Abstract
Latex exploitation-caused latex flow is effective in enhancing latex regeneration in laticifer cells of rubber tree. It should be suitable for screening appropriate reference gene for analysis of the expression of latex regeneration-related genes by quantitative real-time PCR (qRT-PCR). In the present study, the expression stability of 23 candidate reference genes was evaluated on the basis of latex flow by using geNorm and NormFinder algorithms. Ubiquitin-protein ligase 2a (UBC2a) and ubiquitin-protein ligase 2b (UBC2b) were the two most stable genes among the selected candidate references in rubber tree clones with differential duration of latex flow. The two genes were also high-ranked in previous reference gene screening across different tissues and experimental conditions. By contrast, the transcripts of latex regeneration-related genes fluctuated significantly during latex flow. The results suggest that screening reference gene during latex flow should be an efficient and effective clue for selection of reference genes in qRT-PCR.
Keywords: Hevea brasiliensis Muell. Arg., latex flow, reference gene, qRT-PCR, geNorm software, NormFinder software
Introduction
Gene expression analysis provides the potential to explore biological processes. Northern blotting, transcriptome sequencing and qRT-PCR are universal methods that provide reliable and accurate gene expression analysis (Kubista et al., 2006; Josefsen and Nielsen, 2011; Lang et al., 2014; Le et al., 2014). Owing to its technical ease, cost-effectiveness, high specificity and accuracy, qRT-PCR has become the preferred method for detecting and measuring gene expression across different samples (Garson et al., 2005; Kubista et al., 2006). The accuracy of qRT-PCR is influenced by several factors such as the RNA extraction yield, reverse transcription and the efficiency of amplification (Huggett et al., 2005). It is necessary to normalize the expression data of the target genes with gene expression data of reference genes (Vandesompele et al., 2002; Huggett et al., 2005). A reference gene is defined as having a stable expression pattern across different tissues and experimental conditions. Generally, several housekeeping genes such as actin (ACT), eukaryotic elongation factor 1-alpha (eEF-1a), 18S rRNA, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and polyubiquitin (UBQ), have been widely used as reference genes for qRT-PCR in the past decade (Bustin, 2000; Goidin et al., 2001; Kim et al., 2003; Jain et al., 2006). However, accumulating data have shown that the expression levels of these genes are variable across different developmental stages and experimental conditions (Garson et al., 2005). The use of non-validated reference genes could lead to inaccurate results and consequently, erroneous conclusions (Tricarico et al., 2002; Huggett et al., 2005; Remans et al., 2014). The 11 golden rules and Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines are recommended for qRT-PCR performance in the last decade (Udvardi et al., 2008; Bustin et al., 2009, 2013).
Several statistical algorithms, such as geNorm and NormFinder, have been developed for the selection of reference genes for qRT-PCR analysis (Vandesompele et al., 2002; Andersen et al., 2004). The geNorm is a Visual Basic application tool which calculates the expression stability value (M) for all reference genes. The gene with a lower M value suggests its expression is more stable. Additionally, the pairwise variation (Vn/Vn+1) of references can also be estimated by using geNorm. It is recommended that no additional genes are required for normalization when the value of Vn/Vn+1 is below 0.15 (Vandesompele et al., 2002). As another statistical algorithm, the NormFinder ranks the stability of candidate reference genes and calculates not only the candidate reference gene variation but also the variation between subgroup samples in a sample set (Andersen et al., 2004). The lowest variety value indicates the most stable. Application of both algorithms contributes to identify the best stable reference genes for different experimental samples.
The rubber tree (Hevea brasiliensis) is the main source of natural rubber due to the good yield and excellent physical properties of its rubber products (Eschbach and Lacrotte, 1989). Natural rubber biosynthesis takes place in latex, the cytoplasm of highly specialized laticifer cells (Chow et al., 2012; Wang et al., 2013, 2015). In natural rubber production, latex is exploited by successive tappings. Duration of latex flow after tapping is one of crucial factors that determines rubber yield of rubber tree, and easily influenced by multiple factors such as genotypes, age of trees, seasons, etc. (Sethuraj, 1992; Raj et al., 2005; Chao et al., 2015a). Tapping causes rubber tree to be wounded and loose latex (Tang et al., 2010). The loss of latex in turn enhances latex regeneration in laticifer cells (Jacob et al., 1989). Previous studies show that tapping activates the expression of mass of latex metabolism-related genes at transcriptional level, which mainly relay on the polymerase II transcriptional complex that are responsible for eukaryotic mRNA transcription (Hahn, 2004; Hager et al., 2009; Chao et al., 2015a). Such tapping effects should be mainly ascribed to latex flow which results in latex loss from laticifer cells. To screen reference genes on the basis of latex flow may be a more efficient clue than the previously reported methods on the basis of different treatments (Li et al., 2011; Pirrello et al., 2014; Long et al., 2015). In this study, 23 candidate reference genes were evaluated for their expression stability across 18 samples obtained from three stages of latex flow of two rubber tree genotypes with differential duration of latex flow by using geNorm and NormFinder algorithms.
Materials and Methods
Plant Materials
In this study, we used 11-year-old regularly tapped rubber tree clones CATAS7-33-97 and CATAS8-79 with the same circumference. The trees were grown at the Experimental Station of the Rubber Research Institute of the Chinese Academy of Tropical Agricultural Sciences in Danzhou city, Hainan province. After tapping, the latex samples were respectively collected at 1, 30, and 60 min for CATAS7-33-97, and at 1, 80, and 150 min for CATAS8-79, which represented the early, middle, and late stage of latex flow, respectively. Each batch of latex samples were individual collected from three trees for each clone, and stored at -80°C until total RNA extraction.
