interacts directly, or indirectly, through the AT-rich motif.
Figure 4. Sp7 motif analysis and interaction with Dlx5 in osteoblasts.
(A) Summary of de novo motif analysis from Sp7 osteoblast ChIP-seq. Selected motifs are shown in Figure 4A and all enriched motifs are shown in Figure S3A.
(B) Luciferase reporter assay for constructs with twelve tandem copies of the AT-rich motif, or a site-directed variant, in 3T3 cells, and MC3T3E1 cells pre- and post-osteoblast differentiation. Sp7 knockdown (shSp7) lentiviral infection was compared to infection with control (shContol) virus. Data show means ± SDs from triplicate experiments.
(C) Hierarchical clustering comparing relative expression levels of potential AT-binding transcriptional regulators in osteoblast (Ob), chondrocyte (Cho) and MEFs. 47 genes out of 232 potential AT-rich target-binding factors were selected for analysis based on rpkm values > 5 in any of these cell-types. Color scale indicates relative gene expression values.
(D) EMSA analysis of Dlx5 binding to AT-rich motifs in Notch2 intron 2 enhancer. Arrowhead highlights a band shift indicative of a Dlx5-DNA complex.
(E) CisGenome browser screenshots showing Sp7 and Dlx5 associations around Col1a1.
(F) Overlap between Sp7 and Dlx5 FLAG ChIP-seq peaks in transducted MC3T3E1 cells.
(G) de novo motif analysis and centering of AT-rich motifs in each peak set in (F).
(H) Upper: heat map showing Sp7 and Dlx5 peak signals sorted by Sp7 peak ranking in co-associated Sp7-Dlx5 peak set. Lower: scatter plot showing correlation between Sp7 and Dlx5 sequence reads in Sp7-Dlx5 shared peak set. Poisson regression line is shown in red. See also Figures S3 and S4 and Table S4
To test the biological relevance of the AT-rich motif in osteoblast, we generated a reporter containing twelve copies of the AT-rich motif and showed this sequence drove strong, Sp7-dependent reporter activity on osteogenic induction of MC3T3E1 cells (Figure 4B). Further, mutagenesis of each TAATTA region to TGGCTA in the twelve tandem repeat reporter, and the two AT-rich motifs in the Notch2 enhancer reporter, abolished their reporter activations (Figure 4B and Figure S3C). Taken together, these findings suggest the primary site mediating Sp7's regulation of enhancer activity is the AT-rich motif, although it remains to be investigated whether Sp7 interacts directly at this site. The enrichment of other motifs supports the co-engagement of several other transcriptional regulators in Sp7 bound cis-regulatory modules including Runx (motif 6 in Figure S3A and motif 2 in Figure S3B) and NFAT (motif 4 in Figure S3B) supporting published data on their shared interactions (Artigas et al., 2014; Koga et al., 2005).
Dlx3, Dlx5 and Dlx6 are potential transcription factors as Sp7partner proteins
To determine if Sp7 bound the AT-rich motif, we performed EMSA but found no consistent evidence for a direct DNA-binding interaction (Figure 5F), suggesting an indirect interaction and requirement of the partner. To identify potential partners for Sp7, we examined osteoblast-enriched expression for 232 transcription factors annotated in the literature (Berger et al., 2008) and through databases including UniProbe (Hume et al., 2015) and TFClass (Wingender et al., 2013) for their binding to AT-rich target sequences. Several of these showed enriched expression in osteoblasts versus chondrocytes or MEFs (Figure 4C), including three related homeodomain encoding transcriptional regulators, Dlx3, Dlx5 and Dlx6. These Dlx-family members are expressed in bone tissue, bind to a TAATTA homeobox response element well matched to the recovered AT-rich motif, and enhance osteoblast differentiation in vitro (Hume et al., 2015; Wingender et al., 2013; Li et al., 2008). Given that Dlx5 was the most highly expressed of these three factors (Table. S4), and interacts with Dlx5 in vitro (Kawane et al., 2014), we focused on Dlx5 as a candidate for facilitating Sp7 engagement at osteoblast enhancers.
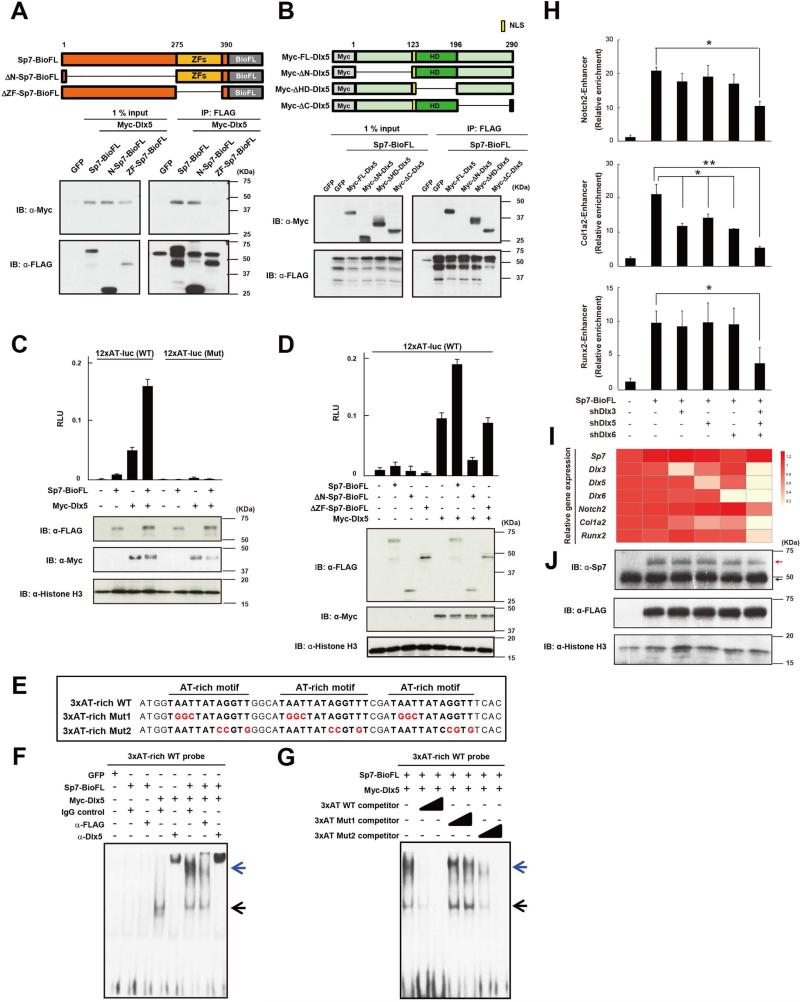
Figure 5. Dlx factors mediate Sp7's regulatory action in osteoblast gene regulation.
