Abstract
The bacterial transcriptional regulator OxyR is known to function as a two-state redox switch. OxyR senses cellular levels of H2O2 via a “sensing cysteine” that switches from the reduced to a disulfide state upon H2O2 exposure, inducing the expression of antioxidant genes. The reduced and disulfide states of OxyR, respectively, bind to extended and compact regions of DNA, where the reduced state blocks and the oxidized state allows transcription and further induces target gene expression by interacting with RNA polymerase. Vibrio vulnificus OxyR2 senses H2O2 with high sensitivity and induces the gene encoding the antioxidant Prx2. In this study, we used mass spectrometry to identify a third redox state of OxyR2, in which the sensing cysteine was overoxidized to S-sulfonated cysteine (Cys-SO3H) by high H2O2 in vitro and in vivo, where the modification deterred the transcription of prx2. The DNA binding preferences of OxyR25CA-C206D, which mimics overoxidized OxyR2, suggested that overoxidized OxyR2 binds to the extended DNA site, masking the −35 region of the prx2 promoter. These combined results demonstrate that OxyR2 functions as a three-state redox switch to tightly regulate the expression of prx2, preventing futile production of Prx2 in cells exposed to high levels of H2O2 sufficient to inactivate Prx2. We further provide evidence that another OxyR homolog, OxyR1, displays similar three-state behavior, inviting further exploration of this phenomenon as a potentially general regulatory mechanism.
Keywords: antioxidant, gene regulation, hydrogen peroxide, peroxiredoxin, reactive oxygen species, OxyR, Vibrio vulnificus, overoxidation, sensing cysteine, three-state redox switch
Introduction
Reactive oxygen species build up as metabolic by-products from oxygen during aerobic respiration processes (1, 2). Of the reactive oxygen species, hydrogen peroxide (H2O2) is toxic to cellular components, especially when it is converted to the hydroxyl radical via the Fenton reaction in the presence of iron or copper ions (3, 4). Pathogenic bacteria inevitably encounter elevated levels of H2O2 imposed by the host immune response system during infection. Therefore, pathogenic bacteria have to cope with H2O2 to survive host environments and in turn to ensure developing illness. Therefore, the bacteria's mechanisms to defense against H2O2 are closely linked to their virulence (5). Bacterial defense against H2O2 relies on a variety of antioxidant defense enzymes, such as peroxiredoxins (Prxs)3 and catalases (6).
OxyR is a central regulator of the antioxidant genes in many bacteria and forms a homotetramer (7). OxyR is a modular structure consisting of an N-terminal DNA binding domain and C-terminal regulatory domain that contains two conserved redox-sensitive cysteines. Typical OxyR, such as Escherichia coli OxyR, has been considered as a “two-state redox switch” that precisely modulates expression of many antioxidant genes in a H2O2-dependent manner (7–9). When H2O2 is low and within safe limits, the redox-sensitive cysteines are present as free thiols separated by an α-helix, resulting in the “reduced state” of OxyR. In the reduced state, the OxyR tetramer adopts an extended conformation (10) and binds to the extended DNA-binding sites to mask the −35 region of the target antioxidant genes (11, 12). Thereby, the reduced OxyR turns off the expressions of target antioxidant genes under conditions of low H2O2 (12–14). When H2O2 increases and exceeds safe limits, one of the redox-sensitive cysteines, named sensing cysteine, senses H2O2 and is rapidly oxidized to Cys-SOH (15). Given the high reactivity, the Cys-SOH readily forms an intramolecular disulfide bond (Cys-S-S-Cys) with the other redox-sensitive cysteine. This “oxidized state or disulfide state” of OxyR adopts compact conformation, binds to more compact DNA-binding sequences, and unmasks the −35 region (11, 12). The oxidized OxyR interacts with the C-terminal domain of RNA polymerase α-subunits and turns on the target antioxidant genes under conditions of high H2O2 (8, 12, 16).
The facultative aerobic pathogen Vibrio vulnificus has two OxyRs, OxyR1 and OxyR2 (11). Amino acid sequences of both OxyR proteins have the conserved residues that are functionally important in OxyR, indicating that OxyR1 and OxyR2 share common functional and structural characteristics with the OxyR homologs of many bacteria. OxyR1 and OxyR2 activate prx1 and prx2, respectively (11). Each of the Prxs scavenges distinct ranges of H2O2 (i.e. working ranges of H2O2 for each Prx). Prx2 has a higher affinity for H2O2 than Prx1 and thus scavenges lower levels of H2O2 more effectively, whereas Prx1 detoxifies higher levels of H2O2 more effectively. It is noteworthy that Prx2 is sensitive, whereas Prx1 is relatively robust, to H2O2. Prx2 is irreversibly inactivated by overoxidation at one of the catalytic cysteines, peroxidatic cysteine, to S-sulfonated cysteine (Cys-SO3H) in cells exposed to 30 μm or higher levels of H2O2 that exceed its working range (17).
This observation led us to raise a question on the redox state and transcriptional activity of OxyR2 in the cells in which cellular H2O2 level is high enough to irreversibly inactivate Prx2. Accordingly, the present study examined the redox state of OxyR2 exposed to various levels of H2O2 in vivo as well as in vitro. MS analysis found that the sensing cysteine is overoxidized to Cys-SO3H by high H2O2 in vitro. The “overoxidized state” is a third redox state in addition to the reduced and disulfide states of OxyR2. We confirmed the presence of the overoxidized state of OxyR2 in V. vulnificus cells exposed to H2O2 exceeding 30 μm. Biochemical and genetic studies demonstrated that the overoxidized OxyR2 deters the production of Prx2 that is no longer functional. The sequences for binding of the overoxidized OxyR2 were determined to explain the mechanism of overoxidized OxyR2 to turn off the prx2 expression. Finally, the possibility that the OxyR1 is also overoxidized to play a role in the regulation of its antioxidant genes was investigated.
Results
Sensing Cysteine of OxyR2 Is Overoxidized at High Levels of H2O2
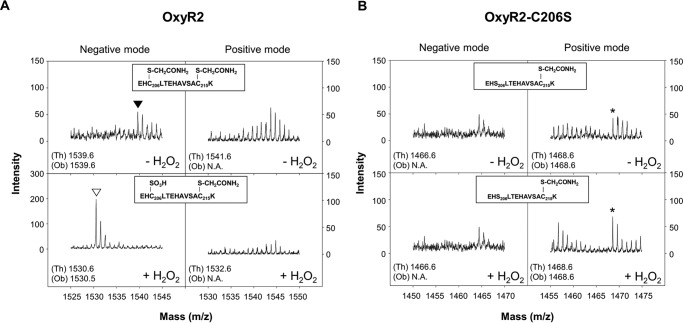
The purified OxyR2 protein was reacted with 500 μm H2O2 in vitro, and the redox state of the sensing cysteine Cys206 was subjected to MALDI-TOF MS analysis in positive and negative ion reflector modes (Fig. 1). Peptides can be detected in positive and/or negative ion reflector modes, depending on the intrinsic properties of the peptides that influence the ionization behavior (18). Mass spectra for the tryptic digests of the OxyR2 protein were analyzed after the free cysteine residues in the sample were alkylated with iodoacetamide to prevent further oxidation during the analysis. As shown in Fig. 1A, the peptide fragment containing both redox-sensitive cysteine residues (EHC206LTEHAVSAC215K) was detectable in the negative ion reflector mode. It remained reduced in the absence of H2O2 and thus produced the doubly alkylated version (Cys-S-CH2CONH2) with a monoisotopic mass [M − H]− of 1539.6 (Fig. 1A, top). However, upon reaction with 500 μm H2O2, the monoisotopic mass [M − H]− of the peptide fragment decreased to 1530.5, suggesting that the peptide fragment contains one S-sulfonated cysteine (Cys-SO3H) and one alkylated cysteine. The peptide fragment with S-sulfinated cysteine (Cys-SO2H; [M − H]− of 1514.6 or [M + H]+ of 1516.6) was not observed under the experimental conditions we tested (data not shown). The EHC206LTEHAVSAC215K peptide fragment was not detectable in the positive ion mode regardless of H2O2 treatment (Fig. 1A, right panels).
