Abstract
Daidzein (DZN) is converted to equol (EQL) by intestinal bacteria. We previously reported that Eggerthella sp. YY7918, which is found in human feces, is an EQL-producing bacterium and analyzed its whole genomic sequence. We found three coding sequences (CDSs) in this bacterium that showed 99% similarity to the EQL-producing enzymes of Lactococcus sp. 20-92. These identified CDSs were designated eqlA, eqlB, and eqlC and thought to encode daidzein reductase (DZNR), dihydrodaidzein reductase (DHDR), and tetrahydrodaidzein reductase (THDR), respectively. These genes were cloned into pColdII. Recombinant plasmids were then introduced into Escherichia coli BL21 (DE3) and DZNR, DHDR, and THDR were expressed and purified by 6×His-Tag chromatography. We confirmed that these three enzymes were involved in the conversion of DZN to EQL. Purified DZNR converted DZN to dihydrodaizein (DHD) in the presence of NADPH. DHDR converted DHD to tetrahydrodaizein (THD) in the presence of NADPH. Neither enzyme showed activities with NADH. THDR converted THD in the absence of cofactors, NAD(P)H, and also produced DHD as a by-product. Thus, we propose that THDR is not a reductase but a new type of dismutase. The GC content of these clusters was 64%, similar to the overall genomic GC content for Eggerthella and Coriobacteriaceae (56–60%), and higher than that for Lactococcus garvieae (39%), even though the gene cluster showed 99% similarity to that in Lactococcus sp. 20-92. Taken together, our results indicate that the gene cluster associated with EQL production evolved in high-GC bacteria including Coriobacteriaceae and was then laterally transferred to Lactococcus sp. 20-92.
Keywords: equol, daidzein, isoflavone, gut-microflora, oxidoreductase, dismutase
INTRODUCTION
Isoflavones are diphenolic compounds that are present in soybean and other Fabaceae. Isoflavones are able to bind to estrogen receptors and exert hormonal effects because of their structural similarity to endogenous estrogens. Major isoflavones, including daidzein (DZN), genistein, and formononetin, can be altered by intestinal bacterial metabolism [1,2,3]. Equol (EQL), a bacterial metabolite of DZN, exhibits higher estrogenic activity than DZN [4]. Therefore, EQL is expected to reduce symptoms of menopausal disorder and prevent hormone-dependent diseases such as osteoporosis and prostate cancer. There is much interest in EQL and its production in the human guts and many researchers are investigating EQL-producing bacteria. Recently, clinical studies using EQL for osteoporosis or menopausal symptoms have been reported [5,6,7,8]. It has been shown that S-EQL has 11 times higher affinity than R-EQL for estrogen receptorβ [9] and that biosynthetic EQL is only present as S-EQL. However, only 30–40% of healthy adult persons are able to produce EQL following ingestion of soy-based food products [10].
It is thought that the microbiological conversion from DZN to S-EQL consists of three steps of enzymatic reactions via dihydrodaidzein (DHD) and tetrahydrodaidzein (THD) [11, 12]. To date, several EQL-producing bacteria have been isolated [13,14,15,16,17,18]. We reported the isolation of Eggerthella sp. YY7918 from the feces of healthy humans, which converts up to 50 μM DZN to S-EQL with almost 100% efficiency [16]. We also analyzed the whole genome sequence of this bacterium [19]. Recently, genes involved in the conversion of DZN to EQL have been identified in three bacterial strains found in the human intestine: Lactococcus sp. strain 20-92 [20, 21], Slackia sp. strain NATTS [22], and Slackia isoflavoniconvertens [23]. In these strains, three genes encoding the enzymes daidzein reductase (DZNR), dihydrodaidzein reductase (DHDR), and tetrahydrodaidzein reductase (THDR) were identified and analyzed for their enzymatic functions.
In this study, we identified three putative genes of Eggerthella sp. YY7918 that shows similarities to those in other EQL-producing bacteria. These three genes were expressed as recombinant enzymes in E. coli and examined for their functions.
MATERIALS AND METHODS
Chemicals
DZN (Tokyo Chemical Industry Co., Ltd., Tokyo, Japan) and (R,S)-Equol (LC Laboratories, Woburn, MA, USA) were dissolved in dimethyl sulfoxide (Wako Pure Chemical Industries, Ltd., Osaka, Japan). DHD and THD were chemically synthesized according to methods described by Wähälä et al. [24]. Resulting products were confirmed by proton nuclear magnetic resonance (1H NMR).
Bacterial strains and culture conditions
Eggerthella sp. YY7918 was cultured under anaerobic conditions at 37°C in GAM medium (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) for 3 days using an anaerobic chamber (Bugbox, Ruskinn Technology, Ltd.). The gas phase was maintained at 80:10:10 (vol. %) for N2:CO2:H2. Recombinant bacteria were grown in 30 ml of Miller’s Luria-Bertani broth (10 g bacto-tryptone, 5 g of bacto-yeast extract, 10 g/l NaCl) containing 100 mg/l of ampicillin (LB-Amp) at 37°C.
