Abstract
MyoD and myogenin are myogenic transcription factors preferentially expressed in adult fast and slow muscles, respectively. Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disorder in which motor neuron loss is accompanied by muscle denervation and paralysis. Studies suggest that muscle phenotype may influence ALS disease progression. Here we demonstrate that myogenin gene transfer into muscle supports spinal cord motor neuron survival and muscle endplate innervation in the G93A SOD1 fALS mice. On the other hand, MyoD gene transfer decreases survival and enhances motor neuron degeneration and muscle denervation. Although an increase in motor neuron count is associated with increased succinic dehydrogenase staining in the muscle, muscle overexpression of PGC-1α does not improve survival or motor function. Our study suggests that postnatal muscle modification influences disease progression and demonstrates that the muscle expression of myogenic and metabolic regulators differentially impact neuropathology associated with disease progression in the G93A SOD1 fALS mouse model.
Introduction
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disorder characterized by the loss of upper and lower motor neurons. Motor neuron losses are accompanied by muscle weakness and paralysis. A number of clinical studies have reported increased vulnerability of fast-fatiguable motor units in ALS patients 1,2. Studies of muscle fiber phenotype 3 and motor units 4 in transgenic mice overexpressing mutant G93A human SOD1 (G93A mice) indicate that fast muscles are preferentially affected. These findings suggest that muscle phenotype may have an influence on ALS neuropathology. However the role of muscle in ALS pathogenesis remains ambiguous. Although muscle specific expression of mutant SOD1 results in motor neuron loss and muscle denervation 5,6, Cre-mediated partial knockdown of mutant SOD1 in the muscle did not have an impact on the disease progression 7. Still, early changes in muscle biochemistry and the effectiveness of muscle-specific expression of either GDNF or IGF-1 in mediating motor neuron rescue and delay in the disease progression in these mice suggest a potentially important and influential role of the muscle on the disease progression 8.
MyoD and myogenin are muscle regulatory factors involved in muscle development and differentiation 9,10. In adult muscle, MyoD is mainly expressed in adult fast muscle fibers 11–13 and regulates fast muscle development 12. On the other hand, myogenin is predominantly expressed in slow muscle fibers 11,13 and its expression increases oxidative metabolism in muscle 14,15. Another factor that has been shown to be a modulator of muscle phenotype is peroxisome proliferator-activated receptor gamma coactivator-1α (PGC-1α). PGC-1α is a potent metabolic regulator of muscle and its expression enhances slow oxidative muscle phenotype 16.
In this study we examined whether postnatal modification of muscle phenotype can modify disease progression in the G93A SOD1 fALS mouse model; adult muscle is highly plastic and can be phenotypically remodeled by intrinsic and extrinsic cues 17,18. We show that postnatal modulation of muscle phenotype as a consequence of exogenous expression of MyoD or myogenin in hindlimb muscles of adult G93A mice modulates motor neuron degeneration, muscle denervation, and survival. On the other hand, muscle overexpression of PGC-1α does not improve survival or motor function in these mice. Our results demonstrate that disease progression in a fALS mouse model can be either improved or worsened through postnatal muscle modification, and demonstrate that myogenic and metabolic factors in the muscle differentially influence the clinical outcome in the G93A mice.
Results
MyoD Gene Transfer Accelerates Disease Progression
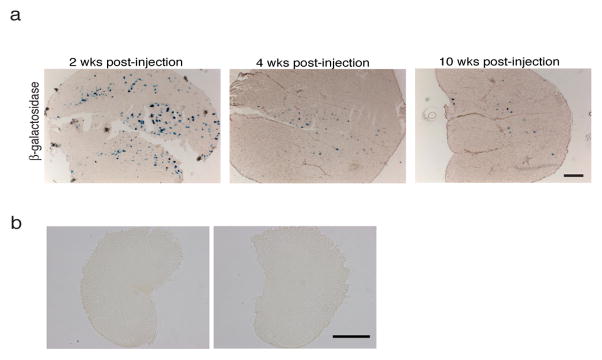
We exogenously expressed MyoD in the hindlimb muscles of adult male G93A mice using adenovirus expressing human MyoD (ad-MyoD) and investigated the effect on motor behavior. Gene transduction efficiency following virus-mediated gene transfer into muscle was determined using adenovirus expressing LacZ. Two-weeks following injection, approximately 16% of the muscle fibers were X-gal positive in the gastrocnemius muscle (Fig. 1a). The number of muscle fibers positive for LacZ stain decreased over time, demonstrating the transient nature of resulting LacZ gene expression (Fig. 1a). Examination of the lumbar portion of spinal cord did not show any X-gal staining suggesting that the virus injected into the hindlimb muscles is not retrogradely taken up by motor neurons in our study (Fig. 1b).
Figure 1. Efficiency of adenovirus-mediated gene transfer into adult muscle was determined in the gastrocnemius muscle using β-galactosidase construct.
(a) A single injection resulted in widely distributed gene expression 2-weeks post injection. Stereological counting showed that approximately 16% of the muscle fibers were visibly stained for b-galactosidase (blue). The gene expression level declined over time showing the transient nature of the b-galactosidase gene expression. (b) Examination of the lumbar portion of the spinal cord at 2-weeks post injection did not show any X-gal staining, demonstrating that virus is not retrogradely transported into motor neurons following virus injection. Scale bar represent 500μm.
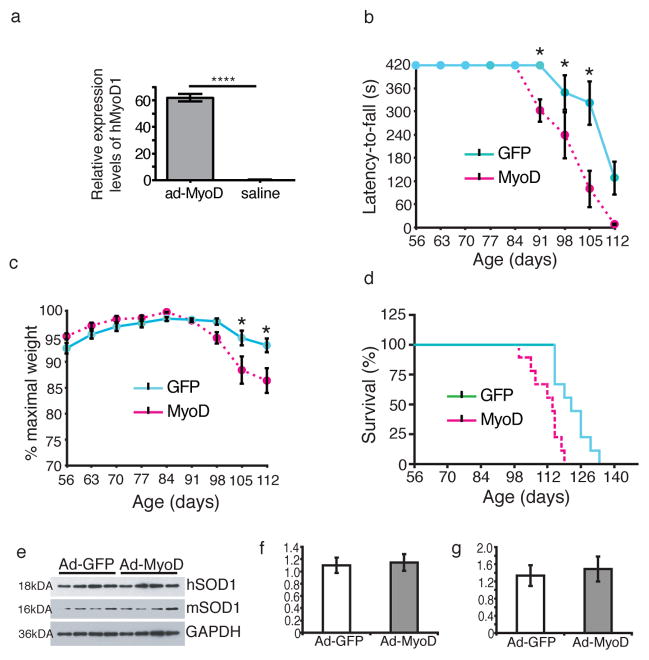
In our first set of experiments, mice were injected with either ad-MyoD or ad-GFP. Expression of human MyoD in the muscle following adenoviral construct injection was confirmed using qPCR (Fig. 2a; t-test, p<0.00001, n=4 samples per group). The behavioral effect of MyoD overexpression in hindlimb muscles of G93A mice was determined through a weekly assessment of Rotarod performance and weight measures starting at 56 days of age. A comparison of the ad-MyoD injection group (n=9) with the ad-GFP injection group (n=9) showed a statistically significant difference in the temporal profile of latency-to-fall on Rotarod (Fig. 2b, two-way ANOVA, F(1,140) = 23.33; p < 0.0001; n=9 mice per group). Bonferroni post-test showed statistically significant difference in the Rotarod performance between the two groups at 91 days (p<0.05), 98 days (p<0.05), and 105 days (p<0.001) of age. Weight loss paralleled the temporal pattern of Rotarod performance. Ad-MyoD injected mice displayed faster weight loss than ad-GFP injected mice (Fig. 2c, two-way ANOVA, F(1,140) = 3.390; p = 0.0014; n=9 mice per group). The differences were statistically significant starting at 105 days (Bonferroni post-test; 105 days, p < 0.05; 112 days, p<0.01; n=9 mice per group). Concomitantly, the survival rate was significantly reduced in ad-MyoD treated mice (Fig. 2d, Kaplan-Meier curve, log ranking, p = 0.0018; n=9 mice per group) and on average reached the endpoint 11% faster (110.56 days vs. 124.57 days, unpaired two-tailed t-test, p = 0.003; n=9 mice per group). Therefore, postnatal expression of MyoD in hindlimb muscles accelerates motor abnormalities as well as weight loss in G93A mice and decreases survival.
Figure 2. MyoD gene-transfer into Adult G93A hindlimb muscle accelerates rotarod deficit and weight loss.