FIGURE 1.
MALDI-TOF mass spectra of OxyR2 and OxyR2-C206S oxidized by 500 μm H2O2in vitro. His6-tagged OxyR2 (A) and OxyR2-C206S (B) proteins were reduced with 100 mm DTT for 1 h and desalted by gel filtration chromatography under anaerobic conditions. Then the OxyR2 proteins were reacted (bottom panels, +H2O2) or unreacted (top panels, −H2O2) with 500 μm H2O2 for 10 min. Each of the OxyR2 proteins was then incubated with iodoacetamide to alkylate reduced cysteine residues, digested with trypsin, and analyzed by MALDI-TOF MS in negative (left panels) and positive (right panels) ion reflector modes. A, mass spectra of peptides containing Cys206 of OxyR2 are indicated. ▾, a peptide with alkylated Cys206 and Cys215; ▿, a peptide with S-sulfonated Cys206 and alkylated Cys215. B, mass spectra of peptides containing Ser206 of OxyR2-C206S are indicated. *, a peptide with alkylated Cys215. The form of each observed peptide is indicated in the box inside each panel. Theoretical (Th) and observed (Ob) monoisotopic masses ([M − H]− for negative mode or [M + H]+ for positive mode) of the peptides are indicated in the bottom left corner of each panel. N.A., the corresponding peptide was not observed.
To determine which cysteine residue was S-sulfonated by H2O2, the same analysis with the OxyR2-C206S variant was performed in the absence and presence of 500 μm H2O2. The tryptic peptide fragment (EHS206LTEHAVSAC215K) was detected in the positive ion reflector mode and had a singly alkylated protein at Cys215 showing the monoisotopic mass [M + H]+ of 1468.6 regardless of the presence of H2O2 (Fig. 1B, right panels). Neither peptide fragment with Cys215-SO3H ([M + H]+ of 1459.6 or [M − H]− of 1457.6) nor with Cys215-SO2H ([M + H]+ of 1443.6 or [M − H]− of 1441.6) was found, indicating that Cys215 did not undergo the overoxidation. The EHS206LTEHAVSAC215K peptide fragment was not detectable in the negative ion mode (Fig. 1B, left panels). Therefore, the combined results demonstrated that the sensing cysteine, Cys206, but not the other cysteine, Cys215, of OxyR2 is overoxidized to Cys-SO3H in the presence of high levels of H2O2 in vitro.
Overoxidation of OxyR2 in the V. vulnificus Cells Exposed to H2O2
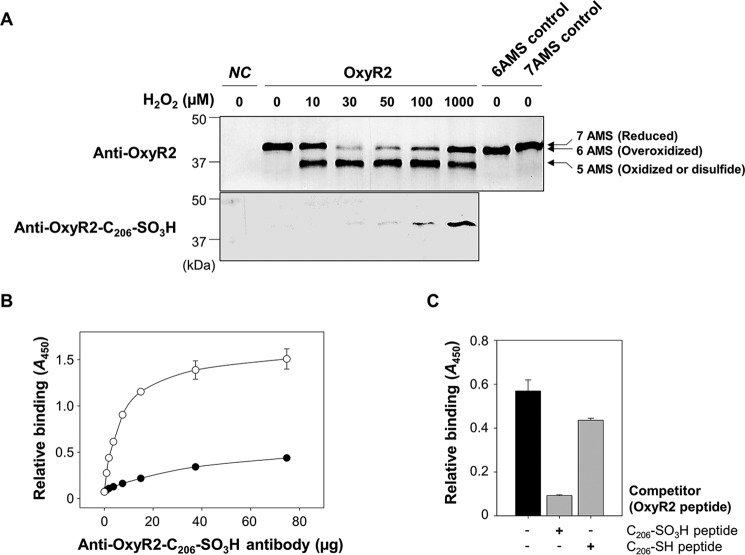
To ascertain whether the overoxidation of OxyR2 at the sensing cysteine indeed occurs by cellular H2O2 in V. vulnificus, the redox state of cellular OxyR2 was analyzed. The oxyR2 mutant OH0703 with pDY1025 expressing oxyR2 was grown anaerobically, exposed to various concentrations of H2O2, and then treated with 0.5-kDa 4-acetamido-4′-maleimidylstilbene-2, 2′-disulfonic acid (AMS; Invitrogen) to alkylate free thiols in the proteins. The total cellular proteins (3.5 μg/well) were resolved on non-reducing SDS-PAGE and immunoblotted with anti-OxyR2 antibody. Alkylation of a free thiol in the proteins with AMS adds 0.5 kDa of molecular mass (Fig. 2A, top). OxyR2 in the cells without exposure to H2O2 existed in the reduced state in which all seven cysteine residues present in OxyR2 were alkylated with AMS (7AMS control). An OxyR2 band containing pentuply alkylated cysteine residues coexisted with the heptuply alkylated OxyR2 band when the cells were exposed to 10 μm H2O2, indicating that a part of OxyR2 was oxidized to form a disulfide bond, as observed in the previous report (11). More importantly, a portion of OxyR2 in the cells exposed to H2O2 exceeding 30 μm appeared to have sextuply alkylated cysteine residues, indicating that one of the seven cysteine residues of OxyR2 had a single modification, such as an oxidation, that could prevent alkylation by AMS. When determined based on the band intensities, relative amounts of the OxyR2 with the single cysteine modification (or sextuply alkylated) were increased gradually by exposure to higher levels of H2O2 (Fig. 2A, top).
FIGURE 2.
