Abstract
Soil bacteria play a key role in the ecological and evolutionary responses of agricultural ecosystems. Domestic herbivore grazing is known to influence soil bacterial community. However, the effects of grazing and its major driving factors on soil bacterial community remain unknown for different plant community compositions under increasing grazing intensity. Thus, to investigate soil bacterial community diversity under five plant community compositions (Grass; Leymus chinensis; Forb; L. chinensis & Forb; and Legume), we performed a four-year field experiment with different grazing intensity treatments (no grazing; light grazing, 4 sheep·ha−1; and heavy grazing, 6 sheep·ha−1) in a grassland in China. Total DNA was obtained from soil samples collected from the plots in August, and polymerase chain reaction (PCR) analysis and denaturing gradient gel electrophoresis (DGGE) fingerprinting were used to investigate soil bacterial community. The results showed that light grazing significantly increased indices of soil bacterial community diversity for the Forb and Legume groups but not the Grass and L. chinensis groups. Heavy grazing significantly reduced these soil bacterial diversity indices, except for the Pielou evenness index in the Legume group. Further analyses revealed that the soil N/P ratio, electrical conductivity (EC), total nitrogen (TN) and pH were the major environmental factors affecting the soil bacterial community. Our study suggests that the soil bacterial community diversity was influenced by grazing intensity and plant community composition in a meadow steppe. The present study provides a baseline assessment of the soil bacterial community diversity in a temperate meadow steppe.
Introduction
Soil biodiversity, a key determinant of the ecological and evolutionary responses of terrestrial ecosystems to current and future environmental change, has become a focus of soil ecological research field [1–2]. Ubiquitous soil bacteria possess enormous metabolic and physiological versatility and are essential to virtually all biogeochemical cycling processes [3]. Soil bacterial community is the basis for nutrient cycling, conversion and utilization, influenced by above- and belowground biota, which can have positive, negative or neutral impacts [4–6]. However, most previous studies have focused on indirect changes to soil bacteria caused by soil, plant and grazing, and scarce data are available on the interactions and mechanisms of aboveground herbivorous animals on bacterial community [7–9].
Soil bacterial communities may be regulated by the physicochemical characteristics of soil, such as texture, pH, water content, and nutrient concentrations (i.e., C, N, C/N), which are key determinants of soil bacterial growth and activity [10]. In general, bacterial diversity has a strong positive correlation with pH, moisture, soil organic carbon and nitrogen, and C/N ratios [11–12], though some experiments have also shown a negative correlation with pH [13]. Plant communities significantly alter microbial community composition through rooting patterns, rhizodeposition, water use, litter chemistry, and subsequent influences on soil properties and microclimate [14]. In recent decades, ecologists have begun to explore the relationships between belowground soil bacterial communities and their functional significance to plant communities and ecosystem processes [15]. The nutrients required by soil bacteria are frequently obtained from plant litter or through root exudates from living plants and root decay [16]. Greater plant production has the potential to lead to greater litter accumulation on the soil surface, and greater C inputs to soil result in greater soil organic C; these changes can have profound effects on soil microbial community composition [17–18].
Although most controlled experiments to date have focused on the responses of plant biomass and species, plant communities can also significantly alter soil bacterial community by changing soil physicochemical characteristics, such as pH, EC, and nutrient content [11, 19]. Additionally, plant root exudates influence the surrounding soil bacteria community, and certain plant species support a highly coevolved soil bacteria community [20]. Experiments have shown that changes in the plant diversity and composition of grassland ecosystems can lead to rapid responses in bacterial activity and diversity [21–22]. Furthermore, previous studies on terrestrial ecosystems have shown that the plant community composition tends to have a greater impact on soil microbial communities compared with plant species richness [19, 22–23]. Nonetheless, few consistent patterns have been detected between plant community composition and soil bacterial community.
The grazing of domestic herbivores significantly affects the vegetation and soil properties of grassland and thereby impacts soil bacterial communities through increased trampling, defoliation, and manure return [24–25]. Indeed, herbivores can alter soil functions by returning carbon and mineral nutrients as dung and urine deposition as well as by trampling, which often reduces soil aeration and moisture [3]. Such activities often modify C and N cycles, potentially changing C and N accumulation in the soil and impacting nitrogen and carbon cycling rates through changes in grassland plant communities [26–28]. Additionally, herbivores alter rhizosphere activity by increasing root exudation via the removal of biomass and effects on the litter breakdown dynamics in plant communities. Most studies have focused on grazing-induced changes in the plant species composition and soil properties of grassland mesocosms [29], whereas relatively few experimental studies have investigated how grazing combined with plant community composition affects soil bacterial community in meadow steppes [30]. Moreover, the associated ecological problems and interaction mechanisms are complex and require investigation through field trials and mesocosm experiments under different grazing intensities. The main objectives of this study were as follows: i. to investigate whether plant community composition affects soil bacterial community composition and diversity; ii. to explore how these changes are affected by intensity of grazing; and iii. to identify the main factors affecting the soil bacterial community in a meadow steppe.
Materials and Methods
Study site
The soil samples used in this study were obtained from an experimental grazing field located at Grassland Ecological Research Station of Northeast Normal University, Jilin Province, China (N 44°32´44"-58," E123°39´52"-40´17"; elevation 130–140 m). The climate is semiarid with a mean annual precipitation of 280–400 mm, with approximately 70% of the rainfall occurring from June to August. The annual mean temperature ranges from 4.6 to 6.4°C [31]. The grassland is a sodic saline meadow steppe with on average 35% clay, 45% silt, and 20% sand; the bulk density is 1.54 g·cm-3, and the average soil pH is approximately 8.7 [32]. The main vegetation type is meadow steppe dominated by the perennial grass L. chinensis (Trin.) Tzvel. Other companion species include Phragmites australis Trin., Calamagrostis epigeios Roth., Chloris virgata Swartz, Lespedeza bicolor Turcz., Melilotus officinalis (Linn.) Pall., Medicago sativa L., Kalimeris integrifolia Turcz., Potentilla flagellaris Willd. and Carex duriuscula C. A. M. [33–34].
Experimental design and animal management
Based on previous research, we first collected evidence for the relationships between aboveground plant community composition and belowground bacterial community diversity to determine whether the correlations are a result of direct associations among the groups of organisms above and below the surface.
For this experiment, 10 ha of grassland that had been mowed for several decades was fenced in 2009 to exclude large herbivores (S1 Fig). For four years, an experiment of different grazing intensities (no grazing: CK; light grazing: LG; heavy grazing: HG) and different plant community composition (Grass; Leymus chinensis; Forb; L. chinensis & Forb; and legume) was performed in a meadow steppe in northeastern China. Three grazing intensities were arranged in a randomized complete block design with three replications, and the stocking rates were 0, 4, and 6 sheep·ha−1, respectively [35]. We analyzed plant diversity in the nine blocks, with a total of 112 plant species. All sampled plant community plots were divided into five compositions: Grass (perennial rhizosphere grass mixture of Phragmites australis, Calamagrostis epigeios, and Chloris virgata); L. chinensis (a clonal dominant species and it played an important role in structuring the plant community); Forb (perennial forb mixture of Kalimeris integrifolia, Potentilla flagellaris, and Carex duriuscula), L. chinensis & Forb (mixture of L. chinensis and perennial forb at a composition of 33 or 67%); and Legume (mixture of Lespedeza daurica, Melilotus officinalis, and Medicago sativa). From 2009 to 2012, the grazing period was approximately 5 months per year from May to September. Grazing occurred twice a day from 6:00 am to 8:00 am and from 4:00 pm to 6:00 pm [36].
