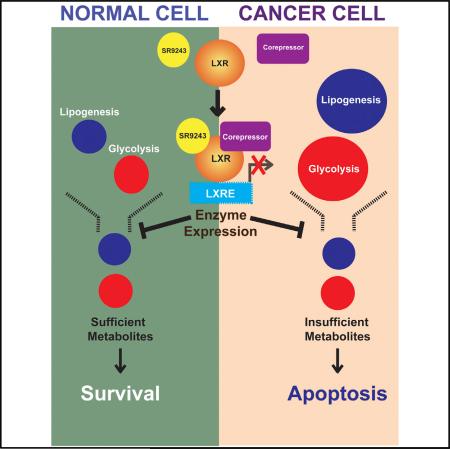
SUMMARY
Malignant cells exhibit aerobic glycolysis (the Warburg effect) and become dependent on de novo lipogenesis, which sustains rapid proliferation and resistance to cellular stress. The nuclear receptor liver-X-receptor (LXR) directly regulates expression of key glycolytic and lipogenic genes. To disrupt these oncogenic metabolism pathways, we designed an LXR inverse agonist SR9243 that induces LXR-corepressor interaction. In cancer cells, SR9243 significantly inhibited the Warburg effect and lipogenesis by reducing glycolytic and lipogenic gene expression. SR9243 induced apoptosis in tumors without inducing weight loss, hepatotoxicity, or inflammation. Our results suggest that LXR inverse agonists may be an effective cancer treatment approach.
Graphical abstract
INTRODUCTION
Metabolism in cancer cells is primarily glycolytic even when oxygen is abundant (Warburg et al., 1927). Aerobic glycolysis or the Warburg effect is well characterized and has been shown to be driven by mitochondrial defects, oncogenic stimuli, hypoxia, and aberrantly enhanced expression of glycolytic enzymes (De-Berardinis et al., 2008; Warburg et al., 1927; Yeung et al., 2008). In particular, elevated glycolytic gene expression is pervasive in cancers of the breast, colon, prostate, and lung. Oncogenes such as mTOR, c-MYC, and hypoxia-inducible factor 1 (HIF-1) promote glycolytic activity by upregulating expression of glycolytic enzymes including phosphofructokinase 1 (PFK1), hexokinases, and pyruvate dehydrogenase kinase-3 (Dang et al., 1997; Jung et al., 2011; Koshiji and Huang, 2004; Lu et al., 2008). In addition, expression of glycolysis-stimulating genes such as PFK1 and phosophofructokinase-2 (PFK2) are strongly associated with highly aggressive and drug-resistant tumor types (DeBerardinis et al., 2008; Phan et al., 2014). Conversely, the tumor suppressor P53 has been shown to block cancer cell growth by suppressing glucose consumption, preventing the downregulation of mitochondrial aerobic respiration, inhibiting NADPH production, and disrupting pentose phosphate synthesis (Yeung et al., 2008). Therefore, the Warburg effect is a central component of the metabolic reprogramming involved in cancer etiology.
Glycolysis is less energy efficient compared to aerobic respiration because it produces significantly fewer molecules of ATP. However, by providing a surplus of metabolic substrates for analplerosis that would be unavailable through normal aerobic respiration, the Warburg effect confers a selective survival advantage to cancer cells. Substrates produced are funneled into other metabolic pathways such as de novo lipid synthesis (lipogenesis), nucleotide production and amino acid synthesis, all of which are indispensable for rapid cancer cell growth. Lactate, produced in abundance in tumors, is instrumental in altering the intracellular redox balance, which promotes cancer cell invasiveness (Bonuccelli et al., 2010; Martinez-Outschoorn et al., 2011; Vander Heiden et al., 2009). Therefore, the Warburg effect functions as the metabolic foundation of oncogenic growth, tumor progression, and tumor resistance to treatment. Despite displaying elevated glycolytic gene expression, cancer cells within the tumor microenvironment can have distinct metabolic profiles depending on pH and oxygen availability (Dang, 2007; Fritz et al., 2010; Huang et al., 2012; Vander Heiden et al., 2009; Yeung et al., 2008). This metabolic plasticity allows cancer cells to evade cell death. Despite the variety of “druggable” targets identified, most glycolysis inhibitors show substantial toxicity in normal tissues and limited therapeutic applications in select cancer types (Pelicano et al., 2006).
The surplus glycolysis metabolites produced by the Warburg effect are integrated into lipogenesis and other metabolic pathways in tumor cells. Glycolysis products are used to synthesize short-, medium-, and long-chain fatty acids that are fundamental building blocks for cell membranes and organelles. Typically, cancer cells show elevated expression of lipogenesis enzymes and endogenous production of lipids, whereas normal cells obtain lipids primarily from exogenous sources (Vander Heiden et al., 2009). Like glycolysis, lipogenic enzyme expression is enhanced in tumors via oncogenic signaling. Although both pathways are linked, compared to tumor glycolysis, lipogenesis is not regulated by changes within the tumor microenvironment such as pH and the availability of oxygen (Blancher and Harris, 1998). Lipids are synthesized by enzymes such as fatty acid synthase (FASN), stearoyl-CoA desaturase (SCD1), and acetyl-CoA carboxylase-1 (ACC1) acting downstream of glycolysis. Lipo-genesis also facilitates immune system evasion and intercellular signaling that promote tumor growth (Phan et al., 2014). Lipid metabolites also provide valuable reducing power within the low nutrient and highly oxidative microenvironment of tumors (Carracedo et al., 2013; Zaytseva et al., 2012). Accordingly, lipogenic gene expression directly correlates with cancer aggressiveness, staging, and drug resistance (Notarnicola et al., 2006, 2012; Ogino et al., 2009; Zaytseva et al., 2012). Increased expression of FASN, SCD1, and ACC1 as well as the sterol-regulatory element binding protein-1c (SREBP1c), a transcription factor that regulates lipogenic gene expression, is associated with numerous forms of cancer (Furuta et al., 2010; Mason et al., 2012). Lipogenesis inhibitors that block FASN, ACC1, SCD1, and SREBP1c activity have been shown to reduce proliferation and induce apoptosis in cancer cells (Chajès et al., 2006; Mason et al., 2012; Notarnicola et al., 2006, 2012; Scaglia et al., 2009). However, clinically viable therapies that effectively block lipogenesis in vivo have not been forthcoming due to adverse side effects such as anorexia and severe weight loss (Clegg et al., 2002; Tu et al., 2005).
The liver-X-receptors, LXRα and LXRβ (NR1H3 and NR1H2, respectively) are nuclear receptors and key regulators of lipid, cholesterol, and carbohydrate metabolism and homeostasis (Kim et al., 2009; Laffitte et al., 2003; Wang et al., 2008; Zhao et al., 2012). LXRβ is ubiquitously expressed, whereas LXRα is expressed in macrophages, liver, adipose, adrenal, intestinal, and lung tissue. Both isoforms form obligate heterodimers with the retinoid-X-receptor (RXR) and bind to endogenous agonists such as the oxysterols 22(R)-hydroxycholesterol and 24(S)-hydroxycholesterol (Baranowski, 2008). LXRs regulate gene expression by directly binding to LXR-responsive elements (LXREs) within the promoter region of LXR-regulated genes. Unliganded LXRs selectively recruit corepressors such as nuclear corepressor 1 and 2 (NCoR1 and SMRT) to form repressor complexes at LXR-target gene promoters (Phelan et al., 2008; Wagner et al., 2003). Through this mechanism, LXRs silence target-gene expression in the absence of ligand activation. Conversely, LXR agonist binding induces dissociation of corepressor complexes and recruitment of LXR coactivators such as thyroid hormone receptor-associated protein (TRAP220/DRIP-2). Coactivator recruitment by ligand-activated LXRs initiates transcription of LXR target genes such as glycolysis enzymes; PFK2 and GCK1 (Kim et al., 2009; Zhao et al., 2012) and lipogenesis genes; SREBP1c, FASN, and SCD1 (Darimont et al., 2006; Joseph et al., 2002; Zhang et al., 2006). Apart from their role in glycolysis and lipogenesis gene regulation, LXRs are also known to attenuate immune function as evidenced by LXR aberrant inflammatory signaling in LXR knockout mice (Jamroz-Wiśniewska et al., 2007; Wójcicka et al., 2007). Moreover, LXR activation stimulates cholesterol efflux via stimulation of activation of ABC transporters (Beyea et al., 2007; Grefhorst et al., 2002). Therefore, LXR has been the focus of a number of studies aimed at developing cholesterol lowering drugs and treatments for atherosclerosis. Unfortunately, LXR agonists are known to promote hepatic steatosis due to enhanced hepatic lipid synthesis, which limits the potential use of LXR agonists as anti-artherogenic drugs in the clinic (Grefhorst et al., 2002; Viennois et al., 2012).
Recent studies have highlighted the emerging role of LXR in cancer metabolism, progression, and immune evasion (Russo, 2011; Villablanca et al., 2010). LXR agonists have been demonstrated to significantly lower intracellular cholesterol levels in cancer cells and therefore exhibit anti-neoplastic activity (Chuu and Lin, 2010; Rough et al., 2010). As a result, LXR agonists have been extensively investigated as pre-clinical anti-cancer drugs. In contrast, tumor cells have been shown to secrete LXR agonists that promote tumor immune evasion and survival (Russo, 2011; Villablanca et al., 2010). Similarly, inhibition of LXR activity also stimulated dendritic cell-mediated tumor cell clearance, enhanced tumor rejection, and prevented tumor recurrence in mice (Jamroz-Wiśniewska et al., 2007; Russo, 2011; Villablanca et al., 2010). Furthermore, other investigations suggest that synthetic LXR agonists may be somewhat antagonistic to chemotherapy treatment (Miller et al., 2011). LXR agonists have been extensively investigated as anti-cancer agents despite the deleterious side effects. However, targeted inhibition of LXR activity to disrupt cancer growth has been left unexplored.