RNA Isolation and cDNA Synthesis
Total RNA was isolated by using the RNAprep Pure Plant Kit (Tiangen, China) and genomic DNA was eliminated according to the manufacturer’s instructions. The concentration and quality of RNA were examined using a NanoDrop 2000 (Thermo Scientific Inc., USA). The RNA integrity of the samples was checked using 1.5% agarose gel electrophoresis. Synthesis of cDNA was performed using a RevertAid TMFirst Strand cDNA Synthesis Kit (Fermentas, Canada) following the manufacturer’s protocol.
qRT-PCR Analysis
A total of 23 housekeeping genes (18S rRNA, ACT, ACT7a, ACT7b, ADF, ADF4, CYP2, eIF2, eIF3, eIF1Aa, eIF1Ab, FP, PTP, RH2a, RH2b, RH8, ROC3, UBC1, UBC2a, UBC2b, UBC3, UBC4, and YLS8), which were evaluated in previous studies (Li et al., 2011; Chao et al., 2015b; Long et al., 2015), were chosen as candidate reference genes in the present study (Supplementary Table S1). Reactions were performed using a SYBR PrimeScript RT-PCR Kit (TaKaRa Biotechnology, Japan) on the CFX96 System (Bio-Rad Laboratories Inc., USA) as follow: 95°C for 3 min followed by 45 cycles at 95°C for 15 s, 56°C for 60 s and 72°C for 30 s, and a melt curve from 55 to 95°C increasing by 0.5°C every 30 s. Each real-time PCR reaction was performed in triplicate. The Bio-Rad CFX96 Manager 3.0 software was used for visualizing and analyzing the data, including the cycle threshold (Ct) values, amplification efficiencies (E) and melting curve of reference genes (Supplementary Table S1; Supplementary Figure S2). The primer efficiency of each gene was evaluated using a standard curve generated by qRT-PCR using a fivefold dilution series of cDNAs (Supplementary Table S1).
Data Analysis
Based on the Ct values for the 23 candidate reference genes, a boxplot was drawn using GraphPad Prism 5 software1 to show the expression variation of each gene. The expression stability of the reference genes was analyzed using the geNorm (version 3.52) and NormFinder (version 0.9533) software packages. To evaluate the gene expression stability, the Ct values of the candidate reference genes were converted into relative quantities using the formula E-ΔCt (ΔCt = Ct value of each sample-the minimum Ct value) (Li et al., 2016), and imported into the software according to the corresponding manuals of the Norm-Finder and geNorm algorithms (Vandesompele et al., 2002; Andersen et al., 2004).
The Expression of Latex Regeneration-Related Genes during Latex Flow
The HbHMGR1 (Genebank accession number: AB294692) and small rubber particle protein (HbSRPP, Genebank accession number: AEH05971) were rubber biosynthesis-related genes (Sando et al., 2008; Guo et al., 2014). HbRpb11 (Genebank accession number: KU301750) and HbTFIIB (Genebank accession number: KU301749) were the genes homologous to RNA polymerase II transcriptional complex members. They were selected to represent latex regeneration-related genes. Specific primers were designed using Primer Premier 5 software4 (Supplementary Table S1) and the amplified efficiency was checked (Supplementary Figure S1). The UBC2b, 18S rRNA and FP were respectively used as reference genes to normalize the transcripts of the target genes. The relative expression level of all of the target genes was evaluated using the 2-ΔΔCt method (Cheng et al., 2015; Hu et al., 2016).
Statistical Analysis
Statistical analysis was performed with SPSS Statistics 17.05 by analysis of variance (ANOVA) based on Duncan’s test (multiple group comparisons) or a t-test (two group comparisons). For Duncan’s test, the capital letter or lowercase represented p < 0.01 or p < 0.05, and the same letter indicated no significant difference among groups. For t-test, ∗∗∗represented p < 0.001.
Results
Determining the Time Intervals for Latex Collection Based on the Different Durations of Latex Flow in the CATAS8-79 and CATAS7-33-97
Latex is usually obtained by tapping the trunk bark of rubber tree every 2-day interval (Figure 1A). The rubber tree clones CATAS8-79 and CATAS7-33-97 exhibited a huge difference in the duration of latex flow and latex production (Figures 1B,C). For CATAS8-79, the latex flow lasted for more than 160 min after tapping, which was significantly longer than that of CATAS7-33-97 (∼70 min) (Figure 1B). As a result, the latex production of CATAS8-79 was notably higher than that of CATAS7-33-97 (Figure 1C). Based on the differential duration of latex flow between the two clones, latex samples were collected at 1, 30, and 60 min after tapping for CATAS7-33-97 and at 1, 80, and 150 min after tapping for CATAS8-79, which represented the early, middle and late stages of latex flow, respectively.
FIGURE 1.
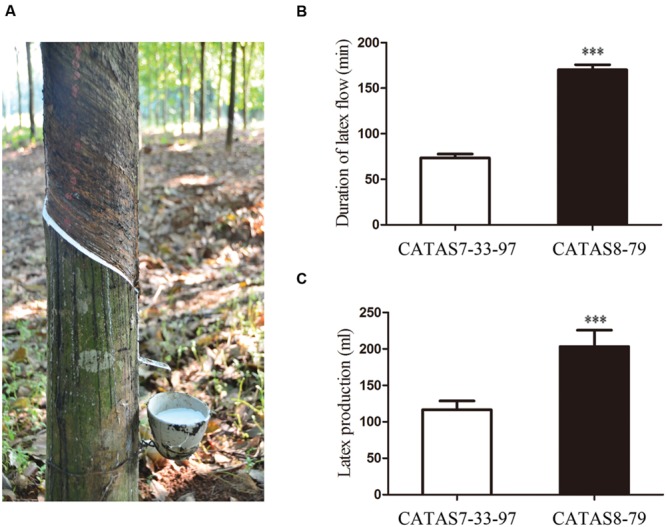
Latex flow of rubber tree (A) and the difference in latex characters (B,C) between CATAS7-33-97 and CATAS8-79. (A) Latex flow of rubber tree after tapping. (B) The difference in duration of latex flow between CATAS7-33-97 and CATAS8-79. (C) The difference in latex production between CATAS7-33-97 and CATAS8-79. Significant difference was indicated by the asterisks above the bars (∗∗∗p < 0.001).
