Abstract
The study of neurological infections by viruses defines the field of neurovirology, which has emerged in the last 30 years and was founded upon the discovery of a number of viruses capable of infecting the human nervous system. Studies have focused on the molecular and biological basis of viral neurological diseases with the aim of revealing new therapeutic options. The first studies of neurovirological infections can be traced back to the discovery that some viruses have an affinity for the nervous system with research into rabies by Louis Pasteur and others in the 1880s. Today, the immense public health impact of neurovirological infections is illustrated by diseases such as neuroAIDS, progressive multifocal leukoencephalopathy, and viral encephalitis. Recent research has seen the development of powerful new techniques for gene editing that promise revolutionary opportunities for the development of novel therapeutic options. In particular, clustered regulatory interspaced short palindromic repeat-associated 9 system provides an effective, highly specific and versatile tool for targeting DNA viruses that are beginning to allow the development of such new approaches. In this short review, we discuss these recent developments, how they pertain to neurological infections, and future prospects.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-016-0439-1) contains supplementary material, which is available to authorized users.
Keywords: Neurological infections, Gene editing, CRISPR/Cas9, NeuroAids, Progressive multifocal encephalopathy
Introduction and Overview
A number of organisms are able to infect the central nervous system (CNS) and can cause diseases with significant morbidity and mortality in the human population. These may be fungal, protozoal, bacterial, or viral in nature. A large number of viruses have been found to be capable of infecting the human nervous system and their study has defined the field of neurovirology, which has emerged as a discipline over the last 30 years [1]. The brain is well protected from viral infections by multiple layers and by the action of the immune system. Nevertheless, it is possible for a number of viruses to invade the CNS, and infections by these viruses usually begin by the virus entering peripheral tissues and then spreading to the CNS either via the peripheral nervous system or by a hematogenous route [2]. By definition, viruses are infective agents that consist of a nucleic acid molecule in a protein coat and are only able to multiply within the living cells of a host. As viral life cycles occur intracellularly, it is common for them to utilize host cellular proteins, making it hard to identify therapeutic targets that are specific for the virus. This is especially true for viruses that cause neurological infections but then enter an inactive persistent/latent state where production of viral proteins does not occur or is limited to a very few proteins. Two examples of viruses that cause neurological infections but can then become inactive are the HIV-1 and polyomavirus JC (JCV). HIV-1 infection in the CNS can cause HIV-associated neurocognitive disorders, which include neurological disorders of various degrees of severity such as HIV-associated dementia, HIV encephalopathy, and mild neurocognitive disorder [3, 4]. JCV causes progressive multifocal leukoencephalopathy (PML), a severe and often fatal, demyelinating disease of the CNS [5–7]. Thus, it would be therapeutically advantageous to develop methodologies that are capable of specifically targeting the nucleotide sequences of these and other CNS viruses. The clustered regulatory interspaced short palindromic repeat (CRISPR)-associated 9 (Cas9) system provides such a methodology and is able to target JCV because it is a DNA virus and HIV-1 because it is an RNA retrovirus that has a DNA proviral replication intermediate. CRISPR/Cas9 provides an effective, highly specific, and versatile tool for targeting DNA viruses or DNA intermediates of RNA viruses. In this review, we will discuss how CRISPR/Cas9 has been used against human viruses that cause neurological infections, including HIV-1 and JCV, and future prospects for therapeutic approaches to these and other neurovirulent viruses.
Viral Diseases of the CNS
A large variety of different viruses of a range of types are able to cause infections of the CNS resulting in disease. As mentioned above, the neurotropic polyomavirus JCV causes PML [5–7]. Interestingly, JCV infects a large proportion of the population during childhood but then enters a persistent/latent state, where viral proteins are not produced, which resides in a variety of tissues, including the glial cells (oligodendrocytes and astrocytes) of the CNS [8]. However, rarely and usually only under conditions of immune dysfunction, JCV reactivates in glia, leading to enlarging lesions of demyelination and the development of PML [7]. HIV-1 infects cells in the CNS and HIV-associated neurocognitive disorders are linked with encephalopathy induced by HIV infection and fueled by immune activation of macrophages and microglia [9, 10]. JCV and HIV-1 are important examples of neurotropic viruses causing neurological infections to which the CRISPR/Cas9 gene editing approach has been applied and thus they will be discussed first [11, 12]. Potentially, the CRISPR/Cas9 could be applied to other DNA viral infections of the human CNS, including herpes simplex encephalitis, which can be caused by herpes simplex type 1 (HSV1) or type 2 (HSV2) [13, 14]; cytomegalovirus (CMV) encephalitis, caused by CMV and seen in patients with AIDS [15, 16]; adenovirus infection-associated CNS dysfunction in children [17]; and varicella zoster encephalitis caused by varicella zoster virus, which is the agent that causes chickenpox in children and shingles in adults [18]. These are also mentioned below. At the present time, there is no such methodology applicable to neurological infections by RNA viruses such as subacute sclerosing panencephalitis (Measles virus), West Nile virus, poliomyelitis, equine encephalitis, rabies, and progressive rubella panencephalitis, or mumps meningoencephalitis. Nevertheless, there may be cellular genes that produce co-factors necessary for the RNA virus life cycle that may provide suitable targets. Neurological viral infections and their amenability to treatments by the CRISPR approach are summarized in Table 1.
Table 1.