(a) Expression of human MyoD1 in the muscle following adenoviral construct injection was confirmed using quantitative real-time PCR.(n=4 per group, t-test, ****: p<0.00001). (b) Rotarod latency-to-fall is decreased at 20 RPM for Ad-MyoD injected mice starting at 91 days of age compared to ad-GFP injected mice (Two-way ANOVA, F(1,140) = 23.33, p < 0.0001; *: Bonferroni post-hoc test, p ≤ 0.05, n=9 mice per group). (c) Weight-loss in ad-MyoD injected mice was also greater starting at 98 days of age compared to ad-GFP injected group (Two-way ANOVA, F(8,140) = 3.390, p = 0.0014; *: Bonferroni test, p ≤ 0.05, n=9 mice per group). (d) Survival is reduced in ad-MyoD injected mice (Kaplan-Meier analysis, log-rank, p = 0.0018). ad-MyoD (dashed line); ad-GFP (solid line); n = 9 miceper group. (e) Western blotting shows that neither the mutant SOD1 (f, hSOD1) nor the mouse SOD1 (g, mSOD1) is altered following MyoD gene transfer (n=4 samples per group). Data are presented as mean±SEM. PLEASE PROVIDE FULL GEL SCANS OF ALL YOUR BLOT DATA IN THE SUPPLEMENTARY INFORMATION AS A SUPPLEMENTARY FIGURE AND CITE IN THE MAIN PAPER.
Recent studies have demonstrated that muscle-specific transgenic expression of mutant SOD1 can cause muscle dysfunction 6 and motor neuron degeneration 5 in mice. Examination of tissue samples by western blotting showed that neither the mutant (Fig. 2e and 2f; full blot images, Supplementary. Fig. 1) nor mouse SOD1 levels (Fig. 2e and 2g; full blot images, Supplementary. Fig. 1) were altered in muscle by MyoD gene transfer, ruling out the possibility of SOD1 modulation in muscle by MyoD as a cause of disease acceleration in G93A mice. Also, the effect observed in G93A mice was disease-specific since similar treatment in non-transgenic littermates did not have any behavioral impact. Weekly assessment of these mice showed consistent weight gain over time (Fig. 3a) and their ability to stay on the Rotarod for full 420 seconds over the course of the testing period (Fig. 3b). These mice remained healthy and did not show any behavioral abnormality even at 6 months following injection, showing that muscle-directed MyoD gene transfer itself was not toxic.
Figure 3. Muscle-directed MyoD gene transfer is not harmful in non-transgenic littermates.
Non-transgenic mice were injected with adenovirus containing MyoD construct (Ad-MyoD) combined with transient CD4+ cell depletion (n=8 mice). These mice were behaviorally observed starting at 56 days of age. (a) Weekly assessment of these mice showed consistent weight gain over time. (b) Weekly Rotarod assessment at 20 RPM showed consistent ability of mice to stay on the rotating beam for full 420 seconds over the duration of testing. After this period the mice were no longer tested but continued being observed. These mice were healthy and did not show any behavioral abnormality even at 6 months following injection. Red dashed line denotes weight at 84 days of age set as 100%. Data are presented as mean±SEM.
Myogenin Gene Transfer Improves Rotarod Performance
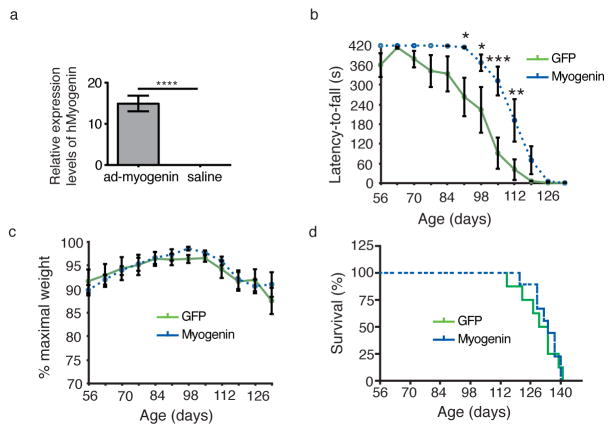
In our second set of experiments, we injected the hindlimb muscles of adult male G93A mice with either ad-myogenin or ad-GFP. Human myogenin expression in the muscle following ad-myogenin injection was confirmed via qPCR (Fig. 4a; t-test, p<0.00001; n=9 mice per group). Comparison of Rotarod performance between ad-myogenin and ad-GFP injected mice showed statistically significant treatment effect (Fig. 4b, Two-way ANOVA, F(1,186) = 30.26; p < 0.0001; n=9 mice per group). Bonferroni post-tests show statistically significant improvement in rotarod performance for ad-myogenin injected mice at 91 (p<0.05),98 (p<0.05), 105 (p<0.001), and 112 (p<0.01) days of age. Myogenin overexpression however did not affect either the weight-loss profile (Fig. 4c) or survival (Fig. 4d). We did not measure the expression levels of hSOD1 in these groups of mice since the study from Cleveland group showed that the knockdown of mutant SOD1 does not affect the disease progression in the fALS mice 7.
Figure 4. Myogenin gene-transfer into Adult G93A hindlimb muscle improves rotarod performance.
(a) Expression of human myogenin in the muscle following adenoviral construct injection was confirmed using quantitative real-time PCR. (n=4 per group, t-test, ****: p<0.00001). (b) Ad-myogenin injected mice are able to remain on the Rotarod for longer duration than ad-GFP injected mice starting at 91-days of age (Two-way ANOVA, F(1,140) = 23.33, p ≤ 0.0001; Bonferroni post-hoc test; *: p ≤ 0.05; **: p<0.01; ***: p<0.001; n=9 mice per group). There was no difference in the weight-loss profile (c) and survival rate (d) between the two treatment groups. Ad-myogenin (dashed line); ad-GFP (solid line); n = 9 mice per group. Data are presented as mean±SEM. Please define ‘**’ and ‘***’
Differential Motor Neuron Degeneration
In our two independent experiments, Rotarod testing showed improvement in motor performance following myogenin gene transfer compared to GFP control mice while MyoD gene transfer worsened motor performance in G93A mice as the disease progressed in comparison to the GFP control group. To determine whether spinal cord motor neuron degeneration is affected by exogenous expression of MyoD and myogenin in muscle, ventral horn motor neurons in the lumbar spinal cord region (L3-L5) were counted stereologically using size exclusion criteria 20. We examined spinal cord samples from 100-day old animals as these mice show significant motor neuron loss starting at 90-days of age 21.
Examination of cresyl violet stained spinal cord sections showed decreased ventral horn staining pattern in ad-MyoD injected mice compared to ad-GFP injected mice (Fig. 5a vs. 5b). On the other hand, ventral horns of ad-myogenin injected mice showed increased staining pattern in the ventral horn compared to ad-GFP injected mice (Fig. 5a vs. 5c). Under higher magnification, ad-MyoD injected mice motor neurons displayed smaller cell body size compared to ad-GFP injected mice (Fig. 5d vs. 5e), suggesting increased atrophy. Spinal cord samples from ad-myogenin mice showed motor neurons that were larger and asymmetrical in shape compared to motor neurons in ad-GFP injected mice, which were smaller and rounder in shape (Fig. 5d vs. 5f).
Figure 5. Muscle gene transfer of either MyoD or myogenin differentially modulates spinal cord motor neuron degeneration in G93A mice.
The impact of MyoD or myogenin muscle gene transfer on spinal cord motor neuron degeneration was determined by analyzing lumbar spinal cord sample from 100-day old mice. Examination of ventral horn motor neurons at lower magnification (10X) showed that compared to ad-GFP injected G93A mice (a), motor neurons in ad-MyoD injected G93A mice were sparsely distributed (b). Ad-myogenin injected mice on the other hand showed greater number of motor neurons (c). Motor neuron morphology also seems to be affected by the treatment. Higher magnification images (20X) showed that in comparison to the ad-GFP injected samples (d), motor neurons in ad-MyoD injected mice were smaller (e). In contrast, motor neurons were less rounded and larger in ad-myogenin injected mice (f). Stereological examination of spinal cord sections showed an impact of muscle directed MyoD and myogenin gene transfer on motor neuron counts using size exclusion criteria (g). Ad-MyoD injected mice showed 26% fewer motor neurons counted compared to ad-GFP injected control mice (one-way ANOVA, p<0.0001; Tukey’s post-hoc test, p<0.05, n=5 samples per group). Ad-myogenin injected mice on the other hand showed 66% more motor neuron counts compared to ad-GFP injected mice (one-way ANOVA, p<0.0001; Tukey’s post-hoc test, p<0.001, n=5 samples per group). Statistically significant differences between the means are represented by different colors. Data are presented as mean±SEM. WT (n=1) was not included in statistical analysis but shown as a benchmark (diagonal pattern). n=5 spinal cord samples per treatments (GFP, MyoD and Myog). Scale bars represent 100μm.
Stereological analysis of ventral horn motor neurons from 2mm portion of lumbar region of spinal cord revealed that muscle directed gene transfer of MyoD or myogenin had a treatment-dependent effect on the motor neuron count based on size exclusion criteria (cross sectional area greater than 200μm2). Ad-MyoD injected mice showed 26% fewer motor neurons counted compared to ad-GFP injected control mice (Fig. 5g, one-way ANOVA, p<0.0001; Tukey’s post-test, p<0.05; n=5 samples per group). On the other hand, ad-myogenin injected mice showed 66% more motor neurons counted compared to ad-GFP injected mice (Fig. 5g, Tukey’s post-test, p<0.001; n=5 samples per group). The motor neuron count in ad-myogenin injected mice is also greater than in ad-MyoD injected mice (Tukey’s post-test, p<0.001; n=5 samples per group). Our results suggest that an exogenous expression of myogenin in muscle helps preserve spinal cord motor neurons while exogenous expression of MyoD in muscle enhances motor neuron degeneration in G93A mice.