Experiment Set Classification and Transcript Abundance Evaluation
The qRT-PCR data from 18 samples were divided into three experiment sets to assess the influence of latex flow on the expression of the candidate reference genes. The first set consisted of nine samples from CATAS7-33-97. The second set was comprised of nine samples from CATAS8-79. The third set included both the first and second sample sets.
The Ct values for 23 candidate reference genes were determined across all of the experimental samples using qRT-PCR (Supplementary Table S1; Supplementary Figure S1). The boxplot analysis was performed using GraphPad Prism 5 software (Figure 2). The results indicated that the selected candidate reference genes displayed a wide range of Ct values, ranging from 9 to 26. The Ct values of the majority of the candidate reference genes ranged from 19 to 22. The 18S rRNA gene was the most abundant reference gene with the lowest mean Ct value (9.85), whereas FP was the least abundant reference gene with the highest mean Ct value (25.94) (Figure 2).
FIGURE 2.
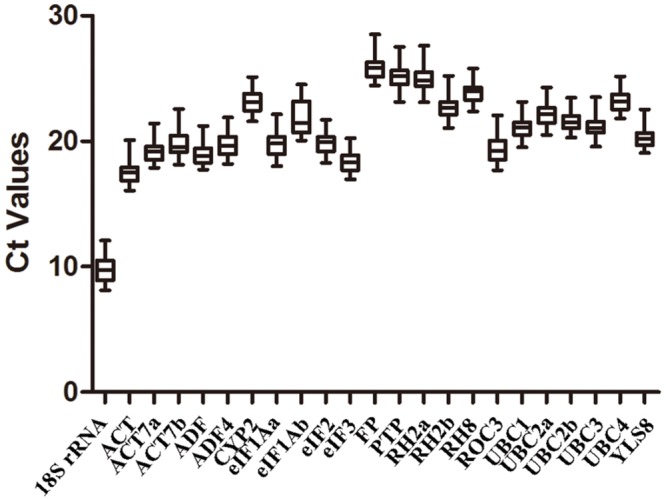
Average cycle threshold (Ct) values for the 23 candidate reference genes upon duration of latex flow.
Evaluation of Expression Stability Using geNorm
geNorm was used to assess the expression stability (M) of each reference gene based on the average pair-wise variation among all of the tested genes (Vandesompele et al., 2002). The lowest M value indicates the most stable expression, whereas a higher M value is indicative of less stable expression. All of the 23 reference genes had an M value below the geNorm threshold of 1.5 and were ranked according to their M values in three sample sets (Figures 3A–C). For CATAS7-33-97 sample set, UBC2a and eIF3 were the most stable genes with the M values of 0.13 (Figure 3A). The seven top-ranked genes were UBC2a/eIF3, UBC2b, RH8, UBC3, eIF1Ab and ROC3. For CATAS8-79 sample set, UBC2a and UBC4 were the most stable genes with the M values of 0.17 (Figure 3B). The seven top-ranked genes were UBC2a/UBC4, YLS8, ADF, ADF4, UBC1 and UBC2b. For total experiment set, RH8 and UBC2b were the most stable genes with the M values of 0.16 (Figure 3C). The seven top-ranked genes were RH8/UBC2b, UBC2a, UBC4, eIF3, UBC1 and ADF4.
FIGURE 3.
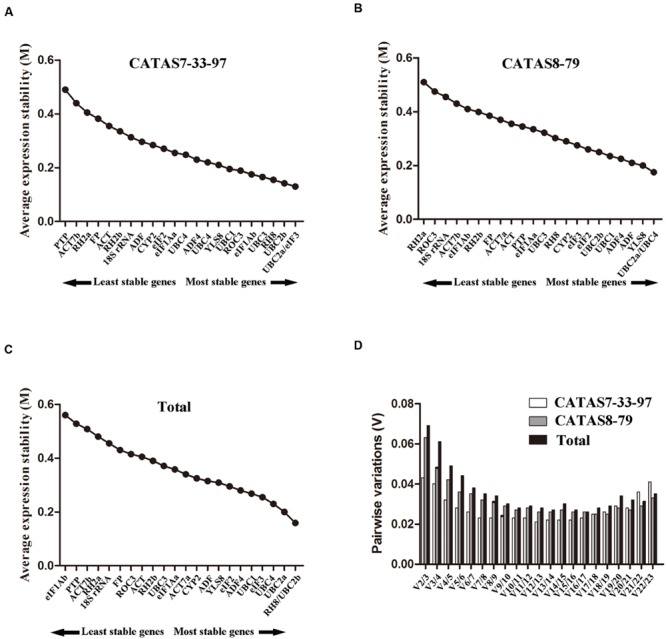
Average gene expression stability (M) and pairwise variation (V) analyses of the candidate reference genes in different sample sets generated by the geNorm software. The ranking of the gene expression stability performed in CATAS7-33-97 (A), CATAS8-79 (B) and total sample (C). The most stable genes are on the right, and the least stable genes are on the left. (D) Pairwise variation calculated to determine the minimum number of reference genes for accurate normalization.
To determine the optimal number of reference genes required for normalization, pairwise variation analysis (Vn/Vn + 1) was performed using the geNorm software (Figure 3D). The cut-off threshold value for Vn/n + 1 is 0.15 and no additional reference gene is required for normalization when the value below this value (Vandesompele et al., 2002). In the present study, the value of V2/3 was below 0.15 under all experimental sets (Figure 3D), which suggests that two reference genes would be sufficient for gene normalization in each experimental set.