Construction of recombinant E. coli expressing DZNR, DHDR, and THDR
Genomic DNA was extracted using an UltraClean® Microbial DNA Isolation Kit (MO BIO Laboratories Inc., Carlsbad, CA, USA). Polymerase chain reactions (PCRs) were used to amplify the open reading frames (ORFs) EGYY15730, 15750, and 15760, which possibly encode DZNR, DHDR, and THDR in Eggerthella sp. YY7918, respectively (Fig. 1). The oligonucleotide primers and the PCR conditions are listed in Table 1. Amplified DNA fragments were introduced into a plasmid, pColdII (Takara Bio Inc., Ohtsu, Japan), between the NdeI and BamHI sites after the N-terminal 6×His-Tag sequence, using an In-Fusion® HD Cloning Kit (Clontech Laboratories Inc., Mountain View, CA, USA). The resultant recombinant plasmids were transformed into E. coli TOP10. The nucleotide sequences were confirmed by sequencing analyses with an ABI PRISM 3130xl DNA sequencing system (Applied Biosystems, Foster City, CA, USA). The recombinant plasmids were designated pColdII-15730, pColdII-15750, and pColdII-15760, and introduced into E. coli BL21 (DE3).
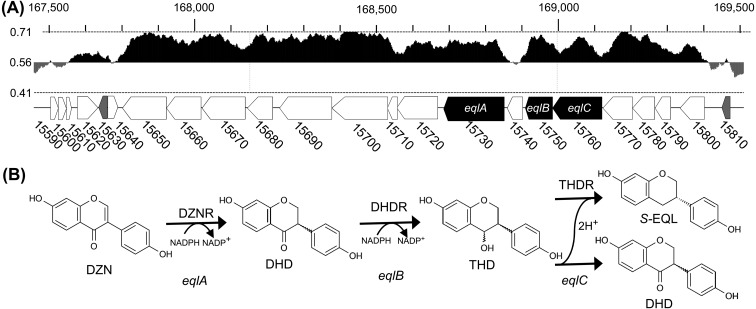
Fig. 1.
A: GC content and the position of coding sequences (CDSs) for genes related to equol (EQL) metabolism in Eggerthella sp. YY7918. The horizontal lines indicate a GC content of 71.2%, 56.2% (the GC content of the whole genome sequence), and 41.2%. The coding regions for daidzein reductase (DZNR), dihydrodaidzein reductase (DHDR), and tetrahydrodaidzein reductase (THDR) commenced at nucleotides 15730, 15750, and 15760, respectively. The eqlA, eqlB, and eqlC genes (all in black) were amplified by polymerase chain reaction using the oligonucleotide primers described in Table 1. B: The metabolic pathway for the conversion of daidzein (DZN) to S-EQL by Eggerthella sp. strain YY7918.
Table 1. Sequence of primers used in this study.
| Exp. | Aim | Primer set | Thermal cycling program | |
|---|---|---|---|---|
| 1 | Const. pColdII-15730 | eqlA Fw | CATCATCATCATCATATGAAGAACAAGTTCTATCC | [98ºC 10 sec – 58ºC 5 sec – 72ºC 10 sec] × 30 cycle |
| amplify EGYY15730 gene | eqlA Rv | GCTTGAATTCGGATCCCTACAGGTTGCAGCCAGCG | ||
| 2 | Const. pColdII-15750 | eqlB Fw | CATCATCATCATCATATGGCACAGGAAGTC | [98ºC 10 sec – 60ºC 5 sec – 72ºC 15 sec ] × 30 cycle |
| amplify EGYY 15750 gene | eqlB Rv | GCTTGAATTCGGATCCTTAGACCTCGATCTCGCCCTG | ||
| 3 | Const. pColdII-15760 | eqlC Fw | CATCATCATCATCATATGGCACAATTCGATGTTGAG | [98ºC 10 sec – 62ºC 5 sec – 72ºC 10 sec ] × 30 cycle |
| amplify EGYY15760 gene | eqlC Rv | GCTTGAATTCGGATCCCTACATAGTGGAGATCGCGTGG | ||
| 4 | Confirm eqlA of strain | seq_eqlA_Nde | GTCCATGAACACGATGC | 98ºC 5 min [98ºC 0 sec – 50ºC 5 sec – 60ºC 4 min ] × 25 cycle |
| YY7918 in pColdII | seq_eqlA_Bam | TCGACCAGACAATCATC | ||
| 5 | Confirm eqlB of strain | seq_eqlB_Nde | ATCATCACCTCCTCCACC | [98ºC 10 sec – 62ºC 5 sec – 72ºC 10 sec ] × 30 cycle |
| YY7918 in pColdII | seq_eqlB_Bam | TAGTCAGACAGGTCGAC | ||
| 6 | Confirm eqlC of strain | seq_eqlC_Nde | AGCTCGTTCTGAATAGAG | 98ºC 5 min [98ºC 0 sec – 50ºC 5 sec – 60ºC 4 min ] × 25 cycle |
| YY7918 in pColdII | seq_eqlC_Bam | ACGACGACGCCTTTATGA | ||
Underlining indicates the homologous 15 bp.