Differential Muscle Denervation
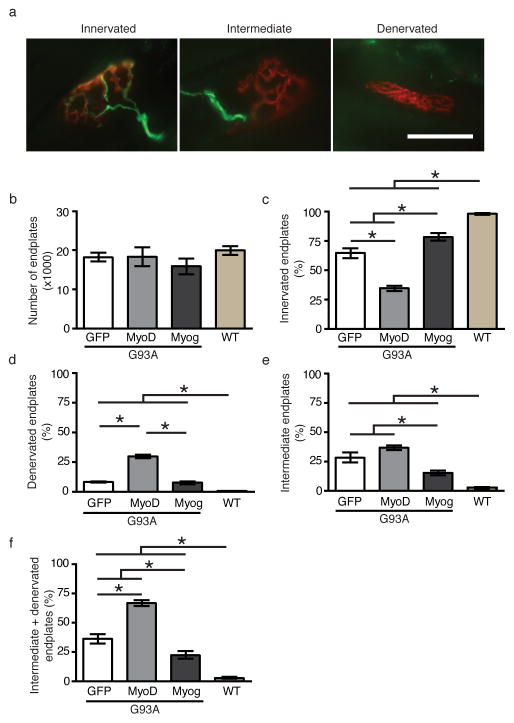
Muscle denervation has been shown to precede motor neuron loss in G93A mice 22,23. Furthermore, Bax deletion-mediated rescue of motor neurons failed to halt muscle endplate denervation 24. To determine whether muscle endplate innervation pattern is modulated following MyoD or myogenin gene transfer, we stereologically examined muscle sections from 100-day old experimental animals. Muscle sections were fluorescently labeled with rhodamine-conjugated α-bungarotoxin along with synaptophysin and neurofilament antibodies (Fig. 6a). Muscle innervation patterns were classified as “innervated”, “intermediate”, or “denervated” according to the staining patterns (Fig. 6a, also described in Methods).
Figure 6. Muscle gene transfer of either MyoD or myogenin differentially modulates muscle endplate innervation in G93A mice.
Muscle endplates were examined in 100-day old mice following muscle-directed gene transfer. (a) NMJ innervation patterns were categorized into one of three distinct patterns based on rhodamine-conjugated α-bungarotoxin (red) and synaptophysin (green) and neurofilament (green) staining patterns. (b) Stereological analysis of exhaustively sectioned muscle samples (n=4 muscles per treatment conditions [GFP, MyoD, Myog, and WT]) did not show any difference in the total number of muscle endplates between groups. However, the proportions of innervated (c, One-way ANOVA, p<0.0001, n= 4 samples per group), denervated (d, One-way ANOVA, p<0.0001, n=4 samples per group), and intermediate (e, One-way ANOVA, p<0.0001, n=4 samples per group) muscle endplates were significantly different between various treatment groups. Muscle innervation was increased in Myogenin gene transfer group whereas MyoD gene transfer decreased the proportion of innervated muscle endplates compared to GFP gene transfer mice (c, Tukey’s post-hoc test; Myog vs. GFP, p<0.05, n= 4 samples per group; MyoD vs. GFP, p<0.001, n=4 samples per group). The proportion of denervated endplates was significantly increased in ad-MyoD injected mice (d, Tukey’s post-hoc test; MyoD vs. GFP, p<0.001, n=4 samples per group) and myogenin gene transfer resulted in reduction of intermediate muscle endplates (e, Tukey’s post-hoc test, p<0.05, n=4 samples per group). When the proportions of denervated and intermediate endplates were combined, there was a clear treatment-dependent effect between all the groups (f, One-way ANOVA, p<0.0001, n=4 samples per group). Following one-way ANOVA, multiple comparison test was performed using Tukey’s test. Different colored bars represent statistically significant differences between the means. Data are presented as mean±SEM. Scale bar represents 25μm.
Stereological analysis of muscle endplates labeled with α-bungarotoxin showed that the total number of muscle endplates per muscle was not different between various treatment groups (Fig. 6b). However, there was a statistically significant treatment-dependent effect on the number of innervated muscle endplates (Fig. 6c, One-way ANOVA, F=44.12, p<0.0001; n=4 samples per group). A greater proportion of muscle endplates were innervated in ad-myogenin injected animals compared to ad-GFP injected mice (Fig. 6c, Tukey’s post-test, p<0.05; n=4 samples per group). On the other hand, there was a significant reduction in the proportion of innervated muscle endplates in ad-MyoD injected mice compared to ad-GFP injected mice (Fig 6c, Tukey’s post-test, p<0.001; n=4 samples per group).
Examination of denervated and intermediate endplates yielded mixed results. A higher proportion of denervated endplates were observed for the ad-MyoD injected animals compared to the ad-GFP and ad-myogenin injected mice. (Fig. 6d, One-way ANOVA, F=112.6, p<0.0001, n=4 samples per group; Tukey’s post-test, MyoD vs. GFP: p<0.001, n=4 samples per group; myogenin vs. MyoD: p<0.001, n=4 samples per group). The denervation pattern was not statistically different between ad-myogenin and ad-GFP injected animals (Fig. 6d). On the other hand, the proportion of intermediate endplates for ad-myogenin injected animals showed statistically significant reduction in comparison to either ad-MyoD or ad-GFP injected mice (Fig. 6e, One-way ANOVA, F=13.54, p<0.0019; Tukey’s post-test, myogenin vs. GFP: p<0.05, n=4 samples per group; myogenin vs. MyoD: p<0.01, n=4 samples per group). There was no statistical difference in intermediate endplate proportions between ad-MyoD and ad-GFP injected mice (Fig. 6e). When the proportions of intermediate and denervated muscle endplates were combined, there was a clear treatment-dependent effect (Fig. 6f, One-way ANOVA, F=43.5, p<0.0001, n=4 samples per group). Compared to the ad-GFP injection group, the proportion of intermediate and denervated endplates was significantly reduced in the ad-myogenin injected animals (Fig. 6f, Tukey’s post-test, myogenin vs. GFP: p<0.05, n=4 samples per group) and was significantly increased in ad-MyoD injected animals (Fig. 6f, Tukey’s post-test MyoD vs. GFP: p<0.001, n=4 samples per group).
Postnatal Gene Expression Alters Muscle Phenotype
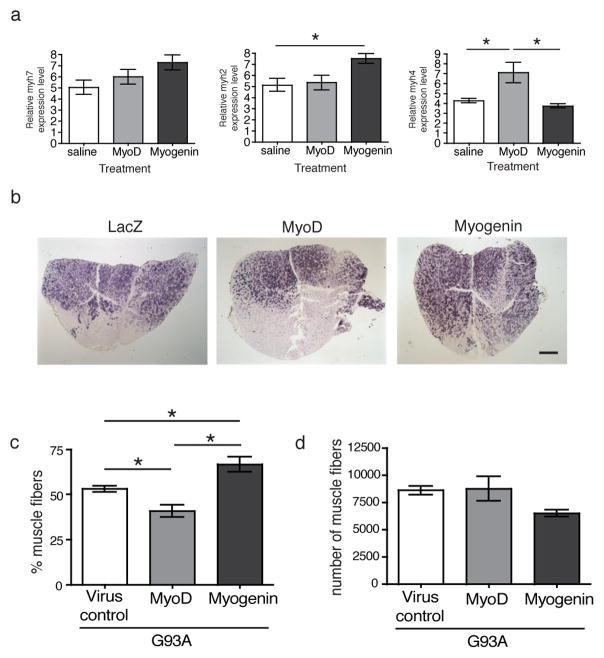
The influence of muscle-directed MyoD and myogenin gene transfer on motor neuron loss and muscle endplate denervation suggests that ensuing modification of muscle properties may influence neuromuscular pathology in G93A mice. It has been shown that myogenin expression induces increased oxidative capacity in pre-existing muscle fibers 14,15 Quantitative real-time PCR (qPCR) shows differential expression of myosin heavy chain isoforms following gene transfer. Two-weeks post-injection, ad-myogenin injected muscle samples show increased expression of myh2 (MyHC IIa gene) (Fig. 7a; one-way ANOVA, p=0.028; Tukey’s post-test, ad-myogenin vs. saline, p<0.05; n=4 samples per group). On the other hand, ad-MyoD injected muscle sample show increased expression of myh4 (MyHC IIb gene) (Fig. 7a; one-way ANOVA, p=0.0083; Tukey’s post-test, ad-MyoD vs saline, p<0.05, n=4 samples per group; ad-MyoD vs. ad-myogenin, p<0.01, n=4 samples per group). The expression levels for myh7 (MyHC I gene) was not statistically different between groups (Fig. 7a). Type IIa fast fibers are more oxidative than type IIb faster fibers. It has been shown that type IIb muscle fibers are preferentially denervated in G93A mice 25. Therefore, oxidative muscle fiber composition in the muscle was assessed using succinic dehydrogenase (SDH) histochemistry.
Figure 7. Oxidative muscle phenotype in G93A mice is reorganized following postnatal gene transfer.
