Abstract
Myxoid liposarcoma has the pathognomonic fusion oncogene FUS-DDIT3 encoding a chimeric transcription factor. Metastatic risk is higher with an increased round cell component and has been linked to aberrations involving the IGFR/PI3K/AKT pathway. These molecular insights have yet to translate to targeted therapies and the lack of experimental models is a major hindrance. We describe the initial in-depth characterization of a new cell line (DL-221) and establishment of a mouse xenograft model.
The cell line DL-221 was derived from a metastatic pleural lesion showing myxoid and round cell histology. This newly established cell line was characterized for phenotypic properties and molecular cytogenetic profile, using PCR, COBRA-FISH and western blot. Next-generation whole exome sequencing was performed to further characterize the cell line and the parent tumor. NOD-SCID-IL2R gamma knockout mice were xenograft hosts.
DL-221 cells grew an adhering monolayer and COBRA-FISH showed an aneuploid karyotype with t(12;16)(q13;p11) and several other rearrangements; RT-PCR demonstrated a FUS-DDIT3 fusion transcript type 1. Both the cell line and the original tumor harbored a TP53 compound heterozygous mutation in exon 4 and 7 and were wild type for PIK3CA. Moreover, among the 1254 variants called by whole exome sequencing, there was 77% concordance between the cell line and parent tumor. The recently described hotspot mutation in the TERT promoter region in myxoid liposarcomas was also found at C228T in DL-221. Xenografts suitable for additional pre-clinical studies were successfully established in mice after subcutaneous injection.
The established DL-221 cell line is the only published available myxoid liposarcoma cell line that underwent spontaneous immortalization, without requiring SV40 transformation. The cell line and its xenograft model are unique and helpful tools to study the biology and novel potential targeted treatment approaches for myxoid liposarcoma.
Keywords: Myxoid liposarcoma, round cell liposarcoma, translocation, FUS-DDIT3, TP53, PIK3CA, HDAC, TERT, CTAG1B, exome sequencing, targeted therapy
Introduction
Myxoid liposarcoma (MLS) is a malignant mesenchymal neoplasm, representing one third of the liposarcomas and accounting for 5% of all adult soft tissue sarcomas.1 Histologically MLS is composed of a variably cellular ovoid cell proliferation with signet-ring lipoblasts set in a myxoid matrix with characteristic plexiform vasculature. Areas of undifferentiated small round cells with scarce cytoplasm and no matrix (round cell variant) are associated with a higher metastatic potential and a worse prognosis.2–8 Other poor prognostic factors are the presence of necrosis, male gender, increasing age and multifocal disease.2;8–10
Localized MLS can be effectively treated with surgery or the combination of surgery and radiation, but 35–50% of patients develop recurrence or metastases. The metastatic pattern is unconventional with spread to sites in soft tissue or bone sometimes occurring before the tumor metastasizes to the lungs.1;6;11 Although MLS is relatively sensitive to radiotherapy and certain chemotherapeutic agents, treatment of extensive disease is challenging.12
MLS is characterized by a reciprocal translocation generating a fusion transcription factor oncogene. In >95% a translocation t(12;16)(q13;p11) is present, fusing FUS (fused in sarcoma; a.k.a. TLS) with DDIT3 (DNA-damage-inducible transcript 3; a.k.a. CHOP).13;14 At least eleven different FUS-DDIT3 fusion types have been described and the fusion type does not impact clinical outcome.2;13;15 Less than 5% of the cases harbour a t(12;22)(q13;q12) leading to an EWSR1-DDIT3 fusion, of which four different transcripts are described.1;15;16
Although the exact mechanism via which the chimeric transcription factor exerts its oncogenic effects remains to be elucidated, it is postulated that it acts as an aberrant transcriptional regulator, stimulating proliferation while inhibiting adipogenic differentiation.17–19 The chimeric product is highly expressed and interferes with heterodimerization of DDIT3 with CCAAT/enhancer-binding protein-β (C/EBPβ). The activity of the transcription factors C/EBPα and PPARγ is inhibited and fat differentiation is blocked.19
Exome sequencing and biomarker analysis of MLS specimens has identified alterations in the IGF/Akt/mTOR axis, implicated in cellular processes such as cell survival, proliferation and growth. Overexpression of the receptor tyrosine kinases AXL, RET and IGF1R, and the ligand IGF1, are negative prognostic biomarkers.8;20;21 Activating mutations in PIK3CA are found in 14–18% of MLS and loss of expression of PTEN is found in 12% of the tumors and is mutually exclusive from PIK3CA mutations.22;23 Increased PI3K/Akt signalling has been demonstrated by high expression of downstream targets like phosphorylated 4EBP1, PRAS40 and S6. The PIK3CA mutation rate, IGFR expression and loss of PTEN were higher in tumors with a round cell component suggesting that this pathway might be involved in round cell transformation and tumor progression. TP53 mutations and reduced protein expression of p16INK4/p14ARF have been identified in a subset of tumors, most frequently in round cell components.24 Hotspot mutations in the TERT (telomerase reverse transcriptase) promoter region were recently reported in MLS (23% to 74%).25;26 These mutations led to increased protein expression of TERT and have been implicated in telomerase dysregulation and the resultant proliferative capability of tumor cells.27 The cancer-testis antigen NY-ESO-1 (a.k.a. CTAG1B) has recently shown to be almost universally expressed in MLS (89–100%).28–30 NY-ESO-1 expression is normally limited to germ cells making it a good cancer immunotherapeutic target.31;32
Over the last few years, we have gained increasing insight into the molecular pathogenesis of MLS; however, translating this knowledge into specific therapies has been challenging. Reliable in vitro and in vivo models are crucial to investigate novel therapies and to study the mechanisms of action. Although MLS is a common sarcoma subtype, so far only two MLS cell lines (402-91 and 1765-92) were established, both immortalized by transfection with the SV40 large T-antigen, and have been made available worldwide.33;34
In this study, we present the establishment and characterization of a new MLS cell line (DL-221), which to our knowledge is the first spontaneously immortalized cell line of this tumor type. This was undertaken as part of the International Myxoid Liposarcoma Consortium and our motivation was to facilitate preclinical research by having representative models to test new therapies.
Materials and Methods
Myxoid liposarcoma primary culture and cell lines
With approval of the Institutional Review Board (IRB) of The University of Texas MD Anderson Cancer Center and with patient’s written informed consent, isolation of tumor cells was performed as described previously.35 Cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (Gemini bio-products, West Sacramento, CA) and 1% penicillin-streptomycin (100U/ml), and placed in a humidified 5% CO2 incubator at 37°C. The previously established human MLS cell lines 402-91 and 1765-92 were cultured in RPMI1640 (Thermo Fisher Scientific, Waltham, MA) supplemented with FBS and antibiotics. The population doubling time was calculated during the exponential growth phase using the algorithm provided by Doubling Time software (http://www.doubling-time.com). All cell lines were tested on a regular basis for mycoplasma infection. Short Tandem Repeat (STR) DNA fingerprinting was performed on the cell lines, the metastatic tumor tissue from which the DL-221 cell line was derived and from xenografted tissue (AmpFLSTR Identifiler PCR Amplification kit; Applied Biosystems, Foster City, CA). The data was analyzed with GeneMapper (v4.0, Applied Biosystems).
