Abstract
Streptococcus agalactiae (group B Streptococcus, GBS) is the predominant cause of early-onset infectious disease in neonates and is responsible for life-threatening infections in elderly and immunocompromised individuals. Clinical manifestations of GBS infection include sepsis, pneumonia, and meningitis. Here, we describe BspA, a deviant antigen I/II family polypeptide that confers adhesive properties linked to pathogenesis in GBS. Heterologous expression of BspA on the surface of the non-adherent bacterium Lactococcus lactis confers adherence to scavenger receptor gp340, human vaginal epithelium, and to the fungus Candida albicans. Complementary crystallographic and biophysical characterization of BspA reveal a novel β-sandwich adhesion domain and unique asparagine-dependent super-helical stalk. Collectively, these findings establish a new bacterial adhesin structure that has in effect been hijacked by a pathogenic Streptococcus species to provide competitive advantage in human mucosal infections.
Keywords: adhesin, bacterial adhesion, bacterial pathogenesis, Candida albicans, protein conformation, Streptococcus, structural model, structure-function, x-ray crystallography, AgI/II family polypeptide
Introduction
Streptococcus agalactiae (group B Streptococcus (GBS)4) is a commensal of the gastrointestinal tract and part of the normal microbiota of the female rectovaginal tract, where it is carried asymptomatically by ∼10–40% of women of childbearing age (1). However, as an opportunistic pathogen, GBS is the leading cause of neonatal meningitis and sepsis in the developed world. The predominant route by which GBS is transmitted to infants is from the mother, either during birth or following breach of the placental barrier in utero. Antenatal GBS screening strategies are therefore commonly used to identify colonized women, who then receive antimicrobial prophylaxis. Nevertheless, GBS remains a major cause of morbidity and mortality in infants worldwide (2).
Because maternal GBS colonization is the primary risk factor for vertical transmission of neonatal infection (3), there has been considerable focus on the mechanisms underlying GBS colonization of the female genitourinary (GU) tract. A number of putative colonization determinants have been identified, including the following: αC protein, mediating entry of GBS into cervical epithelial cells (4); pili, shown to contribute to vaginal attachment (5); and Srr-1, which binds keratin-4 (5) and fibrinogen (6) on the surface of vaginal epithelium. In addition, GBS expresses numerous extracellular matrix-binding proteins, including C5a peptidase (ScpB) and FbsA, which may promote attachment to mucosal tissues of the GU tract (7).
Antigen I/II (AgI/II) family polypeptide adhesins are found widely across the Streptococcus genus and have been best characterized for those streptococci indigenous to the oral cavity (8). In silico analyses have recently revealed the presence of genes encoding AgI/II family polypeptides in GBS, which we have designated here group B Streptococcus surface proteins (Bsp). These proteins conform to a conserved primary structure consisting of seven distinct regions (Fig. 1). The N-terminal region comprises a signal (leader) peptide, an N-terminal domain, and an alanine-rich repeat region (A). The C-terminal region consists of a proline-rich repeat sequence (P) followed by a C-terminal domain (C) and an LPXTG motif required for bacterial cell wall anchorage. The A and P regions flank a central variable region (V), which exhibits the greatest strain-to-strain sequence variation between streptococci and is thought to confer ligand-binding specificity (8). Structural characterization of AgI/II family proteins from Streptococcus mutans (SpaP) and Streptococcus gordonii (SspB) has revealed a distinctive extended fibrillar structure that projects outwards from the wall-proximal C domain (Fig. 1). The stalk is generated by a high affinity interaction between the α-helix and polyproline II (PPII)-helices of the A and P domains, respectively, serving to present the V “head” domain at its tip (9). The N terminus forms a stabilizing scaffold by wrapping behind the base of the stalk (10). Crystal structures of the V regions of SpaP and SspB have revealed a common architecture consisting of a lectin-like fold with a putative binding cleft (11, 12). The structure of the C-terminal subdomains C1, C2, and C3 has also been elucidated by x-ray crystallography and found to consist of β-sandwich domains stabilized by isopeptide bonds (13–16).
FIGURE 1.
A, model structure of Bsp family proteins based on the proposed domain organization of other AgI/II polypeptides (8, 9). This comprises a stalk consisting of the α-helical A domain and the polyproline II (PPII) helical P domain, separating the V domain and the C-terminal domains. The C-terminal domain is followed by the LPXTG motif required for cell wall anchorage. B, amino sequence alignment of BspA, BspB, BspC, and BspD. Structural regions are colored as in A with amino acids conserved in all four proteins highlighted in black.
AgI/II family polypeptides are multifunctional adhesins that serve as important colonization determinants for oral streptococci, mediating binding to host substrata and to other resident microbes. A common receptor for AgI/II polypeptides is glycoprotein-340 (gp340), an innate immunity scavenger receptor produced at mucosal surfaces (17). Fluid-phase gp340, in saliva and other mucosal secretions, agglutinates microbes for clearance from the body, whereas solid-phase gp340 adsorbed onto host surfaces provides a substratum for microbial adherence (18). AgI/II family polypeptides have also been shown to promote microbial community development by binding to oral bacteria Actinomyces oris (19–21) or Porphyromonas gingivalis (22, 23), as well as to the pathogenic fungus Candida albicans (24). More recently, an AgI/II family polypeptide (group A Streptococcus surface protein, AspA) has been described in group A Streptococcus (GAS) (25). The gene encoding AspA is located within an integrative and conjugative element (ICE) designated region of difference 2 (RD2), which is found among GAS serotypes implicated in puerperal fever (26, 27). There is strong evidence that RD2 originated in GBS and was acquired by GAS through horizontal gene transfer. It has been proposed that genes carried within RD2 might contribute to both GAS and GBS pathogenicity (28, 29). Supporting this, AspA has been shown to facilitate GAS biofilm formation on salivary pellicle (25), respiratory infection, and evasion of phagocytosis (30). Given this precedent, we hypothesized that Bsp polypeptides may serve as important host colonization determinants of GBS.
In this study, we introduce four similar, but non-identical, Bsp family proteins that are distributed among GBS of different capsular serotypes. Crystallographic and biophysical characterization of isolated functional domains from one of these proteins (BspA) identifies unique structural features that distinguish it as an AgI/II family deviant. These findings are of significance in directing the development of new vaccines or anti-infective agents that selectively target GBS, while not impacting the commensal microbiota.
Results
Distribution of AgI/II Polypeptides in GBS
In silico analyses were undertaken to determine the prevalence of bsp genes across GBS, using the completed GBS genome sequences available on NCBI. The bsp genes were found in five strains (summarized in Table 1), and each was present on an RD2-like ICE (28). Based upon primary sequence identities, four homologues of the bsp gene were identified, which we designated bspA–D (Fig. 1B). BspA shares 90.35% amino acid (aa) sequence identity with BspB, and both homologues were found in strain NEM316, which carries one copy of bspA and three copies of bspB. The remaining GBS strains carry either bspC or bspD. The proteins encoded by these genes share 99.52–99.76% aa sequence identity, with the only major difference being that BspD is missing a leader peptide (39–41-aa residues) for targeting to the Sec translocation machinery. BspA/B homologues can be separated from BspC/D homologues based upon the presence of two additional sequences in BspA/B homologues, one in the A domain (51-aa residues) and the second in the P domain (25-aa residues). There was no correlation between GBS capsular serotype and bsp homologue distribution.
TABLE 1.
Distribution of AgI/II family polypeptides in GBS
| AgI/II gene | Strain (serotype) | Previous locus tag | New locus tag |
|---|---|---|---|
| bspA | NEM316 (III) | gbs1143 | GBS_RS06095 |
| bspB | NEM316 (III) | gbs0393 | GBS_RS02350 |
| gbs0723 | GBS_RS04015 | ||
| gbs0986 | GBS_RS05325 | ||
| bspC | 515 (Ia) | SAL_2056 | SAL_RS04265 |
| COH1 (III) | SAN_2127 | GBSCOH1_1801 | |
| bspD | 2603V/R (V) | SAG_2021 | |
| 18RS21 (II) | SAJ_1924 |
BspA Promotes Binding to Immobilized gp340
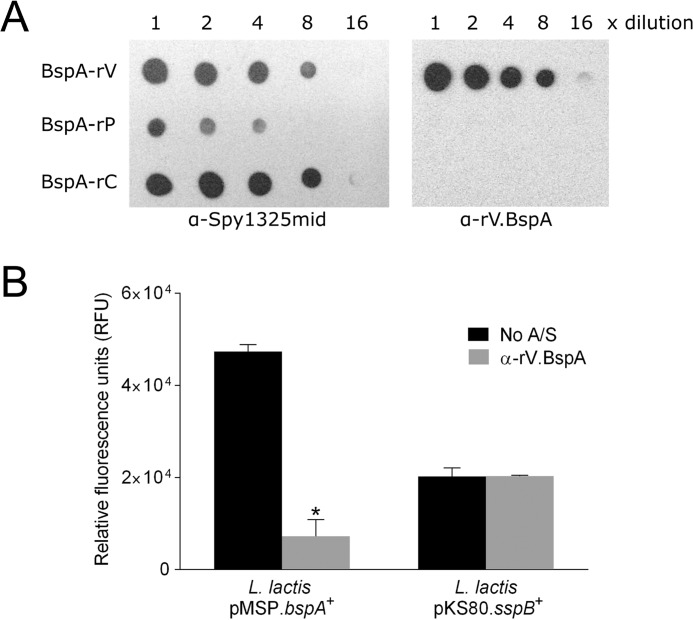
To investigate the functions of BspA as a putative colonization determinant in the absence of confounding problems due to adhesin redundancy in GBS, Lactococcus lactis was utilized as a heterologous expression strain. The bspA gene was cloned under the control of a nisin-inducible promoter into vector pMSP (31), enabling expression of BspA to be regulated by the concentration of nisin added to the growth medium. Expression of BspA on the surface of L. lactis following nisin induction was confirmed by immuno-dot blot analysis (Fig. 2A) with α-Spy1325mid antibodies to Streptococcus pyogenes AspA mid-region (25), which shares 63% aa sequence identity across the corresponding VPC(473–990) region of BspA.
FIGURE 2.
Interactions of BspA with immobilized gp340. A, dot immunoblot to verify the expression of BspA on the surface of L. lactis NZ9800. Lactococci were cultured for 16 h in the absence (uninduced) or presence of 10 or 100 ng/ml nisin to induce BspA expression. Lactococcal suspensions (A600 nm 2.0) were serially 2-fold diluted and spotted onto nitrocellulose membrane. The blot was probed with α-Spy1325 mid antibodies to the V region of AspA (25). B, binding of L. lactis pMSP.bspA+ or vector control (pMSP) to immobilized gp340. Induced or uninduced cells were then incubated in microwells coated with 50 ng of gp340 (black column) or blocking agent BSA (gray column) for 2 h at 37 °C. Nonadherent cells were removed, and total biomass was measured by crystal violet staining. Values given represent mean ± S.D. of three independent experiments performed in triplicate. *, p < 0.005 relative to vector control, calculated using an unpaired Student's t test.