Neurological viral infections and the clustered regulatory interspaced short palindromic repeat (CRISPR) approach
| Virus | Neurological disease | Viral genome | Amenability to CRISPR | Approach used | References |
|---|---|---|---|---|---|
| HIV-1 | NeuroAIDS | RNA | Yes | Yes | [36, 40–42, 44, 45, 49–51] |
| JCV | PML | DNA | Yes | Yes | [11] |
| HSV | Encephalitis | DNA | Yes | Yes | [59] |
| CMV, VZV, etc. | Encephalitis | DNA | Yes | No | |
| Measles, WNV, etc. | Encephalitis | RNA | No | No |
JCV = polyomavirus JC; HSV = herpes simplex virus; CMV = cytomegalovirus; VZV = varicella zoster virus; WNV = West nile virus
Recent Advances in Gene Editing Methodologies
Over the last several years, there have been remarkable advances in the development of gene-editing methodologies with a broad range of important basic science and clinical applications, including the treatment of cancers and viral infections [19, 20]. Three new types of reagents have been developed that specifically target nucleotide sequences within viral genomes and provide important novel antiviral strategies. First, zinc-finger nucleases (ZFN) are fusion proteins of the catalytic DNA cleavage domain of the restriction enzyme FokI with custom-designed specificity-conferring Cys2–His2 zinc-finger domain [21–23]. Second, transcription activator-like effector nucleases are also fusion proteins of FokI, but the specificity-conferring targeting domain in this case is derived from the Xanthomonas transcription activator-like effector proteins [24, 25]. Third, and most powerful, is clustered regulatory interspaced short palindromic repeat (CRISPR)-associated 9 (Cas9), which is effective, highly specific, and incredibly versatile while providing an unprecedented degree of control over genome editing [23, 26–28]. This review will focus on the actual and potential applications of CRISPR/Cas9 system against human neurological viral infections.
CRISPR/Cas9
CRISPR/Cas9 is elegant in its simplicity and ease of use. Because it makes use of a guide RNA (gRNA) to target a specific sequence for cleavage, it is adaptable and flexible to different target nucleotide sequences simply by changing the sequence of the gRNA [29, 30]. CRISPR/Cas nucleases evolved in bacteria as an adaptive immune system, a defense mechanism to deal with attack by viruses, and are found in nearly all Archaea and about half of bacteria [31, 32]. This adaptive immune system has been developed into a very precise gene editing tool, in which a short gRNA is employed to direct the degradation of DNA in a sequence-specific fashion. CRISPR/Cas9 has 2 components: the gRNA and the Cas9 endonuclease. Expression of both the gRNA and Cas9 together in cells results in the DNA target sequence being modified or disrupted. Binding of the 20 base-pair gRNA to the DNA target by Watson–Crick base-pairing recruits the gRNA/Cas9 complex to its target. Additionally to this complementary gRNA guide sequence, Cas9 targeting and subsequent endonuclease cleavage require a trinucleotide protospacer-adjacent motif located immediately 3’ to the target site. The Cas9 endonuclease cuts both strands of the DNA target causing a double-strand break (DSB), which lies 3–4 nucleotides upstream of the protospacer-adjacent motif. The DSB may be repaired by the error-prone nonhomologous end-joining DNA repair pathway, often resulting in the introduction of insertions/deletions at the DSB site. In protein-coding regions, this may lead to frame shifts and/or premature stop codons, which effectively disrupt the open reading frame of the target gene ablating gene function. If 2 gRNAs are employed to different sites on the same DNA, it is possible to delete large segments of DNA that lie between the sites of the DSBs [33]. As opposed to the unpredictable mutations resulting from DSB repair by nonhomologous end-joining, targeted DSBs can be repaired by precise gene editing through homology-directed recombination with an exogenously supplied donor template. Thus, the co-delivery of Cas9/gRNA along with a vector containing DNA homologous to the break site enables high-efficiency homology-directed recombination-based gene editing. Sequence differences present in the donor template can be incorporated into the endogenous locus allowing correction of mutations [23].
As Cas9 is a general endonuclease, all that is required is the gRNA, and this can be generated by chemical synthesis, in vitro transcription, or cellular expression resulting on the production of a highly specific gene editing tool. This ease of use has allowed it to be used in a number of applications in the last 2 years [26, 27]. The relative ease with which new sites can be targeted, by changing the sequence of the short region of the gRNA that determines specificity, has made CRISPR/Cas9 a highly attractive technique for the introduction of site-specific DSBs. In additional, CRISPR/Cas9 protein is highly amenable to multiplexing by using multiple gRNAs to induce DSBs at several loci. Given the occurrence in nature of multiple diverse CRISPR systems, it is probable that new and improved CRISPR gene editing systems will emerge, such as the recently discovered Cpf1 endonuclease and others [34, 35].