Fluorescence in situ hybridization (FISH)
Two color FUS split apart FISH was performed on interphase cells using a probe mix containing bacterial artificial chromosome (BAC) clone RP11–196G11 direct labeled with FITC (FITC-dUTP) (Roche diagnostics GmbH, Mannheim, Germany) and clone RP11–120K18 direct labeled with Cy3 (Roche). Both clones flank the FUS gene and are separated by 120 kbp. Scoring of the split-apart FISH was done by counting at least 100 nuclei; signals were considered split-apart when their distance was larger than the size of a single hybridization signal. Harvested metaphase cells were used for COBRA-FISH. Pictures were taken with a DMRA microscope (Leica, Rijswijk, The Netherlands) and karyotypes analyzed with Colorproc (v2.0, LUMC, Leiden, The Netherlands). A detailed description of the harvesting, hybridization, probe labeling and COBRA-FISH protocols has been published.36;37
FUS-DDIT3 fusion type characterization
Total RNA was extracted from cells using RNeasy Mini Kit (Venio, The Netherlands) as per manufacturer instructions. cDNA was generated using the SuperScript III reverse transcriptase kit (Life Technologies, Carlsbad, CA) and PCR was performed with Taq polymerase (Life Technologies). Sanger sequencing of the amplified product was performed; Chromas (Technelysium, South Brisbane, QLD, Australia) and the BLAST Multiple Alignment tool (NCBI) were used to depict the sequence and analyze the breakpoints, respectively.23 A detailed description of PCR conditions has been published previously (see Table S1 for primers).13
Immunoprecipitation, SDS-PAGE and western blot
Immunoprecipitation was performed on protein lysates from near-confluent cells. The cells were washed twice with ice-cold phosphate-buffered saline (PBS) and harvested by scraping in ice-cold lysis buffer containing 15 mM Tris (pH 8.0), 100 mM NaCl, 1% Triton X-100, supplemented with protease inhibitor cocktail (P2714, Sigma-Aldrich, St. Louis, MO). Lysates were incubated on ice for 30 min, clarified by centrifugation at 4°C, followed by protein estimation using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Overnight immunoprecipitation with anti-DDIT3 (sc-793, Santa Cruz Biotechnology, Dallas, TX) was performed using Dynabeads® Protein G (Life Technologies, Carlsbad, CA) as per manufacturer instructions. Next, SDS-PAGE separation and western blot were performed using anti-FUS (sc-373698, Santa Cruz) and anti-DDIT3 (ab10444, Abcam, Cambridge, UK) for detection. The following Cell Signaling Technology (Danvers, MA) antibodies were also used for western blot: anti-AKT (#2920), anti-phospho-AKT (Ser473) (#9271), anti-AXL (#4566) and PTEN (#9559). Antibodies of Abcam were used for western blot analysis of HDAC1 (ab19845), HDAC2 (ab32117) and HDAC3 (ab32369).38
Molecular characterization: mutation analysis and myxoid liposarcoma associated biomarkers
Mutation analysis was performed for exon 4 to 10 of TP53, exons 9 and 20 of PIK3CA and the TERT promoter region by conventional Sanger sequencing as described elsewhere (Table S1).23 Detailed procedures of NY-ESO-1 immunohistochemistry have been described previously.30
Cell viability assay
In vitro cell viability assays were performed as described previously.39 Briefly, metabolic activity was measured by a WST-1 colorimetric assay as read-out for the cell viability after doxorubicin treatment. Cells were treated with increasing doxorubicin concentrations for 72hr in quadruplicates and experiments were performed 2 to 4 times. GraphPad Prism (v6.05, La Jolla, CA) was used to calculate the IC50-values.
In vivo animal model
All animal procedures and care were approved by the MD Anderson Cancer Center Institutional Animal Care and Usage Committee. Animals received humane care as per the Animal Welfare Act and the NIH “Guide for the Care and Use of Laboratory Animals”. Mouse studies were performed as described previously.40 Briefly, 3×106 cells were injected subcutaneously in the flank of (n=2/cell line) 6 weeks old female NOD-SCID-ILR2 gamma mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, The Jackson Laboratory, Bar Harbour, ME). Tumor growth was assessed twice weekly with a digital caliper (Mitutuyo, Chicago, IL); tumor volume was calculated by using the formula (tumor volume (mm3) = length × width × height × 0.5). Mice injected with DL-221 cells were sacrificed at 79 and 92 days post injection due to ulcerated tumor burden. The tumors were resected, weighed (Ohaus Adventurer® Pro Scale), fixed in formalin and embedded in paraffin. Sections were H&E stained to evaluate tumor histology. FUS split-apart FISH was performed to confirm the presence of the translocation.
Sequencing
Whole exome sequencing was performed on DNA isolated from the DL-221 metastatic tumor tissue and its derivative cell line at passage 33/34. Coding regions were captured with baits from the Agilent SureSelect Human All Exon V4 (Agilent Technologies, Inc., Santa Clara, CA). After selecting for 200 bp median fragment sizes, the samples were run on an Illumina HiSeq 2000 (Illumina Inc., CA) to generate 76 bp paired-end reads. Duplicate reads were removed using Picard and mapping was done using BWA,41 giving an overall mean coverage of 69X for the metastatic tumor and 62X for the derived cell line. The percentages of reads having at least 50X mean coverage were 75% and 65%, respectively.
Variants were called separately for each sample using VarScan142 against the human reference genome. Further comparisons between samples were done by using MuTect43 and Pindel44 with each one of either the tumor or cell line being set as the “reference” with 3% allele frequency cutoff for making calls. To confirm the translocation, Breakdancer45 and Meerkat46 were used. Visual comparisons of allele frequency patterns were made using R: Vioplot47 scatterplot, and 2D kernel density plots. Annotations were obtained using AnnoVar, including protein predictors Mutation Assessor and Polyphen2. Pathway analysis was performed using DAVID.48;49 The exome sequence data has been deposited in the European Genome-phenome Archive (EGA).
Results
Patient history and establishment of the DL-221 cell line
The cell line was generated from a metastatic tumor deposit from the pleura of a 42-year-old male patient (Figure 1A). The patient was originally diagnosed with myxoid liposarcoma of the right anterior thigh that was treated with surgical resection and adjuvant radiation therapy. One year later he was diagnosed with metastatic disease involving the right periorbital area and chest wall (10th rib) for which he received adriamycin/ifosfamide followed by right lateral orbitotomy and chest wall resection. Subsequently, the patient developed multiple metastatic lesions involving the thoracic, lumbar spine and sacrum for which he received multiple chemotherapy regimens. The pleural lesion that was utilized to establish the cell line had been treated with trabectedin prior to resection. The pathology report indicated that the metastatic lesion was found to have a myxoid and sclerotic component. In addition, round cell change was identified. He developed additional metastatic disease involving the lungs, pleura, posterior diaphragmatic region, pelvic muscles and presacral space for which he was again treated with trabectedin followed by single agent doxorubicin. He ultimately succumbed to the disease.