A primary host factor recognized by members of the AgI/II polypeptide family in oral streptococci and GAS is gp340, a mucin-like glycoprotein associated with the surface of mucosal tissues (17). When gp340 becomes adsorbed onto a surface, exposed cryptitopes can provide receptors for microbial adherence. To examine the interaction of BspA with gp340, binding levels of L. lactis expressing BspA (pMSP.bspA+) to immobilized gp340 were compared with L. lactis containing empty vector (pMSP), using a crystal violet spectrophotometric assay. Adhesion levels of L. lactis pMSP.bspA+ were significantly (p < 0.005) higher than those of L. lactis containing empty vector (Fig. 2B). Furthermore, a dose-dependent response in binding to gp340 by BspA was seen with increasing nisin concentration. By contrast, neither L. lactis-expressing BspA nor empty vector adhered to the blocking agent, bovine serum albumin (BSA).
There is evidence that the head (V) regions of SspB, SpaP, and AspA facilitate gp340 binding activity (9, 25, 32). To determine whether the V domain of BspA plays a similar role in mediating binding to immobilized gp340, anti-V domain antibodies were affinity-purified from α-Spy1325mid antiserum (Fig. 3A). These antibodies (designated α-rV.BspA) were then investigated for the capacity to impair adherence of L. lactis pMSP.bspA+ to immobilized gp340. L. lactis expressing the AgI/II family polypeptide SspB from Streptococcus gordonii was included as a control to confirm antiserum specificity. Lactococcal cells were labeled with FITC, and binding to gp340 was measured using a fluorescence plate reader. Adherence of L. lactis pMSP.bspA+ cells to immobilized gp340 was significantly (p < 0.0001) impaired by α-rV.BspA, which reduced binding by 85% relative to the no antiserum control (Fig. 3B). Importantly, this inhibition was specific to the reactivity of α-rV.BspA antibody with BspA, as α-rV.BspA had no significant inhibitory effect on adherence of L. lactis pKS80.sspB+ to immobilized gp340 (Fig. 3B).
FIGURE 3.
Antibody inhibition binding studies. A, affinity purification of α-rV.BspA antibodies. Antibodies reactive against recombinant BspA-V (α-rV.BspA) were affinity-purified from antibodies raised against AspA (α-Spy1325mid). Purified α-rV.BspA antibodies were used to probe dot immunoblots of serially diluted (1:2) recombinant proteins corresponding to BspA-V, -P, and -C domains. Purified antibodies reacted only with BspA-rV. B, effects of affinity-purified antibody α-rV.BspA on adherence of L. lactis expressing BspA to immobilized gp340. L. lactis pMSP.bspA+ cells were FITC-labeled, preincubated without antibody or with 2.4 μg α-rV.BspA, and then incubated in microwells coated with 50 ng of gp340 for 2 h at 37 °C. Nonadherent cells were removed, and relative numbers of attached cells measured in a fluorescence plate reader. L. lactis pKS80.sspB+ expressing S. gordonii SspB was included as a control. Values given represent the mean ± S.D. of two independent experiments performed in duplicate. *, p < 0.0001 relative to no antiserum (A/S) control, calculated using an unpaired Student's t test.
BspA Interacts with C. albicans
Previous studies have shown that SspB expressed by S. gordonii, an integral member of the human oral microbiota, interacts with C. albicans hyphal cell wall Als3 protein, promoting the development of mixed species communities (24). C. albicans co-exists with GBS in the lower female GU tract, and several studies have observed recurrent co-isolation of GBS with C. albicans (33–35). To confirm the capacity for GBS to interact with C. albicans, FITC-labeled GBS NEM316 cells were incubated in suspension with hypha-forming cells of C. albicans SC5314 labeled with calcofluor white, and aggregates were visualized by fluorescence microscopy. As reported for a number of oral streptococci (24, 36–38), GBS NEM316 was shown to intimately interact with candidal cells, preferentially targeting C. albicans hyphal filaments rather than blastospores (Fig. 4A). To determine whether BspA was involved in promoting this association of GBS with C. albicans, the interactions of L. lactis expressing BspA or vector control (pMSP) with C. albicans were compared. L. lactis pMSP cells exhibited few interactions with C. albicans (Fig. 4B). Similarly, uninduced L. lactis pMSP.bspA+ cells associated only sparsely with candidal hyphae (Fig. 4C). By contrast, L. lactis cells induced to express BspA showed high affinity for C. albicans and preferentially bound along the lengths of the hyphal filaments (Fig. 4C).
FIGURE 4.
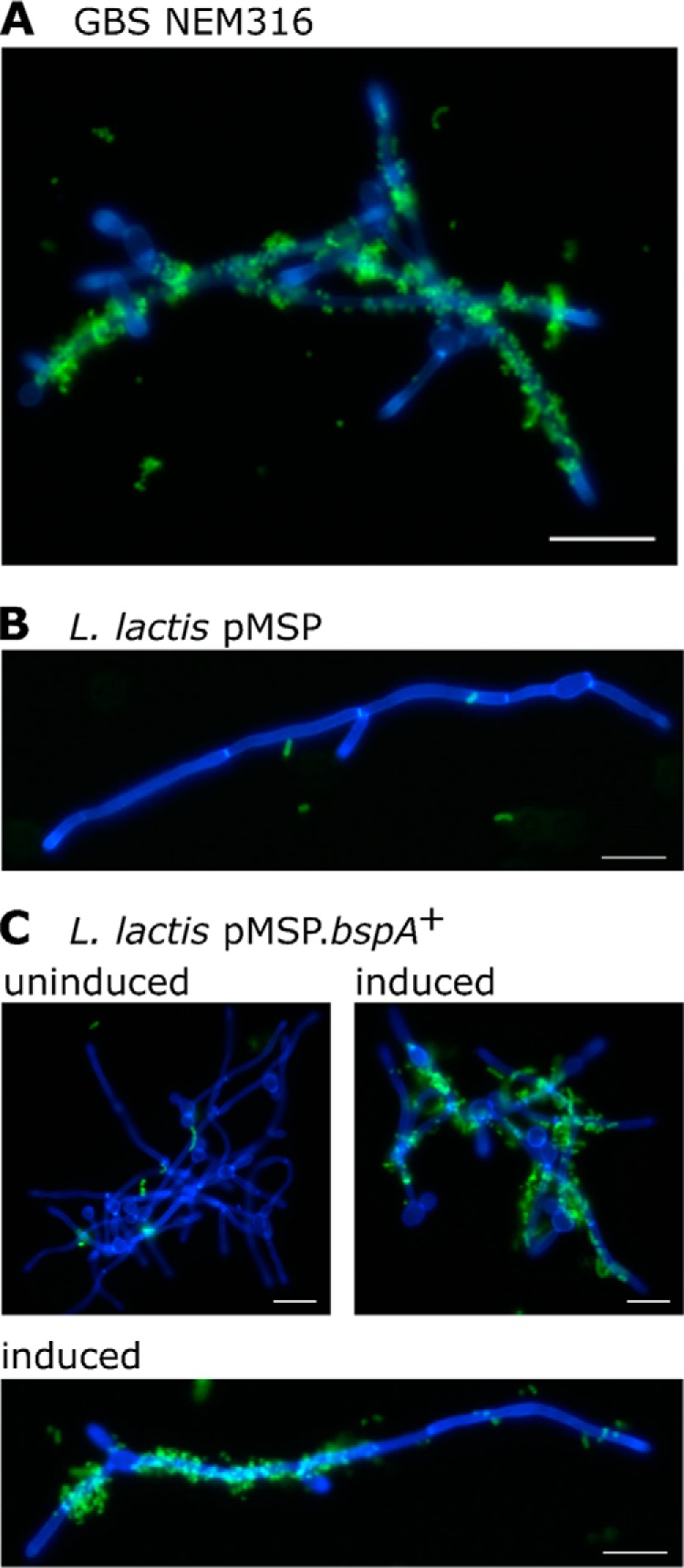
Interactions of GBS or lactococci expressing BspA with C. albicans. A, GBS NEM316 cells; B, L. lactis pMSP vector control; or C, L. lactis pMSP.bspA+. L. lactis strains were cultured for 16 h in the absence (uninduced) or presence of 100 ng/ml nisin to induce BspA expression. GBS or L. lactis cells were FITC-labeled and incubated in suspension with hypha-forming cells of C. albicans stained with calcofluor white. Aggregates were visualized by fluorescence microscopy. Scale bars, 20 μm.
BspA Promotes Adhesion to Human Vaginal Epithelium
GBS colonization of vaginal and cervical epithelia is the primary risk factor for vertical transmission of neonatal infection (3). To determine whether BspA was a candidate for mediating GBS adherence to epithelial cells, binding levels of L. lactis pMSP.bspA+ to a vaginal epithelial cell line, VK2/E6E7, were compared with those of L. lactis pMSP vector control. Lactococci were incubated with VK2/E6E7 monolayers and numbers of associated bacteria enumerated by viable count. Numbers of L. lactis cells expressing BspA associating with VK2/E6E7 monolayers were significantly (p < 0.05) higher than vector control (Fig. 5), implying that BspA mediates adhesion to vaginal epithelial cells.
FIGURE 5.
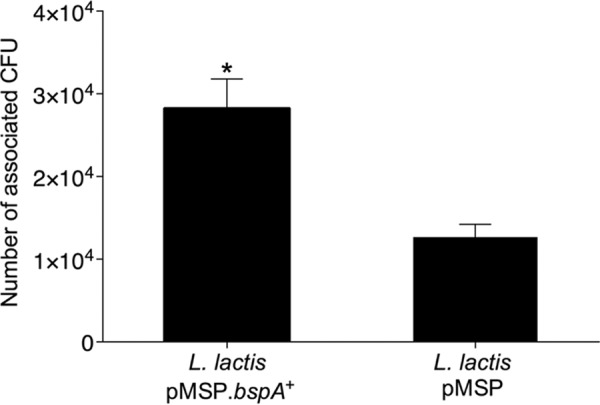
Interactions of L. lactis pMSP.bspA+ or pMSP vector control with VK2/E6E7 vaginal epithelial cell line. Lactococci (multiplicity of infection 5) were incubated with monolayers of VK2/E6E7 cells and numbers of associated CFU enumerated from serial dilutions of recovered cell lysates. Values given represent mean ± S.D. of four independent experiments performed in triplicate. *, p < 0.05 relative to vector control, calculated using a paired Student's t test.
BspA Possesses a Truncated C-terminal Domain That Lacks Bound Metal Ions
To gain further insight into the adhesive properties of BspA, we sought to structurally characterize this protein. Initially, we focused our efforts on the BspA C-terminal domain (BspA-C, 328-aa residues). Although our whole cell binding studies identify the BspA variable domain as being responsible for gp340 binding by this polypeptide, a role for the C-terminal domain of AgI/II family polypeptides in target binding has been suggested by others (9, 15, 39). Despite being significantly shorter (328-aa residues as compared with 502–508 aa in other AgI/II family proteins), the BspA-C domain exhibits a high degree of sequence identity to equivalent regions in other AgI/II family proteins, indicative of analogous function. The crystal structures of BspA-C and a more readily crystallizable point mutant of this protein, BspA-CG744D (both comprising residues 554–881 of full-length BspA), were determined by molecular replacement in the space groups C2122 and P212121 respectively. The structure of BspA-CG744D was determined to 1.8 Å resolution using the SspB-C(1083–1413) crystal structure (PDB code 2WOY, 33% sequence identity) (13) as a molecular replacement search model and refined to an Rwork/Rfree of 19.6:23.4%. The structure of BspA-C domain was determined to 2.2 Å resolution using the BspA-CG744D structure as a molecular replacement search model and refined to a Rwork/Rfree of 21.8:23.9%. For both BspA-CG744D and BspA-C, the asymmetric unit comprises a single monomer of each protein. Crystal packing analysis reveals that Asp-744 of BspA-CG744D forms a salt bridge with Lys-855 in symmetry-related molecules. This interaction is likely to account for the increased propensity of BspA-CG744D to crystallize as compared with BspA–C. There are no significant structural differences between BspA-CG744D and BspA-C structures, with a Cα r.m.s.d. between the two structures of 0.5 Å (255 atoms).