(A, B) Western blotting of co-immunoprecipitates of ectopically expressed indicated Myc-Dlx5 and Sp7-BioFL forms (upper panel) from 293T cell nuclear extracts.
(C, D) 3T3 cell luciferase reporter assay for indicated reporter constructs following transfection with indicated Sp7 and Dlx5 expressions. Data show the means ± SDs from triplicate experiments.
(E, F, G) EMSA of Dlx5 and Sp7 complexes with the AT-rich motif. Black arrow: Dlx5-DNA complex; blue arrow: Sp7/Dlx5-DNA co-complex.
(H, I, J) Sp7-FLAG ChIP-qPCR (H), RT-qPCR (I) and western blotting (J) analysis in MC3T3E1 cells following lentiviral transduction of shRNA-mediated knock-down of Dlx3, 5 and 6 and retroviral transduction of Sp7BioFL expression. The color scale indicates relative gene expression values in the qPCR analysis. Red arrow: Sp7-BioFL protein; black arrow: endogenous Sp7 protein. Data displayed are the means ± SDs from triplicate experiments. *: p < 0.05, **: p < 0.01.
Examining Dlx5 binding by EMSA to an oligonucleotide incorporating the two AT-rich sequences in the Notch2 enhancer sequence demonstrated a robust interaction (Figure 4D). Further, competition analysis demonstrated both sites engage with Dlx5 to maximize enhancer activity and target gene expression (Figure 4D). To directly address Dlx5 interactions in the osteoblast, we infected the MC3T3E1 osteoblast cell line with either FLAG-tagged Dlx5 or BioFL-tagged Sp7, differentiated the cells in osteogenic medium and performed anti-FLAG-mediated ChIP-seq in replicate experiments recovering 24,365 and 5,187 associated regions enriched in FLAG-Dlx5 and Sp7-FLAG datasets, respectively.
To address the biological relevance of these data, we first compared Sp7 peaks identified in vitro in the MC3T3E1 osteoblast cell line with those reported earlier from calvarial osteoblasts. A partial overlap highlighted the significant enrichment of skeletal system-related terms in GREAT GO analysis (Figure S4A). Shared peaks had an higher average ChIP-seq signal (more sequence reads) overall than the entire set of peaks (Figure S4B). These results indicate the in vitro culture system gives a meaningful representation of osteoblast biology.
Comparing the Sp7 and Dlx5 ChIP-seq peak datasets obtained from MC3T3E1cells overexpressing Sp7-BioFL and FLAG-Dlx5, respectively, we observed a striking overlap: about 78% (4,070 out of 5,187) of the Sp7 peaks were shared with Dlx5 peaks; a screen shot of one osteoblast target Col1a1 illustrates their shared interaction signatures (Figure 4E and 4F). Further, the analysis of Sp7-Dlx5 shared peaks included most of those identified intersecting MC3T3E1 cell and calvarial osteoblasts Sp7 ChIP-seq datasets (Figure S4C). Importantly, de novo motif analysis recovered an identical AT-rich sequence within both FLAG-Dlx5 and Sp7-FLAG ChIP-seq data from MC3T3E1 cells to that recovered from Sp7 ChIP-seq in calvarial osteoblasts (Figure 4G). This motif was the most enriched in the top 1,000 Dlx5, Sp7, and shared Sp7-Dlx5 peaks and mapped to peak centers as expected for the primary interaction site (Figure 4G). Heat map and scatterplots for Sp7 and Dlx5 peak signals showed a positive correlation between these datasets (Figure 4H). DAVID analysis identified skeletal system development-related genes as enriched terms in the Sp7-Dlx5 associated regions, while no skeletal-related terms were enriched in those regions bound by FLAG-Dlx5 only (Figure S4D). These data support the conclusion that Sp7 associates with the osteoblast genome via homeodomain containing transcription factor partners such as Dlx5. The observed weaker peak recovery with Sp7-FLAG versus FLAG-Dlx5 (Figure 4H) likely reflects secondary versus primary DNA interactions, respectively, for each transcription factor at the same target sites.
Physical and biological interaction of Sp7 and Dlxs
To obtain a further insight into molecular mechanisms underlying Sp7-Dlx action, we performed a biochemical analysis of their interactions. First, we mapped their protein-protein interaction domains through a series of truncations and co-immunoprecipitation (co-IP) analyses. Co-IP in 293T cells showed that the Sp7 zinc finger domain and Dlx5 N-terminal domain were crucial for protein-protein interaction (Figure 5A and 5B). Next, we performed co-transfection in vitro reporter assays utilizing constructs with twelve copies of the AT-rich motif demonstrating that Sp7 and Dlx5 had synergistic effects on reporter gene activity that was dependent on the predicted homeodomain DNA binding target site within the AT-rich motif (Figure 5C). Deletion of the Sp7 zinc finger domain did not alter Dlx5-mediated reporter activity (Figure 5D), as expected if this form is unable to bind Dlx5 (Figure 5D), In contrast, N-terminal deletion of Sp7 (amino acids 1-274) suppressed Dlx-5 dependent reporter activity (Figure 5D). Sp7's transactivation domain has been localized to this N-terminal region (Nakashima et al., 2002) and this mutant formed physically interacts with Dlx5 (Figure 5A). The N-terminal domain of Dlx5 critical for the interaction with Sp7 has been reported to recruit transactivation co-factors (Masuda et al., 2001). Together these data suggest that the N-terminal deleted form of Sp7 acts in a dominant-negative manner in this context and Sp7 is likely one such co-activator in osteoblasts where the N-terminal region of Sp7 likely functions to activate gene expression in targets bound by a Dlx5-Sp7 regulatory complex. These data provide strong evidence that Sp7 acts through Dlx5, and potentially Dlx3 and Dlx6 as these family members are also specifically enriched in osteoblast progenitors. Sp7-Dlx regulatory complexes engaged at osteoblast enhancers through Dlx binding to AT-rich homeodomain-response elements within osteoblast enhancers are predicted to activate osteoblast target genes.