Because LXR is a key regulator of glycolysis and lipogenesis, enzymes that mediated the Warburg effect and tumor lipogenesis respectively, we decided to target LXR to disrupt cancer cell growth. We designed an LXR inverse agonist, SR9243, to lower the basal transcriptional activity of LXRs and promote suppression of the Warburg effect and lipogenesis. We hypothesized that such a compound would have broad based anti-cancer therapeutic activity and this is investigated in this study.
RESULTS
SR9243 Inhibits LXR Activation by Enhancing LXR-Corepressor Recruitment
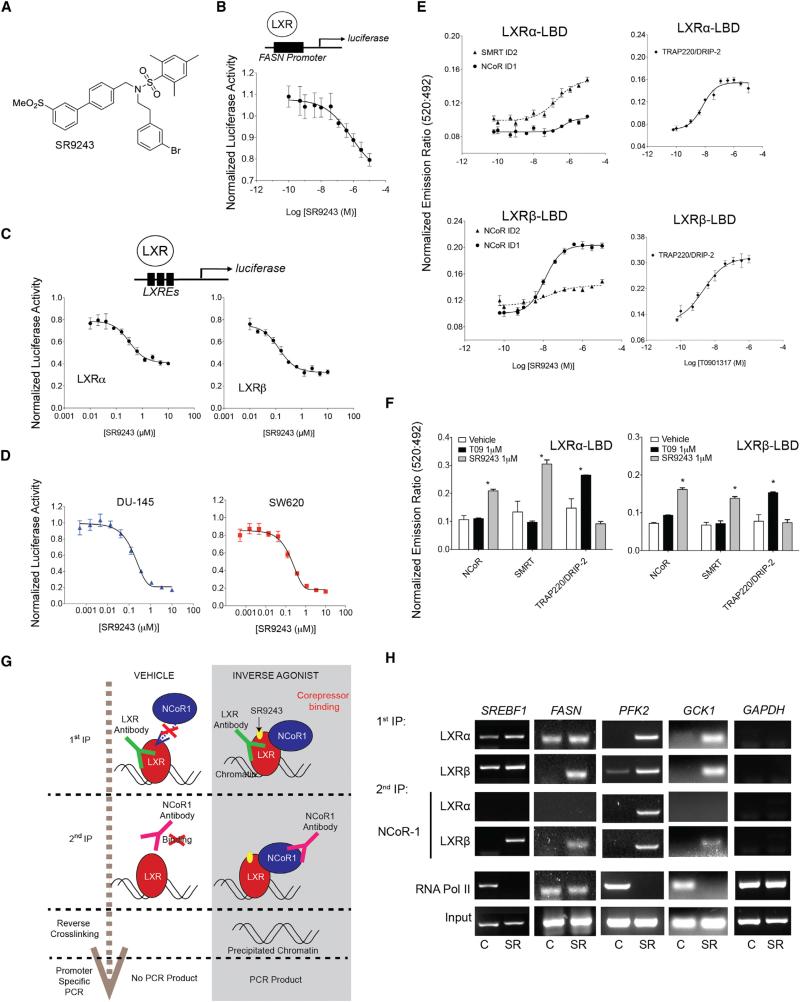
With the goal of targeting the Warburg effect and lipogenesis in cancer cells, we developed an LXR inverse agonist SR9243 (Figure 1A) that specifically targets LXR and downregulates LXR-mediated gene expression to below basal levels (Figures 1B and 1C). SR9243 was designed based on another LXR inverse agonist, SR9238, which we recently described as a “liver-selective” LXR inverse agonist (Griffett et al., 2013). SR9238 contains a rapidly metabolized ester moiety and SR9243 was designed to provide systemic exposure. SR9243 dose-dependently suppressed LXRα- and LXRβ-dependent transcription at nanomolar concentrations (Figures 1B and 1C) in both consensus LXRE-and endogenous (FASN) promoter driven luciferase reporter-assays. In addition, SR9243 potently inhibited LXR-driven luciferase activity in cultured cancer cells (Figure 1D). SR9243 displayed high selectivity for LXR as it failed to significantly influence the activity of any other nuclear receptors (Figure S1A) at 10 μM concentration: a maximally efficacious dose (Figures 1B–1D). Time-resolved fluorescence resonance energy transfer assays (TR-FRET) revealed that LXR interaction with the receptor interaction domains from the nuclear receptor corepressor 1 (NCOR1) and nuclear receptor corepressor 2 (SMRT) was enhanced by SR9243 (Figures 1E and 1F). Interestingly, LXRα ligand binding domains displayed higher affinity for SMRT core-pressor motifs compared to that of NCOR1, which suggests that LXRα may preferentially bind a distinct subset of corepressors compared to LXRβ, which preferred NCOR1 motifs. In contrast to the LXR agonist T0901317, SR9243 failed to enhance recruitment of the LXR coactivators TRAP220/DRIP-2 (Figure 1F). Sequential chromatin immunoprecipitation (Re-ChIP) of LXRα or LXRβ and NCoR1 (Figures 1G, 1H, and S1B–S1D) showed that SR9243 enhanced LXR recruitment of the transcriptional corepressor NCoR1 to the promoters of the LXR-regulated genes SREBF1, FASN, PFK2, and GCK1 (Figures 1H, S1B, and S1C). Interestingly, LXRα and LXRβ isoforms displayed divergent promoter binding characteristics in untreated and SR9243 treated cells. LXRα and LXRβ were bound at the SREBPF promoter in the absence of ligand treatment (Figure 1H). However, unliganded LXRα and LXRβ exclusively occupied the FASN and PFK2 promoters, respectively. Differential promoter occupancy by inactive LXRα and LXRβ has been shown previously and may be responsible for divergent roles each LXR isoform has in metabolism (Wagner et al., 2003). LXRα and β were enriched at FASN, SREBPF, PFK2, and GCK1 promoters upon exposure to SR9243, but not at the GAPDH promoter, suggesting that SR9243 selectively stimulated LXR binding to LXREs (Figures 1H and S1B–S1D). Interestingly, NCoR1 was recruited by LXRβ to the SREBF, FASN, PFK2, and GCK1 promoters, whereas LXRα interacted with NCOR1 at the PFK2 promoter only. In tandem with our results in TR-FRET assays (Figure 1E), this observation suggests that LXRα and LXRβ isoforms differentially recruit corepressors depending on the target promoter context (Figure 1H). Alternatively, these results suggest that LXRβ may be the dominant isoform involved in mediating SR9243-driven gene suppression. Collectively these observations demonstrate that SR9243 induces corepressor recruitment to LXRs at target-gene promoters, leading to suppression of gene expression.
Figure 1. SR9243 Is an LXR Inverse Agonist that Induces Corepressor Recruitment.
(A) Structure of SR9243.
(B) FASN promoter-driven luciferase reporter assay showing SR9243 repression of the basal activity of exogenously expressed full-length LXRα or LXRβ in HEK293 cells. Transfected cells were treated with 0.01–10 μM of SR9243 for 6 hr.
(C) LXRE-driven (3X) luciferase reporter assay showing SR9243 repression of basal activity of endogenously expressed LXRs in HEK293 cells. Cells were treated with 1 nM–10 μM SR9243 for 6 hr.
(D) LXRE-driven luciferase reporter dose-response assays in cancer cell lines expressing exogenous LXRα or LXRβ and treated with SR9243.
(E) TR-FRET assay showing the recruitment of corepressor box peptides NCoR and SMRT to LXRα and LXRβ in response to SR9243.
(F) TR-FRET assay showing relative recruitment of NCoR, SMRT, and TRAP220/DRIP-2 box peptides to LXRα or LXRβ LBDs in response to vehicle of SR9243 or 1 μM of the LXR agonist T0901317.
(G) Diagram illustrating the Re-ChIP experiment shown in (H) PCR results of Re-ChIP assays of LXRα or LXRβ followed by NCOR1 showing SR9243 (SR)-induced corepressor recruitment at the promoters of GCK1, PFK2, SREBF, and FASN. (C, control; SR, SR9243) Representative figure of experiment repeated three times. *p < 0.05, **p < 0.01, ***p < 0.001. Error bars represent ±SEM. See also Figure S1.