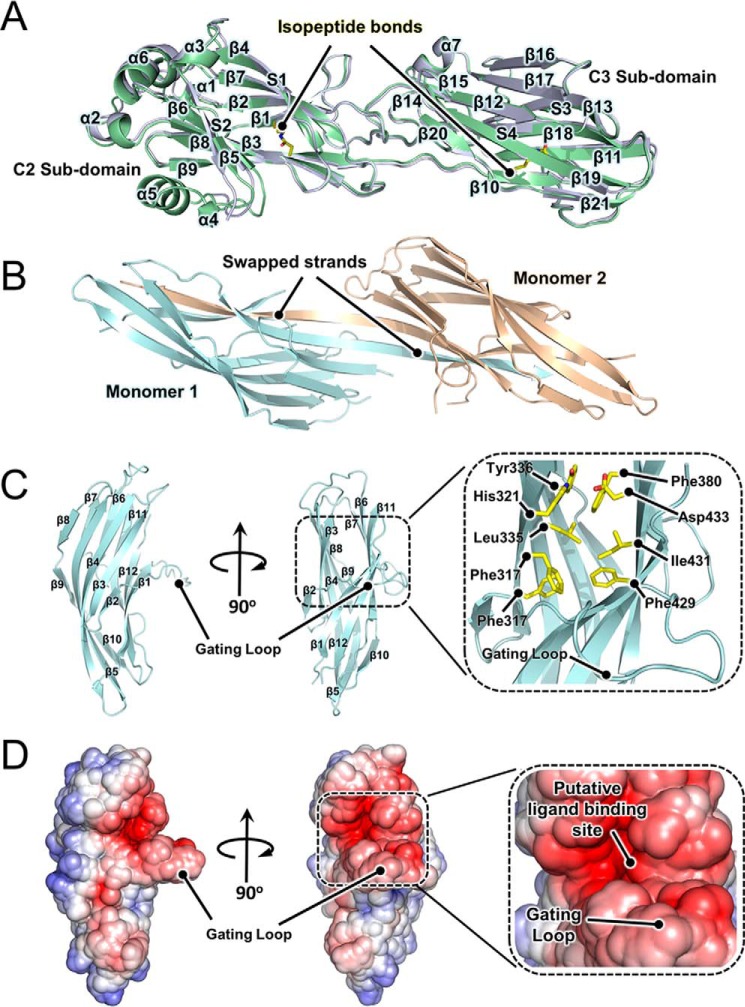
The final models of BspA-CG744D (334-aa residues, including six residues of the N-terminal vector-encoded hexapeptide) and BspA-C (311-aa residues, lacking aa residues 640–657) are presented in Fig. 6A and comprise a pair of compact subdomains (residues 554–725 and 726–881 of full-length BspA) fused by a linker. The two subdomains are equivalent to those termed C2 and C3 in other AgI/II family polypeptides (13–16). The absence of a C1 subdomain, a feature that is present in all other AgI/II family proteins characterized to date, accounts for the size disparity between BspA-C domain and other AgI/II family C domains. The C1-C2 interface region has been suggested as having a role in carbohydrate binding in the S. mutans AgI/II family protein SpaP (15). Its absence in BspA, and by inference from sequence analysis of BspB-D (Fig. 1B), reveals that this interface region is not required for target ligand recognition or binding in Bsp family members.
FIGURE 6.
Crystal structures of BspA-V and -C domains. A, superposition of the crystal structures of BspA-CG744D (green) and BspC (blue). The location of the two isopeptide bonds are highlighted. B, crystal structure of the BspA-V strand-swapped homodimer. C, crystal structure of a BspA-V monomer. The domain adopts a β-sandwich fold, consisting of two anti-parallel sheets made up of five and seven strands, respectively. The N- and C-terminal strands meet to connect the V domain with A and P domains, respectively. A close-up view of the BspA-V target binding pocket is provided as an inset. The putative gating loop is labeled. For clarity, residues within the gating loop for which compelling electron density was not observed (Asp-298 and Asn-299) have been excluded from the structure. D, electrostatic surface representation of BspA-V mirroring the views of the protein shown in C. Electrostatic potentials were calculated using APBS (69).
The two BspA-C subdomains consist of two central β-sheets, S1 and S2 in C2, and S3 and S4 in C3. S1 and S2 comprise four and five strands, respectively. Strands β2 and β3 are connected by two short helices α1 and α2. Helix α6 runs parallel to α1 and α2 and connects β6 and β7. Helix α5 protrudes from the core of the C2 subdomain connecting strands β5 and β6. Similarly to S1 and S2, the β-sheets S3 and S4 contain four and five strands, respectively. Long loops connect strands at the interface of the C2 and C3 subdomains. Helix α7 connects strands β17 and β18. Strands β13 and β16 are connected by two short anti-parallel strands β14 and β15. Connecting loops are shorter on the opposite side of the sandwich, except for a long coiled region connecting β10 and β11. C2 and C3 are connected by a long loop between strands β9 and β10. Protein/protein interactions at the C2-C3 interface region are stabilized by a series of hydrogen bonds. Both C2 and C3 possess a single isopeptide bond (Fig. 6A). In C2 this is formed between the side chains Lys-556 (NZ) and Asn-703 (CG) and fuses S1 to S2. In C3 isopeptide bond formation occurs between Lys-730 (NZ) and Asn-861 (CG), linking S3 to S4.
Previously reported crystal structures of AgI/II family polypeptide C-terminal domains have identified the presence of bound metal ions, which have been suggested to play a role in enhancing stability and conferring adhesive properties (13–16). By contrast, no bound metal ions are observed in the crystal structures of BspA-CG744D or BspA-C. BspA-C2 subdomain does possess a putative unoccupied metal-binding site, equivalently positioned to those reported in SpaP-C2, SspB-C2, and AspA-C2. This site lies at the interface of the loop regions that connect β3 to β4 and β6 to α5 and comprises the residues Gln-607, Pro-608, Val-659 and Glu-661. There is notable sequence disparity in the region of BspA-C3 that corresponds to the SpaP-C3 metal ion-binding site. The residues Asp-1388 and Gln-1391 in SpaP-C3, which act to coordinate a bound calcium ion, are found to be histidines in BspA-C3 (His-777 and His-780). In the AspA-C3 crystal structure (16) a single histidine substitution at the second of these positions (His-1205) appears sufficient to preclude metal ion binding.
The BspA Variable Domain Possesses a β-Sandwich Fold That Is Structurally Distinct from Other AgI/II Family Polypeptides
Having established the role of the BspA variable domain in gp340 binding, we next sought to determine the crystal structure of this region of the protein. The structure of the BspA variable domain (BspA-V, comprising aa residues 285–451 of the full-length protein) was determined in space group C2 using the single anomalous dispersion technique, as applied to crystals of SeMet-labeled BspA-V. The structure was determined to a resolution of 2.4 Å and refined to an Rwork/Rfree of 23.5:26.2%. The asymmetric unit contains two copies of BspA-V arranged in the form of a strand-swapped homodimer (Fig. 6B). BspA dimerization results from the exchange of the C-terminal β-strand (residues 436–606) of each monomer within the dimer. In full-length BspA, the N and C termini of the V domain are fused to their neighboring A and P domains (Fig. 1A), an organization that would preclude BspA-V domain strand-swapping and implies that dimerization is an artifact of crystallization. The positioning of the exchanged C-terminal strands are, however, indicative of their location within the parent monomer. There are no significant structural differences between the monomers within the dimer, with an r.m.s.d. Cα between the two of 0.9 Å (141 atoms). Convincing electron density was not observed for residues 14–15 in monomer 1 and 11–20 and 88–90 in monomer 2, and as such these regions have been excluded from our final model.
BspA-V adopts a fold that is distinct from those reported for other AgI/II family polypeptide V domains (11, 12). This consists of two anti-parallel β-sheets, S1 and S2, comprising 5 and 7 strands, respectively (Fig. 6C). The two sheets are interconnected by four loops, and the strands are connected by five β-hairpins. There are 10 β-bulges in the structure (three G1 bulges, two wide bulges, and five classic β-bulges), of which six occur between strands, and four within β-hairpins. The six inter-strand β-bulges contribute to shaping the long central strands (β4, β5, β11, and β12) of the protein. In its monomeric form, the N- and C-terminal strands of BspA-V (β1 and β12) would converge and be optimally positioned to connect the V domain with its partner A and P domains.
An extended 17-aa residue linker between β1 and β2 projects outward from the surface of the protein exposing a large solvent-accessible pocket (∼500 Å3, as determined by CASTp (40)) located at the S1-S2 interface (Fig. 6, C and D). The pocket is populated by a combination of hydrophobic and negatively charged residues, consistent with a role in binding carbohydrates or glycoproteins, and is peripherally decorated with polar and negatively charged amino acids. Given the absence of any additional cavities, clefts, or highly charged surfaces, it is likely that this pocket constitutes the ligand-binding site of BspA-V domain. The lack of compelling electron density for a number of the residues that constitute the loop linking strands β1 and β2 implies that this region of the protein is flexible, suggesting that it may play a role in gating access to the BspA-V ligand binding pocket (Fig. 6, C and D).
DALI analysis (41) identifies a single functionally analogous structural homologue of BspA-V domain, the carbohydrate-binding protein RbmA from Vibrio cholerae (42). RbmA is a dimeric protein that promotes biofilm scaffolding via protein/carbohydrate interactions. In contrast to BspA-V, however, carbohydrate binding by RbmA is proposed to occur at either the interface of the RbmA dimer or within a positively charged ordered loop present in each monomer. Both these structural features are absent in BspA-V implying that the two proteins employ functionally distinct binding mechanisms.