Evaluation of Expression Stability Using NormFinder
NormFinder is an algorithm tool to identify optimally stable reference genes by determining the expression stability (Andersen et al., 2004). Here, we use NormFinder software to further confirm the results obtained using the geNorm program. The data are listed in Table 1. The seven top-ranked genes were UBC2b, RH8, eIF3, UBC3, UBC2a, eIFAa, and ACT7a in the CATAS7-33-97 experiment set (Table 1), six of which were also present in the top seven genes in the geNorm evaluation (Figure 3A). The seven top-ranked genes were UBC2a, UBC2b, UBC4, ADF4, YLS8, ADF, and RH8 in the CATAS8-79 experiment set (Table 1), six of which were also present in the top seven genes in the geNorm evaluation (Figure 3B). The UBC2a, UBC2b, and RH8 were common between the top seven genes in both experiment sets using both NormFinder and geNorm. Moreover, they were the top three genes in the combined experiment set using both NormFinder (Table 1) and geNorm (Figure 3C).
Table 1.
Expression stability of 23 candidate reference genes as calculated by NormFinder.
| Rank | CATAS7-33-97 |
CATAS8-79 |
Total |
|||
|---|---|---|---|---|---|---|
| Genes | Stability | Genes | Stability | Genes | Stability | |
| 1 | UBC2b | 0.032 | UBC2a | 0.059 | UBC2b | 0.091 |
| 2 | RH8 | 0.096 | UBC2b | 0.097 | UBC2a | 0.096 |
| 3 | eIf3 | 0.101 | UBC4 | 0.109 | RH8 | 0.156 |
| 4 | UBC3 | 0.107 | ADF4 | 0.136 | eIf3 | 0.160 |
| 5 | UBC2a | 0.115 | YLS8 | 0.136 | UBC1 | 0.169 |
| 6 | eIF1Aa | 0.127 | ADF | 0.155 | ADF4 | 0.171 |
| 7 | ACT7a | 0.133 | RH8 | 0.159 | UBC4 | 0.171 |
| 8 | eIF1Ab | 0.152 | UBC1 | 0.162 | ADF | 0.200 |
| 9 | ROC3 | 0.169 | PTP | 0.188 | ACT7a | 0.222 |
| 10 | UBC1 | 0.181 | eIf3 | 0.199 | eIF1Aa | 0.223 |
| 11 | YLS8 | 0.185 | CYP2 | 0.204 | CYP2 | 0.236 |
| 12 | ADF4 | 0.193 | eIF2 | 0.224 | eIF2 | 0.241 |
| 13 | UBC4 | 0.227 | ACT | 0.232 | YLS8 | 0.247 |
| 14 | ADF | 0.228 | eIF1Aa | 0.266 | ACT | 0.250 |
| 15 | RH2b | 0.236 | FP | 0.275 | RH2b | 0.260 |
| 16 | CYP2 | 0.253 | UBC3 | 0.276 | FP | 0.317 |
| 17 | eIF2 | 0.274 | RH2b | 0.280 | UBC3 | 0.317 |
| 18 | 18S rRNA | 0.280 | ACT7a | 0.296 | ROC3 | 0.334 |
| 19 | ACT | 0.281 | eIF1Ab | 0.339 | 18S rRNA | 0.366 |
| 20 | FP | 0.374 | ACT7b | 0.371 | RH2a | 0.446 |
| 21 | RH2a | 0.381 | 18S rRNA | 0.388 | PTP | 0.464 |
| 22 | ACT7b | 0.558 | ROC3 | 0.447 | ACT7b | 0.471 |
| 23 | PTP | 0.649 | RH2a | 0.517 | eIF1Ab | 0.560 |
Influence of Latex Flow on the Expression of Latex Regeneration-Related Genes
The HbHMGR1, HbSRPP1, HbTFIIB and HbRpb11 were selected to analyze the expression of latex regeneration-related gene at different stages of latex flow. The relative expression was respectively normalized against reference genes UBC2b, 18S rRNA and FP. The UBC2b was the most stable reference gene while the traditionally used 18S rRNA and the more stable FP reported by Li et al. (2011) were less stable in the present study. The transcript level of all the four genes were significantly fluctuated during latex flow when normalized by any of the three reference genes (Figure 4). The fluctuating pattern of each target gene was, however, different to some extent among the three normalizations (Figure 4). There was no difference in the transcript level of HbHMGR1 between “middle” and “late” stages of latex flow by using UBC2b normalization in either CATAS7-33-97 or CATAS8-79 (Figure 4A). But it showed significant difference (p < 0.01) by using 18S rRNA normalization in CATAS7-33-97 and difference (p < 0.05) by using FP normalization in CATAS8-79 (Figure 4A). The transcript level of HbSRPP between “middle” and “late” stages showed significant difference (p < 0.01) in CATAS7-33-97 while had no difference in CATAS8-79 by using UBC2b normalization (Figure 4B). It had no difference by using FP normalization while showed significant difference (p < 0.01) by using 18S rRNA in either CATAS7-33-97 or CATAS8-79 (Figure 4B). The relative transcript level of HbRpb11 had no difference between “middle” and “late” stages of latex flow in CATAS7-33-97 by using either UBC2b or FP normalization while significant difference (p < 0.01) by using 18S rRNA normalization (Figure 4C). It showed difference in CATAS8-79 for 18S rRNA normalization and significant difference for both UBC2b and FP normalization (Figure 4C) The UBC2b normalized expression pattern of HbTFIIB in either CATAS7-33-97 or CATAS8-79 was similar with the FP normalized but different from the 18S rRNA normalized expression pattern (Figure 4D).