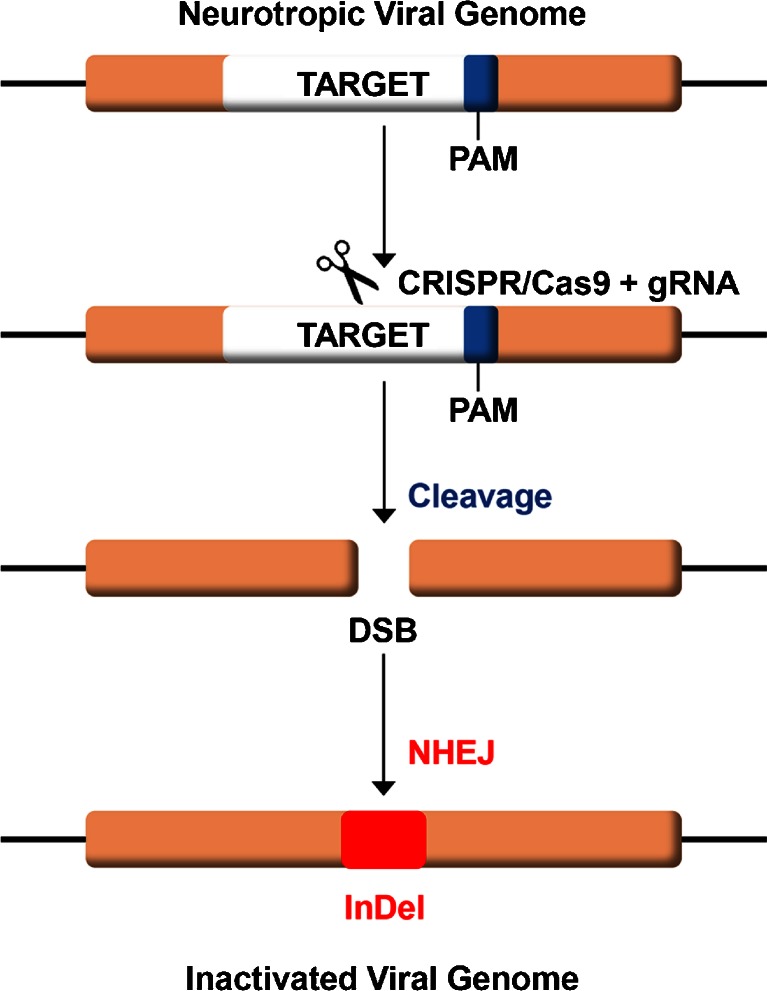
CRISPR/Cas9 also achieves a high degree of specificity and offers virtually exclusive on-target cleavage, which is important for potential clinical applications, given the large size of the human genome. In addition, assays of off-target cleavage have revealed this to be a very rare event [36], and the Cas9 system is remarkably specific. Modifications and improvements to CRISPR/Cas9 continue with respect to genome editing efficiency and reduction of off-target events [37, 38]. Thus, the high specificity of CRISPR/Cas9 means that it has the potential to attack viral DNA in cells without harming the host genome. In principal, eradication of viral neurological infections by CRISPR/Cas9 should be applicable to any DNA virus or RNA virus that has a DNA intermediate in its life cycle [12, 26, 27]. Current progress and future potential for such applications are discussed below. A schematic diagram of CRISPR/Cas9 action on viral DNA is shown in Fig. 1.
Fig. 1.
Schematic of clustered regulatory interspaced short palindromic repeat (CRISPR)/CRISPR-associated 9 (Cas9) action on viral DNA. A guide RNA (gRNA) is designed that is specific for a target sequence immediately adjacent to a 3’ protospacer adjacent motif (PAM) sequence (blue) within the genome of the virus. The gRNA is expressed in host cells together with the Cas9 endonuclease, which are shown schematically as scissors. CRISPR cleaves the viral DNA at the target site resulting in a double-strand break (DSB), which is repaired by nonhomologous end-joining (NHEJ). As NHEJ is error prone, small insertions and deletions (InDel) are found in the repaired DNA that inactivate the target gene
Application of CRISPR/Cas9 to Specific Human Viruses
HIV-1
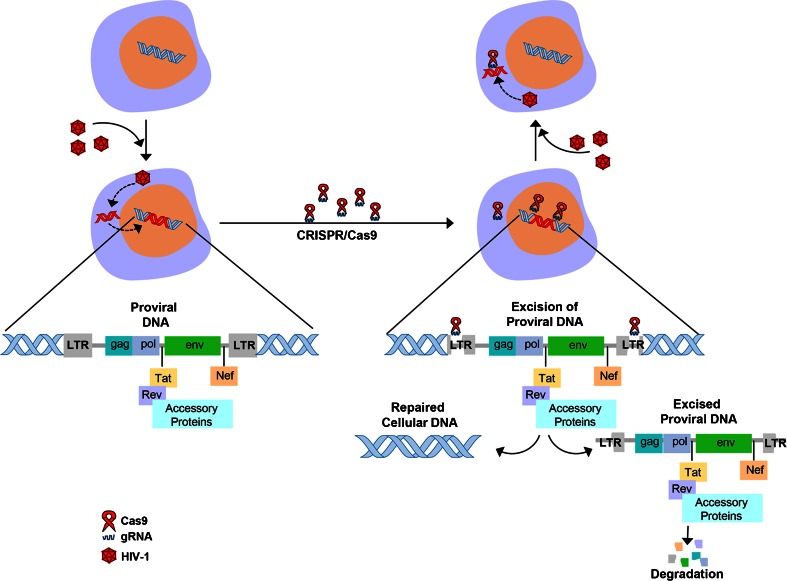
Since its discovery in the 1980s, HIV-1/AIDS remains a significant public health problem worldwide. Despite the development of highly effective treatments that suppress viral replication and robustly reduce viral load in the blood, that is, combination antiretroviral therapy (cART), HIV-1/AIDS remains incurable as HIV-1 is a retrovirus that is able to integrate into the genome of host cells permanently as a DNA copy thereby creating a persistent proviral reservoir from which it can potentially reactivate. This latent HIV-1 reservoir is located in tissues throughout the body, including bone marrow, lymphoid tissue, the genital tract, and the CNS [39]. Thus, HIV-1 remains as a chronic, life-long infection that is invisible to the immune system and not sensitive to anti-HIV drugs, which means there is the risk of viral reactivation, even after effective cART. Therefore, there is a need for HIV-1 genome eradication strategies that give a permanent or “sterile” cure of HIV-1/AIDS. The CRISPR/Cas9 system has been used to target the HIV-1 long terminal repeat (LTR) of integrated proviral DNA in the genome of cells in tissue culture [36, 40]. Hu et al. [36] were able to inactivate viral gene expression and replication in a variety of latently infected cell types, including microglial, promonocytic, and T cells. This is an important first step in a potential therapeutic advance to eliminating all the permanently integrated DNA proviral copies of HIV-1 in an infected individual and indicates the efficacy of CRISPR/Cas9 in cells to which it is delivered independent of cell type. In this study, specific targets in the HIV-1 proviral genome were identified within the HIV-1 LTR U3 region that flank the provirus and allow complete excision of the integrated provirus [36], as shown in Fig. 2.