Soil sampling
Soil samples were collected from 225 plots (five plant community compositions, five replicates in each nine blocks) in mid-August 2011. Five sampling replicates were obtained from the upper 5–20 cm (d = 2.5 cm) using a soil auger; these five cores (0.4 kg fresh weight) were bulked together and divided into two sub-samples after sieving (2 mm2 mesh) to remove coarse roots and stones. All of the samples were stored at 4°C prior to transport to the laboratory. One sub-sample was stored at 4°C for physicochemical analysis, and the other was stored at -20°C for denaturing gradient gel electrophoresis (DGGE) analysis.
Physicochemical analysis
The soil samples (100–200 g wet weight) were air dried and then passed through a 0.14 mm sieve. The soil pH and EC were determined using a soil-water ratio of 1:5. The soil water content (SW) before air drying was obtained by the oven-drying method. The soil organic carbon (SOC) content was measured using the Mebius method and by Walkley-Black acid digestion. The TN content was determined using an autoanalyzer (Foss 2100, FOSS Kjeltec®) with the Kjeldahl method following vitriol digestion. The total phosphorus (TP) content was measured colorimetrically after P extraction by Na2CO3 fusion.
Total DNA extraction from soil samples and DGGE analysis
Total DNA was obtained directly from each soil sample by CTAB-based extraction using a described protocol [37]. In brief, 500 mg of soil was mixed with 250 mg of 0.1 and 0.5 mm (1: 1) zirconia–silica beads, 500 mL of phenol–chloroform–isoamyl alcohol (25: 24: 1; Tris saturated, pH 8.0) and 500 mL of extraction buffer (12.2 mM KH2PO4, 112.8 mM K2HPO4, 5% w/v CTAB, 0.35 M NaCl; pH 8.0). The soil samples were bead-beaten for 30 s at 50 ms-1 and then centrifuged at 10 000 x g for 5 min at 4°C. The supernatant was mixed with 500 mL of chloroform–isoamyl alcohol (24: 1) and centrifuged again at 10 000 x g for 5 min at 4°C. The supernatant was precipitated at room temperature for 2 h with two volumes of a 30% w/v PEG 6000 and 1.6 M NaCl solution. The precipitated nucleic acids were pelleted by centrifugation at 10 000 x g for 10 min at 4°C.
The amount of DNA obtained from the samples varied from 10 to 20 ng·μl−1. The bacterial communities were then assessed by PCR amplification of cDNA templates using the general bacterial primers 16S 341F-GC and 518R. Each reaction mixture contained 50 ng DNA, 1 U Taq DNA Polymerase (Invitrogen®), 1.5 mM MgCl2, 0.2 mM each dNTP, 5 pmol each primer, and 1 μL DMSO to a final volume of 25 μL. The amplification reactions started with an initial denaturation step at 95°C for 5 min, followed by 30 cycles of 30 s at 95°C, 30 s at 55°C and 1 min at 72°C, and a final extension at 72°C for 10 min. Approximately equal amounts of the PCR products were loaded onto 6% (w/v) polyacrylamide gels prepared with 0.5× Tris-acetate-ethylene diamine tetraacetic acid buffer and denaturing gradients ranging from 45% to 65%. Electrophoresis was performed for 16 h at 100 V and 60°C [37–38].
The DGGE gels were scanned using a GS-800 Imaging Densitometer (Bio-Rad), and the bacterial community fingerprints of the DGGE bands were analyzed using Quantity One software (version 4.4.1; Bio-Rad). The images were normalized using markers, and cluster analysis was performed by applying the unweighted pair-group method with mathematical averages (UPGMA) using the similarity matrix generated was based on the Dice coefficient.
Statistical analysis
One-way analysis of variance (ANOVA) and Turkey’s multiple comparison tests were applied for post-hoc analysis of the significant differences among the factors. Two-way ANOVAs were then performed. Soil bacterial diversity indices were calculated, (a) Shannon-Wiener index(H): H = -Σ(Pi×lnPi), was calculated based on gel band intensity; (b) Richness (R): R = S, was calculated based on converting gel image into binary; (c) Pielou evenness index (E): EH = H/Hmax = H/lnS; Pi = Ni /N, where Ni is the height of the peak and N the sum of all peak heights in the densitometric curve, the significance level was set at P < 0.05, all of the data analyses were performed with SAS (Statistical Analysis Software). To evaluate relationships between microbial community composition and environmental variables, we first ordinated the DGGE presence/absence data using nonmetric multidimensional scaling (NMDS) and then looked for significant correlations between the ordination axes and environmental variables, and the statistical differences between soil bacterial fingerprints were measured using ANOSIM (analysis of similarity). The NMDS ordination and ANOSIM test were implemented by R software Version 3.1.1.
Results
Changes in soil properties along a grazing gradient with different plant community compositions
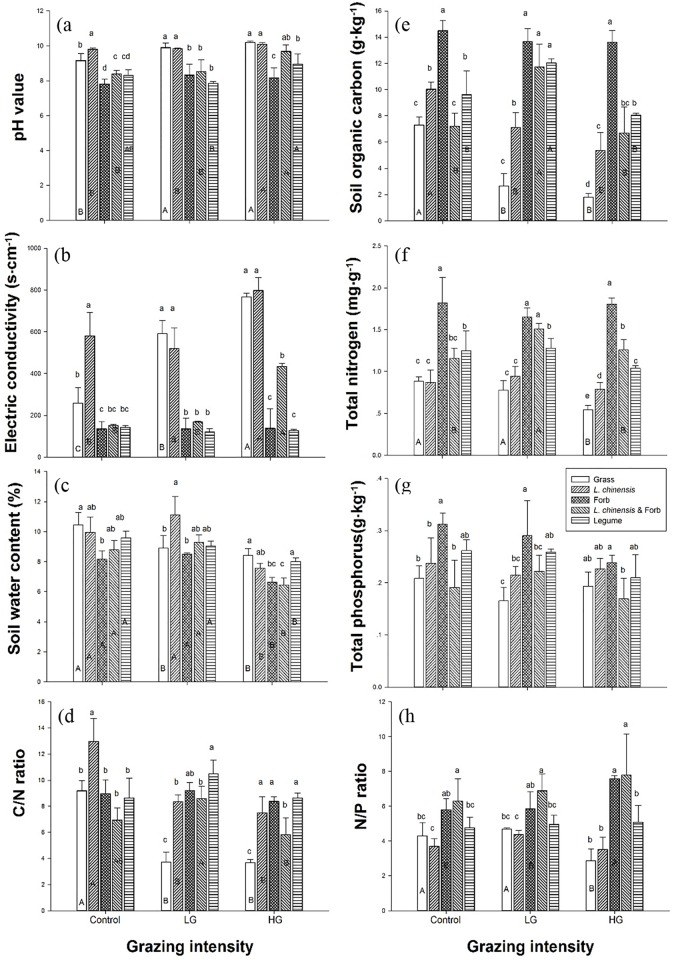
ANOVA detected large and significant differences among the different plant community compositions for all of the soil physicochemical properties along the grazing gradient (Fig 1, Table 1). In general, grazing enhanced the pH value in the Grass, L. chinensis, L. chinensis & Forb, and Legume groups but not in the Forb group (P < 0.001), especially under heavy grazing (Fig 1a). The soil pH was significantly higher in the Grass and L. chinensis groups (P < 0.001). The results for EC were similar to those of pH under the grazing treatments (P < 0.001): EC in the Grass group increased along the grazing gradient, whereas high EC values in the L. chinensis and L. chinensis & Forb groups were only observed with heavy grazing (P < 0.001) (Fig 1b). Grazing reduced SW in the Grass group, but only heavy grazing reduced this parameter in the other plant community compositions (P < 0.001) (Fig 1c). Comparatively, SW was significantly higher in the L. chinensis group and lower in the Forb group in light grazing (P < 0.001) (Fig 1c).
