Abstract
Metallo-β-lactamases catalyze the hydrolysis of most β-lactam antibiotics and hence represent a major clinical concern. The development of inhibitors for these enzymes is complicated by the diversity and flexibility of their substrate-binding sites, motivating research into their structure and function. In this study, we examined the conformational properties of the Bacillus cereus β-lactamase II in the presence of chemical denaturants using a variety of biochemical and biophysical techniques. The apoenzyme was found to unfold cooperatively, with a Gibbs free energy of stabilization (ΔG0) of 32 ± 2 kJ·mol−1. For holoBcII, a first non-cooperative transition leads to multiple interconverting native-like states, in which both zinc atoms remain bound in an apparently unaltered active site, and the protein displays a well organized compact hydrophobic core with structural changes confined to the enzyme surface, but with no catalytic activity. Two-dimensional NMR data revealed that the loss of activity occurs concomitantly with perturbations in two loops that border the enzyme active site. A second cooperative transition, corresponding to global unfolding, is observed at higher denaturant concentrations, with ΔG0 value of 65 ± 1.4 kJ·mol−1. These combined data highlight the importance of the two zinc ions in maintaining structure as well as a relatively well defined conformation for both active site loops to maintain enzymatic activity.
Keywords: antibiotic resistance, circular dichroism (CD), enzyme catalysis, metalloenzyme, nuclear magnetic resonance (NMR), protein folding, zinc, β-lactamase
Introduction
β-Lactamases catalyze hydrolysis of the β-lactam ring of antibiotics belonging to the penicillin family (1–3). Synthesis of these enzymes represents the major cause of bacterial resistance to β-lactam antibiotics (4–7). They are classified into two structural superfamilies (8), namely the active site serine enzymes (both β-lactamases and penicillin-binding proteins) and the metallo-β-lactamases (MBLs).4 The latter, also referred to as class B β-lactamases, require one or two zinc ions for activity (9–11). They show no structural similarity with the active site serine β-lactamases, and they are part of a remarkable set of enzymes (i.e. the zinc metallo-hydrolase family of the β-lactamase fold or, more simply, the MBL superfamily (10–16)) that exhibit a wide variety of functions related to hydrolysis and redox reactions and DNA and RNA metabolism. Seventeen groups of enzymes have been identified on the basis of their biological functions (13), which all share a novel αβ/βα fold. Most of the three-dimensional structures reveal a binuclear center with metal ligands located on loops connecting secondary structure elements (15, 17).
Zinc β-lactamases have been found in many bacterial species, including pathogenic strains (18, 19). Most of them are able to hydrolyze almost all β-lactam antibiotics (20, 21), including carbapenems (i.e. a family of last resort β-lactams that generally escape the activity of the most widespread serine β-lactamases), and they are not sensitive to the classical inactivators of serine β-lactamases, such as clavulanate, sulbactam, and tazobactam (22, 23). Furthermore, these enzymes are often encoded by highly transmissible genetic elements (i.e. plasmids, transposons, and integrons), which allow their spreading among pathogenic bacteria (5, 6, 18, 24). Thus, MBLs have been reported to be of particular concern for public health (18, 19, 24–28), and the development of effective inhibitors of zinc β-lactamases to counteract the ongoing widespread resistance to β-lactam antibiotics is of immediate clinical relevance. The structural diversity of the MBLs and the plasticity of their binding sites, at the level of both the zinc center and the adjacent substrate-binding loops, render the design of such compounds a difficult task (29). This will likely not be possible without a detailed understanding of both the mechanism of action of these enzymes and the interactions that determine the structure-activity relationships among MBL inhibitors (17, 23, 30, 31). This is even more obvious in the light of the challenge of overcoming in vivo toxicity associated with cross-reactivity with human metalloenzymes (32).
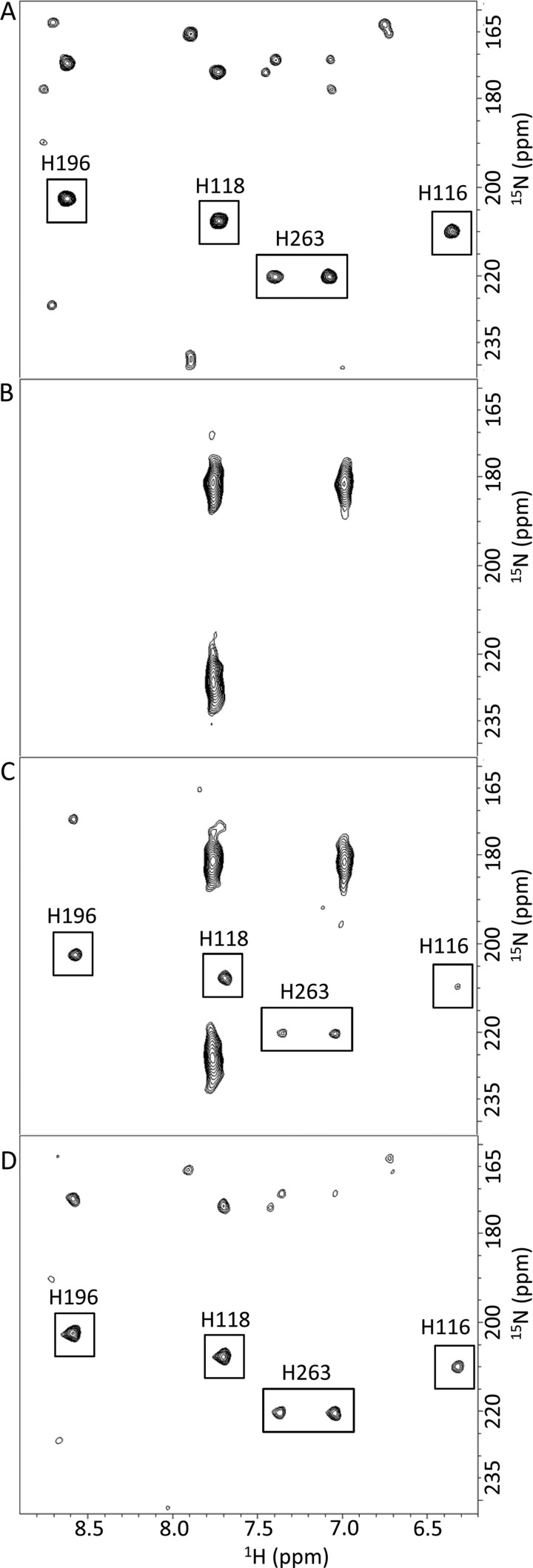
MBLs are grouped according to sequence similarities and zinc coordination into subclasses B1, B2, and B3 (10, 33). Enzymes from each class exhibit specific functional and mechanistic properties (34, 35). In particular, although the B1 and B3 enzymes display maximum activity as dizinc species, the B2 β-lactamases are inhibited upon binding of a second zinc (36). The first class B enzyme was isolated from an innocuous strain of Bacillus cereus (37). This protein, known as BcII, is the archetype, the most extensively studied model of enzymes of the largest ubiquitous and clinically relevant B1 subclass, such as VIM-, IMP-, and NDM-type MBLs (all transferable broad spectrum β-lactamases) (38). BcII consists of 227 residues in the mature form (Mr = 24,960/25,088 in the absence/presence of zinc ions), and its three-dimensional structure (Fig. 1, both x-ray crystal and solution NMR (31, 39–41)) exhibits the classical MBL fold, i.e. a four-layered αβ/βα structure, comprising a central β-sheet sandwich flanked on either side by two α-helices. The active site, with two zinc ions readily accessible to solvent, is located at the bottom of a long wide groove running on the surface of the protein, at one edge of the β-sheet sandwich. The shape of the active site cleft is modulated by conformational changes of two long loops {β3-β4 [residues 32–38(59–66)] and β11-α4 [170–188(223–241)]}. {Residue numbering is presented as follows: number in BcII sequence(number in standard BBL system) (33, 42) throughout the text, and all structural elements are defined according to the solution NMR structure (31)}. Note that these loops have also been widely referred to as L1 and L3, respectively.
FIGURE 1.
Schematic ribbon representation of the structure of BcII 569/H/9 (Protein Data Bank code 1BVT (40)). The zinc ions at the catalytic site are represented as yellow spheres, α-helices and β-strands are shown in red and blue, respectively, and the four tryptophan residues are labeled and colored green. The β3-β4 and β11-α4 loops (residues 32–38(59–66) and 170–188(223–241), respectively) are also indicated. The figure was generated using the open-source molecular graphics system PyMOL (The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC).
BcII has been reported to be active in the presence of either one or two zinc ions at the active site, although maximum activity is observed with two zinc bound (20, 43–45). However, Jacquin et al. (46) demonstrated that at 1:1 [Zn]/[BcII] ratio the only species present were apoenzyme and dizinc enzyme, indicating cooperative binding of the zinc ions and suggesting that the dizinc species is the only relevant form of the enzyme for activity (46). For dizinc MBLs, hydrolysis has been suggested to occur by cleavage of the amide bond of the β-lactam ring via attack of a hydroxide ion on the β-lactam carbonyl carbon, with no formation of covalent adducts (17, 35, 47–49).
The zinc ion in the first binding site (Zn1 or histidine site) is coordinated by four ligands in a tetrahedral geometry as follows: the nitrogen atom of the imidazole groups of three histidine residues (His-86(116), His-88(118), and His-149(196)) and the oxygen atom of a water molecule or hydroxide ion; this oxygen ligand is a bridge to the second zinc ion. The zinc in the second binding site (Zn2 or cysteine site) is coordinated by five ligands as follows: three other amino acid side chains (Asp-90(120), Cys-168(221), and His-210(263)) in a distorted trigonal bipyramidal geometry, an apical water molecule, and the bridging water/hydroxide, which probably acts as the nucleophile in the course of β-lactam hydrolysis (3, 17).
Zinc-bound β-lactamases appear to be more stable than their corresponding metal-depleted forms (9), and removal of the metal is known to induce conformational changes (40, 46, 50). No detailed information on the conformational stability of MBLs has, however, been reported to date (for a brief review of the literature, see under “Discussion”). In this study, the chemical-induced unfolding transitions of both apo- and holoBcII were studied by using a combination of spectroscopic techniques. This analysis gives insights into the role of zinc in stability and provides estimates of the thermodynamic parameters. Furthermore, data obtained at moderate denaturant concentrations indicate population of a heterogeneous ensemble of interconverting partially folded species with two zinc ions bound and modest structural changes at the enzyme surface, particularly in the two active site loops. Observation that these species display no significant changes in the geometry of the active site but are fully inactive demonstrates that both the β3-β4 and β11-α3 loops are important structural elements for catalysis.
This study brings new insights into the role of metal cofactors in protein folding as a whole, in the absence of any clinical implications. Moreover, it highlights structural aspects linked to the catalytic activity of the enzyme, which are relevant to inhibitor discovery.
