Abstract
Actin is the major component of the cytoskeleton playing an essential role in the structure and motility of both muscle and non-muscle cells. It is highly conserved and encoded by a multigene family. α-cardiac actin (αCAA) and α-skeletal actin (αSKA), encoded by two different genes, are the primary actin isoforms expressed in striated muscles. The relative expression levels of αSKA and αCAA have been shown to vary between species and under pathological conditions. In particular, an increased αSKA expression is believed to be a programmed response of a diseased heart. Therefore, it is essential to quantify the relative expression of αSKA and αCAA, which remains challenging due to the high degree of sequence similarity between these isoforms (98.9%). Herein, we developed a top-down liquid chromatography mass spectrometry-based (“LC/MS+”) method for the rapid purification and comprehensive analysis of α-actin extracted from muscle tissues. We thoroughly investigated all of the actin isoforms in healthy human cardiac and skeletal muscles. We found that αSKA is the only isoform expressed in skeletal muscle, whereas αCAA and αSKA are co-expressed in cardiac muscle. We then applied our method to quantify the α-actin isoforms in human healthy hearts and failing hearts with dilated cardiomyopathy (DCM). We found that αSKA is augmented in DCM compared with healthy controls, 43.1 ± 0.9% versus 23.6 ± 1.7%, respectively. As demonstrated, top-down LC/MS+ provides an effective and comprehensive method for the purification, quantification, and characterization of α-actin isoforms, enabling assessment of their clinical potential as cardiac disease markers.
Keywords: actin, muscle, mass spectrometry, myofilament, heart disease
Introduction
Actin is the major component of the cytoskeleton playing an essential role in the structure and motility of both muscle and non-muscle cells.1 In muscle cells, actin is a critical component of the myofilament (the contractile apparatus). Together with the troponin and tropomyosin regulatory complex (Tn/Tm), actin principally makes up the thin filaments in striated (cardiac and skeletal) muscles. Importantly, the interaction between actin thin filaments and myosin thick filaments is responsible for force production during muscle contraction.1-4
Actin is highly conserved, ubiquitous, and encoded by a multigene family.1,5 A total of six actin isoforms, which display tissue specificity and distinct developmental expression patterns, are present in mammalian cells.1,6 α-cardiac actin (αCAA) and α-skeletal actin (αSKA), which are encoded by two different genes: ACTC and ACTA, respectively, are the major actin isoforms expressed in striated muscles.1,5-7 αCAA is the major isoform expressed in cardiac muscle, whereas αSKA is the predominant isoform expressed in skeletal muscle.4,8 Although both αCAA and αSKA are expressed in cardiac muscle, the relative expression levels of αSKA and αCAA have been shown to vary between species, during development, and under pathological conditions.1,4-13 In particular, an increase in the expression of αSKA has been observed in diseased hearts using experimental animal models and human clinical samples.5,7,10-12 Thus, augmented αSKA expression is believed to be a programmed response of a diseased heart and it is essential to determine the αSKA/αCAA isoform ratio for its potential as a cardiac disease marker.5,8
Unfortunately, αCAA and αSKA have very high sequence similarities (98.9%), making analysis of the relative expression level of these two isoforms difficult.5,8,14 Although mRNA expression level has traditionally been used to monitor the relative expression levels,1,6,9,11 the correlation between mRNA level and protein expression has been reported to be notoriously poor.15,16 More recent studies reported the use of αCAA and αSKA specific antibodies to assess the expression of α-actin isoforms by Western blot assays.5,10,17 However, the time-consuming nature and need for standard curve measurements for high-accuracy quantification makes this technique impractical for large cohorts and clinical application.18,19 Therefore, it is essential to develop a fast and reliable method for the quantification of α-actin isoform expression levels for clinical applications.
Mass spectrometry (MS) has become a preferred method for determining protein expression levels because it essentially serves as a “generalized Western blot” without the need to generate antibodies and without the cross-reactivity problem.20 Meanwhile, MS can precisely identify thousands of proteins from complex mixtures, as well as characterize post-translational modifications (PTMs), and protein isoforms arising from genetic variations.20-22 The traditional bottom-up MS approach requires enzymatic digestion of proteins into peptides, so the identification and quantification of proteins depends on the detection of the peptides of interest,21,22 which complicates the quantification of protein isoforms and PTMs (the ‘peptide to protein inference problem’).23 In contrast, top-down MS measures the intact proteins without enzymatic digestion and therefore provides reliable quantification and comprehensive characterization of isoforms and PTMs.23-34
Herein, we aim to develop an integrated top-down liquid chromatography mass spectrometry-based (LC/MS+) method that includes rapid purification, identification, quantification, and characterization of α-actin isoforms extracted from muscles. Specifically, we extracted the myofilament proteins from muscles and collected purified α-actin after reverse-phase LC separation, which provides significantly higher throughput than the traditional time-consuming α-actin purification steps.4,8,35,36 The purified α-actin was analyzed by high-resolution top-down MS, enabling a visualization of all the α-actin isoforms and their PTMs simultaneously in one spectrum for reliable quantification and detailed characterization of the protein sequence and modifications. We then applied this method to identify and quantify isoforms from both human skeletal and cardiac muscles, as well as swine cardiac muscle. We found that αSKA is the only isoform expressed in skeletal muscle, whereas αCAA and αSKA are co-expressed in cardiac muscle. Furthermore, we observed an increase in the expression level of αSKA, relative to αCAA, in end-stage failing hearts from DCM patients compared with healthy controls. This demonstrates the effectiveness and reliability of our integrated LC/MS+ method for assessing α-actin isoforms as a potential disease marker.
Experimental Procedures
Chemicals and Reagents
All chemicals and reagents were purchased from Sigma Chemical Co. (St Louis, MO) and Thermo Fisher Scientific (Pittsburgh, PA) unless noted otherwise. Complete protease inhibitor cocktail tablets were purchased from Roche Diagnostics Corporation (Indianapolis, IN). All solutions were prepared in HPLC grade water (Thermo Fisher Scientific, PA).