(a) Expression levels for various myosin heavy chain isoforms were examined in muscle samples injected with ad-MyoD, ad-myogenin or saline using qPCR. Two-weeks following injection, ad-myogenin injected samples show increased expression for myh2 (one-way ANOVA, p=0.028; Tukey’s post test, ad-myogenin vs saline, p<0.05, n=4 samples per group). The difference in the expression levels for myh7 was not statistically significant between treatments. On the other hand, ad-MyoD injected samples show higher myh4 expression level (one-way ANOVA, p=0.0083; Tukey’s post-test, ad-MyoD vs saline, p<0.05, n=4 samples per group; ad-MyoD vs ad-Myogenin, p<0.01, n=4 samples per group). (B) Transverse muscle sections from 100-day old mice were histochemically stained for SDH in order to identify oxidative muscle fibers. (c) Stereological analysis of muscle samples from 100-day old animals show increased proportion of oxidative muscle fibers (SDH positive) in ad-myogenin injected animals (one-way ANOVA, F=16.6, p<0.0002; Tukey’s post-test, ad-myogenin vs virus control, p<0.5, n= 6 samples per group; ad-myogenin vs. ad-MyoD, p<0.001, n=6 samples per group). On the other hand, ad-MyoD injected muscle sample showed reduction in the proportion of SDH positive oxidative muscle fibers (Tukey’s post-test, ad-MyoD vs virus control, p<0.05, n=6 samples per group). (d) Total number of muscle fibers was not statistically different between treatment groups, suggesting remodeling of muscle oxidative phenotype by MyoD and myogenin gene transfer. Data are presented as mean±SEM. Scale bar represents 500μm.
Histochemical examination of transverse gastrocnemius muscle sections from 100-day old mice shows differences in the distribution of oxidative fibers between the treatment groups. Ad-myogenin treated muscles show a uniform distribution of oxidative muscle fibers as indicated by darkly stained muscle fibers compared to virus control (samples injected with either ad-GFP or ad-Lacz) and MyoD muscle samples (Fig. 7b). Stereological count of SDH stained muscle fibers in 100-day old experimental mice show that the oxidative fibers account for approximately 67% of total muscle fibers in the gastrocnemius injected with ad-myogenin (Fig 7c, n=6). In the virus control muscle samples, SDH positive muscle fibers represent approximately 53% of the total muscle fibers (Fig. 7c, n=6). On the other hand, MyoD treated muscle samples show only 41% of the muscle fibers that are stained positively for SDH, fewer oxidative muscle fibers compared to virus control (Fig 7c, n=6). These values were statistically different from each other (one-way ANOVA, F=16.6, p=0.0002; virus control vs. ad-myogenin, Tukey’s post-test, p<0.05, n=6 samples per group; virus control vs. ad-MyoD, Tukey’s post-test, p<0.05, n=6 samples per group; ad-myogenin vs. ad-MyoD, Tukey’s post-test, p<0.001, n=6 samples per group). The total number of muscle fibers was not statistically different between treatment groups in the 100-day old mice (Fig. 7d).
PGC-1α does not Improve Survival or Motor Performance
To examine whether enhanced mitochondrial biogenesis in muscle by PGC-1α overexpression improves disease progression in the G93A SOD1 fALS mouse model, we crossed G93A mice with transgenic mice overexpressing PGC-1α under the control of muscle creatine kinase (MCK-PCG-1α mice). PGC-1α is a potent metabolic regulator in the muscle and an overexpression of PGC-1α in muscle was reported to cause increased mitochondrial protein and oxidative capacity 16,26.
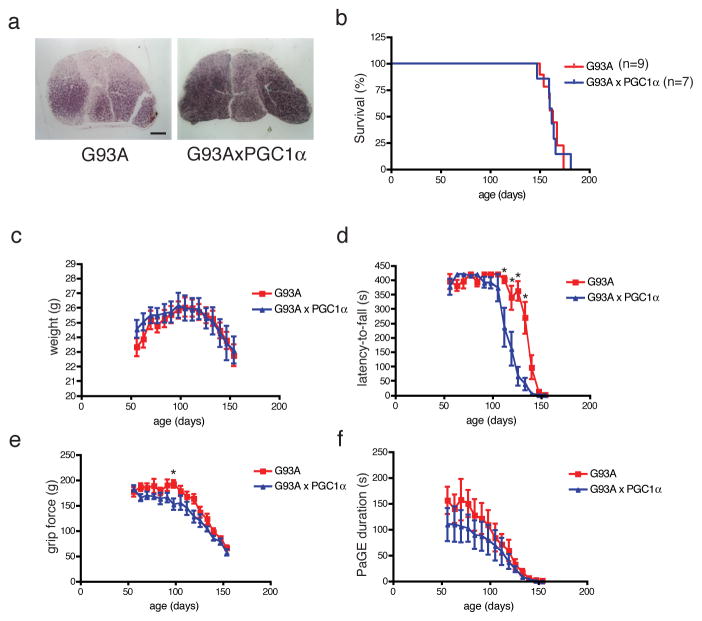
In mice over-expressing PGC-1α in muscle, mitochondrial activity in the gastrocnemius of G93A/MCK-PGC-1α mice was increased as indicated by increased SDH staining in the muscle (Fig. 8a). G93A and G93A/MCK-PGC-1α male mice were longitudinally observed to examine the effect of PGC-1α overexpression in muscle on the disease progression. We did not observe any difference in the survival rate (Fig. 8b, G93A: 163 days vs. G93A/MCK-PGC-1α: 162 days) or progressive weight loss (Fig. 8c) between G93A and G93A/MCK-PGC-1α mice. Motor function was also longitudinally assessed in these mice using various motor function tests. Rotarod analysis demonstrated a genotypic difference between the two genotypic groups (Fig. 8d, two-way ANOVA, p<0.0001; n=9 SOD1; n=7 SOD1/MCK-PGC-1α). Bonferroni post-hoc analysis showed that G93A/MCK-PGC-1α mice performed worse at 112 (p<0.001), 119 (p<0.001), 126 (p<0.001), and 133 (p<0.001) day time-points (Fig. 8d). Paw grip strength was tested using grip strength meter and paw grip endurance (PaGE) test (Fig. 9e and 9f, respectively). Two-way ANOVA showed significant genotype effect on the paw grip strength (Fig. 9e, Two-way ANOVA, p<0.0001, n=9 G93A; n=7 G93A/MCK-PGC-1α). Bonferroni post-hoc analysis showed that G93A/MCK-PGC-1α mice performed worse at 98 day time point (Fig. 8e, p<0.05, n=9 G93A; n=7 G93A/MCK-PGC-1α). PaGE test also showed significant genotype effect (Fig. 8f, Two-way ANOVA, p=0.0082, n=9 G93A; n=7 G93A/MCK-PGC-1α). However there was no statistically significant difference in PaGE between the two genotypes during the any of the time points observed during the course of the disease (Fig. 8f, Bonferroni post-test, n=9 G93A; n=7 G93A/MCK-PGC-1α). Therefore enhancement of mitochondrial biogenesis by constitutive muscle expression of PGC-1α did not affect survival but seemed to have a negative effect on the motor function.
Figure 8. PGC-1α-mediated increase in the muscle oxidative phenotype does not improve survival or motor function in G93A mice.
G93A mice were mated with MCK-PGC-1α mice to examine the effect of enhanced mitochondrial biogenesis in the muscle on disease progression. (a) Gastrocnemius muscle from G93A/MCK-PGC-1α mouse shows higher SDH staining than S93A mouse demonstrating increased mitochondrial activity in the presence of PGC-1α transgene (scale=500μm). (b) Survival analysis and (c) weekly weight measures (Two-way ANOVA, n=9 G93A; n=7 G93A/MCK-PGC-1α) did not show a genotypic difference between the two groups. (c) Motor function was examined in these mice using the Rotarod apparatus. The difference in the rotarod latency-to-fall values between the two groups were statistically significant (Two-way ANOVA, p<0.0001, n=9 G93A; n=7 G93A/MCK-PGC-1α). Bonferroni test shows G93A/MCK-PGC-1α mice performed worse at 112 (p<0.001), 119 (p<0.001), 126 (p<0.001), and 133 (p<0.001) day time-points. Paw grip strength was longitudinally examined using grip strength meter (e) and paw grip endurance test (PaGE, f). Statistical analysis revealed a significant genotype effect for both the grip strength and PaGE (e and f, respectively; Two-way ANOVA; e: p<0.0001; f: p=0.0082, n=9 G93A; n=7 G93A/MCK-PGC-1α). Bonferroni post-test shows that SOD1/PGC1-α mice performed significantly worse at 98-day time-point for the grip strength (e, Bonferroni post-test, p<0.05, n=9 G93A; n=7 G93A/MCK-PGC-1α). However, there was no difference between the genotypes at any of the time-points for the PaGE test (f). Therefore, enhancement of mitochondrial biogenesis in the muscle via PGC-1α overexpression did not improve survival but worsened motor function in G93A mice. Data are presented as mean±SEM.