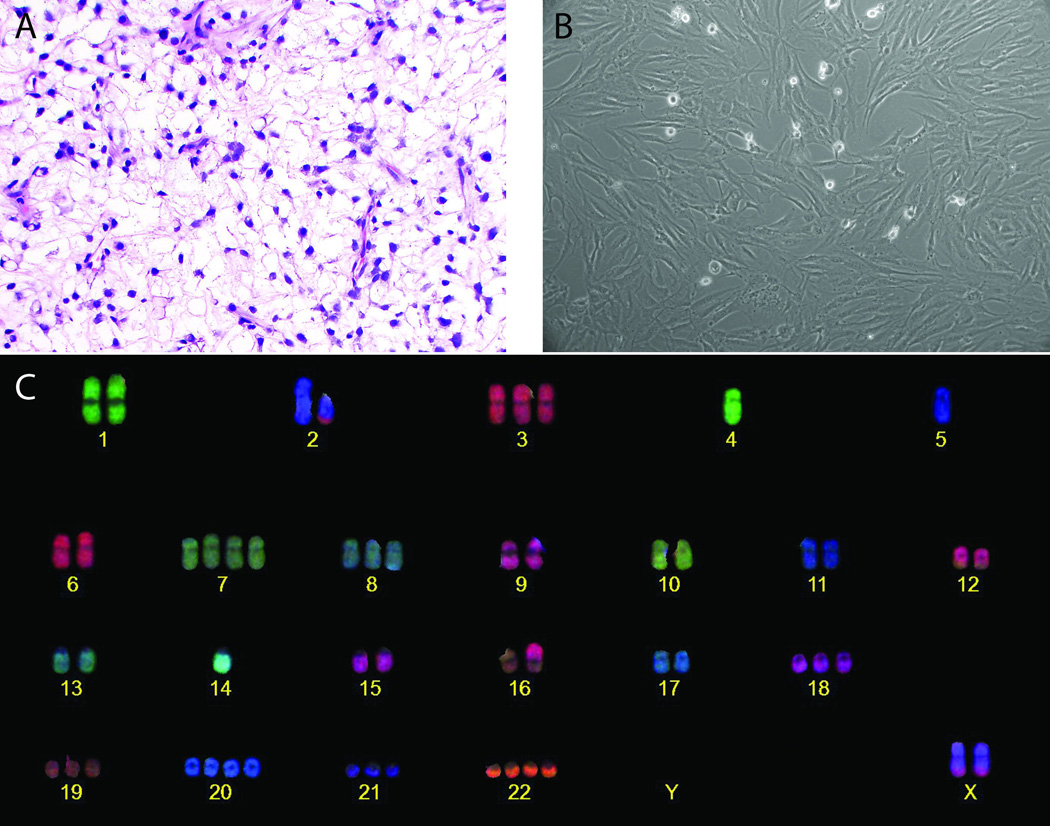
Figure 1. Establishment of the myxoid liposarcoma DL-221 cell line with the characteristic translocation t(12;16)(q13;p11).
(A) Representative H&E-section of the tumor of the pleura of which the tumor cells were isolated for the establishment of the DL-221 cell line (magnification 40×). (B) Tumor cells in culture showing spindle-shaped morphology and nuclei with large nucleoli. (C) COBRA-FISH karyogram of DL-221 cell line showing a near triploid karyotype with several numerical and structural aberrations.
Isolated tumor cells from the metastatic pleura tumor were originally slow growing and passaged once a month, until after 14 months they underwent spontaneous immortalization and a fast growing clone emerged. This clone was isolated and expanded, and after 24 consecutive months of culturing, the cell line has reached over 90 passages. The cell line has a population doubling time of 30 hours calculated at passage 62; this was stable at least through passage 90. Tumor cells grow in an adherent monolayer fashion; phase contrast microscopy revealed spindle cell morphology in monolayer culture (Figure 1B). Cells are mononuclear and the enlarged nuclei contain one or more clearly discernible nucleoli. The cells do not exhibit contact inhibition when reaching confluence. A genetic profile of the cell line was generated by STR typing and was consistent with the metastatic lesion (Table S2).
COBRA-fluorescence in situ hybridization
The presence of the diagnostic FUS-DDIT3 translocation was monitored with FUS split-apart FISH at several times during the culture process to assure a pure culture of tumor cells. The composite karyotype of the DL-221 cell line at passage p31 and p68 was 71~75<3n±>,der(X)t(X;15)(q;q)x2,-Y,der(2)t(X;2)(q;p)t(2;3)(q;p)x2,-4,+5,der(7)t(7;X)(q;q),+i(7)(q10),+7,+8,+8,-9,+11,t(12;16)(q13;p11),-13,+14,-15,der(17)t(10;17)(p;p),+18,+19,+20,+21,idic(22)[10]/ 71~75<3n±>,der(X)t(X;15)(q;q)x2,-Y,der(2)t(X;2)(q;p)t(2;3)(q;p)x2,der(4)t(2;4)(q;p),-4,+5,der(7)t(5;7)(p;q),der(7)t(7;X)(q;q),+8,+8,-9,+10,+11,t(12;16)(q13;p11)x2,-13,+14,-15,+18,+19,+20,+21,+22[6] (Figure 1C). Several additional non-clonal rearrangements were found. The cell line has at least two main subpopulations based on the presence of one or two copies of the translocation t(12;16)(q13;p11) and with two or one copies of chromosome 12 and 16 to complement the triploid genome, respectively. The two different clones are still present in the same proportion, two copies of the translocation versus one copy of the translocation in a ratio of 1.0:1.7, after long term in culture. All chromosomal aberrations that are found at least two times are included in the composite karyotype. The karyotypes are made according to the guidelines of the International System for Human Cytogenetic Nomenclature.50
Fusion-type characterization
The FUS-DDIT3 fusion type was determined with reverse transcriptase-PCR and sequencing. A type 1 fusion was found in the DL-221 cell line, fusing the first seven exons of FUS to the entire reading frame of the DDIT3 gene. The 402-91 and 1765-92 cell lines were included as positive controls in Figure 2A and show as expected type 1 and type 8 fusions, respectively. After electrophoresis the chimeric products reveal a band at the position corresponding to the size of the fusion product. A band was observed around 136 bp in 402-91 and DL-221, consistent with a type 1 fusion product, whereas a larger band at 730 bp was found for 1765-92 due to additional FUS exons in the type 8 fusion product.
Figure 2. Fusion type characterization of the DL-221 cell line and proving the presence of the FUS-DDIT3 chimeric fusion oncoprotein.
(A) RT-PCR was performed on the three myxoid liposarcoma cell lines. This confirms a type 1 fusion product (136 bp) in both DL-221 and 402-91, while the longer fusion product of 730 bp in 1765-92 was consistent with the type 8 fusion product. (B) Immunoprecipitation of FUS-DDIT3 from the myxoid liposarcoma cell lines 402-91, 1765-92 and DL-221 using an anti-DDIT3 antibody. FUS-DDIT3 (arrow) was detected in all three cell lines at around 75 kDa in 402-91 and DL-221 (expressing type 1 FUS-DDIT3) and around 100 kDa (expressing the larger type 8 FUS-DDIT3).
Immunoprecipitation
To prove the presence of the FUS-DDIT3 chimeric fusion oncoprotein in the myxoid liposarcoma cell lines, immunoprecipitation experiments were performed with a DDIT3 antibody on protein lysates from 402-91, 1765-92 and DL-221. Further analysis with SDS-PAGE and immunoblotting with antibodies against DDIT3 and FUS revealed one band for DDIT3 and two bands picked up by the FUS antibody in all samples. The estimated molecular weight of the protein detected with anti-DDIT3 is around 75 kDa in 402-91 and DL-221 and around 100 kDa in 1765-92, corresponding to the estimated weight of the chimeric fusion proteins of 75 kDa and 100 kDa for type 1 and 8 fusion proteins, respectively. Two bands picked up by the anti-FUS antibody were detected in all three cell lines; one band was present at the same level as the DDIT3 band (i.e. FUS-DDIT3) whereas the other band with a weight of 68 kDa represents native FUS protein. The abundant presence of normal FUS is expected based on the high transcriptional activity of this gene; as expected, no expression of the stress-induced native DDIT3 was detected (Figure S1).51 Merging of the immunoblots of FUS (green) and DDIT3 (red) revealed a yellow band in each of the three cell lines, confirming that it represents the fusion oncoprotein (Figure 2B).