To directly examine the role of Dlx factors, we performed biochemical analysis and loss of function studies. EMSA showed a specific DNA interaction of three tandem AT-rich motifs with Dlx5 alone and with an Sp7-Dlx5 complex; as expected no interaction was observed with Sp7 alone (Figure 5E and 5F). The Sp7-Dlx5 interaction was competed by excess wild-type oligomer (3xAT-rich WT, Figure 5E) in a dose-dependent manner (WT in Figure 5G). However, competitive binding was lost when all homeodomain target sites were mutated in the oligomer (Mut2 in Figure 5G), whereas competition was still observed on mutation of flanking nucleotides (Mut1in Figure 5G). Thus, Dlx5 associates directly with the core AT-rich motif and Sp7 forms a ternary complex with Dlx5 bound to its target site. Comparable results were obtained between Sp7 and Sp7BioFL (Figure S4E).
Genetic studies have provided some evidence for Dlx action in osteoblasts; however combinatorial removal of all osteoblast enriched Dlx-factors is not possible with existing alleles due to the combinatorial requirement for Dlx activity for postnatal survival (Robledo et al., 2002). To address the combined roles of Dlx3, Dlx5 and Dlx6 on Sp7 engagement at osteoblast enhancers, we infected MC3T3E1 cells with lentivirus carrying shDlx3, shDlx5 and shDlx6 knock-down cassettes, a retrovirus carrying an Sp7-BioFL expression cassette, and then performed Sp7-FLAG ChIP-qPCR in the presence or absence of Dlx-member knock down (Figure 5H). RT-qPCR analysis demonstrated an 80% reduction of Dlx3, Dlx5 and Dlx6 mRNA levels (Figure 5I), while Sp7 mRNA levels, and Sp7 and Sp7-BioFL protein levels were unaltered (Figure 5I and 5J). In this context, only combinatorial Dlx3, 5, 6 knockdown lead to both a significant reduction in Sp7-FLAG enrichment in Notch2, Col1a2 and Runx2 osteoblast enhancers (Figure 5H) and reduced expression of these target genes (Figure 5I). Together, these results argue for a redundancy amongst Dlx-family members in mediating Sp7's activation of osteoblast enhancers.
Distinct DNA binding action underlies Sp1 and Sp7 actions
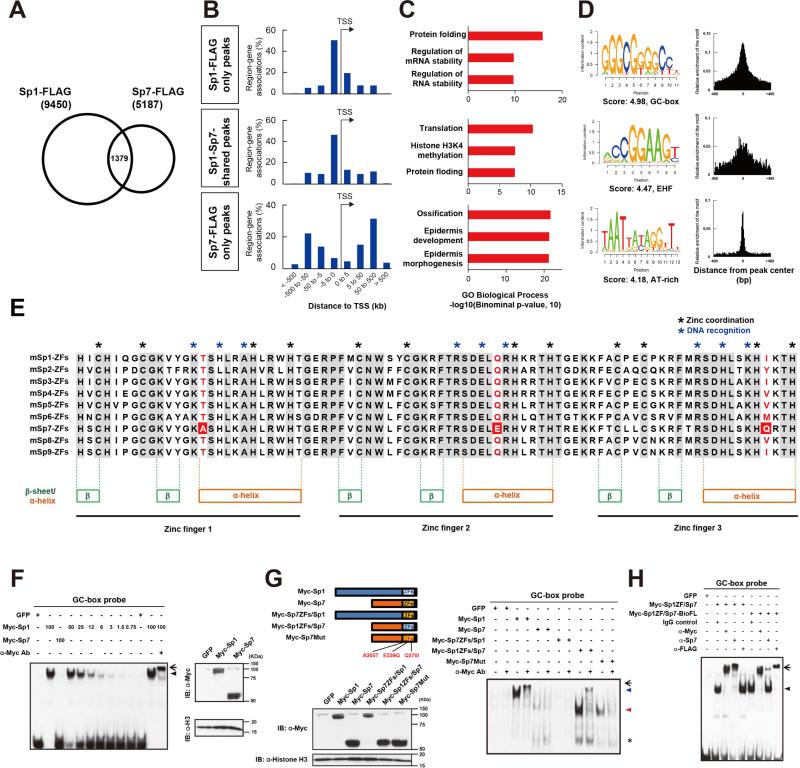
Our results raised the question of whether Sp7 interaction with Dlx factors is distinct for Sp7 or shared by other Sp family members. To address this question, we performed Sp1 ChIP-seq in MC3T3E1 cells overexpressing Sp1-BioFL. Sp1-FLAG ChIP-seq identified 9,450 Sp1binding regions in biological replicates. Less than 30% of Sp7 peaks were shared with this Sp1 peak set (Sp1-Sp7-shared peaks; Figure 6A). A GREAT-GO analysis showed that both Sp1-only and Sp1-Sp7-shared peaks were mainly located promoter proximal regions within 5 kb of the transcriptional start site, coupled to genes related to general cellular processes rather than osteoblast specific programs (Figure 6B and 6C). De novo motif analysis with the top 1,000 Sp1-only peaks identified an expected consensus GC-box, but no AT-rich motif (Figure 6D and Figure S5A). In the shared peaks, an EHF (Ets Homologous Factor) motif was the most enriched site, suggesting EHF factors may partner with both Sp1 and Sp7. However, these peaks also failed to display an osteoblast-specific regulatory signature. Peak intersection revealed that about 25% of Sp1 peaks overlapped with Dlx5 peaks but no correlation in Sp1 and Dlx5 ChIP-seq reads was observed within the shared peak datasets (Figure S5B and S5C) in striking contrast to the analysis of overlapping Sp7 and Dlx5 peaks sets discussed earlier (Figure 4H). These results support distinct modes of action between Sp7 and Sp1 in osteoblast progenitors consistent with the strong Sp7 associated osteoblast specific phenotypes.
Figure 6. Genome-wide comparative analysis of Sp1 and Sp7 DNA association and molecular dissection of their activities.
(A) Overlap between Sp1 and Sp7 FLAG ChIP-seq peaks in transducted MC3T3E1 cells. (B) Genome-wide distribution of intersected Sp1 and Sp7 associated regions relative to TSSs.
(C) GREAT GO annotation for intersected Sp1 and Sp7 peaks showing top three enriched terms.
(D) de novo motif analysis from top 1,000 Sp1 and Sp7 intersected peaks displaying the most enriched motifs and their distribution within peak regions.
