Abstract
Biocomputing nanoplatforms are designed to detect and integrate single or multiple inputs under defined algorithms, such as Boolean logic gates, and generate functionally useful outputs, such as delivery of therapeutics or release of optically detectable signals. Using sensing modules composed of small molecules, polymers, nucleic acids, or proteins/peptides, nanoplatforms have been programmed to detect and process extrinsic stimuli, such as magnetic fields or light, or intrinsic stimuli, such as nucleic acids, enzymes, or pH. Stimulus detection can be transduced by the nanomaterial via three different mechanisms: system assembly, system disassembly, or system transformation. The increasingly sophisticated suite of biocomputing nanoplatforms may be invaluable for a multitude of applications, including medical diagnostics, biomedical imaging, environmental monitoring, and delivery of therapeutics to target cell populations.
Keywords: review, logic gates, Boolean logic, nanoparticle, targeted delivery, gene therapy, drug delivery, imaging agents, stimulus responsive
Graphical abstract
Introduction
Nanoscale platforms designed as drug and gene delivery vectors or diagnostic agents have the potential to dramatically improve the quality of life of patients suffering from a wide range of debilitating diseases, such as cancer, cystic fibrosis, and Alzheimer’s Disease [1]. Delivering drugs, genes, or imaging agents both effectively and specifically to desired target tissues and cells could critically improve biological and clinical understanding of a pathological condition and provide a means for halting or even reversing the disease. Nanoplatforms have also demonstrated utility as detection agents, precisely identifying pathogens and other disease markers, environmental toxins, and hazardous materials. As methods have improved to synthesize a vast array of nanoplatforms with more controllable physicochemical properties, investigators have been pushing the boundaries of what sophisticated functions can be performed by the nanomaterials. In particular, a new breed of nanotechnologies able to perform ‘biocomputation’ is being actively developed. For the purposes of this review, biocomputation is defined as the act of sensing and, importantly, computing biomolecular inputs either through algorithmic trees or continuous functions, and in response generating functional outputs.
The concept of biocomputation, with roots in electrical engineering, has been applied with much success in the field of synthetic biology. Conceptualizing and designing genetic circuits as platforms that compute biomolecular inputs through various methods, such as Boolean logic, have been explored extensively [2–4]. Applying the abstractions of computation to the design of nanoscale platforms, either biologically derived or synthetic, is a relatively new approach to thinking about how these functional nanomaterials can be engineered to improve and expand their capabilities.
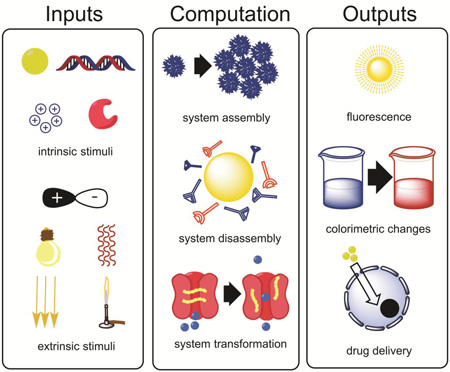
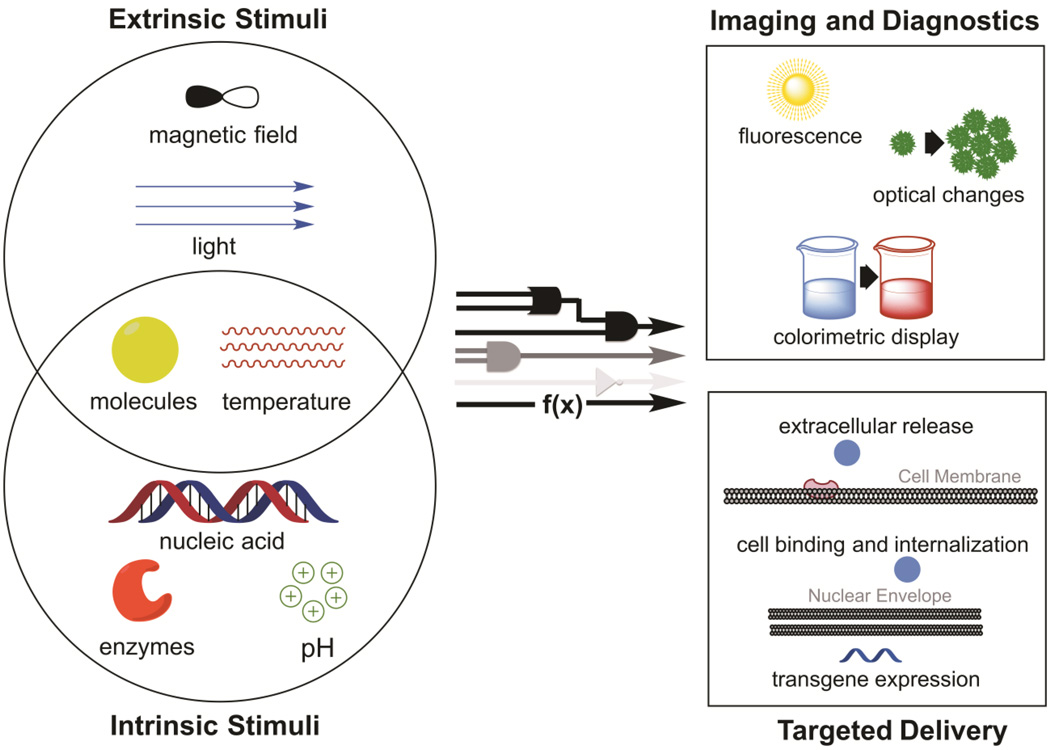
Promisingly, a plethora of inputs and outputs are compatible with nanomaterial-based biocomputation (Figure 1). The biomolecular inputs can be small molecules, enzymes, or nucleic acids. In addition to these intrinsic inputs, a variety of inputs exogenously applied to a biological system, such as light or magnetic fields, can be accepted as stimuli. The inputs can be computed via a number of different logic operators, such as AND, OR, and NOT gates. In response to detecting and computing inputs, the nanoplatforms carry out specific functions, such as delivering transgenes into cells or producing a fluorescence signal. Thus, by enabling nanomaterials to conduct defined computations, platforms can be designed to carry out desired complex tasks, such as delivering desired doses of drugs only to sites of disease characterized by combinations of target biomarkers. Then by characterizing the engineered platforms quantitatively and modeling their behavior mathematically, the engineered biocomputing nanomaterials can become not only controllable but also functionally predictable. This potential to predict the output of a nanoplatform given a set of biomolecular inputs is what makes the biocomputational approach impactful to the future of drug delivery and bioimaging. Much of the work on stimulus-responsive nanomaterials can be easily re-conceptualized through the lens of biocomputation; it would require categorizing the type of computation being conducted by the nanomaterial and quantitatively characterizing the input-output curve under various conditions.
