Abstract
The bacterial messenger cyclic di-GMP (c-di-GMP) binds to a diverse range of effectors to exert its biological effect. Despite the fact that free-standing PilZ proteins are by far the most prevalent c-di-GMP effectors known to date, their physiological function and mechanism of action remain largely unknown. Here we report that the free-standing PilZ protein PA2799 from the opportunistic pathogen Pseudomonas aeruginosa interacts directly with the hybrid histidine kinase SagS. We show that PA2799 (named as HapZ: histidine kinase associated PilZ) binds directly to the phosphoreceiver (REC) domain of SagS, and that the SagS-HapZ interaction is further enhanced at elevated c-di-GMP concentration. We demonstrate that binding of HapZ to SagS inhibits the phosphotransfer between SagS and the downstream protein HptB in a c-di-GMP-dependent manner. In accordance with the role of SagS as a motile-sessile switch and biofilm growth factor, we show that HapZ impacts surface attachment and biofilm formation most likely by regulating the expression of a large number of genes. The observations suggest a previously unknown mechanism whereby c-di-GMP mediates two-component signaling through a PilZ adaptor protein.
Keywords: bacterial signal transduction, biofilm, cyclic di-GMP (c-di-GMP), histidine kinase, Pseudomonas aeruginosa (P. aeruginosa)
Introduction
Cyclic di-GMP (c-di-GMP)4 has emerged as a central regulator in many environmental and pathogenic bacteria. High c-di-GMP concentration generally correlates with the sessile lifestyle and formation of surface-associated biofilm. The cellular concentration of c-di-GMP oscillates with environmental changes through the opposing activities of diguanylate cyclase (DGC) and c-di-GMP phosphodiesterase (PDE) proteins. C-di-GMP exerts its biological effect by binding to riboswitches and a wide variety of protein effectors. PilZ domain proteins are the first discovered c-di-GMP-binding proteins and arguably the most prevalent ones (1). Apart from a small number of atypical PilZ proteins that lack c-di-GMP binding capability, PilZ proteins bind c-di-GMP with a remarkably wide range of binding affinities to regulate different pathways over a wide c-di-GMP concentration range (2, 3). More than half of the PilZ proteins encoded by bacterial genomes are free-standing PilZ proteins that consist of a single PilZ domain of 90∼130 residues. The remaining PilZ proteins are didomain or multidomain proteins that contain a PilZ domain and other functional domains. Recent studies have yielded insight into the function and mechanism of several didomain or multidomain PilZ proteins, including the “backstop brake” YcgR protein from E. coli, the cellulose synthase subunit BcsA from Rhodobacter sphaeroides, the transcriptional regulator MrkH from Klebsiella pneumoniae, the alginate-biosynthesizing protein Alg44 from P. aeruginosa and the chemotaxis receptor Tlp1 from Azospirillum brasiencse (4–12). A common theme emerged from these studies is that the binding of c-di-GMP to the PilZ domain triggers an intra-protein conformational change to modulate the enzymatic activity or binding property of output domains. In contrast, the molecular mechanisms for the free-standing PilZ proteins remain less known despite that several of them have been implicated in bacterial pathogenesis. It was found that the free-standing PilZ proteins from Xanthomonas campestris pv. campestris and Xanthomonas oryzae pv. Oryzae mediate motility, virulence, and extracellular enzyme production (13, 14). It was also found that the sole PilZ protein PlzA in Borrelia burgdorferi regulates motility and virulence by controlling gene expression (15, 16). Although it is widely hypothesized that free-standing PilZ proteins function as c-di-GMP adaptors by binding to their protein partners in a c-di-GMP-dependent fashion, experimental evidence supporting this hypothesis is still lacking. This hypothesis was even challenged when a PilZ protein from X. campestris pv. Campestris was found to interact with its protein partners in a c-di-GMP-independent manner (17).
The opportunistic pathogen P. aeruginosa is notorious for its ability to form multidrug-resistant biofilm on abiotic and biotic surfaces. P. aeruginosa harbors an extensive c-di-GMP signaling network that controls the switch between the motile and sessile life styles and regulates the formation and dispersal of biofilm. The c-di-GMP network of P. aeruginosa PAO1 consists of a large number of DGC and PDE proteins and at least 12 c-di-GMP binding effector proteins. The 12 effectors include FleQ, PelD, FimX, BrlR, PA4395, and seven PilZ-domain-containing proteins. The transcriptional regulator FleQ controls flagellar biosynthesis and the production of extracellular polysaccharides Pel and Psl; whereas the second transcription regulator BrlR is a multidrug transport activator associated with high-level antibiotic resistance (18–22). PelD and FimX control the production of the Pel exopolysaccharide (EPS) and type IV pili biosynthesis, respectively (23–27). The c-di-GMP-binding PA4395, a member of the YajQ family proteins, interacts with a transcriptional regulator for controlling gene transcription and virulence (28). The multi-domain protein Alg44 contains a c-di-GMP-binding PilZ domain to regulate the biosynthesis of the EPS alginate (10). Besides the six proteins with known function, there are six c-di-GMP-binding PilZ proteins whose functions still remain unknown. Among the six PilZ proteins, PA0012, PA2799, PA4324, and PA4608 are free-standing PilZ proteins and PA3353 and PA2989 are didomain proteins, with PA3353 as the homologue of YcgR from Escherichia coli (5). Elucidating the physiological role of the six PilZ proteins will substantially advance our understanding of c-di-GMP signaling in P. aeruginosa. As a step toward this goal, here we report the finding that PA2799 interacts with a sensor histidine kinase in a c-di-GMP-dependent manner. We show that PA2799 plays a role in early and late stages of biofilm formation by regulating the expression of a large number of genes. The results suggest a novel mechanism used by c-di-GMP to regulate two-component signaling through a PilZ adaptor and establish PA2799 as a new regulatory factor in biofilm formation.
Results
PA2799 Interacts with the Phosphoreceiver (REC) Domain of the Hybrid Histidine Kinase SagS
We postulated that free-standing PilZ proteins function as c-di-GMP adaptor proteins by binding to their protein targets. To identify the protein partner of PA2799, a customized bacterial two-hybrid screening system was created with a high-coverage P. aeruginosa genomic DNA library. The P. aeruginosa PAO1 genomic DNA library used for the bacterial two-hybrid screening was constructed by using the pTRG vector and two engineered pTRG vectors (see “Materials and Methods”) to maximize the number of open reading frames (ORFs) covered by the library. The high-coverage genomic library was designed to contain DNA fragments of 1–3 kb in length (supplemental Table S1 and Fig. S1). The bait plasmid was constructed by fusing the PA2799 gene to the bacteriophage λ repressor protein gene. The screening was performed by using PA2799 as the bait to probe for the prey protein encoded by the P. aeruginosa genomic DNA library. After several rounds of optimization to reduce the number of false-positive colonies, the screening yielded consistent results with more than two third of the prey plasmids containing a DNA fragment from the gene PA2824 (Fig. 1, A and B). While most of the other prey plasmids were found to be false positives that contain shifted ORFs, several clones contain a DNA fragment that encodes a portion of the diguanylate cyclase WspR that encompasses the entire GGDEF domain. However, no interaction was detected between PA2799 and the full-length WspR in the subsequent specific two-hybrid binding assay.
FIGURE 1.