FIGURE 4.
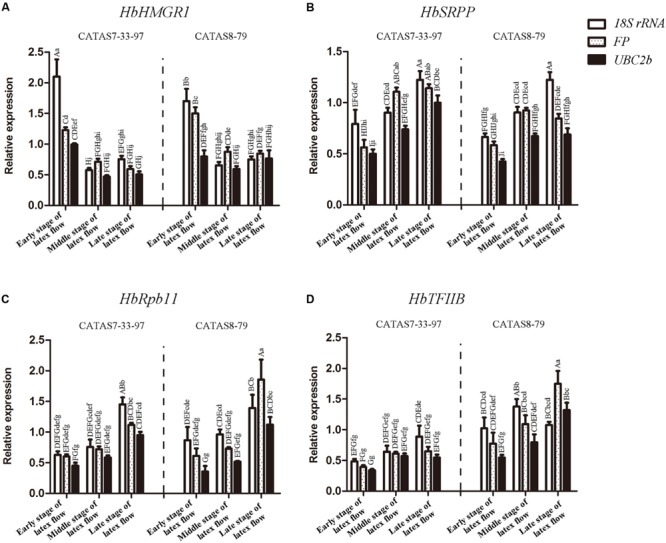
Relative expression of HbHMGR1(A), HbSRPP(B), HbRpb11(C) and HbTFIIB(D) by using 18S rRNA, FP or UBC2b for normalization. Error bars for qRT-PCR showed the standard deviation of three replicates. Different letters indicate statistically significant difference in relative expression of each gene among different tissues. The capital letter represents p < 0.01 while lowercase represents p < 0.05. Analyses were performed with SPSS Statistics 17.0. Bars indicate the standard deviation of three biological replicates in triplicate.
Discussion
Accurate interpretation of the qRT-PCR results mainly depends on the use of stable reference genes for data normalization to minimize non-biological variations among samples (Huggett et al., 2005). However, increasing evidence shows that the best reference gene significantly depends on the experimental conditions (Garson et al., 2005). Therefore, validating suitable reference genes is necessary to obtain accurate gene expression patterns under specific conditions using qRT-PCR (Ji et al., 2014; Wang et al., 2014; Delporte et al., 2015; Kong et al., 2015; Niu et al., 2015).
The rubber tree is one of the most economically important members of the genus Hevea because the milky latex exploited from the tree is the primary source of natural rubber. The latex exploitation is an effective factor that enhances latex regeneration (Jacob et al., 1989). After tapping, the loss of latex from latex flow results in significant changes in turgor pressure of laticifer cells, which may significantly reprogram the transcriptional regulation of latex regeneration-related genes in laticifer cells at the molecular level (Chao et al., 2015a). In the present study, we showed that not only the rubber biosynthesis-related genes such as HbHMGR1 and HbSRPP significantly fluctuated, but also the primary metabolism-related genes such as HbTFIIB and HbRpb11, the homologs of transcriptional complex members, were notably reprogrammed during latex flow (Figure 4). The results indicate that latex regeneration-related genes are strongly influenced in the process of latex flow. This factor is largely ignored when other factors such as tapping and hormone applications are considered in screening reference genes (Li et al., 2011; Pirrello et al., 2014; Long et al., 2015).
The expression stability of 22 candidate reference genes was evaluated based on tapping, genotypes, tissues, and plant hormones (Li et al., 2011). The authors suggested that the top five reference genes were UBC2a, UBC2b, UBC4, eIF3, and FP. Thereafter, 20 of the 22 candidate reference genes were evaluated using successive tapping and the top five reference genes were UBC2a, UBC2b, UBC4, UBC3, and ADF (Long et al., 2015). Three genes were identical between the top five genes. The RH2b, UBC2a, RH8 are the top three best references for abiotic and harvesting stress of rubber tree by screening 11 of the 22 candidate genes and simply evaluating based on the standard deviation and coefficient of variance of Ct value that generated by qRT-PCR (Pirrello et al., 2014). In the present study, the selected 23 candidate reference genes included the 22 candidate reference genes (Li et al., 2011) and an ACT gene (Chao et al., 2015b). The expression stability of all the 23 candidate reference genes was evaluated on the basis of latex flow using geNorm and NormFinder algorithms. The top five genes were UBC2a, UBC2b, UBC4, eIF3, and RH8. Four of the five genes are identical to the genes reported by Li et al. (2011) and three of the five genes are identical to those reported by Long et al. (2015). It is interesting that three UBC family members (UBC2a, UBC2b, and UBC4), are common among these experimental groups of rubber trees (Figure 5).
FIGURE 5.
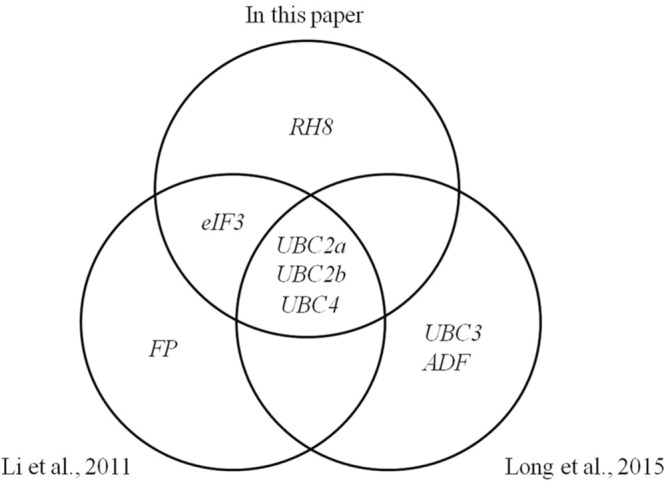
Comparing suitable reference genes screened here with previous studies on rubber tree. The top five best references of each experimental group were analyzed here.