Figure 1.
Biocomputing nanoplatforms accept inputs that are either intrinsic or extrinsic to biological systems, compute the inputs using defined functions, and produce outputs that manipulate the biological environment (therapeutics) or act as an externally detectable readout (diagnostics).
This review will cover some of the recent advances in designing and testing nanoplatforms that conduct biocomputation. The definition of a biocomputing nanoplatform can encompass any nanomaterial or nanoparticle that detects and processes an input signal and outputs a defined response. This review will focus on the more complex biocomputing nanoplatforms developed to date that explicitly incorporate logic gates and often require more than one input for activation. Many different types of nanomaterials are being programmed as biocomputing platforms for a range of applications (Table 1). To date, the main challenges being addressed by biocomputing nanoplatforms are targeted and controlled delivery of drugs or genes, detection of analytes using in vitro diagnostics, and in vivo molecular imaging. The biocomputing approach promises to enable more precise, more selective, and potentially more quantitatively predictable activation of the desired functional output (e.g. delivery of a therapeutic payload or release of a fluorescence signal). The variety of intrinsic and extrinsic input stimuli, output responses, and computational methods used by the different platforms will be discussed.
Table 1.
Applications of Biocomputing Nanoplatforms
| NANOMATERIAL PLATFORM |
APPLICATIONS | REFERENCES |
|---|---|---|
| Polymeric | Inflammatory disease targeting, cancer treatment, targeted imaging | [18,20–31,40,41] |
| Gold-coated | pH sensing, heavy metal sensing, nucleic acid sensing, “lab-on-a-nanoparticle” | [11–14,28,32,33,35] |
| Nucleic acid-based | “Lab-on-a-nanoparticle”, fluorescent and colorimetric biosensing, antibody delivery | [11,12,34,35] |
| Enzyme-based | Detection of biochemical substrates | [31,32] |
| Magnetic | Magnetic resonance imaging, cell sorting | [15,18] |
| Viral | Cancer treatment, toxin sensing, cell targeting | [20,36,39] |
Operators Used by Biocomputing Nanoplatforms
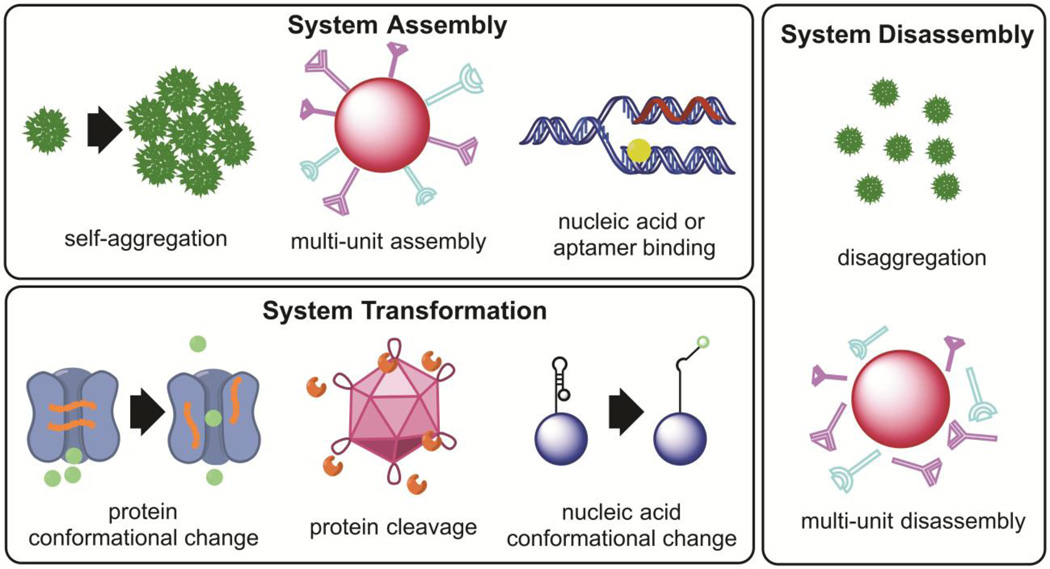
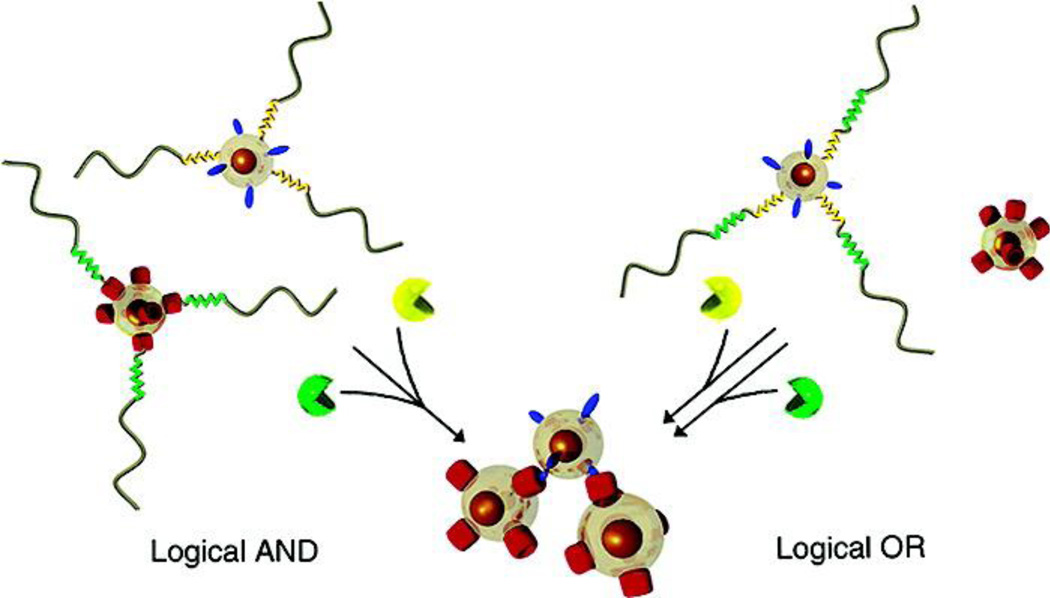
In the most ideal manifestation, nanomaterial-based computation translates inputs into outputs in a quantitatively predictable manner. Researchers have primarily focused on developing systems with binary outputs, similar to the logic gates found in silicon-based computing. These logic gates have been particularly advantageous when applied to biological systems for building complex protein-based signal transduction networks with sophisticated computing-like behavior [5]. These binary systems must be able to detect inputs above a certain threshold level and reject those below the threshold. Many biocomputational nanomaterials have been programmed as AND gates that require detection of two different stimuli to activate. These AND gate nanomaterials can either integrate simultaneous stimuli, or they can utilize consecutive stimuli to activate in phases as they transport to their biological targets. Other biocomputational nanomaterials function as OR gates, NOT gates, or combinations of multiple gates. Some biocomputational nanomaterials behave as tunable devices, where the level of output is determined by the level of input detected. In other words, tunable nanodevices use a continuous mathematical function to connect the level of input signals to the precise level of output, allowing for calibrated responses. The various biocomputing nanoplatforms have different strategies for translating multiple stimuli into a single output, including system assembly, disassembly, and transformation (Figure 2). The remainder of the review will cover examples of these three different strategies.
Figure 2.
Methods of nanoplatform information processing fall into three broad categories: system assembly, system disassembly, and system transformation.
System Assembly as Means of Integrating Inputs