(E) Alignment of zinc finger regions amongst mouse Sp family members highlighting amino acid conservation and amino acids associated with zinc coordination and DNA recognition. Amino acid variants examined for their role in DNA binding specificity are highlighted in red.
(F) GC-box probe EMSA examining nuclear extracts of COS7 cells transfected with N-terminal Myc-tagged Sp1 and Sp7 (left). Similar amounts of nuclear extract were used in lanes 1 to 3 and 11 to 13. GC-box binding of Sp1 was also examined by serial dilution of nuclear extracts (see % of amount used in lane 2) in lanes 4 to 10. Arrowhead: Myc-Sp1 shift band; arrow: supershift product for Myc-Sp1. Protein expression levels were compared by western blotting (right).
(G) GC-box probe EMSA examining nuclear extracts of COS7 cells transfected with the indicated plasmids (right). Blue arrowhead: Myc-Sp1 shift band; Red arrowhead: Myc-Sp1ZFs/Sp7 shift band. Arrow indicates supershift products for Myc-Sp1 and Myc-Sp1ZFs/Sp7 while an asterisk indicates a non-specific band. Protein expression levels were examined by western blotting (left).
(H) GC-box probe EMSA examining nuclear extracts of COS7 cells transfected with the indicated plasmids. Arrowhead: Myc-Sp1ZF/Sp7 or Myc-Sp1ZF/Sp7-BioFL shift products; arrow: Myc-Sp1ZF/Sp7 or Myc-Sp1ZF/Sp7-BioFL supershift products.
See also Figure S5
To directly address these differences in GC-box recognition, previously a canonical property of Sp-family members, we compared Sp1 and Sp7 interaction by EMSA. Sp1's DNA binding activity maps to its ZF domain (Figure 6E; Suske, 1999). The analysis was performed with three different samples; (1) nuclear extracts from in vitro differentiated MC3T3E1 cell-derived osteoblasts (Figure S5D) and antibody supershift assays (Figure S6E), (2) in vitro translated proteins (Figure S5F and S5G), and (3) nuclear extracts from COS7 overexpressing N-terminal Myc-tagged Sp1 and Sp7 (Figure 6F).
We failed to obtain any strong reproducible Sp7 interaction under conditions that gave rise to consistent strong Sp1 interaction with the GC-box target (Figure 6F, S5D and S5F). Substitutions were then performed to introduce the Sp1 zinc finger domain into Sp7 (Sp1ZFs/Sp7) and the Sp7 zinc finger domain into Sp1 (Sp7ZFs/Sp1). EMSA analysis showed the Sp1 ZF conferred GC box binding on the Sp1ZFs/Sp7 protein while DNA binding of Sp1 was lost when the Sp7 ZF containing Sp7ZFs/Sp1 chimeric protein (Figure 6G). Importantly, an Sp1ZFs/Sp7-BioFL variant with the Sp1 ZF substitution in Sp7 and a C-terminal BioFL epitope tag positioned as in the protein produced by targeted Sp7-BioFL allele in mice showed a strong interaction with the GC box motif and a strong supershift in EMSA assay on addition of anti-FLAG antibody (Figure 6H). Thus, it is differences within the ZF binding motif between Sp1 and Sp7, not epitope induced alterations in binding specificity or abrogation of Sp7-BioFL-DNA interactions associated with FLAG antibody association that underlie the failure to recover endogenous Sp7-BioFL interactions at GC motifs in calvarial osteoblasts in vivo.
The amino acid residues interacting with the GC-box recognition are positioned at −1, +3, and +6 of the alpha helical domain of each zinc finger of Sp family members (Figure 6E; Suske, 1999). Interestingly, three amino acids in the alpha helical domain distinguish Sp7 from other Sp family members; A305, E339 and Q370 (Figure 6E). We speculated these differences may contribute to the loss of a GC-box binding preference in Sp7. Consistent with this view, mutating the three variant amino acid to conform to their Sp1 counterparts (A305T, E339Q and Q370I in Sp7) conferred GC-box recognition on the mutated Sp7 protein (Figure 6G). In contrast, mutating just one amino acid to an Sp1-like preference at A305T, a glycosylated amino acid in Sp1; (Chung et al., 2008) did not confer GC-box recognition on Sp7 (Figure S5H and S5I). These results suggest that combinational amino-acid differences within the zinc finger domain of an ancestral Sp family member contributed to the loss of GC-box binding preference in the Sp7 variant of the Sp family.
To determine to what extent these amino acids differences contributed to the distinct mode of Sp7 action, we performed ChIP-seq for the Sp7 mutant carrying A305T, E339Q, and Q370I (Sp7-Mut-FLAG) in MC3T3E1 cells comparing this ChIP-seq signature with those obtained with Sp1 and Sp7. Sp7-Mut-FLAG ChIP-seq identified 8,665 peak regions in biological replicates. While enrichment of the AT-rich motif was still observed in the Sp7 mutant peaks, a weak centering of GC-box at their peak centers was also observed (Figure S5J). Thus, the tested amino acid variants within Sp7's ZF domain contribute, but only partially, to Sp7's distinct mode of action. Further, co-expression of an Sp7ZF/Sp1 chimeric protein encoding construct with Dlx5 did not induce osteoblast marker genes in C3H10T1/2 cells, a stromal cell line with osteogenic potential (Katagiri et al., 1990), in contrast to wild-type Sp7 (Figure S5K). Together, these data suggest that differences in both the N-terminal transactivation domain and ZF domain of Sp7 are likely critical for the acquisition of Sp7's unique osteoblast determining role.