Fig 1. Soil property responses to different plant community composition along a grazing gradient. (a) pH value, (b) electrical conductivity, (c) soil water content, (d) the C/N ratio, (e) soil organic carbon, (f) total nitrogen, (g) total phosphorus and (h) the N/P ratio.
Values represent the means ± SE. Significant differences between different plant community composition levels within a grazing intensity are indicated by lowercase letters (a-c); significant differences between different grazing intensities within a plant community composition level are indicated by capital letters (A-C; P ≤ 0.05). Vertical bars ± 1 SE.
Table 1. Two-way analysis of variance of the effects of grazing and plant community composition on soil pH, electrical conductivity (EC), soil water (SW), soil organic carbon (SOC), total N (TN), total P (TP), C/N and N/P ratios.
| Factors | Grazing intensity (G) | Plant functional group (P) | G × P | |
|---|---|---|---|---|
| d.f. | 2,45 | 4,45 | 8,45 | |
| pH | F | 14.164 | 40.484 | 2.499 |
| P | <0.001 | <0.001 | 0.033 | |
| EC | F | 48.285 | 149.463 | 13.734 |
| P | <0.001 | <0.001 | <0.001 | |
| SW | F | 49.122 | 12.641 | 3.636 |
| P | <0.001 | <0.001 | 0.005 | |
| SOC | F | 24.862 | 96.275 | 9.620 |
| P | <0.001 | <0.001 | <0.001 | |
| TN | F | 4.737 | 80.301 | 2.747 |
| P | 0.016 | <0.001 | 0.021 | |
| TP | F | 3.857 | 10.567 | 1.280 |
| P | 0.032 | <0.001 | 0.291 | |
| C/N | F | 24.094 | 26.352 | 10.450 |
| P | <0.001 | <0.001 | <0.001 | |
| N/P | F | 0.905 | 21.313 | 2.201 |
| P | 0.415 | <0.001 | 0.056 | |
The grazing intensity treatments and plant community compositions significantly affected SOC (P < 0.001), TN (P < 0.001), and TP (P < 0.001) (Fig 1e, 1f and 1g). Grazing decreased SOC in the Grass and L. chinensis groups (P < 0.001), whereas light grazing increased SOC in the L. chinensis & Forb and Legume groups (Fig 1e). Heavy grazing decreased TN in the Grass group, and light grazing increased it in the L. chinensis & Forb group. The SOC, TN and TP contents were highest in the Forb group and lowest in the Grass group. Overall, grazing reduced the C/N ratio (P = 0.016) (Fig 1d) but not the N/P ratio (P = 0.415) (Fig 1h).
PCR-DGGE analysis of soil bacterial communities
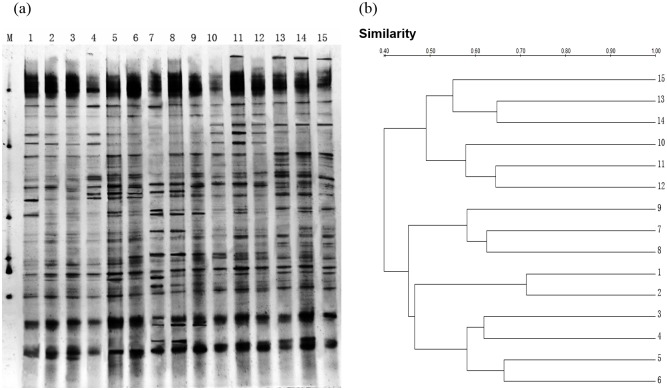
PCR-DGGE analysis of the extracts yielded virtually identical profiles for the fifteen samples analyzed (Fig 2). The diversity of the soil bacterial community was affected by both grazing intensity and plant community composition. The samples were characterized by the presence of a limited number (11–19) of strong bands and a larger number of weak bands; most of the differences were observed in both the strong and weak bands. In Fig 2a, more bands are observed in lanes 1, 4, 8, 11, 13 and 14, indicating more complex bacterial communities in the Grass, L. chinensis and Forb groups under no grazing and in the Grass, Legume, L. chinensis & Forb and Forb groups under light grazing. Conversely, few bands are present in lanes 3, 6 and 12, indicating that heavy grazing decreased the complexity of the bacterial communities in the L. chinensis, Grass and L. chinensis & Forb groups (Fig 2a).
Fig 2. PCR-DGGE analysis of soil bacterial 16S rDNA (a) and dendrogram constructed using the soil bacterial community fingerprints (b) along the grazing gradient with different plant community compositions. M: marker, L. chinensis: 1–3, Grass: 4–6, Legume: 7–9, L. chinensis & Forb: 10–12, Forb: 13–15. Control: 1, 4, 7, 10, 13; light grazing: 2, 5, 8, 11, 14; and heavy grazing: 3, 6, 9, 12, 15.
Difference between the profiles are indicated by the similarity percentage. The dendrogram of the cluster analysis was based on the Dice coefficient and UPGMA clustering algorithm.
UPGMA cluster analysis (Dice coefficient of similarity) of the DGGE patterns confirmed these general observations (Fig 2b). All of the lanes clustered together at a level of approximately 39% similarity. The following lanes were clustered together: lanes 13, 14 and 15; lanes 10, 11 and 12; lanes 7, 8 and 9; lanes 1 and 2; and lanes 3, 4, 5 and 6. The bacterial communities under L. chinensis and Grass were more evidently clustered together. Additionally, a clear trend was observed, in which the same plant community composition resulted in bacterial communities presenting 49% similarity with the other bacterial communities.
Effects of grazing intensity and plant community composition on bacterial diversity indices
The grazing intensity and plant community composition significantly affected bacterial diversity indices. A significant decrease in the Shannon-Wiener index was observed with different grazing intensities under Grass and L. chinensis, especially with heavy grazing. Light grazing significantly increased the Shannon-Wiener index under Forb and Legume, whereas heavy grazing significantly decreased this index. Only heavy grazing significantly decreased the Shannon-Wiener index under L. chinensis & Forb (Fig 3a). The result for richness was similar to that for the Shannon-Wiener index (Fig 3b) under the different grazing intensities. The Shannon-Wiener index (P < 0.001) and richness (P < 0.001) were significantly higher under L. chinensis and lower under L. chinensis & Forb and Legume in the control treatment but were significantly higher under Forb and lower under Grass in the light grazing and heavy grazing treatments (Table 2) (Fig 3a and 3b).
Fig 3. Effects of grazing on bacterial diversity. (a) Shannon-Wiener index, (b) richness, and (c) Pielou evenness index.