Molecular characterization: mutation analysis and myxoid liposarcoma associated biomarkers
Target panel sequencing revealed two different TP53 mutations in the cell line, c.374C>G, p.T125R in exon 4 and c.715A>G, p.N239D in exon 7. The original tumor also harbored these pathogenic variations.52 Mutational analysis of the hotspot regions in exons 9 and 20 of PIK3CA did not identify an aberration in the tumor or cell line, nor any other mutations in this gene as confirmed with whole exome sequencing. No TP53 nor PIK3CA aberrations were found in 402-91 or 1765-92. Mutational analysis of the TERT promoter region revealed a C228T mutation in DL-221 (Figure S2). Next, we confirmed the presence of the previously reported C228T hotspot mutation in 402-91 and 1765-92. None of the cell lines harbored the second reported hotspot mutation in MLS, C250T.26
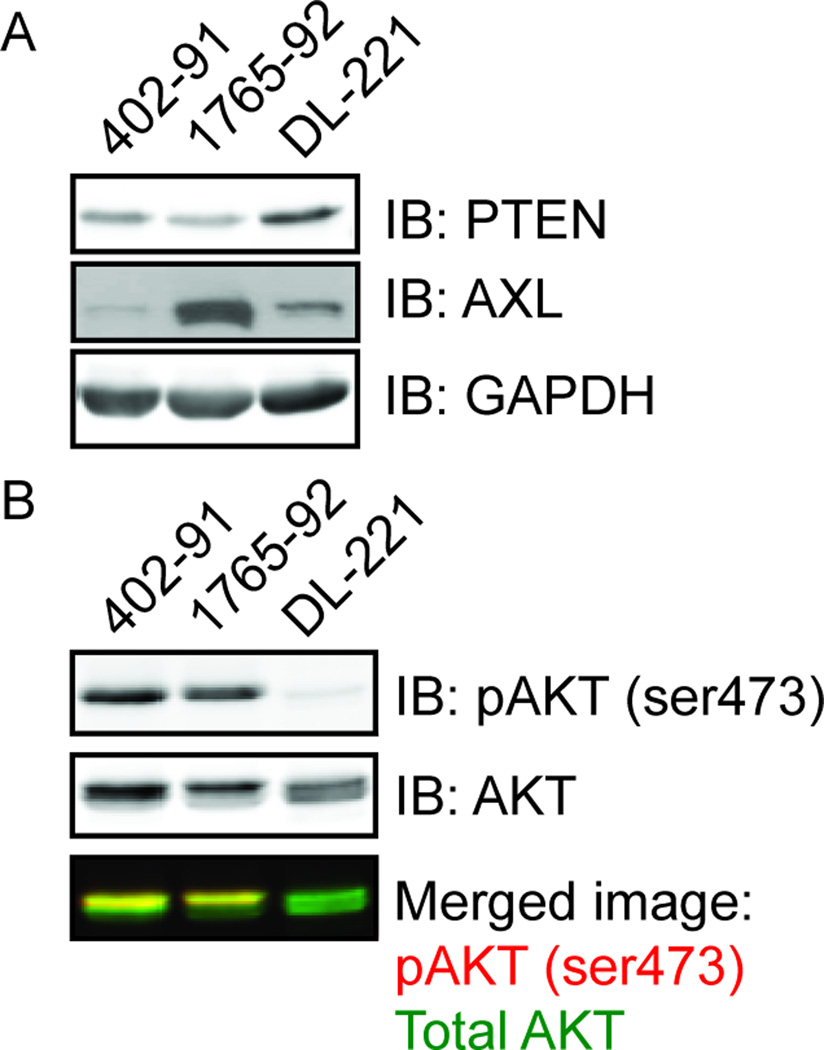
PTEN expression was retained in all three cell lines (Figure 3A). Analysis of the expression of MLS associated biomarkers with western blots showed a strong expression of total Akt in all three cell lines, while phospho-Akt (pAkt-473) was only found in 402-91 and 1765-92 (Figure 3B). The presence of p53 protein in DL-221 was confirmed with western blot analysis, which must be a mutated protein. The p53 protein was abundantly present in 402-91 and 1765-92, but the expression levels are not necessarily relevant in these two cell lines due to the SV40-immortalisation, as reported previously.53 AXL was highly expressed in 1765-92, moderately expressed in DL-221 and minimally expressed in 402-91 (Figure 3A). The protein expression of HDAC-1, −2 and −3 was evaluated. All three histone deacetylase family members were expressed in 402-91, all except HDAC-3 in 1765-92 and, HDAC-2 and −3 in DL-221 (Figure S3). The 402-91 had a moderate immunohistochemical expression of NY-ESO-1; both DL-221 and 1765-92 showed strong expression. The biomarker status of the three MLS cell lines is shown in Table 1.
Figure 3. PTEN and pAKT protein expression.
Western blot analysis of (A) PTEN and AXL expression, with GAPDH expression as a loading control, and (B) pAKT (ser473) expression relative to total AKT expression in the myxoid liposarcoma cell lines. The bottom panel shows the merged image of the pAKT (ser473) signal in red and total AKT signal in green. An overlap of both signals produces a yellow band.
Table 1.
Molecular biomarkers in myxoid liposarcoma cell lines (402-91, 1765-92, DL-221).
| Cell line |
FUS- DDIT3 fusion |
TP53 | P53 (WB) | PIK3CA | TERT | p-AKT473/Total AKT (WB) |
AXL (WB) | HDAC1/2/3 (WB) |
NY-ESO-1 (IHC) |
|---|---|---|---|---|---|---|---|---|---|
| 402-91 | Type 1 | WT | Positive | WT | C228T | +/++ | + | +/++/+ | + |
| 1765-92 | Type 8 | WT | Positive | WT | C228T | +/++ | +++ | +/+/− | ++ |
| DL-221 | Type 1 | T125R, N239D |
Positive | WT | C228T | −/++ | ++ | −/+/+ | ++ |
WB = western blot; WT = wild type; IHC = immunohistochemistry
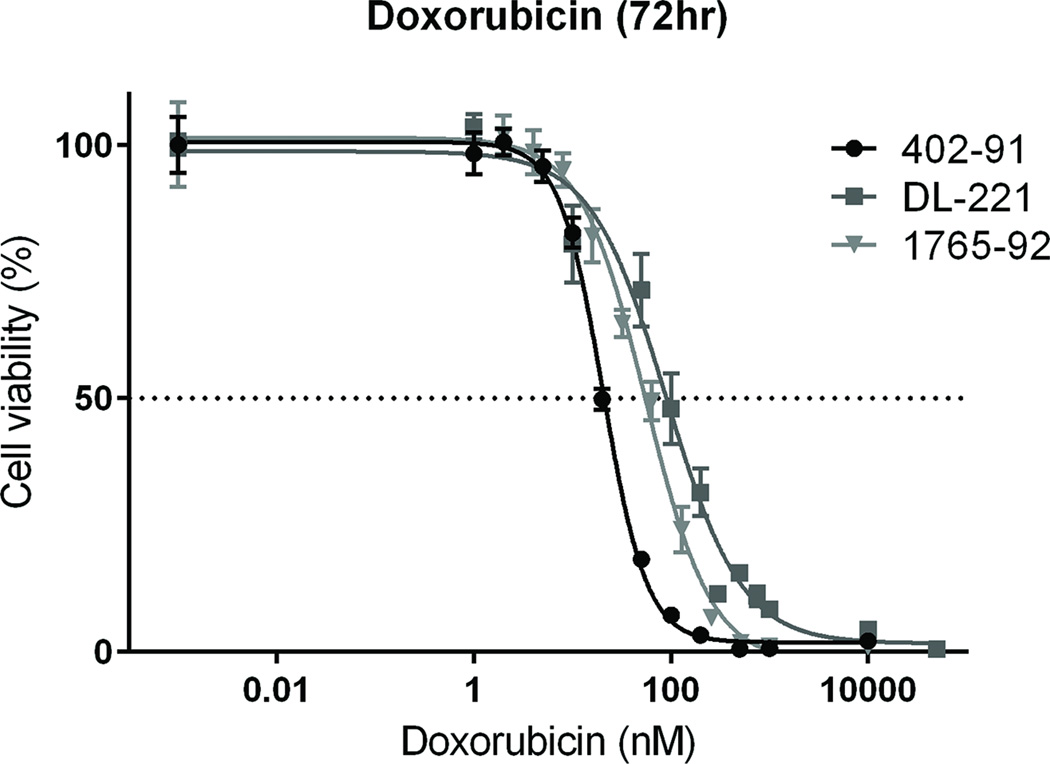
Cytotoxicity experiments with colorimetric assays were performed to determine the effect of treatment on cell viability. The conventional chemotherapeutic doxorubicin was highly toxic for all three MLS cell lines. For DL-221 the IC50 values were in a range of 35 to 89 nM (average 62 nM) and the IC50 values for 402-91 and 1765-92 were 24 nM (range 21–29 nM) and 47 nM (range 31–55 nM) respectively after 72 hour treatment (Figure 4).