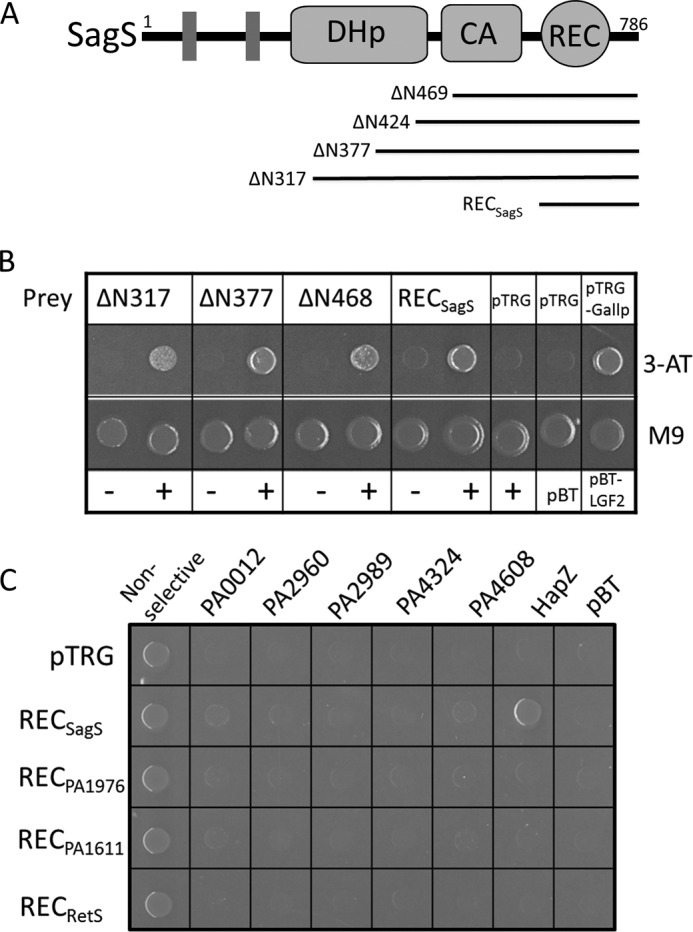
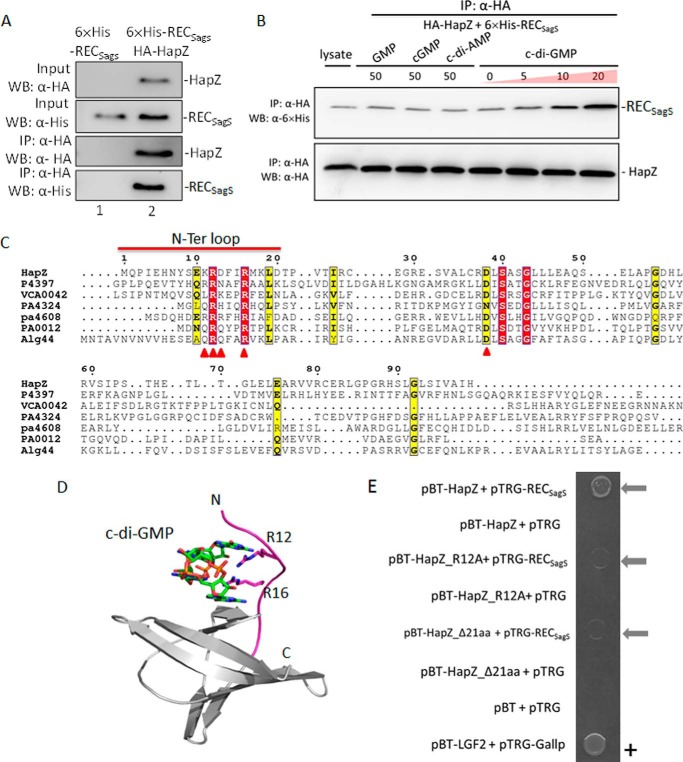
PA2799 (or HapZ) interacts with the phosphoreceiver domain of SagS as demonstrated by bacterial two-hybrid assay. A, domain organization of the hybrid histidine kinase SagS (DHp: dimerization and histidine phosphotransfer domain; CA: catalytic and ATP-binding domain; REC: phosphoreceiver domain). The fragments found to interact with PA2799 are represented by bars. B, colonies from bacterial two-hybrid assay indicate the interaction between PA2799 and various SagS constructs (ΔN317 denotes the construct that lacks the N-terminal 317 residues; − denotes empty pBT vector; + denotes pBT-HapZ vector). C, specific bacterial two-hybrid assays demonstrate the specific interaction between PA2799 (or HapZ) and SagS.
The gene PA2824 codes for the protein SagS, a membrane-anchored hybrid histidine kinase that contains a sensor domain, a dimerization and histidine phosphotransfer (DHp) domain, a catalytic and ATP-binding (CA) domain and a phosphoreceiver (REC) domain (Fig. 1A). To validate the protein-protein interaction and to test whether the C-terminal REC domain of SagS is sufficient for maintaining the interaction with PA2799, we performed specific bacterial two-hybrid binding assays using a construct that contains the free-standing RECSagS domain. As evidenced by the robust growth of the colonies on selective medium (Fig. 1B), the interaction between PA2799 and the RECSagS domain was readily detected to suggest that PA2799 is likely to interact with SagS through the REC domain. As the first PilZ protein found to interact with a histidine kinase, we renamed PA2799 as HapZ (Histidine kinase-associated PilZ protein).
The Protein-Protein Interaction between HapZ and SagS Is Specific
The histidine-containing phosphotransfer protein-B (HptB) is known as one of the downstream and phosphoryl-receiving partners of SagS (29). In addition to SagS, three additional sensor histidine kinases (PA1976, PA1611, and PA4856 or RetS) can also interact with HptB for phospho-transfer (29–32). On the other hand, P. aeruginosa PAO1 contains several PilZ proteins in addition to HapZ. To test the specificity of the interaction between SagS and HapZ, prey plasmids that encode the REC domains of PA1976, PA1611, and PA4856 and bait plasmids that encode the PilZ proteins were constructed for a “cross-binding” assay. Bacterial two-hybrid assays showed that only the interaction between HapZ and RECSagS could be detected (Fig. 1C), suggesting that HapZ is likely the only PilZ protein that interacts with SagS and that the interaction between HapZ and SagS is highly specific. Such specific protein recognition between the PilZ adaptors and its protein partners could be crucial for preventing crosstalk among different signaling pathways.
c-di-GMP Enhances the Protein-Protein Interaction between HapZ and SagS
It is known that HapZ binds c-di-GMP with a dissociation constant (Kd) of 2.0 μm (2). To further validate the interaction between HapZ and RECSagS and examine the effect of c-di-GMP on SagS-HapZ interaction, co-immunoprecipitation (Co-IP) assays were performed in P. aeruginosa PAO1 cell lysate. Plasmids were constructed to express hemagglutinin (HA)-tagged HapZ and 6×His-tagged RECSagS. We found that HA-HapZ could be co-immunoprecipitated with 6×His-RECSagS to confirm the interaction between SagS and HapZ (Fig. 2, A and B). The Co-IP assays also showed that the SagS-HapZ interaction is enhanced by c-di-GMP in a dose-dependent manner within the 0–20 μm range (Fig. 2B). Little enhancement was observed above 20 μm c-di-GMP, which is consistent with the binding affinity (Kd = 2.0 μm) of HapZ for c-di-GMP. The structurally related nucleotides GMP, cGMP, and c-di-AMP had little effect on SagS-HapZ interaction, indicating that the enhancing effect of c-di-GMP is rather specific. The enhancing effect of c-di-GMP on SagS-HapZ interaction is reminiscent of YcgR, a didomain PilZ protein that interacts with the switching complex of flagellar motor in E. coli (4, 5).
FIGURE 2.
The interaction between SagS and HapZ is enhanced by c-di-GMP. A, co-immunoprecipitation of HapZ with RECSagS. The cell lysate obtained from the ΔhapZΔsagS cells that harbor either pUCP-HA-HapZ (lane 1) or pUCP-HA-HapZ/6×His-RECSagS (lane 2) were immunoprecipitated with anti-HA beads at 4 °C (the same batch of total cell lysate was also used for Co-IP in Fig. 2B). The immunoprecipitated proteins and the total cell lysate used for Co-IP were assessed by Western blot analysis using anti-6×His and anti-HA antibodies. B, co-IP assay demonstrates the effect of c-di-GMP, GMP, Cgmp, and c-di-AMP on SagS-HapZ interaction. C, sequence alignment of HapZ, PA0012, PA4608, PA4324, and three structurally characterized PilZ domains. The four key residues for binding c-di-GMP in PA4608 (PDB: 2L74) are indicated by ▵. D, structural model of HapZ in complex with dimeric c-di-GMP. The model was constructed using PA4608 structure as template. C-di-GMP and the conserved R12 and R16 are shown in sticks and the N-terminal loop in magenta. E, specific bacterial two-hybrid assay demonstrates the weakened interaction between RECSagS and the two HapZ mutants that lack Arg12 (HapZ_R12A) or the entire N-terminal loop (HapZΔ21aa).
PilZ proteins bind c-di-GMP by using several conserved residues from the β-barrel and the flexible N-terminal loop (7, 11, 33). Sequence alignment and structural modeling suggests that PA2799 is likely to bind c-di-GMP using the same set of residues from both the β-barrel and the 22-residue N-terminal loop (Fig. 2, C and D). As indicated by the barely visible colony, the protein-protein interaction seemed to be significantly weaker between RECSagS and the HapZ_R12A mutant that lacks the conserved c-di-GMP-binding residue Arg12 (Fig. 2E). Likewise, the interaction also seemed to be weaker between RECSagS and the HapZ_Δ21aa construct that lacks the entire N-terminal loop. The Co-IP and two-hybrid assay results strongly suggest that HapZ and SagS directly interact with each other, and that the binding of c-di-GMP to HapZ enhances the protein-protein interaction.
Unfortunately, our further attempts to characterize the SagS-HapZ interaction and quantify the binding affinity using isothermal titration calorimetry (ITC) and nuclear magnetic resonance (NMR) spectroscopy was not successful because of the tendency of HapZ and its variants to form insoluble aggregate at high concentration (>100 μm), which is required for the in vitro binding assays.
