Abstract
Background
Crohn's disease (CD) is associated with defective sensing of pathogens in genetically susceptible individuals. Nucleotide-binding oligomerization domain containing 2 (NOD2) mutations in coding regions are strongly linked to CD pathogenesis. Our laboratory has reported that microRNAs (miRNAs) are differentially expressed in CD. However, miRNA regulation of NOD2 remains unknown. This study was designed to determine whether miRNAs regulate NOD2 expression as well as downstream nuclear factor kappaB activation and inflammatory responses in colonic epithelial HCT116 cells.
Methods
NOD2 and miRNA expression in stimulated HCT116 cells were assessed by quantitative reverse transcription–polymerase chain reaction. Regulation of NOD2 expression by miRNAs was determined by luciferase reporter construct assays and transfection of specific miRNA mimics. Regulation of NOD2 signaling and immune response by miRNAs was assessed by transfection of mimics followed by muramyl dipeptide stimulation.
Results
Muramyl dipeptide-induced increases in NOD2, interleukin-8, and CXCL3 expression were inversely associated with miRNA expression. Overexpression of miR-192, miR-495, miR-512, and miR-671 suppressed NOD2 expression, muramyl dipeptide-mediated NF-κB activation, and messenger RNA expressions of interleukin-8 and CXCL3 in HCT116 cells. A single-nucleotide polymorphism (rs3135500) located in the NOD2 3′-untranslated region significantly reduced miR-192 effects on NOD2 gene expression.
Conclusions
To our knowledge, this is the first report demonstrating that miRNAs regulate NOD2 and its signaling pathway. Four miRNAs downregulate NOD2 expression, suppress NF-κB activity, and inhibit interleukin-8 and CXCL3 messenger RNA expression. Treatment of CD with miRNAs may represent a potential anti-inflammatory therapeutic strategy in CD patients with and without NOD2 gene mutations.
Keywords: microRNAs, NOD2, MDP, NF-κB
Inflammatory bowel disease (IBD) is a chronic relapsing and remitting inflammatory condition of the gastrointestinal tract. It comprises 2 major disease subtypes, including Crohn's disease (CD) and ulcerative colitis (UC). IBD is characterized by a dysregulation of the inflammatory response to intestinal microbiota in genetically predisposed individuals. Although both forms of IBD share some common features, each disease has its unique clinical course, genetic associations, and gene expression patterns.1 Mucosal inflammation in UC is continuous and only involves the superficial layer of the colon, whereas inflammation in CD is discontinuous and transmural and affects any part of the gastrointestinal tract, with the terminal ileum most commonly involved.2 Since 2005, genome-wide association studies have identified a number of shared and distinct genetic susceptibility loci for CD and UC.3–5 In addition, genome-wide messenger RNA (mRNA) expression profiles have demonstrated that mRNA transcripts are differentially expressed in CD and UC6–8 Furthermore, microRNA (miRNA) expression studies have revealed common and unique miRNA signatures for CD and UC.9–12 Differential protein expression signatures were also identified in CD and UC patients.13
Although the intestinal microbiota are in constant interaction with the host epithelial and immune cells, the intestinal epithelium is the first line of defense against pathogens. Therefore, maintaining intestinal immune homeostasis is critical.14 The detection of invading pathogens by pattern recognition receptors is important in initiating innate immune response. Pattern recognition receptors comprised of cell surface receptors, such as Toll-like receptors (TLRs) and C-type lectin receptors, and intracellular receptors, such as RIG-I-like receptors and nucleotide-binding domain leucine-rich repeat containing receptors.15 Pattern recognition receptors are germline-encoded, evolutionarily conserved receptors that distinguish pathogen-associated molecular patterns. Pathogen-associated molecular patterns include lipopolysaccharide, a major component of outer membrane of gram-negative bacteria, peptidoglycan, a main component of gram-positive bacterial cell wall, flagellin, and microbial nucleic acids.
Nucleotide-binding oligomerization domain containing 2 (NOD2) is a member of the nucleotide-binding domain leucine-rich repeat containing receptors family. It was the first CD susceptibility gene identified.16,17 Individuals who have NOD2 mutations are more susceptible to CD.18 NOD2 is expressed in antigen presenting cells, such as monocytes, macrophages, and dendritic cells, Paneth cells, and numerous epithelial cell types of colonic and pulmonary origin.19 NOD2 confers responsiveness to the ligand muramyl dipeptide (MDP), a degraded bacterial cell wall component, peptidoglycan. Activation of NOD2-dependent pathways leads to the induction of NF-κB as well as the activation of JNK (c-Jun N-terminal kinase) and p38 MAPK. NOD2 activation has been shown to influence the expression and secretion of pro-inflammatory and anti-inflammatory cytokines, including interleukin (IL)-1β, TNFα, IL-6, IL-8, IL-10, and IL-12p40.20–23 Dysfunctional nuclear factor-kappaB (NF-κB) activation impairs production of regulatory cytokines and chemokines and induces death of intestinal epithelial cells and impairment of the intestinal barrier.24
The miRNAs are a group of small (∼22 nucleotide) non-coding RNAs that function as key regulators of mRNA stability, degradation, and translational efficiency.25 miRNAs have been implicated in many biological processes, including development, cell proliferation and differentiation, metabolism, apoptosis, and autophagy.25–28 miRNAs are also involved in the pathogenesis of a number of diseases, such as cancer, metabolic disorders, IBD, and other inflammatory and autoimmune diseases.9–11,26,27,29–32 Our laboratory has previously identified that the IBD-associated miRNA, miR-192, regulates the inflammatory cytokine-induced expression of macrophage inflammatory peptide-2α in intestinal epithelial cells.9 Interestingly, although the regulation of NOD2 by miRNAs has not yet been investigated, in silico analysis of NOD2 3′-untranslated region (3′UTR) reveals 11 putative miRNA binding sites that include 2 putative miR-192 binding sites (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/), implicating miR-192 and other miRNAs in the regulation of NOD2 expression and inflammatory responses.
In this study, we hypothesized that miRNA-mediated repression of NOD2 expression reduces MDP–NOD2 interactions and inhibits NOD2/NF-κB signaling cascade and immune responses in HCT116 cells. We identified that miR-122, miR-192, miR-495, and miR-671 inversely correlated with NOD2 expression in response to MDP stimulation. Ectopic expression of miR-192, miR-495, miR-512, and miR-671 in HCT116 cells suppressed NOD2 expression, NF-κB activation, and immune responses. In addition, mutating the seed regions of both miR-192 binding sites in the 3′UTR of NOD2 influenced NOD2 gene expression. Furthermore, an immune disease–associated single-nucleotide polymorphism (SNP) (rs3135500) in the second miR-192 binding site also resulted in altered NOD2 expression. Our findings suggest that miRNAs may play a critical role in the regulation of NOD2 expression and function.