Overoxidation of OxyR2 Cys206 in V. vulnificus cells exposed to various levels of H2O2. A, OH0703 (pDY1025) was grown anaerobically to an A600 of 0.3 and exposed to various H2O2 concentrations for 3 min. Cellular proteins were precipitated with TCA and alkylated with fresh AMS buffer for 1 h at 37 °C. Proteins (3.5 μg for the top panel and 7 μg for the bottom panel) were resolved by non-reducing SDS-PAGE and immunoblotted using anti-OxyR2 antibody (top) or anti-OxyR2-Cys206-SO3H antibody (bottom). The predicted numbers of AMS that alkylated each OxyR2 molecule and their redox states are indicated at the ends of the gel. Negative control (NC) was OH0703 (pJH0311), 6AMS control was OH0703 (pBANG1416), and 7AMS control was OH0703 (pDY1025). B, the specificity of anti-OxyR2-Cys206-SO3H antibody to overoxidized and reduced OxyR2 was determined using ELISA. The microtiter 96-well plates were coated with 0.1 μg of synthetic peptides corresponding to either OxyR2 active site with overoxidized (S-sulfonated) Cys206 (EKEHC206(SO3H)LTEHA) (○) or with reduced Cys206 (EKEHC206(SH)LTEHA) (●), and the peptides were reacted with various concentrations of the antibody as indicated. C, the S-sulfonated OxyR2 peptide (0.1 μg) was attached to the microtiter 96-well plates and then reacted with 4 μg of anti-OxyR2-Cys206-SO3H antibody. As a binding competitor, either S-sulfonated or reduced OxyR2 peptides (12.5 ng) were added to the reaction with the antibody as indicated. Black bar, control where no competitor was added. Relative binding of the antibody to the specific peptides is presented as A450. All data in B and C represent mean ± S.D. (error bars).
To examine whether the OxyR2 with a single cysteine modification observed in the cells exposed to H2O2 exceeding 30 μm is the OxyR2 with overoxidized Cys206, the total cellular proteins (7.0 μg/well) were resolved on non-reducing SDS-PAGE and immunoblotted with the anti-OxyR2-Cys206-SO3H antibody (Fig. 2A, bottom). The OxyR2 protein with overoxidized Cys206 was specifically detected in cells exposed to H2O2 exceeding 30 μm and increased gradually in the cells exposed to higher levels of H2O2. The results indicated that the sensing cysteine of OxyR2 becomes overoxidized in vivo when the V. vulnificus cells are exposed to high levels of H2O2, as was demonstrated in vitro (Fig. 1).
Concurrently, to determine the specificity of the anti-OxyR2-Cys206-SO3H antibody by ELISA, the antibody was tested to react with the Cys206-SO3H or Cys206-SH peptides that were synthesized and attached to the microtiter 96-well plates. The antibody specifically bound to the Cys206-SO3H peptide (Fig. 2B). Furthermore, the binding of the antibody to the attached Cys206-SO3H peptide was effectively inhibited by the Cys206-SO3H peptide added to the reaction as a competitor (Fig. 2C). The antibody slightly bound to the Cys206-SH peptide (Fig. 2B), and the Cys206-SH peptide competitor could marginally inhibit the antibody reaction to the Cys206-SO3H peptide (Fig. 2C), reflecting that Cys206-SH peptide could be oxidized to Cys206-SO3H peptide in the aerobic conditions we tested. The results indicated that the anti-OxyR2-Cys206-SO3H antibody we used is specific to the OxyR2 with Cys206-SO3H. However, it was not possible to examine the antibody's specificity to Cys206-SO2H. Therefore, the OxyR2 with Cys206-SO2/3H is hereafter designated as the overoxidized state of OxyR2.
Overoxidized State of OxyR2 Turns Off the prx2 Promoter
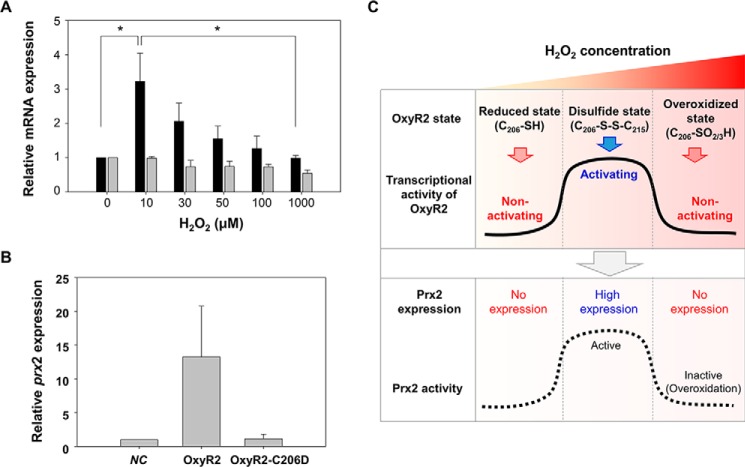
To examine the transcriptional activity of the overoxidized state of OxyR2, the expression of prx2 was monitored in the wild-type V. vulnificus cells. The expression of prx2 was significantly induced when the anaerobically grown cells were exposed to 10 μm H2O2 (Fig. 3A), indicating that OxyR2 was oxidized to the disulfide state under this condition to activate the prx2 promoter (Pprx2) (Fig. 2A). However, the level of prx2 transcript gradually decreased along with increasing concentrations of H2O2 exceeding 30 μm (Fig. 3A), which was in accordance with the increase of the overoxidized state of OxyR2 (Fig. 2A). The changes in prx2 expression were dependent on OxyR2 activity, but not on oxyR2 expression level, because the expression level of oxyR2 was not significantly affected by increasing concentrations of H2O2 below 1 mm (Fig. 3A). The combined results suggested that the overoxidized state of OxyR2 deters further expression of prx2. Because it has been reported that Prx2 is inactivated by H2O2 exceeding 30 μm (17), this deterrence of the prx2 expression may prevent the worthless production of Prx2 in the environments of high H2O2, where Prx2 is no longer functional.
FIGURE 3.
Expression levels of prx2 in response to various levels of H2O2 and a working model for OxyR2 as a three-state redox switch. A, wild-type V. vulnificus was grown anaerobically to an A600 of 0.3 and exposed to various concentrations of H2O2 as indicated for 3 min. Total RNAs were isolated, and the relative levels of prx2 (black bars) and oxyR2 (gray bars) transcripts were determined by quantitative real-time PCR analyses. The level of each transcript from wild type unexposed to H2O2 is presented as 1. *, statistically significant difference (p < 0.05) between groups. B, transcriptional activity of OxyR2-C206D. Cultures were grown aerobically to an A600 of 0.5, total RNAs were isolated, and the relative levels of prx2 transcript were determined by quantitative real-time PCR analyses. The level of transcript from negative control (NC) is presented as 1. Negative control was OH0703 (pJH0311). OxyR2, OH0703 (pDY1025); OxyR2-C206D, OH0703 (pBANG1416). All data in A and B represent mean ± S.D. (error bars). C, OxyR2 shifts to three different redox states, revealing different transcriptional activities in response to various levels of H2O2 in cells (solid line) as indicated. Concurrently, the prx2 expression (dotted line) varies, depending on the redox states of OxyR2. C206-SO2/3H, overoxidized Cys206 with Cys206-SO2H and/or Cys206-SO3H.
However, it was still possible that H2O2 over 30 μm can also detrimentally affect other transcription factors, such as components of RNA polymerase, to deter the prx2 expression. To rule out this possibility, OH0703 containing pBANG1416 expressing OxyR2-C206D was grown aerobically without exogenously added H2O2, and the prx2 expression was compared. It has been reported that the size and the polar properties of aspartic acid are comparable with those of the overoxidized cysteine residues (Cys-SO2H and Cys-SO3H), and therefore OxyR2-C206D is anticipated to mimic the overoxidation state of OxyR2 (10). As shown in Fig. 3B, OH0703 producing wild-type OxyR2 expressed almost 13-fold greater prx2 than the negative control strain lacking OxyR2. This supported our previous observation that wild-type OxyR2 activates the prx2 expression under aerobic conditions (11). In contrast, the prx2 expression level of the strain producing OxyR2-C206D was almost the same as that of the negative control strain (Fig. 3B). The results indicated that the deterrence of the prx2 expression in response to H2O2 exceeding 30 μm is caused mainly, if not solely, by overoxidation of OxyR2 and further confirmed that OxyR2-C206D is functionally similar to the overoxidized OxyR2.