HapZ Blocks the Phosphotransfer between SagS and the Downstream Protein HptB in a c-di-GMP-dependent Manner
It has been known that SagS initiates a phosphorelay cascade in P. aeruginosa with HptB and the bifunctional PA3346 protein as the immediate downstream protein partners (Fig. 3A) (31, 32). PA3346 possesses either kinase or phosphatase activity, depending on the phosphorylation state of the protein. The observed SagS-HapZ interaction hints that HapZ can potentially regulate the SagS two-component signaling pathway by directly binding to SagS. We performed in vitro phosphorylation assay to investigate how HapZ and c-di-GMP affect the phosphotransfer between SagS and HptB. When the 6×His-tagged recombinant proteins SagS246–786 and HptB (1:1 ratio) were incubated in the presence of [γ-32P]ATP, phosphorylation of HptB was observed readily (Fig. 3B). When we included freshly prepared HapZ in the reaction mixture at a SagS/HptB/HapZ ratio of 1:1:1, the phosphorylation of HptB was suppressed, with the final amount of phosphorylated HptB reduced by ∼40%. This observation supports the view that HapZ can interact with SagS even in the absence of c-di-GMP. Nonetheless, addition of c-di-GMP further decreased the phosphorylation level of HptB in a dose-dependent manner till the [c-di-GMP] reached about 50 μm (Fig. 3B). The effect of c-di-GMP is specific because the enhancing effect was not observed for GMP and other nucleotides (Fig. 2C). C-di-GMP did not seem to have a significant effect on HptB phosphorylation when HapZ was replaced by the HapZ_R12A/R16A double mutant that is likely to have a much weaker affinity for c-di-GMP (Fig. 3D), suggesting that c-di-GMP exerts its effect through HapZ. The different effect of HapZ and the double mutant is unlikely due to different protein stability as the recombinant mutant protein seemed to exhibit similar fold efficiency (in E. coli) and in vitro stability (estimated by the decrease of the A280 nm of protein solutions at ambient temperature). Note that although a small sub-family of histidine kinases has been reported to be able to bind c-di-GMP directly (34), SagS is unlikely to bind c-di-GMP due to the absence of c-di-GMP binding residues (supplemental Fig. S2). Taken together, the phosphorylation assays indicate that HapZ can bind to SagS to inhibit phosphorelay, and that the inhibition is likely to be further enhanced at high c-di-GMP concentration.
FIGURE 3.
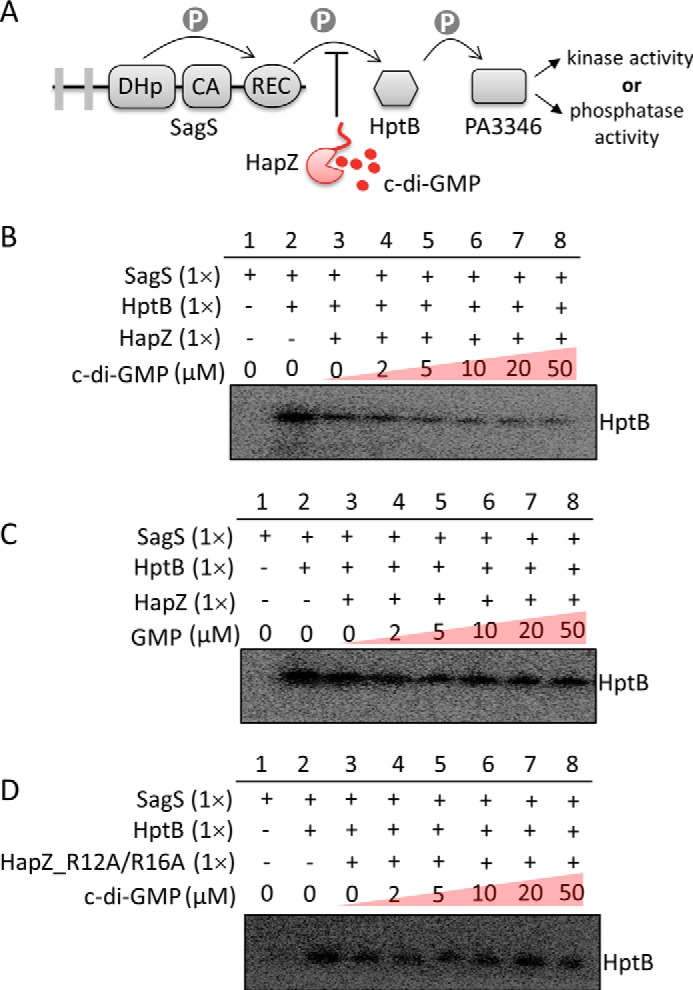
HapZ inhibits the phosphotransfer between SagS and HptB and the inhibition is further enhanced c-di-GMP. A, schematic illustration of the phosphorelay between SagS and downstream signaling proteins HptB and PA3346. B, effect of HapZ and c-di-GMP on the phosphorylation of HptB by SagS. The cytoplasmic portion of SagS (SagS246–786) and [32P]ATP were used for the assays. Detailed reaction conditions are described in “Materials and Methods.” C, effect of GMP on the phosphorylation of HptB. D, effect of c-di-GMP on the phosphorylation of HptB in the presence of the HapZR12A/R16A double mutant instead of HapZ.
HapZ Is Involved in Surface Attachment and Biofilm Formation
P. aeruginosa is notorious for its ability to form antibiotic-resistant biofilm on abiotic and biotic surfaces. SagS, which stands for surface attachment and growth sensor hybrid, is known to control surface attachment and development of P. aeruginosa biofilm (29). If HapZ interacts with SagS in the cell, HapZ is likely to affect surface attachment and biofilm development as well. By using biofilm-growing flow cells and a confocal microscope, we examined how the deletion and overexpression of hapZ affects the formation of P. aeruginosa PAO1 biofilm. As shown in Fig. 4A, PAO1 formed the characteristic mushroom-shaped biofilm after 5 days of growth; whereas the ΔhapZ mutant strain exhibited significant delay in early surface attachment to result in a thin biofilm at the end. On the contrary, the overexpression strain (hapZ+) exhibited enhanced surface attachment at the early stage. However, despite the enhanced surface attachment, the hapZ+ strain forms a flat and fragile biofilm that completely lacks the mushroom structure at the end of five-day growth. The control experiment with PAO1 transformed the empty overexpression plasmid showed that the formation of mushroom-structured biofilm was not affected (data not shown). These observations indicate that HapZ impacts both surface attachment and biofilm development.
FIGURE 4.

HapZ plays a regulatory role in surface attachment and biofilm development. A, flow cell biofilm assay for PAO1 and the two mutants. The biofilms of PAO1, the deletion (ΔhapZ) and overexpression (hapZ+) mutants were grown in flow cells side-by-side and continuously monitored by confocal microscope without disrupting the flows. Confocal scanning laser microcopy micrographs show surface attachment of bacterial cells and formation of biofilm at the end of day 3 and day 5. Scale bars (50 μm). B, confocal scanning laser microcopy micrographs of the biofilm formed by the co-cultured PAO1 and ΔhapZ strain and co-cultured PAO1 and hapZ+ strains.
To further confirm the role of HapZ in initial surface attachment during early biofilm formation, we conducted a competitive biofilm assay with mixed P. aeruginosa populations. When the cyan fluorescence protein (CFP)-tagged PAO1 strain was co-cultured with the YFP-tagged ΔhapZ strain in flow cells, the final biofilm was dominated by PAO1 (Fig. 4B), confirming that the ΔhapZ strain was impaired in surface attachment. In contrast, the matured biofilm was dominated by the hapZ+ strain when PAO1 was co-cultured with YFP-tagged hapZ+ strain, suggesting that the hapZ+ strain is more competent than PAO1 in surface attachment. These observations suggest that HapZ is involved in the processes of surface attachment and biofilm development, which further strengthens the functional relationship between SagS and HapZ.
HapZ Mediates Swarming and Twitching Motility
Flagella-mediated swimming and swarming motility are indispensable for P. aeruginosa biofilm formation. We found that the deletion and overexpression of hapZ affects swarming, but not swimming motility. The ΔhapZ mutant exhibited a hyper-swarming phenotype and the phenotype could be reversed by HapZ overexpression (Fig. 5A). Suppression of swarming by HapZ was also demonstrated by the further subdued swarming activity of hapZ+, suggesting that the swarming phenotype of ΔhapZ is indeed due to the low level of hapZ. The hyper-swarming phenotype of ΔhapZ could not be reversed by the complementation with the hapZ_Δ21 construct that lacks the N-terminal loop, which confirms the importance of c-di-GMP binding for the function of HapZ. A close inspection of the swarming plate revealed large zones of fluid surrounding the swarming tendrils of the ΔhapZ cells. Since the zone of fluid is known to be caused by the production of rhamnolipids (35), we reasoned that the increased swarming for ΔhapZ mutant is, at least partially, caused by an increased production of rhamnolipids. To test this, we quantified the amount of rhamnolipid produced by PAO1 and the two mutants by emulsification activity assay. Indeed, increased rhamnolipid production was observed for ΔhapZ whereas decreased rhamnolipid production was observed hapZ+ (Fig. 5B).