Results
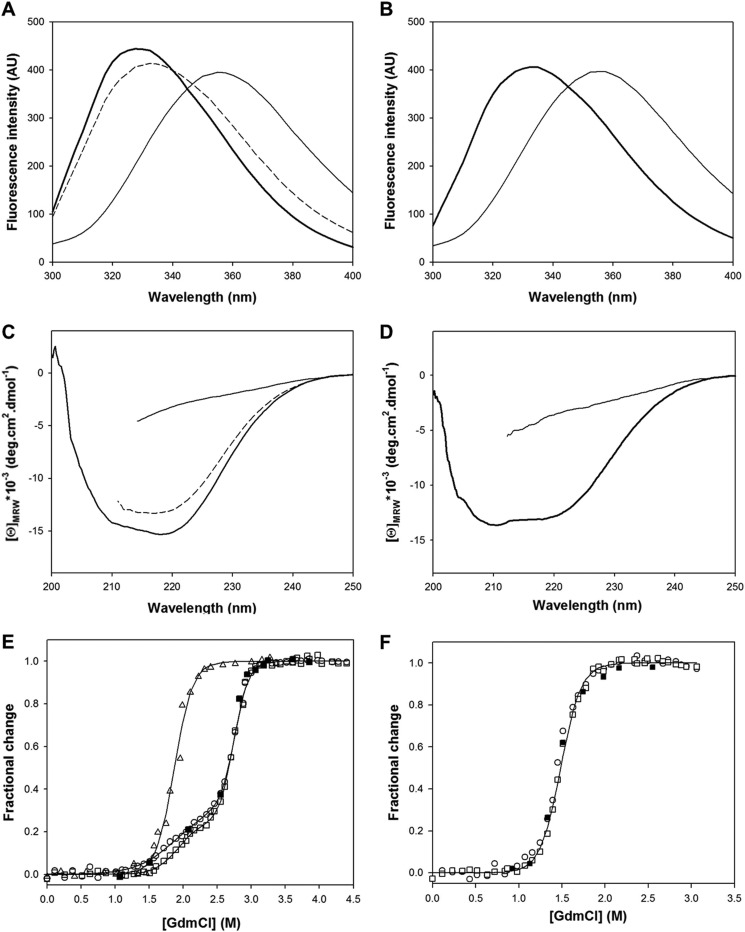
The fluorescence spectra of holo- and apoBcII (Fig. 2, A and B), measured at pH 7.5, 25 °C, are similar and display a broad emission band with maxima at 329 and 334 nm, respectively. These relatively low maximum emission wavelengths (λmax) are consistent with low solvent exposure of the four tryptophan indole side chains, as seen in the enzyme three-dimensional structure (Fig. 1). In 4 m GdmCl, λmax is shifted to 355–356 nm (Fig. 2, A and B), indicating full solvent accessibility of tryptophan indole groups. The near-UV CD spectra (data not shown) of free and metal-bound BcII are also similar, although not identical, indicating a well defined tertiary structure in both cases. The far-UV CD spectra of holo- and apoBcII, shown in Fig. 2, C and D, reveal a significant decrease in helical content of the enzyme in the absence of zinc; this was previously estimated to be around 10% (46). With both apo- and holoenzyme, addition of high concentrations of denaturant led to a dramatic reduction of the CD signal (Fig. 2, C and D), consistent with a major decrease in secondary structure organization.
FIGURE 2.
Fluorescence (A and B) and far-UV CD (C and D) spectra of native ([GdmCl] = 0 m, thick line), partially folded ([GdmCl] = 2.4 m, dashed line), and unfolded ([GdmCl] = 4 m, thin line) forms of holoBcII (A and C) and apoBcII (B and D) are shown. GdmCl-induced equilibrium unfolding transitions of holoBcII (E) and apoBcII (F) BcII at pH 7.5 and 25 °C, monitored by the change in fluorescence intensity at 370 nm (□), CD at 222 nm (○), and enzyme activity (▵; holoBcII only); ■ corresponds to the refolding of BcII after 12 h of incubation in 6 m GdmCl, monitored by the change in fluorescence intensity at 370 nm. Data are shown as the fractional change in signal (99) at each GdmCl concentration; they were analyzed on the basis of two- and three-state models for apoBcII and holoBcII, respectively, and the solid lines represent the best fit calculated using the thermodynamic values in Tables 1 and 2, respectively. The curve for holoBcII unfolding followed by activity measurements is there to guide the eye only.
Incubation of both apo- and holoBcII (0.15 mg·ml−1, i.e. 6 μm) for 12 h in 6 m GdmCl, and subsequent dilution restored fluorescence and far-UV CD spectra that are indistinguishable from those of the native state, indicating that GdmCl-induced unfolding is fully reversible for both forms of the enzyme. With holoBcII, this could be further demonstrated by measuring full recovery of the enzyme activity after unfolding/refolding cycles. Thus, fluorescence and CD spectroscopy were used to follow unfolding transitions of the enzyme by GdmCl and urea.
Unfolding of ApoBcII
With the metal-depleted form of the enzyme, the coincidence of the transition curves obtained by intrinsic fluorescence and far-UV CD measurements (Fig. 2F) indicated that secondary and tertiary structures were destabilized concomitantly. The unfolding curves for the GdmCl-induced denaturation in the absence of zinc can be described by an apparent cooperative two-state model (N ⇌ U), where only the native and unfolded states are significantly populated. Dilution of the fully denatured enzyme (in 6 m GdmCl) into final denaturant concentrations corresponding to the transition zone (i.e. 1–2 m GdmCl) showed that the process was monitored under genuine equilibrium conditions and occurred with full thermodynamic reversibility. Therefore, values of the thermodynamic parameters were calculated assuming a simple two-state model (Table 1). Notably, values obtained for the Gibbs free energy of unfolding (ΔG0NU) were identical, within the error limit, for both optical methods and also for unfolding induced by either GdmCl or urea, providing convincing evidence for a simple two-state unfolding mechanism.
TABLE 1.
Thermodynamic parameters for GdmCl- and urea-induced unfolding transition of apoBcII
| GdmCl |
Urea |
|||||
|---|---|---|---|---|---|---|
| ΔG0NU | −mNU | Cm | ΔG0NU | −mNU | Cm | |
| kJ·mol−1 | kJ·mol−1·m−1 | m | kJ·mol−1 | kJ·mol−1·m−1 | m | |
| Intrinsic fluorescence | 33 ± 1.4 | 21 ± 1 | 1.5 ± 0.2 | 33 ± 1 | 10 ± 0.4 | 3.3 ± 0.3 |
| Far-UV CD | 28 ± 2 | 19.4 ± 1.3 | 1.5 ± 0.2 | 32 ± 1.3 | 9.7 ± 0.4 | 3.3 ± 0.3 |
Unfolding of HoloBcII
By contrast, despite full thermodynamic reversibility, unfolding of holoBcII with both GdmCl (Fig. 2E) and urea (data not shown) displayed non-cooperative behavior, with at least two unfolding transitions. However, because the enzyme was not fully unfolded in 10 m urea (note that no significant unfolding could be observed by both intrinsic fluorescence and far-UV CD measurements at concentrations below 7 m), data obtained using this denaturant were not further analyzed.
With GdmCl, the first and second transitions occurred with midpoint values of 1.9 and 2.8 m, respectively. Both were characterized by changes in intrinsic fluorescence and far-UV CD (Fig. 2E; also near-UV CD, data not shown). These data indicate population of species with intermediate conformations that are in equilibrium with native and fully unfolded enzyme molecules. Optical measurements at 2.4 m GdmCl, i.e. close to the end of the first apparent transition, suggest the existence of a stable intermediate state that retains both a high content of native secondary structure, as deduced from the far-UV CD spectrum shown in Fig. 2C, and a significant protection of the tryptophan residues from the solvent, as indicated by the fluorescence spectrum shown in Fig. 2A. The fluorescence and far-UV CD data shown in Fig. 2E were analyzed separately on the basis of a three-state model ((N ⇌ I ⇌ U), using Equation 3, and the corresponding thermodynamic parameters are given in Table 2. GdmCl-induced unfolding of holoBcII was also followed by activity measurements (Fig. 2E), which indicated that the catalytic activity is lost in a single transition, with a midpoint of 1.9 m, concurrent with the first transition observed by optical methods. The possibility of a denaturant ionic strength effect could be excluded by showing that no significant change in enzymatic activity occurred in the presence of NaCl at concentrations up to 3 m (data not shown). The intermediate state that is apparently populated for the first unfolding transition is thus totally inactive, suggesting a change in specific tertiary and/or secondary structure organization at the enzyme active site, and possibly zinc release.
TABLE 2.
Thermodynamic parameters for GdmCl-induced unfolding transition of holoBcII
| ΔG0NI | −mNI | (Cm)NI | ΔG0IU | −mIU | (Cm)IU | |
|---|---|---|---|---|---|---|
| kJ·mol−1 | kJ·mol−1·m−1 | m | kJ·mol−1 | kJ·mol−1·m−1 | m | |
| Intrinsic fluorescence | 29 ± 6 | 15 ± 3 | 1.9 ± 0.8 | 66 ± 3 | 24 ± 1 | 2.8 ± 0.3 |
| Far-UV CD | 15 ± 3 | 8 ± 2 | 1.9 ± 0.8 | 64 ± 4 | 23 ± 1.4 | 2.7 ± 0.3 |
Finally, GdmCl-induced unfolding was also performed in the presence of the hydrophobic dye ANS; fluorescence measurements at 475 nm revealed no significant enhancement in intensity, and thus no ANS binding, indicating that no species (such as a molten globule (51)) having significant hydrophobic regions exposed to the solvent is significantly populated (52–54).
Characterization of the Intermediate State
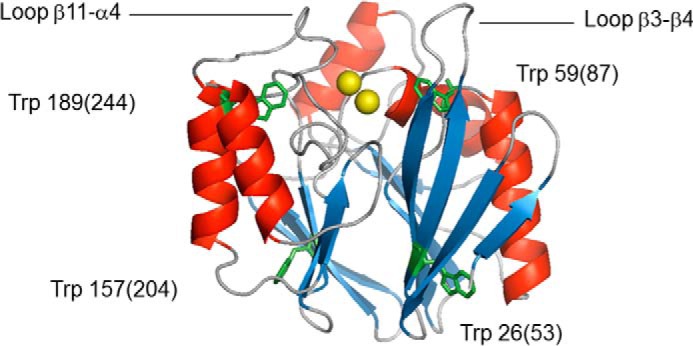
Because the intermediate state is enzymatically inactive, one obvious possibility is that the catalytically essential zinc atoms have dissociated from the enzyme. To gain more insight into the presence of zinc during holoBcII unfolding, the imidazole resonances of the histidine residues involved in the coordination of zinc ions (i.e. His-86(116), His-88(118), and His-149(196) of the histidine site and His-210(263) of the cysteine site) were followed as a function of GdmCl concentration. Fig. 3A shows 1H-15N HSQC spectra focusing on the imidazole signals in the presence (holoBcII) and absence (apoBcII) of zinc, with no GdmCl. The imidazole resonances of the four histidines are well defined and assigned when zinc ions are bound to the catalytic site (55), but not in their absence. The clear difference between holo- and apoBcII in the NMR cross-peak pattern of their imidazole groups allowed detailed analysis of the effect of various GdmCl concentrations on the binding of zinc. At all GdmCl concentrations between 0 and 2.5 m, imidazole signals of the four binding histidines could be distinctly observed (Fig. 3B). Furthermore, the increase of GdmCl concentration from 0 to 2.5 m caused only slight displacements of the imidazole signals (Fig. 3B), close to the experimental error, suggesting little change in the geometry of the zinc-binding sites. These data unambiguously demonstrate that the two zinc ions are bound to their respective sites throughout the first unfolding transition, and therefore, the loss of enzymatic activity is not due to the release of zinc. Furthermore, although there is evidence that the zinc ions have the flexibility to move significantly within the active site (31, 56), the very modest shifts of the imidazole cross-peaks of the zinc ligands suggest that addition of 2.5 m GdmCl not only does not lead to the release of the zinc atoms but in fact has very little effect on the structure of their coordination sites.