Sample Procurement
The human cardiac tissues were obtained from surgical heart transplant specimens in the University of Wisconsin Hospital and Clinics. Only the left ventricles were utilized in this study. All procedures were approved by the University of Wisconsin-Madison Institutional Review Board. Percutaneous skeletal muscle biopsies were taken from the tibialis anterior (predominantly slow) and the vastus lateralis (mixed) from two young healthy women. All procedures have been approved by the ethics committee of the Uppsala University and carried out according to the guidelines of the Declaration of Helsinki. Swine heart tissues were obtained from juvenile Yorkshire domestic swine as approved by the University of Wisconsin Animal Care and Use Committee. The excised tissues were immediately frozen in liquid nitrogen and stored in a −80 °C freezer.
Top-down LC/MS+ analysis of actin
Approximately 5 mg (sufficient for 10 experiments, equivalent to 500 μg per experiment) of cardiac/skeletal muscle tissue was homogenized in HEPES extraction buffer (25 mM HEPES pH 7.5, 50 mM NaF, 0.25 mM Na3VO4, 0.25 mM PMSF (in isopropanol), 2.5 mM EDTA, and a half tablet of Roche protease inhibitor cocktail) using a Teflon homogenizer (1.5 mL tube rounded tip, Scienceware, Pequannock, NJ, USA).28,37 The homogenate was centrifuged at 16.1×1000 rcf at 4 °C for 40 min (Centrifuge 5415R, Eppendorf, Hamburg, Germany), and the supernatant was removed and stored. The pellet was then resuspended and homogenized in TFA extraction buffer (1% TFA, 1 mM TCEP) using the Teflon homogenizer. The homogenate was then centrifuged again at 16.1×1000 rcf at 4 °C for 40 min and the supernatant was collected for further analysis.37
The supernatant collected was mixed with 30% mobile phase B (0.1% formic acid in 50:50 acetonitrile and ethanol), and LC/MS analysis was performed using a 2D-nanoLC system (Eksigent, Dublin, CA) coupled with a linear ion trap (LTQ, Thermo Scientific Inc., Bremen, Germany). Pre-mixed samples were loaded onto the sample loop, and a home-made PLRP column (200 mm ×500 μm, PLRP, 10 μm, 1000 Å, PLRP-S particles were purchased from Varian, Lake Forest, CA) was used for the analysis. All of the myofilament proteins eluted off with a gradient run (12.5 μL/min) in 45 min (mobile phase A: 0.1% formic acid in water, mobile phase B: 0.1% formic acid in 50:50 acetonitrile and ethanol). The gradient started at 30% mobile phase B for 5 min, and then increased to 40% for 10 min, increased to 50% for 7.5 min, again increased to 65% for 7.5 min, and then back to 30%. During the LC/MS analysis, about 5% of the sample went through the tip (25 μm i.d.) and was ionized by electrospray ionization (ESI). In the meantime, the remainder went through the split and was fraction collected and stored on ice for further analysis. The fraction of actin was collected at 28.5-30 minutes retention time. The fraction collected actin samples were analyzed using a 7T linear trap/Fourier transform ion cyclotron resonance (FT-ICR) (LTQ FT Ultra) hybrid mass spectrometer (Thermo Scientific Inc., Bremen, Germany) equipped with an automated chip-based nano ESI source (Triversa NanoMate; Advion BioSciences, Ithaca, NY) as described previously.28,37 The resolving power of the FT-ICR mass analyzer was typically set at 200,000 (at 400 m/z). All the spectra were acquired in full profile mode. For collisionally activated dissociation (CAD) and electron capture dissociation (ECD) fragmentation, individual charge states of protein molecular ions were first isolated and then dissociated using 15-20% of normalized collision energy for CAD or 2.8-3.5% electron energy for ECD with a 70-75 ms duration with no delay. All MS and MS/MS data were processed by an in-house developed software, MASH Suite38 (version 1.2), which uses the THRASH39 algorithm, with S/N threshold of 3 and a fit factor of 60% and manually validated. The fragments in MS/MS spectra were assigned on the basis of the protein sequence of human and swine αCAA and αSKA obtained from Swiss-Prot protein knowledgebase (Unit-ProtKB/Swiss-Prot). Allowance was made for possible PTMs such as removal of N-terminal Cys and Met, acetylation of the N-terminus, and methylation, using 1 Da tolerance for the precursor and product ions, respectively. The assigned ions were manually validated to ensure the accuracy of the assignments. In general, after manual validation using MASH Suite,38 all ions identified have accuracy of less than 10 ppm. All the reported molecular weights (MWs) are the most abundant MWs.
Quantitative analysis and statistics
Quantitative analysis of the relative expression levels of αCAA and αSKA in human heart samples was performed as described previously.27-29 For each detected protein form, the integrated peak heights of the top five isotopomers were used to calculate the relative abundance. The percentages of αCAA and αSKA were defined as the abundances of αCAA and αSKA over the summed abundances of the α-actins. The impact of the number of isotopmers on the calculation of the relative abundances has been investigated using three, five, seven and eleven isotopomers, respectively (Supplementary Table 1). The percent ratio of αSKA over αCAA was used to monitor the changes and data were represented as means ± SEM. Student's T-tests were performed between group comparisons to evaluate statistical significance of variance. Differences among means were considered significant at p < 0.01.