Potential linkage between the Sp7 variant and origin of boney vertebrate evolution
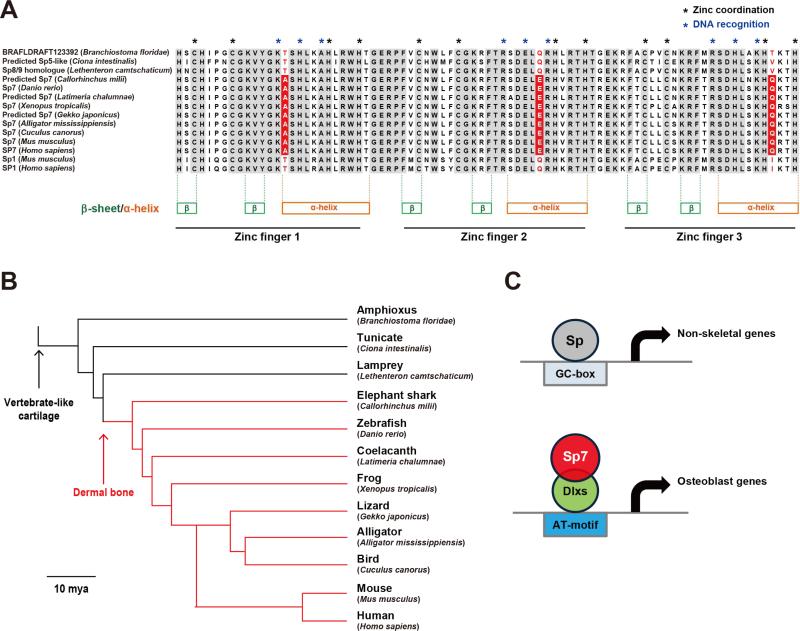
To determine how the appearance of an Sp7-specific zinc finger variant relates to extant bone forming vertebrates, we aligned the most closely related Sp-counterpart to mammalian Sp7 in non bone-forming protochordates, with the Sp7 protein sequence from several bone-forming vertebrates. Interestingly, the protochordates conserve an Sp1-like set of amino acids in the ZF (Figure 7A) suggesting a binding vertebrate Sp1-like action. In contrast, jawed fish, including the basal elephant shark, have dermal bone (Venkatesh et al., 2014) and the bone-associated Sp7 zinc finger variant (Figure 7A). Thus, the acquisition of this distinct Sp-family member may have played a central role in the emergence of bone-forming osteoblasts in the vertebrate evolutionary process (Figure 7B and 7C).
Figure 7. Emergence of Sp7 variant at the origin of boney vertebrate evolution.
(A) Alignment of zinc finger domains in Sp7, Sp1 and the most closely related Sp7 counterparts amongst different chordate species. The three amino acid variants in mammalian Sp7 that correlate with reduced affinities for GC-box DNA binding are highlighted.
(B) A phylogenetic tree of the selected species above in relation to the origin of vertebrate skeletal cell type. Divergence times are from references (Eo and DeWoody, 2010; Larhammar et al., 2009; Venkatesh et al., 2014), Bar: 10 million years ago (10 mya)
(C) Model for Sp7 action in the regulation of osteoblast gene-regulatory programs.
DISCUSSION
Mammalian skeletal development is critically dependent on a small number of transcriptional regulators (Long, 2012). Sox9 is initially required for the establishment of a skeletal primordium and subsequent chondrocyte differentiation, while Sp7 and Runx2 are the two central transcriptional determinants of the vertebrate osteoblast program. The current study gives an insight into the Sp7 regulatory landscape in mammalian osteoblast development and the mechanism by which Sp7 exerts its osteogenic role.
First, long-range genomic interactions (> 5kb) appear to be a predominant feature of the Sp7 regulatory program based on the relationship of cis-regulatory modules and putative promoter targets. Sp7 ChIP-seq identified a number of previously documented Sp7-dependent enhancers and in addition the data predict a large number of osteoblast enhancers several of which demonstrated Sp7-dependent osteoblast activity in cell culture and transgenic mouse assays. The enrichment of predicted motifs bound by Runx2 and other key bone regulatory factors suggests the integration of osteoblast regulatory information through identified cis-regulatory modules. Second, Sp7 targets osteoblast genes that encode osteoblast regulatory factors, signaling components and extracellular matrix proteins known to play important roles in bone development. Third, the data provides strong evidence for an unexpected mode of Sp7 action that distinguishes Sp7 from other Sp members. Whereas Sp1 and other family members bind a consensus GC-box sequence directly (Suske, 1999), we found no evidence of this mechanism in the analysis of Sp7's regulatory action in vivo. Instead, the primary site of engagement present at the center of most ChIP-seq peaks is an AT-rich motif similar to a homeodomain-response element. Fourth, the analysis of comparative ChIP-seq studies, in vivo osteoblast gene expression, and protein-protein and DNA-protein interactions all indicate that Sp7 does not directly engage DNA but interacts with its targets through Dlx-family members directly bound to the recovered AT-rich target sequence. Given the importance of the N-terminal transcriptional activation domain in Sp7 for Sp7-mediated up-regulation of Dlx-targets, we predict Sp7 acts as a transcriptional co-activator in a Dlx-directed process of osteoblast development.
Mode of Sp7 action
In this model, Dlx-members bind through their homeodomain to the core AT-rich motif that is highly enriched in Sp7 and Dlx5 ChIP-seq datasets. Sp7 engages Dlx factors directly through its zinc fingers where several key amino acid substitutions distinguish Sp7 from other Sp-family members. Analysis of three highly conserved amino acid changes, one in each zinc finger, suggest these variants contribute to reducing the affinity of Sp7 binding to the GC-box Sp-family consensus target site. The three amino acids, A305, E339 and Q370, are located adjacent to amino acids directly engaging DNA in alpha helices of the zinc fingers of Sp1, and other family members. These changes may disrupt a secondary structure essential for DNA binding. Interestingly, substituting these three amino acids with the cognate amino acids in Sp1 is sufficient to partially restore GC-box binding in vitro in EMSA assays though these changes are not sufficient to direct an Sp1-like profile of DNA interactions to Sp7 in ChIP-seq studies in an osteoblast cell line. Consequently, other amino-acid changes between Sp7 and Sp1 are critical for distinguishing Sp7's mode of action.