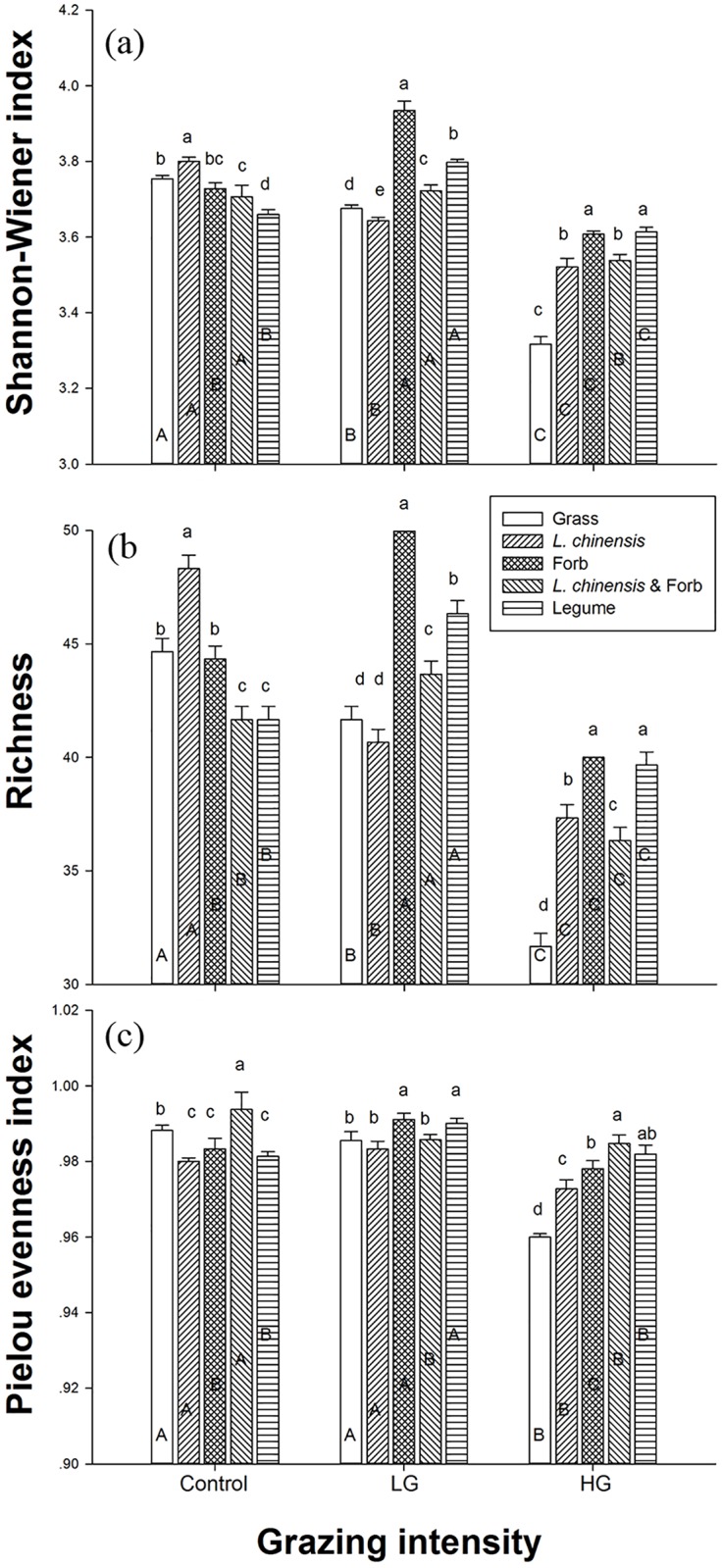
Error bars denote the SE. Significant differences between different plant community composition levels within a grazing intensity are indicated by lowercase letters (a-c); significant differences between different grazing intensities within a plant community composition level are indicated by capital letters (A-C; P ≤ 0.05). Vertical bars ± 1 SE.
Table 2. Two-way analysis of variance of the effects of grazing intensity (G) and plant community composition (P) on the Shannon-Wiener index, richness and Pielou evenness index of the soil bacterial community.
| Factors | Grazing intensity (G) | Plant functional group (P) | G × P | |
|---|---|---|---|---|
| d.f. | 2,45 | 4,45 | 8,45 | |
| Shannon-Wiener index | F | 179.326 | 209.240 | 258.555 |
| P | <0.001 | <0.001 | 0.033 | |
| Richness | F | 821.438 | 150.406 | 106.906 |
| P | <0.001 | <0.001 | <0.001 | |
| Pielou evenness index | F | 123.511 | 34.515 | 26.110 |
| P | <0.001 | <0.001 | <0.001 | |
Heavy grazing significantly decreased the Pielou evenness index under Grass and L. chinensis compared with the control and light grazing treatments. Light grazing significantly increased the Pielou evenness index under Forb, but heavy grazing significantly decreased this index. In addition, grazing decreased the Pielou evenness index under L. chinensis & Forb, though significant changes were not observed between light grazing and heavy grazing. Light grazing significantly increased the Pielou evenness index under Legume (Fig 3c). The heavy grazing and control treatments significantly increased the Pielou evenness index (P < 0.001) under L. chinensis & Forb, though light grazing produced higher index values under Forb and Legume (Table 2) (Fig 3c).
Effects of grazing intensity and plant community composition on the soil bacterial community composition
NMDS revealed the effects of grazing intensity and plant community composition on the soil bacterial community composition (P < 0.001), closer points imply more similar soil bacterial community compositons, Stress = 0.16 for all ordinations, confirming a good correlation between the data and its ordination (Fig 4). Bacterial community compositons differ significantly between the five plant community composition (ANOSIM test, P = 0.001), no significantly between the three grazing intensities (ANOSIM test, P = 0.851), which indicates that plant community composition is the key factor that triggers the variation of soil bacterial community composition.
Fig 4. NMDS ordination of the DGGE profiles of 16S rRNA gene fragments with superimposed vectors derived from soil chemistry data.
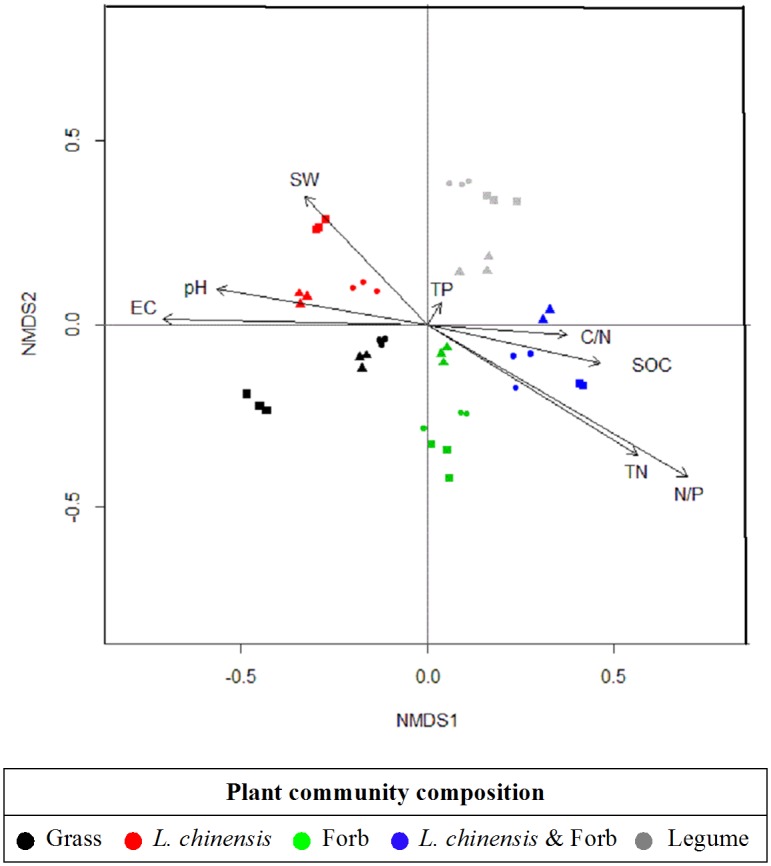
Circles refer to the control, triangles refer to light grazing and squares refer to heavy grazing. The statistical differences between soil bacterial fingerprints were measured using ANOSIM (analysis of similarity).
