Abstract
Purpose
Leber congenital amaurosis (LCA) is an early-onset form of retinal degeneration and six of the 22 known LCA disease genes encode photoreceptor ciliary proteins. Despite the identification of 22 LCA disease genes, the genetic basis of approximately 30% of LCA patients remains unknown. We sought to investigate the cause of disease in the remaining 30% by examining cilia-associated genes.
Methods
Whole-exome sequencing was performed on an LCA cohort of 212 unsolved probands previously screened for mutations in known retinal disease genes. Immunohistochemistry using mouse retinas was used to confirm protein localization and zebrafish were used to perform rescue experiments.
Results
A homozygous nonsynonymous mutation was found in a single proband in CLUAP1, a gene required for ciliogenesis and cilia maintenance. Cluap1 knockout zebrafish exhibit photoreceptor cell death as early as five days post fertilization and rescue experiments revealed that our proband’s mutation is significantly hypomorphic.
Conclusion
Consistent with the knowledge that CLUAP1 plays an important role in cilia function and that cilia are critical to photoreceptor function, our results indicate that hypomorphic mutations in CLUAP1 can result in dysfunctional photoreceptors without systemic abnormalities. This represents the first report linking mutations in CLUAP1 to human disease and establishes CLUAP1 as a candidate LCA gene.
Keywords: Leber congenital amaurosis (LCA), CLUAP1, early-onset retinal disease, photoreceptor connecting cilium, ciliopathy
INTRODUCTION
Leber congenital amaurosis (LCA, http://www.omim.org/phenotypicSeries/PS204000) is the most severe form of nonsyndromic inherited retinal disease with severe visual impairment within the first year of life. LCA is characterized by nystagmus and an extinguished electroretinogram (ERG). LCA affects between 1/30,000 and 1/80,000 individuals differing by population and accounts for 5% of all inherited retinal diseases.1 As of 2015, 22 genes have been linked to LCA; however, conventional techniques for molecular diagnosis find mutations in these genes in only ~70% of LCA patients, highlighting the genetic heterogeneity of LCA.1–5 Gene therapy in the retina of human LCA patients has made significant progress in restoring sight, emphasizing the importance of understanding the genetic etiology of every patient’s disease to allow access to such technologies.6 Genetic counseling based on a patient’s molecular diagnosis also provides the opportunity for mutation screening in offspring and spouses. Discovering the cause of disease in the remaining ~30% of LCA patients is therefore critical to enable treatment for this devastating disease.
An absent ERG, a hallmark of LCA, is indicative of malfunctioning photoreceptor cells which initiate the recorded electrical signal and are responsible for the generation of the a-wave.7 Every identified LCA disease gene has a role relevant to the proper functioning of photoreceptor cells.1,8–14 Photoreceptors are remarkable cells with a highly specialized morphology designed to facilitate photon detection and transduction of visual information. The most striking feature of photoreceptors cells is the elongated outer segment, which contains stacks of membrane discs which house the transmembrane photosensitive opsin proteins that capture photons and activate the visual transduction cascade. The outer segment is linked to the inner segment of photoreceptor cells by a cilium called the connecting cilium from which the outer segment develops; the outer segment in conjunction with the connecting cilium is considered a large modified primary cilium. The inner segment contains the cell’s protein synthesis machinery and therefore every protein with a role in the outer segment must be transported through the connecting cilium for proper function. The outer segment undergoes a massive rate of protein turnover in order to accomplish phototransduction including 10% of the outer segment being shed every day. Shed discs are phagocytized by neighboring retinal pigmented epithelial cells and are then replaced through the voluminous protein transport needed for new disc formation.15 The connecting cilium is therefore homologous to the transition zone of primary cilia found in other cell types and operates as a gatekeeper of protein trafficking to the outer segment.16
Dysfunction of the connecting cilium results in varying degrees of photoreceptor degeneration depending on which component of the connecting cilium is perturbed. In contrast to LCA cilia genes, mutations in MAK, which localizes to the connecting cilium, cause a form of retinal degeneration which onsets at an average age of 20.17 Highlighting the importance of cilia in retinal disease, ~15% of known retinal disease genes are involved in connecting cilium function. Interestingly, this proportion increases to ~25% when only considering known LCA genes (CEP290, IQCB1, LCA5, RPGRIP1, SPATA7, TULP1), supporting the established notion that cilia are critical to early stages of photoreceptor cell development and maintenance.9,18 Combining the knowledge that cilia genes are good retinal disease gene candidates with the knowledge that cilia are important for early photoreceptor development leads to the conclusion that cilia genes are excellent candidate LCA genes.19
Clusterin-associated protein 1 or CLUAP1, is required for ciliogenesis and localizes to the base and tip of cilia in vitro in mouse embryonic fibroblasts.20 CLUAP1 associates with the intraflagellar transport (IFT) complex B group of proteins and undergoes IFT in both invertebrates and vertebrates.21,22 Cluap1−/− mice exhibit mid-gestation lethality due to developmental complications attributed to their lack of primary cilia.20 Interestingly zebrafish with abolished cluap1 expression also display premature mortality due to nonexistent ciliogenesis but can survive until at least 11 days post fertilization (dpf) allowing the examination of retinal tissue. Photoreceptor defects are apparent as early as 3 dpf, the same time point when wildtype zebrafish photoreceptors are undergoing outer segment formation, and rhodopsin in the mutant fish is mislocalized in the photoreceptor layer, an indication of aberrant IFT.22 Aberrant IFT precedes cell death in animal models of LCA caused by mutations in all six known LCA cilia genes.9,23–27 The retinas of cluap1−/− zebrafish lack photoreceptor cells by 5 dpf while the remaining cell layers are intact.28 The CLUAP1 protein contains two major domains, an N-terminal calponin homology-like domain and a C-terminal coiled-coil domain. Both domains are highly conserved from zebrafish to humans and homologous domains can be found in many microtubule binding proteins.29 The two major isoforms of CLUAP1 are expressed in the human retina at moderate levels similar to IFT protein transcripts.30
Based on this evidence we concluded CLUAP1 is a cilia gene important for photoreceptor outer segment formation in vertebrates and therefore an excellent candidate LCA disease gene. In this study of 212 unsettled LCA patients, we found a single proband homozygous for a nonsynonymous amino acid substitution in CLUAP1. Rescue experiments using zebrafish as a model system proved that our mutant allele is hypomorphic.