UBC is a large gene family comprising dozens of members, which widely expands in eukaryotes (Sheng et al., 2012). In the present study, two UBCs, the UBC2a and UBC2b, had the best stability based on latex flow in rubber trees. It is interesting that UBC2a and UBC2b are also stable in laticifer cells and bark tissue of rubber trees. For example, UBC2b was also found to be highly stable across multiple experimental conditions, such as hormone treatments, genotypes and latex regeneration between two tapping intervals (Li et al., 2011; Long et al., 2015). UBC2a was the most stable gene in the bark tissues of rubber trees treated with coronatine (data unpublished). The homolog of UBC2a and UBC2b is also stable in many species such as Brachypodium distachyton (Chambers et al., 2012), Trifolium pretense (Mehdi Khanlou and Van Bockstaele, 2012), Lolium perenne (Huang et al., 2014), and Withania somnifera (Singh et al., 2015) under different experimental conditions. In plants, UBC2a and UBC2b encode ubiquitin-conjugating enzyme subunits that are necessary for the degradation of ubiquitinated proteins by the eukaryotic conserved 26S protein degradation pathway (Stone, 2014). Protein ubiquitination is mediated by the sequential action of E1 (ubiquitin-activating enzyme, UBA), E2 (ubiquitin-conjugating enzyme, UBC), and E3 (ubiquitin-ligating enzyme) (E et al., 2015). E2 UBC enzymes accept activated ubiquitin from E1 through a transthiolation reaction, and then transfer ubiquitin to either a substrate directly aided by E3 or a cysteine of ubiquitin protein ligase through a second transthiolation reaction that then transfers ubiquitin to the substrate (Jue et al., 2015). The housekeeping of UBCs may attribute to the fact that UBCs function as an intermediate step of the ubiquitin-proteasome degradation system to control the abundance of cellular proteins required for plant growth and development (Stone and Callis, 2007; Vierstra, 2009). Taken together, screening reference genes on the basis of latex flow should be an efficient and effective clue for selection of reference genes in qRT-PCR. The UBC2a and UBC2b could be used as suitable reference genes for qRT-PCR analysis of latex regeneration-related genes following different treatments.
Author Contributions
JC designed and carried out the experiment of this study, and wrote the manuscript. SY, YC participated and analyzed data in the experiment. WT planned the study and participated in the design of the experiment. All authors have read and approved the manuscript in its final form.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Fundamental Research Funds for Rubber Research Institute, CATAS (1630022014004) and the earmarked Fund for Modern Agro-industry Technology Research System (CARS-34-GW1).
Abbreviations
- 18S rRNA
18s ribosomal RNA
- ACT7a
Actin (ACTIN7)
- ACT7b
Actin (ACTIN7)
- ADF
Actin depolymerizing factor
- ADF4
Actin depolymerizing factor 4
- Ct
threshold cycle
- eIF1Aa
Eukaryotic translation initiation factor 1A
- eIF1Ab
Eukaryotic translation initiation factor 1A
- eIF2
Eukaryotic translation initiation factor
- eIF3
Eukaryotic translation initiation factor
- FP
F-box family protein
- HMGR1
3-Hydroxy-3-methylglutaryl-coenzyme A reductase
- PTP
Trosine phosphatase
- qRT-PCR
Quantitative real-time PCR
- RH2a
DEAD box RNA helicase, RH2
- RH2b
DEAD box RNA helicase, RH2
- ROC3
Cytosolic cyclophilin (ROC3)
- SRPP
Small rubber particle protein
- TFIIB
Transcription initiation factor IIB
- UBC1
Ubiquitin-protein ligase
- UBC2a
Ubiquitin-protein ligase (ATUBC2)
- UBC2b
Ubiquitin-protein ligase (ATUBC2)
- UBC3
Ubiquitin-protein ligase
- UBC4
Ubiquitin-protein ligase
- YLS8
Mitosis protein YLS8
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01149
The melting curves of the 23 candidate reference genes and four latex regeneration-related genes.
References
- Andersen C. L., Jensen J. L., Orntoft T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64 5245–5250. 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- Bustin S. A. (2000). Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25 169–193. 10.1677/jme.0.0250169 [DOI] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55 611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M. (2013). The need for transparency and good practices in the qPCR literature. Nat. Methods 10 1063–1067. 10.1038/nmeth.2697 [DOI] [PubMed] [Google Scholar]
- Chambers J. P., Behpouri A., Bird A., Ng C. K. (2012). Evaluation of the use of the polyubiquitin genes, Ubi4 and Ubi10 as reference genes for expression studies in Brachypodium distachyon. PLoS ONE 7:e49372 10.1371/journal.pone.0049372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao J., Chen Y., Wu S., Tian W. M. (2015a). Comparative transcriptome analysis of latex from rubber tree clone CATAS8-79 and PR107 reveals new cues for the regulation of latex regeneration and duration of latex flow. BMC Plant Biol. 15:104 10.1186/s12870-015-0488-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao J., Zhang S., Chen Y., Tian W. M. (2015b). Cloning, heterologous expression and characterization of ascorbate peroxidase (APX) gene in laticifer cells of rubber tree (Hevea brasiliensis Muell. Arg.). Plant Physiol. Biochem. 97 331–338. 10.1016/j.plaphy.2015.10.023 [DOI] [PubMed] [Google Scholar]