Materials and Methods
Cell Culture
THP-1, HCT116, HT-29, Caco-2, SW620, and T84 cell lines were previously obtained and used for this study. THP-1 cells were cultured in RPMI-1640 medium. HCT116 cells were cultured in McCoy's 5A medium. HT-29 and Caco-2 cells were cultured in Dulbecco's modified Eagle's medium. SW620 cells were cultured in Leibovitz's L-15 medium. T84 cells were cultured in Dulbecco's modified Eagle's medium/Ham's F12 50/50 medium. All cell lines were supplemented with 10% fetal bovine serum and 1% p enicillin/ streptomycin at 378C in 5% CO2. For experiments in which HCT116 cells were stimulated with MDP, medium alone (control) or medium containing MDP (Sigma–Aldrich, St. Louis, MO) at 50 μg/mL was added and cells were harvested at respective time points for RNA and protein analyses.
Quantitative Reverse Transcription–Polymerase Chain Reaction for mRNA
Total RNA was isolated using TRIzol reagent (Life Technologies, Grand Island, NY) according to the manufacturer's protocol. RNA concentrations were determined using a Nano-Drop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). One microgram of total RNA was converted to complementary DNA (cDNA) using the qScript cDNA SuperMix (Quanta Biosciences, Gaithersburg, MD). Quantitative polymerase chain reaction (qPCR) analysis was performed using QuantiFast SYBR Green PCR kit (Qiagen, Valencia, CA). The qPCR amplifications were performed on the LightCycler 480 real-time PCR system (Roche, Indianapolis, IN). The expression of GAPDH was used as internal control. The primers for NOD2 were 5′-GATTGGCTGCCTTCCTTCTA-3′ (forward) and 5′-GAGCGTCTCTGCTCCATCAT-3′ (reverse). The primers for IL-8 were 5′-CTGCGCCAACACAGAAATTA-3′ (forward) and 5′-TGAATTCTCAGCCCTCTTCAA-3′ (reverse). The primers for CXCL3 were 5′-GAAGTCATAGCCACACTCA-3′ (forward) and 5′-GCTCCCCTTGTTCAGTATCTTT-3′ (reverse). The primers for GAPDH were 5′-CGACCACTTTGTCAAGCTCA-3′ (forward) and 5′-AGGGGAGATTCAGTGTGGTG-3′ (reverse).
Quantitative Reverse Transcription–Polymerase Chain Reaction for miRNA
One microgram of total RNA was converted to cDNA using the NCode VILO miRNA cDNA synthesis kit (Life Technologies) followed by qPCR using the NCode EXPRESS SYBR GreenER miRNA qRT-PCR kit (Life Technologies). The expression of miRNAs was calculated relative to RUNU6B, a ubiquitously expressed small nuclear RNA. The forward primer for miR-20a* was 5′-ACTGCATTATGAGCACTTAAAG-3′. The forward primer for miR-122 was 5′-GGAGTGTGACAATGGTGTTTGTAA-3′ . The forward primer for miR-124 was 5′-TAAGGCACGCGGT-GAATGCC-3′. The forward primer for miR-192 was 5′-CTGACC-TATGAATTGACAGCC-3′. The forward primer for miR-215 was 5′-ATGACCTATGAATTGACAGAC-3′. The forward primer for miR-342-5p was 5′-AGGGGTGCTATCTGTGATTGA-3′. The forward primer for miR-453 was 5′-AGGTTGTCCGTGGT-GAGTTCGCA-3′. The forward primer for miR-495 was 5′-AAACAAACATGGTGCACTTCTTT-3′. The forward primer for miR-512-5p was 5′-TCAGCCTTGAGGGCACTTTCA-3′. The forward primer for miR-671-5p was 5′-AAGCCCTG-GAGGGGCTGGAGGT-3′. The forward primer for RUNU6B was 5′-CGCAAGGATGACACGCAAATTCG-3′. A universal qPCR primer was used as the reverse primer.
Western Blot
Cells were lysed in cold RIPA buffer (Thermo Fisher Scientific, Rockford, IL) supplemented with 1% protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). Cell lysate was suspended in Laemmli sample buffer (Bio-Rad, Hercules, CA) containing 2-mercaptoethanol and boiled for 5 minutes. Protein concentration was determined with BCA protein assay (Thermo Fisher Scientific). After heat denaturation, total protein lysates were subjected to SDS–PAGE and transferred electrophoretically to PVDF membranes. Membranes were blocked in 5% nonfat milk in PBS with 0.1% Tween-20 (PBST) for 1 hour. The blots were incubated with goat anti-NOD2 (Novus Biologicals, Littleton, CO; 1:1000 dilution), rabbit anti-IκBα (Santa Cruz, Dallas, TX; 1:1000 dilution), rabbit anti-p65 subunit of NF-κB (Abcam, Cambridge, MA; 1:500 dilution), rabbit antitubulin (Cell Signaling, Danvers, MA; 1:1000 dilution), and mouse anti-GAPDH (Life Technologies; 1:1000 dilution) overnight at 4°C. After washing with PBST, blots were incubated with Alexa Fluor 680 (Life Technologies) and IRDye 800CW (LI-COR Biosciences, Lincoln, NE) conjugated secondary antibodies (1:10,000 dilution) for 1 hour. The band intensities were quantified using an Odyssey infrared imaging system (LI-COR Biosciences).
NOD2 3′UTR Constructs
The nucleotide sequence of the NOD2 3′UTR spanning 3181 to 4485 (RefSeq # NM_022162.1) was cloned into pmir-GLO dual-luciferase miRNA target expression vector (Promega, Madison, WI) by GenScript. The full-length (WT) pmirGLO-NOD2-3′UTR vector was used as a template to mutate the entire seed region of 1 or both putative miR-192 binding sites or to recreate the rs3135500 SNP. The modified seed sequences of each miR-192 binding site and SNP mutant are outlined in Figure 6A.
Figure 6.
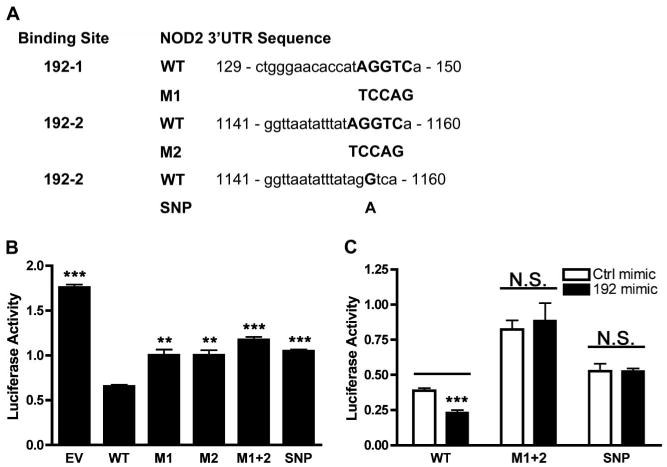
NOD2 miR-192 binding site mutation effects on reporter gene expression. A, Sequence alignments and binding site mutations in the constructs for miR-192 with the proximal putative miR-192 binding site (192-1) and the distal putative miR-192 binding site (192-2). M1 and M2 and SNP are mutant constructs for 192-1 and 192-2 binding sites, respectively. B, Luciferase reporter activity in HCT116 cells transfected with the WT, M1, M2, M1 + 2, and SNP reporter constructs. Results are relative to the WT construct. C, Luciferase reporter activity in HCT116 cells co-transfected with WT, M1 + 2, or SNP reporter constructs and the Ctrl or miR-192 mimics. Results are relative to the Ctrl mimic. Luciferase activity was normalized to Renilla luciferase activity. Results are mean ± standard error, n = 4. **P < 0.01; ***P < 0.001.