Collectively, the data favor a major interaction between Sp7 and Dlx5, and likely the related osteoblast-enriched Dlx-family members, Dlx3 and 6. Sp7 binds Dlx5, and importantly the Sp7 associated genome in MC3T3E1-derived osteoblasts is almost entirely contained within Dlx5 bound target regions. Proving a Dlx-requirement for Sp7 action genetically is difficult given the co-expression of three family members and early lethality of compound mutants, although multiple knockdown of these Dlx members led to a reduction in Sp7 engagement, and Sp7-dependent osteoblast target gene expression, in the MC3T3E1 osteoblast cell line. In the mouse, Dlx5/6 compound mutants do exhibit impaired bone formation; Runx2 is present, as in Sp7 mutants, but Bglap, a mature osteoblast marker is not expressed within the scapula (Robledo et al., 2002). A more definitive insight will likely require conditional approaches to modulate Dlx3, 5 and 6 activities in developing osteoblasts in vivo. Further, we cannot exclude the possibility of other homeodomain transcription factors acting in conjunction with Sp7. Msx1/2, Satb2 and Alx4 are all highly expressed in osteoblasts and may bind to similar homeobox-responsive elements as Dlx's factors in skeletal development (Stains and Civitelli, 2003; Karsenty, 2008)
Sp7-mediated gene regulatory network
The large set of Sp7 associated regions identified in vivo provides an insight into the osteoblast regulatory program. A number of groups have indicated Sp7 binding to GC-boxes or GC-rich sequences similar to the GC-boxes close to the TSS of a number of osteoblast targets including Col1a1 (Koga et al., 2005; Nakashima et al., 2002), Col1a2 (Yano et al., 2014), Col5a1(Wu et al., 2010), Col5a3 (Yun-Feng et al., 2010), Dkk1 (Zhang et al., 2012a), and Mmp13 (Zhang et al., 2012b). However, we failed to recover significant Sp7 binding to any of these regions in osteoblasts in vivo. This could reflect differences in the developmental timing of osteoblasts in our study; our in vivo analysis addressed a mixed population of early osteoblasts present within clavaria of the neonatal mouse. Although our analysis suggests that the epitope tagging strategy did not impact synthetic DNA interactions in Sp1ZF-Sp7 chimeric proteins, we can not rule out that our approach failed to detect a GC-binding subset of Sp7 interactions. However, considering the body of the evidence herein, we think this is unlikely and earlier reports of Sp7 interaction with a GC-box sequence in vitro may not reflect Sp7 interactions in normal osteoblast development.
Evolutionary establishment of osteoblast gene regulatory networks
An evolutionary view provides an insight into the establishment of osteoblast gene regulatory networks. In the mouse, both cartilage and bone formation depend on the early action of Sox9 in skeletal primordia (Long, 2012). SoxE, the ancestor of Sox9, is present in pharyngeal endodermal cells in hemichordates prior to the appearance of cartilage and bone (Fisher and Franz-Odendaal, 2012). These cells synthesize fibrillary collagen. Osteoblast specification upstream of Sp7 action, and mature cartilage programs, depend on Runx2 action in the mouse (Komori et al., 1997). Runx genes likely played an early role in cartilage programs with additional skeletal genes being required to establish the earliest osteoblast gene networks (Fisher and Franz-Odendaal, 2012). In agreement with this view, the cephalochordate Amphioxus forms cartilage but not bone. A single Runx gene is expressed in pharyngeal endoderm of Amphioxus; the potential source of pharyngeal skeletal elements (Fisher and Franz-Odendaal, 2012). Dlx orthologues exist in Drosophila. Multiple paralogues of Dlx genes have arisen in vertebrates through tandem and whole genome duplications and these members regulate a number of developmental processes in addition to skeletal development (Takechi et al., 2013). The appearance of Sp7, favoring protein-protein interaction with Dlx-factors over direct GC box interactions, may have been a key switch in the transition of skeletal progenitors to an osteoblast-like cell type. Comparative analysis of extant species identifies an Sp7-like zinc finger variant in all major vertebrate groups, but none has been identified to date outside of vertebrates. Thus, unlike other osteoblast related regulatory factors, there is a close linkage of Sp7 to bone forming ability. Given pre-existing Dlx-factors, Sp7 may have acted to stabilize or enhance Dlx's transcriptional role on distinct targets within the skeletal regulatory genome. The absence of a documented Sp7 gene in several vertebrate species may reflect a secondary loss of Sp7, and Sp7 independent mechanisms of bone development, or technical issues with sequence coverage/alignment in species with poorly annotated genomes.
Conserved architecture of gene regulatory networks in vitro and in vivo
Our study highlights the strengths and limitation of in vivo and in vitro analysis of gene regulatory networks. The absence of available antibodies and requirement of relatively large number of input cells are challenges to the broad use of factor-specific ChIP-seq on rare cell types, or those that are difficult to isolate such as matrix embedded osteoblasts. Here, we overcame the limitation of Sp7 antibodies by tagging the Sp7 gene through gene-targeting (Yu et al., 2009), which enabled us to generate a decent ChIP-seq dataset. Although the Sp7-BioFL protein level was somewhat reduced relative to wild-type Sp7 protein, several studies support the conclusion that Sp7 tagging did not markedly alter Sp7's osteogenic action. Clearly, recent gene-editing approaches should facilitate more widespread adoption of this approach. ChIP-seq analysis of Sp7 and Dlx5 actions in the MC3T3E1 osteoblast line facilitated rapid hypothesis testing but comparison of data from this cell line with data generated from in vivo purified osteoblasts reveals a less well-delineated osteoblast network. While some differences may be real regulatory differences, reflecting for example the variability in osteoblast makeup between in vitro and in vivo populations, it is likely most differences reflect artificial regulatory interactions induced in vitro by the abnormal environment, or through in vitro propagated genetic changes. This underscores the importance of assessing cells in their normal setting to establish the most biologically meaningful mammalian transcriptional regulatory networks.
EXPERIMENTAL PROCEDURES
Generation of Sp7-BioFL mouse
The targeting strategy used to generate the Biotin-3xFLAG knock-in Sp7 allele is provided in the Supplemental Experimental Procedures. All experiments in this study were carried out in accordance with recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institute of Health, USA. All procedures were approved by the Institutional Animal Care and Use Committee of The University of Southern California. Los Angeles, CA (IACUC #11830 and #11892).
ChIP-seq
ChIP was performed according to Peterson et al., 2012 with minor modifications. The construction of ChIP-seq libraries was performed with a ThruPLEX®-FD Prep Kit (R40012, Rubicon Genomics) according to the manufacturer's instruction. The library was sequenced on Hiseq2000 and NextSeq500 (Illumina) platforms.
ChIP-seq data analysis
DNA-sequence information was aligned to the unmasked mouse genome reference sequence mm9 by bowtie aligner (Langmead et al., 2009). Peak calling was performed by two-sample analysis on CisGenome software (Ji et al., 2008) with a P-value cutoff of 10−5 comparing with the input control without antibody immunoprecipitation. Peaks were incorporated into further analysis displaying an FDR<0.01. For peak distribution in genome and gene ontology analysis, GREAT GO analysis was performed utilizing the online GREAT GO program, version 2.0.2 (McLean et al., 2010). Each peak category was run against whole genome background with assembly mm9. de novo motif analysis were performed using the Gibbs motif sampler provided in the CisGenome package (Ji et al., 2008), and DREME (Bailey, 2011). A 100 bp region surrounding peak center was extracted from mm9 and used for these analyses.