Among all of the soil properties examined, the NMDS plot revealed that the variables explaining the most important factors governing bacterial community composition were N/P ratio (r2 = 0.657, P = 0.001), EC (r2 = 0.503, P = 0.001), and TN (r2 = 0.444, P = 0.001), the other factors were pH (r2 = 0.330, P = 0.002), SW (r2 = 0.231, P = 0.004), SOC (r2 = 0.224, P = 0.006), and C/N ratio (r2 = 0.142, P = 0.053) (S1 Table).
Discussion
The objective of this work was to determine the influence of plant community composition on the diversity of soil microbial communities under grazing intensity treatments and to examine whether these effects are context dependent. Our findings showed that grazing intensity and plant community composition altered the soil bacterial community, and the soil N/P ratio, electrical conductivity (EC), total nitrogen (TN) and pH were the major environmental factors affecting the soil bacterial community. These results provide evidence that partly supports the previously reported changes in ecological processes caused by large herbivore grazing.
Effects of grazing intensity and plant community composition on soil bacterial community diversity
Grazing livestock can play an important role in the microbial ecology of grasslands through a series of specific factors, including plant community composition and biomass, fecal and urine deposition, rhizosphere exudation, and soil texture and physicochemical properties; these factors can have positive, neutral or negative effects on soil bacterial diversity [7,15,39–41]. Our study clearly showed that the diversity of the soil bacterial community varied significantly along the gradient from light to heavy grazing intensity (Fig 3; Table 2). This result is consistent with that of previous studies in which light grazing was shown to markedly increase but heavy grazing to significantly decrease the Shannon-Wiener index, richness, and Pielou evenness index of the soil bacterial community from an upland grassland and an Inner Mongolian dry steppe [42–44]. Previous studies and our results clearly indicate that changes in soil bacterial community diversity are closely related to different grazing intensities [4, 7, 44]. In the present study, light grazing had a limited effect on the pH, EC, SWC and TN of the soil, whereas heavy grazing significantly decreased the parameters of SWC, SOC, TN and the C/N ratio (Fig 1).
Some experiments demonstrated that light or moderate grazing can optimize the aboveground net primary productivity via compensatory growth, which may have occurred due to an increase in nutrient availability (C, N or others), thereby facilitating vegetation regrowth [45–46]. In contrast, heavy grazing may decrease the storage availability of soil C and N and result in a reduction in soil bacterial diversity [47]. Soil pH and moisture are considered to be important factors that affect microbial community composition and diversity, and the soils in the studied grassland are mixed saline and alkaline (pH 8.0–10.0). Other studies have suggested that bacterial diversity declines as the soil pH increases from neutral to alkaline [48]. Indeed, compared with the no grazing treatment, heavy grazing significantly increased pH (from 9.1 to 10.2 under Grass) and EC (from 258.7 to 767.7 under Grass) but decreased SW (from 10.5 to 8.4% under Grass) (Fig 1; Table 1); these changes jointly constrain soil bacterial diversity in nutrient-poor soils [8, 49].
Soil bacteria are mostly heterotrophic and utilize plant exudates or decomposing plant material for growth. Although the reduction in food quantity and changes in food quality caused by alterations to the plant community structure or composition often modify the abundance, activity, and diversity of soil bacterial communities, few studies have focused on it [19, 21, 50]. We found that the diversity of the soil bacterial community differed markedly under the studied plant community composition in the meadow steppe environment. Our results showed that the soil bacterial diversity indices were higher under Grass and L. chinensis than under Forb and Legume in the no grazing plots (Fig 3). Alterations in the plant community composition often result in different structural and chemical compositions of the plant litter that is returned to the soil, subsequently causing different depletion patterns of soil resources [51]. The meadow steppe in our study site is dominated by the perennial grass L. chinensis and other companion grasses, such as P. australis, C. epigejos and C. virgata, a plant composition that is most likely to produce greater litter decomposition and soil exudate profiles under the L. chinensis and Grass groups. Consequently, the organic acid in the root exudates may have reduced the stress of soil salinity and pH and increased soil bacterial diversity at the study site.
Furthermore, the changes in soil bacterial diversity could have been caused by interactions between grazing intensity and plant community composition (Table 2; Fig 3). Our results showed that light grazing significantly increased soil bacterial diversity under the Forb and Legume groups but markedly decreased diversity under the L. chinensis and Grass groups (Fig 3). Previous experiments have shown that light and moderate grazing could lead to overcompensation due to the amounts of litter and root exudates in grassland ecosystems [31, 52–54]. Light grazing increased plant productivity by increasing the quantity of plant litter under the Forb and Legume groups (Fig 3), which may have promoted soil C and N dynamics by accelerating root production and exudation. These conditions are known to favor bacterial growth in soils [55]. However, heavy grazing decreased the Shannon-Wiener index, phylotype richness, and Pielou evenness index of the soil bacterial diversity under all of the plant community compositions (Fig 3). As the forage utilization rate increased, the amount of plant litter returning to the soil decreased with increases in grazing intensity [46]. In particular, heavy grazing led to decreased soil quality, fertility and moisture but promoted the soil pH and EC, which might have decreased the soil bacterial diversity in each plot (Figs 1 and 3). Experiments have also documented the effects of heavy grazing on aboveground plant communities and belowground microbial communities [3–4, 7, 46].
In addition, we observed the highest soil bacterial diversity under Forb and the lowest under Grass with grazing (Fig 3). Forb is composed of a variety of species (perennial forb mixture of Kalimeris integrifolia, Potentilla flagellaris, and Carex duriuscula), and heavy grazing altered the composition of Forb, adding some new species that produce more kinds of root exudates and plant secondary metabolites. This led to the highest SOC, TN, and TP values and the lowest pH under Forb, whereas heavy grazing reduced the Grass biomass, with no change in species composition. Different plant community compositions may create different niche dimensions by varying the litter quantity and quality, consequently inducing changes in groups of soil decomposers [19]. Moreover, trampling by grazers altered the soil texture and physicochemical properties, which could affect soil bacterial community.
Main factors affecting soil bacterial community
Although the results of our study showed that the grazing intensity and plant community composition influenced the soil bacterial community diversity (Fig 3), the latter also significantly affected the soil bacterial community composition (Fig 4), NMDS of the DGGE data revealed that soil N/P ratio (r2 = 0.657), EC (r2 = 0.503), TN (r2 = 0.444), and pH (r2 = 0.330) were the major factors driving soil bacterial community (Fig 4). Previous work has shown that soil bacterial communities are primarily affected by factors such as soil pH, moisture, organic C and N, and aboveground plant species, community composition and diversity in grassland soils [3, 56–59]. In addition, experimental evidence has suggested that soil moisture is a key driving factor in dry steppe and semi-arid steppe ecosystems [60–61], and soil organic C or the C/N ratio plays an important role in controlling bacterial community composition and diversity in alkaline permafrost-affected soils [62–63]. However, these results are not consistent with our observations. The nutrient levels in the studied meadow steppe were low (TN 0.872–1.823 mg·g-1) and precipitation high (280–400 mm), and the soil bacterial diversity of this site was mainly influenced by soil pH and EC, consistent with studies in extreme and/or moderate saline environments [64–66].