FIGURE 3.
Two-dimensional 1H-15N HSQC spectra for histidine imidazole observation. A, superimposition of data obtained for native BcII in the presence (holoBcII, in red) and absence (apoBcII, in blue) of zinc ions, with no GdmCl. B, superimposition of signals observed for holoBcII in the presence of 0 m (blue), 2.25 m (red), and 2.5 m (green) GdmCl. Note that standard BBL numbering system only is used.
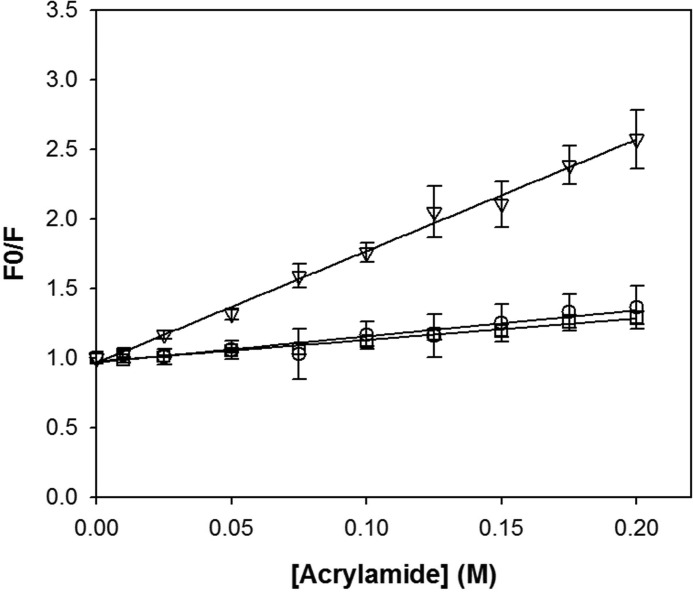
The dynamic accessibility of the tryptophan residues in folded ([GdmCl] = 0 m), intermediate ([GdmCl] = 2.4 m), and unfolded ([GdmCl] = 3.5 m) states of holoBcII was investigated using acrylamide as a fluorescence quencher (57). As the acrylamide concentration was increased, the fluorescence intensity decreased without a noticeable change in spectral shape, and the data were analyzed according to Equation 1 to determine the quenching constant (KSV) (Fig. 4). Complete unfolding of holoBcII in the presence of 3.5 m GdmCl caused an ∼4.5-fold increase of the Stern-Volmer quenching constant, from 1.8 ± 0.1 (in the absence of denaturant) to 8 ± 0.2 m−1, attributable to high solvent exposure of tryptophan side chains in the unfolded protein. By contrast, the Stern-Volmer quenching constant in 2.4 m GdmCl, i.e. KSV = 1.9 ± 0.2 m−1, is identical to that of the folded state, showing no significant change in the solvent accessibility of the tryptophan residues under these conditions.
FIGURE 4.
Stern-Volmer plots of holoBcII tryptophan fluorescence, in the presence of 0.1 mm ZnSO4, quenched by acrylamide at several GdmCl concentrations. Experiments were performed for the native state in the absence of GdmCl (□), the intermediate state in the presence of 2.4 m GdmCl (○), and the unfolded state in the presence of 3.5 m GdmCl (▿). Data are the average of at least three independent experiments, and error bars represent standard deviations.
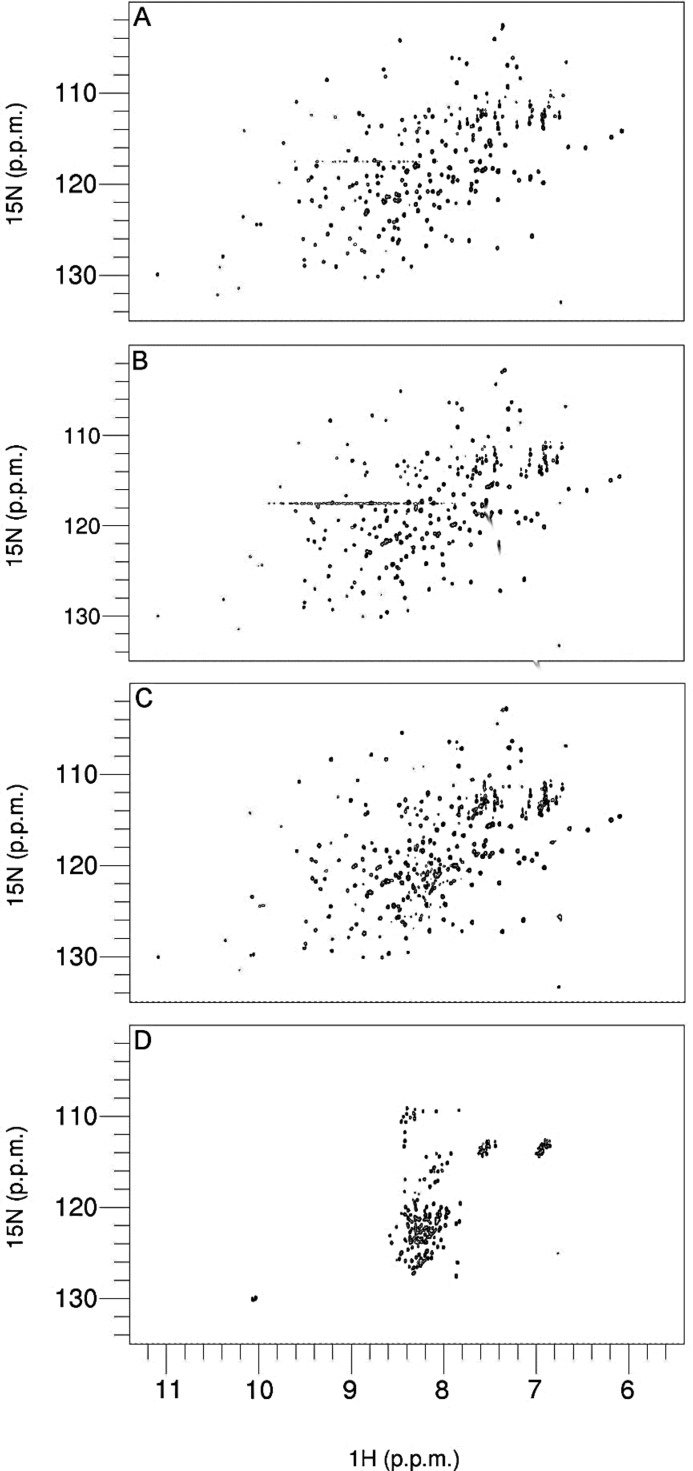
To obtain amino acid-specific information on the unfolding mechanism of BcII, a series of two-dimensional 1H-15N HSQC NMR spectra was recorded under equilibrium conditions in the presence of various GdmCl concentrations in the range from 0 to 4 m. These experiments were performed at pH 6.5, rather than pH 7.5, to slow down intrinsic amide hydrogen exchange. Fig. 5 shows representative spectra of 15N-BcII, collected in the presence of 0, 1.63, 2.34, and 4 m GdmCl. In the absence of denaturant (Fig. 5A), the protein spectrum showed a set of well dispersed peaks characteristic of the folded state (58). At 4 m GdmCl (Fig. 5D), all native enzyme signals disappeared, and the spectrum is typical of an unfolded protein. At intermediate GdmCl concentrations (e.g. 1.63 and 2.34 m), corresponding to the first unfolding transition (see Fig. 2), spectra (Fig. 5, B and C) were similar to that of the folded state (Fig. 5A). Changes in both chemical shift and peak intensity could be observed throughout the entire range of denaturant concentrations. Up to 1.5 m GdmCl, all peaks displayed chemical shifts close to their native state values, although with reduced intensities (probably due to exchange broadening), whereas at higher denaturant concentrations, the disappearance of some peaks occurred. At 2.34 m GdmCl, some peaks corresponding to the unfolded state could be observed (Fig. 6).
FIGURE 5.
Two-dimensional 1H-15N HSQC spectra of holoBcII in the presence of various GdmCl concentrations. A, [GdmCl] = 0 m; B, [GdmCl] = 1.63 m; C, [GdmCl] = 2.34 m; and D, [GdmCl] = 4.0 m. Plots have been drawn using CcpNmr Analysis (97).
FIGURE 6.
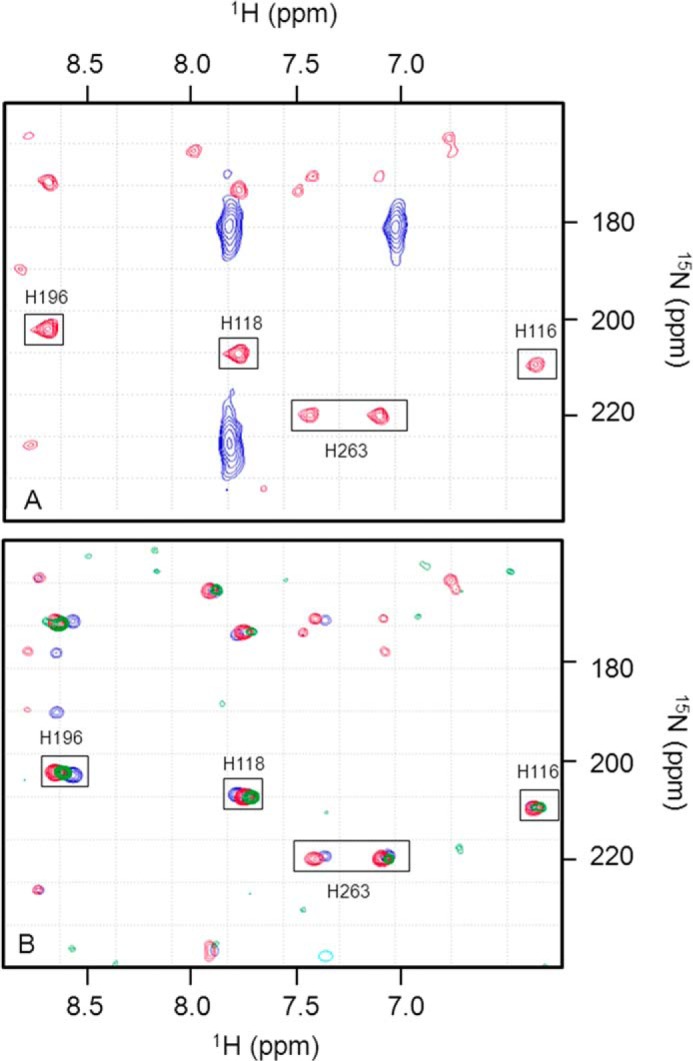
Overlay of two-dimensional 1H-15N HSQC spectra of holoBcII in the presence of various GdmCl concentrations: 0 m (black); 1 m (green); 1.6 m (blue), and 2.34 m (pink). Arrows indicate shifts and their directions; circles are for peaks corresponding to the unfolded protein in 2.34 m GdmCl, and arcs indicate non-linear CSPs. Residues are numbered as in the sequence, i.e. Lys-104(135), Lys-134(181), and Lys-227(291).