miRNA Mimic and Luciferase Construct Transfection
The miRNA mimic negative control (HMC0003) and mimics of miR-122 (HMI1002), miR-124 (HMI0086), miR-192 (HMI0311), miR-495 (HMI0610), miR-512-5p (HMI0640), and miR-671-5p (HMI0901) were obtained from Sigma. An miRNA mimic or a luciferase construct was transfected into HCT116 cells using Lipofectamine 2000 reagent (Life Technologies) according to the manufacturer's guidelines. At 24 hours posttransfection, cells were harvested for RNA and protein analyses or were harvested for measurement of luciferase activities.
Luciferase Reporter Assay
Cells were lysed in passive lysis buffer and then analyzed for the firefly and Renilla luciferase activities using the Dual-Luciferase Reporter Assay System (Promega) on the GloMax-Multi Detection System: Luminometer (Promega) according to the manufacturer's instructions. The firefly luciferase activity was normalized to the renilla luciferase activity.
Statistical Analysis
GraphPad Prism 4.0 (GraphPad Software, La Jolla, CA) was used for the statistical analysis. Statistical significance was determined by standard 2-tailed Student's t test (for comparison of 2 conditions) and 1-way analysis of variance with Dunnett's posttest (for multiple group comparisons). Results were reported as mean ± standard error of the mean. The P value < 0.05 was considered statistically significant.
Results
Endogenous NOD2 is Functional in Colonic Epithelial Cells
We first analyzed the endogenous NOD2 mRNA and protein expression in colonic epithelial cell lines by quantitative reverse transcription–polymerase chain reaction (qRT-PCR) and Western blot, respectively. Specifically, using the THP-1 cell line as a positive control,33 we assessed NOD2 mRNA expression in Caco-2, HCT116, HT-29, SW620, and T84 cells (Fig. 1A). Although NOD2 expression was detected in all cell lines, the HCT116 cells expressed markedly greater NOD2 mRNA as compared with the other cell lines tested. Similarly, we compared NOD2 protein expression in HCT116, HT-29, and Caco-2 cells, with THP-1 cell line as a positive control (Fig. 1B). NOD2 protein expression was markedly greater in HCT116 cells but only minimally expressed in HT-29 and Caco-2 cells.
Figure 1.
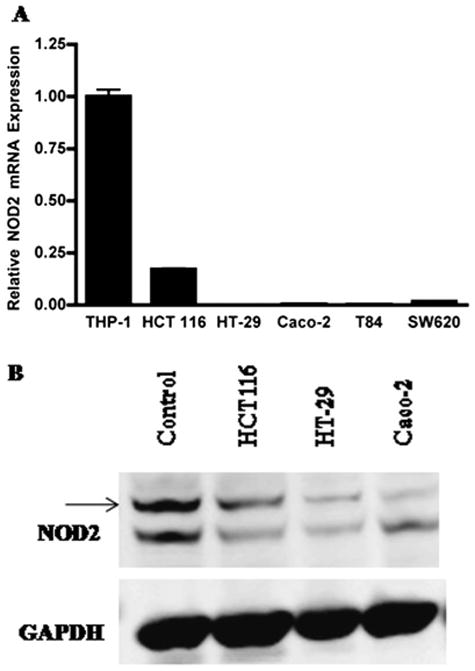
NOD2 mRNA and protein expression in colonic epithelial cell lines. A, NOD2 mRNA expression in various colonic epithelial cell lines (HCT116, HT-29, Caco-2, T84, and SW620), and positive control THP-1 cells was assessed by qRT-PCR. B, NOD2 protein expression in various colonic epithelial cell lines (HCT116, HT-29, and Caco-2) and positive control THP-1 cells was assessed by Western blot analysis.
Previous reports indicate that NOD2 is functional in HCT116 cell.34 First, we confirmed that NOD2 mRNA expression was inducible in HCT116 cells treated with MDP (Fig. 2A). Furthermore, our qRT-PCR analysis indicated that MDP (50 μg/mL) stimulation of NOD2 was time dependent with a maximal 2.4-fold increase in NOD2 mRNA expression observed 6 hours after MDP treatment. Second, we tested whether MDP can activate the NF-κB signaling pathway in HCT116 cells. We induced HCT116 cells with MDP and assessed phosphorylation of NF-κB and degradation of NF-κB inhibitor, alpha (IκBα) at different time points. Our result demonstrated that NF-κB is phosphorylated within the first 30 minutes of MDP induction, whereas IκBα degradation was seen after 60 minutes of MDP stimulation (Fig. 2B). To further study the function of NOD2 in HCT116 cells, we determined the regulatory effects of NOD2 on the inflammatory responses in HCT116 cells. We stimulated the cells with MDP and determined the expression of IL-8 and CXCL3 at different time points. The qRT-PCR analysis demonstrated that IL-8 and CXCL3 mRNA expressions were increased after MDP induction, with a maximal expression 2 hours after induction (Fig. 2C). These results confirmed that NOD2 is functional in HCT116 cells. Because we were able to confirm that NOD2 is highly expressed and functional in HCT116 cells, we used this cell line for our further examination of miRNA regulation of NOD2 in intestinal epithelial cells.
Figure 2.
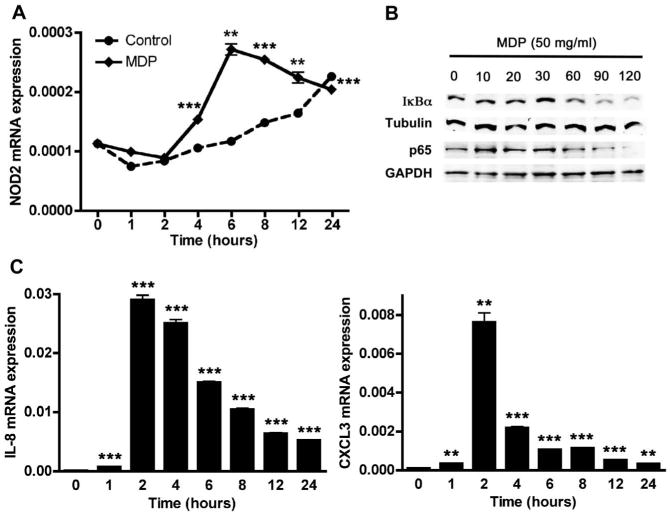
NOD2 is inducible and functional in HCT116 cells. A, Time-dependent NOD2 mRNA expression in MDP (50 μg/mL) treated and untreated (Control) HCT116 cells was assessed by qRT-PCR. B, NF-κB activation in HCT116 cells in response to MDP stimulation for indicated times was assessed via Western blot analysis on whole cell lysates using anti-p65 (for NF-κB phosphorylation), anti-IκBα (for IκBα degradation), anti-tubulin, and anti-GAPDH antibodies. C, Time-dependent IL-8 and CXCL3 mRNA expression in HCT116 cells in response to MDP (50 μg/mL) treatment was assessed by qRT-PCR. Results are mean ± standard error, n = 3. **P < 0.01; ***P < 0.001 compared with 0 hour time point.