Discussion
Our hypothesis was based on experimental evidence suggesting a possible effect of muscle phenotype on the disease progression; fast muscle fibers and motor units are seemingly selectively vulnerable in ALS, while slow muscle fibers, which have greater oxidative capacity, seem to be relatively preserved in the disease 1–4,25. To test our hypothesis, we used viral-mediated postnatal expression of two distinct muscle-specific basic helix-loop-helix transcription factors to directly examine whether muscle phenotype influences disease progression in G93A mouse model of ALS. MyoD and myogenin along with MRF4 and myf5 form a family of myogenic regulatory factors (MRF) that are involved in muscle development and differentiation 27. MyoD and myogenin govern differential muscle gene expression 28. In the adult muscle, MyoD is highly expressed in fast muscle fibers 11,12 and regulates fast muscle development 12. On the other hand, myogenin is highly expressed in slow muscle fibers 11 and its expression increases oxidative metabolism in the muscle 14,15.
In this study, we used adenovirus to exogenously express MyoD and myogenin in the hindlimb muscles of adult G93A mice to examine whether MyoD and myogenin expression in the adult muscle can impact disease progression in an ALS mouse model. Lack of X-gal staining in the spinal cord following injection of adenovirus expressing LacZ into the muscle suggests that that the adenovirus is not retrogradely taken up by the motor neurons and the gene expression is confined to the muscle in our study. Our findings show that exogenous expression of myogenin in adult muscle results in increased motor neuron count, and muscle innervation. On the other hand, exogenous expression of MyoD in the adult muscle results in reduced motor neuron count, muscle innervation and survival. We have previously shown that MyoD expression is decreased in the muscle of G93A mice at 27 days of age 29. Our current finding suggests that this early decrease in MyoD expression may be a protective compensation. We are currently testing this hypothesis by postnatally knocking down MyoD in the G93A mice.
Ad-myogenin injected muscle samples showed higher expression of MyHC IIa gene myh2. On the other hand, ad-MyoD injected muscle samples showed higher expression of MyHC IIb gene myh4. Correspondingly, there was an increase in the proportion of muscle fibers that were strongly stained for SDH in animals injected with ad-myogenin. The total number of muscle fibers between the treatment groups in our study was not statistically different, suggesting an oxidative muscle phenotype conversion by myogenin gene transfer in adult mice. These findings are in agreement with past studies that showed muscle fiber type switching with MyoD and myogenin gene transfer. Myogenin gene transfer has been shown to convert glycolytic muscle fibers to oxidative muscle fibers 14,15. MyoD gene transfer has shown to impart type IIb muscle fiber under denervation condition 13. Our data suggests that MyoD and myogenin gene transfer in the G93A mice at presymptomatic age modulates gene expression and modifies muscle phenotype. The differential effect on neuropathology at 100 days of age in our study suggests that the disease progression can be altered by postnatal muscle phenotype modification. However, the exact physiological changes responsible for mediating the effect in our study remains unclear. More in-depth analysis of physiological changes occurring in the muscle following MyoD and myogenin induction may provide insight into physiological mechanisms affecting NMJ denervation in ALS.
An alternative approach using constitutive overexpression of transgenic PGC-1α to induce an increased oxidative phenotype did not improve survival and worsened motor function in these mice. A similar finding was also reported for the G37R SOD1 mouse model 30. The similar findings between G93A and G37R mouse models of ALS in response to transgenic expression of PGC1-a confirms our methodology and suggests that the MyoD and myogenin findings are unlikely to be due to model differences. The Differential clinical outcome in the fALS mouse model between myogenin- and PGC-1α-mediated muscle remodeling despite the enhanced oxidative phenotype for both factors may be due to differential gene regulation between myogenin and PGC-1α. For example, it has been shown that AICAR-mediated increase in muscle expression of mitochondrial proteins requires PGC-1α 31. However, myogenin expression reamains unchanged despite increased PGC-1α expression in response to AICAR treatment 32. Another possibility is that myogenin expression is virally induced in postnatal muscle whereas PGC-1α is transgenically expressed. Transgenic expression of PGC-1α may induce developmental compensatory gene regulation which may be the reason behind the differential effect on the disease progression in the G93A mice.
It has been shown that muscle denervation precedes motor neuron loss in fALS mouse models and ALS patients 22,23,33. Although, MyoD and myogenin have been shown to activate acetylcholine receptor expression in the muscle 34–37, we did not observe any difference in the total number of alphabungarotoxin bound actetylcholine receptors between the treatment groups. Findings by Williams and colleagues showed that muscle specific microRNA-206 (miR-206), which is also enriched in slow muscle, is important for muscle reinnervation process and developmental knockout of miR-206 in G93A mice accelerates disease progression by delaying muscle reinnervation in these mice 38. Myogenin is a transcriptional regulator of miR-206 39, therefore an increase in myogenin expression could enhance muscle reinnervation process in these mice. However, it is intriguing that muscle denervation is increased in response to MyoD gene transfer since MyoD can also activate miR-206 38. Our finding seems to suggest a differential role of MyoD and myogenin in muscle endplate innervation/denervation process in adult G93A mice and further study will be necessary to determine the role of MyoD and myogenin in the muscle denervation process in the ALS.
Findings from our study provide a potential insight into the role of exercise in ALS. Myogenin and MyoD expression pattern in response to exercise is quite complex and depends on the type of exercise 27,40. For instance, MyoD expression in muscle is highest following high intensity eccentric exercises 40,41. On the other hand, in vivo expression of myogenin is closely correlated with the duration of moderate exercise 40,42,43. However some of the exercise studies show that both the MyoD and myogenin levels are elevated following either resistance or endurance training 27. In cases of strength exercises such as eccentric or resistance exercises that cause muscle damage and repair, MyoD and myogenin are both elevated 27. The expression of MyoD can be quite dynamic as well depending on the type of exercise 40.
The role of exercise has been controversial due to its seemingly conflicting effects on ALS progression reported by various groups. In human subjects, various groups have concluded that exercise is either beneficial or has no benefit 44–47. In contrast, a number of retrospective studies in ALS patients have highlighted strenuous athletics or activities as potential risk factors for ALS 48,49. Studies in ALS mouse models show that moderate exercise delays motor decline 50,51, while high-intensity endurance exercise hastens disease progression 52. Our findings suggest that strenuous exercise or activity that enhances muscle expresssion of MyoD will likely exacerbate ALS disease progression. Additionally, the beneficial exercises for ALS patients would be the ones that enhance myogenin expression in the muscle. It is worth noting that IGF-1 which has shown benefit in G93A mice 8,53 increases myogenin expression in muscle 54,55. Our findings need to be confirmed using other mouse models of fALS to determine whether the observations made in our study may have implications for disease management in ALS patients.
A number of studies have now suggested extraneuronal cells influence motor neuron survival in ALS 56–60. However the potential role of muscle on pathogenesis has been controversial 5–7,30. Our experimental evidence demonstrates that postnatal muscle expression of myogenic factors, MyoD and myogenin, modulates spinal cord motor neuron degeneration, muscle denervation and survival in the G93A SOD1 fALS mouse model. Unlike the beneficial effects of myogenin expression in muscle, constitutive overexpression of PGC1α in the muscle of G93A mice did not improve the outcome in this fALS model, consistent with a recent report. However, this finding does not reflect the potential benefits of postnatal modification of muscle in ALS using other factors, and may reflect pleotropic effects of this transcriptional modifier in addition to increases in muscle oxidative function. Our findings suggest that muscle phenotype moderately influences ALS pathogenesis in the G93A mouse model of ALS. However, this may be due to localized viral injection limited to a small muscle group in our study. A more systemic induction may lead to an increase in therapeutic benefit. Also, it will be necessary to examine other mouse models of fALS to further validate the role of MyoD and myogenin in ALS pathology. Further work in identifying key muscle biochemical pathways by which MyoD and myogenin influence disease progression may lead to effective muscle-directed treatments for ALS.
Methods
Animals
High copy number B6/SJL G93A mice ((male and female mice, 50 days of age) were obtained from Jackson Laboratory (Bar Harbor, ME) and bred and maintained in our animal colony. Mice were maintained on a 12-hour light/dark cycle with free access to food and water. For PGC-1α muscle overexpression study, B6 congenic G93A mice (Jackson Laboratory, Bar Harbor, ME) were crossed to B6 congenic MCK-PGC-1α mice (Jackson Laboratory, Bar Harbor, ME) to generate G93A/MCK-PGC-1α mice. Male mice were used for this study and the genotyping was performed using genomic DNA, as suggested by the Jackson Laboratory genotyping protocol. Mice were killed by decapitation at end-stage and spinal cord and hindlimb muscles were quickly dissected. Dissected samples were either flash-frozen for biochemical analysis or immersion-fixed in 4% paraformaldehyde for immunohistological analysis. Muscle samples were fixed for 2 hours at 4°C and transferred to 30% sucrose solution for 2 hours prior to freezing in OCT (TissueTek). All animal protocols were approved by the Animal Care Committee at the University of British Columbia.