Based on the resonance assignments of native holoBcII (58), assignments of the resonances of the enzyme in the presence of GdmCl concentrations ranging from 0 to 2.34 m could be achieved simply by following chemical shift changes as a function of denaturant concentration. The chemical shifts of 184 out of 227 residues (Fig. 7) were monitored in this way, and for most of them, the changes occurred progressively and linearly with denaturant concentration. For six residues (i.e. Lys-134(181), Lys-147(194), His-149(196), Leu-178(231), and also two unassigned ones), non-linear (i.e. curved; see Fig. 6) chemical shift changes were observed, however, suggesting that more than two states (i.e. in addition to N and I) are involved.
FIGURE 7.
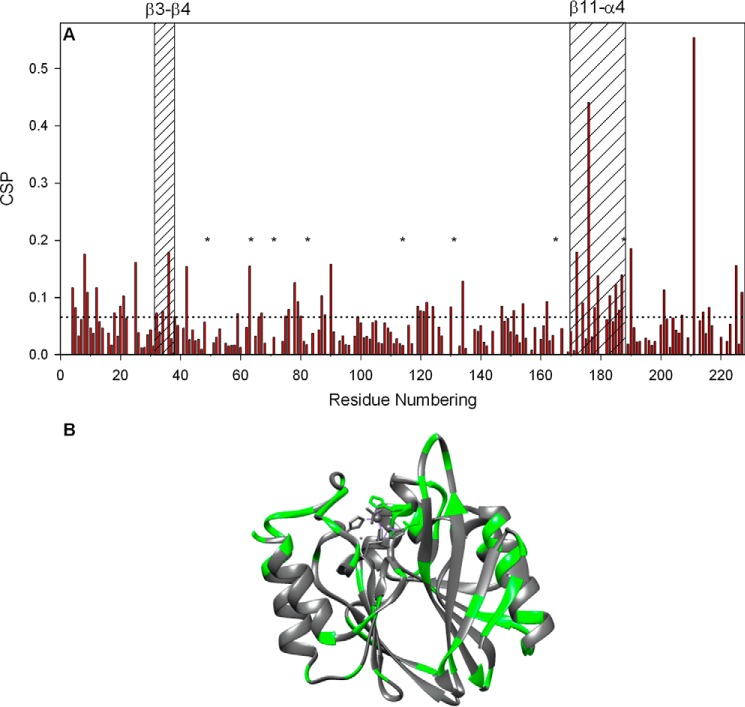
A, combined CSPs between 0 and 2. 34 m GdmCl, calculated using Equation 2 and shown as histograms versus the BcII amino acid sequence. The dashed line represents the threshold (0.064 ppm) for significant chemical shift perturbations calculated as described under “Experimental Procedures.” With residues Ile-83(113), Glu-151(198), Val-165(218), and Lys-176(229), estimation of the CSPs was done by linear extrapolation. The eight residues (Lys-50(78), Thr-64(92), Glu-72(100), Ile-83(113), Ala-115(146), Thr-131(178), Val-165(218), and Glu-188(243)) whose signals disappeared completely as GdmCl concentration was increased above 0.75 m (see text) are indicated by asterisks. Sequence elements corresponding to loops β3-β4 and β11-α4 are indicated; B, schematic ribbon representation of the structure where green areas represent those residues undergoing significant CSPs.
A detailed analysis of the chemical shift data (Fig. 7) showed that 59 residues were significantly perturbed by the denaturant (i.e. CSP > 0.064 ppm between 0 and 2.34 m GdmCl). Among these, the signals of eight residues (Lys-50(78), Thr-64(92), Glu-72(100), Ile-83(113), Ala-115(146), Thr-131(178), Val-165(218), and Glu-188(243)) disappeared completely as GdmCl concentration was increased above 0.75 m, presumably as a result of exchange broadening due to a large shift difference between the native and intermediate states. Those 59 residues, which are all significantly exposed to the solvent (as computed with NACESS (59)), are represented schematically on the structure in Fig. 7B. 32 of them are located in loop regions, although the remaining 27 residues are found essentially at the ends of well organized secondary structure elements, i.e. 16 in β-strands and 11 in α-helices (Fig. 7B). These 11 residues in α-helices correspond to ∼18% of the total number of α-helical residues in the enzyme structure, which is in good agreement with the ∼17% loss in negative ellipticity measured at 222 nm, between 0 and 2.4 m GdmCl (Fig. 2, C and E). These data suggest that the denaturant induces significant conformational changes in the corresponding regions of the polypeptide backbone. We were able to follow specifically the resonances of 50 residues whose backbone amides are fully buried in the enzyme core (as computed with NACCESS (59)), and we observed that they do not undergo any significant shift perturbation (supplemental Table S1), with the exception of Val-25(52), Leu-63(91), Leu-67(95) and Val-207(260), which are in fact positioned close to polar residues that experience significant shift changes. Most importantly, HSQC spectra collected in the presence of urea (data not shown) yielded similar results, thus indicating that no ionic strength effect was observed with GdmCl. In the case of residues located around the active site, these results are in good agreement with the observation that addition of 3 m NaCl had no effect on the enzyme activity (see above).
Fig. 8 shows CSPs of the amide and indole 1H-15N signals of the four tryptophan residues. Progressive and approximately linear shift changes occur with increasing GdmCl concentrations, in good agreement with the behavior of most of the backbone resonances. The side chain of Trp-189(244), which is completely buried in the core of the native protein, showed only minor chemical shift perturbation (0.02 ppm, see Fig. 8) in the presence of the denaturant, indicating the high stability of its immediate environment during the first unfolding transition. Accordingly, residues in the vicinity of Trp-189(244) remained in a native-like state up to at least 2.4 m GdmCl. By contrast, indole groups of Trp-59(87) and Trp-157(204) are more exposed to the solvent, and their environment proved to be a little more sensitive to the denaturant, as shown by CSPs of 0.08 and 0.06 ppm, respectively, in 2.34 m GdmCl (Fig. 8). Finally, Trp26-(53) side chain, which shows very limited solvent accessibility, underwent a dramatic chemical shift perturbation at low GdmCl concentration. Thus, the CSP was 0.14 ppm at 0.5 m GdmCl, and the signal totally disappeared at higher concentrations, probably due to exchange broadening. Because the emission fluorescence intensity of the enzyme measured at 370 nm does not change significantly in the presence of GdmCl concentrations lower than 1.5 m (Fig. 2E), these results suggest that Trp-59(87) and/or Trp-157(204) and not Trp-26(53) are responsible for the increase in intensity observed at higher concentrations (Fig. 2, A and E). For all four tryptophan residues, however, up to 2 m GdmCl the backbone resonance CSP (Fig. 2B) remained below the computed threshold for significant CSP (CSP > 0.064 ppm), in good agreement with the view that the enzyme hydrophobic core remains largely unaffected.
FIGURE 8.
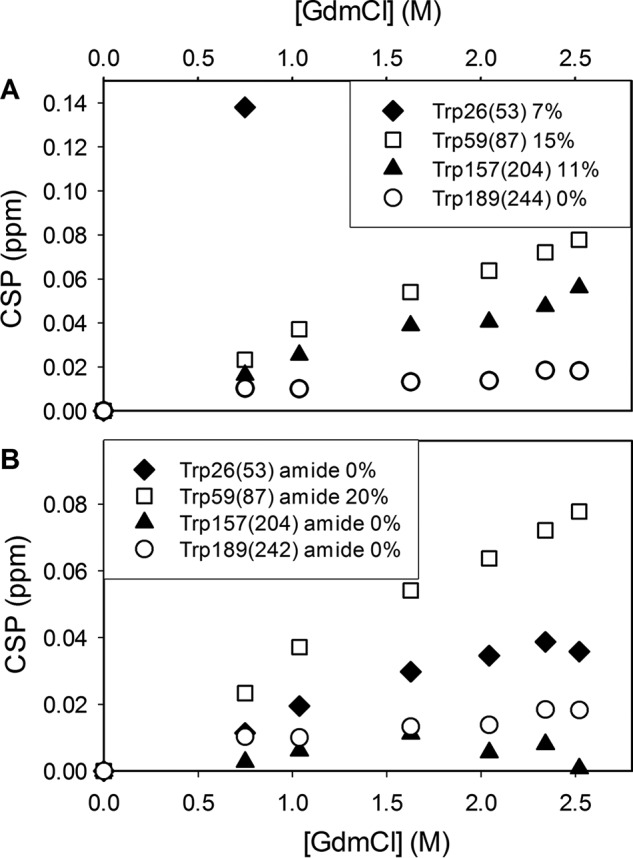
Influence of GdmCl on the indole side chain (A) and backbone amide (B) resonances of the four tryptophan residues of holoBcII. The combined CSPs for side chain Hϵ1-Nϵ1 (A) and backbone H-N peaks (B) are plotted as a function of GdmCl concentration. The solvent accessibility of the corresponding side and main chains, as computed with NACCESS (59), is indicated.
Refolding of ApoBcII by the Addition of Zinc Ions
Transition curves for apo- and holoBcII (Fig. 2, E and F, respectively) highlight significant differences in their conformational stabilities (see also Tables 1 and 2). Thus, at ∼2 m GdmCl, whereas apoBcII is completely unfolded, its dizinc counterpart adopts a native-like, although catalytically inactive, conformation. Using 1H-15N HSQC experiments for selective observation of His imidazole, as described above, we monitored refolding of BcII unfolded in 2 m GdmCl in the absence of zinc, upon addition of 1 and 2 m eq of zinc (Fig. 9). In the presence of denaturant, addition of zinc caused changes in the spectrum of the enzyme, with the concomitant appearance of narrow peaks corresponding to histidine imidazole signals of native holoBcII, and disappearance of the broad peaks characteristic of the histidine imidazole signals of the unfolded protein. At a Zn/BcII molar ratio of 1, the histidine imidazole 1H-15N HSQC spectrum (Fig. 9C) showed a peak distribution consistent with significant population of both unfolded and dizinc-bound BcII. The absence of any signal for a putative monozinc form indicates cooperative binding of the two metal ions (46). Finally, in the presence of 2 eq of zinc (i.e. Zn/BcII molar ratio of 2), complete recovery of a spectrum (Fig. 9D) characteristic of native BcII is observed. These results demonstrate that even in the presence of 2 m GdmCl, zinc binds cooperatively to the enzyme and hence shifts the reversible denaturation by GdmCl from an unfolded state to a native-like intermediate state.
FIGURE 9.
Two-dimensional 1H-15N HSQC spectra for histidine imidazole of BcII in the presence of 2 m GdmCl. A, holoBcII. B, apoBcII. C, zinc/apoBcII molar ratio = 1. D, zinc/apoBcII molar ratio = 2. Note that standard BBL numbering system only is used.
Discussion
As reported before (46, 55, 60), the metal-depleted form of the enzyme is catalytically inactive and displays significant changes in its spectroscopic properties, suggesting both a catalytic and structural role for the metal ions in BcII β-lactamase. The conformational properties of both metal-free and dizinc forms of BcII β-lactamase were probed using chemical denaturants. Full reversibility of the unfolding process was established by both spectroscopic methods (i.e. fluorescence, CD, and NMR) and enzymatic assays. In particular, imidazole-optimized HSQC experiments (Fig. 9) showed that zinc binding to the apoenzyme unfolded in the presence of 2 m GdmCl shifts the equilibrium to the intermediate partially folded state.