Results
Establishment of a top-down LC/MS+ method for the analysis of α-actin isoforms
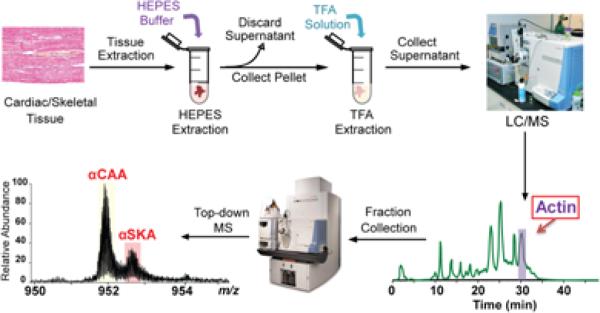
We have developed a top-down LC/MS+ method that allows for the rapid purification, comprehensive characterization, and quantification of α-actin from cardiac and skeletal tissues. Briefly, the method includes the following steps: (i) tissue homogenization in HEPES buffer; (ii) extraction of myofilaments by centrifugation and solubilization of myofilament proteins in TFA solution; (iii) on-line separation of myofilaments by LC; (iv) fraction collection of purified actin concurrent with on-line LC/MS analysis; (v) comprehensive top-down MS analysis of actin isoforms using high-resolution FT-ICR MS (Figure 1, Supplementary Figure 1).
Figure 1. Schematic representation of the integrated top-down LC/MS+-based method for quantification of α-actin isoforms.
Tissues are homogenized in HEPES buffer; the myofilament proteins are extracted by 1% TFA buffer and separated using LC/MS. The α-actin fraction is then collected and analyzed using a high-resolution mass spectrometer.
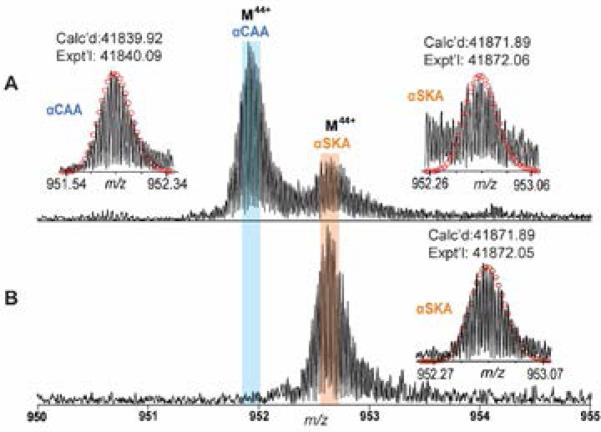
We employed this method to purify α-actin from human cardiac and skeletal tissues and analyzed all detectable isoforms by FT-ICR. A predominant isoform of α-actin, with a MW of 41,840.09, as well as a minor isoform with MW 41,872.06, were present in cardiac muscle (Figure 2A). These two isoforms had a 32 Da mass difference and presumably corresponded to αCAA and αSKA, respectively. As reported previously,5,8 αCAA and αSKA vary by only two juxtaposed amino acids (Asp2Glu3 in αCAA versus Glu2Asp3 in αSKA) and two amino acid substitutions (Met299 and Thr358 in αSKA, versus Leu299 and Ser358 in αCAA) which result in a 32 Da mass difference (Supplementary Figure 2). Moreover, the predominant α-actin peak from skeletal muscle had the same MW of 41,872.05, matching the peak attributed to αSKA in the cardiac sample (Figure 2B). Besides αCAA and αSKA, two minor unknown protein component with the MWs of 18700.69 and 42226.77 were present in the human heart samples (Supplementary Figure 1). The MWs of unknowns do not match any actin isoforms with common modifications. Because of the low S/N of these minor components, it was difficult to obtain sufficient fragmentation ions for further identification.
Figure 2. High-resolution MS for quantitative analysis of α-actin isoforms.
Narrow band FTMS spectrum of α-actin isolated from human (A) heart tissues and (B) skeletal tissues for charge state 44. Insets: isotopically resolved molecular ions of αCAA and αSKA with high accuracy molecular weight measurements. Circles represent the theoretical isotopic abundance distribution of the isotopomer peaks corresponding to the assigned mass. Calc'd, calculated most abundant molecular weight; Expt'l, experimental most abundant molecular weight.
However, the experimental MWs of αCAA (41,840.09) and αSKA (41,872.06) do not match exactly with the theoretical MWs of αCAA (P68032-ACTC_HUMAN, UniProtKB/Swiss-Prot) and αSKA (P68133-ACTS_HUMAN, UniProtKB/Swiss-Prot). Both the experimental MWs of αCAA and αSKA have a mass discrepancy of 177.02 Da from the calculated MWs of 42,016.93 and 42,048.91, respectively, based on the unmodified sequences given in the database. To account for this mass difference, it is reasonable to hypothesize the presence of modifications in the amino acid sequence. After removal of N-terminal Cys and Met, a well-known N-terminal proteolytic cleavage for all actin, and addition of acetylation,14 the calculated MWs (41,825.90 and 41,857.88) still have a mass difference of 14.02 Da from the experimental value. This mass difference is likely due to methylation as the vast majority of α-actin isoforms are post-translationally methylated at His73 to produce N-τ-3-methyl-histidine.40-42 The theoretical most abundant masses (Calc'd) of α-actin, after consideration of N-terminal truncation, acetylation of the new N-terminus and methylation of a histidine residue, are 41,839.92 Da for αCAA and 41,871.89 Da for αSKA, which match the experimental (Expt'l) most abundant mass of αCAA and αSKA (41,840.09 Da and 41,872.06 Da, both with mass errors of 4.1 ppm, respectively) .