FIGURE 5.
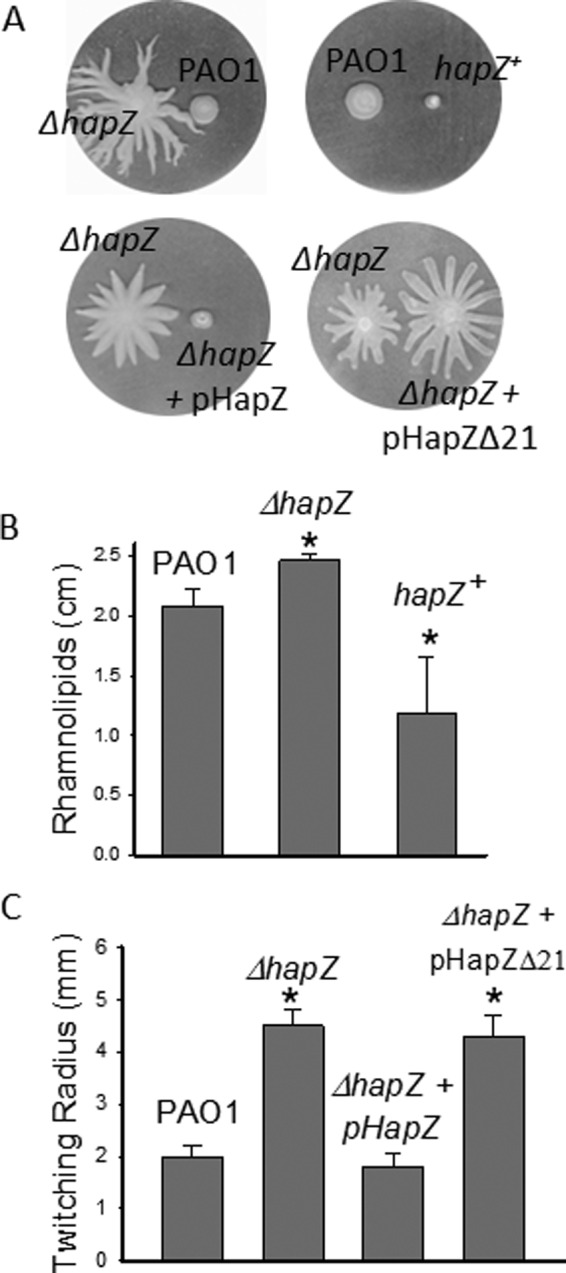
HapZ impacts swarming and twitching motility. A, analysis of swarming motility of PAO1 and mutants on semi-hydrated swarming plates. B, altered production of rhamnolipids for the ΔhapZ and hapZ+ strains as measured by emulsification activity assay. C, analysis of twitching motility of PAO1 and mutant strains by comparing the radius of twitching zones.
Apart from swimming and swarming, type IV pili-dependent twitching mobility is also known to be crucial for the formation of P. aeruginosa biofilm (36). Our twitching motility assay suggests that HapZ affects twitching motility. The ΔhapZ mutant showed increased twitching zone and the enhanced twitching could be suppressed by the overexpression of HapZ, but not by the overexpression of HapZ_Δ21 (Fig. 5C). It is already known that c-di-GMP can regulate twitching motility through the effector FimX in P. aeruginosa (23). The results described here suggest that HapZ is another effector through which c-di-GMP mediates twitching motility.
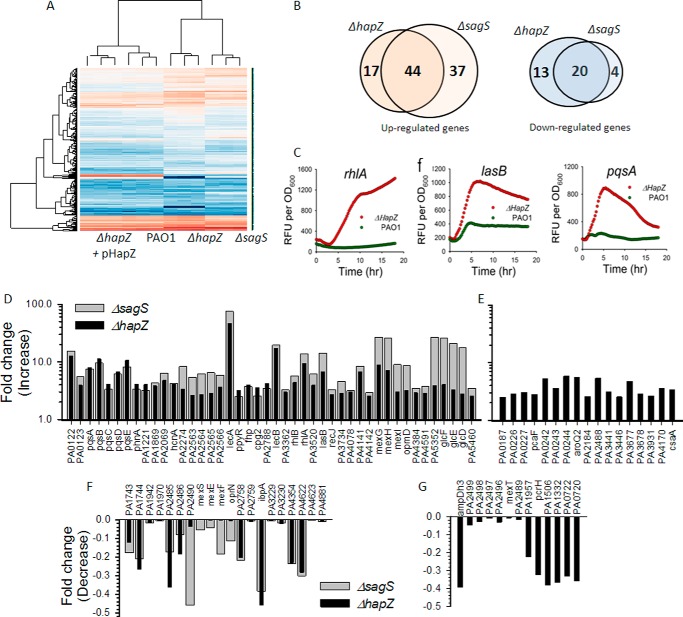
HapZ affects the expression of a large number of genes related to quorum-sensing (QS), motility and other functions(69) SagS and HptB-associated signaling pathways are known to control gene expression (32). We performed transcriptomic analysis by RNA-Seq for PAO1, ΔhapZ, ΔsagS, and the complementation strain (ΔhapZ + pHapZ) to further establish the relationship between SagS and HapZ and to identify the genes under the control of HapZ (32, 37).
For the cells collected at stationary growth stage, the gene expression profile of the ΔhapZ strain differs substantially from that of PAO1 (Fig. 6A). The gene expression pattern could be restored by HapZ complementation, suggesting the differential gene expression is due to a change in HapZ level. Compared with PAO1, a total of 94 genes of ΔhapZ exhibited statistically significant alteration in mRNA level (Fig. 6B). Among the 61 genes showing up-regulation (>2.5 fold-change) (Fig. 6, D and E; supplemental Tables S4 and S6), some are already implicated in motility, virulence and biofilm formation. The greatest increases were observed for the genes lecA (46-fold) and lecB (17-fold), which encodes two lectins that are implicated in cell surface adhesion. LecB is also essential for biofilm development because the lecB-deficient strain is impaired in biofilm formation (38). Pseudomonas quinolone signal (PQS) is a mediator of motility, biofilm formation, population structure and survival of Pseudomonas in hostile environment (39, 40). Several genes (pqsA, B, C, D, and E) involved in the biosynthesis of PQS are up-regulated in the ΔhapZ mutant by 4–11-fold. The expression levels of the rhamnolipid biosynthetic genes rhlA (9.4-fold), rhlB (4.4-fold), and rhlI (2.3-fold) are also up-regulated, which is consistent with the hyper-swarming phenotype and stimulated production of rhamnolipid of ΔhapZ. Elevated expression levels were also observed for the genes that encode the MexG/H/I-OpmD efflux pump (3–8 fold), elastase (lasB, 6.6-fold), and three genes (glcD/E/F) from the central metabolic glycolate pathway. The altered expression levels of the QS-controlled lasB, rhlA and pqsA were further validated for ΔhapZ by using promoter-gfp-fusion reporters. Quantification of GFP fluorescence showed elevated transcription levels for the three QS-controlled genes for ΔhapZ (Fig. 6C). Meanwhile, a total of 33 genes exhibited decreased expression levels in ΔhapZ (Fig. 6, F and G; supplemental Tables S5 and S7). Among the annotated genes, mexE, mexF and oprN code for the MexEF-OprN efflux pump and mexS codes for an associated regulator that has been implicated in biofilm formation, antibiotic resistance, and virulence (41). The expression levels of ibpA and ampDh3, which encode a small heat shock protein and an antibiotic resistance protein, respectively, are also lower in ΔhapZ.
FIGURE 6.
Co-regulation of gene expression by HapZ and SagS. A, heatmaps for the PAO1, ΔhapZ, ΔsagS, and complementation (ΔhapZ + pHapZ) strains obtained from RNA-Seq transcriptome analysis. B, Venn diagrams showing the co-regulated genes by HapZ and SagS. The top genes that exhibited the greatest differences in gene expression are listed in the overlapping regions. C, up-regulated expression of three QS-controlled genes (lasA, rhlA, and pqsA) in ΔhapZ as measured by the gfp-fused reporter assay. D, genes that are up-regulated in both ΔhapZ and ΔsagS. E, genes that are up-regulated in ΔhapZ, but not in ΔsagS. F, genes that are down-regulated in both ΔhapZ and ΔsagS. G, genes that are down-regulated in ΔhapZ, but not in ΔsagS.