Fig. 2.
Schematic of excision of integrated HIV-1 proviral DNA by (CRISPR)/CRISPR-associated 9 (Cas9). A guide RNA (gRNA) specific for the HIV-1 long terminal repeat (LTR) present at each end of the HIV-1 proviral genome is used together with Cas9. This results in cleavage at 2 sites that flank the HIV-1 genome and consequently the excision and subsequent degradation of the proviral DNA
Furthermore, not only can the CRISPR/Cas9 be used to remove provirus present in cells, but also the addition of HIV-1-specific gRNAs and Cas9 to uninfected cells prevents their infection by HIV-1; that is, CRISPR/Cas9 has the potential to be prophylactic, as well as therapeutic [36]. Important properties of the Cas9/gRNAs used in this study are that they caused neither genotoxicity nor off-target editing to the genome of host cells, which was revealed by the use of whole-genome sequence analysis and SURVEYOR assays. Recently, Zhu et al. [41] tested 10 sites in HIV-1 DNA for CRISPR/Cas9 in the JLat10.6 cell line, which is latently infected by HIV-1 and showed that each of the target sites in the HIV-1 proviral DNA was efficiently mutated and that HIV-1 gene expression and virus production were reduced 20-fold. Liao et al. [42] demonstrated that HIV-targeted CRISPR/Cas9 disrupted latently integrated HIV-1 genome and provided long-term adaptive defense against new viral infection, expression, and replication in engineered human-induced pluripotent stem cells, which could then be efficiently differentiated into HIV reservoir cell types and maintained their resistance to challenge with HIV-1.
Another approach to addressing the HIV-1 reservoir problem has been dubbed the “shock and kill” strategy, where latent virus is deliberately reactivated in order to expose it to the action of cART and the immune system [43]. Owing to their poor efficiency and tendencies towards nonspecific and toxic effects, the therapeutic efficacy of the agents currently used in this approach is disappointing. Zhang et al. [44] reported a novel catalytically deficient Cas9-synergistic activation mediator technology to allow more selective and potent reactivation of latent HIV-1. Multiple gRNAs were screened and HIV-1 regions were identified that permit induction of robust reactivation of HIV-1 provirus in latently infected cell lines, including the TZM-bI epithelial HeLa cell derivative, Jurkat T lymphocytes, and CHME5 microglia [44]. Reactivation induced cell death mediated by a toxic build up of viral proteins in the Jurkat and CHME5 cell lines, indicating that the deficient Cas9-synergistic activation mediator system has the potential to serve as a novel therapeutic tool for reversing HIV-1 latency, allowing permanent eradication of latent HIV-1 reservoirs [44]. Limsirichai et al. [45] also showed that engineered transcriptional activation systems based on CRISPR/Cas9 can activate HIV-1 gene expression cell line models of HIV-1 latency and further demonstrated that complementing the Cas9 activators with latency-reversing compounds enhanced HIV-1 transcription. In this study, modulation of the epigenome using CRISPR-based acetyltransferases also promoted HIV-1 gene activation [45].
Saayman et al. [46] used a similar approach and reported activating gRNAs that transcriptionally modulated the HIV-1 latent provirus genome across multiple different in vitro latency cell models. They detected consistent and effective activation of latent virus by this method using gRNAs, whereas latency reversal agents produced variable activation responses. By transcriptomic analysis, it was shown that the transcriptional activation was highly specific, while the well-characterized chemical activator tumor necrosis factor-α induced widespread cellular gene dysregulation [46].
In addition to targeting the HIV-1 genome, an alternative approach is to employ CRISPR/Cas9 to target cellular genes that encode proteins that are required for HIV-1 infection such as C-C chemokine receptor type (CCR) 5, which is a co-receptor for HIV-1 entry [47, 48]. Wang et al. [49] transduced HIV-1 susceptible human CD4+ cells with lentiviral vectors expressing Cas9 and CCR5-specific gRNAs and showed CCR5 gene disruption that became resistant to HIV-1 infection and has no mutations at potential off-target sites, for example CCR2. Hou et al. [50] showed that human C-X-C chemokine receptor type 4 gene disruption by CRISPR/Cas9 led to HIV-1 resistance in human primary CD4(+) T cells and that this was highly specific with negligible off-target effects. Kang et al. [51] reported successful targeting of CCR5 by Cas9/gRNA in green fluorescent protein-marked human induced pluripotent stem cells, resulting in resistance to CCR5-tropic virus challenge.