OxyR2 Regulates prx2 as a Three-state Redox Switch in Response to H2O2
Fig. 3C represents the working model of OxyR2 that regulates the expression of prx2 as a three-state redox switch. OxyR2 adopts the three distinct redox states (reduced state, disulfide state, and overoxidized state) in an H2O2 concentration-dependent manner (Figs. 1 and 2). So far in the widely accepted mechanism of OxyR proteins, OxyR2 shifts its redox state from the reduced state to the disulfide state upon reaction with H2O2 and activates the expression of prx2. However, when exposed to H2O2 exceeding the working range for Prx2, OxyR2 moves to an overoxidized state, a third redox state, and turns off the expression of prx2. It is obvious that OxyR2 is able to prevent production of useless Prx2 and thus save valuable cellular resources by working as a three-state redox switch (Fig. 3C).
Mechanism of the Overoxidized OxyR2 to Turn Off Pprx2
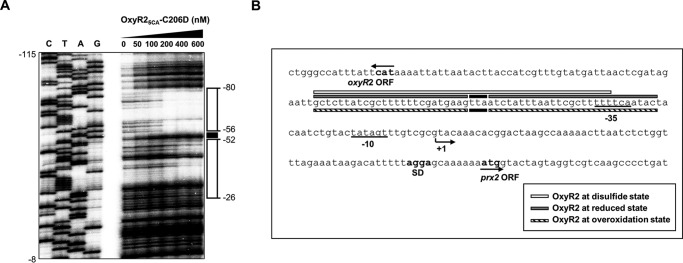
To gain insight into the mechanism by which the overoxidized state of OxyR2 deters the expression of prx2, the sequences for binding of the overoxidized OxyR2 were determined. Purified OxyR25CA-C206D, which mimics overoxidized OxyR2 and is soluble under aerobic conditions, was used in vitro for the DNA footprinting assay. OxyR25CA, in which all five non-catalytic cysteine residues in OxyR2 were replaced with alanine residues, was constructed previously (11). The purified OxyR25CA did not form oligomers in vitro under nonreducing conditions (11). As shown in Fig. 4, the sequences for binding of OxyR25CA-C206D were extended from −80 to −26 relative to the transcription start site of Pprx2. Therefore, one possible mechanism is that the overoxidized OxyR2 presumably turns off the expression of prx2 by masking the −35 region and thereby preventing RNA polymerase binding to Pprx2.
FIGURE 4.
Binding sequences of OxyR25CA-C206D within the prx2 promoter region. A, a 260-bp DNA fragment of the upstream region of Pprx2 was radioactively labeled and then used as probe DNA. The radiolabeled probe DNA (25 nm) was incubated with increasing amounts of the purified OxyR25CA-C206D as indicated and then digested with DNase I. The regions protected by OxyR25CA-C206D are indicated by open boxes, whereas the nucleotides showing enhanced cleavage are indicated by a black box. Lanes C, T, A, and G, nucleotide sequencing ladder of Pprx2. Nucleotide numbers shown are relative to the transcription start site of Pprx2. B, the sequences for binding of OxyR25CA-C206D are indicated below the Pprx2 sequences as diagonal-filled boxes. The sequences for binding of oxidized OxyR2 (white box) and reduced OxyR2 (gray boxes) determined previously (11) are presented above the Pprx2 sequences. The nucleotides showing enhanced cleavage by binding of OxyR25CA-C206D and the reduced OxyR2 are indicated as black boxes. The transcription start site of Pprx2 identified previously (11) is indicated by a bent arrow, and the positions of the putative −10 and −35 regions are underlined. The ATG translation initiation codon and the putative ribosome-binding site (SD) are also indicated in boldface type.
Furthermore, the binding sequences of OxyR25CA-C206D are identical to those of the reduced OxyR2. Enhanced cleavage in several nucleotides (−53 to −55), which was observed in the binding sequences of the reduced OxyR2, was also found in the binding sequences of OxyR25CA-C206D (Fig. 4B). It has been proposed that OxyR tetramer in the reduced state adopts an extended conformation (10) and binds to the elongated DNA-binding sites (11, 12). The combined results suggest that the overoxidized state of OxyR2 turns off the activation of prx2 by adopting a conformation similar to that of the reduced state, binding to the elongated sequences, and preventing RNA polymerase binding to the promoter sequences.
Possible Overoxidation of OxyR1
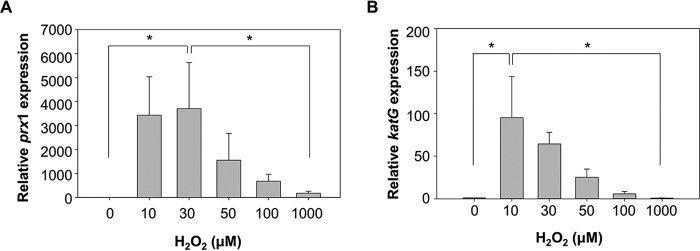
To probe whether other OxyR proteins could function as a three-state redox switch, the expression of prx1 and katG in the V. vulnificus cells exposed to various levels of H2O2 was determined (Fig. 5, A and B). It has been reported that OxyR1, a homolog of E. coli OxyR, directly activates the expression of prx1 and katG (11).4 Expression of both antioxidant genes in V. vulnificus increased along with increasing levels of H2O2 but decreased when H2O2 levels exceeded certain levels; the expression levels of prx1 and katG showed the highest peaks in cells exposed to 30 and 10 μm H2O2, respectively, and then decreased in cells exposed to higher levels of H2O2 (Fig. 5, A and B). This variation of the prx1 and katG expression in response to increasing levels of H2O2 was similar to that of the prx2 expression (Fig. 3A), indicating that OxyR1 was also probably overoxidized to turn off expression of prx1 and katG when the H2O2 levels exceeded the working ranges for Prx1 and KatG. This result suggested that OxyR homologs of other bacteria could function as a three-state redox switch to tightly regulate their target antioxidant genes.
FIGURE 5.
Expression levels of prx1 and katG in response to various levels of H2O2. Wild-type V. vulnificus was grown anaerobically to an A600 of 0.3 and exposed to various concentrations of H2O2 as indicated for 3 min. Total RNAs were isolated, and the relative levels of prx1 (A) and katG (B) transcripts were determined by quantitative real-time PCR analyses. The level of each transcript from wild type unexposed to H2O2 is presented as 1. *, statistically significant difference (p < 0.05) between groups. All data represent mean ± S.D. (error bars).