Expressions of miRNAs that Bind to NOD2 3′UTR in HCT116 Cells
An in silico analysis identified 10 miRNAs with putative binding sites in the 3′UTR of NOD2 (Fig. 3A). We determined the endogenous expression of these 10 miRNAs by qRT-PCR (Fig. 3B). The 4 most highly expressed miRNAs in HCT116 cells were miRs-124, -192, -512, and -671 while the remaining miRNAs had low expression levels. Because we hypothesized that the MDP-induced increase in NOD2 expression may be influenced by a corresponding downregulation of regulatory miRNAs, we then determined which miRNAs exhibited altered expression in response to MDP (Fig. 3C). The results demonstrated that the expression of miRs-122, -192, -495, and -671 were significantly decreased in MDP-stimulated cells compared with unstimulated cells. Our data suggest an inverse correlation between NOD2 expression and the expression of these miRNAs in HCT116 cells, raising the possibility that NOD2 may be regulated by miRNAs and that the effect of MDP on NOD2 expression may result, in part, from a downregulation of these NOD2-associated miRNAs.
Figure 3.
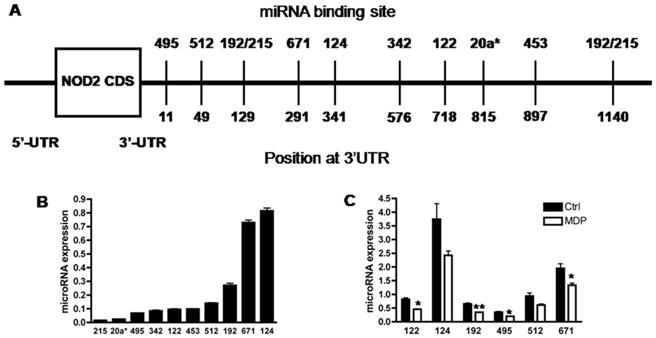
Expression of putative NOD2-associated miRNAs in basal and MDP-treated HCT116 cells. A, Schematic representation of NOD2 mRNA with 10 putative miRNA binding sites. B, Endogenous expression of putative NOD2-associated miRNAs in HCT116 cells was analyzed using qRT-PCR (normalized to U6B). C, Expression of 6 putative NOD2-associated miRNAs in MDP-stimulated (50 μg/mL) and unstimulated (Ctrl) HCT116 cells after 6 hours was assessed using qRT-PCR (normalized to U6B). Results are mean ± standard error, n = 3. *P < 0.05; **P < 0.01 compared with control (Ctrl).
Regulation of NOD2 by miRNAs in Colonic Epithelial Cells
To identify whether miRNAs may regulate NOD2, we first determined whether NOD2 3′UTR may be influenced by endogenous miRNAs. A luciferase reporter construct bearing the NOD2 3′UTR was transfected into unstimulated HCT116 cells. This resulted in a 47% reduction in luciferase activity (Fig. 4A), indicating that the NOD2 3′UTR and endogenous miRNAs can influence NOD2 gene expression (P < 0.01). Next, to determine which miRNAs may influence NOD2 gene expression, we co-transfected HCT116 cells with a luciferase reporter vector containing the NOD2 3′UTR and miRNA mimics (Fig. 4B). Our results demonstrated that miRs-122, -192, -512, and -671 individually suppressed luciferase activity, with miRs-192, -512, and -671 demonstrating the most significant effect. Meanwhile, miR-124 had no demonstrable effect on luciferase activity.
Figure 4.
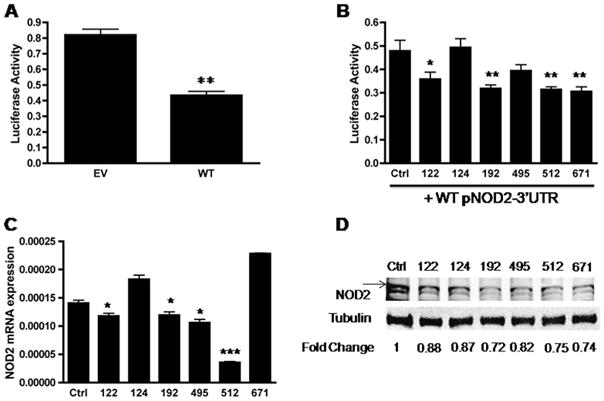
miRNAs modulate NOD2 expression in HCT116 cells. A, Luciferase activity of HCT116 cells transfected with the pmirGLO empty vector (EV) and wild-type pmirGLO-NOD2-3′UTR (WT) reporter constructs. Luciferase activity was normalized to Renilla luciferase activity. B, Luciferase activity of HCT116 cells co-transfected with 50 μg of the WT pNOD2-3′UTR and 50 nM control (Ctrl) mimic or miR-122, miR-124, miR-192, miR-495, miR-512, and miR-671 mimics. Luciferase activity was normalized to Renilla luciferase activity. C, The qRT-PCR quantifying NOD2 mRNA expression levels from HCT116 cells transfected with control (Ctrl) mimic or miR-122, miR-124, miR-192, miR-495, miR-512, and miR-671 mimics. D, Western blot analysis of NOD2 protein expression in HCT116 cells transfected with 50 nM control (Ctrl) mimic or miR-122, miR-124, miR-192, miR-495, miR 512, and miR-671 mimics. Densitometry levels were obtained using an Odyssey infrared imaging system and results are relative to the Ctrl mimic after normalization to the corresponding tubulin. For A, B, C, results are mean ± standard error, n = 3. *P < 0.05; **P < 0.01, ***P < 0.001 compared with EV (A) and Ctrl (B and C).