Comprehensive characterization by MS/MS
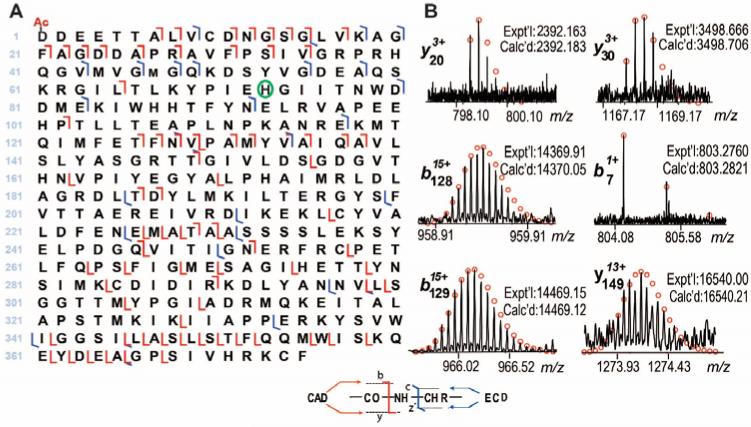
Unlike bottom-up MS, the top-down MS approach allows comprehensive sequence characterization of proteins of interest.23-26 We performed MS/MS to verify the potential modifications in αCAA and αSKA (Supplementary Figure 3). Individual charge states of αCAA and αSKA were isolated in the LTQ/FT mass spectrometer, and fragmented by both ECD and CAD. The representative fragmentation ions and the fragmentation maps for αCAA and αSKA are shown in Figure 3 and Figure 4, respectively. As expected, excluding any protein modifications, only 29 y ions and 12 z· ions, both generated from the C-terminus of the protein, were observed. On the other hand, there was no match for the N-terminus, namely the b and c ions, suggesting the presence of modification at the N-terminus. After removal of the N-terminal Met and Cys, as well as the addition of N-terminal acetylation, 7 b and 19 c ions were generated between 1-72 amino acids, suggesting that the first 72 amino acids from the N-terminus (1-72) do not contain a methylation site. After considering methylation at His73, additional b and c ions were observed. Of note, only methylated forms of c73 were detected, no unmethylated c73 was observed, suggesting that His73 is the only site basally methylated in α-actin. Conversely, if other sites after His73 were methylated (as in the case of positional isomers), one would expect to observe unmethylated c73 ions represented in the ECD/CAD spectrum. After considering all protein modifications, a total of 37 b, 40 y, 16 c and 11 z· ions were detected for human αCAA, as shown in Figure 3A. Representative fragmentation ions are shown in Figure 3B. In the case of αSKA, a total of 25 b, 29 y, 41 c and 12 z· were observed, as shown in Figure 4A. Representative fragmentation ions are shown in Figure 4B.
Figure 3. Comprehensive characterization of α-cardiac actin (αCAA) by top-down high-resolution MS.
(A) The fragmentation map of αCAA based on ECD and CAD spectra, Ac-represents acetylation, and the green circle represents methylation. (B) The representative b and y ions from CAD experiments. Fragmentation assignments were made based on the sequence of human αCAA (P68032, ACTC_HUMAN, UniProtKB/Swiss-Prot) with acetylation at the N-terminus and methylation at His73. Circles represent the theoretical isotopic abundance distribution of the isotopomer peaks corresponding to the assigned mass. Calc'd, calculated molecular weight; Expt'l, experimental molecular weight.
Figure 4. Comprehensive characterization of α-skeletal actin (αSKA) by top-down high-resolution MS.
(A) The fragmentation map of αSKA based on ECD and CAD spectra. Ac-represents acetylation and the green circle represents methylation. (B) Representative b and y ions from CAD data, c and z• ions from ECD data of αSKA. Fragmentation assignments were made based on the sequence of human αSKA (P68133, ACTS_HUMAN, UniProtKB/Swiss-Prot) with acetylation at the N-terminus and methylation at His73. Circles represent the theoretical isotopic abundance distribution of the isotopomer peaks corresponding to the assigned mass. Calc'd, calculated molecular weight; Expt'l, experimental molecular weight.
Investigation of α-actin isoforms in swine heart tissue
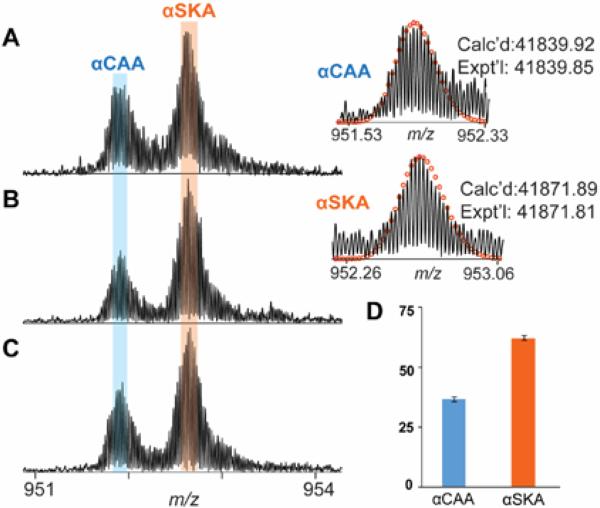
The co-expression of αCAA and αSKA in heart tissue has been shown in various species in addition to humans.4,5,8,13 Given the similarity between swine and human hearts, which include similar size, anatomy, and coronary artery distribution, swine models are preferred in cardiovascular research.43,44 Thus, we applied our method to study the composition of α-actin in swine samples. High-resolution MS of swine actin revealed the presence of two major isoforms of α-actin extracted from cardiac muscle with MWs of 41,840.49 and 41,872.66 and a minor unknown protein component with the MWs of 42048.44 (Figure 5 A-C and Supplementary Figure 4). These correspond to αCAA and αSKA, respectively, and are consistent with the human data (vide supra). Interestingly, unlike healthy human hearts in which αCAA predominates, αSKA is the most abundant α-actin isoform in the swine heart. Next, we measured the relative ratio of αCAA and αSKA in healthy swine heart tissues over three biological replicates. The summed percentages of the αCAA and αSKA over three biological replicates in swine heart with triplicate technical replicates are 37.0 ± 1.0% and 63.0 ± 1.0%, respectively. To address the potential impact of the number of isotopomers on the calculation of the relative abundances, we have used three, five, seven and eleven isotopomers, respectively, and found the impact to be minimal (Supplementary Table 1). Apparently, the ratio of αSKA/αCAA is higher in healthy swine heart than in healthy human heart, which is not surprising since the relative expression levels of αSKA and αCAA have been known to vary between species.4
Figure 5. High-resolution MS for quantitative analysis of α-actin isoforms in swine hearts.