ChIP-qPCR and RT-qPCR
ChIP-qPCR and RT-qPCR were performed with SYBR Premix Ex Taq II (Takara, RR820). Primer sequences are shown in Table S5. Detailed methods and antibodies used are described in the Supplemental Experimental Procedure.
Reporter analysis
The genomic regions used for the reporter analyses are shown in Table S6. G0 transgenic analysis for the Notch2 enhancer was performed using Hsp68-lacZ::nGFP reporter construct as previously described (Peterson et al., 2012). Detailed methods are described in the Supplemental Experimental Procedure.
Immunostaining
Tissues were fixed in 4% paraformaldehyde (PFA)/PBS for 1 hour at 4 °C and soaked in 30% sucrose/PBS overnight at 4 °C. After embedding in OCT compound, cryosections were cut at 12 μm intervals. Sections were blocked with 3% bovine serum albumin (A7960, Sigma-Aldrich) and 1% heat inactivated sheep serum (S2263, Sigma-Aldrich) in PBST. The antibodies used for immunostaining are shown in the Supplemental Experimental Procedure.
EMSA
EMSA was performed using a Light Shift Chemiluminescent EMSA Kit (20148; Fisher Scientific, Hampton, NH), according to the manufacturer's instruction. DNA probes labeled with Biotin were synthesized by Integrated DNA Technologies, Inc. (IDT, Coralville, IA). The sequences of probes are shown in Table S5. Detailed methods and antibodies used are described in the Supplemental Experimental Procedure.
Co-immunoprecipitation, SDS-PAGE and western blotting
Nuclear extracts were obtained with a Nuclear Complex Co-IP Kit (54001, Active motif) according to the manufacturer's instruction. Co-IP was performed using 500 ug of nuclear extract, fifty microliters of Dynabeads M-280 Sheep Anti-Mouse IgG (11201D, Life technologies) and five microgram of Anti-FLAG M2 antibody (F1804, Sigma-Aldrich), followed by SDS-PAGE analysis of protein products. The antibodies used for immunoblotting are shown in the Supplemental Experimental Procedure.
Supplementary Material
Highlights.
Sp7 genome-wide analysis identified osteoblast enhancers in calvarial osteoblasts
Motif recovery and functional analysis indicates Sp7 acts through an AT-rich motif
Sp7 indirectly engages the AT-rich motif through a Dlx-complex
The Sp7 Sp-family variant correlates with the emergence of bone forming vertebrates
ACKNOWLEDGEMENTS
We thank Drs. Henry M. Kronenberg, Clifford J. Tabin, Ung-il Chung, Kevin A. Peterson, and Yuichi Nishi for their helpful inputs; David Butler and Peter Maye for sharing Col2-ECFP mice; Jill McMahon, Charles Nicolet, Selene Tyndale, Helen Truong, Yibu Chen, Meng Li, Seth Ruffins, Gohar Saribekyan, Rie Yonemoto and Nozomi Nagumo for providing technical assistance. H.H. was supported by Research Fellowships of Japan Society for the Promotion of Science (JSPS) and JSPS Postdoctoral Fellowships for Research Abroad. S.O. was funded by Grant-in-Aid for Young Scientists (23689079), Uehara Memorial Foundation Research Grant, and Takeda Science Foundation Research Grant. This study was supported by a grant from the National Institutes of Health to APM (DK056246).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHORS CONTRIBUTIONS
H.H. conceived and performed ChIP-seq, RNA-seq and biochemical experiments; analyzed and interpreted data, and took the lead in preparing the manuscript. S.O. conceived and generated the Sp7-Biotin-3xFLAG knock-in mouse, performed ChIP-seq experiments. X.H. and L.P.L analyzed and interpreted data. A.P.M. conceived and designed the project and interpreted data. All authors participated in the writing of the manuscript.
ACCESSION NUMBERS
ChIP-seq and RNA-seq data reported in this paper are available in the Gene Expression Omnibus (GEO) under accession number GSE76187.
REFERENCES
- Artigas N, Urena C, Rodriguez-Carballo E, Rosa JL, Ventura F. Mitogen-activated protein kinase (MAPK)-regulated interactions between Osterix and Runx2 are critical for the transcriptional osteogenic program. J Biol Chem. 2014;289:27105–27117. doi: 10.1074/jbc.M114.576793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27:1653–1659. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedalov A, Salvatori R, Dodig M, Kronenberg MS, Kapural B, Bogdanovic Z, Kream BE, Woody CO, Clark SH, Mack K, et al. Regulation of COL1A1 expression in type I collagen producing tissues: identification of a 49 base pair region which is required for transgene expression in bone of transgenic mice. J Bone Miner Res. 1995;10:1443–1451. doi: 10.1002/jbmr.5650101004. [DOI] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SS, Kim JH, Park HS, Choi HH, Lee KW, Cho YM, Lee HK, Park KS. Activation of PPARgamma negatively regulates O-GlcNAcylation of Sp1. Biochem Biophys Res Commun. 2008;372:713–718. doi: 10.1016/j.bbrc.2008.05.096. [DOI] [PubMed] [Google Scholar]
- Eo SH, DeWoody JA. Evolutionary rates of mitochondrial genomes correspond to diversification rates and to contemporary species richness in birds and reptiles. Proc Biol Sci. 2010;277:3587–3592. doi: 10.1098/rspb.2010.0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Franz-Odendaal T. Evolution of the bone gene regulatory network. Curr Opin Genet Dev. 2012;22:390–397. doi: 10.1016/j.gde.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Hekmatnejad B, Gauthier C, St-Arnaud R. Control of Fiat (factor inhibiting ATF4-mediated transcription) expression by Sp family transcription factors in osteoblasts. J Cell Biochem. 2013;114:1863–1870. doi: 10.1002/jcb.24528. [DOI] [PubMed] [Google Scholar]
- Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- Hume MA, Barrera LA, Gisselbrecht SS, Bulyk ML. UniPROBE, update 2015: new tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2015;43:D117–122. doi: 10.1093/nar/gku1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga JT, Jones KA, Tjian R. Promoter-specific activation of RNA polymerase II transcription by Sp1. Trends in Biochemical Sciences. 1986;11:20–23. [Google Scholar]