Many biocomputational nanomaterials integrate single or multiple inputs through the assembly of individual components, a process termed ‘system assembly’. System assembly of nanoparticles has been applied in several pioneering studies to monitor biomolecular interactions, as the aggregation of small metallic nanoparticles can influence their optical and magnetic properties [6–10]. Nanoparticles tethered to DNA, proteins, and enzymes can thus be used to optically observe proximity. In this review, we will focus on examples of system assembly where the output is produced based on a logical computation from a set of biomolecular inputs.
Oftentimes, system assembly can be mediated by DNA hybridization. For example, one logic gate-based strategy with DNA nanostructures exploits the hybridization of two different input segments of single-stranded DNA, thus operating as an AND gate, to create a longer near-complementary double-stranded DNA fragment. The combined strand then uses toehold-displacement to assemble onto a gold nanoparticle functionalized with DNA fragments. This unique assembly process displaces a fluorophore-conjugated DNA strand, ultimately resulting in the generation of fluorescence as the output [11]. The platform may be used to detect multiple DNA inputs for achieving DNA-based computation. In another report, similar gold nanoparticles functionalized with single-stranded DNA have been developed to function as target DNA sensors [12]. The detection of target single-stranded DNA facilitates assembly of the nanoparticles into multi-nanoparticle aggregates, a reaction that can be visualized through a solution color change. The platform can be designed to require detection of two different target DNA strands (AND gate), or the detection of one of two DNA segments (OR gate) to signal the presence of pathogen DNA in a sample.
Similarly, DNA aptamers have been attached to the surface of nanoparticles so that aptamer conformational shifts triggered by binding of small molecules can be used to induce the hybridization of near-complementary DNA strands and trigger nanostructure aggregation. For example, one AND gate nanoplatform designed for small molecule sensing used aptamer-modified gold nanoparticles to detect both adenosine and cocaine, inducing nanoparticle aggregation that resulted in a solution color change as the output [13]. Overall, DNA-based aggregation systems utilize the temperature stability and well-characterized hybridization behavior of nucleic acids to create precise logic gate-based nanodevices.
System assembly-based biocomputing nanoplatforms have also been generated without the help of DNA mediators. One nanoplatform relies on spiropyrans to function as an AND-gate, detecting UV light and metal ions as dual inputs [14]. In the presence of the light stimulus, spiropyrans conjugated to the surface of gold nanoparticles can bind metal ions in a multivalent fashion, thus inducing aggregation of the gold nanoparticles and creating a solution color change output. This system can be used to quickly detect a variety of metal ions in solution with a visually detectable change. Another approach utilizes pH and temperature-responsive polymers on the surface of magnetic nanoparticles to relate these stimuli to a continuous range of aggregation outputs [15]. This platform allows cells which contain aggregated nanoparticles to be separated from cells that do not contain aggregated nanoparticles, since only aggregated nanoparticles can be separated with a magnetic field. Other approaches use protease cleavage domains to detect matrix metalloproteinases (MMPs), a class of enzymes elevated in various diseases [16,17]. One platform uses MMP-cleavable peptides on the surface of iron oxide nanoparticles (Figure 3) [18]. These peptides act as AND or OR gates, cleaving in the presence of target MMPs and inducing nanoparticle aggregation to change the magnetic properties of the particles. Such MMP-sensing nanoparticles can be used as in vivo diagnostic agents using magnetic resonance imaging.
Figure 3.
Nanoparticles functionalized with MMP-cleavable peptides can be used to detect MMP activity under defined logic gates. When the peptides are digested by the required combination of MMPs (yellow and green pacmans), binding sites are exposed, allowing the particles to aggregate and trigger an optical change. Figure used with permission [18].