Purification of recombinant proteins
We used 20 mM sodium phosphate buffer (pH 7.0) containing 0.1 mM phenylmethylsulfonyl fluoride (Buffer A) in following purification steps. The transformants were cultivated at 37°C with LB-Amp until the OD660 was approximately 0.3, then the temperature was shifted to 15°C to induce the cold-shock promoters and incubation was continued for 24 hr. The cells were harvested by centrifugation (6,000 × g, 10 min, 4°C) and suspended in 3 ml of Buffer A. One microliterl of 1,000 U/ml recombinant DNase I (Takara Bio Inc. Otsu, Shiga, Japan) was added to each sample, and the samples were then incubated for 15 min at room temperature. Samples were then centrifuged (6,000 × g, 10 min, 4°C), and pellets were resuspended in 1 ml of Buffer A. Cell suspensions were transferred to polypropylene tubes containing 800 mg of glass beads (0.1 mm diameter) and shaken (2,500 rpm, 30 sec, 5 cycles) in a Multi-beads Shocker (Yasui Kikai Corporation, Osaka, Japan). Between each cycle, samples were cooled on ice for 40 sec. Then samples were centrifuged (13,000 × g, 10 min, 4°C). Proteins in the supernatant were purified by Ni-NTA agarose column chromatography (1 mL bed volume, QIAGEN. Hilden, Germany). The proteins were eluted using Buffer A containing 100 mM imidazole and 500 mM NaCl. The concentrations of proteins in samples were determined according to the method described by Bradford et al. [25]. Fractions from each stage of the purification process were subjected to 12.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by coomassie brilliant blue G250 staining.
Enzyme activity assays
Crude enzyme extracts (100 µg) or purified enzymes (5 µg) were subjected to enzyme activity assays. The substrate and buffer had been equilibrated in an anaerobic chamber (80:10:10 [vol. %] for N2:CO2:H2) at 37°C about for 30 min. This was conducted at 37°C in 0.5 ml of 20 mM sodium phosphate buffer containing 50 μM substrate and 1 mM NADPH or NADH. After 15 min of incubation, 500 µl of ethyl acetate was added to stop the reaction. Samples were extracted three times using an equal volume of ethyl acetate, dried under a vacuum, and then dissolved in 0.5 ml of methanol.
Extracted samples (10 µl) were analyzed by high-performance liquid chromatography (HPLC, Hitachi High-Tech Science Corporation, Tokyo, Japan) to quantify levels of DZN and its metabolites (DZN, DHD, THD, and EQL). HPLC analysis was performed using a TSKgel ODS100V reversed-phase column (5 μm, 250 × 4.6 mm i.d.; Tosoh Bioscience, Tokyo, Japan) with an isocratic mobile phase comprised of 45% water:acetic acid (98:2, v/v) in methanol (1 ml/min, 40°C) with detection at 280 nm [18]. The conversion ratio was calculated from the average peak area obtained from three repetitions.
In order to analyze the enantiomeric character of EQL, a SumiChiral OA-7000 (5 μm, 250 mm × 4.6 mm i.d. Sumika Chemicals Analysis Service, Osaka, Japan) was used with a mobile phase composed of 30% 20 mM potassium phosphate, pH 3.0, in acetonitrile [12].
Phylogenetic analysis
A phylogenetic tree of EQL-producing bacteria and related taxa based on the 16S rRNA sequence was generated using the neighbor-joining method [26]. Sequences were obtained from the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/nuccore/). CLUSTAL W 2.1 (http://clustalw.ddbj.nig.ac.jp) was used for sequence alignment [27], and FigTree 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) was used for visualization. Amino acid sequence similarities were determined with BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi) [28].
RESULTS AND DISCUSSION
Putative genes related to isoflavone metabolism
The cording sequences (CDSs) in the Eggerthella sp. YY7918 genome were BLASTP searched with the reported DZNR, DHDR, and THDR of Lactococcus sp. 20-92 (BAJ72750, BAJ72748, BAJ72747), Slackia sp. NATTS (BAL46930, BAL46929, BAL46928), and S. isoflavoniconvertens (AFV15453, AFV15451, AFV15450) as the queries. We identified three CDSs (EGYY15730, EGYY15750, and EGYY15760) with around 99% similarity to DZNR, DHDR, and THDR of Lactococcus sp. 20-92. These genes formed a cluster at nucleotides 1,686,364–1,691,456 in the Eggerthella sp. YY7918 genome (Fig. 1). These identified CDSs were designated putative genes (eqlA, eqlB, and eqlC) of EQL production and thought to encode DZNR, DHDR, and THDR, respectively.
Characteristics of genes
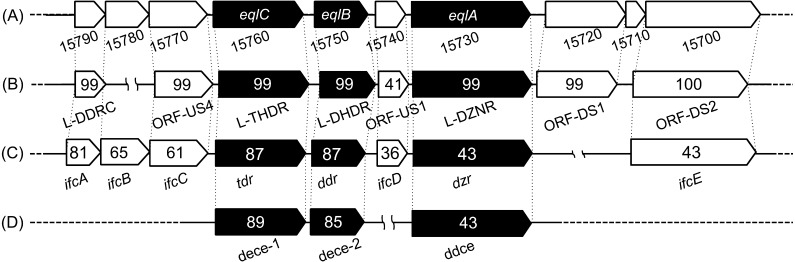
The amino acid sequence of Eggerthella sp. YY7918 DZNR showed 43% identity to that of Slackia sp. NATTS and S. isoflavoniconvertens, while the both DHDR and THDR of Eggerthella sp. YY7918 showed 85–89% identity to the set of Slackia sp. NATTS and S. isoflavoniconvertens (Fig. 6). DZNR belongs to the old yellow enzyme (OYE) family. OYE has been studied for many years. It contains a flavin mononucleotide (FMN) as a prosthetic group, but the biological function of OYE is still unclear. However, some of the family enzymes have been reported to catalyze NAD(P)H-dependent stereoselective redox reactions of the C=C bond [29]. The presence of FMN results in an OYE solution appearing yellow, corresponding to the appearance of our His-Tag-purified recombinant DZNR in solution. It is predicted that DHD, which was converted by DZNR, has chirality, but our partially purified enzyme showed R, S-racemate (data not shown). We believed that further purification is required to confirm this discrepancy.