Discussion
In this study, we present evidence that V. vulnificus OxyR2 regulates the expression of prx2 as a three-state redox switch. When OxyR2 encountered H2O2 exceeding the Prx2 working range, the sensing cysteine of OxyR2 was overoxidized to Cys206-SO3H in vivo as well as in vitro (Figs. 1 and 2). Although MS analysis was unable to detect the peptide fragment of OxyR2 with Cys206-SO2H (Fig. 1A), the S-sulfinated form of OxyR2 was most likely formed as an intermediate in the course of the overoxidation of OxyR2 to the S-sulfonated OxyR2 (19). In addition to the reduced and oxidized (disulfide) states, the overoxidized state is the third redox state of OxyR2, whose function was primarily characterized in this study. The overoxidized OxyR2 actively turns off the transcription of prx2 by binding to elongated sequences masking the −35 region of Pprx2 in the environments of high H2O2, where Prx2 is no longer functional (Figs. 3 and 4) (17). Obviously, the overoxidized OxyR2 can prevent production of useless Prx2 and thus save valuable cellular resources. Therefore, the overoxidized OxyR2 is not simply an expired or perished form of OxyR2 but rather plays an important role in the survival of V. vulnificus during pathogenesis.
It was noteworthy that expression of prx1 and katG was increased, reached a maximum, and then was decreased by exposure of cells to increasing levels of H2O2 in a pattern similar to that of prx2 (Fig. 5, A and B). Both prx1 and katG are regulated by OxyR1, a homolog of E. coli OxyR (11), implying that OxyR2 is not the only OxyR regulating antioxidant genes as a three-state redox switch. OxyR has been primarily thought of as a two-state redox switch that turns the antioxidant genes off and on in an H2O2-dependent manner (7–9). When bacteria encounter manageable ranges of H2O2, OxyR turns on its target genes encoding antioxidants, such as Prxs and catalases. Scavenging the H2O2 using the antioxidants and maintaining metabolic activities to keep growing is beneficial for bacteria. Accordingly, expression of most of the OxyR regulon is governed by the housekeeping σ factor RpoD (σ70) (20). However, when H2O2 exceeds the manageable ranges, maintaining expressions of the antioxidant genes would be worthless because growth and proliferation of bacterial cells under this condition could be reckless. Instead, bacteria rather express many stress tolerance genes governed by the stress-responsive σ factor RpoS (σ38) and shift their global physiology to stationary (or dormant) phase (21, 22). Bacteria can turn off the antioxidant genes using OxyR, as a three-state redox switch, under conditions in which expression of the antioxidants is useless. It is obviously advantageous that bacteria utilize more cellular resources to express stress tolerance genes encoding many additional protective systems (20, 21).
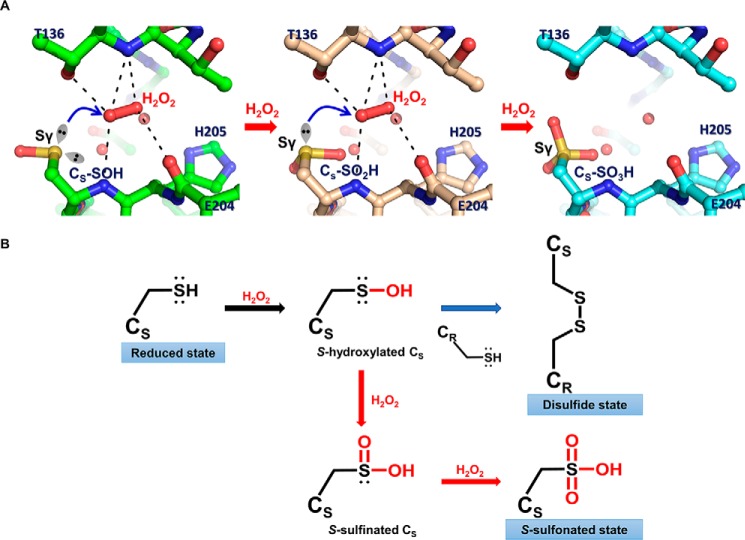
The mechanism for the overoxidation of the sensing cysteine remains unclear. However, structural analysis of the Pseudomonas aeruginosa OxyR-C199D mutant, in which the sensing cysteine residue is replaced with aspartic acid, gives us a hint as to this mechanism (10). The crystal structure OxyR-C199D holds a H2O2 molecule near aspartic acid that is even bulkier than a cysteine residue, implying that H2O2 can also bind to the same site even after the sensing cysteine is oxidized to Cys-SOH or Cys-SO2H. The bound H2O2 can eventually oxidize the cysteine to Cys-SO3H by the same H2O2-driven oxidation mechanism proposed previously (10). We modeled Cys-SOH, Cys-SO2H, and Cys-SO3H forms based on the P. aeruginosa OxyR-C199D structure that contains H2O2 molecules (Fig. 6A). Steric clash was not observed around the modeled cysteine residues with modification. Moreover, the lone pair electrons of the sulfur atoms in the cysteine residues could be facing H2O2 within a reasonable distance for nucleophilic attack on the H2O2 molecule when the -SOH or -SO2H moiety is simply rotated in the model. These findings support the idea that overoxidation at the sensing cysteine residue by H2O2 can occur faster than at other cysteine residues, presumably by the successive H2O2-driven oxidation mechanism.
FIGURE 6.
Proposed overoxidation mechanism of OxyR. A, modeled structures of OxyR, focusing on the sensing cysteine (CS). S-Hydroxylated sensing cysteine (CS-SOH) is oxidized by the bound H2O2 (left), resulting in S-sulfinated sensing cysteine (CS-SO2H; middle), which is in turn further oxidized to S-sulfonated sensing cysteine (CS-SO3H; right). The models were constructed based on the crystal structure of the P. aeruginosa OxyR-C199D variant complexed with a H2O2 molecule. The H2O2 molecule was bound by the polar interactions with the side chain and the backbone amide group of Thr136, the backbone carbonyl group of Glu204, and the backbone amide group of Cys206, as indicated by the dotted lines. The lone pair of electrons are displayed as the sp3 orbital shapes, and the bound water molecules are shown as red spheres. The blue arrows indicate the nucleophilic attack by Sγ of sensing cysteine on an oxygen atom of H2O2. B, schematic reaction mechanism of oxidation of sensing cysteine and the concomitant three redox states of OxyR. Sensing cysteine of OxyR in the reduced state is converted to S-hydroxylated sensing cysteine by H2O2, as a reaction intermediate (black arrow). S-Hydroxylated sensing cysteine rapidly makes a disulfide with the other redox-sensitive cysteine (CR), resulting in the disulfide state of OxyR (blue arrow). Alternatively, S-hydroxylated sensing cysteine is further oxidized consecutively by H2O2 to the final S-sulfonated sensing cysteine in the presence of excess H2O2, resulting in the overoxidized state of OxyR (red arrows).
Three different redox states of OxyR are determined, depending on the cellular levels of H2O2 as depicted in Fig. 6B. At low levels of H2O2 below the sensing level of OxyR, OxyR is maintained in the reduced state. Under the Prx2 working range of H2O2, a kinetic path renders the sensing cysteine in OxyR oxidized to the Cys-SOH intermediate that forms a disulfide bond with the other redox-sensitive cysteine, leading to activation of the target antioxidant genes. However, when H2O2 levels exceed the Prx2 working range, the Cys-SOH intermediate would be rapidly overoxidized to the Cys-SO3H via Cys-SO2H by H2O2 before making the disulfide bond. The resulting overoxidized OxyR2 turns off the production of useless Prx2. We do not yet know what happens to the overoxidized OxyR when the cellular H2O2 level returns to the working range. One plausible mechanism is that the overoxidized OxyR is degraded and new OxyR is synthesized, so that the cell can constantly respond to the changing environment. Given that the cellular amount of OxyR2 is very low (11), the energy needed for this process is not high.