In summary, several biocomputing nanoplatforms use assembly-based strategies, such as DNA hybridization or multi-nanoparticle aggregation mediated by polymers, peptides or DNA. These assembly processes can be triggered by a wide range of stimuli, such as small molecules, enzymes, UV light, temperature and pH. As nanoparticle aggregates often have different fluorescence or absorbance properties as compared to their individual components, the system assembly approach is primarily used to generate diagnostics and other detection tools.
System Disassembly as Means of Integrating Inputs
Biocomputational nanomaterials can also react to input stimuli by either multi-unit nanoparticle disassembly or multi-nanoparticle disaggregation. These platforms often depend on the chemical changes of polymers to induce ‘system disassembly’ [19]. Such disassembly-based platforms have been used primarily for selective targeting of cancer cells for gene therapy and drug delivery. Low pH is one of the most common single inputs for biocomputing nanoplatforms. One report developed a polymer matrix [composed of polyethylene glycol (PEG) and polyethylenimine (PEI)] encapsulating the adeno-associated virus such that upon detection of low pH, the viral vectors are released through triggered chemical expansion [20]. This pH-responsive virus delivery platform may be useful for delivering genes to sites of tumor acidosis. Another interesting platform uses assembly of temperature-responsive block copolymer micelles for drug release [21]. The copolymers have differing low critical solution temperatures (LCST) on each end; thus, depending on the temperature used during particle synthesis, two different types of micelles with differing amounts of loaded cargo can be generated. Each micelle type disassembles under the corresponding temperature stimulus, releasing its drug cargo.
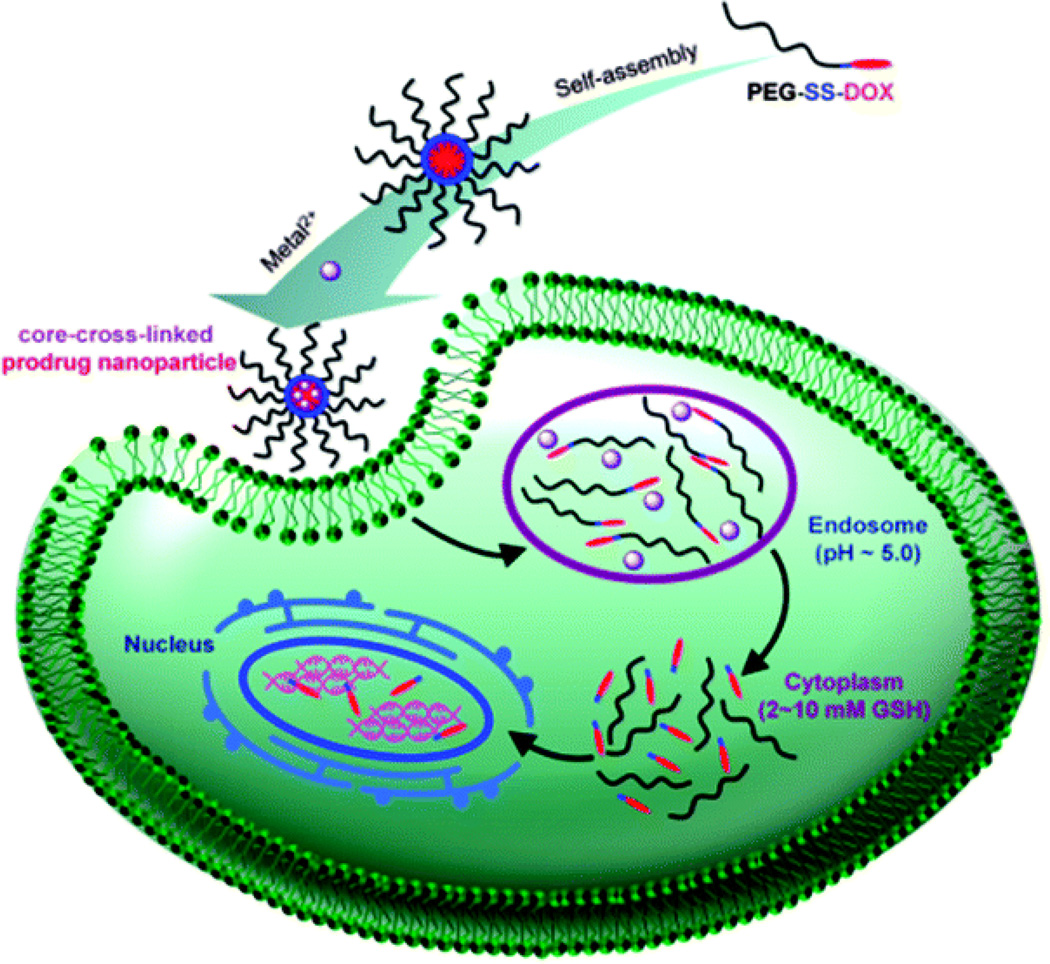
In addition to single input systems, several nanoplatforms have been designed to detect multiple stimuli. One popular approach relies on polymeric micelles that disassemble under the dual stimulus of reducing environment and low pH [22–24]. The micelles encapsulate the drug of interest using polymers that contain both acid-sensitive segments and disulfide bonds. Upon entering an acidic and reducing environment (AND gate inputs), the drug payload is released due to polymer degradation and micelle disassembly (Figure 4). Presence of just one of the inputs is not sufficient to rapidly and completely release the cargo.
Figure 4.
A self-assembling nanoparticle undergoes AND-gate mediated disassembly and drug release during intracellular trafficking. First, acidic pH in the endosome induces nanoparticle disassembly, then the reducing environment of the cytoplasm breaks the disulfide bond linking the cancer drug doxorubicin (DOX) to polyethylene glycol (PEG). Figure used with permission [24].