Fig. 6.
Schematic representation of gene clusters related to EQL production (black boxes) and other flanking proteins against Eggerthella sp. YY7918 with the other EQL-producing human gut bacteria. The gene names are shown under the clusters, and the numbers in the CDSs indicate the amino acid homologies (%). (A) Eggerthella sp. YY7918, (B) Lactococcus sp. 20-92, (C) Slackia isoflavoniconvertens strain, (D) Slackia sp. NATTS.
DHDR belongs to the short-chain dehydrogenase/reductase (SDR) superfamily, and the alignment of the amino acid sequences of the NAD(P)H binding site is conserved [30]. DHDR encoded by eqlB (EGYY 15750) contains an arginine residue (R64) following to GxxKGxG motif (position 49-55), which is observed at the cofactor binding site of the NADPH-specific SDR. It was assumed that DHDR requires NADPH as a cofactor but not NADH because of the presence of the arginine (R64) residue.
THDR also belongs to the SDR superfamily and exhibits similarities to fumarate reductase and succinate dehydrogenase of Sulfurospirillum multivorans [31]. Succinate dehydrogenase catalyzes the oxidation of succinate to fumarate during anaerobic respiration, with fumarate acting as the terminal electron acceptor [32]. The catalytic domain contains a flavin adenine dinucleotide (FAD) group and other iron-sulfur clusters.
Expression and purification of recombinant proteins
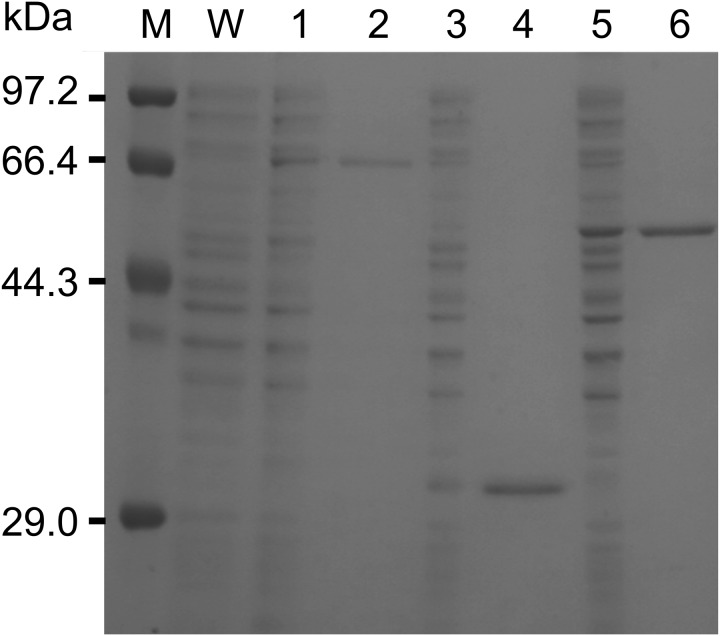
The enzymes encoded by pColdII-15730, pColdII-15750, and pColdII-15760 were expressed in E. coli. These recombinant enzymes were purified by 6×His-Tag affinity column chromatography (Fig. 2). The apparent molecular masses of these three enzymes on SDS-PAGE were 69.7, 30.5, and 52.6 kDa, respectively (Fig. 2), which agree with the theoretical values of EGYY15730 (644 amino acids, 69.0 kDa), EGYY15750 (286 amino acids, 29.8 kDa), and EGYY15760 (486 amino acids, 52.5 kDa), and there was an additional 6×His-tag (+703.8 Da) at the N-termini, respectively.
Fig. 2.
Expression and purification of recombinant enzymes. Lane M, Protein Molecular Weight Marker (Low) (Takara Bio Inc.); W, pColdII/E. coli BL21 (DE3); 1, crude preparation of daidzein reductase (DZNR); 2, purified DZNR; 3, crude preparation of dihydrodaidzein reductase (DHDR); 4, purified DHDR; 5, crude preparation of tetrahydrodaidzein reductase (THDR); and 6, purified THDR.