Antibodies and Reagents
The following antibodies were used in this study: mutant hSOD1 (1:1000) (Novocastra NCL-SOD1, Vector Laboratories; Burlingame, CA); CD4 antibody (100μg/injection) (eBioscience Inc.; San Diego, CA; Cat. No. 16-0041); mouse SOD1 (1:1000) (Biosensis Pty Ltd.; Thebarton, South Australia; Cat. No. R-126-100); GAPDH (1:1000)(Chemicon; Billerica, MA; Cat. No. MAB374); rhodamine conjugated a-bungarotoxin (1:500) (Invitrogen; Cat. No. B13423); synaptophysin (1:500) (Millipore, Billerica, MA; Cat. No. MAB368); neurofilament (1:500) (Covance, Princeton, NJ; Cat. No. SMI-32R). Adenoviral constructs were obtained from Vector Biolabs (Philadelphia, PA). Sodium succinate, sodium phosphate (monbasic and dibasic), nitrobluetetrazolium tablets, and normal goat serum were obtained from Sigma.
Adenoviral Injections
Male mice were injected at 30 days of age with replication incompetent adenovirus (AV) containing GFP, human MyoD or human myogenin construct under the control of cytomegalovirus promoter. Adenoviral constructs were obtained from Vector Biolab (Pennsylvania). 5×108 plaque forming units were injected per site in the hindlimb (gastrocnemius, tibialis anterior, hamstring, and quadriceps) combined with transient depletion of CD4+ cells using commercially available functional grade CD4 antibody (eBioscience) to enhance gene transfer and expression 19. CD4 antibody (100μg) was injected intraperitoneally 3 days and 1 day prior to intramuscular adenoviral injections followed by intraperitoneal injections of CD4 antibody 1 day and 3 days post AV injection. Mice were anesthetized using isofluorane prior to AV injections. Following injections, mice were monitored daily following recovery. None of the treated mice showed any adverse reactions to combined treatments of adenovirus and CD4 antibody.
Behavioral Tests
Mice were trained at 47 days of age on the rotarod (UGO Basile, Italy) for maximum duration of 7 minutes (420 seconds) at 20 RPM. Mice were weighed and tested weekly on the rotarod (UGO Basile, Italy) and grip test (Columbus Instruments) starting at 57 days of age. Mice were tested on the rotarod for maxium duration of 7 minutes (420 seconds). If the animal fell off prior to the maximal duration, they were tested for 4 trials in total and the maximal duration was recorded. Grip strength measurement was repeated 6 times per session and the average value was recorded.
Quantitative Real-Time PCR
Muscle samples were dissected and stored in RNAlater and stored in −20°C until processed (Qiagen, Valencia, CA, USA). RNA extraction was performed using RNeasy Fibrous Tissue Midi Kit (Qiagen, Valencia, CA, USA). cDNAs were generated using SuperScript VILO cDNA Synthesis Kit using 5μg RNA input (Life Technologies, Carlsbad, CA, USA). Quantitative PCR was performed using the StepOne Plus Real-Time PCR system (Life Technologies, Carlsbad, CA, USA) using 5ng cDNA input per well. Fast SYBR Green Master Mix (Life Technologies, Carlsbad, CA, USA) was used for all qPCR. Primer sequences were generated using either Primer Express 3.0 (Life Technologies) or PrimerQuest (IDT, Coralville, IA, USA). The primers were obtained from IDT (Coralville, IA, USA). The sequences for the primers used in the study are listed in Table 1. Gene expression levels are measured by real-time qPCR and expressed relative to the house keeping genes GAPDH and HPRT.
Table 1.
List of primer sequences used for qPCR. Primer sequences are specific for mouse except for those used for MyoD1 and myogenin, which are human specific.
| GAPDH | forward | AACTTTGGCATTGTGGAAGG |
| reverse | ACACATTGGGGGTAGGACA | |
| HPRT | forward | CGTCGTGATTAGCGATGATGA |
| reverse | TCCAAATCCTCGGCATAATGA | |
| Human MyoD1 | forward | TGCCACAACGGACGACTTC |
| reverse | CGGGTCCAGGTCTTCGAA | |
| Human Myogenin | forward | CACTCCCTCACCTCCATCGT |
| reverse | CATCTGGGAAGGCCACAGA | |
| Myh7 (type I) | forward | GTTTCCTTACTTGCTACCCTCAG |
| reverse | TGGATTCTCAAACGTGTCTAGTG | |
| Myh2 (type IIa) | forward | TCCCGAACGAGGCTGAC |
| reverse | AGCTCATGACTGCTGAACTC | |
| Myh4 (type IIb) | forward | GTTGAGAGGTGACAGGAGAA |
| reverse | TCTGCAGAATTTATTTCCGTGA |
Muscle Endplate Immunofluorescence
Muscle sections were outlined using PAP pen (Research Products International) and incubated with rhodamine conjugated a-bungarotoxin (1:500; Invitrogen) for 30 min at room temperature. The sections are rinsed 1X TBS and fixed in ice-cold (−20°C) MeOH for 10 minutes. The sections are rinsed with 1X TBS and blocked with 3% normal goat serum in TBS for 30 min at room temperature. The sections were incubated with synaptophysin (1:500) and neurofilament (1:500) antibodies for 1 hour at room temperature.. The sections are rinsed with 1X TBS and incubated with Alexafluor 488 conjugated secondary (goat anti-mouse, Invitrogen) for 1 hour at room temperature. Sections were dehydrated in 100% EtOH and rinsed in Xylenes to remove PAP pen. The sections were rehydrated and coverslipped using ProlongGold (Invitrogen).
Succinic Dehydrogenase Staining
Succinic dehydrogenase (SDH) staining was done according to SDH staining protocol obtained from Washington University muscle histology lab (URL:http://neuromuscular.wustl.edu/pathol/histol/sdh.htm). Briefly, transverse 12μm muscle sections were mounted on Super Frost microscope slides (Fisher Scientific). Sections were outlined with PAP pen (Research Products International) and incubated in buffer solution (20mM phosphate buffer, 7.5% sucrose, 0.027% sodium succinate, and 10mg nitrobluetetrazoleum) for 45 minutes at RT. The sections were dehydrated briefly rinsed in 30%, 60% and 90% acetone in ddH2O in ascending and descending order, rinsed in H2O, air-dried and coverslipped using VectaMount (Vector Labs).
X-gal Staining
Muscle samples were snap-frozen in liquid N2 and sectioned at 16um thickness and mounted on Superfrost microscope slides (Fisher). Briefly, the muscle sections were outlined with PAP pen and fixed at RT with 0.2% glutaraldehyde in TBS for 10 minutes. Sections were rinsed 3X with TBS and rinsed 1X with LacZ wash buffer (2mM MgCl2, 0.1% sodium deoxycholate, 0.2% Nonidet, in 0.1M phosphate buffer, pH 7.) for 10 minutes. The sections were then stained overnight in LacZ staining solution (LacZ wash buffer, 1mg/ml X-gal, potassium ferrocyanide, potassium ferricyanide). Stained muscle sections were fixed 4% paraformaldehyde in TBS for 10 minutes and rinsed 3X in TBS. Sections were serially dehydrated in EtOH, cleared Xylenes, and coverslipped with DPX mounting medium.
Intact spinal cord samples were immersion fixed in 1% paraformaldehyde/0.2% glutaraldehyde in TBS for 30 minutes at RT. Spinal cords were then rinsed 3X in LacZ wash buffer (15 minutes each) and stained overnight at RT in LacZ staining solution. Stained spinal cord was fixed overnight at 4°C in 4% paraformaldehyde/TBS. The fixed spinal cord was rinsed 3X in TBS (15 minutes each). The spinal cord was then embedded in 1.5% agar and sectioned using vibratome and mounted on Superfrost microscope slides. The mounted sections were dried overnight, cleared in Xylenes and coverslipped with DPX mounting medium.
Cresyl Violet Staining
Briefly, sequential lumbar sections were mounted on slides, dried overnight and stained with 0.5% cresyl violet for 6 min followed by dehydration with a graded series of ethanol solutions (70, 85, 90, 100% for 2 min each). Slides were then treated with CitriSolv (Fisher, Tustin, CA, USA) for 2 min, followed by xylene for 10 min and coverslipped with Cytoseal 60 (Richard-Allan Scientific, Kalamazoo, MI, USA).
Western Blotting
Muscle samples were homogenized in lysis buffer (20mM NaF, 1mM Sodium Vanadate, 0.5% Triton X-100, 0.1% SDS, and protease inhibitor cocktail (10μl/ml; Sigma, P8340) in 1X TBS). Protein quantities were determined using Qubit quantitation platform (Invitrogen). Samples were electrophoresed using the Novex pre-cast gel system (Biorad) and transferred to nitrocellulose (Protran, Schleicher and Schuell, Dassel, Germany), and subjected to immunoblotting. Band density was analyzed using Quantity One software (Biorad, Mississauga, ON). GAPDH (Millipore) was used as loading control. Precision plus protein dual color standards (Biorad) was used to determine the molecular weight sizing of protein bands.
Tissue Imaging
Sections were observed using a Zeiss AxioPlan 2 microscope and images captured using a Coolsnap HQ camera (Photometrics, Tucson, AZ) and MetaMorph software version 6.2r4 (Universal Imaging Corporation, Downingtown, PA).