Thus, the CRISPR/Cas9 system has shown the clear potential to be developed to provide a specific and effective approach to combating HIV-1/AIDS, even when virus is present in a latent state. This approach has been employed both prophylactically and therapeutically in a number of cell types but has not yet been applied to cell types from the CNS, which comprise a significant viral reservoir in individuals infected with HIV-1. An important future goal is to test the feasibility of this approach in a clinical setting.
JCV
JCV is the etiological agent of PML, a severe disorder involving opportunistic infection of the CNS by JCV and usually associated with malfunctions of the immune system [7]. Similar to HIV-1, JCV can establish an asymptomatic persistent infection within the CNS from which it can reactivate to cause PML [8]. The public health significance of PML has increased in recent years with the growth in size of at-risk populations, especially HIV-1/AIDS patients and those receiving new immmunomodulatory therapies for diseases such as multiple sclerosis and rheumatoid arthritis. Although JCV was isolated > 40 years ago [52], there is still no effective therapy for PML [6]. JCV is a has a small (~5.1 Kb), circular DNA genome comprised of 2 coding regions for the early and late proteins, and a noncoding control region with promoter/enhancer elements and the origin of viral DNA replication [53]. The early protein, large T-antigen (T-Ag), accumulates during the early phase, initiates viral DNA replication and is essential for the viral life cycle. A number of treatment options have been applied to PML, including possible viral entry inhibitors and small molecule inhibitors of viral replication, largely without success [6]. Clearly, new therapeutic options are urgently needed for PML.
We have reported the use of CRISPR/Cas9 to eradicate the genome of JCV by targeting regions within the coding region of T-Ag by using different T-Ag gRNAs [11]. Our reasoning was that T-Ag would provide a key target because it is essential for viral DNA replication and, by binding proteins such as p53 and pRb, advancing the cell cycle into S-phase, where the cellular factors required for viral DNA synthesis become available [54, 55]. This latter function means that T-Ag causes the transformation of cells in culture and can promote tumorigenesis in experimental animals [54]. This offered us the possibility of performing functional assays for T-Ag and its disruption by the CRISPR/Cas9 system by in vitro and in vivo experiments. We found that co-expression of Cas9 and JCV T-Ag-specific gRNA in glial cells generated T-Ag deletions and the knockdown of the T-Ag protein expression level [11]. This resulted in loss of T-Ag function in transient transfection assays and JCV infection was inhibited. Derivatives of the T-Ag-transformed HJC-2 cell line, which express doxycycline-inducible Cas9, showed loss of expression of T-Ag upon transduction with a lentiviral vector expressing a JCV T-Ag-specific gRNA and addition of doxycycline to the culture. This correlated with reduced clonogenicity in colony formation assays indicative of the loss of T-Ag function [11]. Off-target events occurring in the most closely related cellular genes to T-Ag in the HJC-2 cells expressing Cas9 and gRNAs were not detected by SURVEYOR assay. Thus, CRISPR/Cas9 is a promising tool with which to eradicate both actively replicating and persistent JCV infection from the human CNS and has the potential to be developed as a therapeutic agent for PML.
HSV
As noted above, HSV encephalitis can be caused by HSV-1 and HSV-2 [13, 14]. As these are DNA viruses, they are, in principle, amenable to targeting by CRISPR/Cas9; indeed, the identity and function of several HSV genes that are essential for the HSV life cycle have been defined [56–58]. On the one hand, to our knowledge, there are no published studies of the use of HSV-specific CRISPR/Cas9 against HSV1 or HSV2. On the other hand, there is one published study using CRISPR/Cas9-directed inactivation against cellular proteins that are necessary for the HSV life cycle. Turner et al. [59] employed CRISPR/Cas9 against the proteins Torsin A and Torsin B, which are involved in nuclear egress, and their stimulatory cofactors lamina associated polypeptide 1 (LAP1) or luminal domain like LAP1 [60, 61]. CRISPR/Cas9 TorsinA/TorsinB double knockout reduced viral production only marginally, but knockout of for luminal domain like LAP1 reduced HSV-1 production by > 1 order of magnitude, while knockout of LAP1 had no effect on viral production [59].
Other Viruses
Other viruses that are capable of causing neurological diseases are amenable to treatment with CRISPR/Cas9 if their genome is DNA, for example, varicella zoster virus or CMV, or they are a retrovirus and have a DNA intermediate in their life cycle. RNA viruses are not amenable to treatment with CRISPR/Cas9 unless a cellular DNA target is available that is necessary for their life cycle, that is, retroviruses. As well as HIV-1, human T-lymphotropic virus-1 (HTLV-1) is another retrovirus that causes human neurological infections. HTLV-1 is the etiological agent of adult T-cell leukemia and can lead to neurological pathologies such as HTLV-1-associated-myelopathy/tropical spastic paraparesis [62]. CRISPR/Cas9 directed against the HTLV-1 DNA provirus may be a potential feasible approach for treating HTLV-1 neurological infections. As far as we are aware, no studies have attempted application of CRISPR/Cas9 for HTLV-1. However, Tanaka et al. [63] designed a ZFN that specifically recognized the conserved region of the HTLV-1 LTR and introduced it into HTLV-1-positive human T-cell lines. The ZFN disrupted the promoter function of the LTR, specifically killed HTLV-1-infected cells, and was effective in an in vivo model of adult T-cell leukemia indicating that gene editing approaches have the potential to form the basis for eradication of HTLV-1 infections.
