Introduction
Methylation defects in the imprinting locus at chromosome 6q24 result in transient neonatal diabetes and small-for-gestational age (SGA) births (1). These phenotypes are primarily ascribed to the overexpression of PLAGL1, a paternally expressed gene on 6q24 that regulates cell cycle and apoptosis (2). Paternal uniparental disomy involving 6q24, as well as copy-number gains of paternal PLAGL1 alleles and epimutations in maternal alleles, have been identified as the causes of hypomethylation at the differentially methylated region (DMR) of PLAGL1 (3, 4).
Recently, Yorifuji et al. reported the identification of 6q24 uniparental disomy in three patients with childhood-onset non-autoimmune diabetes mellitus (5). The three patients were identified through methylation-specific PCR analysis of the PLAGL1 DMR of 113 patients clinically suspected of having maturity-onset diabetes of the young (MODY). These results expanded the phenotypic consequences of 6q24 methylation defects to include MODY-like manifestations without a history of neonatal diabetes. However, the frequency of 6q24 methylation defects among patients with childhood-onset non-autoimmune diabetes remained unknown.
Subjects and Methods
This study was approved by the Institutional Review Board Committee at the National Center for Child Health and Development, and performed in accordance with the Declaration of Helsinki. The study was carried out after obtaining written informed consent from the patients or their parents and from control individuals.
The study population consisted of 58 unrelated Japanese patients with childhood-onset non-autoimmune diabetes who required continuous insulin therapy (22 males and 36 females). The patients were registered with the Japanese Study Group of Insulin Therapy for Childhood and Adolescent Diabetes between March and December 2008. No patients had a history of neonatal diabetes. All patients were assessed as being born appropriate- or large-for-gestational age (birth weight and length > –2.0 SD for gestational age), based on the Japanese neonatal growth charts (6). Birth weight ranged from 2,020 to 4,274 g (mean, 3,100 g). Age at the time of diagnosis ranged from 9 mo to 15 yr (mean, 6 yr) and body mass index SDS ranged from –3.1 to 2.5 (mean, –0.9). Seven patients had a family history of diabetes. As negative controls, we analyzed DNA samples obtained from 49 healthy Japanese volunteers. These samples were purchased from the Human Science Research Resources Bank, Tokyo, Japan (present distributor, National Institute of Biomedical Innovation, Osaka, Japan). To examine the accuracy of our methods, we analyzed samples obtained from three previously reported patients with 6q24 uniparental disomy (5).
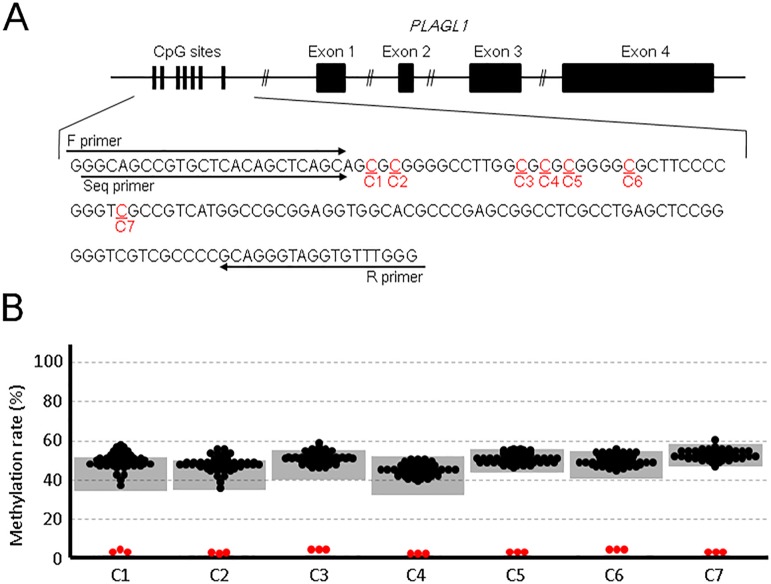
Genomic DNA samples were isolated from the peripheral leukocytes of the patients. The samples were treated with bisulfite. The methylation levels of seven cytosines at the CpG dinucleotides in the DMR (Fig. 1A) were analyzed by pyrosequencing.
Fig. 1.
Methylation analysis of 58 patients with childhood-onset non-autoimmune diabetes. A: Genomic structure of PLAGL1 and its flanking CpG sites. C1–7 represent cytosines at the CpG sites in the differentially methylated region. Forward (F) and reverse (R) primers were used for PCR amplification and the Seq primer was used for pyrosequencing. B: Results of the methylation analysis. Black dots represent the methylation statuses of the 58 patients. Gray shaded areas indicate the reference range obtained from 49 control individuals. The red dots depict the results of three previously reported patients with 6q24 uniparental disomy (5).
Results and Discussion
The methylation statuses of the patients with childhood-onset non-autoimmune diabetes were comparable to those of the control individuals (Fig. 1B). In the present study, we employed pyrosequencing, which is more sensitive than the methylation-specific PCR used by Yorifuji et al. (5, 7). Three previously reported patients with 6q24 uniparental disomy exhibited apparent hypomethylation at all CpG sites examined, confirming the accuracy of our methods. Our findings suggest that 6q24 methylation defects are uncommon among childhood-onset non-autoimmune diabetes patients.