Quantification of Alpha Motor Neurons
Spinal cords were dissected and isolated, immersed in the paraformaldehyde overnight at 4°C and then cryoprotected in 30% sucrose solution in TBS overnight at 4°C. The lumbar spinal cord was then frozen in OCT compound (Sakura Finetek, Torrance, CA) and stored at −80°C until sectioned. The tissue was cut in coronal 50 μm-thick consecutive sections and directly mounted on to Super Frost slides (Fisher Scientific).
Stereological quantification of motorneurons was carried out on a series of cresyl violet stained lumbar sections (every third lumbar section) with a total of 14 sections/mouse used in our calculations. Alpha motorneurons were selected based on morphology and size exclusion criteria (area ≥ 200 mm2). A total estimated number of alpha motorneurons in the ventral horn per spinal cord was obtained using the Optical Fractionator Probe in StereoInvestigator 9.01, a morphometry and stereology software package (MicroBrightField, Colchester, VT, USA). Ventral horns were first delineated with a Zeiss 2.5x objective and then cells counted using a 40x objective on a Zeiss Imager M2 microscope witha motorized computer-controlled stage (Ludl Electronics, Hawthorne, NY) and a Microbrightfield Digital CCD video camera (MicroBrightField, Colchester, VT, USA). All morphometric quantification was conducted with the investigator blind to genotype and treatment.
Quantification of Muscle Endplate Innervation
Left and right gastrocnemius muscles from WT or adenoviral construct (GFP, human MyoD, or human myogenin) injected G93A mice were dissected and fixed in 4% PFA for 20min on ice. Following Incubation in 30% sucrose for 2 hours, muscle was put in OCT and frozen using isopentane on dry ice. Frozen samples were stored at −80°C until sectioned. Muscle samples were sectioned exhaustively sectioned on a cryostat at 40μm thickness, mounted on to Super Frost slides (Fisher Scientific) and let it dry at RT for 10min. Mounted sections were stored at −20°C if short-term and at −80°C if long-term.
Every fourth sections through the whole muscle were fluorescence labeled for a-bungarotoxin, synaptophysin and neurofilament. Each muscle section was delineated using a stereology workstation, consisting of a Zeiss Imager M2 light microscope and a EC Plan Neofluar objective 2.5× (numerical aperture [N.A.], 0.075) and a EC Plan Neofluar objective 40× (N.A., 0.75; Zeiss, Göttingen, Germany) to count. For counting, a motorized specimen stage for automatic sampling (MAX6000; Ludl Electronics, Hawthorne, NY), CCD color video camera (CX9000; Microfire Digital CCD video camera) and stereology software (StereoInvestigator 9.0; MBF Bioscience, Williston, VT) were utilized. Total endplate number was determined from StereoInvestigator 9.0 using the physical fractionator probe with a grid size of 400×400 μm and counting frame of 100×100 μm. The percentage of innervated, intermediate and denervated endplates was determined as total number of endplates divided by the number of innervated (overlapping synaptophysin and alphabungarotoxin staining), intermediate (no overlap but synaptophysin staining was adjacent to alphabungarotoxin) or denervated (no overlap and no apparent synaptophysin staining adjacent to alphabungarotoxin staining) endplates. Overall, the coefficient of error (CE) was less than 0.1. The investigator conducted the stereological analysis blind to genotype and treatment.
Statistics
Two-way ANOVA with Bonferroni post hoc test, One-way ANOVA, two-tailed unpaired t-test, and Kaplan-Meier survival analysis were performed using Prism 4 for Macintosh (Prism v. 4.0, GraphPad Software Inc., San Diego, CA).
Supplementary Material
Acknowledgments
We thank J. Gregg, A. Hill, and C. Lefroy for technical assistance and mouse colony maintenance. This work was supported by Neuromuscular Research Partnership grant (Canadian Institutes of Health Research, ALS Society Canada, and Muscular Dystrophy Canada) to K.H.J.P. and B.R.L.
Footnotes
Author Contributions:
Kevin HJ Park conceived the study; designed and carried out experiments; analyzed data; wrote and edited the manuscript.
Sonia Franciosi: analyzed data.
Blair Leavitt: designed experiments; wrote and edited the manuscript.
All authors reviewed the final manuscript for submission.
The authors declare no competing financial interests.
References
- 1.Dengler R, et al. Amyotrophic lateral sclerosis: macro-EMG and twitch forces of single motor units. Muscle Nerve. 1990;13(6):545–550. doi: 10.1002/mus.880130612. [DOI] [PubMed] [Google Scholar]
- 2.Sanjak M, et al. Quantitative assessment of motor fatigue in amyotrophic lateral sclerosis. J Neurol Sci. 2001;191(1–2):55–59. doi: 10.1016/s0022-510x(01)00624-4. [DOI] [PubMed] [Google Scholar]
- 3.Atkin JD, et al. Properties of slow- and fast-twitch muscle fibres in a mouse model of amyotrophic lateral sclerosis. Neuromuscular Disorders. 2005;15:377–388. doi: 10.1016/j.nmd.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Gordon T, Putman CT, Hegedus J. Amyotrophic lateral sclerosis - evidence of early denervation of fast-twitch muscles. Basic Appl Myology. 2007;17(3–4):141–145. [Google Scholar]
- 5.Wong M, Martin LJ. Skeletal Muscle-Restricted Expression of Human SOD1 Causes Motor Neuron Degeneration in Transgenic Mice. Hum Mol Genet. 2010 doi: 10.1093/hmg/ddq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobrowolny G, et al. Skeletal muscle is a primary target of SOD1G93A-mediated toxicity. Cell Metabolism. 2008;8:425–436. doi: 10.1016/j.cmet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Miller TM, et al. Gene transfer demonstrates that muscle is not a primary target for non-cell-autonomous toxicity in familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2006;103(51):19546–19551. doi: 10.1073/pnas.0609411103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobrowolny G, et al. Muscle expression of a local Igf-1 isoform protects motor neurons in an ALS mouse model. J Cell Biol. 2005;168(2):193–199. doi: 10.1083/jcb.200407021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Megeney L, Rudnicki MA. Determination versus differentation and the MyoD family of transcription factors. Biochem Cell Biol. 1995;73:723–732. doi: 10.1139/o95-080. [DOI] [PubMed] [Google Scholar]
- 10.Perry RL, Rudnicki MA. Molecular mechanisms regulating myogenic determination and differentiation. Front Biosci. 2000;5:D570–D767. doi: 10.2741/perry. [DOI] [PubMed] [Google Scholar]
- 11.Hughes SM, et al. Selective accumulation of myoD and myogenin mRNAs in fast and slow adult skeletal muscle is controlled by innervation and hormones. Dev. 1993;118:1137–1147. doi: 10.1242/dev.118.4.1137. [DOI] [PubMed] [Google Scholar]
- 12.Maves L, et al. Pbx homeodomain proteins direct MyoD activity to promote fast-muscle differentiation. Dev. 2007;134:3371–3382. doi: 10.1242/dev.003905. [DOI] [PubMed] [Google Scholar]
- 13.Ekmark M, Rana ZA, Stewart G, Hardie DG, Gundersen K. De-phosphorylation of MyoD is linking nerve-evoked activity to fast myosin heavy chain expression in rodent adult skeletal muscle. J Physiol. 2007;584(2):637–650. doi: 10.1113/jphysiol.2007.141457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes SM, Chi MM-Y, Lowry OH, Gundersen K. Myogenin induces a shift of enzyme activity from glycolytic to oxidative metabolism in muscles of transgenic mice. J cell Biol. 1999;145(3):633–642. doi: 10.1083/jcb.145.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekmark M, Gr⊘nevik E, Schjerling P, Gundersen K. Myogenin induces higher oxidative capacity in pre-existing mouse muscle fibres after somatic DNA transfer. J Physiol. 2003;548(1):259–269. doi: 10.1113/jphysiol.2002.036228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin J, et al. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 17.Potthoff MJ, Olson EN, Bassel-Duby R. Skeletal muscle remodeling. Curr Opin rheumatol. 2007;19:542–549. doi: 10.1097/BOR.0b013e3282efb761. [DOI] [PubMed] [Google Scholar]
- 18.Goldspink G. Changes in muscle mass and phenotype and the expression of autocrine and systemic growth factors by muscle in response to stretch and overload. J Anat. 1999;194:323–334. doi: 10.1046/j.1469-7580.1999.19430323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Haecker SE, Su Q, Wilson JM. Immunology of gene therapy with adenoviral vectors in mouse skeletal muscle. Hum Mol Genet. 1996;5(11):1703–1712. doi: 10.1093/hmg/5.11.1703. [DOI] [PubMed] [Google Scholar]
- 20.Ciavarro GL, et al. The densitometric physical fractionator for counting neuronal populations: application to a mouse model of familial amyotrophic lateral sclerosis. J Neurosci Methods. 2003;129(1):61–71. doi: 10.1016/s0165-0270(03)00201-2. [DOI] [PubMed] [Google Scholar]