Other types of AND gate nanoplatforms require different input combinations for system disassembly. Low pH and high temperature is another common pair of AND gate input stimuli for nanoparticles aimed at targeting drugs to areas of inflammation or cancer. Both polymer micelles and dendrimers have been developed to function as AND gates requiring both low pH and high temperature, two features observed in the local environment of the target tissues [25,26]. Another similar AND gate system requires a reducing environment as well as UV irradiation for drug delivery [27]. The photosensitive component, a functional nitrobenzyl derivative capable of conformational changes under specific wavelengths of light, is conjugated onto PEG chains that are cross-linked with disulfide bonds. Detection of both UV light by the nitrobenzyl derivative and reduction of the disulfide bonds in the PEG network are required for complete disassembly and release of the encapsulated drug. A more complex example of a multi-stimuli nanoparticle requires pH, temperature, or light to trigger release (i.e. three-input OR gate) [28]. The PEG-based polymeric structure in this platform was designed such that any of these three stimuli is sufficient to trigger disassembly. These reports highlight the potential for functionalized nanoparticles to process many varied stimuli in order to increase targeting specificity. Disassembly-based nanoparticles for drug delivery offer both payload protection throughout transport as well as targeted release.
In addition to therapeutic applications, system disassembly as a method for biocomputing stimuli can be exploited for imaging or diagnostic nanoparticle development. Imaging and diagnostic tools rely on both accurate recognition of the desired target and a detectable visual response. This can be done through changes in fluorescence or absorbance of the nanomaterial-containing solution. For example, the coordination of nucleotide-lanthanide compounds and porphyrin, resulting in a strong fluorescence signal, can be destabilized in the presence of either EDTA or extreme pH values (both low and high) [29,30]. Particle disassembly takes place through ionic porphyrin displacement, altering the fluorescence properties of the complex and releasing its components. The intensity of this fluorescence signal can be regulated by the concentration and type of chemical inputs. This type of nanoplatform may be further engineered to become useful as a simultaneous detection and triggered delivery device.
More complex biocomputing nanoplatforms have been developed for the detection of small molecules. One group is interested in developing in vitro diagnostics to detect the level of metabolic substrates by generating pH-sensitive polymer-functionalized gold nanoparticles that interface with two cellular enzymes [31]. The enzymes, invertase and glucose oxidase, convert sucrose into acidic byproducts, thereby decreasing the solution pH. The low pH then causes the pH-responsive nanoparticles to undergo multinanoparticle disaggregation, resulting in observable absorbance changes. Therefore, this two-tiered information processing platform pairs an AND gate enzyme system with single input (low pH) nanoparticles to yield a visible absorbance change in the solution. Similarly, enzymes covalently attached to the exterior of silicon microparticles have been used to create a local pH change and induce pH-responsive nanoparticle dissolution upon the detection of metabolic substrates in solution [32]. This approach was used to create AND gates responsive to sucrose and oxygen, and OR gates responsive to glucose and ethyl butyrate. Such biocomputing nanoparticles may be used, for example, to image metabolic activity in tissues by quantifying glucose and sucrose availability. Another report developed what could be described as an AND-NOT gated gold nanoparticle platform to detect the simultaneous presence of melamine and the absence of mercury ions [33]. Due to an innate property of gold nanoparticles, the presence of melamine induces the nanoparticles to form multi-nanoparticle aggregates resulting in a blue-colored solution. In the presence of mercury ions, however, the melamine binding sites are blocked by the mercury ions, resulting in a red-colored solution.
In summary, a number of biocomputing nanoplatforms utilize system disassembly, either the disaggregation of multi-nanoparticle structures or the disassembly of nanomaterial components, as the means for integrating stimuli and producing a functional output. The most popular output is the release of therapeutics at target locations, followed by the generation of a detectable fluorescence or absorbance signal for biomedical imaging or diagnostics applications.
System Transformation as Means of Integrating Inputs
Some biocomputing nanoplatforms undergo a more nuanced change upon stimuli detection and are referred to in this review as undergoing a ‘system transformation’ in response to inputs. A promising approach uses DNA segments that can undergo conformational changes. One system is based on DNA-based ‘nanorobots’ with DNA ‘locks’ that hold the box-shaped nanoplatform closed [34]. Each nanoconstruct can have two different DNA locks, which are aptamers binding to specific target cell-associated antigens, allowing for an AND gate operation. When activated, the nanoconstruct can reveal a variety of payloads, including therapeutic antibody fragments.
DNA fragments attached to the surface of gold nanoparticles can utilize system transformation, rather than system assembly/disassembly, to integrate input stimuli. One nanoplatform uses DNA strands to attach fluorophore-quencher pairs to the surface of gold nanoparticles [35]. Upon detection of target DNA sequences in cells, the platform uses toehold displacement to process inputs by altering a fluorophore’s proximity to the quenching gold nanoparticle to induce or quench fluorescence. Conformational changes of DNA structures are a powerful tool for controlling the mechanical availability and proximity of nanoparticle components to create sophisticated downstream behaviors.