Equilibrium folding experiments showed major destabilization of the enzyme in the absence of zinc. Thus, although unfolding of BcII proved to be fully reversible in both the absence and presence of zinc (Figs. 2 and 9), transition curves obtained by measuring fluorescence and CD (Fig. 2, E and F) showed clear differences. Although unfolding of apoBcII starts at 1 m GdmCl and follows a simple two-state cooperative transition (N ⇌ U), with Cm = 1.5 m (i.e. transition midpoint), the metal-bound β-lactamase remains fully active in the presence of 1.5 m GdmCl and, at higher denaturant concentrations, unfolds non-cooperatively in an apparent three-state transition (N ⇌ I ⇌ U), with Cm values of ∼1.9 and 2.8 m. These data are consistent with heat-induced denaturation of the enzyme (61). Thus, unfolding of the metal-bound species was best described by a three-state model, with apparent Tm values of 72 ± 2 and 85 ± 3 °C (61), whereas the apoenzyme was found to unfold in a to a two-state transition, with apparent Tm = 62.3 ± 0.02 °C (61). Thermal unfolding of both enzyme forms proved irreversible, however, and thus no further quantitative analysis could be performed.
Analysis of apo- and holoBcII chemical-induced unfolding curves was performed on the basis of two- and three-state models, respectively, and fitting the corresponding equations to the data in Fig. 2, E and F, yielded values of the thermodynamic parameters displayed in Tables 1 and 2. With both forms, transition curves obtained by intrinsic fluorescence and far-UV CD measurements (also near-UV CD, not shown) indicated that secondary and tertiary structures are destabilized simultaneously, albeit in one and two transitions in the absence and presence of zinc, respectively. This analysis suggests that in the presence of zinc, a partially folded state with reduced fluorescence and CD signal intensities is maximally populated at ∼2.4 m GdmCl. The coincident loss in both signals at all denaturant concentrations, combined with the absence of ANS binding, discards the possibility of a molten globule intermediate (51–54).
For apoBcII, the Gibbs free energy of unfolding (ΔG0NU) was calculated to be 32 ± 2 kJ·mol−1 (Table 1), and the corresponding mUN (= −mNU) values are 20 ± 1 and 9.9 ± 0.2 kJ·mol−1·m−1 for GdmCl and urea, respectively. Interestingly, the experimental mUN values are 10–20% smaller than predicted from the expected change in solvent-accessible surface area upon folding (22.2 and 13 kJ·mol−1·m−1 in GdmCl and urea, respectively; estimated from the size of the enzyme (62)), a finding consistent with an increase of the protein surface exposed to solvent in native apoBcII. On the basis of a three-state model, values of the thermodynamic parameters (i.e. ΔG0NU and mUN) for holoBcII were found to be 80–95 kJ·mol−1 and 30–40 kJ·mol−1·m−1, respectively (Table 2). Although for the second structural transition, fluorescence and CD measurements yielded values that are identical within the error limit (Table 2), major differences between these two measurements appeared for the first transition. In both cases, the global ΔG0UN value (≥80 kJ·mol−1) is significantly greater than the 20–60 kJ·mol−1 found in textbooks (63–66) for most proteins. Furthermore, the corresponding experimental mUN value (≥30 kJ·mol−1 in GdmCl) is also well above that calculated from the size of the protein (22.2 kJ·mol−1 (62)). Thus, values of the thermodynamic parameters in Table 2 suggest that a three-state model might be too simple a description of holoBcII unfolding. This is further supported by the observation that a number of residues, notably Leu-178(231) in the β11-α4 loop, show a non-linear dependence of their amide chemical shifts on GdmCl concentration in the range 1.0–2.34 m (Fig. 6), indicating the presence of more than two states in this concentration range.
Aiming at a better description of the unfolding mechanism of BcII metallo-β-lactamase, we analyzed in detail the structural features of the intermediate state that apparently forms in a non-cooperative manner at moderate GdmCl concentration (Fig. 2, A, C, and E). Although the enzyme was found to lose its activity in the 1.5–2.5 m GdmCl concentration range, optical methods indicated population of a partially folded state, which is not a molten globule and which displays a high content of structural organization, at both the secondary and the tertiary levels. Thus, far-UV CD measurements indicated that the protein retains ∼83% of its α-helical content in the presence of 2.4 m GdmCl, and both fluorescence quenching and ANS binding experiments suggested conservation of a compact and well organized protein core. Two-dimensional NMR experiments allowed the effect of both GdmCl and urea to be monitored at the level of individual residues. In particular, long range 1H–15N HSQC for selective observation of His imidazole resonances demonstrated that the loss of catalytic activity, concomitant with the first transition observed by optical methods, is not due to the loss of zinc from the enzyme active site, which in fact remains occupied by both metal ions up to 2.5 m GdmCl, with no significant change in its geometry. Classical 1H–15N HSQC experiments were performed in the presence of GdmCl and urea to probe the effect on the environment of 184 and 181 residues, respectively, out of 227 in the full-length protein. In the GdmCl and urea concentration ranges of 0 to 2.34 and 0 to 7.9 m, respectively, two-dimensional NMR data showed progressive changes of the chemical shift of many residues, with significant (CSP > 0.064 and CSP > 0.079 ppm, respectively) perturbations for 59 and 40 residues, respectively. These data suggest a gradual transition from the native state to intermediate states with only modest structural changes as indicated by the NMR spectra. These changes are seen to occur at the surface of the enzyme structure and are well localized in loop structures and at the N- and C-terminal extremities of α-helices and β-strands. HSQC experiments thus confirm population of globular species, with a well organized, compact, solvent-protected hydrophobic core. Upon increasing GdmCl concentrations in the range of 1.5 to 2.5 m, the equilibrium between these interconverting folding intermediates is shifted toward the less structured one, characterized by a reduction of ∼17 and 25% in CD (at 222 nm) and fluorescence (at 370 nm) intensity, respectively. Thus, all the data are consistent with the first transition leading to the non-cooperative formation of a heterogeneous ensemble of native-like protein molecules, with two zinc ions bound in the active site but no catalytic activity. A single distinct folding intermediate (e.g. molten globule) is not formed, and a simple three-state model cannot be used to fit the data. In contrast, the second transition from these multiple interconverting native-like species to the fully unfolded state seems to occur with full cooperativity. Interestingly, the corresponding m value (mIN = 23.5 ± 0.7 kJ·mol−1·m−1) is about the same than that calculated for the native enzyme (mUN = 22.2 kJ·mol−1·m−1 (62)), suggesting that the surface that is not accessible to the solvent becomes entirely exposed in this transition. This observation strengthens the conclusion that the first transition leads to the formation of native-like zinc-bound species, with no significant opening of the protein structure and only limited and superficial conformational changes. These are sufficient, however, to cause complete enzyme inactivation.
Finally, our results reveal that global unfolding of the protein takes place over a narrow GdmCl concentration range of 2.5 to 3.0 m, in a cooperative transition characterized by a Gibbs free energy change (ΔG0) of 65 ± 1.4 kJ·mol−1 and a corresponding m value of 23.5 ± 0.7 kJ·mol−1·m−1. These values are compatible with unfolding of the enzyme occurring in a single cooperative transition, from native-like intermediate species to the fully unfolded state. Although holoBcII β-lactamase is remarkably stable, removal of zinc, however, causes a dramatic destabilization of ∼35 kJ·mol−1. This finding is consistent with current knowledge that metal cofactors often stabilize the native state of the bound protein (67, 68). The effect of zinc on the stability of three other MBLs was studied in some detail. With subclass B1 BlaB and VIM-4 β-lactamases, zinc ions were observed to stabilize the enzyme. GdmCl unfolding of BlaB (69) monitored by intrinsic fluorescence measurements occurred at significantly lower denaturant concentration for the apo-form than for the two zinc-bound species. Although the data were interpreted on the basis of a three-state model, the lack of reversibility observed in experiments measuring the restoration of catalytic activity seriously compromises any quantitative analysis. With VIM-4 (70), a dramatic decrease in thermal stability (i.e. a drop of 27 °C in the apparent Tm value) was observed in the absence of zinc. Likewise, with CphA β-lactamase (36), a subclass B2 carbapenemase that is active in the monozinc form only, the catalytic zinc was found to increase the thermal stability by ∼14 °C, whereas the dizinc inactive form was found to be even more resistant to temperature (i.e. a further increase of 4 °C in Tm) than the monozinc enzyme. Finally, study of human glyoxalase II (71), another enzyme of the MBL superfamily, suggested that chemically induced unfolding is a multistep process. Very low GdmCl concentrations (<1 m) were seen to inactivate the enzyme, with no release of zinc and no significant optical changes (both intrinsic fluorescence and CD), an observation that is reminiscent of what is found in this work. A further increase in denaturant concentration led, however, to the formation of a molten globule (with apparent maximum population at 1.2 m (71)), before leading to complete unfolding at concentrations above 3 m.
All the results in this work point to the major structural role of zinc. Up to ∼2.5 m GdmCl, the presence of zinc in the active site appears to lock the protein in an ensemble of native-like structures. The NMR experiments, however, show that there are definite, but limited, structural changes at the enzyme surface at this GdmCl concentration. These limited surface changes must give rise to the observed loss of catalytic activity. In particular, substantial CSPs are seen to occur in the β3-β4 (residues 32–38(59–66)) and β11-α4 (170–188(223–241)) loops that flank the enzyme active site and also for Asp-90(120) in the Zn2 site. It is notable that all these residues also show substantial CSPs on the binding of an inhibitor to the active site (31). The large CSP observed for Asp-90(120) in this work (Fig. 7) most likely results from changes in its environment and not in its position, because observation of imidazole chemical shifts demonstrated that the geometry of both zinc-binding sites remains largely unaffected by GdmCl concentrations below 2.5 m. As for the loops, both have been well documented to be flexible and to modulate, by conformational changes, the shape of the active site (see references below). The NMR solution structure (31) showed that, despite significant flexibility, both structural elements display a relatively well defined conformation. They are generally considered to play a critical role for catalytic activity, especially in the ability of MBLs to hydrolyze β-lactam antibiotics of very diverse structures (11, 17). In particular, the β3-β4 loop (32–38(59–66)) (also known as the “mobile loop” or “flap”), which is conserved in most class B1 MBLs, has been shown to move toward bound inhibitors and substrates and thus to contribute to the formation of a hydrophobic pocket, which is important for ligand binding (31, 40, 72–90). Together with the flap, the β11-α4 loop (170–188(223–241)) on the other side of the active site also contributes to the capacity of MBLs to bind a structurally broad range of substrates and inhibitors (77, 79, 80, 90). In particular, recent crystal structures of NDM-1 MBL (91, 92) have provided further evidence that residues Lys(224) and Asn(233) (BBL numbering; these two residues are conserved in BcII, IMP-1, and CCrA; His(224) and Tyr(224)/Arg(228) are found in VIM-1 and VIM-2, respectively) in the β11-α4 loop play a direct role in substrate binding and hydrolysis. A combination of structural and computational studies of NDM-1 (56) has led to the suggestion that movements of the two loops are important not only in substrate binding but also in the hydration of the active site, including the water molecules that play a central role in the catalytic mechanism. Observation in this work that relatively modest structural changes in the partially folded species, particularly in the two active site loops, lead to complete loss of catalytic activity provides further evidence that these structural elements are key determinants of the enzyme catalytic activity and thus are relevant to the discovery of effective inhibitors.