Enzymatic activity of the recombinant proteins
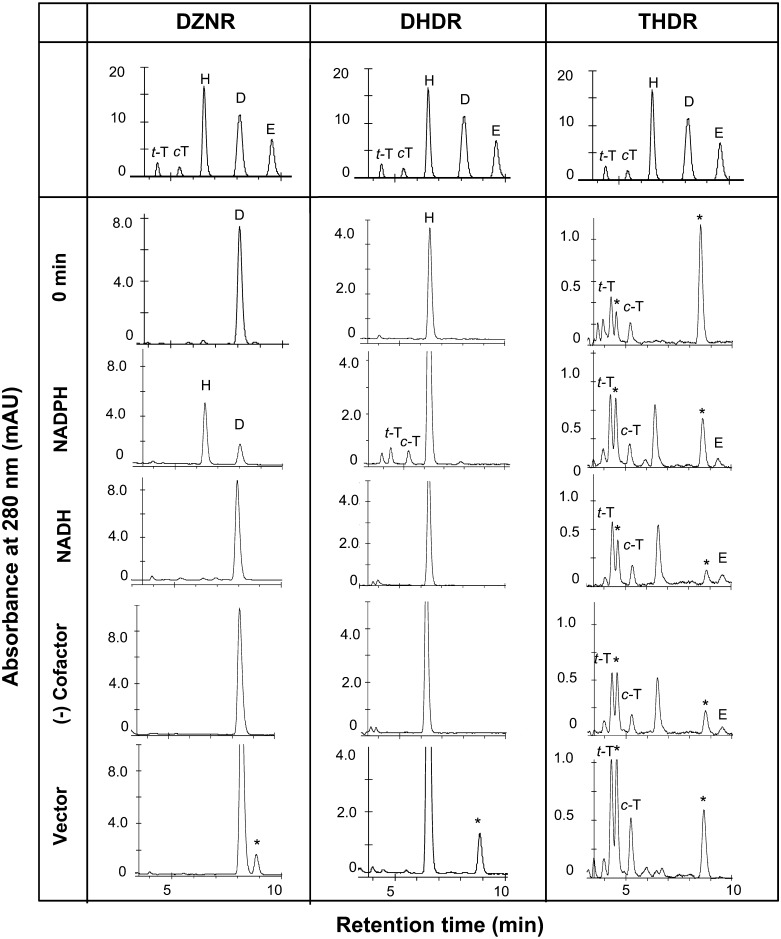
The activities of DZNR and DHDR were detected in purified enzymes of Eggerthella sp. YY7918 as shown in Fig. 3. During incubation for 15 min, 80% of DZN was converted to DHD by 5 µg DZNR in the presence of NADPH under anaerobic condition, while no detectable conversion was observed with NADH. These results suggested that DZNR specifically uses NADPH as a cofactor. The reaction of DHDR was relatively weaker, and only 5% of the DHD was converted to THD in the presence of NADPH in 15 min. No reaction was observed with NADH.
Fig. 3.
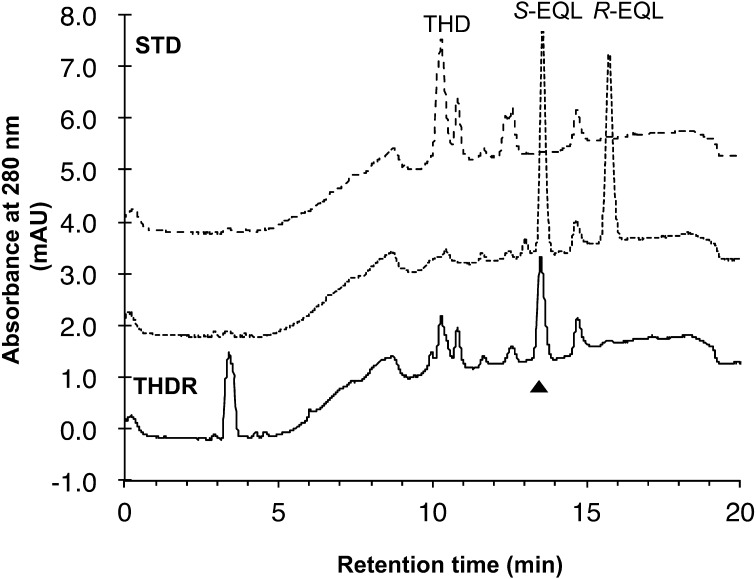
Conversion of DZN, DHD, and THD by purified DZNR, purified DHDR, and crude THDR of Eggerthella sp. YY 7918. Products were analyzed using C18 reversed phase HPLC. STD, elution profiles of reference standards (500 pmol). Samples were assessed after 15 min in the presence of NADPH or NADH. The cell lysates of E.coli BL21(DE3)/pColdII were assessed as the negative control. Asterisks (*) show unknown peaks detected in the case of reaction with E.coli crude cells. t-T: trans-tetrahydrodaidzein, c-T: cis-tetrahydrodaidzein, H: dihydrodaidzein, D: daidzein, E: equol.
The recombinant THDR was unstable. Almost all activity was lost during the purification step. Therefore, we tried to confirm EQL production by using crude enzymes in this study. The conversion of THD to EQL was observed in the presence or absence of either NADPH or NADH using 100 µg crude THDR. The same phenomenon was reported in THDR of Lactococcus sp. 20-92 and S. isoflavoniconvertens [21, 23]. In this reaction, DHD was also detected as a by-product (Fig. 3). These results suggested that THDR converts THD to both DHD and EQL by a kind of disproportionation. We propose that THDR is a new type of dismutase, that takes 2H+ from a THD molecule and produces DHD, and that the 2H+ is then added to another THD to produce EQL (Fig. 1B). To determine the enantiomeric selectivity of the THDR, the product was analyzed by HPLC with chiral column (Fig. 4), with the equol R, S-chiralities being assigned by circular dichroism spectrometry [12, 16]. The data shows that THDR conversed only S-EQL from THD. This result corresponds to biosynthesized EQL in Eggerthella sp. YY7918 [16].
Fig. 4.
HPLC using SumiChiral OA-700 analysis of the products in enantioselective reduction of EQL. The broken line shows chemically synthesized THD and EQL (standard), and the solid line shows the product of THDR. The metabolite has only S-EQL. The broken line shows chemically synthesized EQL, and the solid line shows the product of THDR from THD.