MATERIALS AND METHODS
Patient Recruitment, Clinical Diagnosis, and DNA Preparation
Proband MOGL3628 was recruited and diagnosed with LCA at the Montreal Children’s Hospital at the McGill University Health Centre. This patient was diagnosed with LCA after we found nystamus and very poor visual fixation at 6 weeks of age. An ERG was non-detectable and the retina appeared normal. There were no systemic abnormalities. This study was approved by the McGill University Health Center Research Institute Research Ethics Board and adhered to the tenets of the declaration of Helsinki. Blood samples were collected from the proband and both parents after obtaining informed consent; DNA was extracted using the Qiagen blood genomic DNA extraction kit.
WES and Next-Generation Sequencing (NGS) Data Processing
1μg of total patient DNA was sheared into 300–500bp fragments. Fragments were end-repaired and a 3′ adenosine base added. Illumina Y-shaped adapters were ligated to DNA fragments and 10 cycles of PCR were used to amplify the library with a unique barcode. Library DNA concentration was quantified using Life Technologies picogreen assay and the library was pooled together with five other libraries. 3μg of pooled DNA was capture enriched using the NimbleGen SeqCap EZ Human Exome Library v2.0 Hybridization and Wash Kit. Exome captured libraries were quantified and multiplexed sequenced on an Illumina HiSeq 2000.
NGS reads were mapped to the hg19 human reference genome using BWA-MEM, duplicate reads were removed using Picard, local realignments were performed using GATK, and variants were called using Atlas2. A population frequency threshold of 0.5% was used to filter out common variants which occur too frequently to be the cause of a rare Mendelian disease. Four NGS cohort databases were used to determine allele frequencies. The functional consequence of remaining rare variants was annotated using ANNOVAR, and dbNSFP was used to compile in silico predictions about the deleteriousness of nonsynonymous variants. UGENE was used to perform the multiple sequence alignment using the MUSCLE alignment algorithm.
Please see the Supplementary Methods for references and details.
Sanger Sequencing
Sanger sequencing was used to confirm the authenticity of the variant identified by NGS, to confirm the variant properly segregated in the proband’s parents, and to screen five additional LCA probands for mutations in CLUAP1. A PCR primer pair was designed for each exon of interest. After PCR amplification, the amplicons were sequenced on an Applied Biosystems 3730XL or 3500XL Genetic Analyzer.
Immunohistochemistry
In vivo immunohistochemistry was performed on adult mouse retinal sections. Anti-CLUAP1 and anti-acetylated α-tubulin were used for primary antibodies while DAPI was used as a counterstain. In vitro immunohistochemistry was performed on hTERT-RPE1 cells transiently transfected to overexpress human FLAG-tagged CLUAP1 under control of a CMV promoter. CLUAP1 cDNA was mutagenized to recreate the proband’s mutation. Anti-FLAG tag and anti-acetylated α-tubulin were used for primary antibodies while DAPI was used as a counterstain.
Please see the Supplementary Methods for experimental details, reagent sources, and reagent concentrations.
Zebrafish Functional Experiments
The proband’s mutation was recreated at the homologous zebrafish cDNA residue. Capped mRNA was amplified, template DNA degraded, and mRNA purified. Embryos were lysed following a previously published protocol.31 Day 0 embryos were lysed at 8~9 hours post fertilization (hpf), and day 1 embryos were lysed at 24 hpf. Western blotting was performed using anti-GFP. Anti-β-tubulin was used as a loading control. Zebrafish rescue experiments were performed by injecting embryos from cluap1−/+ incrosses at the one cell stage with varying concentrations of zebrafish wildtype and mutant cluap1 mRNA tagged with GFP. GFP mRNA was injected as a negative control. An average of 96.5 embryos were injected per allele per concentration as well as for controls. The percent of phenotypic zebrafish was quantified at 3 dpf. Rescue experiments were performed twice thus reported data is the average of two experiments. P-values were obtained using a student’s t-test. Error bars represent the standard deviation of the two independent experiments.