- 21.Chiu AY, et al. Age-dependent penetrance of disease in a transgenic mouse model of familial amyotrophic lateral sclerosis. Mol Cell Neurosci. 1995;6:349–362. doi: 10.1006/mcne.1995.1027. [DOI] [PubMed] [Google Scholar]
- 22.Frey D, et al. Early and selective loss of neuromuscular synapse subtypes with low sprouting competence in motoneuron diseases. J Neurosci. 2000;20(7):2534–2542. doi: 10.1523/JNEUROSCI.20-07-02534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer LR, et al. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Experimental Neurology. 2004;185:232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Gould TW, et al. Complete dissociation of motor neuron death from motor dysfunction by Bax deletion in a mouse model of ALS. J Neurosci. 2006;26(34):8774–8786. doi: 10.1523/JNEUROSCI.2315-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hegedus J, Putman C, Tyreman N, Gordon T. Preferential motor unit loss in the SOD1 G93A transgenic mouse model of amyotrophic lateral sclerosis. J Physiol. 2008;586(14):2227–2251. doi: 10.1113/jphysiol.2007.149286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finck B, Kelly D. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116(3):615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gundersen K. Excitation-transcription coupling in skeletal muscle: the molecular pathways of exercise. Biol Rev Camb Philos Soc. 2011;86(3):564–600. doi: 10.1111/j.1469-185X.2010.00161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davie JK, et al. Target gene selectivity of the myogenic basic helix-loop-helix transcription factor myogenin in embryonic muscle. Dev Biol. 2007;311:650–664. doi: 10.1016/j.ydbio.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 29.Park KH, Vincent I. Presymptomatic biochemical changes in hindlimb muscle of G93A human Cu/Zn superoxide dismutase 1 transgenic mouse model of amyotrophic lateral sclerosis. Biochim Biophys Acta. 2008;1782(7–8):462–468. doi: 10.1016/j.bbadis.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Da Cruz S, et al. Elevated PGC-1α activity sustains mitochondrial biogenesis and muscle function without extending survival in a mouse model of inherited ALS. Cell Metab. 2012;15(5):778–786. doi: 10.1016/j.cmet.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leick L, et al. PGC-1{alpha} is required for AICAR-induced expression of GLUT4 and mitochondrial proteins in mouse skeletal muscle. Am J Physiol Endocrinol Metab. 2010;299(3):E456–E465. doi: 10.1152/ajpendo.00648.2009. [DOI] [PubMed] [Google Scholar]
- 32.Suwa M, Nakao H, Kumagai S. Effects of chronic AICAR treatment on fiber composition, enzyme activity, UCP3, and PGC-1 in rat muscles. J Appl Physiol. 2003;95:960–968. doi: 10.1152/japplphysiol.00349.2003. [DOI] [PubMed] [Google Scholar]
- 33.Kennel PF, Finiels F, Revah F, Mallet J. Neuromuscular function impairment is not caused by motor neurone loss in FALS mice: an electromyographic study. Neuroreport. 1996;7(8):1427–1431. doi: 10.1097/00001756-199605310-00021. [DOI] [PubMed] [Google Scholar]
- 34.Merlie JP, Mudd J, cheng T-C, Olson EN. Myogenin and acetylcholine receptor α gene promoters mediate transcriptional regulation in response to motor innervation. J Biol Chem. 1994;269(4):2461–2467. [PubMed] [Google Scholar]
- 35.Prody CA, Merlie JP. A developmental and tissue-specific enhancer in the mouse skeletal muscle acetylcholine receptor α-subunit gene regulated by myogenic factors. J Biol Chem. 1991;33:22588–22596. [PubMed] [Google Scholar]
- 36.Gilmour BP, Fanger GR, Newton C, Evans SM, Gardener PD. Multiple binding sites for myogenic regulatory factors are required for expression of the acetylcholine receptor γ-subunit gene. J Biol Chem. 1991;266(30):19871–19874. [PubMed] [Google Scholar]
- 37.Piette J, Bessereau J-L, Huchet M, Changeaux J-P. Two adjacent myoD1-binding sites regulate expression of the acetylcholine receptor α-subunit gene. Nature. 1990;345:353–355. doi: 10.1038/345353a0. [DOI] [PubMed] [Google Scholar]
- 38.Williams AH, et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moresi V, et al. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Legerlotz K, Smith HK. Role of MyoD in denervated, disused and exercised muscle. Muscle Nerve. 2008;38:1087–1100. doi: 10.1002/mus.21087. [DOI] [PubMed] [Google Scholar]
- 41.Peters D, et al. Asynchronous functional, cellular and transcriptional changes after a bout of eccentric exercise in the rat. J Physiol. 2003;553(Pt 3):947–957. doi: 10.1113/jphysiol.2003.048462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flynn JM, Meadows E, Fiorotto M, Klein WH. Myogenin regulates exercise capacity and skeletal muscle metabolism in the adult mouse. PLoS One. 2010;5(10):e13535. doi: 10.1371/journal.pone.0013535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siu PM, Donley DA, Bryner RW, Alway SE. Myogenin and oxidative enzyme gene expression levels are elevated in rat soleus muscles after endurance training. J Appl Physiol. 2004;97(1):277–285. doi: 10.1152/japplphysiol.00534.2004. [DOI] [PubMed] [Google Scholar]
- 44.Liebetanz D, Hagermann K, von Lewinski F, Kahler E, Paulus W. Extensive exercise is not harmful in amyotrophic lateral sclerosis. Eur J Neurosci. 2004;20:3115–3120. doi: 10.1111/j.1460-9568.2004.03769.x. [DOI] [PubMed] [Google Scholar]
- 45.McCrate ME, Kaspar BK. Physical activity and neuroprotection in amyotrophic lateral sclerosis. Neuromol Med. 2008 doi: 10.1007/s12017-008-8030-5. [DOI] [PubMed] [Google Scholar]
- 46.Chen A, Montes J, Mitsumoto H. The role of exercise in amyotrophic lateral sclerosis. Phys Med Rehabil Clin N Am. 2008;19:545–557. doi: 10.1016/j.pmr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 47.Bello-Haas V, et al. A randomized controlled trial of resistance exercise in individuals with ALS. Neurology. 2007;68(23):2003–2007. doi: 10.1212/01.wnl.0000264418.92308.a4. [DOI] [PubMed] [Google Scholar]
- 48.Wicks P, et al. Three soccer playing friends with simultaneous amyotrophic lateral sclerosis. Amyotrophic Lateral Sclerosis. 2007;8:177–179. doi: 10.1080/17482960701195220. [DOI] [PubMed] [Google Scholar]
- 49.Chiò A, Benzi G, Dossena M, Mutani R, Mora G. Severely increased risk of amyotrophic lateral sclerosis among Italian professional football players. Brain. 2005;128:472–476. doi: 10.1093/brain/awh373. [DOI] [PubMed] [Google Scholar]
- 50.Carreras I, et al. Moderate exercise delays the motor performance decline in a transgenic model of ALS. Brain Res. 2009;1313:192–201. doi: 10.1016/j.brainres.2009.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kirkinezos IG, Hernandez D, Bradley WG, Moraes CT. Regular exercise is beneficial to a mouse model of amyotrophic lateal sclerosis. Ann Neurol. 2003;53:804–807. doi: 10.1002/ana.10597. [DOI] [PubMed] [Google Scholar]
- 52.Mahoney DJ, Rodriguez C, Devries M, Yasuda N, Tarnopolsky MA. Effects of high-intensity endurance exercise training in the G93A mouse model of amyotrophic lateral sclerosis. Muscle Nerve. 2004;29:656–662. doi: 10.1002/mus.20004. [DOI] [PubMed] [Google Scholar]
- 53.Kaspar BK, Llado J, Sherkat N, Rothstein JD, Gage FH. Retrograde viral delivery of IGF-1 prolongs survival in a mouse ALS model. Science. 2003;301:839–842. doi: 10.1126/science.1086137. [DOI] [PubMed] [Google Scholar]
- 54.Tureckova J, Wilson EM, Cappalonga JL, Rotwein P. Insulin-like growth factor-mediated muscle differentiation: collaboration between phosphatidylinositol 3-kinase-Akt-signaling pathways and myogenin. J Biol Chem. 2001;276(42):39264–39270. doi: 10.1074/jbc.M104991200. [DOI] [PubMed] [Google Scholar]
- 55.Sumitani S, Goya K, Testa JR, Kouhara H, Kasayama S. Akt1 and Akt2 differently regulate muscle creatine kinase and myogenin gene transcription in insulin-induced differentiation of C2C12 myoblasts. Endocrinology. 2002;143(3):820–828. doi: 10.1210/endo.143.3.8687. [DOI] [PubMed] [Google Scholar]
- 56.Boillée S, et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 57.Boillée S, Vande Velde C, Cleveland DW. ALS: a disease of motor neurons and their non-neuronal neighbors. Neuron. 2006;52(1):39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 58.Clement AM, et al. Wild-type nonneuronal cells extend survival of SOD1 mutant motor neurons in ALS mice. Science. 2003;302(5642):113–117. doi: 10.1126/science.1086071. [DOI] [PubMed] [Google Scholar]
- 59.Lobsinger CS, et al. Schwann cells expressing dismutase active mutant SOD1 unexpectedly slow disease progression in ALS mice. PNAS. 2009 doi: 10.1073/pnas.0813339106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lobsinger CS, Cleveland DW. Glial cells as intrinsic components of non-cell-autonomous neurodegenerative disease. Nat Neurosci. 2007;10(11):1355–1360. doi: 10.1038/nn1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.