Based on the above results, we tested whether miRNA mimics could directly influence endogenous NOD2 mRNA and protein expression. Similar to our luciferase assay results, we observed that miRs-122, -192, -495, and -512 mimic transfection significantly decreased endogenous NOD2 mRNA expression (Fig. 4C), with the miR-512 mimic transfection resulting in the greatest reduction in NOD2 mRNA expression and miR-124 mimic transfection having no effect on NOD2 mRNA expression. Interestingly, miR-671 mimic transfection had no effect on endogenous NOD2 mRNA expression. When we examined miRNA mimic transfection effects on endogenous NOD2 protein expression by Western blot analysis, our densitometry analysis demonstrated that transfection of the miRs-192, -512, and -671, and to a lesser extent miR-495, mimics resulted in a marked decreased NOD2 protein expression (Fig. 4D). The effect of miRs-122 and -124 mimic transfection on NOD2 protein expression seemed less robust. Similarly, when we assessed the effect of miRNA mimic transfection on MDP-stimulated NOD2 expression after 6 hours, our results demonstrated that miRs-192, -495, -512, and -671 inhibited MDP-stimulated NOD2 protein expression as well (see Figure, Supplemental Digital Content 1, http://links.lww.com/IBD/A346). Overall, our results indicated that miRs-192, -512, and -671, consistently influence NOD2 expression. Results for miR-495 and miR-122 were less convincing. The miR-122 mimic transfection significantly reduced luciferase activity when co-transfected with the luciferase reporter vector containing the NOD2-3′UTR, influenced endogenous NOD2 mRNA expression and seemed to modestly reduce NOD2 protein expression. The miR-495 mimic transfection did not significantly reduce luciferase activity when co-transfected with the luciferase reporter vector containing the NOD2-3′UTR. However, the miR-495 mimic seemed to modestly reduce endogenous NOD2 mRNA and protein expression. Our results with the miR-124 mimic indicated that it, most likely, does not directly influence NOD2 mRNA or protein expression.
miRNAs Attenuate Innate Immune Responses via Suppression of NOD2 Signaling Pathway
Because miRNAs were demonstrated to influence NOD2 protein expression, we hypothesized that these miRNAs may influence downstream NOD2 function. To test this hypothesis, we examined the effect of NOD2-associated miRNAs on MDP-induced NF-κB activity, transfected HCT116 cells with miRNA mimics, and stimulated cells with MDP. We assessed phosphorylation of NF-κB at 30 minutes (Fig. 5A) and degradation of IκBα at 60 minutes (Fig 5B). Our results demonstrated that mimic transfection of all 4 miRNAs with the greatest effect on NOD2 protein expression, miRs-192, -495, -512 and -671, resulted in reduced NF-κB phosphorylation (Fig. 5A) and reduced IκBα degradation (Fig. 5B). Meanwhile, transfection of a miR-124 mimic, with the least effect on NOD2 expression, resulted in only modest alterations in IκBα degradation and no effect in NF-κB phosphorylation. Interestingly, transfection of the miR-122 mimic, while only having modest effects on NOD2 protein expression, had significant effects on NF-κB phosphorylation and IκBα degradation.
Figure 5.
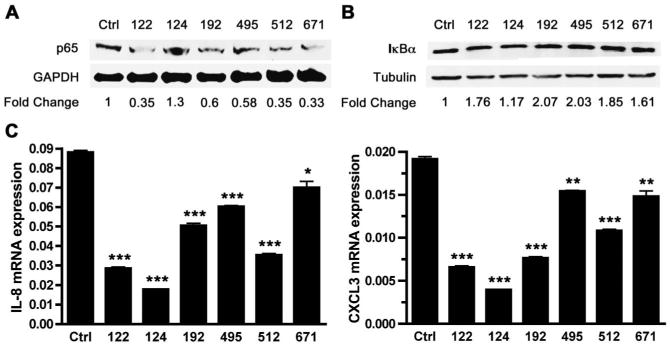
Decreased NF-κB phosphorylation and degradation of IkBa in HCT116 cells transfected with miRNA mimics. A, Western blot analysis for NF-κB phosphorylation in HCT116 cells transfected with miRNA mimics for 24 hours followed by MDP stimulation (50 μg/mL) for 30 minutes using anti-phospho-NF-κB (p65) and anti-GAPDH antibodies. Densitometry levels were obtained using an Odyssey infrared imaging system and results are relative to the Ctrl mimic after normalization to individual GAPDH levels. B, Western blot analysis for IκBα degradation in HCT116 cells transfected with miRNA mimics for 24 hours followed by MDP stimulation (50 μg/mL) for 60 minutes using anti-IκBα and anti-tubulin antibodies. Densitometry levels were obtained using an Odyssey infrared imaging system and results are relative the Ctrl mimic after normalization to individual tubulin levels. C, The qRT-PCR analysis of IL-8 and CXCL3 mRNA expression in HCT116 cells transfected with 50 nM Ctrl, miR-122, miR-124, miR-192, miR-495, miR-512, and miR-671 mimics for 24 hours followed by 2 hours of MDP stimulation (50 μg/mL). Results are mean ± standard error, n = 3. *P < 0.05; **P < 0.01; ***P < 0.001 compared with Ctrl mimic.
Next, we examined whether the NOD2-associated miRNA mimic transfection influenced downstream IL-8 and CXCL3 mRNA expression (Fig. 5C). Our results indicated that the transfection of mimics of miRs-192, -495, -512, and -671 all, to varying degrees, significantly reduced both CXCL3 and IL-8 mRNA expression. Interestingly, we found that both miR-122 and miR-124 mimic transfection also reduced CXCL3 and IL-8 mRNA expression.
Mutating the miR-192 Binding Sites on NOD2 Increase Luciferase Activities
Of the miRNAs that were found to influence NOD2 expression and function, miR-192 was of particular interest for 3 reasons: first, the differential expression of miR-192 has been noted in IBD9; second, there are 2 predicted miR-192 binding sites within the NOD2 3′UTR; third, an SNP (rs3135500) in the NOD2 3′UTR was found to be associated with ileal CD35 and other immune-related diseases36 and corresponds to the seed region of 1 of the 2 putative miR-192 binding sites. We next examined whether 1 or both putative miR-192 binding sites influenced NOD2 expression and whether the SNP within the putative miR-192 binding site could influence NOD2 gene expression. The sequence alignments of the 2 putative miR-192 binding sites and the SNP are shown in Figure 6A. To determine which miR-192 binding site may be influenced by endogenous miR-192, 2 constructs containing mutations in each of the putative miR-192 binding sites, a construct containing mutations in both binding sites and an additional construct containing the SNP (rs3135500) in the second putative miR-192 binding site were created (Fig 6A). Transfecting mutant constructs into HCT116 cells resulted in a restoration of luciferase activity (Fig. 6B). The results indicated that the first miR-192 binding site (nucleotides 129–150) (P < 0.01) and the second miR-192 binding site (nucleotides 1141-1160) (P < 0.01) were equally influential, resulting in a 32% restoration in luciferase activity. Of note, the construct containing mutations of both miR-192 binding sites (M1 + 2) exerted the greatest influence on the NOD2 expression in HCT116 cells (47% restoration in luciferase activity), indicating that both miR-192 bindings sites are functional and the cumulative effects of both miR-192 binding sites are additive.