SR9243 Reduces Cancer Cell Viability and Induces Apoptotic Cell Death
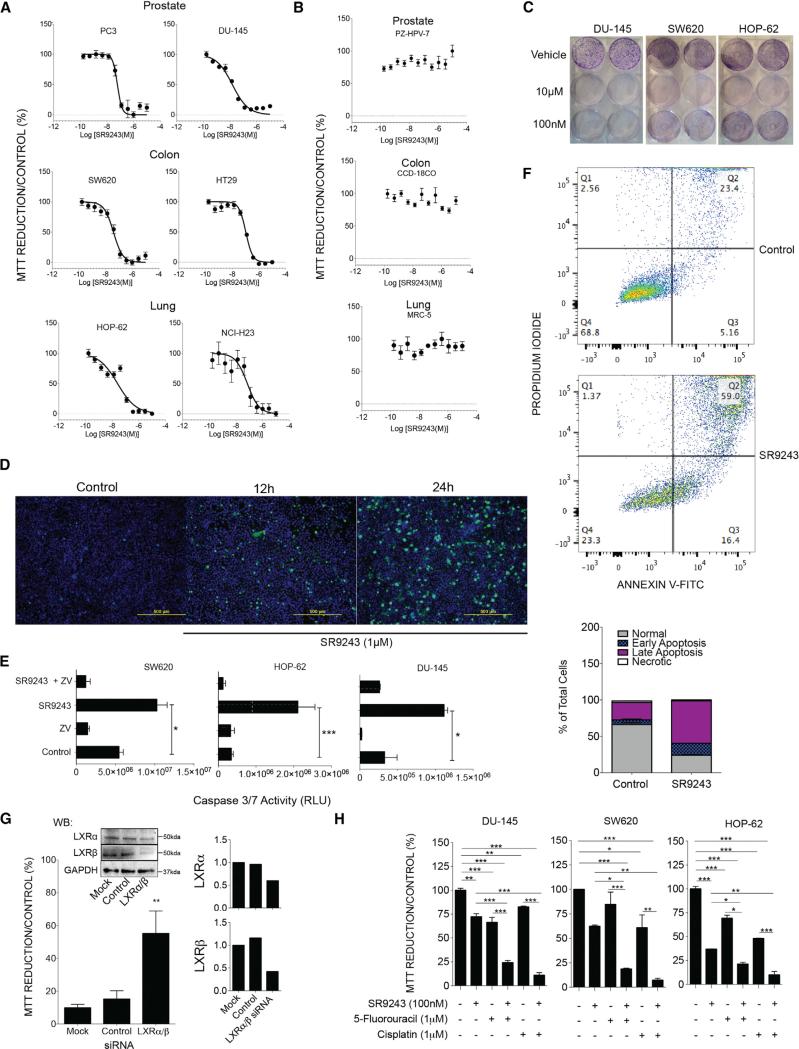
Most cancer cells are highly dependent on the Warburg effect and are often forced into apoptotic cell death when subjected to glycolytic blockade. First, to determine the efficacy of SR9243 as an anti-tumor agent, we assessed the effect of SR9243 on cancer cell viability in a variety of cancer cell types. SR9243 potently reduced cancer cell viability at nanomolar concentrations (half-maximal inhibitory concentration [IC50] ~15–104 nM) in MTT reduction assays in prostate (PC3 and!DU-145), colorectal (SW620 and HT29), and lung (HOP-62 and NCI-H23) cancer cell lines (Table 1 and Figure 2A). Conversely, SR9243 did not reduce the viability of non-malignant cells that originate from the same organs—prostate (PZ-HPV-7), colon (CCD-18Co), and lung (MRC-5)—which express similar levels of LXRs (Figures 2B and S2A). The colony-forming capacity of cancer cells was also significantly lowered by SR9243 in a dose-dependent manner (Figure 2C). Cancer cell death occurred as early as 12 hr following SR9243 treatment (Figure 2D), which coincided with a robust increase in caspase-3/7 activation (Figure 2E) and induction of apoptotic cell death (Figure 2F). LXRs have been shown to modulate cell cycle regulation genes, and LXR agonists have been shown to inhibit proliferation by blocking cell cycle progression (Meng et al., 2009; Nguyen-Vu et al., 2013; Vedin et al., 2012). However, SR9243-treated cells showed no significant changes in the expression a number of key cell cycle arrest genes:P21, P15, P27, MDM2, P52, and GADD45 (Figure S2B); or cell cycle progression genes: CCNE, CDC25, CCNB1, CCND1, and CCNA2 (Figure S2C), suggesting that SR9243-induced apoptotic cell death was not due to LXR-mediated cell cycle arrest. Like the inverse agonist SR9243, the LXR agonist (GW3965) has been shown to inhibit cancer cell growth. However, GW3965 as well as other LXR agonists mediate their anti-neoplastic effects via reduction of intracellular cholesterol levels and induction of cell cycle arrest (Lo Sasso et al., 2013; Nguyen-Vu et al., 2013; Vedin et al., 2012). Co-treatment of cancer cells with GW3965 and SR9243 shows reciprocal modulation of LXR activity (Figure S2D). In addition, GW3965 dose-dependently reduced the toxicity of SR9243 in cancer cells, demonstrating that both types of ligands target the LXR and modulate LXR activity in quantifiably opposite directions (Figure S2E). This shows that LXR agonists and inverse agonists disrupt cancer cell growth by modulating LXR gene regulated pathways through distinct mechanisms.
Table 1.
Calculated IC50 of SR9243 on the Viability of Cancer Cells from Different Tissue Origins
| Cancer Cell Line | Tissue | SR9243 IC50 (nM) |
|---|---|---|
| SW620 | colon | 40 |
| HT-29 | colon | 104 |
| DU-145 | prostate | 15 |
| PC-3 | prostate | 61 |
| NCI-H23 | lung | 72 |
| HOP-62 | lung | 26 |
| MIAPACA-2 | pancreatic | 18 |
Figure 2. SR9243 Induces Apoptotic Cell Death in Multiple Cancer Cell Types in an LXR-Dependent Manner.
(A) MTT assay showing the viability of cancer cells from the prostate (PC3, DU-145), colon (SW620, HT-29), and lung (HOP-62, NCI-H23) treated with increasing doses of SR9243.
(B) MTT assay of non-malignant cells from the prostate (PZ-HPV-7), colon (CCD-18Co), and lung (MRC5) treated with SR9243.
(C) Colony formation assay of cancer cells treated with vehicle, or 100 nM or 10 μM SR9243.
(D) Fluorescent cell viability/apoptosis assay showing the viable cells (blue) and or the apoptotic cells (green) from SW620 cells treated with 1 μM SR9243 for 12 and 24 hr.
(E) Luminescence-based caspase 3/7 activity of SW620, HOP62, and DU-145 cells treated with vehicle, 1 μM SR9243, and or the caspase inhibitor (Z-VAD-FMK) for 24 hr.
(F) FACs sorting of Annexin V FITC/PI-stained SW620 cells treated with SR9243 (100 nM) for 24 hr. The percentage of apoptotic, necrotic and live cell populations are graphically represented in lower image.
(G) MTT reduction assay showing rescue of cell viability of SW620 colon cancer cells transfected with LXRα and LXRβ-specific siRNAs compared to control or mock transfected cells. LXR expression in SW620 cells treated with LXR siRNAs (right).
(H) MTT reduction assay showing the viability of DU-145, SW620, and HOP-62 cancer cells in response to SR9243 alone or in combination with 5′-fluorouracil or cisplatin. Experiments were repeated three times. *p < 0.05, **p < 0.01, ***p < 0.001. Error bars represent ±SEM. See also Figure S2.
To determine whether cancer cell death and gene suppression by SR9243 was LXR dependent, we decided to knock down the expression of LXR in cancer cells using LXRα and β specific siRNAs. We postulated that if the activity of SR9243 is LXR-dependent, depleting LXR levels through siRNA knockdown would diminish the anti-neoplastic and gene suppressive effects of SR9243. Indeed, downregulation of LXR expression reduced SR9243-mediated repression of LXR target genes (Figure S2F) and rescued cancer cell viability in SR9243-treated cells (Figure 2G). Collectively, these results demonstrate that the effects of SR9243 on cancer cell viability are LXR dependent and non-toxic to normal cells.
SR9243 Sensitizes Cancer Cells to Chemotherapeutic Treatments
As SR9243 effectively reduced cancer cell viability, we investigated whether SR9243 could be a complementary treatment to cytotoxic chemotherapeutic drugs when used in combination. SR9243 treatment profoundly enhanced the efficacy of 5′-fluorouracil or cisplatin in all cancer cells tested (Figure 2H). This collectively highlights that SR9243 could be a viable cancer treatment approach either administered alone or as a part of combination therapy.
SR9243 Disrupts the Warburg Effect in Cancer Cells
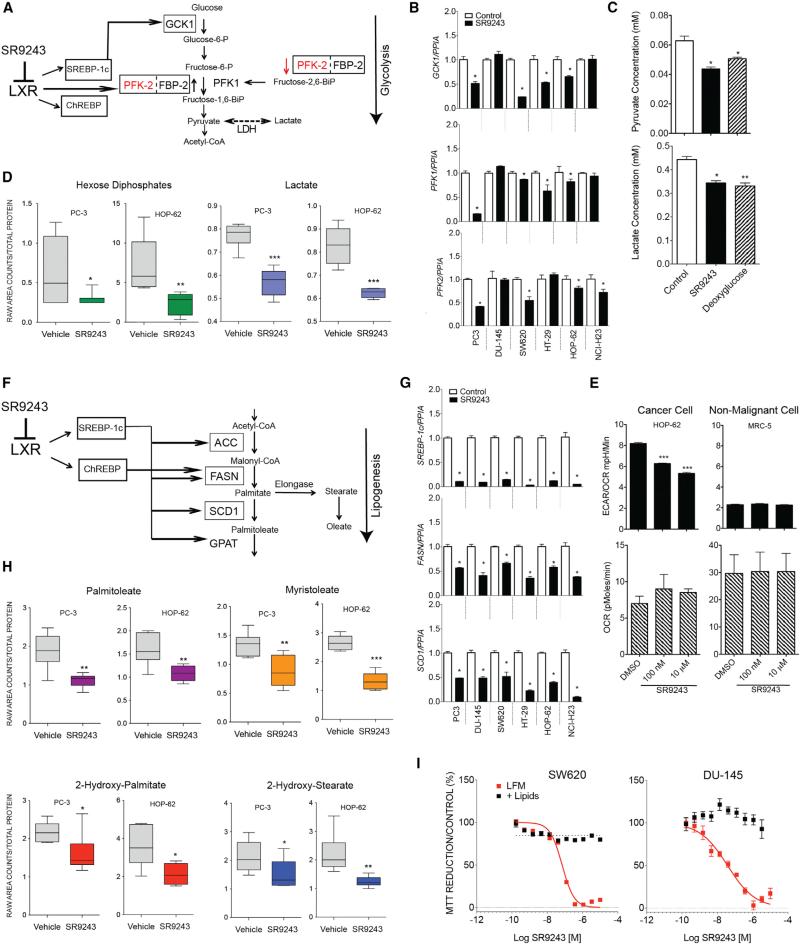
LXR directly regulates multiple components of the glycolysis pathway (Figure 3A). Considering the potent selective effect SR9243 has on cancer cell viability, we sought to investigate the effects SR923 on LXR-regulated gene expression specifically in cancer cells. First, we assessed whether SR9243 suppressed expression of enzymes that are known to mediate the Warburg effect in cancer cells. Cells treated with SR9243 showed significantly suppressed glycolytic gene expression (GCK1, PFK2, PFK1, and LDH) (Figures 3B and S3A). To truly inhibit the Warburg effect in cancer cells, SR9243 must reduce the intracellular concentrations of glycolytic metabolites. Biochemical assessment and gas chromatography/mass spectrometry (GC/MS) analysis revealed that intracellular levels of pyruvate, hexose phosphates, and lactate were significantly reduced by SR9243 (Figures 3C and 3D). Notably, SR9243 reduced the levels of lactate and pyruvate to that of cells treated with 2-deoxyglucose (Figure 3D), a potent hexokinase inhibitor that has reached phase II clinical cancer-drug trials (Mohanti et al., 1996). These results suggest that SR9243 mediated a substantial decrease in glycolytic enzyme expression that significantly affected glycolytic output in cancer cells.