RESULTS
WES Identifies Homozygous Variants in CLUAP1 as Candidate Pathogenic Mutations in an LCA Proband
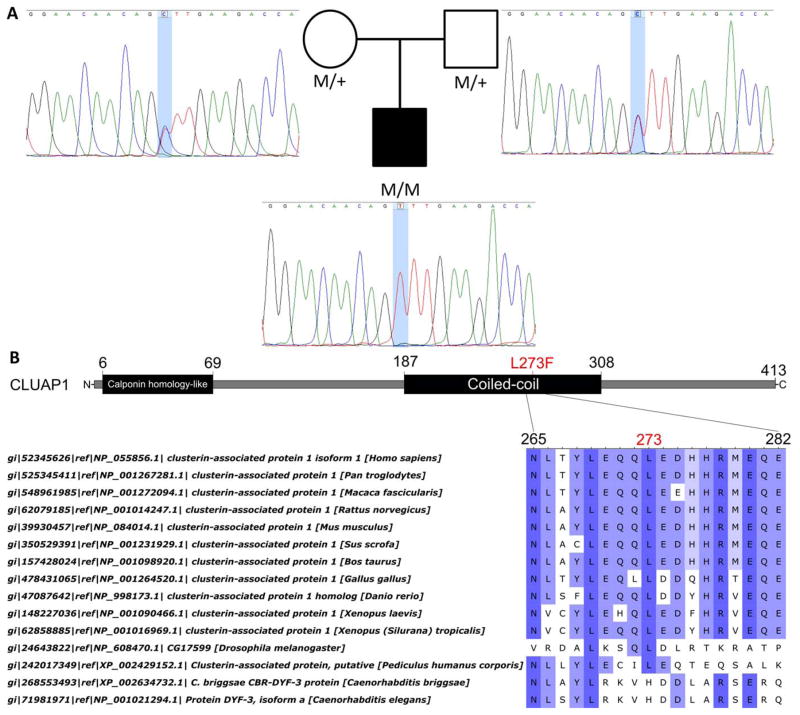
Saudi Arabian proband MOGL3628, currently age 5, exhibited severe visual function limited to light perception by 6 weeks of age accompanied by nystagmus, the oculo-digit sign, and an extinguished ERG (Table 1). The proband’s fundi appeared relatively normal (Supplementary Figure S1). Whole exome sequencing identified 349 rare protein altering variants (Supplementary Table S1). No causal mutations were found among the 8 variants located in known retinal disease genes (Supplementary Table S2) so a list of all potentially biallelic variants was created assuming the recessive inheritance pattern usually seen in LCA patients (Supplementary Table S3). Due to the importance of cilia associated genes in retinal disease, a list of cilia genes (Supplementary Methods) was compared to the genes present in the biallelic variant list and the cilia gene CLUAP1 was identified as containing a homozygous nonsynonymous mutation which was used as reasoning for further analysis of this proband. All 35 potentially biallelic variants in 21 genes were subsequently subjected to a systematic candidate disease gene prioritization strategy in order to exclude or prioritize each gene for further testing. Both gene-level and variant-level information were leveraged during the prioritization process and the primary reasons behind gene exclusion or prioritization are included in Supplementary Table S3. A description of the process is found in the Supplementary Methods. The mutations in CLUAP1 were deemed the most likely candidates for the cause of disease and so their authenticity was validated and proper segregation of the mutations confirmed by Sanger sequencing (Figure 1A).
Table 1.
Proband MOGL3628 information
| Age | Age of onset | Best corrected visual acuity (both eyes) | Nystagmus | Oculodigit sign | Electroretinogram |
|---|---|---|---|---|---|
| 5 | First 6 weeks | Light Perception | Yes | Yes | Extinguished |
Figure 1.
Pedigree, Sanger sequencing traces, and 2D CLUAP1 protein structure with an MSA of the affected residue. (A) The proband’s pedigree and Sanger sequencing traces showing proper segregation of the mutant allele. (B) A 2D graphical representation of the long isoform of CLUAP1 containing two characterized functional domains with the location of the proband’s mutation displayed in red. A protein multiple sequence alignment surrounding the amino acid affected by the proband’s mutation showing the leucine at position 273 is conserved in sighted organisms. Aligned columns are colored a darker blue with increasing conservation at that position.
CLUAP1 has two isoforms, which are both expressed in the human retina, a long isoform (NM_015041, NP_055856) which encodes 413 amino acids and a short isoform (NM_024793, NP_079069) which encodes 247 amino acids. The mutation occurs in the eighth exon of CLUAP1 and affects the cDNA sequence of both isoforms, c.817C>T long, c.319C>T short (Table 2). This mutation results in a leucine to phenylalanine substitution p.L273F long and p.L107F short (Table 2), which is predicted to be either damaging or have a functional impact by 11/12 of the in silico prediction algorithms queried in dbNSFP (Table 2). The mutation is extraordinarily rare and is found only in the Exome Aggregation Consortium database at a frequency of 1/121,086 chromosomes or 0.0008%, although Saudi Arabians are underrepresented in the examined databases and the single individual seen to be a carrier for the allele is of reported European descent (Table 2).
Table 2.