Figure 4. Dose-response curves of MLS cell lines to doxorubicin treatment in vitro.
All three cell lines were treated with doxorubicin for 72 hour. For each cell line, one representative experiment of consecutive experiments is depicted.
Mouse studies
DL-221 cells from passage 15 were injected subcutaneously in the flank of two NOD-SCID-ILR2 gamma mice. Tumor development was observed in both mice and they were sacrificed at day 79 and 92 post-injection, due to ulcerated tumor burden (Figure 5A). No metastatic lesions were found. Histological evaluation of the xenografts revealed a similar morphology, consisting of a moderately cell-rich, myxoid lesion with short spindle-shaped, moderately pleomorphic tumor cells. Marked nuclear atypia with hyperchromatic and heterogeneous nuclei was present and large nucleoli were visible in the majority of the cells (Figure 5B). A FUS-split apart FISH assay was performed on FFPE sections from both xenografts and revealed a segregation of the signals, consistent with the expected rearrangement at the FUS locus (Figure 5C). Cells obtained from both first generation resections were successfully passaged. The STR profile of the xenografts revealed the same profile as the original tumor and the cell line (Table S2). Injection of the two other available MLS cell lines, 402-91 and 1765-92, did not lead to tumor formation in vivo in 16 months.
Figure 5. Xenograft model of DL-221 cell line.
(A) tumor growth in two mice, (B) H&E picture of xenograft tumor (magnification 40×) and (C) FUS split-apart FISH reveals a break in the FUS gene in the tumor cells in mice, the gain of the green signal was consistent with the karyotype image shown in figure 1.
Next generation sequencing
The metastatic tumor and its derivative cell line both underwent whole exome sequencing to an average mean coverage of 60X. Using Varscan1 to compare each sample to the hg19 reference genome, a total of 1254 exonic variants were called (1230 nonsynonymous, 23 stopgains, 1 stoploss), 960 of which were shared between the two samples. Among those shared, 38 genes overlap with the Sanger Cancer Gene Census, 10 of which were “Probably damaging” by Polyphen2 and 7 with “Medium Impact” by Mutation Assessor. Analysis of these alleles using DAVID showed significant p-values from both Panther (p = 5.2 × 10−5) and KEGG (p = 4.8 × 10−4) for the Wnt signaling pathway, which includes Polyphen2-predicted “probably damaging” variants in APC (p.R99W), APC2 (p.G323R), and TP53 (p.N239D, p.T125R) (Table S3). Ninety percent (863/960) of the alleles retained insignificant allele frequency changes of <0.2 between the metastatic tumor and cell line (Figure S4). Driver mutations such as the two TP53 missense mutations p.T125R and p.N239D that were characterized in the primary tumor were confirmed in both samples and retained consistent allele frequencies (metastatic p.T125R 0.43 allele freq, depth = 54, cell line 0.46 allele freq, depth = 54; p.N239D metastatic 0.4 allele freq, depth = 37, cell line 0.32 allele freq, depth = 50). These two variants are not found in any of the population databases ESP6500 and 1000G. As well, the FUS-DDIT3 translocation was confirmed from split reads found using Meerkat and a call found with Breakdancer that was ~1 kb away from the actual breakpoint (Table S4 and S5).46
By MuTect, 63 exonic variants were found in the metastatic tumor that were absent from the cell line, while 32 exonic variants were present in the cell line but not in the tumor. A small group of 28 nonsynonymous alleles were selected for during cell line establishment, as evidenced by increases in allele frequencies of >0.5 (Table S6). There are several possible explanations for these frequency changes. First, all of these alleles come from just three chromosomes: chr2 (9/28), chr4 (6/28), and chr9 (13/28), which could be due to copy number events. The losses of both chromosomes 4 and 9 seen by FISH karyotyping of the cell line would explain the loss of heterozygosity effects that resulted in allele frequencies of 1.00 in the cell line for these two chromosomes. Other possibilities are that clonal selection or purification of the tumor from normal cell contamination resulted in higher allele frequencies, which may explain those alleles from chromosome 2. Of these 28 alleles, five are predicted “Probably damaging” by Polyphen2 (FAT4 p.P1313S; DSPP p.D1152N; SP140L p.C181R; IFIH1 p.L125M; and IPPK p.R130W).
Discussion
Myxoid liposarcoma is a relatively common sarcoma subtype, yet there are only two previously established cell lines that are widely available.33 Representative models for preclinical testing, including cell lines and functioning xenografts, are essential to understanding the underlying molecular determinants of round cell and metastatic progression in this disease and to identify and develop targeted therapeutic strategies.
Here we report the establishment of a new MLS cell line (DL-221) and demonstrate that this cell line can be successfully used in a xenograft model. The cell line was grown from isolated myxoid liposarcoma cells from a metastatic pleura lesion. After 14 months of culturing, a fast growing clone emerged and was expanded. DL-221 has been cultured for more than 24 months and shows a stable doubling time of around 30 hours. In contrast to the two other established MLS cell lines, 402-91 and 1765-92, which were both immortalized by induction of the large T-cell SV40 antigen,33 DL-221 is the first published MLS cell line that underwent spontaneous immortalization. SV40 suppresses the transcriptional activity of the tumor suppressor p53 and hence affects the p53 pathway in the cells. SV40 immortalization also involves the inactivation of other growth suppressors as pRB and SEN6.54 This has to be kept in mind when studying the in vitro effect of new treatment options in these cell lines, as it is not fully representative of the oncogenic process that occurs in human patient tumors. One of the reasons for the limited MLS models could be the difficulty of culturing them in vitro, which has been our experience as well. This might be related to a lack of necessary cytokines, growth factors, adhesion molecules or endothelial cells in the extracellular environment, difficulty in adjusting to a monolayer culture or a combination of these factors. Most primary cultures from MLS tumor tissues slow down after several weeks or months in culture, and finally result in terminal fat differentiation or senescence.
Exome sequencing of the original tumor and the DL-221 cell line was performed to search for new genetic aberrations that are common between the cell line and original tumor and those that arise in the cell line specific to the stress of growing in culture. The FUS-DDIT3 translocation and the TP53 mutations were present in both samples.