Delivery of CRISPR/Cas9
The greatest challenge for utilizing the CRISPR/Cas9 is obtaining efficient delivery to the cells of the nervous system. There are various available means of delivery and these include viral transduction with adenovirus, adeno-associated virus (AAV) or lentiviruses [64–66], as well as physical methods [67]. Adenovirus vectors are useful but restricted in application because they are immunogenic [64]. However, lentiviral vectors, which are frequently derived from HIV-1, are capable of effecting permanent genetic alterations to their DNA targets but are limited by off-target events if the presence of the transduced nuclease is not transient. Therefore, integrase-defective lentiviruses or self-inactivating replication-incompetent lentiviruses are preferable because they give transient expression and have the ability to transduce both dividing and nondividing cells [65, 66]. The feasibility of lentiviral CRISPR/Cas9 delivery vectors is illustrated by many studies, including the eradication of latent infection by HIV-1 and JCV [11, 12]. AAV lacks an integration machinery and so, unlike lentiviruses, does not integrate and remains mostly in an episomal state. Further advantages of recombinant AAV vectors are low pathogenicity and low immunogenicity, and the capacity to transduce both dividing and nondividing cells without integrating. However, AAV is limited by the small size of the transgene that can be accommodated. There are a number of approaches to this problem, including delivering Cas9 and gRNAs in separate AAV vectors [68], splitting the Cas9 enzyme into halves that are delivered separately [69], or using a split-Cas9 with split-inteins and intein-mediated trans-splicing reconstitution of the full-length Cas9 protein [70]. Ran et al. [71] reported that Cas9 from Staphylococcus aureus (SaCas9) is able to edit with similar efficiency to Cas9 from Streptococcus pyogenes but is > 1 kb shorter.
The same general considerations outlined above apply to the suitability of viral vectors for gene delivery specifically to the CNS. Because the potential benefits of gene therapy for neurological diseases such as Alzheimer's, Parkinson's, and amyotrophic lateral sclerosis are enormous, significant effort has been made in developing vectors capable of delivering therapeutic genes to the CNS. In most applications, AAV is presently the most appropriate vector for the delivery of therapeutic genes to the CNS [72].
Conclusions and Future Directions
The CRISPR/Cas9 is a new and powerful tool for specific, straightforward, and facile genetic manipulation of any DNA target and is applicable to the eradication of DNA viruses, including those that cause neurological infections. As noted in the previous section, the major challenge to adopting CRISPR/Cas9 for the therapy of neurological infections is achieving efficient delivery to the cells of the nervous system. Although CRISPR/Cas9 has only been available for use in eukaryotic cells for about 3 years, it has already shown huge potential for the development of therapeutic applications to human diseases. Surely this is only the beginning and the rapid pace of advancement in this field will continue in the future.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 668 kb)
Acknowledgments
We thank past and present members of the Department of Neuroscience for their continued support and insightful discussions. We also acknowledge the intellectual contributions of the Temple University School of Medicine Comprehensive NeuroAIDS Center (Basic Science Cores I and II). We are grateful to Cynthia Papaleo for editorial assistance. This work was supported by grants R01NS087971 (KK and WH), R01AI077460 (MKW) P30MH092177 and P01DA037830 (KK) awarded by the National Institutes of Health.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
References
- 1.Johnson RT. Neurovirology: evolution of a new discipline. J Neurovirol. 1995;1:2–4. doi: 10.3109/13550289509111005. [DOI] [PubMed] [Google Scholar]
- 2.Koyuncu OO, Hogue IB, Enquist LW. Virus infections in the nervous system. Cell Host Microbe. 2013;13:379–393. doi: 10.1016/j.chom.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Del Valle L, Piña-Oviedo S. HIV disorders of the brain: pathology and pathogenesis. Front Biosci. 2006;11:718–732. doi: 10.2741/1830. [DOI] [PubMed] [Google Scholar]
- 4.Kovalevich J, Langford D. Neuronal toxicity in HIV CNS disease. Future Virol. 2012;7:687–698. doi: 10.2217/fvl.12.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger JR. The clinical features of PML. Cleve Clin J Med. 2011;78(Suppl 2):S8–S12. doi: 10.3949/ccjm.78.s2.03. [DOI] [PubMed] [Google Scholar]
- 6.Tavazzi E, White MK, Khalili K. Progressive multifocal leukoencephalopathy: clinical and molecular aspects. Rev Med Virol. 2012;22:8–32. doi: 10.1002/rmv.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wollebo HS, White MK, Gordon J, Berger JR, Khalili K. Persistence and pathogenesis of the neurotropic polyomavirus JC. Ann Neurol. 2015;77:560–570. doi: 10.1002/ana.24371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White MK, Khalili K. Pathogenesis of progressive multifocal leukoencephalopathy—revisited. J Infect Dis. 2011;203:578–586. doi: 10.1093/infdis/jiq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anthony IC, Bell JE. The neuropathology of HIV/AIDS. Int Rev Psychiatry. 2008;20:15–24. doi: 10.1080/09540260701862037. [DOI] [PubMed] [Google Scholar]