We next examined whether an alteration of a single nucleotide in the second miR-192 binding site, corresponding to the rs3135500 SNP, could regulate NOD2 expression. Transfection of the SNP construct into HCT116 cells significantly restored luciferase activity (P < 0.05) (Fig. 6B) and this restoration of luciferase activity was as robust as the mutation of the entire seed region of the second miR-192 binding site. To further examine the influence of the SNP on miR-192 regulation of NOD2 expression, we co-transfected a miR-192 mimic with wild-type, M1 + 2, or SNP constructs into HCT116 cells. The result demonstrated that miR-192 mimic transfection reduced luciferase activity in cells transfected with wild-type construct. However, the ability of miR-192 mimic transfection to reduce luciferase activity was abolished in the HCT116 cells transfected with the M1 + 2 and SNP constructs (Fig. 6C), providing additional evidence that miR-192 regulates NOD2 expression and the rs3135500 SNP influences the ability of miR-192 to regulate NOD2 expression.
Discussion
Multiple regulatory mechanisms have been implicated in the regulation of NOD2 gene expression. These include regulatory elements in the promoter region of the NOD2 gene, promoter methylation, and ubiquitination pathways.37–39 Specifically, NF-κB regulatory elements have been shown to significantly influence intestinal epithelial NOD2 gene expression.40,41 In addition, it has been demonstrated that NOD2 gene expression was reactivated by the demethylating agent 5-aza-2′ deoxycytidine in the phytohemaglutinin-stimulated peripheral blood mononuclear cells.39 Recently, it has been reported that NOD2 can be degraded by TRIM27 through ubiquitination and proteasomal degradation.37 Finally, environmental stimuli, such as cigarette smoking, have been shown to delay TNFα-induced NOD2 mRNA expression.43
In this report, we present the first evidence that miRNAs also play a significant role in regulating NOD2 gene expression. Specifically, we demonstrated that miRs-192, -495, -512, and -671 influenced NOD2 gene expression as well as its downstream NF-κB activity and immune responses in intestinal epithelial cells.
Modulation of miRNA expression in response to various pro-inflammatory components in primary human monocytes was previously investigated.44 miR-129-5p was upregulated in response to MDP activation.44 Thirteen biological processes targeted by miRNAs also respond to MDP stimulation.44 Modulation of miRNA expression by TLR stimulation has been shown in several studies. Specifically, miR-155, miR-146, miR-132, miR-147, miR-9, miR-21, and miR-34645–48 are increased in response to TLR2 and TLR4 activation while miR-19a/b are decreased in response to TLR2 activation.45 However, miRNAs can regulate TLR expression and its signaling pathway. Specifically, let-7i suppresses TLR4 expression in human cholangiocytes.49
In our study, we identified miRs-192, -495, -512, and -671 as having direct influences on NOD2 gene expression. Of these miRNAs, only miR-192 has been associated with TLR expression changes. It has been shown that TLR-1, TLR-3, and TLR4a expressions were altered after overexpression of miR-192 in lipopolysaccharide-induced zebrafish liver (ZFL) cell line.50 However, each of these miRNAs has been shown to influence other genes. Specifically, upregulation of miR-495 has been found to be associated with reduced methionine adenosyltransferase 1A (MAT1A) expression in hepatocellular carcinoma.51 Increased miR-512 expression has been found to be associated with cervical neoplasm,52 whereas miR-671 has been found to be associated with nonalcoholic fatty liver disease.53
Of particular interest is the finding that miR-192 regulates NOD2 expression and function. We previously demonstrated that miR-192 is dysregulated in intestinal epithelial cells of patients with IBD and that miR-192 influences intestinal epithelial cell innate immunity via the direct regulation of macrophage inflammatory peptide 2α expression.9 Here, we demonstrated that miR-192 significantly altered NOD2 mRNA and protein expression. In addition, a miR-192 mimic significantly reduced NF-κB phosphorylation and IκBα degradation and downstream IL-8 and CXCL3 expression. These results suggest that miR-192 is a key miRNA in the pathogenesis of IBD and in the regulation of intestinal epithelial cell innate immune responses.
Multiple genetic analyses have implicated NOD2 in the pathogenesis of a number of chronic inflammatory diseases and autoimmune diseases, including CD,16,17 psoriasis,42,54 psoriatic arthritis,54,55 asthma,36 sarcoidosis,56 atopic dermatitis,57 allergic rhinitis,57 and Blau syndrome.58 A number of studies have shown that SNPs in miRNA binding sites are associated with diseases32,59,60 and may significantly alter miRNA binding to associated gene transcripts.61,62 In particular, 8 SNPs in the NOD2 gene were recently analyzed for associations with atopic phenotypes in a large German adult population (n = 1875).36 Of particular interest in this report is that an SNP with an A allele at rs3135500 (G4384A) is located in the NOD2 putative miRNA binding site for miR-192. The G4384A SNP was identified to be associated with an increased risk of asthma (odds ratio = 1.374, P = 0.023) and predicted to significantly alter miRNA binding to the NOD2 transcript.36 Here, we experimentally confirmed that the rs3135500 variant is a loss-of-function SNP by luciferase assay in HCT116 cells. Specifically, transfecting the NOD2 3′UTR vector possessing the rs3135500 SNP into HCT116 cells resulted in a higher baseline luciferase activity when compared with the WT vector and conferred the loss of response to miR-192 mimic transfection. It may be hypothesized that the rs3135500 variant may physiologically contribute to altered NOD2 expression in disease. Further studies are necessary to determine whether epithelial cells or other immune cells harboring the rs3135500 SNP demonstrate altered NOD2 expression and immune responses, contributing to the chronicity of inflammation as seen in CD and atopic disease.
Of note, although our data demonstrated that the transfection of mimics of miRs-192, -495, -512, and -671 influenced NOD2 expression, the proportional effects of these mimics on downstream NF-κB activity and immune responses in intestinal epithelial cells did not correlate solely to NOD2 expression levels. There was much greater correlation to miRNA mimic effects on reducing NF-κB phosphorylation and IκBα degradation than downstream IL-8 and CXCL3 expression. It has been suggested that a single miRNA can target hundreds of mRNAs while a mRNA can be regulated by multiple miRNA.63 It has also been demonstrated that several miRNAs react to 1 stimulus.44 It is quite possible that each miRNA has multiple targets in the cytokine signaling pathways examined. This is notable in the case of IL-8, which has putative miR-124 binding site in the 3′UTR. The down-regulation of IL-8 in miR-124 overexpressed cells may be due to the direct effect of miR-124 on 3′UTR of IL-8 rather than indirect effect from NOD2 signaling. Furthermore, the potential role of miRNAs with putative binding sites in the NOD2 3′UTR regulating other immune functions is indicated by the results obtained with the miR-124 mimic transfection, in which we observed very little direct effect on NOD2 expression but significant regulation of MDP-stimulated IL-8 and CXCL3 gene expression. Further studies are necessary to dissect all the gene targets of these NOD2-regulatory miRNAs.