Experimental Procedures
Enzyme and Chemicals
Ultrapure GdmCl and benzylpenicillin were purchased from Sigma. All other chemicals were of reagent grade.
Both the recombinant (46) and uniformly 15N isotopically enriched (55) β-lactamase II from B. cereus 569/H/9 (BcII) were expressed and purified as described. The enzyme concentration was determined by absorbance measurements at 280 nm, using a molar extinction coefficient value of 30,500 m−1·cm−1 (43).
The metal-free BcII enzyme (apoBcII) was prepared following the procedure described in Jacquin et al. (46). All final preparations were stored at −20 °C.
Buffers
Unless otherwise mentioned, experiments were performed in 10 mm Hepes buffer, pH 7.5, with 300 mm NaCl in the presence (holoBcII) or absence (apoBcII) of 0.1 mm ZnSO4. GdmCl and acrylamide solutions were prepared in the same buffer, and, if necessary, pH was adjusted to 7.5 with HCl or NaOH.
Enzyme Activity Measurements
Hydrolysis of benzylpenicillin was monitored by measuring absorbance changes at 235 nm using a Specord 200 spectrophotometer (Analytikjena) equipped with a thermostatically controlled cell holder. Values of the kinetic parameters were determined at 30 °C in 10 mm Hepes, pH 7.5, as described (93).
Chemical-induced Unfolding Transitions
Unfolding was studied at 25 °C. Samples at various denaturant (GdmCl and urea) concentrations were allowed to equilibrate for at least 12 h (under these conditions, equilibrium is reached throughout the transition, see “Results”). Unfolding and refolding transitions were obtained by monitoring the changes in intrinsic fluorescence emission (λex = 280 nm; λem = 370 nm) and circular dichroism (CD) at 222 nm, using a Varian Cary Eclipse spectrofluorimeter and a Jasco J-810 spectropolarimeter, respectively, both equipped with a thermostatically controlled cell holder. With all samples, the data were corrected for the contribution of the solution (buffer + denaturant). Unfolding of holoBcII was also monitored by measuring enzyme activity, as described above. Denaturant concentrations in the samples were determined from refractive index measurements (94) using a R5000 hand-held refractometer from Atago (Japan). Protein concentrations of ∼0.15 mg·ml−1 (6.0 μm) were used throughout.
Quenching of Tryptophan Fluorescence by Acrylamide
All fluorescence experiments were performed at 25 °C using a protein concentration of 0.10 mg·ml−1 (4 μm). The enzyme was diluted with acrylamide concentrations ranging from 0 to 0.2 m and tryptophan fluorescence emission spectra were recorded from 305 to 450 nm, following excitation at 295 nm. Each spectrum was the average of five acquisitions and was corrected for the contribution of the solution. The solvent accessibility of tryptophan residues was estimated according to the Stern-Volmer Equation 1 (95),
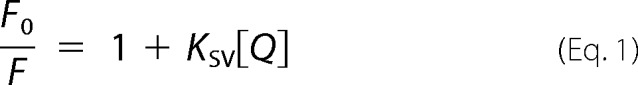 |
where F0 and F are the tryptophan fluorescence intensities (computed as areas below the curves for the tryptophan emission fluorescence spectra) in the absence and presence of acrylamide, respectively, KSV is the Stern-Volmer quenching constant and [Q] is the molar concentration of acrylamide.
Nuclear Magnetic Resonance (NMR)
Freeze-dried 15N-labeled holoBcII was dissolved in 10 mm Hepes buffer, pH 6.5, containing 300 mm NaCl and 0.1 mm ZnSO4, to give a final enzyme concentration of 10 mg·ml−1 (400 μm) in the presence of GdmCl or urea concentrations ranging from 0 to 4 m or 0 to 7.9 m, respectively. NMR spectra were collected using a home-built 750 MHz NMR spectrometer, controlled with GE/Omega software and equipped with an Oxford Instruments Co. magnet and a home-built triple-resonance pulse-field-gradient probe head. Spectral widths of 10204.08 and 2659.57 Hz in F2 (1H) and F1 (15N) respectively, were used for two-dimensional 1H-15N HSQC spectra. All spectra were collected at 25 °C, processed with NMRPipe (96), and analyzed with CcpNmr Analysis (97).
For chemical-induced unfolding under equilibrium conditions, the concomitant population of folded and unfolded species in the transition region was accompanied by changes in the NMR spectrum, both in terms of peak intensity and chemical shifts. The combined CSP was calculated as shown in Equation 2 (98),
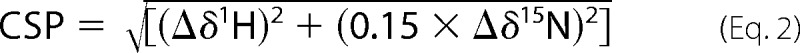 |
where δ represents the chemical shift in ppm.
To determine significant shift changes, we defined a threshold value, as follows. In a first step, all shift distances were considered and averaged (〈CSP〉), with errors calculated at the 95% confidence limit (i.e. twice the standard deviation, σ). Then, the highest shift changes (CSP ≥ 〈CSP〉 + 2σ) were removed from the data and a new 〈CSP〉, with standard deviation, was calculated. This operation was repeated until all data lie within 2σ values of the average T and the final 〈CSP〉 + 2σ value corresponds to the threshold below which the residues are not significantly perturbed.
The two-dimensional 1H-15N HSQC experiments for the observation of histidine imidazole groups were carried out with a total dephasing delay of 16.67 ms, at 25 °C, on a Bruker Avance 600 MHz NMR spectrometer equipped with a cryoprobe. The apoprotein concentration was 12.5 mg·ml−1 (500 μm) in 10 mm Hepes, 300 mm NaCl, pH 7.5. 1 and 2 m eq of zinc ions were added to the same sample.
Data Analysis
Unfolding curves were analyzed on the basis of a two-state (N ⇌ U) or a three-state (N ⇌ I ⇌ U) model, according to Vandenameele et al. (99) and Equation 3 and 4, respectively. This analysis is based on the assumption that the differences in Gibbs free energy between various species exhibit a linear dependence on denaturant concentration (100, 101),
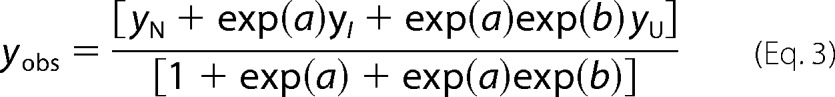 |
where
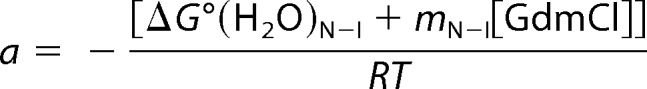 |
and
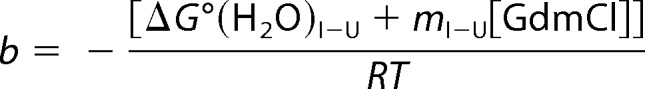 |
yobs is the measured parameter at a given denaturant concentration, and yN, yI, and yU are the values of this parameter for the folded, partially unfolded (intermediate), and unfolded states, respectively, at the same denaturant concentration. The observed linear dependence of this parameter on denaturant concentration was taken into account as described (102). ΔG0(H2O)N-I and ΔG0(H2O)I-U represent the unfolding free energy on going from N to I and from I to U, respectively, in the absence of denaturant; mN-I and mI-U are the slopes, δ(ΔG0)/δ[GdmCl], of the corresponding linear plots of the Gibbs free energy against denaturant concentration. R is the gas constant, and T is the absolute temperature. The midpoints of the transition, i.e. the denaturant concentrations at which [I]/[N] and [U]/[I] are equal to unity, are given by Cm = −ΔG0(H2O)/m.
The program GraFit 5.0.10 (Erithacus Software Ltd.) was used for non-linear least squares analysis of the data. Unless otherwise stated, errors are reported as standard deviations throughout.
Author Contributions
C. M. and M. N. contributed equally to this work. A. M. conceived and coordinated the study and wrote the paper. C. M. and M. N. contributed substantially to conception and design, acquisition, and analysis of data and also to writing the article. O. J., N. W., M. D., and A. I. K. contributed significantly to acquisition of data. G. C. K. R., C. D., and C. R. contributed decidedly to design, analysis, and interpretation of NMR experiments and also to drafting the article. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We acknowledge Prof. Jean-Marie Frère for critical reading of the manuscript and Prof. Roger H. Pain for his suggestion of a “punchy” title. Also, we are grateful to Dr. Catherine Goodman (acting on behalf of the Journal of Biological Chemistry) for valuable suggestions to improve the manuscript. Some NMR experiments in Oxford, United Kingdom, were collected with access provided by the European Commission's Framework Program 7 (FP7) East-NMR (Contract 228461) and BioNMR projects (Contract 261863).
This work was supported in part by Fonds de la Recherche Fondamentale et Collective Contracts 2.4550.05 and 2.4530.09 and by the Belgian Program of Interuniversity Attraction Poles initiated by the Federal Office for Scientific Technical and Cultural Affairs Grants P5/33, P6/19, and P7/44). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1.
- MBL
- metallo β-lactamase
- BcII
- β-lactamase II from B. cereus 569/H/9
- GdmCl
- guanidinium chloride
- ANS
- 1-anilino-8-naphthalenesulfonate
- HSQC
- heteronuclear single quantum coherence
- CSP
- combined chemical shift perturbation.