A-C) Representative spectrum of α-actin isolated from three different biological samples. D) The bar graph represents the ratio of αCAA and αSKA in pig hearts. Circles represent the theoretical isotopic abundance distribution of the isotopomer peaks corresponding to the assigned mass. Calc'd, calculated most abundant molecular weight; Expt'l, experimental most abundant molecular weight.
Investigation of disease-related changes in α-actin isoforms in human heart tissues
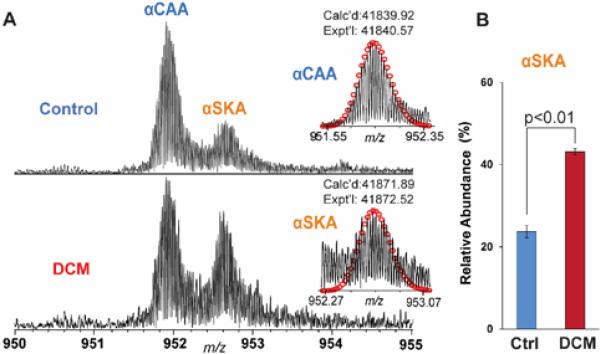
To assess the potential of α-actin isoforms as a cardiac disease maker, we investigated the differences of α-actin expression levels in healthy and diseased human hearts using top-down MS. We utilized three healthy human donor hearts as control (Ctrl) and three dilated cardiomyopathy (DCM) human hearts to study the disease-related changes in α-actin isoforms in human cardiac tissues. αCAA remains the major isoform in both Ctrl and DCM samples although the relative abundance of αSKA is much higher in DCM than Ctrl (Figure 6). The individual values of relative abundances of αCAA and αSKA from each human heart were listed in Supplementary Table 2. The summed percentages of the αCAA and αSKA over three biological replicates (each with three technical replicates) in healthy heart were 76.3±1.7% and 23.7±0.9%, respectively (Figure 6A). Strikingly, the percentage of αSKA increased to 43.1 ± 0.9% (Figure 6B) in failing heart samples with DCM. This clearly shows the level of αSKA is augmented in DCM compared to healthy heart tissue.
Figure 6. Quantification of the relative abundance of αSKA from control and DCM human hearts.
A) Representative high-resolution MS spectra of human α-actin purified from healthy human heart (top) and end-stage failing human heart with DCM (bottom). Calc'd, calculated molecular weight; Expt'l, experimental molecular weight. B) The bar graph represents αSKA ratio to total α-actin. All measurements were performed with three biological replicates per group. The data are represented as the mean peak area ± S.E.M. Ctrl, control.
Discussion
Top-down LC/MS+ method for the quantification of α-actin isoforms
In this study, we developed an integrated top-down LC/MS+ method for the rapid purification, accurate quantification, and comprehensive sequence characterization of α-actin isoforms from tissue samples. The key advantages of our method over existing methods for the quantification and analysis of α-actin isoforms include: i) rapid purification of actin; ii) a minimal amount of tissue needed (~500 μg per LC/MS analysis); iii) high reproducibility and reliable quantification of actin isoforms; and iv) comprehensive characterization of the sequences and modifications of all actin isoforms.
The traditional methods for the quantification of α-actin isoforms include mRNA expression1,6,9,11 and Western blot assays;5,10,17 however, both of these methods have intrinsic limitations. It is known that the correlation between mRNA level and protein expression, especially in complex samples, is far from perfect due to multiple parameters such as RNA secondary structure, regulatory proteins and sRNAs, codon bias and adaptation index, ribosomal density and occupancy, protein half-lives, and other biological factors influencing mRNA-protein correlation.15 Although Western blot has been the primary tool used to quantify proteins over the last three decades, there are numerous potential pitfalls, including unequal loading of proteins, transfer problems, the amount of substrate for chemiluminescence, and nonlinearity of the signal, which makes the quantification of Western blot very challenging.19 To improve protein quantification by Western blot, Copeland et al. applied a standard curve method by making serial dilutions of purified αSKA to quantitate αSKA expression in human heart tissues.5 However, the method is very time consuming and is impractical for analysis of large cohorts.
Recently, MS has become a preferred method for determining protein expression because it essentially serves as a “generalized Western blot”.20 Ravenscroft et al. successfully applied a multiple reaction monitoring (MRM)-based targeted MS approach to quantify α-actin isoforms.13 However, their method requires SDS-PAGE separation of the muscle tissue and extraction of peptides from the gel. Consequently, instead of measuring the protein itself, the quantification depends on peptides generated from digestion, which complicates the quantification of isoforms and PTMs (the ‘peptide to protein inference problem’).23 The major caveats of any peptide-based quantitative method include: i) not all peptides can be analyzed by MS since some may not be favorably retained on LC column and larger peptides may not be detected by MS; ii) The propagation of quantitative errors may result from processing steps including digestion and LC/MS of peptides.45 Alternatively, Bergen et al. investigated the expression of α-actin isoforms by measuring intact protein masses by FT-ICR.8 Although this method eliminates the needs for digestion of proteins and the quantification relies on the abundance of the protein not the peptide, it requires 2-3 days of laborious α-actin purification from a large amount of muscle making it impractical for analysis of α-actin expression in normal and pathological human heart tissues.
In contrast, our method provides fast, reliable α-actin isoform analysis with high-accuracy from a minimal amount of tissue. Rather than the time-consuming traditional biochemical α-actin purification,8,35,36 we can extract the myofilament subproteome in less than two hours and then purify actin by reverse phase LC/MS gradient run in ~45 min. The purified protein is obtained directly from HPLC separation and is ready for MS analysis without the need for desalting. Thus, the α-actin proteins could be ready for high-resolution MS analysis in less than 3 hours, compared to the traditional actin purification which requires more than two days of work.4,8,35,36 Furthermore, a minimal amount of heart and skeletal muscle tissue (equivalent to 500 μg per LC injection) is enough for LC/MS separation. The reason we started with 5 mg was due to the difficulty in measuring 500 μg accurately. Such advantages make this a promising method for future clinical applications.