We also tried to produce of EQL or THD using a combination of these 3 enzymes. To produce EQL from DZN, we added 5 µg each of DZNR, DHDR, and THDR. The conversion of DZN to THD was confirmed, but EQL was not detected (data not shown). As shown above, we confirmed the activities of DZNR, DHDR, and THDR of Eggerthella sp. YY7918.
However, the enzymatic reactions of these three enzymes were weak and not stable enough to perform more critical enzymatic characterization. More refined purification and stabilization methods are needed for these enzymes to perform further enzymatic studies. It is possible other accessory proteins contribute to the enzymatic reactions of three enzymes.
Shimada et al. reported that dihydrodaidzein racemase (L-DDRC) transformed R-dihydrodaidzein to S-dihydrodaidzein in Lactococcus sp. 20-92, which increased EQL formation [33]. In Eggerthella sp. YY7918, EGYY15790 showed high homology to the CDS of L-DDRC. For the production of EQL, another factor(s) may be needed, such as DDRC.
Regarding this point, the detailed reaction mechanism of EQL production from DZN has not been agreed upon among research groups, including such things as the stereoselectivity of DZNR and DHDR, importance of the racemase, and the enzymatic function of THDR. Further critical researches is necessary to resolve such issues.
Phylogenetic analysis
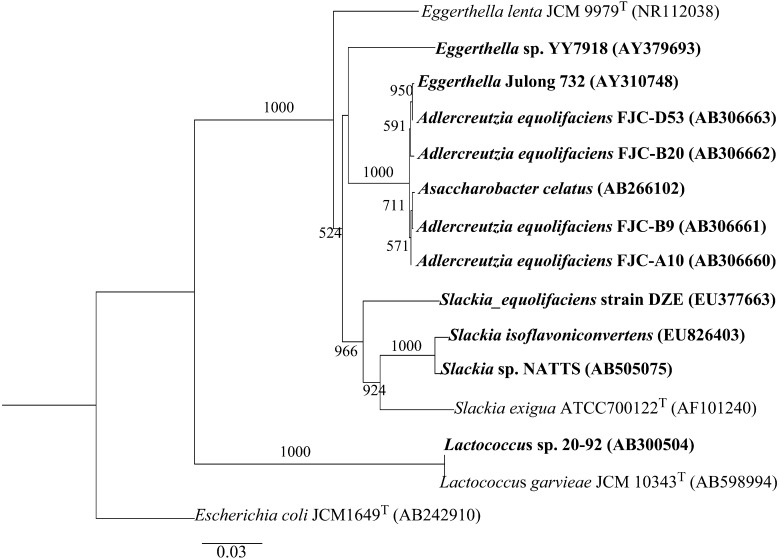
We performed a phylogenetic analysis of 16S rRNA sequences from EQL-producing bacteria. The type strains and E. coli as an out-group were also added to the analysis (Fig. 5). All EQL-producing bacteria, except for Lactococcus sp. 20-92, belong to the Coriobacteriaceae family. The amino acid sequences of DZNR, DHDR, and THDR exhibited high levels of identity (around 99%) with the corresponding amino acid sequences of Lactococcus sp. 20-92. The sequence identity of DZNR (43%) was lower than that for the other two enzymes (85–89%) in a comparation with Slackia sp. NATTS and S. isoflavoniconvertens. Eggerthella sp. YY7918 converted DZN and DHD into S-EQL but failed to metabolize glysitein or genistein [17]. In contrast, S. isoflavoniconvertens was able to reduce DZN and genistein [16]. DZNRs in bacterial species are diverse, exhibiting different enzymatic characteristics, such as substrate specificity and kinetics.
Fig. 5.
Neighbor-joining phylogenetic tree based on the 16S rRNA gene sequences of EQL-producing bacteria (bold) and related EQL-non-producing type strains of the Coriobacteriaceae and Streptococcaceae. A GenBank accession numbers are presented in parentheses. E. coli JCM1649 was used as an out-group. The type strains are represented by a superscript T. Numbers at branch points are values based on 1,000 bootstraps replicates.
The GC content of this gene cluster was 64%, consistent with that for the flanking region (Fig. 1), and somewhat higher than that for the entire genome (56.2%) of Eggerthella sp. YY7918. BLASTP analyses revealed that the homologous genes exist in the franking region with a conserved orientation (Fig. 6). This conserved gene-cluster of EQL-producing bacteria consists of DDRC (EGYY15790) [33], putative electron transfer flavoprotein (EGYY15770) and glutamate synthase (EGYY15700). Schröder et al. showed that these proteins were concurrently expressed in S. isoflavonicovertens using a proteome analysis [23]. It is possible that these conserved clusters play an unknown physiological role in these EQL-producing bacteria, in addition to the DZN-EQL pathway.
The BLASTP search against CDSs (EGYY15550-15820) revealed that only the genes of high-GC contents bacteria, such as Slackia, Adlercreutzia, Bifidobacterium, or Gordonibacter had high score.