Mutation information
| Genome-level consequence | Gene | Gene-level consequence | |||
|---|---|---|---|---|---|
| NC_000016.9:g.3573261C>T | CLUAP1 |
NM_024793:c.319C>T:p.L107F NM_015041:c.817C>T:p.L273F |
|||
| In silico nonsynonymous mutation deleteriousness prediction algorithms and corresponding predictions | |||||
| SIFT | Polyphen2 HDIV | Polyphen2 HVAR | LRT | MutationTaster | MutationAssessor |
| Damaging | Damaging | Damaging | Damaging | Damaging | Medium impact |
| FATHMM | MetaSVM | MetaLR | VEST3 | PROVEAN | CADD |
| Tolerated | Damaging | Damaging | Damaging | Damaging | Damaging |
| Frequency of variant in large-scale population studies (mutant allele count/total alleles) | |||||
| 1000 Genomes Project | Human Genetic Variation Database | Exome Aggregation Consortium | CHARGE | ||
| 0 (0/5,008) | 0 (0/2,416) | 0.0008% (1/121,086) | 0 (0/21,880) | ||
Our proband’s mutation falls in CLUAP1’s coiled-coil domain, one of two characterized functional domains in the CLUAP1 protein. Both the coiled-coil and calponin homology-like domains have been reported to be involved in microtubule binding activity as shown in IFT proteins with homologous domains. The coiled-coil domain is also associated with facilitating protein-protein interactions, and it is within this region that our proband’s mutation lies (Figure 1B). A protein multiple sequence alignment of human CLUAP1 and its orthologs reveals that the affected leucine residue is conserved in all sighted organisms, including the fruitfly D. melanogaster and the body louse P. humanus corporis whose protein sequences display much less overall conservation compared to analyzed vertebrates. Similar to CLUAP1, the worm ortholog DYF-3 has been shown to be required for neuronal sensory cilium formation in C. elegans yet the specific residue is not conserved in either worm aligned (Figure 1C).32
To ascertain additional patients with CLUAP1 mutations, we specifically corresponded with our LCA expert clinician-scientist collaborator (R.K.K) who was able to identify five additional unsolved LCA probands for whom homozygosity mapping had mapped a significant homozygous interval surrounding and containing CLUAP1. Direct Sanger sequencing of the CLUAP1 gene in these individuals did not identify any candidate mutations. We additionally contacted members of the European Retinal Disease Consortium, of which R.K.K. is a member, as well as submitted our interest in CLUAP1 to GeneMatcher, a component of the Matchmaker Exchange network, but were unable to identify potential collaborators having retinal disease patients with CLUAP1 mutations.
CLUAP1 is Localized at the Connecting Cilium of Photoreceptor Cells
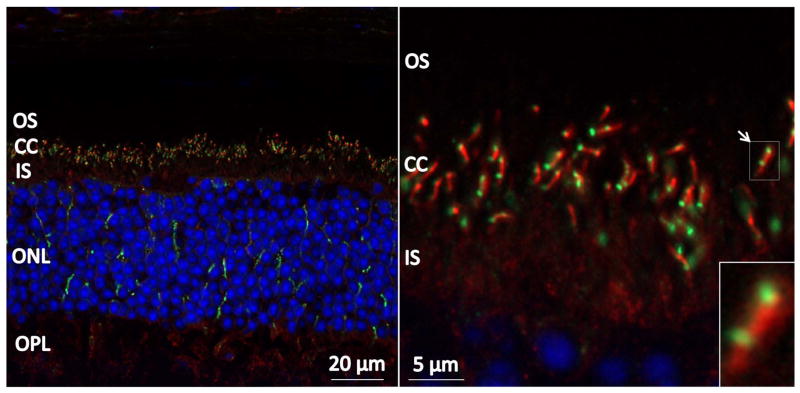
Although CLUAP1 is proposed to be a ciliary protein based on functional studies, its precise localization in the retina is unknown. Given our proband’s phenotype, we hypothesize that CLUAP1 is likely to be expressed in photoreceptor cells. To test this, we performed immunohistochemistry on sections of mouse retinas using an anti-CLUAP1 antibody as well as anti-acetylated α-tubulin and DAPI. CLUAP1 localizes to the connecting cilium of photoreceptor cells located between the inner and outer segment layers of the retina (Figure 2). An inconsistent CLUAP1 signal is also seen in certain areas of the outer nuclear layer but this may be the result of non-specific antibody binding.
Figure 2.
CLUAP1 localizes to the connecting cilium in adult mouse retinas. Mouse immunohistochemistry staining using anti-CLUAP1 (green), anti-acetylated α-tubulin (red), and DAPI (blue). CLUAP1 can be seen to localize to specific puncti between the inner segment (IS) and outer segment (OS) layers of photoreceptors cells. CLUAP1 localization overlaps with the tip and base of the acetylated α-tubulin staining corresponding to the tip and base of connecting cilia (CC). ONL = outer nuclear layer, OPL = outer plexiform layer.
The Proband’s Missense Mutation Negatively Impacts CLUAP1 Function
To test if the proband’s mutation affects CLUAP1 function by altering its localization we performed immunofluorescence on hTERT-RPE1 cells transiently overexpressing either FLAG-CLUAP1 or FLAG-CLUAP1Mut. CLUAP1 harboring the proband’s mutation was observed to localize to the base of cilia similarly to wildtype (Supplementary Figure S2). To test if the proband’s mutation is detrimental to CLUAP1 function in vivo, mutagenesis of wild type zebrafish cluap1 cDNA was performed to generate a cDNA construct containing a mutation at the zebrafish residue that is homologous to the human variant. Taking advantage of the cluap1 knockout zebrafish model, which shows developmental defects typical for a ciliopathy, a functional analysis of the mutant cluap1 was performed using rescue experiments. As described in the methods section, GFP-tagged wild type and mutant mRNA were injected into zebrafish embryos resulting from cluap1+/− incrosses.