Compared to the 94 genes of ΔhapZ that showed different expression from PAO1, a total of 105 genes exhibited altered expression levels in ΔsagS (supplemental Tables S4, S5, S8, and S9). The regulons of SagS and HapZ overlap with a total of 64 co-regulated genes (Fig. 6B). Notably, among the 64 genes, 44 are up-regulated and 20 are down-regulated, and none of the genes showed opposite change in ΔsagS and ΔhapZ. The 44 up-regulated genes shared by ΔsagS and ΔhapZ include lasB, lecA/B rhlA/B, mexG/H/I, opmD, pqsA-E, glcD/E/F, and other unannotated genes (Fig. 6D). The most notable down-regulated genes shared by the ΔsagS and ΔhapZ are the efflux pump genes mexE/F, oprN, and several other unannotated genes (Fig. 6F). Sauer et al. recently reported that inactivation of sagS altered the expression of mexA/B, oprM, mexE/F and oprN genes (20). The observed changes in the expression level for these genes for the ΔsagS strain are consistent with their observations. Taken together, the gene expression studies show that both SagS and HapZ are involved in gene regulation and they share a large number of co-regulated genes.
Discussion
In P. aeruginosa, c-di-GMP exerts its effect by binding to several PilZ domain proteins to regulate pathways that are not yet known. Elucidating the function of the PilZ effectors will significantly advance our understanding of the role of c-di-GMP in P. aeruginosa. To this end, our studies yield insight into the function of one PilZ effector (HapZ) by identifying the hybrid histidine kinase SagS as its potential physiological partner. Our biochemical studies suggest that although HapZ can interact directly with SagS in the absence of c-di-GMP, the SagS-HapZ interaction is enhanced in the presence of c-di-GMP. We demonstrated that HapZ mediates bacterial motility and biofilm formation, and that the deletion of hapZ had a pleiotropic effect on gene expression. These results suggest a previously unknown mechanism used by c-di-GMP to control two-component signaling through a discrete PilZ adaptor.
An important finding of our study is that HapZ plays an important role in biofilm formation. Given the importance of biofilm in P. aeruginosa chronic infection (42), HapZ may play an active role in the in vivo survival and pathogenesis of P. aeruginosa and contribute to the persistence of chronic infection. Formation of biofilm involves many regulatory factors and several stages that include a reversible attachment stage, an irreversible attachment stage and two maturation or development stages (43, 44). As a master regulator of biofilm formation in P. aeruginosa, c-di-GMP controls different stages of biofilm formation by regulating a myriad of cellular functions (45). It has been suggested that SagS acts as a motile switch during early stage of biofilm formation to control surface attachment (29, 37). In accordance with the role of SagS, we found that HapZ affects the ability of P. aeruginosa cells in surface attachment. Ability to swarm on solid or semi-solid surface is one of the major factors that affect surface attachment; and suppression of swarming motility to create a non-motile subpopulation is crucial for initial surface attachment during biofilm formation (46, 47). Our study indicates that swarming motility is likely one of the functions controlled by HapZ to impact surface attachment. The thin biofilm formed by the ΔhapZ mutant is partially due to enhanced swarming motility that prevents cells from initiating surface attachment. The ability of P. aeruginosa to swarm is dependent on many factors (35, 48). The HapZ-dependent pathway is likely to mediate swarming motility by affecting the production of QS signals, rhamnolipids and likely other factors.
In addition to its role in surface attachment, our data suggests that HapZ is actively involved in the process of biofilm development or maturation. Development of robust P. aeruginosa biofilm microcolonies following surface attachment requires phenotypic changes in swarming and twitching motility, EPS biosynthesis and export, QS activity, rhamnolipid production and central metabolism. The transition from surface attachment stage to maturation stage during biofilm formation is accompanied by extensive shift in gene expression (46). Such shift in gene expression is vital for biofilm maturation as the surface-attached cells need to build extracellular matrix and grow microcolonies vertically. The lack of microcolonies for the hapZ+ mutant that is highly efficient in surface attachment indicates that a high level of HapZ impedes the shift from a “surface attachment phenotype” to a “maturation phenotype” in biofilm formation. HapZ and its associated pathways are likely to contribute to biofilm development by controlling multiple factors and pathways. Most notably, our data suggest that HapZ controls the expression of many genes that are under the control of the three QS systems (Las, Rhl, and PQS) of P. aeruginosa. As the three QS systems have all been implicated in biofilm development (49–51), it is likely for HapZ to impact biofilm development by regulating quorum-sensing activity. Secondly, SagS is a key regulator in the formation of highly antimicrobial tolerant biofilm microcolonies (20). HapZ is likely to contribute to the development of resistant biofilm because it mediates the expression of genes that encode several major efflux pumps and metabolic enzymes. Thirdly, as twitching motility is essential for the formation of structured biofilm microcolonies (52), HapZ may also participate in biofilm development by controlling twitching motility.
SagS is part of a highly complex two-component signaling network composed of multiple sensor kinases, response regulators, connectors, and transcriptional regulators. SagS can initiate phosphorelay cascades through both HptB-dependent and -independent pathways (29, 31, 32, 37, 53). Our current understanding of this intricate two-component signaling network is far from complete. What is clear now is that this complex two-component signaling network controls the expression of a large number of genes to coordinate the timing of surface attachment and biofilm development, with the pathways activated or deactivated at different stages during biofilm formation (37, 54–57). Integration of HapZ in the two-component signaling network presumably allows the phosphorelay cascades to be regulated by additional inputs sensed by c-di-GMP pathways. It was presumed that the input signal sensed by the sensor domain of SagS triggers the SagS-controlled motile-to-sessile switching. The current study raises the possibility that the switching is triggered by a change in cellular c-di-GMP level instead. Moreover, SagS has been suggested to regulate gene expression by interacting with the histidine kinase BfiS and controlling its phosphorylation (37). It is plausible that c-di-GMP and HapZ may also mediate this HptB-independent pathway to impact gene expression. This may be the reason why the deletion of sagS and hapZ did not have opposite effect on gene expression, as one would infer from the observation that HapB inhibits the phosphotransfer between SagS and HptB. As a step toward fully unraveling this intricate two-component signaling network, the current study unveils the unexpected involvement of c-di-GMP and HapZ and sets the stage for investigating how c-di-GMP controls various stages of biofilm formation through this signaling network.
The involvement of c-di-GMP in two-component signaling is a common phenomenon, as evidenced by the fact that many bacterial genomes contain a large number of genes that encode response regulators containing c-di-GMP-synthesizing (GGDEF) and degrading (EAL or HD-GYP) domains (58–63). By using the c-di-GMP-synthesizing or degrading response regulators, the environmental inputs sensed by histidine kinases can be translated into oscillation of cellular c-di-GMP concentration. One of the findings from recent studies is that c-di-GMP can also control the activity and function of histidine kinases to regulate two-component signaling. For example, the bifunctional histidine kinase CckA involved in cell cycle control in C. crescentus binds c-di-GMP directly in the catalytic- and ATP-binding domain for regulating the kinase and phosphatase activities (34). Another example is the orphan hybrid histidine kinase SgmT from Myxococcus xanthus. SgmT has a C-terminal non-enzymatic GGDEF domain that functions as a c-di-GMP-binding effector. The binding of c-di-GMP to the GGDEF domain is crucial for the spatial sequestration, and likely cellular function of SgmT (64). The current study suggests that another way for c-di-GMP to control histidine kinase is by using a discrete PilZ adaptor. In principle, the use of PilZ adaptors has the advantage of allowing c-di-GMP to control multiple pathways through highly specific kinase-adaptor interaction. Because PilZ proteins exhibit a wide range of binding affinity for c-di-GMP (23), the effect of c-di-GMP on a specific pathway can be “felt” only when the c-di-GMP concentration reaches a certain threshold. It is also conceivable that evolving a discrete PilZ adaptor to regulate histidine kinase is easier than creating a c-di-GMP-binding site in the catalytic domain of histidine kinases because of fewer evolutionary constricts. The ability to rapidly evolve new c-di-GMP-binding PilZ adaptors for two-component signaling pathways may be crucial for the survival of such highly adaptable bacteria as P. aeruginosa.
Materials and Methods
Construction of P. aeruginosa Genomic DNA Libraries
For genomic library construction, the BamHI site was used for vector digestion and Sau3AI site for gDNA partial digestion. The vector pTRG of the BacterioMatch II Two-Hybrid system (Stratagene) was used for prey library construction. Three independent genomic DNA libraries were constructed by using the vector pTRG, and the engineered pTRG-derived pTRG-A2 and pTRG-AC1. The plasmids pTRG-A2 and pTRG-AC1 were constructed by inserting the nucleotides “A” and “AC” between the 3′-end of the RNAP-α (992 bp) gene and the BamHI site (993 bp) of, respectively. The insertion resulted in a shift of the polylinker site by +1 or +2 nucleotides. The vectors pTRG, pTRG-A2 and pTRG-AC1 were digested by BamHI and dephosphorylated with phosphatase. The vector digestion mixture was extracted with phenol: chloroform, precipitated, and resuspended into 1× TE buffer. The genomic DNA of P. aeruginosa strain PAO1 was isolated using the chloroform-cetyltrimethylammonium bromide extraction method, and partially digested by Sau3AI. The randomly digested fragments ranging in size from 1,000 to 3,000 bp were purified using NucleoSpin® Gel and PCR Clean-up kit (MACHEREY-NEGEL). Gel-purified DNA fragments were ligated to the linearized and dephosphorylated pTRG, pTRG-A2, or pTRG-AC1 vectors. The resulting ligation mixtures were precipitated and resuspended into 1× TE buffer, followed by transformation into E. coli XL1-Blue MRF' Kan competent cells using a standard electroporation procedure. The three libraries were collected and pooled separately as the prey libraries and stored in −80 °C freezer.