BspA-A and -P Domains Form an Anti-parallel Coiled-coil Super-helix Stabilized through an Asparagine-dependent Hydrogen Bond Network
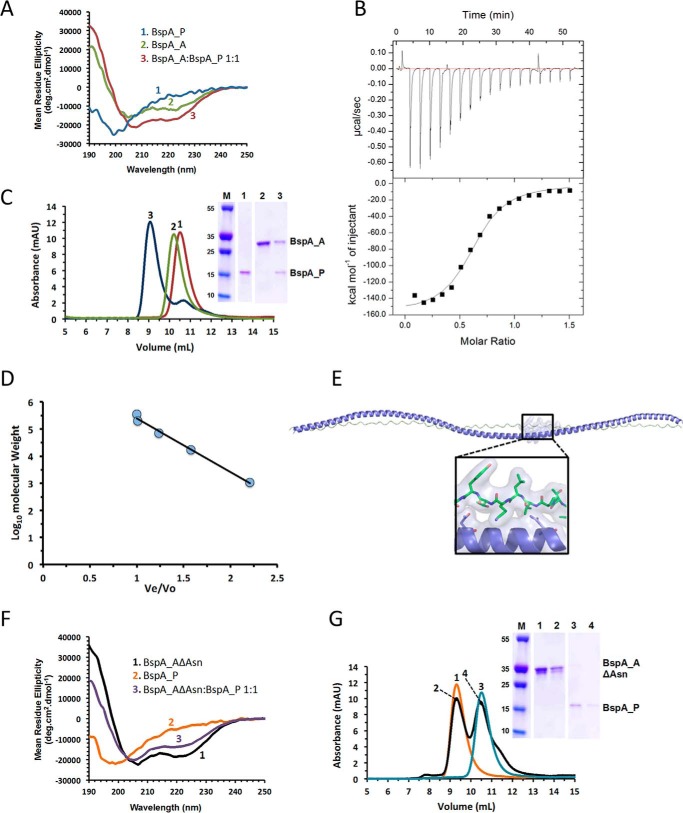
Having elucidated the molecular structures of the BspA-V and -C domains, we next turned our attention to the A and P domains of this protein. A common structural feature of AgI/II family polypeptides is the presence of an extended antiparallel coiled-coil super-helix resulting from association of their A and P domains (9). This stalk-like structure serves to project the V domain of the protein away from the bacterial cell surface, where the N and C domains are located, and in previously characterized AgI/II proteins arises due to the complementary “knobs-into-holes” packing of hydrophobic residues within the alanine-rich A domain and PXXP motifs present in the polyproline P domain (9). Bioinformatic analysis of the A and P domains from Bsp proteins demonstrates that these regions are significantly shorter than those of other AgI/II family polypeptides (117–168 residues for Bsp-A domains and 77–102 residues for Bsp-P domains) and differ considerably in amino acid composition. Most notably Bsp-A domains are largely devoid of the AXYXA(LV) heptad motif identified in SpaP and conserved in AspA and SspB (8). In an effort to reconcile these features, the A and P domains of BspA were produced recombinantly, and their ability to associate in vitro was examined. Initially, circular dichroism (CD) spectroscopy was used to monitor BspA-A·P association and assess changes in helicity resulting from coiled-coil formation. The CD spectrum of BspA-A is consistent with this domain adopting a predominantly α-helical structure, with >55% helicity as predicted using CDSSTR (43–47). BspA-P domain exhibits a CD spectrum consistent with an unfolded protein. Sequential titration of BspA-A into BspA-P, as monitored by CD spectroscopy, demonstrates a stepwise recovery in helicity, consistent with the formation of a coiled-coil structure (Fig. 7A). To further characterize the BspA-A·P interaction, isothermal titration calorimetry (ITC) was used to monitor the thermodynamic parameters of BspA-A·P formation. Experiments titrating BspA-A into BspA-P were performed (Fig. 7B). These analyses demonstrate that the BspA-A·P interaction is predominantly enthalpically driven with ΔG = −38 ± 4 kJ/mol, Ka = 4.3 ± 0.7 μm−1, Kd = 270 ± 3 nm, ΔH = −660 ± 18 kJ/mol, ΔS = −2.1 kJ/mol/K, and n = 0.62 ± 0.01, at a temperature of 298 K.
FIGURE 7.
Biophysical characterization of BspA-A·P complex formation. A, CD spectroscopy of BspA-A, BspA-P, and an equimolar mixture of both proteins. B, ITC analysis monitoring the titration of BspA-A into BspA-P. C, SEC elution profiles of BspA-A, BspA-P, and the BspA-A·P complex. D, SEC column calibration curve calculated using Bio-Rad protein standards (thyroglobulin, 670 kDa; γ-globulin, 158 kDa; ovalbumin, 44 kDa; myoglobin, 17 kDa; vitamin B12, 1.35 kDa). E, molecular model of the BspA-A·P complex highlighting the role of BspA-A domain (blue) asparagine residues in mediating interaction with BspA-P (green). F, CD spectroscopy of BspA-AΔAsn, BspA-P, and an equimolar mixture of both proteins. G, SEC elution profiles of BspA-AΔAsn, BspA-P, and an equimolar mixture of both proteins.
Next, we sought to demonstrate that BspA-A·P domain mixing results in the formation of a single tightly associated A·P complex. To investigate this, the hydrodynamic properties of BspA-A, BspA-P, and BspA-A·P domains were assessed using SEC (Fig. 7C). Comparisons of the elution profiles of recombinant BspA-A and BspA-P with those of proteins of known molecular mass (Fig. 7D) are consistent with masses of >30 and >20 kDa, respectively, significantly larger than those established by SDS-PAGE (Fig. 7C). However, given that SEC elution behavior is dictated by Stokes radius rather than molecular mass (48), the elution behaviors of BspA-A and BspA-P are consistent with both proteins being single species of extended conformation in solution. A BspA-A·P complex, generated by mixing BspA-A and BspA-P in near-equimolar quantities, yields a single species that elutes prior to both BspA-A and BspA-P domains. SDS-PAGE analysis of this species confirms the presence of both BspA-A and BspA-P domains in this species (Fig. 7C). Together our spectroscopic, thermodynamic, and hydrodynamic data are fully consistent with the ability of BspA-A and BspA-P domains to self-assemble to form a super-helical structure.
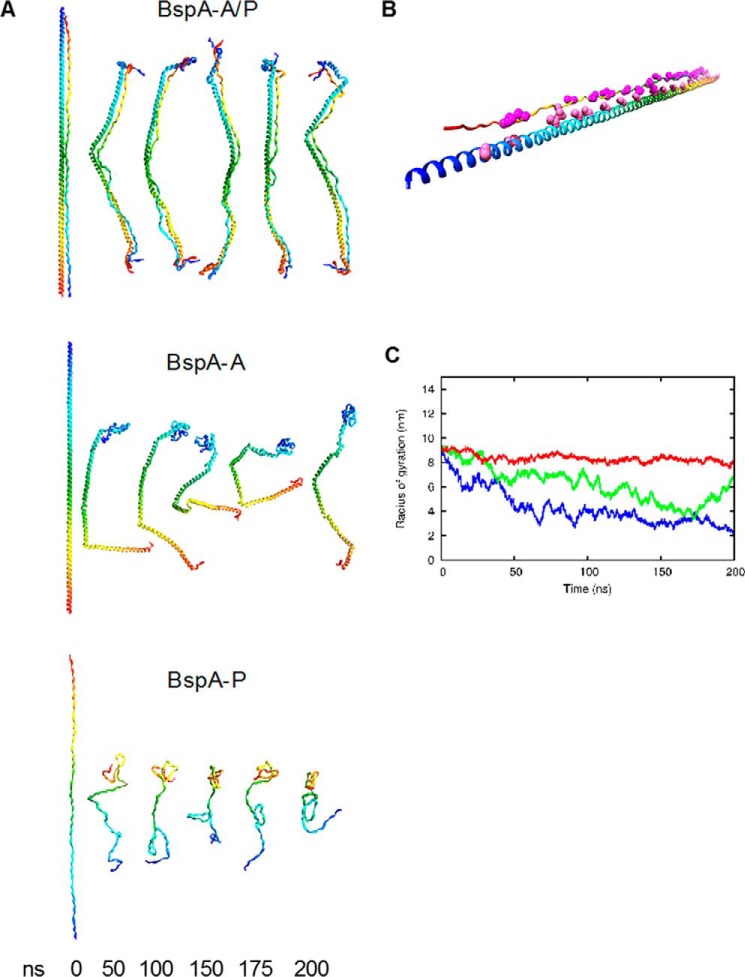
In an effort to investigate the molecular basis of BspA-A·P complex formation, a molecular model of this assembly was generated (Fig. 7E). Inspection of this model suggests the BspA-A·P helical assembly is mediated largely via an asparagine seam, present on the A domain, which forms a series of complementary hydrogen bond interactions to main chain oxygen and nitrogen atoms within BspA-P domain. The BspA-A·P model remains extended over 200 ns of molecular dynamics simulations (Fig. 8). To test the validity of our model, an asparagine-less mutant of BspA-A domain was generated (BspA-AΔAsn) within which all 19 asparagines were substituted to alanines. BspA-AΔAsn exhibits a CD spectrum nearly identical to that of BspA-A, demonstrating that the helical structure of BspA-AΔAsn is not perturbed by the Asp-Ala substitutions (Fig. 7F). Titration of BspA-AΔAsn into BspA-P, monitored using CD spectroscopy, reveals no increase in helicity. Similarly, ITC (data not shown) and SEC analyses (Fig. 7G) provide no evidence of BspA-AΔAsn·P interaction or complex formation, illustrating the critical role of the BspA-A domain asparagine seam in BspA-A·P assembly.
FIGURE 8.
Molecular dynamics simulations of the model of BspA-A·P, BspA-A, and BspA-P. A, structures at time points during the simulations. The dimeric BspA-A·P model behaves as a semi-rigid rod during the simulation, whereas the BspA-A α-helix shows kinking and partial unfolding of the helix. The PPII BspA-P rapidly collapses into a random coil. Ribbons are rainbow colored blue N terminus through red C terminus. B, initial model. The alignment of most of the asparagine residues in BspA-A toward the backbone of BspA-P is shown. Asn residues, pink spheres; Pro residues, magenta spheres. C, plots of radius of gyration with respect to time. BspA-A·P, red; BspA-A, green; BspA-P, blue.
Discussion
GBS is a leading cause of severe invasive bacterial infection and represents a significant global health and economic burden (2). Studies of the machineries employed by this bacterium to invade and colonize human hosts are thus of both broad fundamental interest and potential significance for informing the design of new therapeutic agents or interventions. AgI/II family polypeptides are important colonization determinants of oral viridans streptococci and have been identified in GBS (49, 50). Our bioinformatic analyses demonstrate that five GBS strains, representing four capsular serotypes, carry genes encoding AgI/II family polypeptides, which we here designate as Bsp proteins. These data support a recent study in which the presence of ICEs carrying a bsp gene was shown to be prevalent across a collection of 303 GBS strains derived from 11 host species, including humans, cattle, and fish (50). Given the capacity for ICEs to undergo sequence deletions during mobilization, the frequency with which an intact bsp gene has been acquired by these strains supports the hypothesis and our findings here that Bsp proteins convey an important competitive advantage to GBS. Analysis of bsp gene sequences suggests that they conform to the canonical AgI/II family N-A-V-P-C domain architecture. Nonetheless, the significant sequence variations exhibited by Bsp proteins, as compared with other AgI/II family polypeptides (8), imply a disparity in both domain size and architecture and hint at a diversification of function.
AgI/II family polypeptides have been shown to play important roles in host colonization by facilitating binding interactions with epithelial cell surfaces (30, 51, 52). The ability of the archetypal Bsp family member BspA to facilitate binding to vaginal epithelial cells when presented on the surface of non-adherent bacterium L. lactis suggests a role for BspA, and perhaps other Bsp proteins, in facilitating host cell adherence and colonization. This has implications for disease risk associated with GBS carriage. Furthermore, a potential target for mucosal tissue colonization is identified as innate immune component gp340. BspA was shown to bind immobilized gp340 via a mechanism that appeared to be dependent upon the BspA-V domain. Vaginal epithelial cells express high levels of cell-associated gp340, and gp340 targeting as a bridging molecule to support epithelial cell interactions has already been shown for HIV (53). Likewise, attachment of GBS pili to immobilized gp340 within the oral cavity has been proposed as a potential mechanism to promote GBS colonization of the oropharynx of adults and neonates, an initial step in progression of invasive diseases (25, 54). BspA-mediated interactions with immobilized gp340 may therefore represent an important mechanism for facilitating GBS colonization of infection-relevant sites.