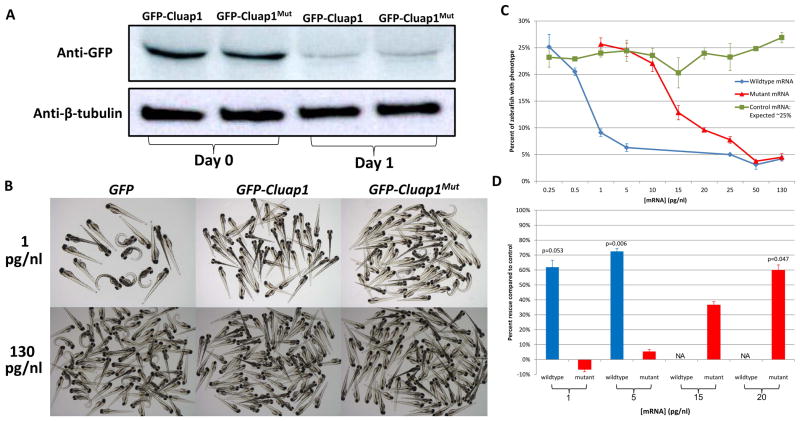
To test if the mutation affects cluap1 expression, we first performed Western blots using embryo lysates. Mutant Cluap1 was expressed at similar levels to wildtype leading to the conclusion that the mutation does not affect protein expression or degradation (Figure 3A). To assess the dysfunction of our mutant Cluap1, we screened for the ventral spine curvature feature present in cluap1−/− knockout zebrafish at an early age. The percent of phenotypic embryos was compared to the expected ~25% observed after control injections of GFP. To determine if mutant Cluap1 could rescue the phenotype of cluap1−/− knockout zebrafish, low and high concentrations (1 and 130 pg/nl) of mutant mRNA were tested along with wildtype. At the lower concentration of 1 pg/nl, no significant rescue effect was observed from mutant cluap1 mRNA injected embryos (25.7% phenotypic progeny, p=0.118, Figure 3B and Supplementary Table S4) while wildtype cluap1 mRNA displayed a borderline significant rescue (9.1% phenotypic progeny, p=0.053, Figure 3B and Supplementary Table S4). At the higher concentration of 130 pg/nl, mutant cluap1 performed similar to wildtype and exhibited a clearly significant rescue effect (4.5% phenotypic progeny, p=0.007, Figure 3B and Supplementary Table S4).
Figure 3.
Mutant Cluap1 is expressed at levels similar to wildtype but requires 20 times the amount in order to achieve an equivalent rescue effect. (A) Zebrafish embryos injected with 25 pg/nl GFP-cluap1 or GFP-cluap1Mut mRNA express similar amounts of Cluap1 as verified by western blotting. (B) Embryos resulting from cluap1−/+ incrosses injected with control GFP mRNA exhibit the expected 25% phenotypic proportion. At a low concentration (1 pg/nl), wildtype cluap1 mRNA noticeably rescues the spine curvature feature of cluap1−/− zebrafish embryos while mutant cluap1 mRNA does not. At a high concentration (130 pg/nl), mutant cluap1 mRNA rescues the spine curvature feature comparable to wildtype cluap1 mRNA revealing a dose-dependent response. (C) Dose response curves of rescue experiments. Zebrafish injected with control GFP mRNA (green) showed approximately the expected 25% Mendelian ratio. Zebrafish injected with GFP-cluap1 mRNA (blue) reached a borderline significant (p=0.053) rescue at 1 pg/nl. Zebrafish injected with GFP-cluap1Mut mRNA (red) significantly (p=0.047) rescued the mutant phenotype at a concentration of 20 pg/nl. (D) The percent of progeny rescued as compared to control injections at the same concentration highlights the 20-fold increase in the amount of mutant cluap1 mRNA required to achieve a level of rescue significance comparable to wildtype cluap1. Errors bars in (C) and (D) represent the standard deviation from two independent experiments.
Since a high concentration of mutant cluap1 rescued the spine curvature feature, we next quantified the functionality of the mutant protein by varying the concentration of cluap1 mRNA injections from 0.25 – 130 pg/nl. Graphing the dose response curve of these rescue experiments revealed that the proband’s mutation does indeed interfere with Cluap1 function. While wildtype cluap1 mRNA can reach a borderline significant rescue effect at the previously mentioned concentration of 1 pg/nl, mutant cluap1 mRNA requires a concentration of 20 pg/nl in order to achieve a similar level of rescue significance (9.6% phenotypic progeny, p=0.047, Figure 3C and Supplementary Table S4). To visualize this data in an alternate form, we compared the percent of progeny that were expected to be phenotypic based on control injections to the percent actually observed after mutant and wildtype cluap1 mRNA injections to determine the percent of progeny that were rescued which highlights the hypomorphic nature of our proband’s allele (Figure 3D). Based on these functional rescue assays Cluap1 containing our proband’s mutation has as little as 5% of its remaining activity as judged from the 20 fold increase in mutant Cluap1 concentration required to achieve a rescue significance comparable to wildtype.
DISCUSSION
We conclude that since CLUAP1 is required for photoreceptor function in other vertebrates, and our LCA proband’s allele is an extremely hypomorphic CLUAP1 allele, that CLUAP1 is very likely to be necessary for photoreceptor function in humans. This therefore represents the first report linking hypomorphic mutations in CLUAP1 to human disease and establishes CLUAP1 as a candidate LCA gene. Identifying CLUAP1 as a novel candidate LCA gene is important both for understanding human biology and for unlocking the future molecular diagnosis of patients affected by mutations in this gene. CLUAP1 can now be studied in the context of being a human ciliopathy gene and establishing a new candidate disease gene allows for quicker and less expensive molecular diagnosis of future patients since geneticists can target the CLUAP1 locus for examination. CLUAP1 mutations appear to be a rare cause of LCA so further studies screening additional patient cohorts are needed to determine the true proportion of LCA patients with CLUAP1 mutations.