Bacterial Two-hybrid Screening
The plasmid pBT-PA2799 was used as the bait to probe the genomic DNA libraries using the BacterioMatch II Two-Hybrid system (Stratagene). The plasmids extracted from the cells harboring the library fragments were co-transformed into the reporter strain with the bait plasmid by electroporation. The co-transformed cells were grown on M9+ His-deficient medium containing 5 mm 3-AT for 48–72 h at 30 °C. Colonies that grew on these plates were selected as positive colonies. The equivalent of 1 × 106 colonies were screened, as assessed by plating the transformed cells on M9+ His-deficient medium. Positive colonies were subsequently picked and re-streaked on M9+ His-deficient medium containing 5 mm 3-AT and 12.5 μg/ml streptomycin. The colonies that grew on the double selection medium were cultured in liquid medium, and used for colony PCR with pTRG F/R pairs to amplify the insert from the library plasmid. The positive colonies were grown for the preparation of plasmids for sequencing. For the specific two-hybrid binding assay, the bait and prey plasmids were co-transformed and the co-transformed mixtures were plated onto non-selective medium and incubated at 37 °C overnight. The growth of the selected co-transformed clones was verified by dotting on the selective medium by 37 °C overnight incubation. Normal growth on the selective screening medium was considered as an indicator of positive protein-protein interaction.
Co-immunoprecipitation (Co-IP) Assay
To validate the interaction between HapZ and RECSags in P. aeruginosa by Co-IP assay, the two plasmids pUCP-HA-HapZ, pUCP-HA-HapZ/6×His-RECSagS (supplemental Table S2) were constructed and transformed into PAO1. The strain harboring pUCP-HA-HapZ consistently expressed protein HA-HapZ while the stain harboring pUCP-HA-HapZ/6×His-RECSagS produced both HA-HapZ and 6×His-RECSagS. The bacterial strains were cultured in LB containing 250 μg/ml carbenicillin at 37 °C overnight with shaking and subsequently sub-cultured to an OD600 of 1.0 at 37 °C. Cells were harvested by centrifugation (13,000 × g for 10 min) and lysed with ice-cold cell lysis 1× PBS buffer (10 mm NaPi (pH 7.4), 137 mm NaCl, and 2.7 mm KCl, 0.5% Nonidet P-40, 10% glycerol, 1 mm EDTA and protease inhibitor mixture (Roche)). The total cell lysates were clarified by centrifugation at 15,000 × g for 10 min, and the supernatant was pre-clarified with protein G beads after being filtered by Minisart® high flow Syringe Filters. Lysates was subsequently immunoprecipitated with the protein A-Sepharose beads (EZview™ Red Anti-HA Affinity Gel, Sigma-Aldrich). Each immunoprecipitation sample containing 800 μl of lysate, 20 μl of the prewashed beads with different concentrations of nucleotides (c-di-GMP, GMP or cGMP) was incubated for 1 h at 4 °C. The beads were collected by centrifugation and washed six times with lysis buffer at 4 °C, and then the immunoprecipitated proteins were diluted with 2× sample buffer and subjected to SDS-PAGE and Western blotting with HA antibody.
Cloning, Expression, and Purification of Recombinant Proteins
The recombinant proteins HapZ, HptB, and SagS246–786 were expressed using E. coli Rosetta competent cells. Transformed cells were grown at 37 °C to OD600 of 0.6 in LB medium containing 50 μg/ml kanamycin and induced for protein expression. Protein expression was induced using 0.5 mm isopropyl β-d-thiogalactopyranoside (IPTG) (16 °C). After 16 h, cells were harvested by centrifugation and resuspended in phosphate buffer, pH 8.0 and lysed using French Press homogenizer at 15,000 psi. The cells were then centrifuged at 25,000 rpm for 30 min, and supernatants were filtered and loaded onto a nickel-charged column equilibrated with KPi buffer (pH 8.0). The column was washed sequentially using W1 (KPi buffer, pH 8.0 + 20 mm imidazole) and W2 (KPi buffer, pH 8.0 + 50 mm imidazole) buffer. The 6×His-tagged proteins were eluted from the column using elution buffers (KPi buffer, pH 8.0) containing increasing concentrations of imidazole (100, 200, and 300 mm) and were analyzed using SDS-PAGE. The fractions containing the desired proteins were pooled with a purity > 95%, desalted using PD-10 column (GE Healthcare) and concentrated using Amicon concentrator (10 kDa or HptB and SagS246–786 and 5 kDa for HapZ) at 4,000rpm at 4 °C. The concentrations of the proteins were determined by Bradford method and were aliquot and stored at −80 °C with the exception of HapZ, which must be used without storage due to its propensity to aggregation.
In Vitro Phosphorylation Assay
C-di-GMP was prepared using a thermophilic diguanylate cyclase as described previously (65, 66). Phosphorelay between SagS and HptB was examined by performing in vitro phosphorylation assay with 6×His-tagged recombinant proteins. The reaction mixture that contains SagS246–786 and HptB (1:1 ratio), [32P]ATP, and MgCl2 was incubated at 25 °C for 30 min in reaction buffer, which is composed of 100 mm Tris-HCl (pH 8.0), 50 mm KCl, 5 mm MgCl2, 1 mm DTT, 1 μCi of [γ-32P]ATP (about 111 TBq/mmol, 10 mCi/ml, 3.33 μm, 3,000 Ci/mmol). Phosphorylation of HptB was visualized by running 15% SDS-polyacrylamide gel electrophoresis and autoradiography. The effect of HapZ and c-di-GMP on HptB phosphorylation was examined by incubating the corresponding components with the reaction mixture.
Biofilm Formation in Flow-cell
The P. aeruginosa strains were tagged with GFP reporter by inserting the mini-Tn7-eGFP-Gmr, mini-Tn7-eCFP-Strepr and mini-Tn7-eYFP-Strepr cassette. Biofilms were grown in flow chambers with individual channel dimensions of 1 × 4×40 mm. The flow chambers were inoculated by injecting 350 μl overnight culture diluted to an OD600 nm of 0.01 into each flow channel using a small syringe. After inoculation, the flow channels were left without flow for 1 h, after which medium flow was started using a Watson Marlow 205S peristaltic pump. The mean flow velocity in the flow-chambers was 0.2 mm s−1. All microscopic observations and image acquisitions were done with a Zeiss LSM780 confocal laser scanning microscope (CLSM). Data were analyzed to generate the simulated three-dimensional images using the IMARIS software package (Bitplane AG) (67).
Transcriptome Analysis by RNA-Seq
Colonies of P. aeruginosa PAO1 and the three mutant strains were grown overnight in ABTGC medium at 37 °C. The cultures were diluted to a starting OD600 nm of 0.01, and 1 ml of culture was added to a well of a 24-well plate. The plate was sealed with parafilm and incubated at 37 °C to reach stationary phase. The cultures were mixed immediately with 2 volumes of RNAprotect® Bacteria Reagent (Qiagen). After a 5-min incubation at room temperature, samples were centrifuged, and the pellets were stored at −80 °C. Bacterial cells were treated with lysozyme and total RNA was extracted with RNeasy Mini Purification kit (Qiagen). Removal of DNA was carried out by on-column DNase digestion with the RNase-free DNase Set (Qiagen). The integrity of total RNA and DNA contamination was assessed and the 16S, 23S, and 5S rRNAs were removed by using the Ribo-Zero™ Magnetic Kit (Epicenter). Gene expression analysis was performed in duplicate by RNA sequencing. The rRNA-depleted RNA was fragmented to 200–300-bp fragments, and the first and second strand cDNA were synthesized, followed by end repair and adaptor ligation. The libraries were sequenced using the Illumina HiSeq2000 platform with a paired-end protocol and read lengths of 100 nt. The RNA-Seq data are available in the NCBI GEO Short Read Archives (SRA No: SAMN02369270). The sequence reads were assembled and analyzed in RNA-Seq and the expression analysis application of CLC genomics Workbench 6.0 (CLC Bio, Aarhus, Denmark). The following criteria were used to filter the unique sequence reads: minimum length fraction of 0.9, minimum similarity fraction of 0.8, and maximum number of two mismatches. Data were normalized by calculating the reads per kb per million mapped reads for each gene (68). ANOVA and t test were performed on transformed data to identify the genes with significant changes in expression (p < 0.05, fold change > 2.5).