In summary, V. vulnificus OxyR2 is primarily characterized as a three-state redox switch. A thiol-based sensor protein OxyR2 senses H2O2 and shifts to a third redox state, the overoxidized state, as a result of overoxidation of sensing cysteine residue by H2O2. Overoxidized OxyR2 prevents production of Prx2 where Prx2 is no longer functional and thereby saves valuable cellular resources. The DNA sequences for binding of OxyR25CA-C206D suggested that overoxidized OxyR2 adopts an extended conformation and turns off Pprx2 by binding to elongated sequences masking the −35 region. Expression patterns of prx1 and katG in response to various H2O2 levels suggested that OxyR1, a homolog of E. coli OxyR, also functions as a three-state redox switch, as does OxyR2.
Experimental Procedures
Bacterial Strains, Plasmids, and Culture Media
The strains and plasmids used in this study are listed in Table 1. The V. vulnificus strains were grown in LB medium supplemented with 2.0% (w/v) NaCl (LBS) at 30 °C. Anaerobic conditions were obtained using an anaerobic chamber with an atmosphere of 90% N2, 5% CO2, and 5% H2 (Coy Laboratory Products, Grass Lake, MI). For anaerobic culture, the media were preincubated to remove dissolved O2 in the anaerobic chamber, which was verified by adding 0.00001% (w/v) resazurin salt (Sigma) to the media as described previously (11).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains or plasmids | Relevant characteristicsa | Reference or source |
|---|---|---|
| Bacterial strains | ||
| V. vulnificus | ||
| MO6-24/O | Clinical isolate; virulent | Laboratory collection |
| OH0703 | MO6-24/O with oxyR2::nptI; Kmr | Ref. 11 |
| E. coli | ||
| SM10 λ pir | thi thr leu tonA lacY supE recA::RP4–2-Tc::Mu λpir; Kmr; host for -requiring plasmids; conjugal donor | Ref. 23 |
| BL21 (DE3) | F− ompT hsdS (rB−mB−) gal (DE3) | Laboratory collection |
| Plasmids | ||
| pJH0311 | 0.3-kb MCS of pUC19 cloned into pCOS5; Apr, Cmr | Ref. 26 |
| pDY1025 | pJH0311 with wild-type oxyR2; Apr, Cmr | Ref. 11 |
| pBANG1416 | pJH0311 with the mutant oxyR2 encoding OxyR2-C206D; Apr, Cmr | This study |
| pET-28a(+) | His6-tag fusion protein expression vector; Kmr | Novagen |
| pDY1001 | pET-28a (+) with wild-type oxyR2; Kmr | Ref. 11 |
| pDY1104 | pET-28a (+) with the mutant oxyR2 encoding OxyR2-C206S; Kmr | This study |
| pDY1014 | pET-28a (+) with the mutant oxyR2 encoding OxyR25CA; Kmr | Ref. 11 |
| pBANG1501 | pET-28a (+) with the mutant oxyR2 encoding OxyR25CA-C206D; Kmr | This study |
a Apr, ampicillin-resistant; Cmr, chloramphenicol-resistant; Kmr, kanamycin-resistant.
Site-directed Mutagenesis of oxyR2 and Purification of Mutant OxyR2 Proteins
A mutant OxyR2 in which Cys206 was replaced with aspartic acid was constructed using the QuikChange® site-directed mutagenesis kit (Agilent Technologies, Loveland, CO) as described previously (11). The complementary mutagenic primers listed in Table 2 were used in conjunction with the plasmid pDY1025 (oxyR2 cloned into a broad host-range vector pJH0311 under the lac promoter) to create pBANG1416 (oxyR2-C206D on pJH0311) (Table 1). The mutation was confirmed by DNA sequencing. E. coli SM10 λ pir, tra (23) harboring pJH0311, pDY1025, or pBANG1416 was used as a conjugal donor to the oxyR2 mutant (OH0703). The conjugation was conducted as described previously (11).
TABLE 2.
Oligonucleotides used in this study
| Name | Oligonucleotide Sequence (5′→3′)a,b | Use |
|---|---|---|
| For mutagenesis | ||
| OXYR2C206D-F | GGAAAAAGAGCATGATCTGACTGAACACGCGGTGTCGG | Construction of OxyR2-C206D mutant |
| OXYR2C206D-R | CCGACACCGCGTGTTCAGTCAGATCATGCTCTTTTTCC | |
| OXYR2C206S-F | GGAAAAAGAGCATAGTCTGACTGAACAC GCGGTGTCGGCC | Construction of OxyR2-C206S mutant |
| OXYR2C206S-R | GGCCGACACCGCGTGTTCAGTCAGACTATGCTCTTTTTCC | |
| For quantitative real-time PCR | ||
| PRX2QRT-F | GTTGCTTTCCGTGGCTCTTTCC | Expression of prx2 |
| PRX2QRT-R | TACTTCGCCGTGCTTCTGGTG | |
| OXYR2QRT2-F | GCCCAGTCTGAAGCAGTTACACTATC | Expression of oxyR2 |
| OXYR2QRT2-R | GAGAGGACAGCCGATCAACTCTTC | |
| PRX1QRT-F | TACCCAGCGGACTTCACTTTCG | Expression of prx1 |
| PRX1QRT-R | TGACACTGAGTACACTTCCACACC | |
| KATGQRT-F | CGTTCCGTTTGCAGCAGGTC | Expression of katG |
| KATGQRT-R | CCAGTTGCGGAAGCCATCTG | |
| For DNA footprinting | ||
| PRX2P02-F | GCAGAGTAATAAGATAGTGTAACTGC | Amplification of prx2 upstream region |
| PRX2P02-R | CCTACTAGTACCATTTTTTTGCTCC | |
Mutant OxyR2 proteins, OxyR2-C206S and OxyR25CA-C206D, were constructed and purified for MALDI-TOF MS analysis and DNA footprinting assay, respectively. Cys206 of OxyR2 was replaced with serine using pDY1001 (oxyR2 on pET-28a(+) as a template) and the complementary mutagenic primers (Table 2) to result in pDY1104 (oxyR2-C206S on pET-28a(+)). Similarly, Cys206 of OxyR25CA was replaced with serine using pDY1014 (oxyR25CA on pET-28a(+) as a template) to result in pBANG1501 (oxyR25CA-C206D on pET-28a(+)) (Table 1). The resulting His6-tagged mutant OxyR2 proteins were produced in E. coli BL21 (DE3) and purified by affinity chromatography using Ni2+-nitrilotriacetic acid resin (Qiagen, Valencia, CA) as described previously (11).