Near-neutral soils present a greater availability of nutrients to support copiotrophic bacteria, whereas high pH and EC conditions significantly reduce utilization of soil nutrients in alkaline-saline regions [67]. In general, near-neutral pH levels might be regarded as a proxy for the physiological availability of a variety of nutrients. The internal pH of bacterial cells is normally close to neutral, and an external pH environment similar to this intracellular value may suggest a reduction in the energy expenditure required to maintain this internal pH and fewer specialized adaptations [63, 68]. However, high pH could be considered a “stressful” environment because of grazing-induced disturbances and changes in plant community traits; such environments might require specialized adaptations that relatively few taxa have been able to acquire [67]. At our study site, heavy grazing significantly increased pH (from 9.1 to 10.2) and EC (from 258.7 to 767.7) and decreased TN (from 0.88 mg·g-1 to 0.55 mg·g-1), thereby resulting in a reduction in soil bacterial diversity (Shannon-Wiener index, richness and Pielou evenness index) (Figs 1 and 3). Thus, it is clear that multiple stresses due to herbivore grazing lead to additive negative effects on soil bacteria diversity through changes in soil physical and chemical properties in meadow steppe ecosystems.
Conclusions
The experimental results presented here suggest that grazing intensity and plant community composition as well as their interactions markedly influence the Shannon-Wiener index, phylotype richness, and Pielou evenness index of the soil bacterial diversity. Light grazing significantly increased the diversity of the soil bacterial community under the Forb and Legume groups but notably decreased diversity under the L. chinensis and Grass groups. In contrast, heavy grazing greatly decreased the diversity of the soil bacterial community diversity for all of the plant community compositions. Our study suggests that the soil bacterial community diversity is influenced by grazing intensity and plant community composition, soil N/P, EC, TN, and pH to be the main driving forces affecting soil bacterial community in this meadow steppe at a regional scale. The results of this study reveal that the diversity of the soil bacterial community is influenced by grazing intensity, plant community composition and soil physicochemical properties. The present study provides a baseline assessment of the soil bacterial community in a temperate meadow steppe and could be a useful tool for future assessments of the effects of grassland management.
Supporting Information
(TIF)
(DOC)
Acknowledgments
We thank Liang Jin, Jun Liang and Jushan Liu for providing valuable comments on the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This project was supported by National Natural Science Foundation of China (31230012) to DW and (34201839) to ZY, the State Basic Research Program (2012FY111900-3) to DW, the Program for Introducing Talents to Universities (B16011) to DW, and the China Postdoctoral Science Foundation Project (20110491286) to TQ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Prosser JI, Bohannan BJM, Curtis TP, Ellis RJ, Firestone MK, Freckleton RP, et al. The role of ecological theory in microbial ecology. Nat Rev Microbiol 2007; 5(5): 384–392. 10.1038/nrmicro1643 . [DOI] [PubMed] [Google Scholar]
- 2.Bardgett RD, Putten WHVD. Belowground biodiversity and ecosystem functioning. Nature 2014; 515 (7528): 505–511. 10.1038/nature13855 . [DOI] [PubMed] [Google Scholar]
- 3.Clegg CD. Impact of cattle grazing and inorganic fertiliser additions to managed grasslands on the microbial community composition of soils. Appl Soil Ecol 2006; 31 (1–2): 73–82. 10.1016/j.apsoil.2005.04.003 [DOI] [Google Scholar]
- 4.Wardle DA, Bardgett RD, Klironomos JN, Setala H. Ecological linkages between aboveground and belowground biota. Science 2004; 304(5677): 1629–33. . [DOI] [PubMed] [Google Scholar]
- 5.Singh BK, Dawson LA, Macdonald CA, Buckland SM. Impact of biotic and abiotic interaction on soil microbial communities and functions: A field study. Appl Soil Ecol 2009; 41(3): 239–248. 10.1016/j.apsoil.2008.10.003 [DOI] [Google Scholar]
- 6.Birkhofer K, Schoning I, Alt F, Herold N, Klarner B, Maraun M, et al. General relationships between abiotic soil properties and soil biota across spatial scales and different land-use types. PloS ONE 2012; 7 (8): 1–8. 10.1371/journal.pone.0043292 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bardgett RD, Jones AC, Jones DL, Kemmitt SJ, Cook R, Hobbs PJ. Soil microbial community patterns related to the history and intensity of grazing in sub-montane ecosystems. Soil Biol Biochem 2001; 33: 1653–1664. 10.1016/S0038-0717(01)00086-4 [DOI] [Google Scholar]
- 8.Braun B, Böckelmann U, Grohmann E, Szewzyk U. Bacterial soil communities affected by water-repellency. Geoderma 2010; 158 (3–4): 343–351. 10.1016/j.geoderma.2010.06.001 [DOI] [Google Scholar]
- 9.Marshall CB, McLaren JR, Turkington R. Soil microbial communities resistant to changes in plant functional group composition. Soil Biol Biochem 2011; 43 (1): 78–85. 10.1016/j.soilbio.2010.09.016 [DOI] [Google Scholar]
- 10.Angassa A, Sheleme B, Oba G, Treydte AC, Linstädter A, Sauerborn J. Savanna land use and its effect on soil characteristics in southern Ethiopia. J Arid Environ 2012; 81: 67–76. 10.1016/j.jaridenv.2012.01.006 [DOI] [Google Scholar]
- 11.Douterelo I, Goulder R, Lillie M. Soil microbial community response to land-management and depth, related to the degradation of organic matter in English wetlands: Implications for the in situ preservation of archaeological remains. Appl Soil Ecol 2010; 44 (3): 219–227. 10.1016/j.apsoil.2009.12.009 [DOI] [Google Scholar]
- 12.Nautiyal CS, Chauhan PS, Bhatia CR. Changes in soil physico-chemical properties and microbial functional diversity due to 14 years of conversion of grassland to organic agriculture in semi-arid agroecosystem. Soil Till Res 2010; 109 (2): 55–60. 10.1016/j.still.2010.04.008 [DOI] [Google Scholar]
- 13.Acosta-Martínez V, Dowd SE, Sun Y, Wester D, Allen V. Pyrosequencing analysis for characterization of soil bacterial populations as affected by an integrated livestock-cotton production system. Appl Soil Ecol 2010; 45 (1): 13–25. 10.1016/j.apsoil.2010.01.005 [DOI] [Google Scholar]
- 14.Fichtner A, Oheimb GV, Härdtle W, Wilken C, Gutknecht JLM. Effects of anthropogenic disturbances on soil microbial communities in oak forests persist for more than 100 years. Soil Biol Biochem 2014; 70: 79–87. 10.1016/j.soilbio.2013.12.015 [DOI] [Google Scholar]