Swarming and Twitching Motility Assay
Swarming motility was analyzed on 0.5% agar M8 salt plates supplemented with 0.2% glucose, 2 mm MgSO4, and 0.5% casamino acid. 1.5 μl of cell culture grown at 37 °C overnight in LB medium were dotted on freshly prepared swarming plates and incubated for 16 h at 37 °C. The PAO1 strain was included as a control on each swarming plate. For twitching motility assay, PAO1 cells were stab inoculated with a toothpick through a thin Difco LB agar (3 mm, 1% Difco granulated agar) layer to the bottom of the Petri dish. After overnight growth at 37 °C, the zone of twitching was visualized by staining with Coomassie Brilliant Blue R250, and the radius of the zone was measured.
Quantification of Rhamnolipid Production
The emulsification activity assay was modified from as previously described (69). 1 ml of filtered overnight culture of the different P. aeruginosa strains was mixed with 1 ml of hexadecane in a glass tube. The mixture was vortexed rigorously for 2 min, and was allowed to stand for 2 h in room temperature. Emulsification activity was measured as the height of emulsion layer divided by the total height of the mixture. Experiments were performed in triplicate, and the results are shown as the mean ± S.D.
Gene Expression Reporter Fusion Assay
Overnight cultures of P. aeruginosa PAO1 and ΔhapZ strains carrying the different QS report fusions (PlasB-gfp (69), PrhlA-gfp (70), and PpqsA-gfp (71)) were diluted 100 times in ABTGC medium. 200 μl of each diluted culture was added to each wells of a 96-well plate. GFP fluorescence and observance at 600 nm was monitored overnight using a microplate reader (Tecan Infinite 2000). Experiments were performed in six replicates, and the results are shown as the mean ± S.D.
Author Contributions
Z. X. L. designed the study and wrote the paper with the assistance of L. X., P. V., and Y. L. The two-hybrid screening, colony morphology, and motility experiments were performed by L. X., P. V., Y. D., L. X., R. Y., L. Z. The in vitro phosphorylation and recombinant protein preparation were performed by L. X., P. V., and G. L. Y. The flow-cell biofilm, RNA-Seq, 3D-SIM, and fluorescent reporter assays were designed and performed by L. Y., Y. L., and Y. D.
Supplementary Material
Acknowledgment
The Pseudomonas mutant strains were obtained from the mutant library supported by Grant NIH P30 DK089507.
This work was supported by a Tier II ARC grant (to Z. X. L.) from the Ministry of Education of Singapore. This work is also supported by the National Basic Research Program of China (973 Program, grant number 2015CB150600) and the Introduction of Innovative R&D Team Program of Guangdong Province (No.2013S034), China. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables S1–S9 and Figs. S1-S2.
- c-di-GMP
- cyclic di-GMP
- DGC
- diguanylate cyclase
- PDE
- c-di-GMP phosphodiesterase
- HapZ
- histidine kinase-associated PilZ
- REC
- phosphoreceiver.
References
- 1. Amikam D., and Galperin M. Y. (2006) PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22, 3–6 [DOI] [PubMed] [Google Scholar]
- 2. Pultz I. S., Christen M., Kulasekara H. D., Kennard A., Kulasekara B., and Miller S. I. (2012) The response threshold of Salmonella PilZ domain proteins is determined by their binding affinities for c-di-GMP. Mol. Microbiol. 86, 1424–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ho C. L., Chong K. S., Oppong J. A., Chuah M. L., Tan S. M., and Liang Z. X. (2013) Visualizing the perturbation of cellular cyclic di-GMP levels in bacterial cells. J. Am. Chem. Soc. 135, 566–569 [DOI] [PubMed] [Google Scholar]
- 4. Boehm A., Kaiser M., Li H., Spangler C., Kasper C. A., Ackermann M., Kaever V., Sourjik V., Roth V., and Jenal U. (2010) Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141, 107–116 [DOI] [PubMed] [Google Scholar]
- 5. Paul K., Nieto V., Carlquist W. C., Blair D. F., and Harshey R. M. (2010) The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “Backstop Brake” mechanism. Mol. Cell 38, 128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morgan J. L. W., Strumillo J., and Zimmer J. (2013) Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493, 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morgan J. L. W., McNamara J. T., and Zimmer J. (2014) Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat. Struct. Mol. Biol. 21, 489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Russell M. H., Bible A. N., Fang X., Gooding J. R., Campagna S. R., Gomelsky M., and Alexandre G. (2013) Integration of the Second Messenger c-di-GMP into the Chemotactic Signaling Pathway. mBio 4, e00001–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fang X., and Gomelsky M. (2010) A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 76, 1295–1305 [DOI] [PubMed] [Google Scholar]
- 10. Merighi M., Lee V. T., Hyodo M., Hayakawa Y., and Lory S. (2007) The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65, 876–895 [DOI] [PubMed] [Google Scholar]
- 11. Whitney J. C., Whitfield G. B., Marmont L. S., Yip P., Neculai A. M., Lobsanov Y. D., Robinson H., Ohman D. E., and Howell P. L. (2015) Dimeric c-di-GMP is required for post-translational regulation of alginate production in Pseudomonas aeruginosa. J. Biol. Chem. 290, 12451–12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wilksch J. J., Yang J., Clements A., Gabbe J. L., Short K. R., Cao H., Cavaliere R., James C. E., Whitchurch C. B., Schembri M. A., Chuah M. L., Liang Z. X., Wijburg O. L., Jenney A. W., Lithgow T., and Strugnell R. A. (2011) MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Path. 7, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCarthy Y., Ryan R. P., O'Donovan K., He Y. Q., Jiang B. L., Feng J. X., Tang J. L., and Dow J. M. (2008) The role of PilZ domain proteins in the virulence of Xanthomonas campestris pv. campestris. Mol. Plant Pathol. 9, 819–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang F., Tian F., Chen H., Hutchins W., Yang C.-H., and He C. (2015) The Xanthomonas oryzae pv. oryzae PilZ domain proteins function differentially in cyclic di-GMP binding and regulation of virulence and motility. App. Environ. Microbiol. 81, 4358–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pitzer J. E., Sultan S. Z., Hayakawa Y., Hobbs G., Miller M. R., and Motaleb M. A. (2011) Analysis of the Borrelia burgdorferi cyclic-di-GMP-binding protein PlzA reveals a role in motility and virulence. Infect. Immun. 79, 1815–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. He M., Zhang J.-J., Ye M., Lou Y., and Yang X. F. (2014) Cyclic di-GMP receptor PlzA controls virulence gene expression through RpoS in Borrelia burgdorferi. Infect. Immun. 82, 445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ryan R. P., McCarthy Y., Kiely P. A., O'Connor R., Farah C. S., Armitage J. P., and Dow J. M. (2012) Dynamic complex formation between HD-GYP, GGDEF and PilZ domain proteins regulates motility in Xanthomonas campestris. Mol. Microbiol. 86, 557–567 [DOI] [PubMed] [Google Scholar]
- 18. Hickman J. W., and Harwood C. S. (2008) Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69, 376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baraquet C., Murakami K., Parsek M. R., and Harwood C. S. (2012) The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 40, 7207–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gupta K., Marques C. N. H., Petrova O. E., and Sauer K. (2013) Antimicrobial tolerance of Pseudomonas aeruginosa biofilms is activated during an early developmental stage and requires the two-component hybrid SagS. J. Bacteriol. 195, 4975–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liao J., and Sauer K. (2012) The MerR-like transcriptional regulator BrlR contributes to Pseudomonas aeruginosa biofilm tolerance. J. Bacteriol. 194, 4823–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chambers J. R., Liao J., Schurr M. J., and Sauer K. (2014) BrlR from Pseudomonas aeruginosa is a c-di-GMP-responsive transcription factor. Mol. Microbiol. 92, 471–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang B., Whitchurch C. B., and Mattick J. S. (2003) FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 185, 7068–7076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Navarro M. V., De N., Bae N., Wang Q., and Sondermann H. (2009) Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure 17, 1104–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qi Y., Chuah M. L. C., Dong X., Xie K., Luo Z., Tang K., and Liang Z.-X. (2011) Binding of C-di-GMP in the non-catalytic EAL domain of FimX induces a long-range conformational change. J. Biol. Chem. 286, 2910–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Z., Chen J. H., Hao Y., and Nair S. K. (2012) Structures of the PelD cyclic diguanylate effector involved in pellicle formation in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 287, 30191–30204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Whitney J. C., Colvin K. M., Marmont L. S., Robinson H., Parsek M. R., and Howell P. L. (2012) Structure of the cytoplasmic region of PelD, a degenerate diguanylate cyclase receptor that regulates exopolysaccharide production in Pseudomonas aeruginosa. J. Biol. Chem. 287, 23582–23593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. An S.-Q., Caly D. L., McCarthy Y., Murdoch S. L., Ward J., Febrer M., Dow J. M., and Ryan R. P. (2014) Novel cyclic di-GMP effectors of the YajQ protein family control bacterial virulence. PLoS Pathog. 10, e1004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hsu J. L., Chen H. C., Peng H. L., and Chang H. Y. (2008) Characterization of the histidine-containing phosphotransfer protein B-mediated multistep phosphorelay system in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 283, 9933–9944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin C. T., Huang Y. J., Chu P. H., Hsu J. L., Huang C. H., and Peng H. L. (2006) Identification of an HptB-mediated multi-step phosphorelay in Pseudomonas aeruginosa PAO1. Res. Microbiol. 157, 169–175 [DOI] [PubMed] [Google Scholar]