The AgI/II family polypeptides of oral streptococci have been shown to promote bacterial cell interactions with other resident microbes, a process fundamental to the accretion of oral microbial biofilm communities. Likewise, colonization of the GU tract by the resident microbiota results from the formation of polymicrobial communities on the vaginal epithelium (55). This study provides the first evidence of interkingdom interactions between GBS and C. albicans. Given the reported co-isolation of these two species (33–35), candidal/GBS interactions may serve as an important mechanism to promote co-colonization of the GU tract. BspA-expressing L. lactis cells were shown to interact with C. albicans, binding preferentially to hyphal filaments. AgI/II family polypeptide SspB promotes S. gordonii binding to C. albicans hyphae via recognition of the hypha-specific adhesin Als3 (24). A similar molecular mechanism may underpin the targeting of BspA to C. albicans hyphae.
Our demonstration of the potential role of Bsp proteins in facilitating GBS colonization encouraged us to probe in greater detail the molecular determinants of target recognition by Bsp polypeptides. Although they retain the same general architecture as other AgI/II family proteins, the aa sequences of Bsp proteins suggested functional divergence. Our structural studies provide evidence for a mechanism of target binding that is distinct from other AgI/II family adhesins. The C-terminal domain of BspA lacks a discrete C1 subdomain, proposed to enable the binding of carbohydrates or glycoproteins via the C1-C2 interface. Furthermore, the absence of bound metal ions, suggested in other AgI/II family polypeptide C-terminal domains to affect stability and adhesive properties (13–16), may indicate altered binding behavior. Although it is not possible to discount metal binding capability entirely based on our crystallographic studies, variations in aa composition within the C2 and C3 metal-binding sites are consistent with loss of this function. As Bsp-V domain antibodies block BspA binding to gp340, it appears unlikely that the Bsp-C domain makes any significant contribution to mediate this interaction.
Our proposed role of the BspA-V domain in target binding is consistent with studies of other AgI/II family polypeptides, where the V domain has been identified as a major determinant of adherence (8). The crystal structure of the BspA-V domain demonstrates that this domain adopts a fold that is distinct from those reported for other AgI/II family polypeptides, comprising an elaborated β-sandwich core, which exposes a sizeable binding pocket at the interface of its two constituent β-sheets. The BspA-V domain has no structural precedent among AgI/II family proteins or bacterial adhesins in general. The binding pocket of BspA is formed by a series of hydrophobic and negatively charged residues, consistent with a role in binding carbohydrates or glycosylated proteins. gp340 is a highly complex molecule designed to recognize many microbial ligands. As such, gp340 contains numerous N- and O-linked glycosylation sites together with multiple domains (SRCR and CUB) within the protein backbone (56). Given the architecture of the BspA-V domain, it is likely that gp340 recognition occurs at a site distinct to those targeted by other AgI/II family polypeptides. The role of Bsp proteins as multifunctional adhesins correlates with our findings that BspA targets gp340, vaginal epithelium and C. albicans, and is further supported by the recent evidence that BspC may contribute to biofilm formation (50).
Like other AgI/II family polypeptides, Bsp proteins retain a coiled-coil super-helical A·P domain stalk structure. In vitro assembly of the A·P domain complex unambiguously demonstrates that recombinant BspA-A and BspA-P domains self-assemble to form a discrete BspA-A·P species, consistent with an elongated super-helix. However, Bsp proteins lack the defining sequence features observed in SpaP, SspB, and AspA (9) that are involved in stabilizing the stalk via hydrophobic interactions. Our modeling studies highlight that the proposed assembly mechanism of SpaP and SspB A·P super-helices is not applicable to BspA. By contrast, BspA-A·P assembly is facilitated through a hydrogen bond network primarily contributed to by asparagine residues present in the BspA-A domain. The key role of these residues is further evidenced by the inability of an asparagine-less BspA-A domain mutant to associate with BspA-P domain in vitro. As a result of shorter A·P stalk length and the absence of the C1 domain, BspA is predicted to be significantly shorter (25–35 nm) than SpaP (>50 nm) (9). AgI/II family polypeptides in oral streptococcal species have been proposed to act as longer distance adhesins, making initial contacts with host surfaces (8). The shorter length of BspA may reflect that in GBS a two-step process occurs, whereby longer appendages carrying adhesins, e.g. pili, establish primary host contacts, enabling BspA to make secondary interactions over shorter distances.
In summary, this study identifies BspA as an important adhesin of GBS that could play a pivotal role in facilitating host colonization. BspA binds to epithelial cell surfaces and to another opportunistic member of the human microbiota, C. albicans. The prototypical Bsp family member, BspA, is shown to possess a novel molecular architecture distinct from other AgI/II family proteins, including a unique functional variable domain with no structural precedent. This divergent structural configuration could be a key to assisting adhesion and survival of GBS in the presence of commensal streptococci or fungi. Moreover, the structural and functional information that we describe for BspA can be developed toward designing novel therapeutics, not only against GBS but also with potential to impact colonization by C. albicans. Vaccines or pharmaceuticals that worked efficiently by targeting this conserved antigen specific to GBS might therefore restrict host colonization or control invasive disease by these two opportunistic pathogens.
Experimental Procedures
Bacterial Strains and Growth Conditions
Strains used in this study are outlined in Table 2. Escherichia coli K12 MG1655 was used for molecular cloning experiments to generate pMSP7517-based constructs (abbreviated pMSP), and StellarTM competent cells were used to generate pOPINF-based constructs, and E. coli BL21(DE3) was used for expression of recombinant proteins. The methionine auxotroph E. coli B834(DE3) was employed for expression of selenomethionine (SeMet)-labeled proteins. All E. coli strains were cultured in Luria Bertani (LB) medium at 37 °C, with shaking at 220 rpm. Streptococcus strains were cultured in Todd-Hewitt broth containing 0.5% yeast extract (THY). All Streptococcus strains were grown at 37 °C in an atmosphere of 5% CO2. L. lactis strains were cultured in M17 broth supplemented with 0.5% glucose (GM17) in a candle jar at 30 °C. Lactococci carrying nisin-inducible pMSP-based constructs were subcultured into GM17 medium containing 10–100 ng/ml nisin. C. albicans SC5314 was cultivated on Sabouraud dextrose agar (SAB) at 37 °C for 48 h. Single colonies were used to inoculate YPD broth (2% yeast extract, 4% mycological peptone, and 4% glucose), and cells were cultured at 37 °C with shaking at 220 rpm. Media were supplemented when necessary with 100 μg/ml ampicillin (Amp100) or 300 μg/ml erythromycin (Erm300) for E. coli; and 5 μg/ml erythromycin (Erm5) for lactococci.
TABLE 2.
Microbial strains used in study
| Strain | Characteristics | Source |
|---|---|---|
| S. agalactiae | ||
| NEM316 | Wild-type (serotype III) | Laboratory stock |
| L. lactis | ||
| NZ9800 | Wild type | Laboratory stock |
| UB2658 | pMSP.bspA+ | This study |
| UB2635 | Empty pMSP | This study |
| C. albicans | ||
| SC5314 | Wild-type | Gillum et al. (70) |
| E. coli | ||
| K12 MG1655 | F− λ− rph-1 | Laboratory stock |
| StellarTM | F′ endA1 supE44 thi-1 recA1 relA1 gyrA96 phoA Φ80d lacZΔ M15 Δ (lacZYA-argF) U169 Δ (mrr-hsdRMS-mcrBC) ΔmcrA, λ− | Clontech |
| B834 (DE3) | F− ompT hsdSB (rB− mB−) gal dcm met (DE3) | Novagen |
| BL21 (DE3) | F− ompT hsdSB (rB− mB−) gal dcm (DE3) | Novagen |
| UB2878 | pOPINF.bspA+; full-length; BL21 | This study |
| UB2879 | pOPINF.bspA-V+; BL21 | This study |
| UB2880 | pOPINF.bspA-C+; 2231G>A; BL21 | This study |
| UB2881 | pOPINF.bspA-C+; BL21 | This study |
| UB2882 | pOPINF.bspA-A+; BL21 | This study |
| UB2883 | pOPINF.bspA-AΔAsn+; BL21 | This study |
| UB2884 | pOPINF.bspA-P+; BL21 | This study |
Generation of L. lactis Heterologous Expression Strains
Primers were designed to amplify locus GBS_RS06095 corresponding to full-length BspA (Table 3). NcoI and XhoI restriction sites were incorporated at the termini of forward and reverse primers, respectively. PCR products were subcloned into nisin-inducible expression vector pMSP (31) via NcoI and XhoI sites, generating plasmid pMSP.bspA+. The construct was transformed directly into electrocompetent L. lactis NZ9800, prepared by culturing in GM17G broth (GM17 broth supplemented with 2.5% glycine) at 30 °C to A600 nm 0.5–0.6. Cultures were incubated on ice for 10 min, harvested by centrifugation (3000 × g, 4 °C, 10 min), and suspended in wash buffer (0.5 m glucose, 10% glycerol). Cells were electroporated in chilled electroporation cuvettes (2-mm electrode gap) using a Gene Pulser (Bio-Rad) set at 2.5 kV voltage, 25 microfarad capacitance, and 400 ohm resistance. Transformants were recovered in SGM17MC medium (GM17 supplemented with 0.5 m sucrose, 20 mm MgCl2, and 2 mm CaCl2), plated onto GM17 agar supplemented with 5 μg/ml erythromycin and incubated for 48 h at 30 °C in a candle jar. Transformants were confirmed by plasmid isolation and PCR, and expression of BspA in L. lactis was verified by dot immunoblot.
TABLE 3.
Primers used in study
| Protein product | Primer name | Sequencea |
|---|---|---|
| BspA(1–932) | pMSP.bspA.F | CATGCCATGGAGGAGGAAACAATATGAATTCAC |
| pMSP.bspA.R | CCGCTCGAGGCAACCCGATTATGAGAGG | |
| BspA(39–881) | bspA-N.F | AAGTTCTGTTTCAGGGCCCGGCTGATCAAGTTACAACTCAAGCA |
| bspA-C.R | ATGGTCTAGAAAGCTTTACTATTGCTCTCCTGTGGATGGTAA | |
| BspA-V(285–451) | bspA-V.F | AAGTTCTGTTTCAGGGCCCGAATGTTGCTTTTGACATTAAAGCC |
| bspA-V.R | ATGGTCTAGAAAGCTTTACTAATTATTATTAACATAAGACACTAAAGC | |
| BspA-C(554–881) | bspA-C.F | AAGTTCTGTTTCAGGGCCCGCCAACTAAGAAAGTTCTAGATGAA |
| bspA-C.R | ATGGTCTAGAAAGCTTTACTATTGCTCTCCTGTGGATGGTAA | |
| BspA-A(117–284) | bspA-A.F | AAGTTCTGTTTCAGGGCCCGATTAAAGATGCTACTGCTGAAAAT |
| bspA-A.R | ATGGTCTAGAAAGTCTTACTAATTAATAGACTGTTTATCTGCTTC | |
| BspA-P(452–553) | bspP-A.F | AAGTTCTGTTTCAGGGCCCGGATGCTGTTCCTAATGTGGTC |
| bspP-A.R | ATGGTCTAGAAAGCTTTACTATGTGGCCTTTGGTATCTGTAC | |
| BspA-AΔAsn | bspA-AΔN-F | AAGTTCTGTTTCAGGGCCCGATGGCGAAGAAAGCGGCT |
| bspA-AΔN-R | ATGGTCTAGAAAGCTTTATCAGGCAATCGACTGTTTATCGGCTTC |
a Restriction endonuclease sites are underlined. Sequences in bold indicate additional bases required for cloning into the ligation-independent expression vector pOPINF.