The importance of cilia to many areas of the body and the importance of CLUAP1 to cilia raises a concern about the clinical presentation of the presented proband and why syndromic features have not been observed. We considered addressing this question by sectioning the retinas of embryos rescued with mutant cluap1 mRNA to verify that a retinal phenotype persists after the correction of spine curvature. Unfortunately due to the rapid decay of the injected mRNA and the exogenous protein being minimally detectable at 1 dpf (Figure 3A), it is unlikely that this approach would be informative as photoreceptor development isn’t complete until around 5 dpf. However, three reasonable explanations exist for this observation. The first is that our proband does indeed have a syndromic disease, but additional features have yet to manifest due to the young age. This would not be surprising as multiple syndromic ciliopathies such as Joubert syndrome, Senior-Løken syndrome, and Alström syndrome are known to present a retinal degeneration feature preceding complications in other tissues. These syndromes can in fact be misdiagnosed as LCA because of the lack of additional symptoms at an affected individual’s initial clinical presentation.1
The second explanation is that the specific mutation that our proband harbors is only detrimental to a retinal specific function, and therefore only a retinal phenotype is observed. The connecting cilium in photoreceptors is a unique form of cilia containing proteins not found in most other ciliated cell types in the body. The presence of retinal specific proteins at the connecting cilium means that proteins like CLUAP1 whose expression is not restricted to the retina conceivably have a purpose specialized to the connecting cilium involving interactions with retinal specific proteins. This explanation is supported by the fact that the proband’s mutation causes a nonsynonymous amino acid substitution and that the mutation resides in CLUAP1’s coiled-coil domain known to accommodate protein-protein interactions. The residue that our proband’s mutation affects is not conserved in C. elegans and C. briggsae, implying the residue is not essential for ciliogenesis but instead is important for some function specific to the organisms in which it is strictly conserved. The absence of eyes is one of the defining features separating these worms from the other species in the multiple sequence alignment and so a safe hypothesis appears to be that the conserved leucine is required for a function involved in vision.
The third explanation is that our proband’s mutation results in a general hypomorphic form of CLUAP1 and that the retina is more sensitive to this decrease in CLUAP1 function than other tissue types possibly comparable to the ubiquitously expressed LCA gene NMNAT1.8 The variability in phenotype caused by mutations in the same gene can range drastically, especially in retinal disease, depending on the exact nature of the patients’ alleles. It is possible that a mutation residing in CLUAP1’s calponin homology-like domain would confer a different phenotype impacting other tissues, and it is likely the complete loss of function of CLUAP1 would not be compatible with human life as seen in other vertebrates. The function and expression of genes are also not always conserved across species and human CLUAP1 is not detected in the human kidney by northern blotting which could explain the lack of kidney defects observed in the original zebrafish polycystic kidney disease screen.33,34
In 2008 known LCA genes were reported to account for 70% of LCA cases, based on our experience in 2015 this percentage can vary up to 75% but for the most part has unappreciatively changed despite the identification of an additional eight genes. At least six of the reported LCA disease genes including the two most recently published genes account for a remarkably small portion (~1%) of all LCA cases.11–13,35–37 CLUAP1 adds to this trend accounting for 1/212 LCA patients in our cohort (0.47%). The more than a quarter of all LCA patients waiting for a molecular diagnosis inspires two interesting hypotheses regarding future directions for studying the genetics of LCA. The first is the big fish hypothesis which can be summarized in that any one gene responsible for a significant proportion of patients’ disease (a “big fish”) will likely have already been discovered at this stage in human genetics research. This means that only “little fish” are left, which is why we see new LCA disease genes accounting for a minor fraction of patients. It is therefore possible that the source of disease representing the 30% of unsolved LCA cases will be composed of >25 novel disease genes each accounting for an ever smaller fraction of patients. The second alternative hypothesis is that the cause of disease resides in known LCA genes but modern genotyping techniques cannot identify the causal variation. For most genetic disorders whole exome sequencing is widely considered the standard method for molecular diagnosis but whole exome sequencing is currently unsuitable for identifying potentially causative variation in introns, promoters, enhancers, nor large structural variations. Even in regions captured by whole exome sequencing there is data we do not fully know how to interpret including variation in UTRs, synonymous variation and intronic variation close to an exon but outside of the 2bp splicing consensus sequence. Understanding variation in these atypical regions is imperative in the research of human disease because groups may focus efforts on finding novel disease genes when the disease-causing mutations they seek are “right under their noses”. The truth is most likely a combination of these two hypotheses leaving numerous possibilities for the future of LCA research.
Supplementary Material
The fundi of proband MOGL3628 appear normal. The right and left eyes are shown in these colored retinal photos illustrating normal appearing discs, fovea, and maculae. The retinal arteries are very subtly narrowed.
The proband’s mutation in CLUAP1 does not prevent its ciliary localization when transiently overexpressed in hTERT-RPE1 cells. Immunohistochemistry analysis was performed using anti-FLAG (red), anti-acetylated α-tubulin (green), and DAPI (blue). (A) FLAG-CLUAP1 and (B) FLAG-CLUAP1Mut are both observed weakly throughout the cell with the strongest signal at the base of the cilium.
Supplementary Table S1: 349 rare protein-altering variants in 335 genes
Acknowledgments
We would like to thank the proband and their family for participating in this study. This work is supported by the National Institutes of Health [grant numbers R01-DK092808, 1P30-DK090744] to Z.Sun., [shared grant 1S10RR026550] to R.C.; the National Eye Institute [grant numbers R01EY022356, R01EY018571, P30EY002520] to R.C.; the Retina Research Foundation; the Foundation Fighting Blindness [grant number BR-GE-0613-0618-BCM] to R.C.; R.K.K. acknowledges the Foundation Fighting Blindness Canada, the Canadian Institutes of Health Research, and Fonds de recherché Santé Quebéc et Réseau Vision all of whom have supported this work. Z. Soens is supported by National Eye Institute training grant 5T32EY007001-38.