- Cheng H. M., Chern Y., Chen I. H., Liu C., Li S. H., Chun S. J., et al. (2015). Effects on murine behavior and lifespan of selectively decreasing expression of mutant huntingtin allele by supt4h knockdown. PLoS Genet. 11:e1005043 10.1371/journal.pgen.1005043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow K. S., Mat-Isa M. N., Bahari A., Ghazali A. K., Alias H., Mohd-Zainuddin Z., et al. (2012). Metabolic routes affecting rubber biosynthesis in Hevea brasiliensis latex. J. Exp. Bot. 63 1863–1871. 10.1093/jxb/err363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delporte M., Legrand G., Hilbert J. L., Gagneul D. (2015). Selection and validation of reference genes for quantitative real-time PCR analysis of gene expression in Cichorium intybus. Front. Plant Sci. 6:651 10.3389/fpls.2015.00651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- E Z., Zhang Y., Li T., Wang L., Zhao H. (2015). Characterization of the ubiquitin-conjugating enzyme gene family in rice and evaluation of expression profiles under abiotic stresses and hormone treatments. PLoS ONE 10:e0122621 10.1371/journal.pone.0122621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschbach J. M., Lacrotte R. (1989). “Factors influencing response to hormonal yield stimulation: limits of this stimulation,” in Physiology of Rubber Tree Latex eds d’Auzac J., Jacob J. L., Chrestin H. (Boca Raton, FL: CRC Press; ) 321–342. [Google Scholar]
- Garson J. A., Grant P. R., Ayliffe U., Ferns R. B., Tedder R. S. (2005). Real-time PCR quantitation of hepatitis B virus DNA using automated sample preparation and murine cytomegalovirus internal control. J. Virol Methods 126 207–213. 10.1016/j.jviromet.2005.03.001 [DOI] [PubMed] [Google Scholar]
- Goidin D., Mamessier A., Staquet M. J., Schmitt D., Berthier-Vergnes O. (2001). Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal. Biochem. 295 17–21. 10.1006/abio.2001.5171 [DOI] [PubMed] [Google Scholar]
- Guo D., Li H. L., Tang X., Peng S. Q. (2014). Molecular and functional characterization of the HbSRPP promoter in response to hormones and abiotic stresses. Transgenic Res. 23 331–340. 10.1007/s11248-013-9753-0 [DOI] [PubMed] [Google Scholar]
- Hager G. L., McNally J. G., Misteli T. (2009). Transcription dynamics. Mol. Cell 35 741–753. 10.1016/j.molcel.2009.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S. (2004). Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11 394–403. 10.1038/nsmb763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Hu W., Xia Z., Zhou X., Wang W. (2016). Validation of reference genes for relative quantitative gene expression studies in cassava (Manihot esculenta Crantz) by using quantitative real-time PCR. Front. Plant Sci. 7:680 10.3389/fpls.2016.00680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Yan H., Jiang X., Yin G., Zhang X., Qi X., et al. (2014). Identification of candidate reference genes in perennial ryegrass for quantitative RT-PCR under various abiotic stress conditions. PLoS ONE 9:e93724 10.1371/journal.pone.0093724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J., Dheda K., Bustin S., Zumla A. (2005). Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 6 279–284. 10.1038/sj.gene.6364190 [DOI] [PubMed] [Google Scholar]
- Jacob J. L., Prevot J. C., Roussel D., Lacrotte R., Serres E., d’Auzac J., et al. (1989). “Yield limiting factors, latex physiological parameters, latex diagnosis, and clonal typology,” in Physiology of Rubber Tree Latex eds d’Auzac J., Jacob J. L., Chrestin H. (Boca Raton, FL: CRC Press; ) 345–382. [Google Scholar]
- Jain M., Nijhawan A., Tyagi A. K., Khurana J. P. (2006). Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 345 646–651. 10.1016/j.bbrc.2006.04.140 [DOI] [PubMed] [Google Scholar]
- Ji Y., Tu P., Wang K., Gao F., Yang W., Zhu Y., et al. (2014). Defining reference genes for quantitative real-time PCR analysis of anther development in rice. Acta Biochim. Biophys. Sin. (Shanghai) 46 305–312. 10.1093/abbs/gmu002 [DOI] [PubMed] [Google Scholar]
- Josefsen K., Nielsen H. (2011). Northern blotting analysis. Methods Mol. Biol. 703 87–105. 10.1007/978-1-59745-248-9_7 [DOI] [PubMed] [Google Scholar]
- Jue D., Sang X., Lu S., Dong C., Zhao Q., Chen H., et al. (2015). Genome-wide identification, phylogenetic and expression analyses of the ubiquitin-conjugating enzyme gene family in maize. PLoS ONE 10:e0143488 10.1371/journal.pone.0143488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. R., Nam H. Y., Kim S. U., Kim S. I., Chang Y. J. (2003). Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnol. Lett. 25 1869–1872. 10.1023/A:1026298032009 [DOI] [PubMed] [Google Scholar]
- Kong Q., Yuan J., Gao L., Zhao L., Cheng F., Huang Y., et al. (2015). Evaluation of appropriate reference genes for gene expression normalization during watermelon fruit development. PLoS ONE 10:e0130865 10.1371/journal.pone.0130865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubista M., Andrade J. M., Bengtsson M., Forootan A., Jonak J., Lind K., et al. (2006). The real-time polymerase chain reaction. Mol. Aspects Med. 27 95–125. 10.1016/j.mam.2005.12.007 [DOI] [PubMed] [Google Scholar]
- Lang V., Usadel B., Obermeyer G. (2014). De novo sequencing and analysis of the lily pollen transcriptome: an open access data source for an orphan plant species. Plant Mol. Biol. 87 69–80. 10.1007/s11103-014-0261-2 [DOI] [PubMed] [Google Scholar]
- Le M. Q., Pagter M., Hincha D. K. (2014). Global changes in gene expression, assayed by microarray hybridization and quantitative RT-PCR, during acclimation of three Arabidopsis thaliana accessions to sub-zero temperatures after cold acclimation. Plant Mol. Biol. 87 1–15. 10.1007/s11103-014-0256-z [DOI] [PubMed] [Google Scholar]