The DL-221 cell line was regularly tested for the presence of the chimeric fusion oncogene, which was confirmed by FUS-split apart FISH on interphase cells. COBRA-FISH karyotyping of metaphase cells revealed a near triploid karyotype with several numerical and structural aberrations including the characteristic balanced t(12;16)(q13;p11). The tumor cell population consists of two clones, which differ in the number of copies per cell of this translocation. The most common clonal cell population contains one copy of the translocation and the other clone two (with the ratio of the clones being 1.7:1.0), and this was stable across multiple passages. It is as yet unknown if this has any consequences on the level of FUS-DDIT3 fusion product in the different clones. Fusion type characterization revealed a type 1 product, fusing exon 7 of FUS to the complete coding sequence of DDIT3. This is the same fusion as in the cell line 402-91; while the 1765-92 cell line carries the longer type 8 fusion. No cell line is available with the most common (65%) type 2 fusion; by comparison type 1 fusions are encountered in 22% of patients, whereas type 8 fusions are rare.2;13
MLS can contain several additional molecular genetic alterations, like TP53, PIK3CA and TERT mutations, which directly influence tumor cell biology and might be involved in round cell transformation, migration capacity and differential response to drugs. Alterations of the p53 pathway have been described in several studies on MLS and the percentage of cases with an alteration varies from <5% to 83%.2;8;24;55;56 Whole exome sequencing of the DL-221 cell line revealed two TP53 missense mutations, T125R in exon 4 and N239D in exon 7; both mutations lead to the substitution of an amino acid of a different class and are expected to be deleterious. Sanger sequencing of exon 4 to 10 did confirm the mutations in exons 4 and 7. TP53 alterations are most frequently encountered in the hotspot exons 5 to 8; these two mutations have both been reported before in carcinomas (cosmic database). PIK3CA mutations have been identified in 14–18% of the MLS cases and are associated with round cell progression and shorter disease-specific survival.22;23 Loss of PTEN expression, which is mutually exclusive with PIK3CA mutations, and IGF1R overexpression are other mechanisms to activate the PI3K/Akt pathway. Immunohistochemical expression of the downstream pathway member, phosphorylated 4EBP1, is significantly correlated to pathway activating events, and p-4EBP1 is highly expressed in 60% of MLS. All three cell lines are wildtype for PIK3CA and retain PTEN expression. Total-Akt was expressed in all three cell lines, whereas p-Akt473 was only present in 402-91 and 1765-92, possibly implying that the PI3K pathway is activated in the 402-91 and 1765-92 cell lines but not in the DL-221 cell line. A minor response has been reported in two MLS patients treated with mTOR inhibitors (everolimus and temsirolimus), a downstream target of the PI3K pathway.57 These findings warrant testing of the several new inhibitors of PI3K, Akt and mTOR in the preclinical and clinical setting for MLS. Expression of the receptor tyrosine kinase AXL is associated with metastasis and worse outcome in several malignancies, and has been identified as a prognosticator of disease-specific survival in MLS patients.8 AXL was present in DL-221 and 1765-92 but is only weakly expressed in 402-91. This might indicate that AXL inhibition is a candidate treatment in a subset of MLS. Point mutations in the telomerase reverse transcriptase (TERT) promotor region have been identified in 23% (3/13) to 74% (29/39) of MLS.25;26 This broad range in the mutation rate warrants investigation of more patients to elucidate the role of TERT mutations in MLS. The mutation creates a new transcription factor-binding site leading to an upregulation of TERT and finally lengthening of the telomeres. DL-221 carries the C228T mutation, as do both the other cell lines we tested. NY-ESO-1, a cancer testis antigen expressed in the majority of the myxoid liposarcomas,30 is strongly expressed in 1765-92 and DL-221 cells. The patients from whom these cell lines originated could have been candidates for NY-ESO-1 directed immunotherapeutic approaches and the cell lines might serve as good models to test its efficacy.58
HDAC inhibitors have shown inhibitory and cytotoxic effects on several soft tissue sarcoma tumor cell models grown in vitro and in vivo, including the 402-91 cell line.59 The MLS fusion gene can affect gene transcription processes by interacting with C/EBP family members; in addition, the EWSR1-DDIT3 fusion gene has shown to act on at an epigenetic level and enhance histone deacetylation and DNA methylation.60 Epigenetic alterations might be involved in the transcriptional regulation of MLS. Expression of HDAC members −1, −2 and −3 was investigated. HDAC-2 was the only one showing moderate to strong expression in all three cell lines, thereby making this a potential target for treatment.61 HDAC-1 and −3 were moderately expressed in 2 out of 3 cell lines. A recent phase II trial with a pan-HDAC inhibitor SB939 as a single agent tested in 22 translocation associated sarcoma patients (5 of whom had MLS) revealed no objective responses but 2 out of 4 evaluable MLS patients achieved stable disease.62 Those patients with high HDAC-2 expression appeared to benefit the most;61–63 however extensive pre-clinical selection of the most promising HDAC-inhibitors might be worthwhile in specific soft tissue sarcoma subtypes. DL-221, similar to the other two cell lines displays sensitivity to the commonly used chemotherapeutic doxorubicin; we are in the process of performing a high throughput drug screen to look for activity with novel targeted agents.
In summary, DL-221, a new spontaneously immortalized myxoid liposarcoma cell line and generated xenograft, represents a much-needed addition to the in vitro and in vivo models available to study this disease. We also report whole exome sequencing to reflect the concordant abnormalities between the cell line and original tumor, data informative for preclinical testing and selecting novel targeted therapeutics that might be translated to the clinic. We hope this cell line and xenograft model will serve to accelerate the pace of preclinical and translational research in MLS, leading to new treatment options to prevent or treat metastatic disease.
Supplementary Material
Myxoid liposarcoma cell lines 402-91, 1765-92 and DL-221 do not express the 29 kDa native DDIT3 under regular culture conditions. Tunicamycin treatment at 5 mg/ml was included as a positive control to cause endoplasmic reticulum stress, and induce DDIT3 protein expression. The higher molecular weight FUS-DDIT3 fusion proteins were also observed in all three cell lines at a higher level of expression compared to native DDIT3.
Mutational analysis for promoter hotspot mutations in TERT, showing the C228T mutation in DL-221.
Expression levels of the HDAC-1, −2 and −3 proteins in the three myxoid liposarcoma cell lines.
(A) The R package vioplot was used to graph violin density plots of all VarScan1 calls from both the metastatic tumor and the derivative cell line. The similarity of the plots indicates high identity between the two samples. 960/1255 variants are shared between the two, with 63 exonic variants present in the metastatic tumor but not in the cell line and 32 exonic variants present in the cell line but not in the metastatic tumor. (B) 2D allele frequency plot showing that 90% of changes in allele frequency between the metastatic lesion and the cell line are small (<0.2). The two samples have a high correlation in variants (r2 = 0.623).
Acknowledgments
Funding: This work is supported by the Liddy Shriver Sarcoma Initiative.
Abbreviations
- AXL
AXL receptor tyrosine kinase
- BAC
Bacterial artificial chromosome
- C/EBP
CCAAT-enhancer-binding protein
- CHOP
C/EBP homologous protein
- COBRA-FISH
Combined binary ratio labeling fluorescence in situ hybridization
- CTAGB1
Cancer testis antigen 1B, gene name of NY-ESO-1 (New York Esophageal Squamous Cell Carcinoma 1)
- DDIT3
DNA damage-inducible transcript 3
- EWSR1
Ewing sarcoma RNA-binding protein 1
- FFPE
Formalin-fixed paraffin-embedded
- FUS
Fused in Sarcoma
- HDAC
Histone deacetylase
- H&E
Haematoxylin and eosin
- IC50
half maximal inhibitory concentration
- IGF1R
Insulin-like growth factor 1 receptor
- MLS
Myxoid liposarcoma
- mTOR
mechanistic target of rapamycin
- PI3K
Phosphatidylinositol 3-kinase
- PIK3CA
Phosphatidylinositol-4,5-Bisphosphate 3-Kinase, Catalytic Subunit Alpha
- PCR
Polymerase chain reaction
- PPARγ
Peroxisome proliferator activated receptor gamma
- PTEN
Phosphatase and tensin homolog
- RET
Rearranged during transfection proto-oncogene
- RT-PCR
Reverse transcriptase polymerase chain reaction
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- STR-typing
Short tandem repeat typing
- SV40
Simian vacuolating virus 40
- TERT
Telomerase reverse transcriptase
- TLS
Translocated in Sarcoma
Footnotes
Disclosures
The authors have no conflicts of interest to declare.