Figure 3. SR9243 Inhibits Glycolytic Lipogenic Enzymes in Cancer Cells.
(A) Schematic showing glycolysis genes regulated by LXRs and associated metabolites. Gene names outlined in boxes are directly regulated by LXRs.
(B) RT-PCR showing expression of LXR-regulated glycolysis genes GCK1, PFK1, and PFK2 after 6 hr treatment with SR9243 (10 μM).
(C) Biochemically determined cellular levels of lactate and pyruvate in HOP-62 cancer cells treated with SR9243 for 24 hr.
(D) GC/MS determined levels of lactate and hexose phosphates in PC3 and HOP62 cancer cells.
(E) Relative glycolytic rate (ECAR/OCR) and oxygen consumption rate (OCR) of HOP-62 lung cancer cells or the corresponding non-tumorigenic lung epithelial cells (MRC5) treated with SR9243 (100 nM or 10 μM). Cells were assessed for extracellular acidification rate and OCR using the Seahorse XF extracellular flux analyzer.
(F) Schematic showing the lipogenesis genes regulated by LXR and their cognate substrates and products. RT-PCR showing expression profile of LXR-regulated lipogenesis genes SREBP1c, FASN, and SCD1 in cancer cells treated with SR9243 (10 μM) for 6 hr.
(G and H) (G) GC/MS showing cellular levels of the long-chain fatty acids palmitoleate and myristoleate in SR9243 treated cancer cells and (H) the short-chain fatty acids (2-hydroxy-palmitate and 2-hydroxy-stearate) in SW620 and HOP-62 cells treated with SR9243. Samples were normalized by total protein concentration.
(I) MTT assay of DU-145 and SW620 cells treated with increasing amounts of SR9243 in LFM (lipid-free media) or positive lipid media (supplemented with 25 nM oleate, palmitate, and stearate) for 96 hr. Data were analyzed using Welch's t tests and/or Wilcoxon's rank sum tests. *p < 0.05, *p < 0.05, **p < 0.01, ***p < 0.001. Error bars represent ±SEM. See also Figure S3.
SR9243 is Non-toxic to Normal Cells
Normal cells require glycolysis for energy production. Therefore glycolytic enzyme inhibitors often produce undesired toxic effects in non-tumor tissues because they also inadvertently disrupt the activity of glycolysis enzymes required for sustaining respiration in normal cells. Whether SR9243 truly had a selectively pernicious effect on the elevated rate of glycolysis in cancer cells was uncertain. SR9243 did not suppress glycolytic gene expression or glycolysis metabolite concentrations in non-malignant cells (Figures S3B and S3C). To further investigate this selective effect of SR9243 on cancer cell glycolytic output, the extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) of cancer cells and non-malignant cells treated with SR9243 was measured. As expected, due to the Warburg effect, the cancer cells assayed displayed a much higher ECAR compared to normal cells. Interestingly, SR9243 treatment selectively reduced the relatively high glycolytic output of cancer cells to levels comparable to that of non-malignant cells (Figure 3E). Notably, SR9243 did not significantly suppress glycolysis in non-malignant cells (Figure 3E). In addition, SR9243 failed to significantly disrupt OCR in both cancer cells and non-malignant cells (Figure 3E, bottom). These results were consistent with the selective effect SR9243 had on cancer cell viability compared to the innocuous effect SR9243 had on non-malignant cells (Figures 2A and 2B). Further, these observations suggest that SR9243-mediated glycolytic gene suppression preferentially disrupts elevated glycolytic output in cancer cells without adversely affecting respiration in normal cells.
SR9243 Suppresses Lipogenesis Gene Expression and Lipid Production in Cancer Cells
Similar to LXR-regulated glycolysis genes, SR9243 repressed the lipogenesis genes directly regulated by LXR: FASN, SREBP1-c, and SCD1 (Figures 3F and 3G). SR9243 substantially disrupted lipogenic gene expression in all the cancer cell lines tested (Figure 3F). In particular, NCI-H23 and DU-145 cells showed potent reduction in FASN, SREBP1c, and SCD1 expression in contrast to the modest effect on glycolysis enzyme expression observed in these cell lines (Figures 3B and 3F; Figure S3A). SR9243 reduced the expression of the lipogenic genes FASN and SCD1 (Figure S3D) and intriguingly did not inhibit the expression of SREBP1c in normal cell lines in contrast to the profound effect on SREBP1c expression observed in cancer cells (Figures S3D and 3G). SR9243 reduced expression of the cholesterol transport gene ABCA1 in both normal and malignant cell lines (Figures S3E and S3F), which suggested that SR9243 selective activity was not due to enhanced cholesterol export. Within cancer cells the intracellular levels of major end products of lipogenesis—palmitate, stearate, palmitoleate, and myristoleate (Figure 3H)—were significantly reduced by SR9243. Select fatty acids like oleate have been shown to rescue cell viability in cancer cells treated with lipogenesis enzyme-specific inhibitors (e.g., TOFA), which target upstream lipogenesis enzymes like SCD1 (Mason et al., 2012). Interestingly culture media supplemented with individual fatty acids, such as oleate, stearate, and palmitate, could not significantly rescue the viability of SR9243-treated cancer cells (Figure S3G). Further, combined supplementation of cancer cell media with oleate, stearate, and palmitate in combination completely rescued cancer cell viability in cancer cells (Figure 3I). This suggests that multiple points in the lipogenesis pathway are targeted by SR9243 treatment. Fatty acid supplementation also rescued the viability of SW620 cells in which glycolysis was substantially disrupted (Figure 3I). This observation is supported by previous studies that have similarly shown that lipid uptake by cancer cells can temporarily sustain cancer cell viability when energy substrates are limited (Carracedo et al., 2013; Dang et al., 1997). As discussed previously, non-malignant cells are not typically dependent on lipogenesis for normal function. Therefore, these findings suggest that SR9243 is a potent inhibitor of lipogenic gene expression that selectively kills cancer cells by depleting intracellular lipids. Based on these results, it is predicted therefore that SR9243 should display therapeutic activity in both highly glycolytic and or lipogenic tumor cell subtypes.