The high amino acid sequence identity (99%) between the gene clusters of Eggerthella sp. YY7918 and Lactococcus sp. 20-92 suggested that these clusters are likely closely related in termes of evolution. In comparison with the GC content of the Lactococcus genome (< 40%), Eggerthella sp. YY7918 could be considered to have an extraordinarily high GC content (56%). No other Lactobacilli or Firmicutes have been reported to contain EQL-producing genes. In contrast, the GC content of the three EQL-producing genes of Lactococcus sp. 20-92 was 68%, while the genomic GC content was around 39% in other Lactococcus strains, such as L. garvieae [34, 35]. These results suggest that this region was horizontally transferred from high-GC bacteria, including Coriobacteriaceae, to Lactococcus species via horizontal transmission, as noted by Schröder et al. [22].
According to our findings, the gene cluster associated with EQL-production likely evolved in the Coriobacteriaceae, which are often found in the guts of animals. In our hypothesis, three independent oxidoreductase genes that encode the prototypes of DZNR, DHDR, and THDR and the flanking genes have gathered and evolved into the metabolic system of isoflavone and its derivatives DZN, DHD, THD, and EQL. The physiological function of these enzymes and their metabolites in these bacteria remain unclear. However, under some specific conditions, the identified gene cluster might provide some survival advantage to the bacteria and/or its host animals.
Acknowledgments
The authors thank Drs. M. Hattori and K. Oshima (The University of Tokyo) for their help with the genomic analysis. This work was supported by JSPS KAKENHI Grant Number 25450098, and a grant from the Tojuro Iijima Foundation for Food Science and Technology (2014).
REFERENCES
- 1.Bowey E, Adlercreutz H, Rowland I. 2003. Metabolism of isoflavones and lignans by the gut microflora: a study in germ-free and human flora associated rats. Food Chem Toxicol 41: 631–636. [DOI] [PubMed] [Google Scholar]
- 2.Joannou GE, Kelly GE, Reeder AY, Waring M, Nelson C. 1995. A urinary profile study of dietary phytoestrogens. The identification and mode of metabolism of new isoflavonoids. J Steroid Biochem Mol Biol 54: 167–184. [DOI] [PubMed] [Google Scholar]
- 3.Hur H, Rafii F. 2000. Biotransformation of the isoflavonoids biochanin A, formononetin, and glycitein by Eubacterium limosum. FEMS Microbiol Lett 192: 21–25. [DOI] [PubMed] [Google Scholar]
- 4.Willard ST, Frawley LS. 1998. Phytoestrogens have agonistic and combinatorial effects on estrogen-responsive gene expression in MCF-7 human breast cancer cells. Endocrine 8: 117–121. [DOI] [PubMed] [Google Scholar]
- 5.Tousen Y, Ezaki J, Fujii Y, Ueno T, Nishimuta M, Ishimi Y. 2011. Natural S-equol decreases bone resorption in postmenopausal, non-equol-producing Japanese women: a pilot randomized, placebo-controlled trial. Menopause 18: 563–574. [DOI] [PubMed] [Google Scholar]
- 6.Jackson RL, Greiwe JS, Desai PB, Schwen RJ. 2011. Single-dose and steady-state pharmacokinetic studies of S-equol, a potent nonhormonal, estrogen receptor β-agonist being developed for the treatment of menopausal symptoms. Menopause 18: 185–193. [PubMed] [Google Scholar]
- 7.Aso T, Uchiyama S, Matsumura Y, Taguchi M, Nozaki M, Takamatsu K, Ishizuka B, Kubota T, Mizunuma H, Ohta H. 2012. A natural S-equol supplement alleviates hot flushes and other menopausal symptoms in equol nonproducing postmenopausal Japanese women. J Womens Health (Larchmt) 21: 92–100. [DOI] [PubMed] [Google Scholar]
- 8.Utian WH, Jones M, Setchell KDR. 2015. S-equol: a potential nonhormonal agent for menopause-related symptom relief. J Womens Health (Larchmt) 24: 200–208. [DOI] [PubMed] [Google Scholar]
- 9.Muthyala RS, Ju YH, Sheng S, Williams LD, Doerge DR, Katzenellenbogen BS, Helferich WG, Katzenellenbogen JA. 2004. Equol, a natural estrogenic metabolite from soy isoflavones: convenient preparation and resolution of R- and S-equols and their differing binding and biological activity through estrogen receptors alpha and beta. Bioorg Med Chem 12: 1559–1567. [DOI] [PubMed] [Google Scholar]
- 10.Arai Y, Uehara M, Sato Y, Kimira M, Eboshida A, Adlercreutz H, Watanabe S. 2000. Comparison of isoflavones among dietary intake, plasma concentration and urinary excretion for accurate estimation of phytoestrogen intake. J Epidemiol 10: 127–135. [DOI] [PubMed] [Google Scholar]
- 11.Joannou GE, Kelly GE, Reeder AY, Waring M, Nelson C. 1995. A urinary profile study of dietary phytoestrogens. The identification and mode of metabolism of new isoflavonoids. J Steroid Biochem Mol Biol 54: 167–184. [DOI] [PubMed] [Google Scholar]
- 12.Wang XL, Hur HG, Lee JH, Kim KT, Kim SI. 2005. Enantioselective synthesis of S-equol from dihydrodaidzein by a newly isolated anaerobic human intestinal bacterium. Appl Environ Microbiol 71: 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minamida K, Tanaka M, Abe A, Sone T, Tomita F, Hara H, Asano K. 2006. Production of equol from daidzein by gram-positive rod-shaped bacterium isolated from rat intestine. J Biosci Bioeng 102: 247–250. [DOI] [PubMed] [Google Scholar]