The purified α-actin was analyzed by high-resolution top-down MS, enabling visualization and extensive characterization of all α-actin isoforms together with their PTMs. A total of six actin isoforms resulting from different genes are known to be expressed in a tissue-specific way. 46 In addition to the ubiquitous β- and γ-cytoplasmic actins, four muscle specific actins, αCAA, αSKA, α-smooth muscle actin (αSMA), and γ-smooth muscle actin (γSMA). Here only αCAA, αSKA, but neither αSMA nor γSMA were detected in the adult cardiac and skeletal muscle tissues. This is not surprising since αSMA is known to be transiently expressed in the embryonic cardiac and skeletal muscles.46 A previous study demonstrated that αSMA is not expressed in the healthy and pathologic cardiac tissues.7 γSMA is known to be absent in the myocardium and the re-expression of γSMA in the transgenic mouse heart resulted in a hypodynamic and hypertrophied heart.46 Characterization of α-actin isoforms revealed that N-terminal modifications and histidine methylation are the only modifications of αCAA and αSKA, which is in agreement with previous reports.40-42 Note that we employed a “LC/MS+” platform which allowed fraction collection concurrent with on-line LC/MS,28,37 which is also known as “data-directed top-down” reported by Whitelegge and co-workers.47,48 Here, the low-resolution LTQ/MS is used as a detector to monitor the elution of the proteins in order to find the specific fraction that contains actin for offline high-resolution top-down MS analysis. The advantage of fraction collection is the generation of the purified proteins for further comprehensive high-resolution MS analysis off-line and other biochemical assays if desired. Nonetheless, the off-line fraction collection undoubtedly compromised the throughput. With the development of the high sensitivity of high-resolution instruments,49,50 we expect to perform the high-resolution MS analysis of actin isoforms on-line on a chromatographic scale, which will further increase the throughput of this method.
The αSKA/ αCAA relative expression ratio as a potential cardiac biomarker
High-resolution FT-ICR MS spectra of purified actin from healthy human heart tissue exhibited the co-expression of αCAA and αSKA, 76.3 ± 1.7% and 23.7 ± 0.9% of total α-actin, respectively. This is in close agreement with the previously reported 21 ± 2% αSKA ratio in healthy human heart tissues determined by Western blot analysis.5 Moreover, we found α-actin purified from human skeletal tissue exhibited only αSKA with no detectable αCAA (Figure 2B), which is also in agreement with previous reports.4,8 Interestingly, the healthy swine heart tissue expressed high levels of αSKA (62%) (Figure 5) compared to healthy human heart tissue (Figure 6). This value is a bit higher than the percentage reported by Ravenscroft et al. using MRM-based quantification, which measured 52% αSKA in swine hearts.13 The variation could be due to ‘peptide to protein inference’23 between the peptide-based MRM quantification in comparison to protein-based top-down MS. The top-down MS approach has the advantage for relative quantification of isoforms since the ionization efficiency is much less affected by the small variation in the protein sequence (in this case two amino acid substitutions, Met299 and Thr358 in αSKA, versus Leu299 and Ser358 in αCAA, which result in a 32 Da mass difference) in comparison to peptides in the bottom-up approach.51,52 It may also be due to differences in the breeds of animals used.5
The higher level of αSKA in pathological conditions such as heart failure and hypertrophy has been shown by mRNA assays and αSKA-specific antibody application.5,7,10,11 To further validate our method, we investigated the relative expression of αSKA in healthy and end-stage failing human hearts with DCM (Figure 6). In failing heart samples, the percentage of αSKA increased from 23.6 ± 1.7% (Ctrl) to 43.1 ± 0.9 % (DCM). Although the percentage of αSKA in healthy human hearts is similar to the previous report (21 ± 2%), the value in DCM hearts is slightly less than the previously reported value of 53 ± 5%, determined by quantitative Western blot in end-stage failing hearts.5 It is likely that these variations reflect differences in the pathological conditions (our study used tissue samples from patients with DCM), or differences caused by varying medical treatments received for each individual. Further investigation of αSKA expression levels in large groups of patients with diverse hypertrophic disease conditions could provide better insight into pathology-specific αSKA expression.
The functional significance of the varying expression levels of αSKA in the heart muscle during differentiation, development, and disease is incompletely understood.5,7,12 Nevertheless, a previous study by Robbins and co-workers in BALB/C mouse heart demonstrated a significant functional correlation between α-actin isoform content and cardiac contractile function, suggesting that αSKA is associated with increased contractility.1 Therefore, increased αSKA might be part of a compensation mechanism to maintain cardiac contractility and serve as a programmed response of a disease-compromised heart which is unable to meet the demand.53 Recently, the Marston group employed an in vitro motility assay which observed basic sliding speed and response to Ca2+ concentrations to examine the contractile properties of αSKA.5 They found no functional differences in direct comparison of reconstituted thin filaments containing 20% αSKA versus 60% αSKA. Thus it has been suggested that accumulation of αSKA might not be the sole factor for compensation of contractility.5 However it could be a part of the gene reprogramming known as the “fetal gene program”54, which includes the re-expression of certain fetal genes and the down-regulation of multiple adult genes, such as β-myosin heavy chain, atrial natriuretic factor, and αSKA.55,56 Conceivably, re-expression of αSKA could serve as a marker for dedifferentiation and an important molecular indicator of heart failure.12
Conclusion
We have developed a LC/MS+ method for the rapid purification, reliable quantification, and comprehensive characterization of α-actin isoforms due to genetic variations together with PTMs. We have applied this method to assess the α-actin isoforms in both cardiac and skeletal tissues. We confirmed both αCAA and αSKA are expressed in cardiac muscle, whereas αSKA is the only isoform expressed in skeletal muscle. Furthermore, our data revealed that αSKA/αCAA ratio is disturbed in end-stage failing hearts with DCM compared with healthy hearts. Importantly, our LC/MS+ method only requires small amounts of samples which makes it feasible to analyze biopsy samples and is practical to be used in large human cohorts in clinical applications. Moreover, it can be easily developed into a high throughput method using a high-sensitivity and high-resolution mass spectrometer.