SR9243 Inhibits LXR-Dependent Gene Expression and Tumor Growth In Vivo
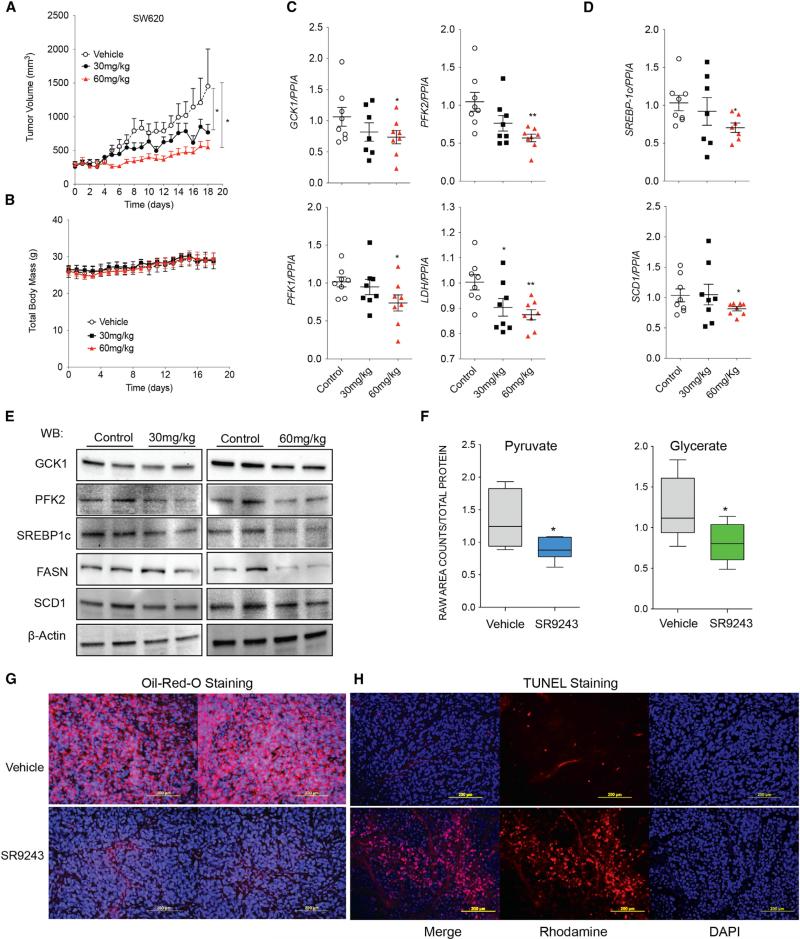
To examine whether SR9243 could elicit a similarly potent down-regulation of LXR activity in vivo, we first tested the ability of SR9243 to inhibit lipogenesis in mice. To accomplish this, we test the effects of SR9243 on Ob/Ob mice fed a high-fat diet, an established mouse model of elevated lipogenic signaling and enzyme expression. In this mouse model, SR9243 was able to suppress hepatic steatosis in vivo (Figure S4A). We then went on to test the anti-cancer properties of SR9243 in xenograft cancer models to determine if SR9243 could impede tumor growth in vivo. Colon cancer tumor xenograft growth (SW620) was substantially reduced by SR9243 treatment (Figure 4A) in a dose-dependent manner. A number of lipogenesis inhibitors have been tested in vivo that promote rapid weight loss and loss of appetite, making them unviable clinical therapies (Clegg et al., 2002; Tu et al., 2005). However, SR9243-treated tumor-bearing mice did not display any weight loss (Figure 4B). Notably, SR9243 treatment did not significantly modulate LXR receptor levels (Figures S4B and S4C). SR9243 also significantly and dose-dependently reduced glycolytic (GCK1, PFK2, PFK1, and LDH; Figures 4C and 4E) and lipogenic (SCD1, FASN, and SREBP1c) enzyme expression in colon tumor xenografts (Figures 4D and 4E). In addition, GC/MS analysis showed that treated mice had reduced levels of the key metabolite markers of glycolysis (pyruvate and glycerate; Figure 4F) and a substantial reduction in total tumor lipid content (Figure 4G). In situ TUNEL assays also revealed that SR9243-treated tumors contained significantly more apoptotic cells (Figure 4H). These results indicate that SR9243 was able to profoundly inhibit tumor glycolysis, lipogenesis, and induce apoptotic cancer cell death without promoting weight loss in vivo. Further, these results suggest that LXR inverse agonists unlike previous lipogenesis inhibitors may not adversely affect lipid homeostasis or food intake.
Figure 4. SR9243 Reduces Tumor Growth, Glycolytic and Lipogenic Enzyme Expression in Tumors In Vivo.
(A) Volume of SW620 colon cancer xenografts in athymic mice treated with vehicle (n = 9), 30 mg/kg (n = 9), or 60 mg/kg (n = 9) SR9243.
(B) Total body weight of mice from (A).
(C and D) RT-PCR analysis of the (C) glycolytic genes GCK1, PFK1, and PFK2 and (D) the lipogenesis genes SREBP1c and SCD1 in tumors of mice treated with 30 and 60 mg/kg SR9243. RT-PCR data were analyzed using Student's t test. *p < 0.05.
(E) Immunoblot showing expression levels of SREBP1c, FASN, SCD1, GCK1, and PFK2 protein in SW620 tumors treated with 30 or 60 mg/kg SR9243.
(F) GC/MS determined concentration of pyruvate and glycerate in SW620 xenografts in mice treated with 30 mg/kg SR9243. Data were analyzed using Welch's t tests and/or Wilcoxon's rank sum tests. *p < 0.05. Error bars represent ±SEM.
(G) Oil red O staining showing lipid content of SW620 tumors in mice treated with 60 mg/kg SR9243 or vehicle.
(H) Fluorescence based in situ TUNEL assay showing apoptotic cells in tumors from mice treated with SR9243 (60 mg/kg) or vehicle control. See also Figure S4.
SR9243-Mediated Lipogenic Enzyme Expression Is Sufficient for Inhibition of Tumor Growth In Vivo
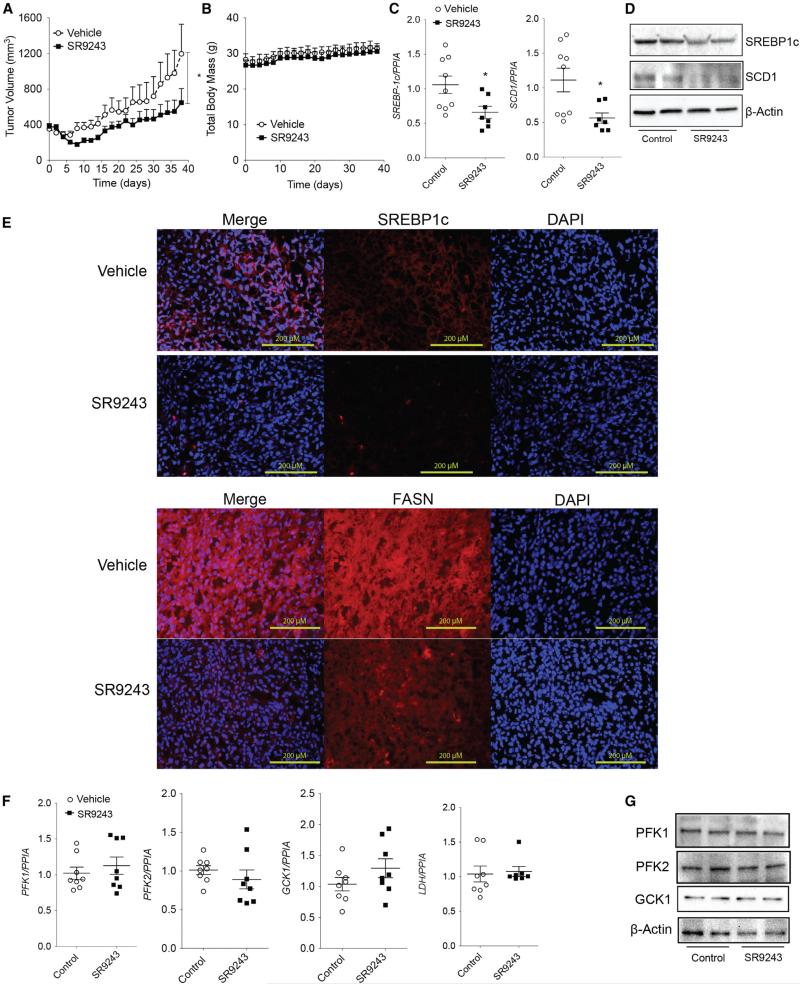
As mentioned earlier, cancer cells within the tumor microenvironment can display metabolic plasticity and temporarily reduced dependence on glycolytic metabolite production depending on pH and oxygen availability. In tumor cells, glycolytic metabolites are funneled into lipogenic pathways where they are converted into short-, medium-, and long-chain fatty acids that can be incorporated into plasma cell and organelle membranes or used as signaling molecules. Unlike glycolysis, tumor lipogenesis is less dependent on tumor microenvironment changes. Therefore, effectively blocking both glycolytic and lipogenic pathways exploited by cancer cells may be especially useful for targeting tumor cell types with distinct metabolic profiles. We observed that SR9243 dually inhibited glycolytic and lipogenic enzyme expression in vitro and in vivo. Interestingly, SR9243 failed to reduce the expression of multiple glycolytic enzymes. Specifically in DU-145 and NCI-H23 cells (Figures 3B and S3A), only LDH and PFK2 expression were modestly reduced. These mild effects on glycolysis gene expression did not fully correlate with the significant disruption of cell growth observed in these cell lines (Figure 2A). Considering this, we tested whether SR9243 could similarly ablate DU-145 tumor xenograft growth in vivo without substantially inhibiting glycolytic gene expression. SR9243 potently reduced DU-145 tumor growth (Figure 5A). As observed in colon cancer tumor xenograft models, SR9243 did not cause any reduction in total body weight (Figure 5B). In these tumors SREBP-1c, SCD1, and FASN expression was markedly suppressed (Figures 5C–5E). Consistent with our observations in vitro, SR9243 did not reduce the expression of GCK1, PFK2, or PFK1 (Figure 5F). Interestingly, SR9243 also had no effect on LDH expression levels in DU-145 tumors as observed in cultured cells (Figures 5F and S3A). In addition, SR9243 did not influence pyruvate or lactic acid levels within tumors (Figure S5B). However, SR9243 significantly reduced DU-145 fatty acid metabolite content (Figure S5B). Therefore, these results demonstrated that SR9243 inhibits tumor growth without profoundly repressing glycolytic gene expression. This implies that because of its dual-pathway activity, SR9243 should have efficacy in cancer cells that display variable glycolytic activity within the metabolically heterogeneous tumor microenvironment.
Figure 5. SR9243 Treatment Inhibits Lipogenic Enzyme Expression to Suppress Tumor Growth.
(A) Volume of prostate cancer (DU-145)-xenografts in nude mice treated with 60 mg/kg SR9243 (n = 8) or vehicle (n = 7).
(B) Total body weight of mice treated in (A).
(C) RT-PCR showing expression of SREBP1c and SCD1 in tumors from mice treated with SR9243 or vehicle. Data were analyzed using Student's t test followed by Bonferroni post-test. *p < 0.05. Error bars represent ±SEM).
(D) Immunoblot showing protein expression of SREBP1c and SCD1 in control and SR9243-treated tumors.
(E) Fluorescence-based immunohistochemistry SREBP1c and FASN expression in tumors from vehicle and SR9243-treated mice.