- Karsenty G. Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- Katagiri T, Yamaguchi A, Ikeda T, Yoshiki S, Wozney JM, Rosen V, Wang EA, Tanaka H, Omura S, Suda T. The non-osteogenic mouse pluripotent cell line, C3H10T1/2, is induced to differentiate into osteoblastic cells by recombinant human bone morphogenetic protein-2. Biochem Biophys Res Commun. 1990;172:295–299. doi: 10.1016/s0006-291x(05)80208-6. [DOI] [PubMed] [Google Scholar]
- Kawane T, Komori H, Liu W, Moriishi T, Miyazaki T, Mori M, Matsuo Y, Takada Y, Izumi S, Jiang Q, et al. Dlx5 and mef2 regulate a novel runx2 enhancer for osteoblast-specific expression. J Bone Miner Res. 2014;29:1960–1969. doi: 10.1002/jbmr.2240. [DOI] [PubMed] [Google Scholar]
- Koga T, Matsui Y, Asagiri M, Kodama T, de Crombrugghe B, Nakashima K, Takayanagi H. NFAT and Osterix cooperatively regulate bone formation. Nat Med. 2005;11:880–885. doi: 10.1038/nm1270. [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapunzina P, Aglan M, Temtamy S, Caparros-Martin JA, Valencia M, Leton R, Martinez-Glez V, Elhossini R, Amr K, Vilaboa N, et al. Identification of a frameshift mutation in Osterix in a patient with recessive osteogenesis imperfecta. Am J Hum Genet. 2010;87:110–114. doi: 10.1016/j.ajhg.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larhammar D, Nordstrom K, Larsson TA. Evolution of vertebrate rod and cone phototransduction genes. Philos Trans R Soc Lond B Biol Sci. 2009;364:2867–2880. doi: 10.1098/rstb.2009.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Marijanovic I, Kronenberg MS, Erceg I, Stover ML, Velonis D, Mina M, Heinrich JG, Harris SE, Upholt WB, et al. Expression and function of Dlx genes in the osteoblast lineage. Dev Biol. 2008;316:458–470. doi: 10.1016/j.ydbio.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2012;13:27–38. doi: 10.1038/nrm3254. [DOI] [PubMed] [Google Scholar]
- Masuda Y, Sasaki A, Shibuya H, Ueno N, Ikeda K, Watanabe K. Dlxin-1, a novel protein that binds Dlx5 and regulates its transcriptional function. J Biol Chem. 2001;276:5331–5338. doi: 10.1074/jbc.M008590200. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Benkusky NA, Onal M, Pike JW. Selective regulation of Mmp13 by 1,25(OH)D, PTH, and Osterix through distal enhancers. J Steroid Biochem Mol Biol. 2015 doi: 10.1016/j.jsbmb.2015.09.001. In Press, doi:10.1016/j.jsbmb.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Olsen BR, Reginato AM, Wang W. Bone development. Annu Rev Cell Dev Biol. 2000;16:191–220. doi: 10.1146/annurev.cellbio.16.1.191. [DOI] [PubMed] [Google Scholar]
- Peterson KA, Nishi Y, Ma W, Vedenko A, Shokri L, Zhang X, McFarlane M, Baizabal JM, Junker JP, van Oudenaarden A, et al. Neural-specific Sox2 input and differential Gli-binding affinity provide context and positional information in Shh-directed neural patterning. Genes Dev. 2012;26:2802–2816. doi: 10.1101/gad.207142.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo RF, Rajan L, Li X, Lufkin T. The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev. 2002;16:1089–1101. doi: 10.1101/gad.988402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- Stains JP, Civitelli R. Genomic approaches to identifying transcriptional regulators of osteoblast differentiation. Genome Biol. 2003;4:222. doi: 10.1186/gb-2003-4-7-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- Takechi M, Adachi N, Hirai T, Kuratani S, Kuraku S. The Dlx genes as clues to vertebrate genomics and craniofacial evolution. Semin Cell Dev Biol. 2013;24:110–118. doi: 10.1016/j.semcdb.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Tang W, Yang F, Li Y, de Crombrugghe B, Jiao H, Xiao G, Zhang C. Transcriptional regulation of Vascular Endothelial Growth Factor (VEGF) by osteoblast-specific transcription factor Osterix (Osx) in osteoblasts. J Biol Chem. 2012;287:1671–1678. doi: 10.1074/jbc.M111.288472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB, Ohta Y, Flajnik MF, Sutoh Y, Kasahara M, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505:174–179. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Schoeps T, Donitz J. TFClass: an expandable hierarchical classification of human transcription factors. Nucleic Acids Res. 2013;41:D165–170. doi: 10.1093/nar/gks1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YF, Matsuo N, Sumiyoshi H, Yoshioka H. Sp7/Osterix is involved in the up-regulation of the mouse pro-alpha1(V) collagen gene (Col5a1) in osteoblastic cells. Matrix Biol. 2010;29:701–706. doi: 10.1016/j.matbio.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Yano H, Hamanaka R, Nakamura-Ota M, Adachi S, Zhang JJ, Matsuo N, Yoshioka H. Sp7/Osterix induces the mouse pro-alpha2(I) collagen gene (Col1a2) expression via the proximal promoter in osteoblastic cells. Biochem Biophys Res Commun. 2014;452:531–536. doi: 10.1016/j.bbrc.2014.08.100. [DOI] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun-Feng W, Matsuo N, Sumiyoshi H, Yoshioka H. Sp7/Osterix up-regulates the mouse pro-alpha3(V) collagen gene (Col5a3) during the osteoblast differentiation. Biochem Biophys Res Commun. 2010;394:503–508. doi: 10.1016/j.bbrc.2010.02.171. [DOI] [PubMed] [Google Scholar]
- Zhang C, Dai H, de Crombrugghe B. Characterization of Dkk1 gene regulation by the osteoblast-specific transcription factor Osx. Biochem Biophys Res Commun. 2012a;420:782–786. doi: 10.1016/j.bbrc.2012.03.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Tang W, Li Y. Matrix metalloproteinase 13 (MMP13) is a direct target of osteoblast-specific transcription factor osterix (Osx) in osteoblasts. PLoS One. 2012b;7:e50525. doi: 10.1371/journal.pone.0050525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zhang Z, Feng JQ, Dusevich VM, Sinha K, Zhang H, Darnay BG, de Crombrugghe B. Multiple functions of Osterix are required for bone growth and homeostasis in postnatal mice. Proc Natl Acad Sci U S A. 2010;107:12919–12924. doi: 10.1073/pnas.0912855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.