- Li H., Qin Y., Xiao X., Tang C. (2011). Screening of valid reference genes for real-time RT-PCR data normalization in Hevea brasiliensis and expression validation of a sucrose transporter gene HbSUT3. Plant Sci. 181 132–139. 10.1016/j.plantsci.2011.04.014 [DOI] [PubMed] [Google Scholar]
- Li J., Han J., Hu Y., Yang J. (2016). Selection of reference genes for quantitative real-time PCR during flower development in tree Peony (Paeonia suffruticosa Andr.). Front. Plant Sci. 7:516 10.3389/fpls.2016.00516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X., He B., Gao X., Qin Y., Yang J., Fang Y., et al. (2015). Validation of reference genes for quantitative real-time PCR during latex regeneration in rubber tree. Gene 563 190–195. 10.1016/j.gene.2015.03.026 [DOI] [PubMed] [Google Scholar]
- Mehdi Khanlou K., Van Bockstaele E. (2012). A critique of widely used normalization software tools and an alternative method to identify reliable reference genes in red clover (Trifolium pratense L.). Planta 236 1381–1393. 10.1007/s00425-012-1682-2 [DOI] [PubMed] [Google Scholar]
- Niu X., Qi J., Zhang G., Xu J., Tao A., Fang P., et al. (2015). Selection of reliable reference genes for quantitative real-time PCR gene expression analysis in Jute (Corchorus capsularis) under stress treatments. Front. Plant Sci. 6:848 10.3389/fpls.2015.00848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrello J., Leclercq J., Dessailly F., Rio M., Piyatrakul P., Kuswanhadi K., et al. (2014). Transcriptional and post-transcriptional regulation of the jasmonate signalling pathway in response to abiotic and harvesting stress in Hevea brasiliensis. BMC Plant Biol. 14:341 10.1186/s12870-014-0341-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj S., Das G., Pothen J., Dey S. K. (2005). Relationship between latex yield of Hevea brasiliensis and antecedent environmental parameters. Int. J. Biometeorol. 49 189–196. 10.1007/s00484-004-0222-6 [DOI] [PubMed] [Google Scholar]
- Remans T., Keunen E., Bex G. J., Smeets K., Vangronsveld J., Cuypers A. (2014). Reliable gene expression analysis by reverse transcription-quantitative PCR: reporting and minimizing the uncertainty in data accuracy. Plant Cell 26 3829–3837. 10.1105/tpc.114.130641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sando T., Takaoka C., Mukai Y., Yamashita A., Hattori M., Ogasawara N., et al. (2008). Cloning and characterization of mevalonate pathway genes in a natural rubber producing plant, Hevea brasiliensis. Biosci. Biotechnol. Biochem. 72 2049–2060. 10.1271/bbb.80165 [DOI] [PubMed] [Google Scholar]
- Sethuraj M. R. (1992). “Yield components in Hevea Brasiliensis,” in Natural Rubber: Biology, Cultivation and Technology eds Sethuraj M. R., Mathew N. M. (Amsterdam: Elsevier Press; ) 137–163. [Google Scholar]
- Sheng Y., Hong J. H., Doherty R., Srikumar T., Shloush J., Avvakumov G. V., et al. (2012). A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen. Mol. Cell Proteomics 11 329–341. 10.1074/mcp.O111.013706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V., Kaul S. C., Wadhwa R., Pati P. K. (2015). Evaluation and selection of candidate reference genes for normalization of quantitative RT-PCR in Withania somnifera (L.) Dunal. PLoS ONE 10:e0118860 10.1371/journal.pone.0118860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S. L. (2014). The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front. Plant Sci. 5:135 10.3389/fpls.2014.00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S. L., Callis J. (2007). Ubiquitin ligases mediate growth and development by promoting protein death. Curr. Opin. Plant Biol. 10 624–632. 10.1016/j.pbi.2007.07.010 [DOI] [PubMed] [Google Scholar]
- Tang C., Huang D., Yang J., Liu S., Sakr S., Li H., et al. (2010). The sucrose transporter HbSUT3 plays an active role in sucrose loading to laticifer and rubber productivity in exploited trees of Hevea brasiliensis (para rubber tree). Plant Cell Environ. 33 1708–1720. 10.1111/j.1365-3040.2010.02175.x [DOI] [PubMed] [Google Scholar]
- Tricarico C., Pinzani P., Bianchi S., Paglierani M., Distante V., Pazzagli M., et al. (2002). Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal. Biochem. 309 293–300. 10.1016/S0003-2697(02)00311-1 [DOI] [PubMed] [Google Scholar]
- Udvardi M. K., Czechowski T., Scheible W. R. (2008). Eleven golden rules of quantitative RT-PCR. Plant Cell 20 1736–1737. 10.1105/tpc.108.061143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3 1–11. 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R. D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10 385–397. 10.1038/nrm2688 [DOI] [PubMed] [Google Scholar]
- Wang H., Wang J., Jiang J., Chen S., Guan Z., Liao Y., et al. (2014). Reference genes for normalizing transcription in diploid and tetraploid Arabidopsis. Sci. Rep. 4:6781 10.1038/srep06781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Shi M., Wang D., Chen Y., Cai F., Zhang S., et al. (2013). Comparative proteomics of primary and secondary lutoids reveals that chitinase and glucanase play a crucial combined role in rubber particle aggregation in Hevea brasiliensis. J. Proteome Res. 12 5146–5159. 10.1021/pr400378c [DOI] [PubMed] [Google Scholar]
- Wang X., Wang D., Sun Y., Yang Q., Chang L., Wang L., et al. (2015). Comprehensive proteomics analysis of laticifer latex reveals new insights into ethylene stimulation of natural rubber production. Sci. Rep. 5:13778 10.1038/srep13778 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The melting curves of the 23 candidate reference genes and four latex regeneration-related genes.