References
- 1. Waley S. G. (1992) in The Chemistry of β-Lactams (Page M. I., ed) pp. 198–228, Blackie, London, UK [Google Scholar]
- 2. Frère J. M. (1995) β-Lactamases and bacterial resistance to antibiotics. Mol. Microbiol. 16, 385–395 [DOI] [PubMed] [Google Scholar]
- 3. Fink A. L., and Page M. I. (2012) in β-Lactamases (Frère J. M., ed) pp. 41–77, Nova Science Publishers, Inc., New York [Google Scholar]
- 4. Poole K. (2004) Resistance to β-lactam antibiotics. Cell. Mol. Life Sci. 61, 2200–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jacoby G. A., and Munoz-Price L. S. (2005) The new β-lactamases. N. Engl. J. Med. 352, 380–391 [DOI] [PubMed] [Google Scholar]
- 6. Rossolini G. M., and Docquier J. D. (2006) New β-lactamases: a paradigm for the rapid response of bacterial evolution in clinical setting. Future Microbiol. 1, 295–308 [DOI] [PubMed] [Google Scholar]
- 7. Frère J. M., (ed) (2012) β-Lactamases, Nova Science Publishers, Inc., New York [Google Scholar]
- 8. Matagne A., Dubus A., Galleni M., and Frère J. M. (1999) The β-lactamase cycle: a tale of selective pressure and bacterial ingenuity. Nat. Prod. Rep. 16, 1–19 [DOI] [PubMed] [Google Scholar]
- 9. Heinz U., and Adolph H. W. (2004) Metallo-β-lactamases: two binding sites for one catalytic metal ion? Cell. Mol. Life Sci. 61, 2827–2839 [DOI] [PubMed] [Google Scholar]
- 10. Bebrone C. (2007) Metallo-β-lactamases (Classification, Activity, Genetic Organization, Structure, Zinc Coordination) and their superfamily. Biochem. Pharmacol. 74, 1686–1701 [DOI] [PubMed] [Google Scholar]
- 11. Palzkill T. (2013) Metallo-β-lactamase structure and function. Ann. N.Y. Acad. Sci. 1277, 91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Neuwald A. F., Liu J. S., Lipman D. J., and Lawrence C. E. (1997) Extracting protein alignment models from the sequence database. Nucleic Acids Res. 25, 1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daiyasu H., Osaka K., Ishino Y., and Toh H. (2001) Expansion of the zinc metallo-hydrolase family of the β-lactamase fold. FEBS Lett. 503, 1–6 [DOI] [PubMed] [Google Scholar]
- 14. Herzberg O., and Fitzgerald M. D. (2004) in Handbook of Metalloproteins (Messerschmidt A., Bode W., and Gygler M., eds) Vol. 3, pp. 217–234, John Wiley & Sons, Ltd., New York [Google Scholar]
- 15. Bebrone C., Garau G., Garcia-Saez I., Chantalat L., Carfi A., and Dideberg O. (2012) in β-Lactamases (Frère J. M., ed) pp. 41–77, Nova Science Publishers, Inc., New York [Google Scholar]
- 16. Baier F., and Tokuriki N. (2014) Connectivity between catalytic landscapes of the metallo-β-lactamase superfamily. J. Mol. Biol. 426, 2442–2456 [DOI] [PubMed] [Google Scholar]
- 17. Karsisiotis A. I., Damblon C. F., and Roberts G. C. (2014) A variety of roles for versatile zinc in metallo-β-lactamases. Metallomics 6, 1181–1197 [DOI] [PubMed] [Google Scholar]
- 18. Walsh T. R., Toleman M. A., Poirel L., and Nordmann P. (2005) Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18, 306–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cornaglia G., Giamarellou H., and Rossolini G. M. (2011) Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect. Dis. 11, 381–393 [DOI] [PubMed] [Google Scholar]
- 20. Felici A., Amicosante G., Oratore A., Strom R., Ledent P., Joris B., Fanuel L., and Frère J. M. (1993) An overview of the kinetic parameters of class B β-lactamases. Biochem. J. 291, 151–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rasmussen B. A., and Bush K. (1997) Carbapenem-hydrolyzing β-lactamases. Antimicrob. Agents Chemother. 41, 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spencer J., and Walsh T. R. (2006) A new approach to the inhibition of metallo-β-lactamases. Angew. Chem. Int. Ed. Engl. 45, 1022–1026 [DOI] [PubMed] [Google Scholar]
- 23. Drawz S. M., and Bonomo R. A. (2010) Three decades of β-lactamase inhibitors. Clin. Microbiol. Rev. 23, 160–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Payne D. J. (1993) Metallo-β-lactamases: a new therapeutic challenge. J. Med. Microbiol. 39, 93–99 [DOI] [PubMed] [Google Scholar]
- 25. Livermore D. M., and Woodford N. (2000) Carbapenemases: a problem in waiting? Curr. Opin. Microbiol. 3, 489–495 [DOI] [PubMed] [Google Scholar]
- 26. Oelschlaeger P., Ai N., Duprez K. T., Welsh W. J., and Toney J. H. (2010) Evolving carbapenemases: can medicinal chemists advance one step ahead of the coming storm? J. Med. Chem. 53, 3013–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Patel G., and Bonomo R. A. (2013) “Stormy waters ahead”: global emergence of carbapenemases. Front. Microbiol. 4, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bush K. (2013) Proliferation and significance of clinically relevant β-lactamases. Ann. N.Y. Acad. Sci. 1277, 84–90 [DOI] [PubMed] [Google Scholar]
- 29. Fast W., and Sutton L. D. (2013) Metallo-β-lactamase: inhibitors and reporter substrates. Biochim. Biophys. Acta 1834, 1648–1659 [DOI] [PubMed] [Google Scholar]
- 30. Crowder M. W., Spencer J., and Vila A. J. (2006) Metallo-β-lactamases: novel weaponry for antibiotic resistance in bacteria. Acc. Chem. Res. 39, 721–728 [DOI] [PubMed] [Google Scholar]
- 31. Karsisiotis A. I., Damblon C. F., and Roberts G. C. (2013) Solution structures of the Bacillus cereus metallo-β-lactamase BcII and its complex with the broad spectrum inhibitor R-thiomandelic acid. Biochem. J. 456, 397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. King A. M., Reid-Yu S. A., Wang W., King D. T., De Pascale G., Strynadka N. C., Walsh T. R., Coombes B. K., and Wright G. D. (2014) Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance. Nature 510, 503–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Galleni M., Lamotte-Brasseur J., Rossolini G. M., Spencer J., Dideberg O., Frère J. M., and Metallo-beta-lactamases Working Group. (2001) Standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 45, 660–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Frère J. M., Galleni M., Bush K., and Dideberg O. (2005) Is it necessary to change the classification of β-lactamases? J. Antimicrob. Chemother. 55, 1051–1053 [DOI] [PubMed] [Google Scholar]
- 35. Page M. I., and Badarau A. (2008) The mechanisms of catalysis by metallo-β-lactamases. Bioinorg. Chem. Appl. 2008, 576297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hernandez Valladares M., Felici A., Weber G., Adolph H. W., Zeppezauer M., Rossolini G. M., Amicosante G., Frère J. M., and Galleni M. (1997) Zn(II) dependence of the Aeromonas hydrophila AE036 metallo-β-lactamase activity and stability. Biochemistry 36, 11534–11541 [DOI] [PubMed] [Google Scholar]
- 37. Sabath L. D., and Abraham E. P. (1966) Zinc as a cofactor for cephalosporinase from Bacillus cereus 569. Biochem. J. 98, 11C–13C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meini M. R., Llarrull L. I., and Vila A. J. (2014) Evolution of metallo-β-lactamases: trends revealed by natural diversity and in vitro evolution. Antibiotics 3, 285–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carfi A., Pares S., Duée E., Galleni M., Duez C., Frère J. M., and Dideberg O. (1995) The 3-D structure of a zinc metallo-β-lactamase from Bacillus cereus reveals a new-type of protein fold. EMBO J. 14, 4914–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carfi A., Duée E., Galleni M., Frère J. M., and Dideberg O. (1998) 1.85 Å resolution structure of zinc β-lactamase from Bacillus cereus. Acta Crystallogr. D Biol. Crystallogr. 54, 313–323 [DOI] [PubMed] [Google Scholar]
- 41. Fabiane S. M., Sohi M. K., Wan T., Payne D. J., Bateson J. H., Mitchell T., and Sutton B. J. (1998) Crystal structure of the zinc-dependent β-lactamase from Bacillus cereus at 1.9 Å resolution: binuclear active site with features of a mononuclear enzyme. Biochemistry 37, 12404–12411 [DOI] [PubMed] [Google Scholar]
- 42. Garau G., García-Sáez I., Bebrone C., Anne C., Mercuri P., Galleni M., Frère J. M., and Dideberg O. (2004) Update of the standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 48, 2347–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Paul-Soto R., Bauer R., Frère J. M., Galleni M., Meyer-Klaucke W., Nolting H., Rossolini G. M., de Seny D., Hernandez-Valladares M., Zeppezauer M., and Adolph H. W. (1999) Mono and binuclear Zn2+-β-lactamase. Role of the conserved cysteine in the catalytic mechanism. J. Biol. Chem. 274, 13242–13249 [DOI] [PubMed] [Google Scholar]
- 44. Wommer S., Rival S., Heinz U., Galleni M., Frere J. M., Franceschini N., Amicosante G., Rasmussen B., Bauer R., and Adolph H. W. (2002) Substrate-activated zinc binding of metallo-β-lactamases–physiological importance of the mononuclear enzymes. J. Biol. Chem. 277, 24142–24147 [DOI] [PubMed] [Google Scholar]
- 45. Rasia R. M., and Vila A. J. (2002) Exploring the role and the binding affinity of a second zinc equivalent in B. cereus metallo-β-lactamase. Biochemistry 41, 1853–1860 [DOI] [PubMed] [Google Scholar]
- 46. Jacquin O., Balbeur D., Damblon C., Marchot P., De Pauw E., Roberts G. C., Frère J. M., and Matagne A. (2009) Positively cooperative binding of zinc ions to Bacillus cereus 569/H/9 β-lactamase II suggests that the binuclear enzyme is the only relevant form for catalysis. J. Mol. Biol. 392, 1278–1291 [DOI] [PubMed] [Google Scholar]
- 47. Wang Z., Fast W., and Benkovic S. J. (1999) On the mechanism of the metallo-β-lactamase from Bacteroides fragilis. Biochemistry 38, 10013–10023 [DOI] [PubMed] [Google Scholar]
- 48. Wang Z., Fast W., Valentine A. M., and Benkovic S. J. (1999) Metallo-β-lactamase: structure and mechanism. Curr. Opin. Chem. Biol. 3, 614–622 [DOI] [PubMed] [Google Scholar]
- 49. Frère J. M. (ed) (2012) in β-Lactamases, pp. 41–77, Nova Science Publishers, Inc., New York [Google Scholar]
- 50. Hemmingsen L., Damblon C., Antony J., Jensen M., Adolph H. W., Wommer S., Roberts G. C., and Bauer R. (2001) Dynamics of mononuclear cadmium β-lactamase revealed by the combination of NMR and PAC spectroscopy. J. Am. Chem. Soc. 123, 10329–10335 [DOI] [PubMed] [Google Scholar]
- 51. Redfield C. (1999) Molten globules. Curr. Biol. 9, R313. [DOI] [PubMed] [Google Scholar]
- 52. Kuwajima K. (1989) The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Protein Struct. Funct. Gen. 6, 87–103 [DOI] [PubMed] [Google Scholar]
- 53. Ptitsyn O. B., Pain R. H., Semisotnov G. V., Zerovnik E., and Razgulyaev O. I. (1990) Evidence for a molten globule state as a general intermediate in protein folding. FEBS Lett. 262, 20–24 [DOI] [PubMed] [Google Scholar]
- 54. Semisotnov G. V., Rodionova N. A., Razgulyaev O. I., Uversky V. N., Gripas' A. F., and Gilmanshin R. I. (1991) Study of the “Molten Globule” intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers 31, 119–128 [DOI] [PubMed] [Google Scholar]