The differences in the results between the prior and present studies likely reflect the differences in the inclusion criteria. The study population reported on by Yorifuji et al. consisted of 113 patients with MODY-like phenotypes including 11 SGA cases (5), whereas none of our 58 patients were born SGA. Notably, all three patients with 6q24 uniparental disomy identified by Yorifuji et al. had a history of SGA (5). This is consistent with the observation that overexpression of paternally expressed genes usually results in prenatal and postnatal growth failure (1, 2). Our data imply that a history of SGA appears is an essential marker for diabetes due to 6q24 methylation defects. Since molecular diagnoses of methylation defects likely serve to improve the clinical management of the patients (5), methylation analyses should be considered for childhood-onset non-autoimmune diabetes patients with a history of SGA. Further studies are necessary to clarify the precise frequency and phenotypic spectrum of diabetes due to 6q24 methylation defects.
Conflict of interests
T.U. received honoraria from Novo Nordisk and Sanofi as a speaker and for attendance at advisory boards. No other authors have nothing to declare.
Acknowledgements
The work was supported by the Grants from the Ministry of Health, Labor and Welfare, the Japan Agency for Medical Research and Development, the National Center for Child Health and Development, the Takeda Foundation, the Japan Diabetes Foundation, and the Manpei Suzuki Diabetes Foundation.
The Japanese Study Group of Insulin Therapy for Childhood and Adolescent Diabetes (JSGIT): Koji Takemoto, Ehime University School of Medicine; Noriyuki Takubo, Kitasato University School of Medicine; Kohji Tsubouchi, Chuno Kosei Hospital; Reiko Horikawa, National Center for Child Health and Development; Kisho Kobayashi, Yamanashi University School of Medicine; Yoshihito Kasahara, Kanazawa University School of Medicine; Akemi Koike, Koike Child Clinic; Takahiro Mochizuki, Osaka City General Medical Center; Kanshi Minamitani, Teikyo University Chiba Medical Center, Ryuzo Takaya, Osaka Medical College; Hiroshi Mochizuki, Saitama Children’s Medical Center; Aki Nishii, JR Sendai Hospital; Zenro Kizaki, Japanese Red Cross Kyoto Daiichi Hospital; Tetsuo Mori, Nagano Red Cross Hospital; Naoto Shimura, Dokkyo Medical University; Tokuo Mukai, Asahikawa Medical University; Nobuo Matsuura, Teine Keijinkai Hospital; Takao Fujisawa, National Mie Hospital; Kenji Ihara, Kyushu University School of Medicine; Kitaro Kosaka, Kyoto Prefectural University of Medicine; Rika Kizu, Yokosuka Kyosai Hospital; Toshikazu Takahashi, Takahashi Clinic; Satoshi Matsuo, Matsuo Child Clinic; Keiichi Hanaki, Tottori Prefectural Kousei Hospital; Yutaka Igarashi, Igarashi Children’s Clinic; Goro Sasaki, Tokyo Dental College Ichikawa General Hospital; Shun Soneda, St. Marianna University School of Medicine; Shinichi Teno, Teno Clinic; Susumu Kanzaki, Tottori University Faculty of Medicine.
References
- 1.Temple IK, Shield JPH. 6q24 transient neonatal diabetes. Rev Endocr Metab Disord 2010;11: 199–204. doi: 10.1007/s11154-010-9150-4 [DOI] [PubMed] [Google Scholar]
- 2.Abdollahi A. LOT1 (ZAC1/PLAGL1) and its family members: mechanisms and functions. J Cell Physiol 2007;210: 16–25. doi: 10.1002/jcp.20835 [DOI] [PubMed] [Google Scholar]
- 3.Mackay DJ, Temple IK, Shield JPH, Robinson DO. Bisulphite sequencing of the transient neonatal diabetes mellitus DMR facilitates a novel diagnostic test but reveals no methylation anomalies in patients of unknown aetiology. Hum Genet 2005;116: 255–61. doi: 10.1007/s00439-004-1236-1 [DOI] [PubMed] [Google Scholar]
- 4.Boonen SE, Mackay DJ, Hahnemann JM, Docherty L, Grønskov K, Lehmann A, et al. Transient neonatal diabetes, ZFP57, and hypomethylation of multiple imprinted loci: a detailed follow-up. Diabetes Care 2013;36: 505–12. doi: 10.2337/dc12-0700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yorifuji T, Matsubara K, Sakakibara A, Hashimoto Y, Kawakita R, Hosokawa Y, et al. Abnormalities in chromosome 6q24 as a cause of early-onset, non-obese, non-autoimmune diabetes mellitus without history of neonatal diabetes. Diabet Med 2015;32: 963–7. doi: 10.1111/dme.12758 [DOI] [PubMed] [Google Scholar]
- 6.Itabashi K, Miura F, Uehara R, Nakamura Y. New Japanese neonatal anthropometric charts for gestational age at birth. Pediatr Int 2014;56: 702–8. doi: 10.1111/ped.12331 [DOI] [PubMed] [Google Scholar]
- 7.Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by Pyrosequencing. Biotechniques 2003;35: 152–6. [DOI] [PubMed] [Google Scholar]