In summary, we have observed that MDP enhances NOD2 expression and suppresses the expression of miRNAs with putative NOD2 3′UTR binding sites in HCT116 cells. We also demonstrated that miR-192, miR-495, miR-512, and miR-671 are capable of regulating NOD2 expression in different degrees by binding to the putative complementary binding sites within the 3′UTR of NOD2 mRNA. This raises the possibility that overexpression of more than 1 miRNA may increase the efficiency of the regulation of NOD2 expression. In addition, we have shown that these miRNAs inhibit NOD2 signaling pathway and expression of IL-8 and CXCL3 in response to MDP stimulation. Taken together, we have identified a novel mechanism in which MDP-mediated down-regulation of miRNAs leads to overexpression of NOD2, excessive NF-κB activation, and amplification of innate immune responses and susceptibility to chronic inflammatory and autoimmune diseases. HCT116 findings observed in our study may not be true in other cell lines or primary cells.
Supplementary Material
Supplemental Figure. MDP-induced NOD2 protein expression is inhibited by miRNAs. Western blot analysis of NOD2 protein expression in HCT116 cells transfected with 50 nM of control (Ctrl) mimic or miR-122, miR-124, miR-192, miR-495, miR-512 and miR-671 mimics for 24 hours and then stimulated with MDP for 6 hours. Densitometry levels were obtained using an Odyssey infrared imaging system and results are relative to the Ctrl mimic after normalization to individual GAPDH.
Acknowledgments
Supported by National Institute of Health (NIH; K08 DK078046 to J. H. Kwon; T32 DK07074 to Z. Zhai) and the NIH DDRCC Core Grant (P30 DK42086).
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.ibdjournal.org).
The authors have no conflicts of interest to disclose.
References
- 1.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–429. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 2.Baumgart DC, Sandborn WJ. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet. 2007;369:1641–1657. doi: 10.1016/S0140-6736(07)60751-X. [DOI] [PubMed] [Google Scholar]
- 3.Budarf ML, Labbe C, David G, et al. GWA studies: rewriting the story of IBD. Trends Genet. 2009;25:137–146. doi: 10.1016/j.tig.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGovern DP, Gardet A, Torkvist L, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 2010;42:332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costello CM, Mah N, Hasler R, et al. Dissection of the inflammatory bowel disease transcriptome using genome-wide cDNA microarrays. PLoS Med. 2005;2:e199. doi: 10.1371/journal.pmed.0020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawrance IC, Fiocchi C, Chakravarti S. Ulcerative colitis and Crohn's disease: distinctive gene expression profiles and novel susceptibility candidate genes. Hum Mol Genet. 2001;10:445–456. doi: 10.1093/hmg/10.5.445. [DOI] [PubMed] [Google Scholar]
- 8.Wu F, Dassopoulos T, Cope L, et al. Genome-wide gene expression differences in Crohn's disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm Bowel Dis. 2007;13:807–821. doi: 10.1002/ibd.20110. [DOI] [PubMed] [Google Scholar]
- 9.Wu F, Zikusoka M, Trindade A, et al. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624–1635e24. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 10.Wu F, Guo NJ, Tian H, et al. Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn's disease. Inflamm Bowel Dis. 2011;17:241–250. doi: 10.1002/ibd.21450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu F, Zhang S, Dassopoulos T, et al. Identification of microRNAs associated with ileal and colonic Crohn's disease. Inflamm Bowel Dis. 2010;16:1729–1738. doi: 10.1002/ibd.21267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fasseu M, Treton X, Guichard C, et al. Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS One. 2010;5:e13160. doi: 10.1371/journal.pone.0013160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shkoda A, Werner T, Daniel H, et al. Differential protein expression profile in the intestinal epithelium from patients with inflammatory bowel disease. J Proteome Res. 2007;6:1114–1125. doi: 10.1021/pr060433m. [DOI] [PubMed] [Google Scholar]
- 14.Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol. 2004;12:129–134. doi: 10.1016/j.tim.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 16.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 17.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 18.Maeda S, Hsu LC, Liu H, et al. Nod2 mutation in Crohn's disease potentiates NF-kappaB activity and IL-1beta processing. Science. 2005;307:734–738. doi: 10.1126/science.1103685. [DOI] [PubMed] [Google Scholar]
- 19.Franchi L, Warner N, Viani K, et al. Function of Nod-like receptors in microbial recognition and host defense. Immunol Rev. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Moran T, Swanson E, et al. Regulation of IL-8 and IL-1beta expression in Crohn's disease associated NOD2/CARD15 mutations. Hum Mol Genet. 2004;13:1715–1725. doi: 10.1093/hmg/ddh182. [DOI] [PubMed] [Google Scholar]
- 21.Xu ZJ, Zhao GQ, Wang Q, et al. Nucleotide oligomerization domain 2 contributes to the innate immune response in THCE cells stimulated by Aspergillus fumigatus conidia. Int J Ophthalmol. 2012;5:409–414. doi: 10.3980/j.issn.2222-3959.2012.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noguchi E, Homma Y, Kang X, et al. A Crohn's disease-associated NOD2 mutation suppresses transcription of human IL10 by inhibiting activity of the nuclear ribonucleoprotein hnRNP-A1. Nat Immunol. 2009;10:471–479. doi: 10.1038/ni.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beynon V, Cotofana S, Brand S, et al. NOD2/CARD15 genotype influences MDP-induced cytokine release and basal IL-12p40 levels in primary isolated peripheral blood monocytes. Inflamm Bowel Dis. 2008;14:1033–1040. doi: 10.1002/ibd.20441. [DOI] [PubMed] [Google Scholar]
- 24.Gutierrez O, Pipaon C, Inohara N, et al. Induction of Nod2 in myelomo-nocytic and intestinal epithelial cells via nuclear factor-kappa B activation. J Biol Chem. 2002;277:41701–41705. doi: 10.1074/jbc.M206473200. [DOI] [PubMed] [Google Scholar]
- 25.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 26.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 27.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frankel LB, Lund AH. MicroRNA regulation of autophagy. Carcinogenesis. 2012;33:2018–2025. doi: 10.1093/carcin/bgs266. [DOI] [PubMed] [Google Scholar]
- 29.Dalal SR, Kwon JH. The role of microRNA in inflammatory bowel disease. Gastroenterol Hepatol (N Y) 2010;6:714–722. [PMC free article] [PubMed] [Google Scholar]
- 30.Sonkoly E, Wei T, Janson PC, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanczyk J, Pedrioli DM, Brentano F, et al. Altered expression of micro-RNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 32.Tan Z, Randall G, Fan J, et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Till A, Rosenstiel P, Brautigam K, et al. A role for membrane-bound CD147 in NOD2-mediated recognition of bacterial cytoinvasion. J Cell Sci. 2008;121:487–495. doi: 10.1242/jcs.016980. [DOI] [PubMed] [Google Scholar]