Footnotes
Supplementary information is available at the Genetics in Medicine website (http://www.nature.com/gim).
References
- 1.den Hollander AI, Roepman R, Koenekoop RK, Cremers FP. Leber congenital amaurosis: genes, proteins and disease mechanisms. Progress in retinal and eye research. 2008 Jul;27(4):391–419. doi: 10.1016/j.preteyeres.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 2.RetNet Retinal Information Network. University of Texas-Houston Health Science Center; Houston, TX: [accessed 2015]. http://www.sph.uth.tmc.edu/RetNet. [Google Scholar]
- 3.Wang X, Wang H, Sun V, et al. Comprehensive molecular diagnosis of 179 Leber congenital amaurosis and juvenile retinitis pigmentosa patients by targeted next generation sequencing. Journal of medical genetics. 2013 Oct;50(10):674–688. doi: 10.1136/jmedgenet-2013-101558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Wang H, Cao M, et al. Whole-exome sequencing identifies ALMS1, IQCB1, CNGA3, and MYO7A mutations in patients with Leber congenital amaurosis. Human mutation. 2011 Dec;32(12):1450–1459. doi: 10.1002/humu.21587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo Y, Prokudin I, Yu C, et al. Advantage of Whole Exome Sequencing over Allele-specific and Targeted Segment Sequencing, in Detection of Novel TULP1 Mutation in Leber Congenital Amaurosis. Ophthalmic genetics. 2014 Feb 19; doi: 10.3109/13816810.2014.886269. [DOI] [PubMed] [Google Scholar]
- 6.Hufnagel RB, Ahmed ZM, Correa ZM, Sisk RA. Gene therapy for Leber congenital amaurosis: advances and future directions. Graefe’s archive for clinical and experimental ophthalmology = Albrecht von Graefes Archiv fur klinische und experimentelle Ophthalmologie. 2012 Aug;250(8):1117–1128. doi: 10.1007/s00417-012-2028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robson JG, Frishman LJ. The rod-driven a-wave of the dark-adapted mammalian electroretinogram. Progress in retinal and eye research. 2014 Mar;39:1–22. doi: 10.1016/j.preteyeres.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koenekoop RK, Wang H, Majewski J, et al. Mutations in NMNAT1 cause Leber congenital amaurosis and identify a new disease pathway for retinal degeneration. Nature genetics. 2012 Sep;44(9):1035–1039. doi: 10.1038/ng.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eblimit A, Nguyen TM, Chen Y, et al. Spata7 is a retinal ciliopathy gene critical for correct RPGRIP1 localization and protein trafficking in the retina. Human molecular genetics. 2014 Nov 14; doi: 10.1093/hmg/ddu573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aldahmesh MA, Al-Owain M, Alqahtani F, Hazzaa S, Alkuraya FS. A null mutation in CABP4 causes Leber’s congenital amaurosis-like phenotype. Molecular vision. 2010;16:207–212. [PMC free article] [PubMed] [Google Scholar]
- 11.Sergouniotis PI, Davidson AE, Mackay DS, et al. Recessive mutations in KCNJ13, encoding an inwardly rectifying potassium channel subunit, cause leber congenital amaurosis. American journal of human genetics. 2011 Jul 15;89(1):183–190. doi: 10.1016/j.ajhg.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henderson RH, Williamson KA, Kennedy JS, et al. A rare de novo nonsense mutation in OTX2 causes early onset retinal dystrophy and pituitary dysfunction. Molecular vision. 2009;15:2442–2447. [PMC free article] [PubMed] [Google Scholar]
- 13.Asai-Coakwell M, March L, Dai XH, et al. Contribution of growth differentiation factor 6-dependent cell survival to early-onset retinal dystrophies. Human molecular genetics. 2013 Apr 1;22(7):1432–1442. doi: 10.1093/hmg/dds560. [DOI] [PubMed] [Google Scholar]
- 14.Estrada-Cuzcano A, Koenekoop RK, Coppieters F, et al. IQCB1 mutations in patients with leber congenital amaurosis. Investigative ophthalmology & visual science. 2011 Feb;52(2):834–839. doi: 10.1167/iovs.10-5221. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen-Legros J, Hicks D. Renewal of photoreceptor outer segments and their phagocytosis by the retinal pigment epithelium. International review of cytology. 2000;196:245–313. doi: 10.1016/s0074-7696(00)96006-6. [DOI] [PubMed] [Google Scholar]
- 16.Wheway G, Parry DA, Johnson CA. The role of primary cilia in the development and disease of the retina. Organogenesis. 2014 Jan 1;10(1):69–85. doi: 10.4161/org.26710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Huet RA, Siemiatkowska AM, Ozgul RK, et al. Retinitis pigmentosa caused by mutations in the ciliary MAK gene is relatively mild and is not associated with apparent extra-ocular features. Acta ophthalmologica. 2015 Feb;93(1):83–94. doi: 10.1111/aos.12500. [DOI] [PubMed] [Google Scholar]
- 18.Rachel RA, Li T, Swaroop A. Photoreceptor sensory cilia and ciliopathies: focus on CEP290, RPGR and their interacting proteins. Cilia. 2012;1(1):22. doi: 10.1186/2046-2530-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Q, Zhang Q, Pierce EA. Photoreceptor sensory cilia and inherited retinal degeneration. Advances in experimental medicine and biology. 2010;664:223–232. doi: 10.1007/978-1-4419-1399-9_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Botilde Y, Yoshiba S, Shinohara K, et al. Cluap1 localizes preferentially to the base and tip of cilia and is required for ciliogenesis in the mouse embryo. Developmental biology. 2013 Sep 1;381(1):203–212. doi: 10.1016/j.ydbio.2013.05.024. [DOI] [PubMed] [Google Scholar]