Supplementary Material
Acknowledgments
We would like to thank Zachery Gregorich for critical reading of this manuscript. Financial support was kindly provided by NIH R01 HL109810 and R01 HL096971 (to Y.G.).
Footnotes
Supporting Information Available:
Additional information as noted in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hewett TE, Grupp IL, Grupp G, Robbins J. Circ Res. 1994;74:740–746. doi: 10.1161/01.res.74.4.740. [DOI] [PubMed] [Google Scholar]
- 2.Moss RL, Razumova M, Fitzsimons DP. Circ.Res. 2004;94:1290–1300. doi: 10.1161/01.RES.0000127125.61647.4F. [DOI] [PubMed] [Google Scholar]
- 3.Kabsch W, Vandekerckhove J. Annu. Rev. Biophys. Biomol. Struct. 1992;21:49–76. doi: 10.1146/annurev.bb.21.060192.000405. [DOI] [PubMed] [Google Scholar]
- 4.Vandekerckhove J, Bugaisky G, Buckingham MJ. Biol. Chem. 1986;261:1838–1843. [PubMed] [Google Scholar]
- 5.Copeland O, Nowak KJ, Laing NG, Ravenscroft G, Messer AE, Bayliss CR, Marston SB. J. Muscle Res. Cell. Motil. 2010;31:207–214. doi: 10.1007/s10974-010-9224-7. [DOI] [PubMed] [Google Scholar]
- 6.Clement S, Chaponnier C, Gabbiani G. Circ. Res. 1999;85:e51–58. doi: 10.1161/01.res.85.10.e51. [DOI] [PubMed] [Google Scholar]
- 7.Suurmeijer AJ, Clement S, Francesconi A, Bocchi L, Angelini A, Van Veldhuisen DJ, Spagnoli LG, Gabbiani G, Orlandi A. J. Pathol. 2003;199:387–397. doi: 10.1002/path.1311. [DOI] [PubMed] [Google Scholar]
- 8.Bergen HR, 3rd, Ajtai K, Burghardt TP, Nepomuceno AI, Muddiman DC. Rapid Commun. Mass Spectrom. 2003;17:1467–1471. doi: 10.1002/rcm.1075. [DOI] [PubMed] [Google Scholar]
- 9.Bennetts BH, Burnett L, dos Remedios CG. J. Mol. Cell. Cardiol. 1986;18:993–996. doi: 10.1016/s0022-2828(86)80013-x. [DOI] [PubMed] [Google Scholar]
- 10.Berni R, Savi M, Bocchi L, Delucchi F, Musso E, Chaponnier C, Gabbiani G, Clement S, Stilli D. Am. J. Physiol. Heart Circ. Physiol. 2009;296:H1625–1632. doi: 10.1152/ajpheart.01057.2008. [DOI] [PubMed] [Google Scholar]
- 11.Boheler KR, Carrier L, de la Bastie D, Allen PD, Komajda M, Mercadier JJ, Schwartz K. J. Clin. Invest. 1991;88:323–330. doi: 10.1172/JCI115295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Driesen RB, Verheyen FK, Debie W, Blaauw E, Babiker FA, Cornelussen RN, Ausma J, Lenders MH, Borgers M, Chaponnier C, Ramaekers FC. J. Cell. Mol. Med. 2009;13:896–908. doi: 10.1111/j.1582-4934.2008.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ravenscroft G, Colley SM, Walker KR, Clement S, Bringans S, Lipscombe R, Fabian VA, Laing NG, Nowak KJ. Neuromuscul. Disord. 2008;18:953–958. doi: 10.1016/j.nmd.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Vandekerckhove J, Weber K. Differentiation. 1979;14:123–133. doi: 10.1111/j.1432-0436.1979.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 15.Maier T, Güell M, Serrano L. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 16.Gygi SP, Rochon Y, Franza BR, Aebersold R. Mol. Cell. Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stilli D, Bocchi L, Berni R, Zaniboni M, Cacciani F, Chaponnier C, Musso E, Gabbiani G, Clement S. Exp. Physiol. 2006;91:571–580. doi: 10.1113/expphysiol.2005.032607. [DOI] [PubMed] [Google Scholar]
- 18.Kurien BT, Scofield RH. Methods. 2006;38:283–293. doi: 10.1016/j.ymeth.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Taylor SC, Berkelman T, Yadav G, Hammond M. Mol Biotechnol. 2013;55:217–226. doi: 10.1007/s12033-013-9672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chait BT. Kornberg RD, Raetz CRH, Rothman JE, Thorner JW, editors. Ann. Rev. Biochem. 2011;80:239–246. doi: 10.1146/annurev-biochem-110810-095744. [DOI] [PubMed] [Google Scholar]
- 21.Mann M, Jensen ON. Nat. Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 22.Mann M, Hendrickson RC, Pandey A. Annu. Rev. Biochem. 2001;70:437–473. doi: 10.1146/annurev.biochem.70.1.437. [DOI] [PubMed] [Google Scholar]
- 23.Kelleher NL, Thomas PM, Ntai I, Compton PD, LeDuc RD. Expert Rev Proteomic. 2014;11:649–651. doi: 10.1586/14789450.2014.976559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gregorich ZR, Ge Y. Proteomics. 2014;14:1195–1210. doi: 10.1002/pmic.201300432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siuti N, Kelleher NL. Nat Methods. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Ge Y. Circ.-Cardiovasc. Genet. 2011;4:711. doi: 10.1161/CIRCGENETICS.110.957829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng Y, Chen X, Zhang H, Xu Q, Hacker TA, Ge Y. J Proteome Res. 2013;12:187–198. doi: 10.1021/pr301054n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng Y, Gregorich ZR, Valeja SG, Zhang H, Cai W, Chen YC, Guner H, Chen AJ, Schwahn DJ, Hacker TA, Liu X, Ge Y. Mol Cell Proteomics. 