Dot Immunoblot
Expression of BspA on the surface of L. lactis was induced by culturing cells in the presence of 10–100 ng nisin/ml. Suspensions were adjusted to A600 nm 2.0 and spotted (2 μl) onto nitrocellulose membrane. The membrane was blocked with TBS (50 mm Tris-HCl, pH 7.2, 0.15 m NaCl) containing 10% (w/v) milk powder and subsequently probed with rabbit α-Spy1325mid (25) followed by swine anti-rabbit IgG-horseradish peroxidase (HRP) conjugate (Dako), both diluted 1:1000 into TBS supplemented with 0.1% Tween 20 and 1% milk powder. The membrane was then developed using Amersham BiosciencesTM ECLTM Western blotting analysis system (GE Healthcare) according to the manufacturer's instructions.
In Vitro gp340 Binding Assays
gp340 was prepared from parotid saliva samples pooled from multiple donors using a multistep procedure, including adsorption onto S. mutans cells, as described previously (18). Adherence of L. lactis cells expressing BspA to immobilized gp340 was performed by crystal violet assay and data converted to cell numbers as described previously (25).
Antibody Inhibition Assays
Antibodies reactive against BspA-V (α-rV.BspA) were affinity-purified from antibodies raised against AspA (α-Spy1325mid) (25) by a protocol adapted from Jenkinson and Easingwood (57). Nitrocellulose membrane disks (4 cm diameter) were soaked in 250 μl of recombinant BspA-V (25 mg/ml) and left to dry for 10 min. Disks were blocked by incubating in TBS containing 10% (w/v) milk (TBSm) for 1 h, then washed with TBS, and incubated for 3.5 h with 3.5 ml of α-Spy1325mid diluted 1:10 in TBSm. Residual antibody was collected for analysis, and disks were washed three times in TBS. The disks were incubated in 3.5 ml of 0.2 m glycine hydrochloride for 8 min at 25 °C and then neutralized with 1 m K2HPO4. Dialysis into PBS (50 mm Na2HPO4·NaH2PO4, pH 7.2, 0.15 m NaCl) was performed by concentrating antibodies with a 10-kDa concentrator (Vivaspin, GE Healthcare). Antibodies were stored at 4 °C. To check antibody specificity, recombinant proteins (see below) corresponding to BspA-V, BspA-P, or BspA-C domains (0.6 mg/ml) were 2-fold serially diluted and spotted onto nitrocellulose membrane. The blots were probed with antiserum α-Spy1325mid (1:1000) or affinity-purified α-rV.BspA (0.1 mg) and developed as described above.
Coating of microtiter plate wells with gp340 was as described previously (25). Lactococci were harvested (5000 × g, 7 min), washed in TBSC (TBS containing 5 mm CaCl2), resuspended in 1.5 mm fluorescein isothiocyanate (FITC) in carbonate buffer (50 mm Na2CO3, 100 mm NaCl), and incubated for 1 h at 22 °C in the dark with gentle agitation. Cells were harvested (5000 × g, 7 min), washed three times in carbonate buffer, and suspended in TBSC at A600 nm 1.0. FITC-labeled L. lactis cells were incubated with or without 12.5 μg of α-rV.BspA for 30 min at 25 °C. Suspensions (50 μl) were transferred to appropriate wells and incubated at 37 °C for 2 h. Wells were washed twice with TBS and fixed by adding 100 μl of 25% formaldehyde for 30 min at 25 °C. Formaldehyde was removed, and wells were washed twice with TBS before detection of binding levels using a fluorescence plate reader (excitation wavelength 485 nm and emission wavelength 535 nm).
C. albicans Binding Assays
After 16 h of growth in YPD, C. albicans cells were harvested (5000 × g, 5 min), washed, and resuspended in YNBPT (20 mm Na2HPO4·KH2PO4, pH 7.0, 1× yeast nitrogen base (YNB), 0.02% tryptone) at A600 nm 1.0. Adjusted cell suspensions (200 μl; 2 × 105 cells) were transferred to glass bijoux bottles containing 1.8 ml of YNBPTG (YNBPT supplemented with 0.4% glucose) and incubated at 37 °C, 220 rpm for 3 h to induce filamentation. Lactococci or GBS cells were harvested (5000 × g, 7 min), washed in YNBPT, FITC-labeled as described above, and suspended in YNBPTG at A600 nm 0.2. FITC-labeled bacteria (1 ml; 1 × 108 cells) were added to each bijoux containing hypha-producing C. albicans and incubated at 37 °C, 220 rpm for a further 1 h. Freshly prepared calcofluor white (1 μg/ml final concentration) was added to each suspension just prior to depositing co-cultures (10 μl) onto microscope slides and visualizing by fluorescence microscopy.
VK2/E6E7 Binding Assays
Immortalized human vaginal (VK2/E6E7) epithelial cell lines were obtained from American Type Culture Collection (ATCC CRL-2616) and maintained at 37 °C with 5% CO2 in keratinocyte serum-free medium (KSFM; Gibco®) supplemented with 0.1 ng/ml human recombinant epidermal growth factor, 0.05 mg/ml bovine pituitary extract, and 0.4 nm CaCl2. Bacterial association assays were performed as described by Sheen et al. (5), with minor modifications. In brief, VK2/E6E7 cells (105) were seeded into 24-well plates and grown to confluence. Monolayers were then washed and incubated with L. lactis suspensions at a multiplicity of infection of 5 for 2 h at 37 °C. Nonadherent bacteria were removed by washing, and the monolayers were dispersed with trypsin/EDTA (0.1 ml) and then lysed with 0.025% Triton X-100 in water (0.4 ml). Serial dilutions of this lysate were plated onto GM17 agar plates to enumerate associated bacterial colony-forming units (CFU).
Protein Expression and Purification
Primers were designed to amplify regions of locus GBS_RS06095 corresponding to full-length BspA (BspA(39–881)), V domain (BspA-V(285–451)), C-terminal domain (BspA-C(554–881)), A domain (BspA-A(117–284)), and P domain (BspA-P(452–553)), and also a synthetic gene (MWG Eurofins) encoding a variant of the BspA-A domain within which all asparagine residues were substituted to alanine residues (BspA-AΔAsn) (Table 3). PCR products from amplification of GBS strain NEM316 chromosomal DNA were cloned into the pOPINF expression vector using the In-FusionTM recombinase (Clontech), incorporating a His6 tag at the N terminus of the cloned product. pOPINF was linearized as described previously (58). pOPINF and PCR amplicons were purified using a NucleoSpin PCR clean-up kit (Clontech) and ligated as instructed by the manufacturer's protocol. All constructs were verified by DNA sequencing. A nucleotide substitution (2231G→A) was identified in bspA-C, resulting in a point mutation of G744D. This protein was designated BspA-CG744D. A new construct was subsequently generated lacking this mutation, which was designated BspA-C.
Resulting plasmids were transformed into competent E. coli BL21(DE3) cells. Expression strains were grown in 1 liter of LB medium to A600 nm 0.4–0.6, at which time isopropyl 1-thio-β-d-galactopyranoside (0.2 mm final concentration for BspA-V and BspA-C; 1 mm for BspA, BspA-A, BspA-AΔAsn, and BspA-P) was added, and the culture was incubated at 20 °C, 220 rpm for a further 16 h. Cells were harvested by centrifugation (5000 × g, 15 min, 4 °C), washed in His-Load buffer (20 mm imidazole, 50 mm Tris, 150 mm NaCl, pH 7.5), and cell pellets frozen at −80 °C.
For SeMet-substituted BspA-V, pOPINF-bspA-V was transformed into competent E. coli B834(DE3) cells. Cells were grown to A600 nm 0.4–0.6, harvested by centrifugation (5000 × g, 15 min, 4 °C), washed with distilled water to remove residual methionine, and resuspended in 5 ml of 50 mm Tris-HCl, pH 7.5. This suspension was then used to inoculate 1 liter of pre-warmed M9 medium containing 2.2% (w/v) SeMet Medium Base, 10.2% (w/v) Nutrient Mix, and 0.014% (w/v) SeMet (all Molecular Dimensions Ltd.). The culture was grown to A600 nm 0.6, and isopropyl 1-thio-β-d-galactopyranoside (0.2 mm final concentration) was added, and the culture was incubated at 37 °C with shaking for a further 16 h. Cells were harvested by centrifugation (5000 × g, 15 min, 4 °C) and washed in His-Load buffer supplemented with 1 mm tris(2-carboxyethyl)phosphine-HCl, pH 7.5, and the cell pellet was frozen at −80 °C.
Frozen cell pellets were resuspended in His-load buffer supplemented with Pierce Protease inhibitor mini-tablets (Thermo Scientific). Cells were lysed by sonication, and cellular debris was removed by centrifugation (20,000 × g, 30 min, 4 °C). Lysates were purified by nickel affinity chromatography using a HiTrap IMAC column (GE Healthcare), followed by SEC using either a High Load 16/60 Superdex 75 column (BspA-V, BspA-C; BspA-A, BspA-AΔAsn, BspA-P) or a Superdex 200 column (BspA) (both from GE Healthcare).
Protein Crystallization, Data Collection, and Processing
BspA-CG744D, BspA-C, and SeMet-labeled BspA-V were concentrated to 17, 23, and 25 mg/ml, respectively, in buffer consisting of 20 mm Tris, 150 mm NaCl, pH 7.5. SeMet-labeled BspA-V crystals were grown in 0.1 m Buffer System 1 at pH 6.5 (MES, imidazole), 0.1 m amino acids, and 37.5% MPD_P1K_P3350 (Morpheus Screen, Molecular Dimensions Ltd.). BspA-CG744D crystals were grown in 0.1 m MMT buffer (1:2:2 l-malic acid, MES, Tris-HCl, pH 6.0) and 25% (w/v) PEG 1500. BspA-C crystals were grown in 0.02 m sodium/potassium phosphate and 20% (w/v) PEG 3350. X-ray diffraction data were collected on beamlines I03 or I04 at Diamond Light Source, UK. Data for BspA-V and BspA-C were processed with MOSFLM (59). Data for BspA-CG744D were processed with XDS (60). All data collection and refinement statistics are provided in Table 4.
TABLE 4.
Crystallization, data collection, and refinement statistics
| BspA-VSe-Met, PDB code 5DZ8 | BspA-CG744D, PDB code 5DZ9 | BspA-CWT, PDB code 5DZA | |
|---|---|---|---|
| Data collection | |||
| Space group | C121 | P212121 | C2221 |
| Cell dimensions | |||
| a, b, c (Å) | 148.1, 34.4, 92.6 | 38.1, 48.2, 234.1 | 34.1, 206.2, 121.4 |
| α, β, γ (°) | 90.0, 127.3, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Wavelength Å | 0.979 | 0.979 | 0.920 |
| Resolution range (Å)a | 73.7–2.4 (2.5–2.4) | 58.5–1.9 (1.9–1.8) | 39.1–2.2 (2.3–2.2) |
| Rmerge | 0.06 (0.14) | 0.11 (0.84) | 0.09 (0.45) |
| I/σI | 19.9 (8.6) | 17.2 (3.4) | 9.7 (1.9) |
| CC1/2 (%) | 99.8 (98.8) | 99.7 (78.6) | 99.6 (76.4) |
| Completeness (%) | 99.6 (97.2) | 99.4 (96.4) | 92.3 (94.3) |
| Redundancy | 5.9 (4.8) | 12.3 (9.9) | 3.4 (3.5) |
| No. of selenium sites | 4 | ||
| Figure of merit | 0.25 | ||
| Refinement | |||
| Resolution (Å) | 73.7–2.4 | 55.5–1.9 | 102.9–2.2 |
| No. of reflections (working set/test set) | 87, 362/13, 981 | 436, 280/33, 734 | 69, 391/19, 496 |
| Rwork/Rfree | 23.5/26.2 | 19.6/23.4 | 21.8/23.9 |
| No. of atoms | 2566 | 2944 | 2505 |
| Protein | 2487 | 2624 | 2437 |
| Ligand/ion | 15 | 0 | 15 |
| Water | 64 | 320 | 53 |
| B-factors Å2 | |||
| Protein | 25 | 22 | 24 |
| Ligand | 30 | 31 | |
| Water | 29 | 32 | 24 |
| r.m.s.d. | |||
| Bond lengths (Å) | 0.02 | 0.02 | 0.02 |
| Bond angles (°) | 1.7 | 1.8 | 1.8 |
a Values in parentheses are for highest resolution shell.