- 55. Damblon C., Prosperi C., Lian L. Y., Barsukov I., Paul-Soto R., Galleni M., Frère J. M., and Roberts G. C. (1999) 1H–15N HMQC for the identification of metal-bound histidines in 113Cd substituted Bacillus cereus zinc β-lactamase. J. Am. Chem. Soc. 121, 11575–11576 [Google Scholar]
- 56. Kim Y., Cunningham M. A., Mire J., Tesar C., Sacchettini J., and Joachimiak A. (2013) NDM-1, the ultimate promiscuous enzyme: substrate recognition and catalytic mechanism. FASEB J. 27, 1917–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Eftink M., and Ghiron C. A. (1981) Fluorescence quenching studies with proteins. Anal. Biochem. 114, 199–227 [DOI] [PubMed] [Google Scholar]
- 58. Karsisiotis A. I., Damblon C., and Roberts G. C. (2014) Complete 1H, 15N, and 13C resonance assignments of Bacillus cereus metallo-β-lactamase and its complex with the inhibitor R-thiomandelic acid. Biomol. NMR Assign. 8, 313–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hubbard S. J., and Thornton J. M. (1993) NACCESS, Version 2.1.1, University College; London [Google Scholar]
- 60. Rasia R. M., and Vila A. J. (2004) Structural determinants of substrate binding to Bacillus cereus metallo-β-lactamase. J. Biol. Chem. 279, 26046–26051 [DOI] [PubMed] [Google Scholar]
- 61. Jacquin O. (2011) Etude des Propriétés de Repliement et de Fixation du Zinc de la Métallo-β-lactamase BcII de Bacillus cereus 569/H/9. Ph.D. thesis, University of Liège [Google Scholar]
- 62. Myers J. K., Pace C. N., and Scholtz J. M. (1995) Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 4, 2138–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pfeil W. (1998) Protein Stability and Folding: A Collection of Thermodynamic Data, Springer-Verlag, Berlin [Google Scholar]
- 64. Whitford D. (2005) Proteins: Structure and Function, John Wiley & Sons, Ltd., Chichester, UK [Google Scholar]
- 65. Creighton T. E. (2010) The Biophysical Chemistry of Nucleic Acids and Proteins, Helvetian Press [Google Scholar]
- 66. Lehninger A. L., Nelson D. L., and Cox M. M. (2013) Principles of Biochemistry, 6th Ed., W. H. Freeman & Co., New York [Google Scholar]
- 67. Palm-Espling M. E., Niemiec M. S., and Wittung-Stafshede P. (2012) Role of metal in folding and stability of copper proteins in vitro. Biochim. Biophys. Acta 1823, 1594–1603 [DOI] [PubMed] [Google Scholar]
- 68. Wittung-Stafshede P. (2002) Role of cofactors in protein folding. Acc. Chem. Res. 35, 201–208 [DOI] [PubMed] [Google Scholar]
- 69. Selevsek N., Rival S., Tholey A., Heinzle E., Heinz U., Hemmingsen L., and Adolph H. W. (2009) Zinc ion-induced domain organization in metallo-β-lactamases. J. Biol. Chem. 284, 16419–16431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lassaux P., Traoré D. A., Loisel E., Favier A., Docquier J. D., Sohier J. S., Laurent C., Bebrone C., Frère J. M., Ferrer J. L., and Galleni M. (2011) Biochemical and structural characterization of the subclass B1 metallo-β-lactamase VIM-4. Antimicrob. Agents Chemother. 55, 1248–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dragani B., Cocco R., Ridderström M., Stenberg G., Mannervik B., and Aceto A. (1999) Unfolding and refolding of human glyoxalase II and its single-tryptophan mutants. J. Mol. Biol. 291, 481–490 [DOI] [PubMed] [Google Scholar]
- 72. Concha N. O., Rasmussen B. A., Bush K., and Herzberg O. (1996) Crystal structure of the wide-spectrum binuclear zinc β-lactamase from Bacteroides fragilis. Structure 4, 823–836 [DOI] [PubMed] [Google Scholar]
- 73. Concha N. O., Janson C. A., Rowling P., Pearson S., Cheever C. A., Clarke B. P., Lewis C., Galleni M., Frère J. M., Payne D. J., Bateson J. H., and Abdel-Meguid S. S. (2000) Crystal structure of the IMP-1 metallo β-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor. Biochemistry 39, 4288–4298 [DOI] [PubMed] [Google Scholar]
- 74. Fitzgerald P. M., Wu J. K., and Toney J. H. (1998) Unanticipated inhibition of the metallo-β-lactamase from Bacteroides fragilis by 4-morpholineethanesulfonic acid (MES): a crystallographic study at 1.85-A resolution. Biochemistry 37, 6791–6800 [DOI] [PubMed] [Google Scholar]
- 75. Toney J. H., Fitzgerald P. M., Grover-Sharma N., Olson S. H., May W. J., Sundelof J. G., Vanderwall D. E., Cleary K. A., Grant S. K., Wu J. K., Kozarich J. W., Pompliano D. L., and Hammond G. G. (1998) Antibiotic sensitization using biphenyl tetrazoles as potent inhibitors of Bacteroides fragilis metallo-β-lactamase. Chem. Biol. 5, 185–196 [DOI] [PubMed] [Google Scholar]
- 76. Toney J. H., Hammond G. G., Fitzgerald P. M., Sharma N., Balkovec J. M., Rouen G. P., Olson S. H., Hammond M. L., Greenlee M. L., and Gao Y. D. (2001) Succinic acids as potent inhibitors of plasmid-borne IMP-1 metallo-β-lactamase. J. Biol. Chem. 276, 31913–31918 [DOI] [PubMed] [Google Scholar]
- 77. Scrofani S. D., Chung J., Huntley J. J., Benkovic S. J., Wright P. E., and Dyson H. J. (1999) NMR characterization of the metallo-β-lactamase from Bacteroides fragilis and its interaction with a tight-binding inhibitor: role of an active site loop. Biochemistry 38, 14507–14514 [DOI] [PubMed] [Google Scholar]
- 78. Yang Y., Keeney D., Tang X., Canfield N., and Rasmussen B. A. (1999) Kinetic properties and metal content of the metallo-β-lactamase CcrA harboring selective amino acid substitutions. J. Biol. Chem. 274, 15706–15711 [DOI] [PubMed] [Google Scholar]
- 79. Huntley J. J., Scrofani S. D., Osborne M. J., Wright P. E., and Dyson H. J. (2000) Dynamics of the metallo-β-lactamase from Bacteroides fragilis in the presence and absence of a tight-binding inhibitor. Biochemistry 39, 13356–13364 [DOI] [PubMed] [Google Scholar]
- 80. Huntley J. J., Fast W., Benkovic S. J., Wright P. E., and Dyson H. J. (2003) Role of a solvent-exposed tryptophan in the recognition and binding of antibiotic substrates for a metallo-β-lactamase. Protein Sci. 12, 1368–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mollard C., Moali C., Papamicael C., Damblon C., Vessilier S., Amicosante G., Schofield C. J., Galleni M., Frere J. M., and Roberts G. C. (2001) Thiomandelic acid, a broad spectrum inhibitor of zinc β-lactamases. J. Biol. Chem. 276, 45015–45023 [DOI] [PubMed] [Google Scholar]
- 82. Payne D. J., Hueso-Rodríguez J. A., Boyd H., Concha N. O., Janson C. A., Gilpin M., Bateson J. H., Cheever C., Niconovich N. L., Pearson S., Rittenhouse S., Tew D., Díez E., Pérez P., De La Fuente J., et al. (2002) Identification of a series of tricyclic natural products as potent broad-spectrum inhibitors of metallo-β-lactamases. Antimicrob. Agents Chemother. 46, 1880–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Docquier J. D., Lamotte-Brasseur J., Galleni M., Amicosante G., Frère J. M., and Rossolini G. M. (2003) On functional and structural heterogeneity of VIM-type metallo-β-lactamases. J. Antimicrob. Chemother. 51, 257–266 [DOI] [PubMed] [Google Scholar]
- 84. Moali C., Anne C., Lamotte-Brasseur J., Groslambert S., Devreese B., Van Beeumen J., Galleni M., and Frère J. M. (2003) Analysis of the importance of the metallo-β-lactamase active site loop in substrate binding and catalysis. (2003) Chem. Biol. 10, 319–329 [DOI] [PubMed] [Google Scholar]
- 85. Krauss M., Gresh N., and Antony J. (2003) Binding and hydrolysis of ampicillin in the active site of a zinc lactamase. J. Phys. Chem. B 107, 1215–1229 [Google Scholar]
- 86. Tomatis P. E., Fabiane S. M., Simona F., Carloni P., Sutton B. J., and Vila A. J. (2008) Adaptive protein evolution grants organismal fitness by improving catalysis and flexibility. Proc. Natl. Acad. Sci. U.S.A. 105, 20605–20610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Salsbury F. R. Jr., Crowder M. W., Kingsmore S. F., and Huntley J. J. (2009) Molecular dynamic simulations of the metallo-β-lactamase from Bacteroides fragilis in the presence and absence of a tight-binding inhibitor. J. Mol. Model. 15, 133–145 [DOI] [PubMed] [Google Scholar]
- 88. González J. M., Buschiazzo A., and Vila A. J. (2010) Evidence of adaptability in metal coordination geometry and active site loop conformation among B1 metallo-β-lactamases. Biochemistry 49, 7930–7938 [DOI] [PubMed] [Google Scholar]
- 89. Valdez C. E., Sparta M., and Alexandrova A. N. (2013) The role of the flexible L43-S54 protein loop in the CcrA metallo-β-lactamase in binding structurally dissimilar β-lactam antibiotics. J. Chem. Theory Comput. 9, 730–737 [DOI] [PubMed] [Google Scholar]
- 90. Rydzik A. M., Brem J., van Berkel S. S., Pfeffer I., Makena A., Claridge T. D., and Schofield C. J. (2014) Monitoring conformational changes in the NDM-1 metallo-β-lactamase by 19F NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 53, 3129–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang H., and Hao Q. (2011) Crystal structure of NDM-1 reveals a common β-lactam hydrolysis ;Mechanism. FASEB J. 25, 2574–2582 [DOI] [PubMed] [Google Scholar]
- 92. King D. T., Worrall L. J., Gruninger R., and Strynadka N. C. (2012) New Delhi metallo-β-lactamase: structural insights into β-lactam recognition and inhibition. J. Am. Chem. Soc. 134, 11362–11365 [DOI] [PubMed] [Google Scholar]
- 93. Matagne A., Misselyn-Bauduin A. M., Joris B., Erpicum T., Granier B., and Frère J. M. (1990) The diversity of the catalytic properties of class A β-lactamases. Biochem. J. 265, 131–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nozaki Y. (1972) The preparation of guanidine hydrochloride. Method Enzymol. 26, 43–50 [DOI] [PubMed] [Google Scholar]
- 95. Lakowicz J. R. (1999) Principles of Fluorescence Spectroscopy, 2nd Ed., pp. 237–265, Kluwert Academic/Plenum Publishers, New York [Google Scholar]
- 96. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 97. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., and Laue E. D. (2005) The CCPN Data Model for NMR Spectroscopy: development of a software pipeline. Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 98. Mulder F. A., Schipper D., Bott R., and Boelens R. (1999) Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol. 292, 111–123 [DOI] [PubMed] [Google Scholar]
- 99. Vandenameele J., Lejeune A., Di Paolo A., Brans A., Frère J. M., Schmid F. X., and Matagne A. (2010) Folding of class A β-lactamase is rate-limited by peptide bond isomerization and occurs via parallel pathways. Biochemistry 49, 4264–4275 [DOI] [PubMed] [Google Scholar]
- 100. Santoro M. M., and Bolen D. W. (1988) Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl α-chymotrypsin using different denaturants. Biochemistry 27, 8063–8068 [DOI] [PubMed] [Google Scholar]
- 101. Pace C. N. (1990) Measuring and increasing protein stability. Trends Biotechnol. 8, 93–98 [DOI] [PubMed] [Google Scholar]
- 102. Dumoulin M., Conrath K., Van Meirhaeghe A., Meersman F., Heremans K., Frenken L. G., Muyldermans S., Wyns L., and Matagne A. (2002) Single-domain antibody fragments with high conformational stability. Protein Sci. 11, 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.