- 34.Zhao L, Lee JY, Hwang DH. The phosphatidylinositol 3-kinase/Akt pathway negatively regulates Nod2-mediated NF-kappaB pathway. Biochem Pharmacol. 2008;75:1515–1525. doi: 10.1016/j.bcp.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 35.Lee YH, Song GG. Pathway analysis of a genome-wide association study of ileal Crohn's disease. DNA Cell Biol. 2012;31:1549–1554. doi: 10.1089/dna.2012.1605. [DOI] [PubMed] [Google Scholar]
- 36.Weidinger S, Klopp N, Rummler L, et al. Association of CARD15 polymorphisms with atopy-related traits in a population-based cohort of Caucasian adults. Clin Exp Allergy. 2005;35:866–872. doi: 10.1111/j.1365-2222.2005.02269.x. [DOI] [PubMed] [Google Scholar]
- 37.Zurek B, Schoultz I, Neerincx A, et al. TRIM27 negatively regulates NOD2 by ubiquitination and proteasomal degradation. PLoS One. 2012;7:e41255. doi: 10.1371/journal.pone.0041255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu C, Sun L, Hu Y, et al. Functional characterization of the NF-kappaB binding site in the human NOD2 promoter. Cell Mol Immunol. 2010;7:288–295. doi: 10.1038/cmi.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kerkel K, Schupf N, Hatta K, et al. Altered DNA methylation in leukocytes with trisomy 21. PLoS Genet. 2010;6:e1001212. doi: 10.1371/journal.pgen.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stronati L, Negroni A, Merola P, et al. Mucosal NOD2 expression and NF-kappaB activation in pediatric Crohn's disease. Inflamm Bowel Dis. 2008;14:295–302. doi: 10.1002/ibd.20332. [DOI] [PubMed] [Google Scholar]
- 41.Rosenstiel P, Fantini M, Brautigam K, et al. TNF-alpha and IFN-gamma regulate the expression of the NOD2 (CARD15) gene in human intestinal epithelial cells. Gastroenterology. 2003;124:1001–1009. doi: 10.1053/gast.2003.50157. [DOI] [PubMed] [Google Scholar]
- 42.Nair RP, Henseler T, Jenisch S, et al. Evidence for two psoriasis susceptibility loci (HLA and 17q) and two novel candidate regions (16q and 20p) by genome-wide scan. Hum Mol Genet. 1997;6:1349–1356. doi: 10.1093/hmg/6.8.1349. [DOI] [PubMed] [Google Scholar]
- 43.Aldhous MC, Soo K, Stark LA, et al. Cigarette smoke extract (CSE) delays NOD2 expression and affects NOD2/RIPK2 interactions in intestinal epithelial cells. PLoS One. 2011;6:e24715. doi: 10.1371/journal.pone.0024715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hasler R, Jacobs G, Till A, et al. Microbial pattern recognition causes distinct functional micro-RNA signatures in primary human monocytes. PLoS One. 2012;7:e31151. doi: 10.1371/journal.pone.0031151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Philippe L, Alsaleh G, Suffert G, et al. TLR2 expression is regulated by microRNA miR-19 in rheumatoid fibroblast-like synoviocytes. J Immunol. 2012;188:454–461. doi: 10.4049/jimmunol.1102348. [DOI] [PubMed] [Google Scholar]
- 46.O'Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of Tolllike receptor signalling. Nat Rev Immunol. 2011;11:163–175. doi: 10.1038/nri2957. [DOI] [PubMed] [Google Scholar]
- 47.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 48.Taganov KD, Boldin MP, Chang KJ, et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen XM, Splinter PL, O'Hara SP, et al. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. 2007;282:28929–28938. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu TH, Pan CY, Lin MC, et al. In vivo screening of zebrafish microRNA responses to bacterial infection and their possible roles in regulating immune response genes after lipopolysaccharide stimulation. Fish Physiol Biochem. 2012;38:1299–1310. doi: 10.1007/s10695-012-9617-1. [DOI] [PubMed] [Google Scholar]
- 51.Yang H, Cho ME, Li TW, et al. MicroRNAs regulate methionine adeno-syltransferase 1A expression in hepatocellular carcinoma. J Clin Invest. 2013;123:285–298. doi: 10.1172/JCI63861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheung TH, Man KN, Yu MY, et al. Dysregulated microRNAs in the pathogenesis and progression of cervical neoplasm. Cell Cycle. 2012;11:2876–2884. doi: 10.4161/cc.21278. [DOI] [PubMed] [Google Scholar]
- 53.Estep M, Armistead D, Hossain N, et al. Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2010;32:487–497. doi: 10.1111/j.1365-2036.2010.04366.x. [DOI] [PubMed] [Google Scholar]
- 54.Li WQ, Han JL, Chan AT, et al. Psoriasis, psoriatic arthritis and increased risk of incident Crohn's disease in US women. Ann Rheum Dis. 2013;72:1200–1205. doi: 10.1136/annrheumdis-2012-202143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karason A, Gudjonsson JE, Upmanyu R, et al. A susceptibility gene for psoriatic arthritis maps to chromosome 16q: evidence for imprinting. Am J Hum Genet. 2003;72:125–131. doi: 10.1086/345646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanazawa N, Okafuji I, Kambe N, et al. Early-onset sarcoidosis and CARD15 mutations with constitutive nuclear factor-kappaB activation: common genetic etiology with Blau syndrome. Blood. 2005;105:1195–1197. doi: 10.1182/blood-2004-07-2972. [DOI] [PubMed] [Google Scholar]
- 57.Kabesch M, Peters W, Carr D, et al. Association between polymorphisms in caspase recruitment domain containing protein 15 and allergy in two German populations. J Allergy Clin Immunol. 2003;111:813–817. doi: 10.1067/mai.2003.1336. [DOI] [PubMed] [Google Scholar]
- 58.Miceli-Richard C, Lesage S, Rybojad M, et al. CARD15 mutations in Blau syndrome. Nat Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 59.Zwiers A, Kraal L, van de Pouw Kraan TC, et al. Cutting edge: a variant of the IL-23R gene associated with inflammatory bowel disease induces loss of microRNA regulation and enhanced protein production. J Immunol. 2012;188:1573–1577. doi: 10.4049/jimmunol.1101494. [DOI] [PubMed] [Google Scholar]
- 60.Landi D, Gemignani F, Naccarati A, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis. 2008;29:579–584. doi: 10.1093/carcin/bgm304. [DOI] [PubMed] [Google Scholar]
- 61.Wang K, Li J, Guo H, et al. MiR-196a binding-site SNP regulates RAP1A expression contributing to esophageal squamous cell carcinoma risk and metastasis. Carcinogenesis. 2012;33:2147–2154. doi: 10.1093/carcin/bgs259. [DOI] [PubMed] [Google Scholar]
- 62.Luo J, Cai Q, Wang W, et al. A microRNA-7 binding site polymorphism in HOXB5 leads to differential gene expression in bladder cancer. PLoS One. 2012;7:e40127. doi: 10.1371/journal.pone.0040127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure. MDP-induced NOD2 protein expression is inhibited by miRNAs. Western blot analysis of NOD2 protein expression in HCT116 cells transfected with 50 nM of control (Ctrl) mimic or miR-122, miR-124, miR-192, miR-495, miR-512 and miR-671 mimics for 24 hours and then stimulated with MDP for 6 hours. Densitometry levels were obtained using an Odyssey infrared imaging system and results are relative to the Ctrl mimic after normalization to individual GAPDH.