(F) RT-PCR showing expression of PFK1, PFK2, GCK1, and LDH in tumors treated with SR9243 or control. Data were analyzed using Student's t test followed by Bonferroni post-test. *p < 0.05.
(G) Immunoblot showing protein expression of SREBP1c and SCD1 in control and SR9243-treated tumors and SCD1 in tumors from mice treated with SR9243 or vehicle. See also Figure S5.
SR9243 Blocks Tumor Growth without Causing Immune or Hepatic Toxicity In Vivo
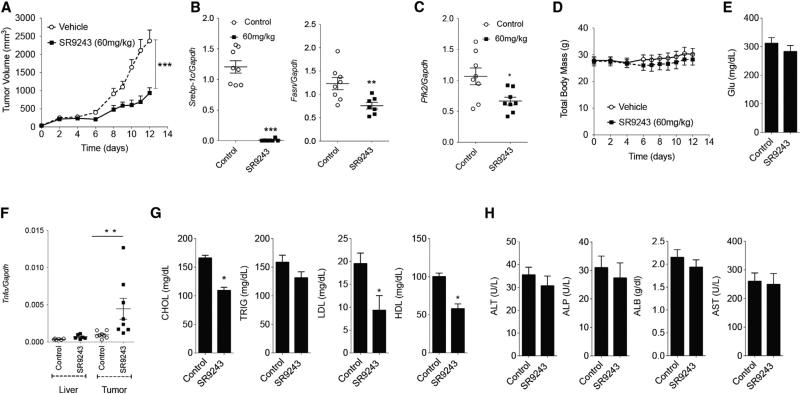
We have shown previously that long-term treatment with an LXR inverse agonist did not enhance inflammatory cytokines in the liver and inhibited liver inflammation in a mouse model of fatty liver disease (Griffett et al., 2013). We assessed whether administration of SR9243 to immune competent (C57BL6J) tumor-bearing mice stimulated inflammation or hepatic toxicity. Similar to the effect of SR9243 on tumor growth in nude mice, SR9243 profoundly inhibited Lewis lung carcinoma (LLC1) tumor growth in C57BL6J mice (Figure 6A). LXR-regulated lipogenic and glycolytic gene expression was also substantially reduced (Figures 6B and 6C) without promoting weight loss (Figure 6D). Biochemical profiling of mouse plasma revealed that plasma glucose levels was unchanged (Figure 6E). Interestingly, SR9243 also significantly increased Tnfα levels within tumors, without elevating expression levels of Tnfα, IL6, or IL5 in the liver (Figures 6F, S6A, and S6B). Thus, immune activation in SR9243-treated mice was isolated to tumors and relatively absent in normal tissue. Previous studies have shown that tumors secrete LXR agonists that suppress dendritic cell activation and promote immune silencing or tumor “masking.” This tumor-specific elevation of Tnfα suggests that SR9243 may be inducing tumor “unmasking” in treated mice. SR9243 also reduced total cholesterol and significantly reduced both low-density and high-density lipoprotein (LDL and HDL) plasma levels (Figure 6G) in SR9243-treated mice, consistent with previous observations in mice treated with LXR inverse agonists (Griffett et al., 2013). SR9243 treatments mediated these effects without inducing liver toxicity markers (Figure 6H). Together, these results indicate that SR9243 inhibits tumor lipogenesis and glycolysis and induces apoptotic cancer cell death with no evidence of hepatic toxicity or pro-inflammatory effects.
Figure 6. SR9243 Inhibits Tumor Growth and Lipogenesis without Hepatotoxicity or Inflammation.
(A) Volume of mouse Lewis lung carcinoma (LLC1) syngeneic tumors implanted in C57BL6/J mice treated with vehicle (n = 8) or 60 mg/kg SR9243 (n = 8).
(B and C) RT-PCR analysis of (B) Srebp1c, Fasn, and (C) Pfk2 expression in tumors from mice treated with SR9243 or vehicle.
(D) Total body weight of tumor-bearing mice treated with SR9243.
(E) Serological analysis of blood glucose levels in vehicle and SR9243-treated mice.
(F) RT-PCR analysis showing expression of the cytokine Tnfα in livers and LLC1 tumors of C57BL6J mice treated with SR9243. See also Figure S5.
(G) Plasma levels of total cholesterol (CHOL), triglycerides (TRIG), low-density lipoproteins (LDL), and high-density lipoproteins (HDL) in vehicle and SR9243 (60 mg//kg)-treated mice.
(H) Plasma levels of the liver transaminases aspartate transaminase (AST), alkaline phosphatase (ALP), and alanine aminotransferase (ALT) as well as total albumin (ALB) in vehicle and SR9243-treated tumor-bearing mice. All RT-PCR and serological data were analyzed using a two-tailed t test followed by Bonferroni post-test. *p < 0.05. Error bars represent ±SEM. See also Figure S6.
DISCUSSION
The metabolic profiles of cancer cells are distinct from those of normal cells due to the Warburg effect and lipogenesis. These are key metabolic pathways that drive cancer progression, growth, survival, immune evasion, resistance to treatment, and disease recurrence (Fritz et al., 2010; Huang et al., 2012; Vander Heiden et al., 2009; Yeung et al., 2008). Therefore, by targeting glycolysis and lipogenesis, a broad range of cancers can be treated. This work establishes that targeted repression of glycolysis and lipogenesis can be achieved via suppression of LXR target-gene expression. Notably, LXR signaling has not been shown to initiate the Warburg effect in tumor cells. LXRs, however, directly regulate a number of key glycolytic and lipogenic enzymes that facilitate the Warburg effect and tumor lipogenesis, respectively. Therefore, we used an LXR inverse agonist to “hijack” the unliganded LXR and promote corepressor recruitment and formation of repressor complexes at the promoters of LXR-regulated genes. This approach reduced LXR transcriptional activity to below basal levels and therefore suppressed lipogenic and glycolytic gene expression, thereby inhibiting tumor growth.
The past few decades the Warburg effect and lipogenesis has been extensively studied with the goal of identifying targeted therapies that are selectively cytotoxic to cancer cells. A number of enzyme-specific inhibitors that target glycolysis enzymes and lipogenesis enzymes have been tested in rodent cancer models. Despite these efforts, no clinically viable cancer metabolism inhibitors for solid tumors have been forthcoming and only one glycolysis inhibitor, 2-deoxy-glucose, has made it to phase II clinical trials (Mohanti et al., 1996). The challenges of using enzyme-specific glycolysis and lipogenesis inhibitors for treating solid tumors may be inherently flawed due to their mode of action. Ideally, to effectively inhibit glycolytic and lipogenic enzyme activity, enzyme inhibitors must block the catalytic activity of overexpressed enzymes in tumor tissues. Cancer cells have a surplus pool of catalytically active enzyme molecules relative to normal cells. Therefore, to effectively disrupt tumor metabolism, enzyme-specific inhibitors are expected to selectively target and disrupt metabolic processes in relatively more metabolically active tumor cells while sparing less active non-malignant cells. Therefore, the dosage of enzyme inhibitor required to obtain the desired therapeutic effects in vivo also adversely affects the metabolic functions of normal cells. These limitations restrict the “therapeutic window” significantly for enzyme-specific inhibitors as clinical treatments for solid tumors.
Interestingly, in this study we observed that the effect of SR9243 on cancer cell glycolytic gene expression was not conserved across all cell types. The prostate cancer cell line DU-145 and non-small cell lung carcinoma cell line NCI-H23 both displayed limited repression of glycolytic genes in response to SR9243 but were highly responsive to SR9243 treatment. The underlying mechanism responsible for the differential effect SR9243 had on glycolysis gene expression in these cell types may be due to a number of factors. These cancer cells may exhibit cell type specific deregulation of glycolytic gene expression that limit LXR-mediated gene repression. Despite directly regulating the expression of key glycolytic genes, LXRs have a shared role in glycolytic gene regulation along with other nuclear receptors (e.g., PPARs and TRα) and oncogenes such (e.g., HIF and MYC). Therefore, resistance to LXR-mediated glycolytic disruption could be due to metabolic compensation driven by other receptor signaling pathways or oncogenes. Interestingly, NCI-H23 and DU-145 cells lack functional LKB1, a tumor suppressor that inhibits the Warburg effect through regulation of AMPK and downstream regulation of glycolysis enzyme expression. However, SR9243 was able to repress glycolysis gene expression in cancer cells that expressed mutant LKB1 (data not shown). This suggests that the underlying mechanisms for resistance to glycolysis gene repression may be more complex. Nonetheless, identification of the factors that may contribute to resistance to SR9243 mediated repression of glycolytic gene expression should be the subject of further inquiry. Our investigation did demonstrate, however, that LXR repression of lipogenic gene expression was sufficient to induce tumor cell death.
The favorable safety profile and lack of toxicity SR9243 displayed in non-malignant cells and tissues may be inherent to the mechanism of action of LXR-mediated glycolytic and lipogenic gene suppression. Our observations suggest that receptor-mediated downregulation of glycolytic and lipogenic enzyme expression using an LXR inverse agonist may be a more selective therapeutic strategy for disrupting the Warburg effect and lipogenesis than targeted enzyme inhibition. Our observations suggest that downregulation of key enzymes that drive the Warburg effect and/or lipogenesis in cancer cells produces a metabolic environment that is unable to support cancer cell growth but sufficient for the function of normal cells. Therefore, we propose that SR9243 facilitates the reprogramming of cancer cell metabolism to “normal” levels that cannot sustain cancer cell growth. SR9243 therefore is able to promote apoptotic cancer cell death without adversely affecting non-malignant cells. However, our observations notwithstanding, the underlying mechanism driving SR9243 cancer cell selectivity and low toxicity is still uncertain and should be the subject of further investigation.