- 10.Rappaport J, Volsky DJ. Role of the macrophage in HIV-associated neurocognitive disorders and other comorbidities in patients on effective antiretroviral treatment. J Neurovirol. 2015;21:235–241. doi: 10.1007/s13365-015-0346-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wollebo HS, Bellizzi A, Kaminski R, Hu W, White MK, Khalili K. CRISPR/Cas9 System as an agent for eliminating polyomavirus JC infection. PLoS One. 2015;10 doi: 10.1371/journal.pone.0136046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khalili K, Kaminski R, Gordon J, Cosentino L, Hu W. Genome editing strategies: potential tools for eradicating HIV-1/AIDS. J Neurovirol. 2015;21:310–321. doi: 10.1007/s13365-014-0308-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pires de Mello CP, Bloom DC, Paixão IC. Herpes simplex virus type-1: replication, latency, reactivation and its antiviral targets. Antivir Ther 2016 Jan 4 [Epub ahead of print]. [DOI] [PubMed]
- 14.Berger A, Shahar T, Margalit N. Post-craniotomy herpes simplex type 2 encephalitis: case report and literature review. World Neurosurg 2015;pii:S1878-8750(15)01724-6. [DOI] [PubMed]
- 15.Morgello S, Cho ES, Nielsen S, Devinsky O, Petito CK. Cytomegalovirus encephalitis in patients with acquired immunodeficiency syndrome: an autopsy study of 30 cases and a review of the literature. Hum Pathol. 1987;18:289–297. doi: 10.1016/S0046-8177(87)80012-6. [DOI] [PubMed] [Google Scholar]
- 16.Mamidi A, DeSimone JA, Pomerantz RJ. Central nervous system infections in individuals with HIV-1 infection. J Neurovirol. 2002;8:158–167. doi: 10.1080/13550280290049723. [DOI] [PubMed] [Google Scholar]
- 17.Huang YC, Huang SL, Chen SP, et al. Adenovirus infection associated with central nervous system dysfunction in children. J Clin Virol. 2013;57:300–304. doi: 10.1016/j.jcv.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 18.Grahn A, Studahl M. Varicella-zoster virus infections of the central nervous system—prognosis, diagnostics and treatment. J Infect. 2015;71:281–293. doi: 10.1016/j.jinf.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 19.White MK, Khalili K. CRISPR/Cas9 and cancer targets: future possibilities and present challenges. Oncotarget 2016 Jan 31 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 20.White MK, Hu W, Khalili K. The CRISPR/Cas9 genome editing methodology as a weapon against human viruses. Discov Med. 2015;19:255–262. [PMC free article] [PubMed] [Google Scholar]
- 21.Kim YG, Li L, Chandrasegaran S. Insertion and deletion mutants of FokI restriction endonuclease. J Biol Chem. 1994;269:31978–31982. [PubMed] [Google Scholar]
- 22.Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF. Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat Methods. 2012;9:805–807. doi: 10.1038/nmeth.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeder ML, Gersbach CA. Genome editing technologies for gene and cell therapy. Mol Ther. 2016;24:430–446. doi: 10.1038/mt.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright DA, Li T, Yang B, Spalding MH. TALEN-mediated genome editing: prospects and perspectives. Biochem J. 2014;462:15–24. doi: 10.1042/BJ20140295. [DOI] [PubMed] [Google Scholar]
- 25.Ousterout DG, Gersbach CA. The development of TALE nucleases for biotechnology. Methods Mol Biol. 2016;1338:27–42. doi: 10.1007/978-1-4939-2932-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014;346:1258096. [DOI] [PubMed]
- 27.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolukbasi MF, Gupta A, Wolfe SA. Creating and evaluating accurate CRISPR-Cas9 scalpels for genomic surgery. Nat Methods. 2015;13:41–50. doi: 10.1038/nmeth.3684. [DOI] [PubMed] [Google Scholar]
- 29.Ran FA, Hsu PD, Lin CY, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 32.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–997. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 33.Canver MC, Bauer DE, Dass A, et al. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J Biol Chem. 2014;289:21312–21324. doi: 10.1074/jbc.M114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shmakov S, Abudayyeh OO, Makarova KS, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu W, Kaminski R, Yang F, et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci USA. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fois AF, Brew BJ. The potential of the CNS as a reservoir for HIV-1 infection: Implications for HIV eradication. Curr HIV/AIDS Rep. 2015;12:299–303. doi: 10.1007/s11904-015-0257-9. [DOI] [PubMed] [Google Scholar]
- 40.Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu W, Lei R, Le Duff Y, et al. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology. 2015;12:22. doi: 10.1186/s12977-015-0150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao HK, Gu Y, Diaz A, Marlett J, et al. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun. 2015;6:6413. doi: 10.1038/ncomms7413. [DOI] [PubMed] [Google Scholar]