Structure Determination
The structure of the BspA-V domain was determined using the single wavelength anomalous dispersion method as applied to SeMet-labeled crystals of BspA-V. Identification of heavy atom sites and the resulting initial phase calculation was carried out using PEHINIX AUTOSOL (61). This was followed by iterative rounds of manual model building and refinement using COOT (62) and REFMAC (63), the latter as implemented in the CCP4 suite of programs (64). The final model of BspA-V domain comprises two copies of the protein, each encompassing residues 285–451 of the native sequence, 64 water molecules, and 1 molecule each of acetate, diethylene glycol, and ethylene glycol, and 96.44% of residues are in the allowed or favored region of the Ramachandran plot with five outliers (Lys-295, Asp-325, Val-327, Asp-348, and Gln-370).
The structure of BspA-CG744D was determined by molecular replacement using PHASER (65) employing the S. gordonii SspB-C(1083–1413) (13) as a search model (PDB code 2WOY, 33% identity). The initial structure was subjected to iterative rounds of manual model building and refinement using COOT and REFMAC, respectively. The final model of BspA-CG744D includes one copy of the protein and 320 water molecules. 98.50% of residues are in the allowed or favored region of the Ramachandran plot; there were no outliers.
The structure of BspA-C domain was determined by molecular replacement using MOLREP (66) employing BspA-CG744D as a search model. Model building and refinement were performed as detailed above for BspA-CG744D. The final model of BspA-C includes one copy of the protein, 53 water molecules, 1 molecule of diethylene glycol, and 2 molecules of ethylene glycol; 96.10% of residues are in the allowed or favored region of the Ramachandran plot with one outlier (Asn-743).
Circular Dichroism Spectroscopy
CD spectra were collected using a Jasco 810 spectropolarimeter (Jasco, Tokyo, Japan) fitted with a Peltier temperature controller. All spectra were collected from 190 to 260 nm, using a 1-mm path length cuvette, at 20 °C, in buffer containing 20 mm sodium phosphate, 50 mm sodium fluoride, pH 7.5. All proteins were dialyzed into this buffer prior to experimentation. Titration experiments were conducted by the sequential addition of BspA-P in 9.1-μl increments to BspA-A or BspA-AΔAsn, to a final concentration of 12.3 μm BspA-A and 11.7 μm BspA-P, respectively. Final spectra were generated as averages of eight repeat scans, with appropriate protein-free buffer spectra subtracted. All data were plotted without smoothing using Excel. Spectra were analyzed using the DICHROWEB suite of programs (46, 47). Secondary structure predictions were performed using the CDSSTR method (43–47).
Isothermal Titration Calorimetry
Calorimetric titrations were performed using a MicroCal iTC200 system (GE Healthcare). The reaction cell (250 μl) contained BspA-A, BspA-AΔAsn, or BspA-P domains at concentrations ranging from 4 to 12 μm. The syringe (40 μl) contained BspA-A, BspA-AΔAsn, or BspA-P at concentrations ranging from 48 to 130 μm. Experiments were carried out in reaction buffer containing 20 mm sodium phosphate, 150 mm sodium chloride, pH 7.5, at 25 °C with a stirring speed of 800 rpm. All proteins were dialyzed into reaction buffer prior to experimentation. An initial injection of 0.4 μl (excluded from data fitting) was followed by 19 injections of 2.0 μl, separated by 180-s intervals. Data were fitted with a single binding site model using Origin software (version 7 with MicroCal ITC analysis module) permitting calculation of the thermodynamic properties ΔG, ΔH, ΔS, Ka, and stoichiometries (n) of the binding interactions. The enthalpy of dilution of the various titrants were measured by titration into buffer and found to be insignificant.
Analytical Size Exclusion Chromatography
Analytical SEC was performed using a Superdex 75 10/300 GL column (GE Healthcare) in buffer comprising 20 mm sodium phosphate, 150 mm NaCl, pH 7.5. 200-μl aliquots of each sample (20 μm) were loaded onto the column and eluted at 0.5 ml/min for 30 min. The absorbance of the column eluant was monitored at 280 nm throughout.
Molecular Modeling
The only template for homology modeling the BspA-A·P domain is the S. mutans adhesin SpaP (PDB code 3IOX). In this structure, the seam between the α-helical (A) and PPII (P) components is formed by stretches of Asn side chains interacting with the P backbone and stretches of Tyr residues interdigitating P proline side chains. The BspA-A·P domain is about three times longer than the corresponding region in the 3IOX structure, and the A domain is rich in Asn (10%) but poor in Tyr (2%). Mapping the sequence of BspA-A·P domain onto a template derived from three copies of the 3IOX A·P domain failed to capture many Asn to Pro backbone interactions due to supercoiling in the domain. Hence, an alternative approach was sought, and an appropriate template was constructed in the following manner. First, a regular α-helix was built with the sequence BspA-A Ala-71–Asn-288 and flanked by two turns of glycine residues, resulting in a linear seam, comprising the majority of Asn residues, along one face of the helix. Copies of a short Asn-rich section of the 3IOX A·P domain (Ala-420–Lys-446 and Pro-848–Val-861) were superimposed along the template helix, providing a guide to model the P domain from three copies of the 3IOX P domain with minimal adjustments of backbone torsion angles. The P domain sequence Asp-452–Thr-553 was also flanked by glycine residues, and 10 registers of this P sequence were threaded onto the template, side chains repacked with SCWRL (67), and the interaction energy of each thread evaluated with BUDE (68). The best energy model was prepared for energy calculations under the AMBER99-SB-ildn force field using the GROMACS tools to add hydrogen atoms consistent with pH 7, and surrounding the protein in a box of TIP3P waters containing 0.15 m sodium chloride. Molecular dynamics simulations (200 ns) were performed under periodic boundary conditions using PME electrostatics at 278 K and 1 bar as an NTP ensemble. The two components (A and P) were treated in the same way. Setup, simulations, and analysis were performed using GROMACS 4.6.7, structure processing and imaging with Chimera 1.9 and VMD 1.9.1.
Statistical Analysis
All data are reported as mean ± S.D. unless otherwise indicated. Significance between samples was determined using the two-tailed Student's t test, and a value of p < 0.05 was accepted as indicating significance. Data were analyzed with GraphPad Prism version 6 software.
Author Contributions
H. F. J., R. J. L., P. R. R., and A. H. N. conceived and designed the study. S. R., T. J. H., G. R. P., M. T., A. R., R. B. S., H. F. J., P. R. R., and A. H. N. performed experiments and analyzed data. S. R., H. F. J., R. J. L., P. R. R., and A. H. N. wrote the manuscript.
Acknowledgments
We thank Jane Brittan and Lindsay Dutton for technical assistance and Catherine Back for advice and discussions. We thank Nicklas Strömberg for provision of gp340, Shaynoor Dramsi for GBS strains, and Gary Dunny for plasmid pMSP7517.
This work was funded in part by Biotechnology and Biological Sciences Research Council Grant BB/I006478/I (to P. R. R.), National Institutes of Health Grants DE016690 (to H. F. J. and R. J. L.) and DE012505 (to R. J. L.), and Royal Society University Research Fellowship Award UF080534 (to P. R. R.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (codes 5DZ8, 5DZ9, and 5DZA) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GBS
- group B Streptococcus
- GAS
- group A Streptococcus
- Bsp
- group B Streptococcus surface protein
- PPII
- polyproline II
- gp340
- glycoprotein-340
- RD2
- region of difference 2
- ICE
- integrative conjugative element
- SEC
- size exclusion chromatography
- SeMet
- selenomethionine
- r.m.s.d.
- root-mean-square deviation
- ITC
- isothermal titration calorimetry
- PDB
- Protein Data Bank
- aa
- amino acid
- GU
- genitourinary
- AgI/II
- antigen I/II.