A number of recently published studies suggest that LXR activation may be a key pathway mediating cancer cell immune evasion through silencing of dendritic cell activity (Russo, 2011; Villablanca et al., 2010). In our investigations, we observed that SR9243 specifically induced substantial inflammation in the tumors without causing systemic inflammation. These observations may be due to enhanced immune cell infiltration into tumors due to SR9243 treatment. Therefore, SR9243 may mediate tumor “unmasking” via downregulation of the immune-suppressive effects of LXR ligands within the tumor microenvironment. Preliminary experiments have suggested that SR9243 is able to induce dendritic cell activity that is suppressed by tumor-produced LXR ligands (data not shown). However, whether SR9243 can promote immune-mediated tumor clearance is unclear and should be the subject of further investigation and may highlight a promising avenue of exploration into the role of LXR as a therapeutic cancer target.
EXPERIMENTAL PROCEDURES
Animals
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by Institutional Animal Care and Use Committee of the Saint Louis University School of Medicine. Nu/Nu (Charles River) C57BL/6J and Ob/Ob mice (Jackson Laboratories) were housed in sterile ventilated cages, fed a standard diet unless otherwise specified, and provided water ad libitum. Mice were killed using CO2 followed by cervical dislocation.
Synthesis and Purification of SR9243
Detailed synthesis of SR9243 can be found in the Supplemental Experimental Procedures.
Chromatin Immunoprecipitation Assay
HepG2 were treated with DMSO vehicle or SR9243 (10 μM) for 48 hr fixed using Formaldehyde (1%) for 10 min. Sequential chromatin immunoprecipitation (Re-ChIP) was performed using the Re-ChIP IT kit (Active Motif) as per manufacturer's instruction.
Tumor Xenograft Experiments
Cancer cell lines were harvested using Trypsin/EDTA washed with PBS and re-suspended in PBS containing Matrigel. SW620 (5 × 106 cells), DU-145 (5 × 106 cells), and LLC cells (1 × 106) cells were implanted subcutaneously in the lower right flank of 5-week-old Nu/Nu (Charles River) or C57BL6J (LLC1) mice. All tumor xenografts were allowed to reach a volume of 100 mm3 before treatment commenced.
Statistical Analysis
All data were subjected to either ANOVA or t tests where specified with Tukey's post hoc test. *p < 0.05 is considered statistically significant. *p < 0.05, **p < 0.01 ***p < 0.001.
Supplementary Material
Highlights.
Glycolysis and lipogenesis are key metabolic pathways in cancer
LXRs directly regulate glycolysis and lipogenesis enzyme expression
The LXR inverse agonist SR9243 inhibits glycolysis and lipogenesis in cancer cells
SR9243 induces cancer cell death but is non-toxic to non-malignant cells
SR9243 has therapeutic effects without weight loss, liver toxicity, or inflammation
Significance.
A number of small molecules that target the Warburg effect and lipogenesis have been developed but none have become clinical treatments because of off-target effects such as excessive weight loss, anorexia, high toxicity, and low efficacy in vivo. Here we describe the anti-cancer properties of an LXR inverse agonist SR9243. Unlike previously developed targeted treatments, SR9243 selectively induces apoptosis in cancer cells but spares non-malignant tissues, exhibiting significant anti-tumor activity without overt toxicity, inflammation, or weight loss. The favorable safety profile of SR9243 demonstrates that LXR inverse agonists hold significant promise as prospective clinical treatments.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures and six figures and can be found with this article online at http://dx.doi.org/10.1016/j.ccell.2015.05.007.
REFERENCES
- Baranowski M. Biological role of liver X receptors. J. Physiol. Pharmacol. 2008;59(Suppl 7):31–55. [PubMed] [Google Scholar]
- Beyea MM, Heslop CL, Sawyez CG, Edwards JY, Markle JG, Hegele RA, Huff MW. Selective up-regulation of LXR-regulated genes ABCA1, ABCG1, and APOE in macrophages through increased endogenous synthesis of 24(S),25-epoxycholesterol. J. Biol. Chem. 2007;282:5207–5216. doi: 10.1074/jbc.M611063200. [DOI] [PubMed] [Google Scholar]
- Blancher C, Harris AL. The molecular basis of the hypoxia response pathway: tumour hypoxia as a therapy target. Cancer Metastasis Rev. 1998;17:187–194. doi: 10.1023/a:1006002419244. [DOI] [PubMed] [Google Scholar]
- Bonuccelli G, Tsirigos A, Whitaker-Menezes D, Pavlides S, Pestell RG, Chiavarina B, Frank PG, Flomenberg N, Howell A, Martinez-Outschoorn UE, et al. Ketones and lactate “fuel” tumor growth and metastasis: Evidence that epithelial cancer cells use oxidative mitochondrial metabolism. Cell Cycle. 2010;9:3506–3514. doi: 10.4161/cc.9.17.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat. Rev. Cancer. 2013;13:227–232. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chajès V, Cambot M, Moreau K, Lenoir GM, Joulin V. Acetyl-CoA carboxylase alpha is essential to breast cancer cell survival. Cancer Res. 2006;66:5287–5294. doi: 10.1158/0008-5472.CAN-05-1489. [DOI] [PubMed] [Google Scholar]
- Chuu CP, Lin HP. Antiproliferative effect of LXR agonists T0901317 and 22(R)-hydroxycholesterol on multiple human cancer cell lines. Anticancer Res. 2010;30:3643–3648. [PubMed] [Google Scholar]
- Clegg DJ, Wortman MD, Benoit SC, McOsker CC, Seeley RJ. Comparison of central and peripheral administration of C75 on food intake, body weight, and conditioned taste aversion. Diabetes. 2002;51:3196–3201. doi: 10.2337/diabetes.51.11.3196. [DOI] [PubMed] [Google Scholar]
- Dang CV. The interplay between MYC and HIF in the Warburg effect. Ernst Schering Found Symp Proc. 2007:35–53. doi: 10.1007/2789_2008_088. [DOI] [PubMed] [Google Scholar]
- Dang CV, Lewis BC, Dolde C, Dang G, Shim H. Oncogenes in tumor metabolism, tumorigenesis, and apoptosis. J. Bioenerg. Biomembr. 1997;29:345–354. doi: 10.1023/a:1022446730452. [DOI] [PubMed] [Google Scholar]
- Darimont C, Avanti O, Zbinden I, Leone-Vautravers P, Mansourian R, Giusti V, Macé K. Liver X receptor preferentially activates de novo lipogenesis in human preadipocytes. Biochimie. 2006;88:309–318. doi: 10.1016/j.biochi.2005.08.010. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Fritz V, Benfodda Z, Rodier G, Henriquet C, Iborra F, Avancès C, Allory Y, de la Taille A, Culine S, Blancou H, et al. Abrogation of de novo lipogenesis by stearoyl-CoA desaturase 1 inhibition interferes with oncogenic signaling and blocks prostate cancer progression in mice. Mol. Cancer Ther. 2010;9:1740–1754. doi: 10.1158/1535-7163.MCT-09-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta E, Okuda H, Kobayashi A, Watabe K. Metabolic genes in cancer: their roles in tumor progression and clinical implications. Biochim. Biophys. Acta. 2010;1805:141–152. doi: 10.1016/j.bbcan.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefhorst A, Elzinga BM, Voshol PJ, Plösch T, Kok T, Bloks VW, van der Sluijs FH, Havekes LM, Romijn JA, Verkade HJ, Kuipers F. Stimulation of lipogenesis by pharmacological activation of the liver X receptor leads to production of large, triglyceride-rich very low density lipoprotein particles. J. Biol. Chem. 2002;277:34182–34190. doi: 10.1074/jbc.M204887200. [DOI] [PubMed] [Google Scholar]
- Griffett K, Solt LA, El-Gendy Bel.-D., Kamenecka TM, Burris TP. A liver-selective LXR inverse agonist that suppresses hepatic steatosis. ACS Chem. Biol. 2013;8:559–567. doi: 10.1021/cb300541g. [DOI] [PubMed] [Google Scholar]
- Huang WC, Li X, Liu J, Lin J, Chung LW. Activation of androgen receptor, lipogenesis, and oxidative stress converged by SREBP-1 is responsible for regulating growth and progression of prostate cancer cells. Mol. Cancer Res. 2012;10:133–142. doi: 10.1158/1541-7786.MCR-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamroz-Wiśniewska A, Wójcicka G, Horoszewicz K, Be1towski J. Liver X receptors (LXRs). Part II: non-lipid effects, role in pathology, and therapeutic implications. Postepy Hig Med Dosw (Online) 2007;61:760–785. [PubMed] [Google Scholar]
- Joseph SB, Laffitte BA, Patel PH, Watson MA, Matsukuma KE, Walczak R, Collins JL, Osborne TF, Tontonoz P. Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. J. Biol. Chem. 2002;277:11019–11025. doi: 10.1074/jbc.M111041200. [DOI] [PubMed] [Google Scholar]
- Jung SY, Song HS, Park SY, Chung SH, Kim YJ. Pyruvate promotes tumor angiogenesis through HIF-1-dependent PAI-1 expression. Int. J. Oncol. 2011;38:571–576. doi: 10.3892/ijo.2010.859. [DOI] [PubMed] [Google Scholar]