- 21.Omori Y, Zhao C, Saras A, et al. Elipsa is an early determinant of ciliogenesis that links the IFT particle to membrane-associated small GTPase Rab8. Nature cell biology. 2008 Apr;10(4):437–444. doi: 10.1038/ncb1706. [DOI] [PubMed] [Google Scholar]
- 22.Lee C, Wallingford JB, Gross JM. Cluap1 is essential for ciliogenesis and photoreceptor maintenance in the vertebrate eye. Investigative ophthalmology & visual science. 2014 Jul;55(7):4585–4592. doi: 10.1167/iovs.14-14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boldt K, Mans DA, Won J, et al. Disruption of intraflagellar protein transport in photoreceptor cilia causes Leber congenital amaurosis in humans and mice. The Journal of clinical investigation. 2011 Jun;121(6):2169–2180. doi: 10.1172/JCI45627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anand M, Khanna H. Ciliary transition zone (TZ) proteins RPGR and CEP290: role in photoreceptor cilia and degenerative diseases. Expert opinion on therapeutic targets. 2012 Jun;16(6):541–551. doi: 10.1517/14728222.2012.680956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ronquillo CC, Bernstein PS, Baehr W. Senior-Loken syndrome: a syndromic form of retinal dystrophy associated with nephronophthisis. Vision research. 2012 Dec 15;75:88–97. doi: 10.1016/j.visres.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grossman GH, Watson RF, Pauer GJ, Bollinger K, Hagstrom SA. Immunocytochemical evidence of Tulp1-dependent outer segment protein transport pathways in photoreceptor cells. Experimental eye research. 2011 Nov;93(5):658–668. doi: 10.1016/j.exer.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Won J, Gifford E, Smith RS, et al. RPGRIP1 is essential for normal rod photoreceptor outer segment elaboration and morphogenesis. Human molecular genetics. 2009 Nov 15;18(22):4329–4339. doi: 10.1093/hmg/ddp385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Sun Z. Qilin is essential for cilia assembly and normal kidney development in zebrafish. PloS one. 2011;6(11):e27365. doi: 10.1371/journal.pone.0027365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schou KB, Andersen JS, Pedersen LB. A divergent calponin homology (NN-CH) domain defines a novel family: implications for evolution of ciliary IFT complex B proteins. Bioinformatics. 2014 Apr 1;30(7):899–902. doi: 10.1093/bioinformatics/btt661. [DOI] [PubMed] [Google Scholar]
- 30.Farkas MH, Grant GR, White JA, Sousa ME, Consugar MB, Pierce EA. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC genomics. 2013;14:486. doi: 10.1186/1471-2164-14-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zamora LY, Lu Z. Alcohol-induced morphological deficits in the development of octavolateral organs of the zebrafish (Danio rerio) Zebrafish. 2013 Mar;10(1):52–61. doi: 10.1089/zeb.2012.0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murayama T, Toh Y, Ohshima Y, Koga M. The dyf-3 gene encodes a novel protein required for sensory cilium formation in Caenorhabditis elegans. Journal of molecular biology. 2005 Feb 25;346(3):677–687. doi: 10.1016/j.jmb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi M, Lin YM, Nakamura Y, Furukawa Y. Isolation and characterization of a novel gene CLUAP1 whose expression is frequently upregulated in colon cancer. Oncogene. 2004 Dec 9;23(57):9289–9294. doi: 10.1038/sj.onc.1208100. [DOI] [PubMed] [Google Scholar]
- 34.Sun Z, Amsterdam A, Pazour GJ, Cole DG, Miller MS, Hopkins N. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 2004 Aug;131(16):4085–4093. doi: 10.1242/dev.01240. [DOI] [PubMed] [Google Scholar]
- 35.Abu-Safieh L, Alrashed M, Anazi S, et al. Autozygome-guided exome sequencing in retinal dystrophy patients reveals pathogenetic mutations and novel candidate disease genes. Genome research. 2013 Feb;23(2):236–247. doi: 10.1101/gr.144105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson DA, Li Y, McHenry CL, et al. Mutations in the gene encoding lecithin retinol acyltransferase are associated with early-onset severe retinal dystrophy. Nature genetics. 2001 Jun;28(2):123–124. doi: 10.1038/88828. [DOI] [PubMed] [Google Scholar]
- 37.Mackay DS, Henderson RH, Sergouniotis PI, et al. Novel mutations in MERTK associated with childhood onset rod-cone dystrophy. Molecular vision. 2010;16:369–377. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The fundi of proband MOGL3628 appear normal. The right and left eyes are shown in these colored retinal photos illustrating normal appearing discs, fovea, and maculae. The retinal arteries are very subtly narrowed.
The proband’s mutation in CLUAP1 does not prevent its ciliary localization when transiently overexpressed in hTERT-RPE1 cells. Immunohistochemistry analysis was performed using anti-FLAG (red), anti-acetylated α-tubulin (green), and DAPI (blue). (A) FLAG-CLUAP1 and (B) FLAG-CLUAP1Mut are both observed weakly throughout the cell with the strongest signal at the base of the cilium.
Supplementary Table S1: 349 rare protein-altering variants in 335 genes