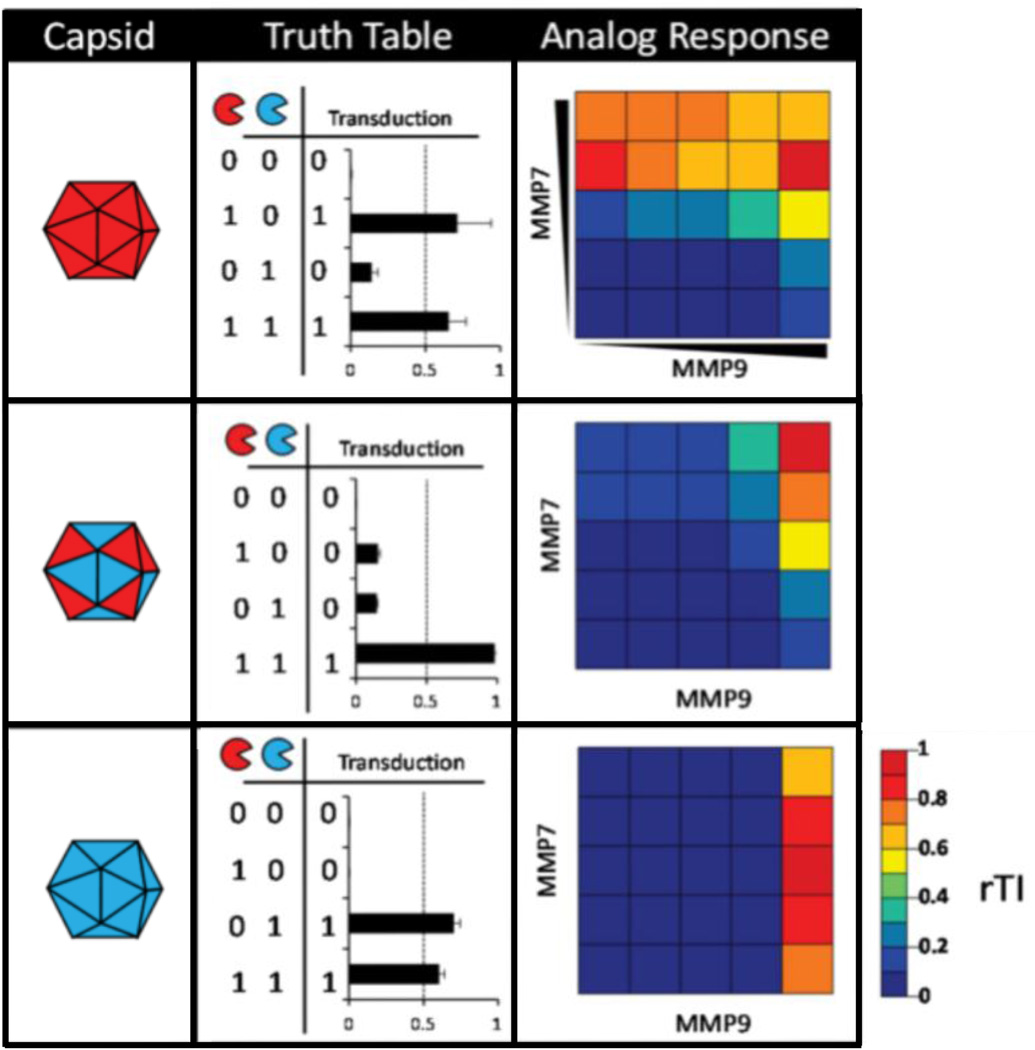
Another example of using system transformation to integrate biomolecular information is the incorporation of MMP-cleavable masking motifs on the surface of nanoplatforms. For example, masking motifs have been genetically incorporated into the cell surface receptor binding pocket of adeno-associated virus (AAV) nanoparticles such that when the motifs are in place, the vector cannot transduce cells [36]. This system can act as an AND gate by combining two different masking motifs, each susceptible to different MMPs, on the same virus capsid (Figure 5). In order for the AAV nanoparticles to successfully transduce cells, both types of masking motifs need to be cleaved from the capsid. Moreover, the protease-activatable virus nanoparticle was modeled mathematically as a multi-input node, enabling its functional output to be quantitatively predictable. Protease detection systems are particularly promising candidates for targeted disease therapy since MMPs are often upregulated in a variety of disease sites [37].
Figure 5.
Virus nanoparticles designed to be activated by two different MMPs for controlled gene delivery. Tunable virus nanoparticles are generated from the combination of two capsid proteins with different masking motifs (red or blue in schematic), each sensitive to a different MMP (red or blue pacmans). The mixed ‘mosaic capsid’ (middle row) demonstrates an AND-gate response to stimulation with the two different MMPs. The ‘analog response’ column displays heatmaps of the relative transduction index (rTI - the percent of fluorescence positive cells multiplied by the mean fluorescence intensity, normalized to control) of the given capsids in response to various concentration combinations of MMP inputs. Figure adapted and used with permission [36].
Nanoparticles have also used various biologically derived materials which exhibit altered physical behaviors, such as motility or swelling, in response to external stimuli. One such system uses two molecular motors to mechanically induce the release of a payload from a hollow nanoparticle for targeted drug delivery [38]. The platform combines light-activated ‘nanoimpellers’ and pH-activated ‘nanovalves’ into an AND gate configuration that requires detection of both light and pH stimuli to allow the payload to escape the nanoparticle. Another system transformation-based approach uses phage capsid proteins, which undergo a structural change upon binding to their unique environmental targets [39]. Two different phages can be combined into one phage-bundle nanostructure to create an AND gate. Thus, detection of two different nitroaromatic molecules results in swelling of the nanostructure, leading to a change in reflected color based on the arrangement of fibers. Thus, these phage nanostructures can be used to detect the presence of explosives by identifying multiple signature chemicals.
In summary, biocomputing nanoplatforms that utilize system transformation harness a more subtle change of the nanomaterial as the means for sensing inputs and generating outputs. Many of these platforms are built from naturally occurring materials, such as DNA, viruses, and proteins.
Conclusions and Perspectives
The future of successful therapeutic and diagnostic nanoplatforms hinges on new and improved mechanisms for connecting inputs (both intrinsic and extrinsic) to predictable and reliable output responses. The wide range of platforms discussed here represents a promising foundation to achieving this vision. System assembly-based biocomputing nanoplatforms show promise in many diagnostics applications, as generation of multi-nanoparticle aggregates can lead to changes in absorbance or fluorescence of the nanoparticle solution. Nanoparticle aggregation or multi-subunit assembly allows for signal integration through the coming together of many signal-detecting components. Biocomputing platforms based on system disassembly have shown more promise in the field of drug delivery. Multi-nanoparticle disaggregation or multi-subunit disassembly allows for the release of therapeutic cargo only at the site of input detection. Finally, system transformation uses more nuanced changes to the nanomaterials to yield desired functional outcomes, and shows promise in imaging and diagnostics, as well as for drug delivery.
In order for more sophisticated multi-tiered algorithmic trees to be built from individual nanoparticle logic gates, the individual nanoparticles should be better characterized quantitatively. Based on experimental data, the ‘transfer function’ of each nanoparticle should be determined under various parameters. These quantitative functions can help predict how a system would function given a certain set of conditions. Thus, approaching nanoparticle design with quantitative biocomputation in mind should lead to the generation of more predictable nanoplatforms.