- Kim TH, Kim H, Park JM, Im SS, Bae JS, Kim MY, Yoon HG, Cha JY, Kim KS, Ahn YH. Interrelationship between liver X receptor alpha, sterol regulatory element-binding protein-1c, peroxisome proliferator-activated receptor gamma, and small heterodimer partner in the transcriptional regulation of glucokinase gene expression in liver. J. Biol. Chem. 2009;284:15071–15083. doi: 10.1074/jbc.M109.006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiji M, Huang LE. Dynamic balancing of the dual nature of HIF-1alpha for cell survival. Cell Cycle. 2004;3:853–854. doi: 10.4161/cc.3.7.990. [DOI] [PubMed] [Google Scholar]
- Laffitte BA, Chao LC, Li J, Walczak R, Hummasti S, Joseph SB, Castrillo A, Wilpitz DC, Mangelsdorf DJ, Collins JL, et al. Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue. Proc. Natl. Acad. Sci. USA. 2003;100:5419–5424. doi: 10.1073/pnas.0830671100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Sasso G, Bovenga F, Murzilli S, Salvatore L, Di Tullio G, Martelli N, D'Orazio A, Rainaldi S, Vacca M, Mangia A, et al. Liver X receptors inhibit proliferation of human colorectal cancer cells and growth of intestinal tumors in mice. Gastroenterology. 2013;144:1497–1507. doi: 10.1053/j.gastro.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Lu CW, Lin SC, Chen KF, Lai YY, Tsai SJ. Induction of pyruvate dehydrogenase kinase-3 by hypoxia-inducible factor-1 promotes metabolic switch and drug resistance. J. Biol. Chem. 2008;283:28106–28114. doi: 10.1074/jbc.M803508200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Outschoorn UE, Prisco M, Ertel A, Tsirigos A, Lin Z, Pavlides S, Wang C, Flomenberg N, Knudsen ES, Howell A, et al. Ketones and lactate increase cancer cell “stemness,” driving recurrence, metastasis and poor clinical outcome in breast cancer: achieving personalized medicine via Metabolo-Genomics. Cell Cycle. 2011;10:1271–1286. doi: 10.4161/cc.10.8.15330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P, Liang B, Li L, Fremgen T, Murphy E, Quinn A, Madden SL, Biemann HP, Wang B, Cohen A, et al. SCD1 inhibition causes cancer cell death by depleting mono-unsaturated fatty acids. PLoS ONE. 2012;7:e33823. doi: 10.1371/journal.pone.0033823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng ZX, Nie J, Ling JJ, Sun JX, Zhu YX, Gao L, Lv JH, Zhu DY, Sun YJ, Han X. Activation of liver X receptors inhibits pancreatic islet beta cell proliferation through cell cycle arrest. Diabetologia. 2009;52:125–135. doi: 10.1007/s00125-008-1174-x. [DOI] [PubMed] [Google Scholar]
- Miller DH, Fischer AK, Chu KF, Burr R, Hillenmeyer S, Brard L, Brodsky AS. T0901317 inhibits cisplatin-induced apoptosis in ovarian cancer cells [corrected]. Int. J. Gynecol. Cancer. 2011;21:1350–1356. doi: 10.1097/IGC.0b013e318228f558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanti BK, Rath GK, Anantha N, Kannan V, Das BS, Chandramouli BA, Banerjee AK, Das S, Jena A, Ravichandran R, et al. Improving cancer radiotherapy with 2-deoxy-D-glucose: phase I/II clinical trials on human cerebral gliomas. Int. J. Radiat. Oncol. Biol. Phys. 1996;35:103–111. doi: 10.1016/s0360-3016(96)85017-6. [DOI] [PubMed] [Google Scholar]
- Nguyen-Vu T, Vedin LL, Liu K, Jonsson P, Lin JZ, Candelaria NR, Candelaria LP, Addanki S, Williams C, Gustafsson JA, et al. Liver x receptor ligands disrupt breast cancer cell proliferation through an E2F-mediated mechanism. Breast Cancer Res. 2013;15:R51. doi: 10.1186/bcr3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notarnicola M, Altomare DF, Calvani M, Orlando A, Bifulco M, D'Attoma B, Caruso MG. Fatty acid synthase hyperactivation in human colorectal cancer: relationship with tumor side and sex. Oncology. 2006;71:327–332. doi: 10.1159/000107106. [DOI] [PubMed] [Google Scholar]
- Notarnicola M, Tutino V, Calvani M, Lorusso D, Guerra V, Caruso MG. Serum levels of fatty acid synthase in colorectal cancer patients are associated with tumor stage. J Gastrointest Cancer. 2012;43:508–511. doi: 10.1007/s12029-011-9300-2. [DOI] [PubMed] [Google Scholar]
- Ogino S, Shima K, Nosho K, Irahara N, Baba Y, Wolpin BM, Giovannucci EL, Meyerhardt JA, Fuchs CS. A cohort study of p27 localization in colon cancer, body mass index, and patient survival. Cancer Epidemiol. Biomarkers Prev. 2009;18:1849–1858. doi: 10.1158/1055-9965.EPI-09-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25:4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- Phan LM, Yeung SC, Lee MH. Cancer metabolic reprogramming: importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol. Med. 2014;11:1–19. doi: 10.7497/j.issn.2095-3941.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan CA, Weaver JM, Steger DJ, Joshi S, Maslany JT, Collins JL, Zuercher WJ, Willson TM, Walker M, Jaye M, Lazar MA. Selective partial agonism of liver X receptor alpha is related to differential core-pressor recruitment. Mol. Endocrinol. 2008;22:2241–2249. doi: 10.1210/me.2008-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rough JJ, Monroy MA, Yerrum S, Daly JM. Anti-proliferative effect of LXR agonist T0901317 in ovarian carcinoma cells. J Ovarian Res. 2010;3:13. doi: 10.1186/1757-2215-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo V. Metabolism, LXR/LXR ligands, and tumor immune escape. J. Leukoc. Biol. 2011;90:673–679. doi: 10.1189/jlb.0411198. [DOI] [PubMed] [Google Scholar]
- Scaglia N, Chisholm JW, Igal RA. Inhibition of stearoylCoA desaturase-1 inactivates acetyl-CoA carboxylase and impairs proliferation in cancer cells: role of AMPK. PLoS ONE. 2009;4:e6812. doi: 10.1371/journal.pone.0006812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y, Thupari JN, Kim EK, Pinn ML, Moran TH, Ronnett GV, Kuhajda FP. C75 alters central and peripheral gene expression to reduce food intake and increase energy expenditure. Endocrinology. 2005;146:486–493. doi: 10.1210/en.2004-0976. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedin LL, Gustafsson JA, Steffensen KR. The oxysterol receptors LXRalpha and LXRbeta suppress proliferation in the colon. Mol. Carcinog. 2012 doi: 10.1002/mc.21924. [DOI] [PubMed] [Google Scholar]
- Viennois E, Mouzat K, Dufour J, Morel L, Lobaccaro JM, Baron S. Selective liver X receptor modulators (SLiMs): what use in human health? Mol. Cell. Endocrinol. 2012;351:129–141. doi: 10.1016/j.mce.2011.08.036. [DOI] [PubMed] [Google Scholar]
- Villablanca EJ, Raccosta L, Zhou D, Fontana R, Maggioni D, Negro A, Sanvito F, Ponzoni M, Valentinis B, Bregni M, et al. Tumor-mediated liver X receptor-alpha activation inhibits CC chemokine receptor-7 expression on dendritic cells and dampens antitumor responses. Nat. Med. 2010;16:98–105. doi: 10.1038/nm.2074. [DOI] [PubMed] [Google Scholar]
- Wagner BL, Valledor AF, Shao G, Daige CL, Bischoff ED, Petrowski M, Jepsen K, Baek SH, Heyman RA, Rosenfeld MG, et al. Promoter-specific roles for liver X receptor/corepressor complexes in the regulation of ABCA1 and SREBP1 gene expression. Mol. Cell. Biol. 2003;23:5780–5789. doi: 10.1128/MCB.23.16.5780-5789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Rogers PM, Su C, Varga G, Stayrook KR, Burris TP. Regulation of cholesterologenesis by the oxysterol receptor, LXRalpha. J. Biol. Chem. 2008;283:26332–26339. doi: 10.1074/jbc.M804808200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. J. Gen. Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wójcicka G, Jamroz-Wiśniewska A, Horoszewicz K, Be1towski J. Liver X receptors (LXRs). Part I: structure, function, regulation of activity, and role in lipid metabolism. Postepy Hig Med Dosw (Online) 2007;61:736–759. [PubMed] [Google Scholar]
- Yeung SJ, Pan J, Lee MH. Roles of p53, MYC and HIF-1 in regulating glycolysis - the seventh hallmark of cancer. Cell. Mol. Life Sci. 2008;65:3981–3999. doi: 10.1007/s00018-008-8224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytseva YY, Rychahou PG, Gulhati P, Elliott VA, Mustain WC, O'Connor K, Morris AJ, Sunkara M, Weiss HL, Lee EY, Evers BM. Inhibition of fatty acid synthase attenuates CD44-associated signaling and reduces metastasis in colorectal cancer. Cancer Res. 2012;72:1504–1517. doi: 10.1158/0008-5472.CAN-11-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang X, Chen L, Wu J, Su D, Lu WJ, Hwang MT, Yang G, Li S, Wei M, et al. Liver X receptor agonist TO-901317 upregulates SCD1 expression in renal proximal straight tubule. Am. J. Physiol. Renal Physiol. 2006;290:F1065–F1073. doi: 10.1152/ajprenal.00131.2005. [DOI] [PubMed] [Google Scholar]
- Zhao LF, Iwasaki Y, Nishiyama M, Taguchi T, Tsugita M, Okazaki M, Nakayama S, Kambayashi M, Fujimoto S, Hashimoto K, et al. Liver X receptor α is involved in the transcriptional regulation of the 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase gene. Diabetes. 2012;61:1062–1071. doi: 10.2337/db11-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.