MALDI-TOF MS Analysis
To investigate the redox state of Cys206 in OxyR2 in vitro, the full-length His6-tagged OxyR2 and OxyR2-C206S proteins were analyzed by MALDI-TOF MS, as described previously (11, 17). Briefly, to reduce the proteins, 20 μg of proteins in a reaction buffer (20 mm Tris (pH 7.4), 0.3 m KCl, 5 mm MgCl2, 0.5 mm EDTA, and 10% (v/v) glycerol) were treated with 100 mm DTT for 1 h, and then the DTT was removed by gel filtration chromatography under anaerobic conditions. To prepare oxidized proteins, reduced OxyR2 proteins were reacted with 500 μm H2O2 for 10 min after the DTT removal. Subsequently, the reduced cysteine residues of each reduced and overoxidized OxyR2 protein were alkylated with 50 mm iodoacetamide for 1.5 h in the dark under anaerobic conditions. Alkylated OxyR2 proteins were resolved on non-reducing SDS-PAGE, and the protein bands were excised and in-gel digested with trypsin (Sigma). Peptides were extracted from the gel pieces with 0.1% trifluoroacetic acid in 50% acetonitrile, concentrated to volumes of 10 μl using a SpeedVac concentrator (Savant Instruments Inc., Farmingdale, NY), and desalted using Zip-Tip C18 reverse phase peptide separation matrix (Millipore, Billerica, MA).
MALDI-TOF MS analyses were carried out on a Voyager-DETM STR biospectrometry work station (Applied Biosystems Inc., Foster City, CA) operating in positive and negative ion reflector modes. α-Cyano-4-hydroxycinnamic acid (10 mg/ml) in 50% acetonitrile and 0.1% trifluoroacetic acid was used as a matrix. The theoretical monoisotopic masses ([M − H]− for the deprotonated form in the negative ion reflector mode or [M + H]+ for the protonated form in the positive ion reflector mode) of the cleavage peptides were determined using PeptideMass software from the ExPASy proteomics server. The masses from the positive ion reflector mode were calibrated internally with known masses of autolytic trypsin peptides. However, the masses from the negative ion reflector mode were uncalibrated because the trypsin peaks were not detectable in the negative ion mode.
In Vivo Alkylation of OxyR2 and Western Blotting Analysis
The oxyR2 mutant OH0703 with pDY1025 expressing oxyR2 was used for Western blotting analysis of OxyR2 as described previously (11). OH0703 (pDY1025) was grown anaerobically to an A600 of 0.3, aliquoted to the same volume, and exposed to various concentrations of H2O2. To alkylate free thiols in the proteins with AMS, the cells were immediately precipitated with ice-cold TCA, and then the resulting pellets were dissolved in 50 μl of the fresh AMS buffer (15 mm AMS, 1 m Tris, 1 mm EDTA, 0.1% (w/v) SDS, pH 8.0) (11).
After incubation at 37 °C for 1 h, the same amounts of pelleted total protein were resolved on SDS-PAGE under non-reducing conditions and immunoblotted with either anti-OxyR2 or anti-OxyR2-Cys206-SO3H antibody as described previously (11). The anti-OxyR2 polyclonal antibody was prepared previously in a rabbit by using the purified His6-tagged OxyR2 as an antigen (11). An S-sulfonated peptide corresponding to the active site of OxyR2, EKEHC206(SO3H)LTEHA, was synthesized and conjugated with keyhole limpet hemocyanin and then used to raise rabbit anti-OxyR2-Cys206-SO3H polyclonal antibody (AbFrontier, Seoul, South Korea).
ELISA
The Cys206-SH peptide EKEHC206(SH)LTEHA and its S-sulfonated form, Cys206-SO3H peptide EKEHC206(SO3H)LTEHA, were synthesized (AbFrontier) and used to analyze the binding specificity of anti-OxyR2-Cys206-SO3H antibody. The microtiter 96-well plates were coated with 100 μl of each of the synthetic peptides (1 μg/ml) for overnight at 4 °C and blocked with the blocking buffer (2% skim milk in PBS with 20 mm 2-mercaptoethanol) for 2 h to prevent a nonspecific binding. After the blocking buffer was disposed, various concentrations of anti-OxyR2-Cys206-SO3H antibody in 100 μl of the blocking buffer were added to each well and reacted for 2 h. When needed, the peptides were added to the reaction along with the antibody as binding competitors.
Then the wells were reacted with HRP-conjugated goat anti-rabbit IgG (0.2 μg/ml) (AbFrontier) for 2 h. For the colorimetric development of HRP, 3,3′,5,5′-tetramethylbenzidine solution (0.0075% (w/v), Sigma) was added and reacted for 1 min. The reaction was stopped by adding 100 μl of 2 m H2SO4. Absorbance at 450 nm (A450) was recorded to measure the amount of anti-OxyR2-Cys206-SO3H antibody in each well using an InfiniteTM M200 microplate reader (Tecan, Männedorf, Switzerland). The wells were washed between each step of the reaction either with TBST-M (50 mm Tris, 150 mm NaCl, 0.1% Tween 20, 20 mm 2-mercaptoethanol) before anti-OxyR2-Cys206-SO3H antibody was added or with TBST (50 mm Tris, 150 mm NaCl, 0.1% Tween 20) after the antibody was added. All reactions were carried out at room temperature.
RNA Purification and Transcript Analysis
Total cellular RNAs from the cultures grown anaerobically to an A600 of 0.3 or aerobically to an A600 of 0.5 were isolated using RNAprotect® bacteria reagent and an RNeasy® minikit (Qiagen). When necessary, the cultures were exposed to H2O2 for 3 min and then harvested. For quantitative real-time PCR, cDNA was synthesized using the iScriptTM cDNA synthesis kit (Bio-Rad), and real-time PCR amplification of the cDNA was performed using the Chromo 4 real-time PCR detection system (Bio-Rad) with pairs of primers listed in Table 2. Relative expression levels of the specific transcripts were calculated using the 16S rRNA expression level as the internal reference for normalization (24).
DNA Footprinting Assay
DNA footprinting assays were conducted as described previously (11). The 260-bp upstream region of Pprx2, extending from −246 to +14, was amplified by PCR using [γ-32P]ATP-labeled PRX2P02-R and unlabeled PRX2P02-F primers (Table 2). The labeled 260-bp DNA (25 nm) probe was incubated with various concentrations of purified OxyR25CA-C206D for 30 min at 30 °C in a 20-μl reaction mixture containing 1× binding buffer (25 mm Tris-Cl (pH 8.0), 25 mm KCl, 6 mm MgCl2, 0.5 mm EDTA, 10% (v/v) glycerol, 0.05% (v/v) Tween 20, and 50 μg/ml BSA). The DNA-protein complexes were digested with 0.1 units of DNase I for 30 s at 30 °C. After ethanol precipitation, the digested DNA products were resolved on a sequencing gel alongside sequencing ladders of the same 260-bp upstream region of Pprx2. The gels were visualized using a phosphor imager analyzer (BAS1500, Fuji Photo Film Co. Ltd., Tokyo, Japan).
Modeling of OxyR2 Structures
To construct models for an overoxidized OxyR2 structure containing a H2O2 molecule, we used the coordinates of the full-length OxyR-C199D variant from P. aeruginosa (Protein Data Bank code 4X6G), where an H2O2 molecule is bound near the mutated Asp199 residue corresponding to Cys206 of OxyR2. The Asp199 residue in the structure was changed to Cys-SOH, Cys-SO2H, or Cys-SO3H using the program COOT (25). Then -SOH, -SO2H, or -SO3H was rotated to be suitable for the nucleophilic attack on the bound H2O2 molecule.