- 15.Bardgett RD, Wardle DA. Herbivore-mediated linkages between aboveground and belowground communities. Ecology. 2003; 84:2258–2268. 10.1890/02-0274 [DOI] [Google Scholar]
- 16.Bhullar GS, Edwards PJ, Olde VH. Influence of different plant species on methane emissions from soil in a restored Swiss wetland. PloS ONE 2014; 9 (2): e89588 10.1371/journal.pone.0089588.t001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allison VJ, Yermakov Z, Miller RM, Jastrow JD, Matamala R. Using landscape and depth gradients to decouple the impact of correlated environmental variables on soil microbial community composition. Soil Biol. Biochem 2007; 39 (2): 505–516. 10.1016/j.soilbio.2006.08.021 [DOI] [Google Scholar]
- 18.Buyer JS, Zuberer DA, Nichols KA, Franzluebbers AJ. Soil microbial community function, structure, and glomalin in response to tall fescue endophyte infection. Plant Soil 2010; 339 (1–2): 401–412. 10.1007/s11104-010-0592-y [DOI] [Google Scholar]
- 19.Fry EL, Manning P, Allen DGP, Hurst A, Everwand G, Rimmler M, et al. Plant functional group composition modifies the effects of precipitation change on grassland ecosystem function. PloS ONE 2013; 8 (2): 1–14. 10.1371/journal.pone.0057027 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Badri DV, Quintana N, Kassis EG, Kim HK, Choi YH, Sugiyama A, et al. An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol 2009; 151 (4): 2006–2017. 10.1104/pp.109.147462 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loranger-Merciris G, Barthes L, Gastine A, Leadley P. Rapid effects of plant species diversity and identity on soil microbial communities in experimental grassland ecosystems. Soil Biol Biochem 2006; 38(8): 2336–2343. 10.1016/j.soilbio.2006.02.009 [DOI] [Google Scholar]
- 22.Kooyman R, Rossetto M. Definition of plant functional groups for informing implementation scenarios in resource-limited multi-species recovery planning. Biodiversity Conserv 2008; 17(12): 2917–2937. 10.1007/s10531-008-9405-5 [DOI] [Google Scholar]
- 23.Ledeganck P, Nijs I, Beyens L. Plant functional group diversity promotes soil protist diversity. Protist 2003; 154: 239–249. 10.1078/143446103322166536 . [DOI] [PubMed] [Google Scholar]
- 24.Kohler F, Hamelin J, Gillet F, Gobat JM, Buttler A. Soil microbial community changes in wooded mountain pastures due to simulated effects of cattle grazing. Plant Soil 2005; 278 (1–2): 327–340. 10.1007/s11104-005-8809-1 [DOI] [Google Scholar]
- 25.Wang L, Wang DL, Liu J, Huang Y, Hodgkinson KC. Diet selection variation of a large herbivore in a feeding experiment with increasing species numbers and different plant functional group combinations. Acta Oecol 2011; 37(3): 263–268. 10.1016/j.actao.2011.02.010 [DOI] [Google Scholar]
- 26.Piñeiro G, Paruelo JM, Oesterheld M. Potential long-term impacts of livestock introduction on carbon and nitrogen cycling in grasslands of southern South America. Global Change Biol 2006; 12(7): 1267–1284. 10.1111/j.1365-2486.2006.01173.x [DOI] [Google Scholar]
- 27.Patra AK, Roux XL. Effects of grazing on microbial functional groups involved in soil N dynamics. Ecol Monogr 2005; 75 (1): 65–80. 10.1890/03-0837 [DOI] [Google Scholar]
- 28.Olsen YS, Dausse A, Garbutt A, Ford H, Thomas DN, Jones DL. Cattle grazing drives nitrogen and carbon cycling in a temperate salt marsh. Soil Biol Biochem 2011; 43(3): 531–541. 10.1016/j.soilbio.2010.11.018 [DOI] [Google Scholar]
- 29.Semmartin M, Bella C, Salamone IG. Grazing-induced changes in plant species composition affect plant and soil properties of grassland mesocosms. Plant Soil 2009; 328(1–2): 471–481. 10.1007/s11104-009-0126-7 [DOI] [Google Scholar]
- 30.Roux XL, Poly F, Currey P, Commeaux C, Hai B, Nicol GW, et al. Effects of aboveground grazing on coupling among nitrifier activity, abundance and community structure. ISME J 2008; 2(2): 221–232. 10.1038/ismej.2007.109 . [DOI] [PubMed] [Google Scholar]
- 31.Liu J, Wang L, Wang DL, Bonser SP, Sun F, Zhou YF, et al. Plants can benefit from herbivory: stimulatory effects of sheep saliva on growth of Leymus chinensis. PLoS ONE 2012; 7(1): 29–59. 10.1371/journal.pone.0029259 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang RZ, Ripley EA. Effects of grazing on a Leymus chinensis grassland on the Songnen plain of northeastern China. J Ari Environ 1997; 36: 307–318. 10.1006/jare.1996.0214 [DOI] [Google Scholar]
- 33.Ba L, Ning J, Wang DL, Facelli E, Facelli JM, Yang YN, et al. The relationship between the diversity of arbuscular mycorrhizal fungi and grazing in a meadow steppe. Plant Soil 2012; 352(1–2): 143–156. 10.1007/s11104-011-0985-6 [DOI] [Google Scholar]
- 34.Gao Y, Wang D, Xing F, Liu J, Wang L. Combined effects of resource heterogeneity and simulated herbivory on plasticity of clonal integration in a rhizomatous perennial herb. Plant Biol 2014; 16: 774–782. 10.1111/plb.12122 . [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Wang D, Han S, Wang X. Effect of grazing intensity on the regrowth capability in Leymus chinensis grassland. Acta Pratacultural Science 2004; 13 (6): 39–44. [Google Scholar]
- 36.Zhu H, Wang DL, Guo QF, Liu J, Wang L. Interactive effects of large herbivores and plant diversity on insect abundance in a meadow steppe in China. Agric Ecosyst Environ 2015; 212: 245–252. 10.1016/j.agee.2015.07.008 [DOI] [Google Scholar]
- 37.Yergeau E, Bokhorst S, Huiskes AH, Boschker HT, Aerts R, Kowalchuk GA. Size and structure of bacterial, fungal and nematode communities along an antarctic environmental gradient. FEMS Microbiology Ecology 2007; 59(2): 436–451. 10.1111/j.1574-6941.2006.00200.x [DOI] [PubMed] [Google Scholar]
- 38.Dini-Andreote F, Andreote FD, Costa R, Taketani RG, Elsas JK, Araújo WL. Bacterial soil community in a Brazilian sugarcane field. Plant Soil 2010; 336(1–2): 337–349. 10.1007/s11104-010-0486-z [DOI] [Google Scholar]
- 39.Dorrough J, Ash J, Mcintyre S. Plant responses to livestock grazing frequency in an australian temperate grassland. Ecography 2004; 27(6): 798–810. 10.1111/j.0906-7590.2004.04004.x [DOI] [Google Scholar]
- 40.Ritz K, McNicol JW, Nunan N, Grayston S, Millard P, Atkinson D, et al. Spatial structure in soil chemical and microbiological properties in an upland grassland. FEMS Microbiol Ecol 2004; 49: 191–205. 10.1016/j.femsec.2004.03.005 . [DOI] [PubMed] [Google Scholar]