2014;13:2752–2764. doi: 10.1074/mcp.M114.040675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong XT, Sumandea CA, Chen YC, Garcia-Cazarin ML, Zhang J, Balke CW, Sumandea MP, Ge Y. J Biol Chem. 2012;287:848–857. doi: 10.1074/jbc.M111.293258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ge Y, Rybakova IN, Xu Q, Moss RL. Proc. Natl. Acad. Sci. U.S. A. 2009;106:12658–12663. doi: 10.1073/pnas.0813369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Guy MJ, Norman HS, Chen YC, Xu QG, Dong XT, Guner H, Wang SJ, Kohmoto T, Young KH, Moss RL, Ge Y. J Proteome Res. 2011;10:4054–4065. doi: 10.1021/pr200258m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chamot-Rooke J, Mikaty G, Malosse C, Soyer M, Dumont A, Gault J, Imhaus AF, Martin P, Trellet M, Clary G, Chafey P, Camoin L, Nilges M, Nassif X, Dumenil G. Science. 2011;331:778–782. doi: 10.1126/science.1200729. [DOI] [PubMed] [Google Scholar]
- 33.Ryan CM, Souda P, Bassilian S, Ujwal R, Zhang J, Abramson J, Ping P, Durazo A, Bowie JU, Hasan SS, Baniulis D, Cramer WA, Faull KF, Whitelegge JP. Mol. Cell. Proteomics. 2010;9:791–803. doi: 10.1074/mcp.M900516-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tran JC, Zamdborg L, Ahlf DR, Lee JE, Catherman AD, Durbin KR, Tipton JD, Vellaichamy A, Kellie JF, Li M, Wu C, Sweet SM, Early BP, Siuti N, LeDuc RD, Compton PD, Thomas PM, Kelleher NL. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katz AM, Hall EJ. Circ. Res. 1963;13:187–198. doi: 10.1161/01.res.13.3.187. [DOI] [PubMed] [Google Scholar]
- 36.Pardee JD, Spudich JA. Methods Enzymol. 1982;85(Pt B):164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y-C, Sumandea M, Larsson L, Moss R, Ge Y. J. Muscle Res. Cell. Motil. 2015:1–13. doi: 10.1007/s10974-015-9404-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guner H, Close PL, Cai W, Zhang H, Peng Y, Gregorich ZR, Ge Y. J. Am. Soc. Mass Spectrom. 2014;25:464–470. doi: 10.1007/s13361-013-0789-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horn DM, Zubarev RA, McLafferty FW. J Am Soc Mass Spectrom. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 40.Asatoor AM, Armstrong MD. Biochem. Biophys. Res. Commun. 1967;26:168–174. doi: 10.1016/0006-291x(67)90229-x. [DOI] [PubMed] [Google Scholar]
- 41.Johnson P, Harris CI, Perry SV. Biochem J. 1967;105:361–370. doi: 10.1042/bj1050361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nyman T, Schuler H, Korenbaum E, Schutt CE, Karlsson R, Lindberg U. J. Mol. Biol. 2002;317:577–589. doi: 10.1006/jmbi.2002.5436. [DOI] [PubMed] [Google Scholar]
- 43.Ye L, Chang Y-H, Xiong Q, Zhang P, Zhang L, Somasundaram P, Lepley M, Swingen C, Su L, Wendel JS, Guo J, Jang A, Rosenbush D, Greder L, Dutton JR, Zhang J, Kamp TJ, Kaufman DS, Ge Y, Zhang J. Cell Stem Cell. 2014;15:750–761. doi: 10.1016/j.stem.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Dong X, Hacker TA, Ge Y. J. Am. Soc. Mass Spectrom. 2010;21:940–948. doi: 10.1016/j.jasms.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ong SE, Mann M. Nat. Chem. Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 46.Clément S, Stouffs M, Bettiol E, Kampf S, Krause K-H, Chaponnier C, Jaconi M. J. Cell Sci. 2007;120:229–238. doi: 10.1242/jcs.03340. [DOI] [PubMed] [Google Scholar]
- 47.Whitelegge JP, Zhang HM, Aguilera R, Taylor RM, Cramer WA. Mol Cell Proteomics. 2002;1:816–827. doi: 10.1074/mcp.m200045-mcp200. [DOI] [PubMed] [Google Scholar]
- 48.Thangaraj B, Ryan CM, Souda P, Krause K, Faull KF, Weber APM, Fromme P, Whitelegge JP. Proteomics. 2010;10:3644–3656. doi: 10.1002/pmic.201000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kostyukevich Y, Vladimirov G, Nikolaev E. J. Am. Soc. Mass Spectrom. 2012;23:2198–2207. doi: 10.1007/s13361-012-0480-1. [DOI] [PubMed] [Google Scholar]
- 50.Makarov A, Denisov E, Lange O. J. Am. Soc, Mass Spectrom. 2009;20:1391–1396. doi: 10.1016/j.jasms.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 51.Zabrouskov V, Ge Y, Schwartz J, Walker JW. Mol. Cell. Proteomics. 2008;7:1838–1849. doi: 10.1074/mcp.M700524-MCP200. [DOI] [PubMed] [Google Scholar]
- 52.Pesavento JJ, Mizzen CA, Kelleher NL. Anal. Chem. 2006;78:4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- 53.Lim D-S, Roberts R, Marian AJ. J. Am. Coll. Cardiol. 2001;38:1175–1180. doi: 10.1016/s0735-1097(01)01509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rajabi M, Kassiotis C, Razeghi P, Taegtmeyer H. Heart Fail. Rev. 2007;12:331–343. doi: 10.1007/s10741-007-9034-1. [DOI] [PubMed] [Google Scholar]
- 55.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 56.Izumo S, Nadal-Ginard B, Mahdavi V. Proc. Natl. Acad. Sci. U. S. A. 1988;85:339–343. doi: 10.1073/pnas.85.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.