- 14.Uchiyama S, Ueno T, Suzuki T. 2007. Identification of a newly isolated equol-producing lactic acid bacterium from the human feces. J Intest Microbiol 21: 217–220(in Japanese). [Google Scholar]
- 15.Maruo T, Sakamoto M, Ito C, Toda T, Benno Y. 2008. Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus Eggerthella. Int J Syst Evol Microbiol 58: 1221–1227. [DOI] [PubMed] [Google Scholar]
- 16.Yokoyama S, Suzuki T. 2008. Isolation and characterization of a novel equol-producing bacterium from human feces. Biosci Biotechnol Biochem 72: 2660–2666. [DOI] [PubMed] [Google Scholar]
- 17.Tsuji H, Moriyama K, Nomoto K, Miyanaga N, Akaza H. 2010. Isolation and characterization of the equol-producing bacterium Slackia sp. strain NATTS. Arch Microbiol 192: 279–287. [DOI] [PubMed] [Google Scholar]
- 18.Matthies A, Clavel T, Gütschow M, Engst W, Haller D, Blaut M, Braune A. 2008. Conversion of daidzein and genistein by an anaerobic bacterium newly isolated from the mouse intestine. Appl Environ Microbiol 74: 4847–4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yokoyama S, Oshima K, Nomura I, Hattori M, Suzuki T. 2011. Complete genomic sequence of the equol-producing bacterium Eggerthella sp. strain YY7918, isolated from adult human intestine. J Bacteriol 193: 5570–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimada Y, Yasuda S, Takahashi M, Hayashi T, Miyazawa N, Sato I, Abiru Y, Uchiyama S, Hishigaki H. 2010. Cloning and expression of a novel NADP(H)-dependent daidzein reductase, an enzyme involved in the metabolism of daidzein, from equol-producing Lactococcus strain 20-92. Appl Environ Microbiol 76: 5892–5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimada Y, Takahashi M, Miyazawa N, Ohtani T, Abiru Y, Uchiyama S, Hishigaki H. 2011. Identification of two novel reductases involved in equol biosynthesis in Lactococcus strain 20-92. J Mol Microbiol Biotechnol 21: 160–172. [DOI] [PubMed] [Google Scholar]
- 22.Tsuji H, Moriyama K, Nomoto K, Akaza H. 2012. Identification of an enzyme system for daidzein-to-equol conversion in Slackia sp. strain NATTS. Appl Environ Microbiol 78: 1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schröder C, Matthies A, Engst W, Blaut M, Braune A. 2013. Identification and expression of genes involved in the conversion of daidzein and genistein by the equol-forming bacterium Slackia isoflavoniconvertens. Appl Environ Microbiol 79: 3494–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wähälä K, Koskimies JK, Mesilaakso M, Salakka A, Leino TK, Adlercreutz H. 1997. The synthesis, structure, and anticancer activity of cis - and trans-4ʹ, 7-dihydroxyisoflavan-4-ols. J Org Chem 62: 7690–7693. [Google Scholar]
- 25.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 26.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 27.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall M, Stueckler C, Ehammer H, Pointner E, Oberdorfer G, Gruber K, Hauer B, Stuermer R, Kroutil W, Macheroux P, Fabe K. 2008. Asymmetric bioreduction of C=C bonds using enoate reductases OPR1, OPR3 and YqjM: enzyme-based stereocontro. Adv Synth Catal 350: 411–418. [Google Scholar]
- 30.Tanaka N, Nonaka T, Nakanishi M, Deyashiki Y, Hara A, Mitsui Y. 1996. Crystal structure of the ternary complex of mouse lung carbonyl reductase at 1.8 A resolution: the structural origin of coenzyme specificity in the short-chain dehydrogenase/reductase family. Structure 4: 33–45. [DOI] [PubMed] [Google Scholar]
- 31.Goris T, Schubert T, Gadkari J, Wubet T, Tarkka M, Buscot F, Adrian L, Diekert G. 2014. Insights into organohalide respiration and the versatile catabolism of Sulfurospirillum multivorans gained from comparative genomics and physiological studies. Environ Microbiol 16: 3562–3580. [DOI] [PubMed] [Google Scholar]
- 32.Lancaster CRD, Simon J. 2002. Succinate:quinone oxidoreductases from ε-proteobacteria. Biochim Biophys Acta 1553: 84–101. [DOI] [PubMed] [Google Scholar]
- 33.Shimada Y, Takahashi M, Miyazawa N, Abiru Y, Uchiyama S, Hishigaki H. 2012. Identification of a novel dihydrodaidzein racemase essential for biosynthesis of equol from daidzein in Lactococcus sp. strain 20-92. Appl Environ Microbiol 78: 4902–4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reimundo P, Pignatelli M, Alcaraz LD, D’Auria G, Moya A, Guijarro JA. 2011. Genome sequence of Lactococcus garvieae UNIUD074, isolated in Italy from a lactococcosis outbreak. J Bacteriol 193: 3684–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morita H, Toh H, Oshima K, Yoshizaki M, Kawanishi M, Nakaya K, Suzuki T, Miyauchi E, Ishii Y, Tanabe S, Murakami M, Hattori M. 2011. Complete genome sequence and comparative analysis of the fish pathogen Lactococcus garvieae. PLoS ONE 6: e23184. [DOI] [PMC free article] [PubMed] [Google Scholar]