- 41.Guitian R, Bardgett RD. Plant and soil microbial responses to defoliation in temperate semi-natural grassland. Plant Soil 2000; 220: 271–277. 10.1023/a:1004787710886 [DOI] [Google Scholar]
- 42.Clegg CD, Ritz K, Griffiths BS. %G+C profiling and cross hybridization of microbial DNA reveals great variation in below-ground community structure in UK upland grasslands. Appl Soil Ecol 2000; 14: 125–134. 10.1016/S0929-1393(00)00045-7 [DOI] [Google Scholar]
- 43.Grayston SJ, Griffith GS, Mawdsley JL, Campbell CD, Bardgett RD. Accounting for variability in soil microbial communities of temperate upland grassland ecosystems. Soil Biol Biochem 2001; 33: 533–551. 10.1016/S0038-0717(00)00194-2 [DOI] [Google Scholar]
- 44.Zhou X, Wang J, Hao Y, Wang Y. Intermediate grazing intensities by sheep increase soil bacterial diversities in an Inner Mongolian steppe. Biol Fert Soils 2010; 46(8): 817–824. 10.1007/s00374-010-0487-3 [DOI] [Google Scholar]
- 45.Ruess RW, Mcnaughton SJ. Grazing and the dynamics of nutrient and energy regulated microbial processes in the serengeti grasslands. Oikos 1987; 49(1): 101–110. 10.2307/3565559 [DOI] [Google Scholar]
- 46.Han GD, Hao XY, Zhao ML, Wang MJ, Ellert BH, Willms W, et al. Effect of grazing intensity on carbon and nitrogen in a meadow steppe in Inner Mongolia. Agric Ecosyst Environ 2008; 125: 21–32. 10.1016/j.agee.2007.11.009 [DOI] [Google Scholar]
- 47.Steffens M, Kölbl A, Kai UT, Kögel-Knabner I. Grazing effects on soil chemical and physical properties in a semiarid steppe of Inner Mongolia (P.R. China). Geoderma 2008; 143: 63–72. 10.1016/j.geoderma.2007.09.004 [DOI] [Google Scholar]
- 48.Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. PNAS 2006; 103: 626–631. 10.1073/pnas.0507535103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dimitriu PA, Grayston SJ. Relationship between soil properties and patterns of bacterial B-diversity across reclaimed and natural boreal forest soils. Microb Ecol 2010; 59(3): 563–573. 10.1007/s00248-009-9590-0 [DOI] [PubMed] [Google Scholar]
- 50.Lamb EG, Kennedy N, Siciliano SD. Effects of plant species richness and evenness on soil microbial community diversity and function. Plant Soil 2011; 338: 483–495. 10.1007/s11104-010-0560-6 [DOI] [Google Scholar]
- 51.Wardle DA, Barker GM, Yeates GW, Bonner KI, Ghani A. Introduced browsing mammals in New Zealand natural forests: aboveground and belowground consequences. Ecolo Monogr 2001; 71(4): 587–614. [Google Scholar]
- 52.Aarssen LW. Hypotheses for the evolution of apical dominance in plants: implications for the interpretation of overcompensation. Oikos 1995; 74: 149–156. 10.2307/3545684 [DOI] [Google Scholar]
- 53.Huhta AP, Hellstrӧm K, Rautio P, Tuomi J. Grazing tolerance of Gentianella amarelle and other monocarpic herbs: why is tolerance highest at low damage levels? Plant Ecol 2003; 166: 49–61. 10.1023/A:1023278502972 [DOI] [Google Scholar]
- 54.Gao Y, Xing F, Jin Y, Nie D, Wang Y. Foraging responses of clonal plants to multi-patch environmental heterogeneity: spatial preference and temporal reversibility. Plant Soil 2012; 359: 137–147. 10.1007/s11104-012-1148-0 [DOI] [Google Scholar]
- 55.Kawasaki A, Watson ER, Kertesz MA. Indirect effects of polycyclic aromatic hydrocarbon contamination on microbial communities in legume and grass rhizospheres. Plant Soil 2011; 358(1–2): 169–182. 10.1007/s11104-011-1089-z [DOI] [Google Scholar]
- 56.Kowalchuk GA, Buma DS, Boer WD, Klinkhamer PGL, Veen JAV. Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Antonie Leeuwenhoek 2002; 8: 509–520. 10.1023/A:1020565523615 . [DOI] [PubMed] [Google Scholar]
- 57.Nunan N, Daniell TJ, Singh BK, Papert A, McNicol JW, Prosser JI. Links between plant and rhizosplane bacterial communities in grassland soils, characterized using molecular techniques. Appl Environ Microbiol 2005; 71: 6784–6792. 10.1128/AEM.71.11.6784-6792.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Macdonald CA, Thomas N, Robinson L, Tate KR, Ross DJ, Dando J, et al. Physiological, biochemical and molecular responses of the soil microbial community after afforestation of pastures with Pinus radiata. Soil Biol Biochem 2009; 41: 1642–1651. 10.1016/j.soilbio.2009.05.003 [DOI] [Google Scholar]
- 59.Zhu H, Wang DL, Wang L, Bai YG, Fang J, Liu J. The effects of large herbivore grazing on meadow steppe plant and insect diversity. J Appl Ecol 2012; 49(5): 1075–1083. 10.1111/j.1365-2664.2012.02195.x [DOI] [Google Scholar]
- 60.Lin Y, Hong M, Han G, Zhao M, Bai Y, Chang SX. Grazing intensity affected spatial patterns of vegetation and soil fertility in a desert steppe. Agric Ecosyst Environ 2010; 138 (3–4): 282–292. 10.1016/j.agee.2010.05.013 [DOI] [Google Scholar]
- 61.Reszkowska A, Krümmelbein J, Peth S, Horn R, Zhao Y, Gan L. Influence of grazing on hydraulic and mechanical properties of semiarid steppe soils under different vegetation type in Inner Mongolia, China. Plant Soil 2010; 340 (1–2): 59–72. 10.1007/s11104-010-0405-3 [DOI] [Google Scholar]
- 62.Zhang X, Xu S, Li C, Zhao L, Feng H, Yue G, et al. The soil carbon/nitrogen ratio and moisture affect microbial community structures in alkaline permafrost-affected soils with different vegetation types on the Tibetan plateau. Res Microbiol 2014; 165(2): 128–139. 10.1016/j.resmic.2014.01.002 . [DOI] [PubMed] [Google Scholar]
- 63.Chu H, Fierer N, Lauber CL, Caporaso JG, Knight R, Grogan P. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol 2010; 12: 2998–3006. 10.1111/j.1462-2920.2010.02277.x [DOI] [PubMed] [Google Scholar]
- 64.Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 2009; 75: 5111–5120. 10.1128/AEM.00335-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rousk J, Bååth E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 2010; 4: 1340–1351. 10.1038/ismej.2010.58 . [DOI] [PubMed] [Google Scholar]
- 66.Cerritos R, Eguiarte LE, Avitia M, Siefert J, Travisano M, Rodriguez-Verdugo A, et al. Diversity of culturable thermo-resistant aquatic bacteria along an environmental gradient in Cuatro Cienegas, Coahuila, Mexico. Antonie van Leeuwenhoek 2011; 99 (2): 303–318. 10.1007/s10482-010-9490-9 . [DOI] [PubMed] [Google Scholar]
- 67.Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology 2007; 88: 1354–64. 10.1890/05-1839 . [DOI] [PubMed] [Google Scholar]
- 68.Tripathi BM, Kim M, Singh D, Lee-Cruz L, Lai-Hoe A, Ainuddin AN, et al. Tropical soil bacterial communities in Malaysia: pH dominates in the equatorial tropics too. Microb Ecol 2012; 64(2): 474–484. 10.1007/s00248-012-0028-8 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.