References
- 1.Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F. World Health Organisation Classification of Tumours. Pathology and Genetics of Tumours of Soft Tissue and Bone. 4th. Lyon: IARC Press; 2013. [Google Scholar]
- 2.Antonescu CR, Tschernyavsky SJ, Decuseara R, et al. Prognostic impact of P53 status, TLS-CHOP fusion transcript structure, and histological grade in myxoid liposarcoma: a molecular and clinicopathologic study of 82 cases. Clin Cancer Res. 2001;7:3977–3987. [PubMed] [Google Scholar]
- 3.Fiore M, Grosso F, Lo VS, et al. Myxoid/round cell and pleomorphic liposarcomas: prognostic factors and survival in a series of patients treated at a single institution. Cancer. 2007;109:2522–2531. doi: 10.1002/cncr.22720. [DOI] [PubMed] [Google Scholar]
- 4.Haniball J, Sumathi VP, Kindblom LG, et al. Prognostic factors and metastatic patterns in primary myxoid/round-cell liposarcoma. Sarcoma. 2011;2011:538085. doi: 10.1155/2011/538085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kilpatrick SE, Doyon J, Choong PF, et al. The clinicopathologic spectrum of myxoid and round cell liposarcoma. A study of 95 cases. Cancer. 1996;77:1450–1458. doi: 10.1002/(SICI)1097-0142(19960415)77:8<1450::AID-CNCR5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 6.Moreau LC, Turcotte R, Ferguson P, et al. Myxoid\round cell liposarcoma (MRCLS) revisited: an analysis of 418 primarily managed cases. Ann Surg Oncol. 2012;19:1081–1088. doi: 10.1245/s10434-011-2127-z. [DOI] [PubMed] [Google Scholar]
- 7.ten Heuvel SE, Hoekstra HJ, van Ginkel RJ, et al. Clinicopathologic prognostic factors in myxoid liposarcoma: a retrospective study of 49 patients with long-term follow-up. Ann Surg Oncol. 2007;14:222–229. doi: 10.1245/s10434-006-9043-7. [DOI] [PubMed] [Google Scholar]
- 8.Hoffman A, Ghadimi MP, Demicco EG, et al. Localized and metastatic myxoid/round cell liposarcoma: clinical and molecular observations. Cancer. 2013;119:1868–1877. doi: 10.1002/cncr.27847. [DOI] [PubMed] [Google Scholar]
- 9.Nishida Y, Tsukushi S, Nakashima H, et al. Clinicopathologic prognostic factors of pure myxoid liposarcoma of the extremities and trunk wall. Clin Orthop Relat Res. 2010;468:3041–3046. doi: 10.1007/s11999-010-1396-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Vreeze R, de Jong D, Nederlof P, et al. Multifocal myxoid liposarcoma-metastasis or second primary tumor? A molecular biological analysis. J Mol Diagn. 2010;12:238–243. doi: 10.2353/jmoldx.2010.090117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuglo HM, Maretty-Nielsen K, Hovgaard D, et al. Metastatic pattern, local relapse, and survival of patients with myxoid liposarcoma: a retrospective study of 45 patients. Sarcoma. 2013;2013:548628. doi: 10.1155/2013/548628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katz D, Boonsirikamchai P, Choi H, et al. Efficacy of first-line doxorubicin and ifosfamide in myxoid liposarcoma. Clin Sarcoma Res. 2012;2:2. doi: 10.1186/2045-3329-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powers MP, Wang WL, Hernandez VS, et al. Detection of myxoid liposarcoma-associated FUS-DDIT3 rearrangement variants including a newly identified breakpoint using an optimized RT-PCR assay. Mod Pathol. 2010;23:1307–1315. doi: 10.1038/modpathol.2010.118. [DOI] [PubMed] [Google Scholar]
- 14.Crozat A, Aman P, Mandahl N, et al. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 15.Bode-Lesniewska B, Frigerio S, Exner U, et al. Relevance of translocation type in myxoid liposarcoma and identification of a novel EWSR1-DDIT3 fusion. Genes Chromosomes Cancer. 2007;46:961–971. doi: 10.1002/gcc.20478. [DOI] [PubMed] [Google Scholar]
- 16.Sandberg AA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: liposarcoma. Cancer Genet Cytogenet. 2004;155:1–24. doi: 10.1016/j.cancergencyto.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Tan AY, Manley JL. The TET family of proteins: functions and roles in disease. J Mol Cell Biol. 2009;1:82–92. doi: 10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goransson M, Andersson MK, Forni C, et al. The myxoid liposarcoma FUS-DDIT3 fusion oncoprotein deregulates NF-kappaB target genes by interaction with NFKBIZ. Oncogene. 2009;28:270–278. doi: 10.1038/onc.2008.378. [DOI] [PubMed] [Google Scholar]
- 19.Perez-Mancera PA, Bermejo-Rodriguez C, Sanchez-Martin M, et al. FUS-DDIT3 prevents the development of adipocytic precursors in liposarcoma by repressing PPARgamma and C/EBPalpha and activating eIF4E. PLoS One. 2008;3:e2569. doi: 10.1371/journal.pone.0002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng H, Dodge J, Mehl E, et al. Validation of immature adipogenic status and identification of prognostic biomarkers in myxoid liposarcoma using tissue microarrays. Hum Pathol. 2009;40:1244–1251. doi: 10.1016/j.humpath.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Negri T, Virdis E, Brich S, et al. Functional mapping of receptor tyrosine kinases in myxoid liposarcoma. Clin Cancer Res. 2010;16:3581–3593. doi: 10.1158/1078-0432.CCR-09-2912. [DOI] [PubMed] [Google Scholar]
- 22.Barretina J, Taylor BS, Banerji S, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010;42:715–721. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demicco EG, Torres KE, Ghadimi MP, et al. Involvement of the PI3K/Akt pathway in myxoid/round cell liposarcoma. Mod Pathol. 2012;25:212–221. doi: 10.1038/modpathol.2011.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oda Y, Yamamoto H, Takahira T, et al. Frequent alteration of p16(INK4a)/p14(ARF) and p53 pathways in the round cell component of myxoid/round cell liposarcoma: p53 gene alterations and reduced p14(ARF) expression both correlate with poor prognosis. J Pathol. 2005;207:410–421. doi: 10.1002/path.1848. [DOI] [PubMed] [Google Scholar]
- 25.Saito T, Akaike K, Kurisaki-Arakawa A, et al. TERT promoter mutations are rare in bone and soft tissue sarcomas of Japanese patients. Mol Clin Oncol. 2016;4:61–64. doi: 10.3892/mco.2015.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koelsche C, Renner M, Hartmann W, et al. TERT promoter hotspot mutations are recurrent in myxoid liposarcomas but rare in other soft tissue sarcoma entities. J Exp Clin Cancer Res. 2014;33:33. doi: 10.1186/1756-9966-33-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vinagre J, Almeida A, Populo H, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4:2185. doi: 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- 28.Hemminger JA, Ewart TA, Scharschmidt TJ, et al. The cancer-testis antigen NY-ESO-1 is highly expressed in myxoid and round cell subset of liposarcomas. Mod Pathol. 2013;26:282–288. doi: 10.1038/modpathol.2012.133. [DOI] [PubMed] [Google Scholar]
- 29.Pollack SM, Jungbluth AA, Hoch BL, et al. NY-ESO-1 is a ubiquitous immunotherapeutic target antigen for patients with myxoid/round cell liposarcoma. Cancer. 2012;118:4564–4570. doi: 10.1002/cncr.27446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Endo M, de Graaff MA, Ingram DR, et al. NY-ESO-1 (CTAG1B) expression in mesenchymal tumors. Mod Pathol. 2015;28:587–595. doi: 10.1038/modpathol.2014.155. [DOI] [PubMed] [Google Scholar]
- 31.Chen YT, Boyer AD, Viars CS, et al. Genomic cloning and localization of CTAG, a gene encoding an autoimmunogenic cancer-testis antigen NY-ESO-1, to human chromosome Xq28. Cytogenet Cell Genet. 1997;79:237–240. doi: 10.1159/000134734. [DOI] [PubMed] [Google Scholar]