Data Analyses
Mean and S.D. were calculated from at least three independent experiments. To analyze H2O2 concentration-dependent expression of prx2, oxyR2, prx1, and katG, data were assessed for normal distribution and homogeneity of variance by Shapiro-Wilks' test. Data that had normal distribution were analyzed by one-way analysis of variance, and data that did not have normal distribution were analyzed by Kruskal-Wallis one-way analysis of variance on ranks. Statistical significance was assessed by the Student-Newman-Keuls method as a post hoc test using SigmaPlot version 13. Significance of differences between experimental groups was accepted at a p value of <0.05.
Author Contributions
Y.-J. B., Z.-W. L., D. K., I. J., N.-C. H., and S. H. C. designed the research; Y.-J. B., Z.-W. L., D. K., and I. J. performed the research; Y.-J. B., I. J., N.-C. H., and S. H. C. analyzed the data; and Y.-J. B., I. J., N.-C. H., and S. H. C. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
This work was supported by Mid-career Researcher Program Grant 2015R1A2A1A13001654 through the National Research Foundation, funded by the Ministry of Science, ICT, and Future Planning (to S. H. C.) and the R&D Convergence Center Support Program of the Ministry of Agriculture, Food, and Rural Affairs (to S. H. C. and N.-C. H.), Republic of Korea. The authors declare that they have no conflicts of interest with the contents of this article.
S. H. Choi, unpublished observation.
- Prx
- peroxiredoxin
- AMS
- 4-acetamido-4′-maleimidylstilbene-2, 2′-disulfonic acid.
References
- 1. Storz G., and Imlay J. A. (1999) Oxidative stress. Curr. Opin. Microbiol. 2, 188–194 [DOI] [PubMed] [Google Scholar]
- 2. Stadtman E. R. (2001) Protein oxidation in aging and age-related diseases. Ann. N.Y. Acad. Sci. 928, 22–38 [DOI] [PubMed] [Google Scholar]
- 3. Lemire J. A., Harrison J. J., and Turner R. J. (2013) Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 11, 371–384 [DOI] [PubMed] [Google Scholar]
- 4. Cooke M. S., Evans M. D., Dizdaroglu M., and Lunec J. (2003) Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 17, 1195–1214 [DOI] [PubMed] [Google Scholar]
- 5. Miller R. A., and Britigan B. E. (1997) Role of oxidants in microbial pathophysiology. Clin. Microbiol. Rev. 10, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Storz G., and Zheng M. (2000) Oxidative stress. in Bacterial Stress Responses (Storz G., and Hengge-Aronis R. eds) pp. 44–59, American Society for Microbiology Press, Washington, D. C. [Google Scholar]
- 7. Dubbs J. M., and Mongkolsuk S. (2012) Peroxide-sensing transcriptional regulators in bacteria. J. Bacteriol. 194, 5495–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi H., Kim S., Mukhopadhyay P., Cho S., Woo J., Storz G., and Ryu S. E. (2001) Structural basis of the redox switch in the OxyR transcription factor. Cell 105, 103–113 [DOI] [PubMed] [Google Scholar]
- 9. Zheng M., Aslund F., and Storz G. (1998) Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279, 1718–1721 [DOI] [PubMed] [Google Scholar]
- 10. Jo I., Chung I. Y., Bae H. W., Kim J. S., Song S., Cho Y. H., and Ha N. C. (2015) Structural details of the OxyR peroxide-sensing mechanism. Proc. Natl. Acad. Sci. U.S.A. 112, 6443–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim S., Bang Y. J., Kim D., Lim J. G., Oh M. H., and Choi S. H. (2014) Distinct characteristics of OxyR2, a new OxyR-type regulator, ensuring expression of Peroxiredoxin 2 detoxifying low levels of hydrogen peroxide in Vibrio vulnificus. Mol. Microbiol. 93, 992–1009 [DOI] [PubMed] [Google Scholar]
- 12. Toledano M. B., Kullik I., Trinh F., Baird P. T., Schneider T. D., and Storz G. (1994) Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78, 897–909 [DOI] [PubMed] [Google Scholar]
- 13. Heo Y. J., Chung I. Y., Cho W. J., Lee B. Y., Kim J. H., Choi K. H., Lee J. W., Hassett D. J., and Cho Y. H. (2010) The major catalase gene (katA) of Pseudomonas aeruginosa PA14 is under both positive and negative control of the global transactivator OxyR in response to hydrogen peroxide. J. Bacteriol. 192, 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tseng H. J., McEwan A. G., Apicella M. A., and Jennings M. P. (2003) OxyR acts as a repressor of catalase expression in Neisseria gonorrhoeae. Infect. Immun. 71, 550–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee C., Lee S. M., Mukhopadhyay P., Kim S. J., Lee S. C., Ahn W. S., Yu M. H., Storz G., and Ryu S. E. (2004) Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat. Struct. Mol. Biol. 11, 1179–1185 [DOI] [PubMed] [Google Scholar]
- 16. Tao K., Fujita N., and Ishihama A. (1993) Involvement of the RNA polymerase α subunit C-terminal region in co-operative interaction and transcriptional activation with OxyR protein. Mol. Microbiol. 7, 859–864 [DOI] [PubMed] [Google Scholar]
- 17. Bang Y. J., Oh M. H., and Choi S. H. (2012) Distinct characteristics of two 2-Cys peroxiredoxins of Vibrio vulnificus suggesting differential roles in detoxifying oxidative stress. J. Biol. Chem. 287, 42516–42524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Janek K., Wenschuh H., Bienert M., and Krause E. (2001) Phosphopeptide analysis by positive and negative ion matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 15, 1593–1599 [DOI] [PubMed] [Google Scholar]
- 19. Groitl B., and Jakob U. (2014) Thiol-based redox switches. Biochim. Biophys. Acta 1844, 1335–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seo S. W., Kim D., Szubin R., and Palsson B. O. (2015) Genome-wide reconstruction of OxyR and SoxRS transcriptional regulatory networks under oxidative stress in Escherichia coli K-12 MG1655. Cell Rep. 12, 1289–1299 [DOI] [PubMed] [Google Scholar]
- 21. Klauck E., and Hengge R. (2012) σS-controlling networks in Escherichia coli. in Bacterial Regulatory Networks (Filloux A., ed) pp. 1–26, Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 22. Dong T., Joyce C., and Schellhorn H. E. (2008) The role of RpoS in bacterial adaptation. in Bacterial Physiology: A Molecular Approach (El-Sharoud W., ed) pp. 313–337, Springer, Berlin [Google Scholar]
- 23. Miller V. L., and Mekalanos J. J. (1988) A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. bacteriol. 170, 2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lim J. G., Bang Y. J., and Choi S. H. (2014) Characterization of the Vibrio vulnificus 1-Cys peroxiredoxin Prx3 and regulation of its expression by the Fe-S cluster regulator IscR in response to oxidative stress and iron starvation. J. Biol. Chem. 289, 36263–36274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 26. Goo S. Y., Lee H. J., Kim W. H., Han K. L., Park D. K., Lee H. J., Kim S. M., Kim K. S., Lee K. H., and Park S. J. (2006) Identification of OmpU of Vibrio vulnificus as a fibronectin-binding protein and its role in bacterial pathogenesis. Infect. Immun. 74, 5586–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]