- 31. Bordi C., Lamy M. C., Ventre I., Termine E., Hachani A., Fillet S., Roche B., Bleves S., Méjean V., Lazdunski A., and Filloux A. (2010) Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol. Microbiol. 76, 1427–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bhuwan M., Lee H. J., Peng H. L., and Chang H. Y. (2012) Histidine-containing phosphotransfer protein-B (HptB) regulates swarming motility through partner-switching system in Pseudomonas aeruginosa PAO1 strain. J. Biol. Chem. 287, 1903–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Habazettl J., Allan M. G., Jenal U., and Grzesiek S. (2011) Solution structure of the PilZ domain protein PA4608 complex with cyclic di-GMP identifies charge clustering as molecular readout. J. Biol. Chem. 286, 14304–14314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lori C., Ozaki S., Steiner S., Böhm R., Abel S., Dubey B. N., Schirmer T., Hiller S., and Jenal U. (2015) Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 523, 236–239 [DOI] [PubMed] [Google Scholar]
- 35. Caiazza N. C., Shanks R. M. Q., and O'Toole G. A. (2005) Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 187, 7351–7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barken K. B., Pamp S. J., Yang L., Gjermansen M., Bertrand J. J., Klausen M., Givskov M., Whitchurch C. B., Engel J. N., and Tolker-Nielsen T. (2008) Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10, 2331–2343 [DOI] [PubMed] [Google Scholar]
- 37. Petrova O. E., and Sauer K. (2011) SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation. J. Bacteriol. 193, 6614–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tielker D., Hacker S., Loris R., Strathmann M., Wingender J., Wilhelm S., Rosenau F., and Jaeger K. E. (2005) Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology 151, 1313–1323 [DOI] [PubMed] [Google Scholar]
- 39. Ha D. G., Merritt J. H., Hampton T. H., Hodgkinson J. T., Janecek M., Spring D. R., Welch M., and O'Toole G. A. (2011) 2-Heptyl-4-quinolone, a precursor of the Pseudomonas quinolone signal molecule, modulates swarming motility in Pseudomonas aeruginosa. J. Bacteriol. 193, 6770–6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Häussler S., and Becker T. (2008) The Pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog. 4, e1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tian Z. X., Mac Aogáin M., O'Connor H. F., Fargier E., Mooij M. J., Adams C., Wang Y. P., and O'Gara F. (2009) MexT modulates virulence determinants in Pseudomonas aeruginosa independent of the MexEF-OprN efflux pump. Microbial Pathogenesis 47, 237–241 [DOI] [PubMed] [Google Scholar]
- 42. Costerton J. W. (2001) Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol. 9, 50–52 [DOI] [PubMed] [Google Scholar]
- 43. Sauer K., Camper A. K., Ehrlich G. D., Costerton J. W., and Davies D. G. (2002) Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184, 1140–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O'Toole G., Kaplan H. B., and Kolter R. (2000) Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79 [DOI] [PubMed] [Google Scholar]
- 45. Boyd C. D., and O'Toole G. A. (2012) Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu. Rev. Cell Dev. Biol. 28, 439–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kuchma S. L., Brothers K. M., Merritt J. H., Liberati N. T., Ausubel F. M., and O'Toole G. A. (2007) BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189, 8165–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Caiazza N. C., Merritt J. H., Brothers K. M., and O'Toole G. A. (2007) Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189, 3603–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shrout J. D., Chopp D. L., Just C. L., Hentzer M., Givskov M., and Parsek M. R. (2006) The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62, 1264–1277 [DOI] [PubMed] [Google Scholar]
- 49. Sakuragi Y., and Kolter R. (2007) Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J. Bacteriol. 189, 5383–5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ueda A., and Wood T. K. (2009) Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog. 5, e1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gupta R., and Schuster M. (2012) Quorum sensing modulates colony morphology through alkyl quinolones in Pseudomonas aeruginosa. BMC Microbiology 12, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. O'Toole G. A., and Kolter R. (1998) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304 [DOI] [PubMed] [Google Scholar]
- 53. Petrova O. E., and Sauer K. (2009) A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. Plos Pathogens 5, e1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mikkelsen H., Sivaneson M., and Filloux A. (2011) Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ. Microbiol. 13, 1666–1681 [DOI] [PubMed] [Google Scholar]
- 55. Petrova O. E., and Sauer K. (2012) Sticky situations: key components that control bacterial surface attachment. J. Bacteriol. 194, 2413–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Petrova O. E., and Sauer K. (2010) The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J. Bacteriol. 192, 5275–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kong W. N., Chen L., Zhao J. Q., Shen T., Surette M. G., Shen L. X., and Duan K. M. (2013) Hybrid sensor kinase PA1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol. Microbiol. 88, 784–797 [DOI] [PubMed] [Google Scholar]
- 58. Galperin M. Y. (2006) Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 188, 4169–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Aldridge P., Paul R., Goymer P., Rainey P., and Jenal U. (2003) Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 47, 1695–1708 [DOI] [PubMed] [Google Scholar]
- 60. De N., Navarro M. V., Raghavan R. V., and Sondermann H. (2009) Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J. Mol. Biol. 393, 619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kulasekara H. D., Ventre I., Kulasekara B. R., Lazdunski A., Filloux A., and Lory S. (2005) A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55, 368–380 [DOI] [PubMed] [Google Scholar]
- 62. Chen M. W., Kotaka M., Vonrhein C., Bricogne G., Rao F., Chuah M. L., Svergun D., Schneider G., Liang Z. X., and Lescar J. (2012) Structural insights into the regulatory mechanism of the response regulator RocR from Pseudomonas aeruginosa in cyclic Di-GMP signaling. J. Bacteriol. 194, 4837–4846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ryan R. P., Fouhy Y., Lucey J. F., Crossman L. C., Spiro S., He Y.-W., Zhang L.-H., Heeb S., Cámara M., Williams P., and Dow J. M. (2006) Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. U.S.A. 103, 6712–6717 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64. Petters T., Zhang X., Nesper J., Treuner-Lange A., Gomez-Santos N., Hoppert M., Jenal U., and Søgaard-Andersen L. (2012) The orphan histidine protein kinase SgmT is a c-di-GMP receptor and regulates composition of the extracellular matrix together with the orphan DNA binding response regulator DigR in Myxococcus xanthus. Mol. Microbiol. 84, 147–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rao F., Pasunooti S., Ng Y., Zhuo W., Lim L., Liu W., and Liang Z.-X. (2009) Enzymatic synthesis of c-di-GMP using a thermophilic diguanylate cyclase. Anal. Biochem. 389, 138–142 [DOI] [PubMed] [Google Scholar]
- 66. Pasunooti S., Surya W., Tan S. N., and Liang Z. X. (2010) Sol-gel immobilization of a thermophilic diguanylate cyclase for enzymatic production of cyclic-di-GMP. J. Mol. Catal. B: Enz. 67, 98–103 [Google Scholar]
- 67. Tolker-Nielsen T., and Sternberg C. (2011) Growing and analyzing biofilms in flow chambers. Curr. Protoc. Microbiol. 21, 1B.2.1–1B.2.17 [DOI] [PubMed] [Google Scholar]
- 68. Mortazavi A., Williams B. A., McCue K., Schaeffer L., and Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods 5, 621–628 [DOI] [PubMed] [Google Scholar]
- 69. Yang L., Rybtke M. T., Jakobsen T. H., Hentzer M., Bjarnsholt T., Givskov M., and Tolker-Nielsen T. (2009) Computer-Aided Identification of Recognized Drugs as Pseudomonas aeruginosa Quorum-Sensing Inhibitors. Antimicrob. Agents Chemother. 53, 2432–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rasmussen T. B., Bjarnsholt T., Skindersoe M. E., Hentzer M., Kristoffersen P., Köte M., Nielsen J., Eberl L., and Givskov M. (2005) Screening for Quorum-Sensing Inhibitors (QSI) by Use of a Novel Genetic System, the QSI Selector. J. Bacteriol. 187, 1799–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yang L., Barken K. B., Skindersoe M. E., Christensen A. B., Givskov M., and Tolker-Nielsen T. (2007) Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 153, 1318–1328 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.