References
- 1. Berner R. (2002) Group B streptococci during pregnancy and infancy. Curr. Opin. Infect. Dis. 15, 307–313 [DOI] [PubMed] [Google Scholar]
- 2. Le Doare K., and Heath P. T. (2013) An overview of global GBS epidemiology. Vaccine 31, D7–D12 [DOI] [PubMed] [Google Scholar]
- 3. Verani J. R., McGee L., and Schrag S. J. (2010) Prevention of perinatal group B streptococcal disease-revised guidelines from CDC. MMWR Recomm. Rep. 59, 1–36 [PubMed] [Google Scholar]
- 4. Baron M. J., Bolduc G. R., Goldberg M. B., Aupérin T. C., and Madoff L. C. (2004) αC protein of group B Streptococcus binds host cell surface glycosaminoglycan and enters cells by an actin-dependent mechanism. J. Biol. Chem. 279, 24714–24723 [DOI] [PubMed] [Google Scholar]
- 5. Sheen T. R., Jimenez A., Wang N. Y., Banerjee A., van Sorge N. M., and Doran K. S. (2011) Serine-rich repeat proteins and pili promote Streptococcus agalactiae colonization of the vaginal tract. J. Bacteriol. 193, 6834–6842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang N. Y., Patras K. A., Seo H. S., Cavaco C. K., Rösler B., Neely M. N., Sullam P. M., and Doran K. S. (2014) Group B streptococcal serine-rich repeat proteins promote interaction with fibrinogen and vaginal colonization. J. Infect. Dis. 210, 982–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doran K. S., and Nizet V. (2004) Molecular pathogenesis of neonatal group B streptococcal infection: no longer in its infancy. Mol. Microbiol. 54, 23–31 [DOI] [PubMed] [Google Scholar]
- 8. Brady L. J., Maddocks S. E., Larson M. R., Forsgren N., Persson K., Deivanayagam C. C., and Jenkinson H. F. (2010) The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol. Microbiol. 77, 276–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Larson M. R., Rajashankar K. R., Patel M. H., Robinette R. A., Crowley P. J., Michalek S., Brady L. J., and Deivanayagam C. (2010) Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of α- and PPII-helices. Proc. Natl. Acad. Sci. U.S.A. 107, 5983–5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heim K. P., Crowley P. J., Long J. R., Kailasan S., McKenna R., and Brady L. J. (2014) An intramolecular lock facilitates folding and stabilizes the tertiary structure of Streptococcus mutans adhesin P1. Proc. Natl. Acad. Sci. U.S.A. 111, 15746–15751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Troffer-Charlier N., Ogier J., Moras D., and Cavarelli J. (2002) Crystal structure of the V-region of Streptococcus mutans antigen I/II at 2.4 Å resolution suggests a sugar preformed binding site. J. Mol. Biol. 318, 179–188 [DOI] [PubMed] [Google Scholar]
- 12. Forsgren N., Lamont R. J., and Persson K. (2009) Crystal structure of the variable domain of the Streptococcus gordonii surface protein SspB. Protein Sci. 18, 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Forsgren N., Lamont R. J., and Persson K. (2010) Two intramolecular isopeptide bonds are identified in the crystal structure of the Streptococcus gordonii SspB C-terminal domain. J. Mol. Biol. 397, 740–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nylander A., Forsgren N., and Persson K. (2011) Structure of the C-terminal domain of the surface antigen SpaP from the caries pathogen Streptococcus mutans. Acta Crystallogr Sect. F Struct. Biol. Cryst. Commun. 67, 23–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Larson M. R., Rajashankar K. R., Crowley P. J., Kelly C., Mitchell T. J., Brady L. J., and Deivanayagam C. (2011) Crystal structure of the C-terminal region of Streptococcus mutans antigen I/II and characterization of salivary agglutinin adherence domains. J. Biol. Chem. 286, 21657–21666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hall M., Nylander S., Jenkinson H. F., and Persson K. (2014) Structure of the C-terminal domain of AspA (antigen I/II-family) protein from Streptococcus pyogenes. FEBS Open Bio. 4, 283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Madsen J., Mollenhauer J., and Holmskov U. (2010) Review: Gp-340/DMBT1 in mucosal innate immunity. Innate. Immun. 16, 160–167 [DOI] [PubMed] [Google Scholar]
- 18. Loimaranta V., Jakubovics N. S., Hytönen J., Finne J., Jenkinson H. F., and Strömberg N. (2005) Fluid- or surface-phase human salivary scavenger protein gp340 exposes different bacterial recognition properties. Infect. Immun. 73, 2245–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Egland P. G., Dû L. D., and Kolenbrander P. E. (2001) Identification of independent Streptococcus gordonii SspA and SspB functions in coaggregation with Actinomyces naeslundii. Infect. Immun. 69, 7512–7516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jakubovics N. S., Strömberg N., van Dolleweerd C. J., Kelly C. G., and Jenkinson H. F. (2005) Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Mol. Microbiol. 55, 1591–1605 [DOI] [PubMed] [Google Scholar]
- 21. Back C. R., Douglas S. K., Emerson J. E., Nobbs A. H., and Jenkinson H. F. (2015) Streptococcus gordonii DL1 adhesin SspB V-region mediates coaggregation via receptor polysaccharide of Actinomyces oris T14V. Mol. Oral. Microbiol. 30, 411–424 [DOI] [PubMed] [Google Scholar]
- 22. Lamont R. J., El-Sabaeny A., Park Y., Cook G. S., Costerton J. W., and Demuth D. R. (2002) Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148, 1627–1636 [DOI] [PubMed] [Google Scholar]
- 23. Park Y., Simionato M. R., Sekiya K., Murakami Y., James D., Chen W., Hackett M., Yoshimura F., Demuth D. R., and Lamont R. J. (2005) Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect. Immun. 73, 3983–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silverman R. J., Nobbs A. H., Vickerman M. M., Barbour M. E., and Jenkinson H. F. (2010) Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect. Immun. 78, 4644–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maddocks S. E., Wright C. J., Nobbs A. H., Brittan J. L., Franklin L., Strömberg N., Kadioglu A., Jepson M. A., and Jenkinson H. F. (2011) Streptococcus pyogenes antigen I/II-family polypeptide AspA shows differential ligand-binding properties and mediates biofilm formation. Mol. Microbiol. 81, 1034–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Green N. M., Zhang S., Porcella S. F., Nagiec M. J., Barbian K. D., Beres S. B., LeFebvre R. B., and Musser J. M. (2005) Genome sequence of a serotype M28 strain of group A Streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J. Infect. Dis. 192, 760–770 [DOI] [PubMed] [Google Scholar]
- 27. Brochet M., Couvé E., Glaser P., Guédon G., and Payot S. (2008) Integrative conjugative elements and related elements are major contributors to the genome diversity of Streptococcus agalactiae. J. Bacteriol. 190, 6913–6917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang S., Green N. M., Sitkiewicz I., Lefebvre R. B., and Musser J. M. (2006) Identification and characterization of an antigen I/II family protein produced by group A Streptococcus. Infect. Immun. 74, 4200–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sitkiewicz I., Green N. M., Guo N., Mereghetti L., and Musser J. M. (2011) Lateral gene transfer of streptococcal ICE element RD2 (region of difference 2) encoding secreted proteins. BMC Microbiol. 11, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Franklin L., Nobbs A. H., Bricio-Moreno L., Wright C. J., Maddocks S. E., Sahota J. S., Ralph J., O'Connor M., Jenkinson H. F., and Kadioglu A. (2013) The AgI/II family adhesin AspA is required for respiratory infection by Streptococcus pyogenes. PLoS ONE 8, e62433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hirt H., Erlandsen S. L., and Dunny G. M. (2000) Heterologous inducible expression of Enterococcus faecalis pCF10 aggregation substance asc10 in Lactococcus lactis and Streptococcus gordonii contributes to cell hydrophobicity and adhesion to fibrin. J. Bacteriol. 182, 2299–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Purushotham S., and Deivanayagam C. (2014) The calcium-induced conformation and glycosylation of scavenger-rich cysteine repeat (SRCR) domains of glycoprotein 340 influence the high affinity interaction with antigen I/II homologs. J. Biol. Chem. 289, 21877–21887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Monif G. R., and Carson H. J. (1998) Female genital tract bacterial coisolates with Candida albicans in patients without clinical vaginitis. Infect. Dis. Obstet. Gynecol. 6, 52–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bayó M., Berlanga M., and Agut M. (2002) Vaginal microbiota in healthy pregnant women and prenatal screening of group B streptococci (GBS). Int. Microbiol. 5, 87–90 [DOI] [PubMed] [Google Scholar]
- 35. Cools P., Jespers V., Hardy L., Crucitti T., Delany-Moretlwe S., Mwaura M., Ndayisaba G. F., van de Wijgert J. H., and Vaneechoutte M. (2016) A multi-country cross-sectional study of vaginal carriage of group B streptococci (GBS) and Escherichia coli in resource-poor settings: prevalences and risk factors. PLoS ONE 11, e0148052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jenkinson H. F., Lala H. C., and Shepherd M. G. (1990) Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect. Immun. 58, 1429–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kolenbrander P. E., Ganeshkumar N., Cassels F. J., and Hughes C. V. (1993) Coaggregation: specific adherence among human oral plaque bacteria. FASEB J. 7, 406–413 [DOI] [PubMed] [Google Scholar]
- 38. Bamford C. V., d'Mello A., Nobbs A. H., Dutton L. C., Vickerman M. M., and Jenkinson H. F. (2009) Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect. Immun. 77, 3696–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kelly C. G., and Younson J. S. (2000) Anti-adhesive strategies in the prevention of infectious disease at mucosal surfaces. Expert Opin. Investig. Drugs 9, 1711–1721 [DOI] [PubMed] [Google Scholar]
- 40. Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., and Liang J. (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 34, W116–W118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Giglio K. M., Fong J. C., Yildiz F. H., and Sondermann H. (2013) Structural basis for biofilm formation via the Vibrio cholerae matrix protein RbmA. J. Bacteriol. 195, 3277–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Compton L. A., and Johnson W. C. Jr. (1986) Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal. Biochem. 155, 155–167 [DOI] [PubMed] [Google Scholar]
- 44. Manavalan P., and Johnson W. C. Jr. (1987) Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal. Biochem. 167, 76–85 [DOI] [PubMed] [Google Scholar]
- 45. Sreerama N., and Woody R. W. (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 46. Whitmore L., and Wallace B. A. (2004) DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Whitmore L., and Wallace B. A. (2008) Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89, 392–400 [DOI] [PubMed] [Google Scholar]
- 48. Siegel L. M., and Monty K. J. (1966) Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta 112, 346–362 [DOI] [PubMed] [Google Scholar]
- 49. Tettelin H., Masignani V., Cieslewicz M. J., Donati C., Medini D., Ward N. L., Angiuoli S. V., Crabtree J., Jones A. L., Durkin A. S., Deboy R. T., Davidsen T. M., Mora M., Scarselli M., Margarit y Ros I., et al. (2005) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U.S.A. 102, 13950–13955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chuzeville S., Dramsi S., Madec J. Y., Haenni M., and Payot S. (2015) Antigen I/II encoded by integrative and conjugative elements of Streptococcus agalactiae and role in biofilm formation. Microb. Pathog. 88, 1–9 [DOI] [PubMed] [Google Scholar]
- 51. Al-Okla S., Chatenay-Rivauday C., Klein J. P., and Wachsmann D. (1999) Involvement of α5β1 integrins in interleukin 8 production induced by oral viridans streptococcal protein I/IIf in cultured endothelial cells. Cell. Microbiol. 1, 157–168 [DOI] [PubMed] [Google Scholar]
- 52. Nobbs A. H., Shearer B. H., Drobni M., Jepson M. A., and Jenkinson H. F. (2007) Adherence and internalization of Streptococcus gordonii by epithelial cells involves β1 integrin recognition by SspA and SspB (antigen I/II family) polypeptides. Cell. Microbiol. 9, 65–83 [DOI] [PubMed] [Google Scholar]
- 53. Stoddard E., Cannon G., Ni H., Karikó K., Capodici J., Malamud D., and Weissman D. (2007) gp340 expressed on human genital epithelia binds HIV-1 envelope protein and facilitates viral transmission. J. Immunol. 179, 3126–3132 [DOI] [PubMed] [Google Scholar]
- 54. Brittan J. L., and Nobbs A. H. (2015) Group B Streptococcus pili mediate adherence to salivary glycoproteins. Microbes Infect. 17, 360–368 [DOI] [PubMed] [Google Scholar]
- 55. Verstraelen H., and Swidsinski A. (2013) The biofilm in bacterial vaginosis: implications for epidemiology, diagnosis and treatment. Curr. Opin. Infect. Dis. 26, 86–89 [DOI] [PubMed] [Google Scholar]
- 56. Ligtenberg A. J., Veerman E. C., Nieuw Amerongen A. V., and Mollenhauer J. (2007) Salivary agglutinin/glycoprotein-340/DMBT1: a single molecule with variable composition and with different functions in infection, inflammation and cancer. Biol. Chem. 388, 1275–1289 [DOI] [PubMed] [Google Scholar]
- 57. Jenkinson H. F., and Easingwood R. A. (1990) Insertional inactivation of the gene encoding a 76-kilodalton cell surface polypeptide in Streptococcus gordonii Challis has a pleiotropic effect on cell surface composition and properties. Infect. Immun. 58, 3689–3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Berrow N. S., Alderton D., Sainsbury S., Nettleship J., Assenberg R., Rahman N., Stuart D. I., and Owens R. J. (2007) A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 35, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., and Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 62. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 63. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vagin A., and Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 67. Krivov G. G., Shapovalov M. V., and Dunbrack R. L. Jr. (2009) Improved prediction of protein side-chain conformations with SCWRL4. Proteins 77, 778–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. McIntosh-Smith S., Price J., Sessions R. B., and Ibarra A. A. (2015) High performance in silico virtual drug screening on many-core processors. Int. J. High Perform. Comput. Appl. 29, 119–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Baker N. A., Sept D., Joseph S., Holst M. J., and McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gillum A. M., Tsay E. Y., and Kirsch D. R. (1984) Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198, 179–182 [DOI] [PubMed] [Google Scholar]