- 43.Archin NM, Liberty AL, Kashuba AD, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Yin C, Zhang T, Li F, et al. CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci Rep. 2015;5:16277. doi: 10.1038/srep16277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Limsirichai P, Gaj T, Schaffer DV. CRISPR-mediated activation of latent HIV-1 expression. Mol Ther. 2016;24:499–507. doi: 10.1038/mt.2015.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saayman SM, Lazar DC, Scott TA, et al. Potent and targeted activation of latent HIV-1 using the CRISPR/dCas9 activator complex. Mol Ther. 2016;24:488–498. doi: 10.1038/mt.2015.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manjunath N, Yi G, Dang Y, Shankar P. Newer gene editing technologies toward HIV gene therapy. Viruses. 2013;5:2748–2766. doi: 10.3390/v5112748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stone D, Kiem HP, Jerome KR. Targeted gene disruption to cure HIV. Curr Opin HIV AIDS. 2013;8:217–223. doi: 10.1097/COH.0b013e32835f736c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang W, Ye C, Liu J, Zhang D, Kimata JT, Zhou P. CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hou P, Chen S, Wang S, et al. Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Sci Rep. 2015;5:15577. doi: 10.1038/srep15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang H, Minder P, Park MA, Mesquitta WT, Torbett BE, Slukvin II. CCR5 disruption in induced pluripotent stem cells using CRISPR/Cas9 provides selective resistance of immune cells to CCR5-tropic HIV-1 virus. Mol Ther Nucleic Acids. 2015;4 doi: 10.1038/mtna.2015.42. [DOI] [PubMed] [Google Scholar]
- 52.Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet. 1971;1:1257–1260. doi: 10.1016/S0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- 53.Frisque RJ, Bream GL, Cannella MT. Human polyomavirus JC virus genome. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White MK, Khalili K. Polyomaviruses and human cancer: molecular mechanisms underlying patterns of tumorigenesis. Virology. 2004;324:1–16. doi: 10.1016/j.virol.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 55.White MK, Khalili K. Interaction of retinoblastoma protein family members with large T-antigen of primate polyomaviruses. Oncogene. 2006;25:5286–5293. doi: 10.1038/sj.onc.1209618. [DOI] [PubMed] [Google Scholar]
- 56.Reynolds AE, Ryckman BJ, Baines JD, Zhou Y, Liang L, Roller RJ. U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J Virol. 2001;75:8803–8817. doi: 10.1128/JVI.75.18.8803-8817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reynolds AE, Wills EG, Roller RJ, Ryckman BJ, Baines JD. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J Virol. 2002;76:8939–8952. doi: 10.1128/JVI.76.17.8939-8952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simpson-Holley M, Baines J, Roller R, Knipe DM. Herpes simplex virus 1 U(L)31 and U(L)34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J Virol. 2004;78:5591–5600. doi: 10.1128/JVI.78.11.5591-5600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turner EM, Brown RS, Laudermilch E, Tsai PL, Schlieker C. The Torsin activator LULL1 Is required for efficient growth of Herpes Simplex Virus 1. J Virol. 2015;89:8444–8452. doi: 10.1128/JVI.01143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao C, Brown RS, Chase AR, Eisele MR, Schlieker C. Regulation of Torsin ATPases by LAP1 and LULL1. Proc Natl Acad Sci U S A. 2013;110:E1545–E1554. doi: 10.1073/pnas.1300676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rose AE, Zhao C, Turner EM, Steyer AM, Schlieker C. Arresting a Torsin ATPase reshapes the endoplasmic reticulum. J Biol Chem. 2014;289:552–564. doi: 10.1074/jbc.M113.515791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaplan JE, Osame M, Kubota H, et al. The risk of development of HTLV-I-associated myelopathy/tropical spastic paraparesis among persons infected with HTLV-I. J Acquir Immune Defic Syndr. 1990;3:1096–1101. [PubMed] [Google Scholar]
- 63.Tanaka A, Takeda S, Kariya R, et al. A novel therapeutic molecule against HTLV-1 infection targeting provirus. Leukemia. 2013;27:1621–1627. doi: 10.1038/leu.2013.46. [DOI] [PubMed] [Google Scholar]
- 64.Wang D, Mou H, Li S, et al. Adenovirus-mediated somatic genome editing of Pten by CRISPR/Cas9 in mouse liver in spite of Cas9-specific immune responses. Hum Gene Ther. 2015;26:432–442. doi: 10.1089/hum.2015.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen X, Gonçalves MA. Engineered viruses as genome editing devices. Mol Ther. 2016;24:447–457. doi: 10.1038/mt.2015.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gori JL, Hsu PD, Maeder ML, Shen S, Welstead GG, Bumcrot D. Delivery and specificity of CRISPR-Cas9 genome editing technologies for human gene therapy. Hum Gene Ther. 2015;26:443–451. doi: 10.1089/hum.2015.074. [DOI] [PubMed] [Google Scholar]
- 67.67 Li L, He ZY, Wei XW, Gao GP, Wei YQ. Challenges in CRISPR/CAS9 Delivery: Potential Roles of Nonviral Vectors. Hum Gene Ther 2015;26:452-462. [DOI] [PubMed]
- 68.Swiech L, Heidenreich M, Banerjee A, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wright AV, Sternberg SH, Taylor DW, et al. Rational design of a split-Cas9 enzyme complex. Proc Natl Acad Sci U S A. 2015;112:2984–2989. doi: 10.1073/pnas.1501698112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Truong DJ, Kühner K, Kühn R, et al. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res. 2015;43:6450–6458. doi: 10.1093/nar/gkv601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ran FA, Cong L, Yan WX, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lentz TB, Gray SJ, Samulski RJ. Viral vectors for gene delivery to the central nervous system. Neurobiol Dis. 2012;48:179–188. doi: 10.1016/j.nbd.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 668 kb)