Additionally, if therapeutic biocomputing nanoplatforms are to be used in complicated biological contexts, such as inside cells or in intact animals, the concentrations of various input stimuli need to be quantified in relevant biological environments, such as diseased cells and tissues. This information is being curated by various databases, but patient-to-patient variability may require personalized measurements of the target biomarker(s) either via biopsies or through the use of companion diagnostics. The nanoplatforms must then be optimized for the gradients of stimuli they will encounter in the target biological environments.
In conclusion, the field of biocomputing nanomaterials is expanding to encompass many more complex systems than ever before. By conceptualizing the design process as one analogous to building electrical circuits, engineered nanomaterials may be programmed to carry out sophisticated, multi-step computations that can result in better targeted and more efficient drugs, or the detection of complex mixtures of desired analytes.
Acknowledgments
This material is based upon work supported by the National Science Foundation under grant number 0955536, Cancer Prevention Research Institute of Texas under grant number RP130455, the American Heart Association under grant number 15GRNT23070007, the National Institutes of Health under grant numbers R21CA187316 and R21HL126053, and a John S. Dunn Foundation award to J.S, and a National Science Foundation Graduate Research Fellowship to A.C.E.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ganta S, Devalapally H, Shahiwala A, Amiji M. A review of stimuli-responsive nanocarriers for drug and gene delivery. Journal of Controlled Release. 2008;126:187–204. doi: 10.1016/j.jconrel.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 2.Kis Z, Pereira HS, Homma T, Pedrigi RM, Krams R. Mammalian synthetic biology: emerging medical applications. J R Soc Interface. 2015;12:20141000–20141000. doi: 10.1098/rsif.2014.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye H, Fussenegger M. Synthetic therapeutic gene circuits in mammalian cells. FEBS Lett. 2014;588:2537–2544. doi: 10.1016/j.febslet.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat. Rev. Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lienert F, Lohmueller JJ, Garg A, Silver PA. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat. Rev. Mol. Cell Biol. 2014;15:95–107. doi: 10.1038/nrm3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JY, Lee JS. Synthesis and thermally reversible assembly of DNA - Gold nanoparticle cluster conjugates. Nano Lett. 2009;9:4564–4569. doi: 10.1021/nl9030709. [DOI] [PubMed] [Google Scholar]
- 7.Qian X, Li J, Nie S. Stimuli-responsive SERS nanoparticles: conformational control of plasmonic coupling and surface Raman enhancement. J. Am. Chem. Soc. 2009;131:7540–7541. doi: 10.1021/ja902226z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian X, Zhou X, Nie S. Surface-enhanced raman nanoparticle beacons based on bioconjugated gold nanocrystals and long range plasmonic coupling. J. Am. Chem. Soc. 2008;130:14934–14935. doi: 10.1021/ja8062502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez JM, Josephson L, O'Loughlin T, Högemann D, Weissleder R. Magnetic relaxation switches capable of sensing molecular interactions. Nature Biotechnology. 2002;20:816–820. doi: 10.1038/nbt720. [DOI] [PubMed] [Google Scholar]
- 10.Sonnichsen C, Reinhard BM, Liphard J, Alivisatos AP. A molecular ruler based on plasmon coupling of single gold and silver nanoparticles. Nature Biotechnology. 2005;23:741–745. doi: 10.1038/nbt1100. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Dong C, Dong Y, Liu S, Pan L, Zhang C. Logic nanoparticle beacon triggered by the binding-induced effect of multiple inputs. ACS Appl Mater Interfaces. 2014;6:14486–14492. doi: 10.1021/am5036994. [DOI] [PubMed] [Google Scholar]
- 12.Zhang C, Yang J, Xu J. Molecular logic computing model based on self-assembly of DNA nanoparticles. Chin. Sci. Bull. 2011;56:3566–3571. [Google Scholar]
- 13.Xu X, Zhang J, Yang F, Yang X. Colorimetric logic gates for small molecules using split/integrated aptamers and unmodified gold nanoparticles. Chemical Communications. 2011;47:9435–9437. doi: 10.1039/c1cc13459k. [DOI] [PubMed] [Google Scholar]
- 14.Liu D, Chen W, Sun K, Deng K, Zhang W, Wang Z, et al. Resettable, multi-readout logic gates based on controllably reversible aggregation of gold nanoparticles. Angewandte Chemie International Edition. 2011;50:4103–4107. doi: 10.1002/anie.201008198. [DOI] [PubMed] [Google Scholar]
- 15.Majewski AP, Schallon A, Jérôme V, Freitag R, Müller AHE, Schmalz H. Dual-responsive magnetic core-shell nanoparticles for nonviral gene delivery and cell separation. Biomacromolecules. 2012;13:857–866. doi: 10.1021/bm2017756. [DOI] [PubMed] [Google Scholar]
- 16.Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circulation Research. 2003;92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- 17.Birkedal-Hansen H, Moore WG, Bodden MK, Windsor LJ, Birkedal-Hansen B, DeCarlo A, et al. Matrix metalloproteinases: a review. Crobm. 1993;4:197–250. doi: 10.1177/10454411930040020401. [DOI] [PubMed] [Google Scholar]
- 18.von Maltzahn G, Harris TJ, Park J-H, Min D-H, Schmidt AJ, Sailor MJ, et al. Nanoparticle self-assembly gated by logical proteolytic triggers. J. Am. Chem. Soc. 2007;129:6064–6065. doi: 10.1021/ja070461l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikitin MP, Shipunova VO, Deyev SM, Nikitin PI. Biocomputing based on particle disassembly. Nat Nanotechnol. 2014;9:716–722. doi: 10.1038/nnano.2014.156. [DOI] [PubMed] [Google Scholar]