- 32.Jungbluth AA, Antonescu CR, Busam KJ, et al. Monophasic and biphasic synovial sarcomas abundantly express cancer/testis antigen NY-ESO-1 but not MAGE-A1 or CT7. Int J Cancer. 2001;94:252–256. doi: 10.1002/ijc.1451. [DOI] [PubMed] [Google Scholar]
- 33.Aman P, Ron D, Mandahl N, et al. Rearrangement of the transcription factor gene CHOP in myxoid liposarcomas with t(12;16)(q13;p11) Genes Chromosomes Cancer. 1992;5:278–285. doi: 10.1002/gcc.2870050403. [DOI] [PubMed] [Google Scholar]
- 34.Thelin-Jarnum S, Lassen C, Panagopoulos I, et al. Identification of genes differentially expressed in TLS-CHOP carrying myxoid liposarcomas. Int J Cancer. 1999;83:30–33. doi: 10.1002/(sici)1097-0215(19990924)83:1<30::aid-ijc6>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 35.Lahat G, Zhu QS, Huang KL, et al. Vimentin is a novel anti-cancer therapeutic target; insights from in vitro and in vivo mice xenograft studies. PLoS One. 2010;5:e10105. doi: 10.1371/journal.pone.0010105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Szuhai K, Bezrookove V, Wiegant J, et al. Simultaneous molecular karyotyping and mapping of viral DNA integration sites by 25-color COBRA-FISH. Genes Chromosomes Cancer. 2000;28:92–97. doi: 10.1002/(sici)1098-2264(200005)28:1<92::aid-gcc11>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 37.Szuhai K, Tanke HJ. COBRA: combined binary ratio labeling of nucleic-acid probes for multi-color fluorescence in situ hybridization karyotyping. Nat Protoc. 2006;1:264–275. doi: 10.1038/nprot.2006.41. [DOI] [PubMed] [Google Scholar]
- 38.Lopez G, Song Y, Lam R, et al. HDAC Inhibition for the Treatment of Epithelioid Sarcoma: Novel Cross Talk Between Epigenetic Components. Mol Cancer Res. 2016;14:35–43. doi: 10.1158/1541-7786.MCR-15-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willems SM, Schrage YM, Bruijn IH, et al. Kinome profiling of myxoid liposarcoma reveals NF-kappaB-pathway kinase activity and casein kinase II inhibition as a potential treatment option. Mol Cancer. 2010;9:257. doi: 10.1186/1476-4598-9-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin Z, Lahat G, Korchin B, et al. Midkine enhances soft-tissue sarcoma growth: a possible novel therapeutic target. Clin Cancer Res. 2008;14:5033–5042. doi: 10.1158/1078-0432.CCR-08-0092. [DOI] [PubMed] [Google Scholar]
- 41.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koboldt DC, Chen K, Wylie T, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye K, Schulz MH, Long Q, et al. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen K, Wallis JW, McLellan MD, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang L, Luquette LJ, Gehlenborg N, et al. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell. 2013;153:919–929. doi: 10.1016/j.cell.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hintze JL, Nelson RD. Violin Plots: A Box Plot-Density Trace Synergism. The American Statistician. 1998;52:181–184. [Google Scholar]
- 48.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 50.Shaffer LG, McGowan-Jordan J, Schmid M. ISCN (2013): An International System for Human Cytogenetic Nomenclature. Basel: S. Karger; 2013. [Google Scholar]
- 51.Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 52.Shihab HA, Gough J, Cooper DN, et al. Predicting the functional consequences of cancer-associated amino acid substitutions. Bioinformatics. 2013;29:1504–1510. doi: 10.1093/bioinformatics/btt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stahlberg A, Kabjorn GC, Engtrom K, et al. Normal and functional TP53 in genetically stable myxoid/round cell liposarcoma. PLoS One. 2014;9:e113110. doi: 10.1371/journal.pone.0113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jha KK, Banga S, Palejwala V, et al. SV40-Mediated immortalization. Exp Cell Res. 1998;245:1–7. doi: 10.1006/excr.1998.4272. [DOI] [PubMed] [Google Scholar]
- 55.Pilotti S, Lavarino C, Mezzelani A, et al. Limited role of TP53 and TP53-related genes in myxoid liposarcoma. Tumori. 1998;84:571–577. doi: 10.1177/030089169808400512. [DOI] [PubMed] [Google Scholar]
- 56.Dei Tos AP, Piccinin S, Doglioni C, et al. Molecular aberrations of the G1-S checkpoint in myxoid and round cell liposarcoma. Am J Pathol. 1997;151:1531–1539. [PMC free article] [PubMed] [Google Scholar]
- 57.Sanfilippo R, Dei Tos AP, Casali PG. Myxoid liposarcoma and the mammalian target of rapamycin pathway. Curr Opin Oncol. 2013;25:379–383. doi: 10.1097/CCO.0b013e32836227ac. [DOI] [PubMed] [Google Scholar]
- 58.Robbins PF, Kassim SH, Tran TL, et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin Cancer Res. 2015;21:1019–1027. doi: 10.1158/1078-0432.CCR-14-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu S, Cheng H, Kwan W, et al. Histone deacetylase inhibitors induce growth arrest, apoptosis, and differentiation in clear cell sarcoma models. Mol Cancer Ther. 2008;7:1751–1761. doi: 10.1158/1535-7163.MCT-07-0560. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki K, Matsui Y, Higashimoto M, et al. Myxoid liposarcoma-associated EWSR1-DDIT3 selectively represses osteoblastic and chondrocytic transcription in multipotent mesenchymal cells. PLoS One. 2012;7:e36682. doi: 10.1371/journal.pone.0036682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pacheco M, Nielsen TO. Histone deacetylase 1 and 2 in mesenchymal tumors. Mod Pathol. 2012;25:222–230. doi: 10.1038/modpathol.2011.157. [DOI] [PubMed] [Google Scholar]
- 62.Chu QS, Nielsen TO, Alcindor T, et al. A phase II study of SB939, a novel pan-histone deacetylase inhibitor, in patients with translocation-associated recurrent/metastatic sarcomas-NCIC-CTG IND 200dagger. Ann Oncol. 2015;26:973–981. doi: 10.1093/annonc/mdv033. [DOI] [PubMed] [Google Scholar]
- 63.Cassier PA, Lefranc A, Amela EY, et al. A phase IItrial of panobinostat in patients with advanced pretreated soft tissue sarcoma. A study from the French Sarcoma Group. Br J Cancer. 2013;109:909–914. doi: 10.1038/bjc.2013.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Myxoid liposarcoma cell lines 402-91, 1765-92 and DL-221 do not express the 29 kDa native DDIT3 under regular culture conditions. Tunicamycin treatment at 5 mg/ml was included as a positive control to cause endoplasmic reticulum stress, and induce DDIT3 protein expression. The higher molecular weight FUS-DDIT3 fusion proteins were also observed in all three cell lines at a higher level of expression compared to native DDIT3.
Mutational analysis for promoter hotspot mutations in TERT, showing the C228T mutation in DL-221.
Expression levels of the HDAC-1, −2 and −3 proteins in the three myxoid liposarcoma cell lines.
(A) The R package vioplot was used to graph violin density plots of all VarScan1 calls from both the metastatic tumor and the derivative cell line. The similarity of the plots indicates high identity between the two samples. 960/1255 variants are shared between the two, with 63 exonic variants present in the metastatic tumor but not in the cell line and 32 exonic variants present in the cell line but not in the metastatic tumor. (B) 2D allele frequency plot showing that 90% of changes in allele frequency between the metastatic lesion and the cell line are small (<0.2). The two samples have a high correlation in variants (r2 = 0.623).