- 20.Tseng S-J, Kempson IM, Peng S-F, Ke B-H, Chen H-H, Chen P-F, et al. Environment acidity triggers release of recombinant adeno-associated virus serotype 2 from a tunable matrix. J Control Release. 2013;170:252–258. doi: 10.1016/j.jconrel.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Kotsuchibashi Y, Ebara M, Hoffman AS, Narain R, Aoyagi T. Temperature-responsive mixed core nanoparticle properties determined by the composition of statistical and block copolymers in the core. Polymer Chemistry. 2015;6:1693–1697. [Google Scholar]
- 22.Cheng R, Meng F, Deng C, Klok H-A, Zhong Z. Dual and multi-stimuli responsive polymeric nanoparticles for programmed site-specific drug delivery. Biomaterials. 2013;34:3647–3657. doi: 10.1016/j.biomaterials.2013.01.084. [DOI] [PubMed] [Google Scholar]
- 23.Mahmoud EA, Sankaranarayanan J, Morachis JM, Kim G, Almutairi A. Inflammation responsive logic gate nanoparticles for the delivery of proteins. Bioconjug. Chem. 2011;22:1416–1421. doi: 10.1021/bc200141h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bai L, Wang X-H, Song F, Wang X-L, Wang Y-Z. “AND” logic gate regulated pH and reduction dual-responsive prodrug nanoparticles for efficient intracellular anticancer drug delivery. Chem. Commun. (Camb.) 2015;51:93–96. doi: 10.1039/c4cc07012g. [DOI] [PubMed] [Google Scholar]
- 25.Shen Y, Ma X, Zhang B, Zhou Z, Sun Q, Jin E, et al. Degradable dual pH- and temperature-responsive photoluminescent dendrimers. Chemistry - a European Journal. 2011;17:5319–5326. doi: 10.1002/chem.201003495. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y-C, Liao L-C, Lu P-L, Lo C-L, Tsai H-C, Huang C-Y, et al. The accumulation of dual pH and temperature responsive micelles in tumors. Biomaterials. 2012;33:4576–4588. doi: 10.1016/j.biomaterials.2012.02.059. [DOI] [PubMed] [Google Scholar]
- 27.Huang Q, Liu T, Bao C, Lin Q, Ma M, Zhu L. Light and reductive dual stimuli-responsive PEI nanoparticles: “AND” logic response and controllable release. Journal of Materials Chemistry B. 2014;2:3333–3339. doi: 10.1039/c4tb00087k. [DOI] [PubMed] [Google Scholar]
- 28.Oishi M, Nakamura T, Jinji Y, Matsuishi K, Nagasaki Y. Multi-stimuli-triggered release of charged dye from smart PEGylated nanogels containing gold nanoparticles to regulate fluorescence signals. Journal of Materials Chemistry. 2009;19:5909–5904. [Google Scholar]
- 29.Pu F, Ju E, Ren J, Qu X. Multiconfigurable logic gates based on fluorescence switching in adaptive coordination polymer nanoparticles. Advanced Materials. 2014;26:1111–1117. doi: 10.1002/adma.201304109. [DOI] [PubMed] [Google Scholar]
- 30.Pu F, Ren J, Qu X. “Plug and play” logic gates based on fluorescence switching regulated by self-assembly of nucleotide and lanthanide ions. ACS Appl Mater Interfaces. 2014;6:9557–9562. doi: 10.1021/am501949t. [DOI] [PubMed] [Google Scholar]
- 31.Motornov M, Zhou J, Pita M, Gopishetty V, Tokarev I, Katz E, et al. “Chemical transformers” from nanoparticle ensembles operated with logic. Nano Lett. 2008;8:2993–2997. doi: 10.1021/nl802059m. [DOI] [PubMed] [Google Scholar]
- 32.Pita M, Krämer M, Zhou J, Poghossian A, Schöning MJ, Fernández VM, et al. Optoelectronic properties of nanostructured ensembles controlled by biomolecular logic systems. ACS Nano. 2008;2:2160–2166. doi: 10.1021/nn8004558. [DOI] [PubMed] [Google Scholar]
- 33.Du J, Yin S, Jiang L, Ma B, Chen X. A colorimetric logic gate based on free gold nanoparticles and the coordination strategy between melamine and mercury ions. Chemical Communications. 2013;49:4196–4198. doi: 10.1039/c2cc36955a. [DOI] [PubMed] [Google Scholar]
- 34.Douglas SM, Bachelet I, Church GM. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 35.Yang B, Zhang X-B, Kang L-P, Huang Z-M, Shen G-L, Yu R-Q, et al. Intelligent layered nanoflare: “lab-on-a-nanoparticle” for multiple DNA logic gate operations and efficient intracellular delivery. Nanoscale. 2014;6:8990–8996. doi: 10.1039/c4nr01676a. [DOI] [PubMed] [Google Scholar]
- 36.Judd J, Ho ML, Tiwari A, Gomez EJ, Dempsey C, Van Vliet K. Tunable protease-activatable virus nanonodes. ACS Nano. 2014;8:4740–4746. doi: 10.1021/nn500550q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vartak DG, Gemeinhart RA. Matrix metalloproteases: Underutilized targets for drug delivery. Journal of Drug Targeting. 2008;15:1–20. doi: 10.1080/10611860600968967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angelos S, Yang Y-W, Khashab NM, Stoddart JF, Zink JI. Dual-controlled nanoparticles exhibiting AND logic. J. Am. Chem. Soc. 2009;131:11344–11346. doi: 10.1021/ja9042752. [DOI] [PubMed] [Google Scholar]
- 39.Oh J-W, Chung W-J, Heo K, Jin H-E, Lee BY, Wang E, et al. Biomimetic virusbased colourimetric sensors. Nat Commun. 2014;5:3043. doi: 10.1038/ncomms4043. [DOI] [PubMed] [Google Scholar]
- 40.Wei C, Guo J, Wang C. Dual stimuli-responsive polymeric micelles exhibiting “AND” logic gate for controlled release of Adriamycin. Macromolecular Rapid Communications. 2011;32:451–455. doi: 10.1002/marc.201000708. [DOI] [PubMed] [Google Scholar]
- 41.Nagase K, Kobayashi J, Okano T. Temperature-responsive intelligent interfaces for biomolecular separation and cell sheet engineering. J R Soc Interface. 2009;6 doi: 10.1098/rsif.2008.0499.focus. rsif.2008.0499.focus–S309. [DOI] [PMC free article] [PubMed] [Google Scholar]