Abstract
Synaptogyrin-2 is a non-neuronal member of the synaptogyrin family involved in synaptic vesicle biogenesis and trafficking. Little is known about the function of synaptogyrin-2. Severe fever with thrombocytopenia syndrome (SFTS) is an emerging infectious disease characterized by high fever, thrombocytopenia, and leukocytopenia with high mortality, caused by a novel tick-borne phlebovirus in the family Bunyaviridae. Our previous studies have shown that the viral nonstructural protein NSs forms inclusion bodies (IBs) that are involved in viral immune evasion, as well as viral RNA replication. In this study, we sought to elucidate the mechanism by which NSs formed the IBs, a lipid droplet-based structure confirmed by NSs co-localization with perilipin A and adipose differentiation-related protein (ADRP). Through a high throughput screening, we identified synaptogyrin-2 to be highly up-regulated in response to SFTS bunyavirus (SFTSV) infection and to be a promoter of viral replication. We demonstrated that synaptogyrin-2 interacted with NSs and was translocated into the IBs, which were reconstructed from lipid droplets into large structures in infection. Viral RNA replication decreased, and infectious virus titers were lowered significantly when synaptogyrin-2 was silenced in specific shRNA-expressing cells, which correlated with the reduced number of the large IBs restructured from regular lipid droplets. We hypothesize that synaptogyrin-2 is essential to promoting the formation of the IBs to become virus factories for viral RNA replication through its interaction with NSs. These findings unveil the function of synaptogyrin-2 as an enhancer in viral infection.
Keywords: lipid droplet, negative-strand RNA virus, protein-protein interaction, viral protein, viral replication
Introduction
Synaptogyrin-2 (SYNGR2)2 is a tetraspan vesicle membrane protein (1–3) and a member of the synaptogyrin family. Synaptogyrin-1 and -3, abundantly present in synaptic vesicles, are believed to be involved in various aspects in the synaptic vesicle cycle, including vesicle biogenesis, exocytosis and endocytotic recycling, and neurotransmission (2, 4). They are encoded by multigene classes in mammals and evolutionarily conserved throughout the animal kingdom (5). The synaptogyrin-2 gene transcript of 1.6 kb is expressed at high levels in all tissues, except brain, encoding a protein of 224 amino acids in length (5). However, studies on synaptogyrin-2 have been limited, and the function of this protein is largely unknown.
Emerging tick-borne bunyaviruses have been recognized to be a serious threat to public health and include severe fever with thrombocytopenia syndrome virus (SFTSV) (6). Since first identified in 2010 in China (7), SFTSV has caused over 3000 reported cases with mortalities ranging from 5 to 15% and has quickly spread to more than 13 provinces (8, 9). Although the symptoms of SFTS are mainly nonspecific, drastic loss of white blood cells and platelets are reported in most cases, and multiorgan failure can occur in severe cases, which usually are fatal (10, 11). SFTSV has also been isolated from patients in Korea and Japan since 2013 (12–15). The closely related Heartland virus was characterized after its isolation from two febrile patients in Missouri (16–18) and has been blamed for at least eight cases of Heartland disease with two fatalities in three more states since 2012. Patients infected with Heartland virus presented with similar symptoms. A few species of ticks are reported to be the vectors for this group of arboviruses, which infect various species of agricultural and wildlife animals (7, 19).
SFTSV and Heartland virus are enveloped RNA viruses with three single-stranded RNA genome segments (7, 20). The L and M segments are of negative polarity, encoding an RNA-dependent RNA polymerase and precursor of glycoproteins Gn and Gc, respectively. The S segment uses an ambisense strategy to encode a nucleoprotein (N) in antisense orientation and a nonstructural protein (NSs) in sense orientation, separated by an intergenic region (7, 20). Studies have unveiled the function of NSs in its suppression of antiviral interferon induction. We and other groups have identified a unique strategy of viral immune evasion used by NSs to suppress IFN signaling; that is, that NSs sequesters TBK1, as well as IKKϵ and IRF3, into viral inclusion bodies (IBs) formed by NSs itself, blocking the IRFs from translocation into the nucleus and resulting in an attenuated induction of IFN (21, 22). Interestingly, we found that NSs-formed IBs is lipid droplet-based, and viral dsRNA, the intermediate form of viral RNA during its replication, is also present in IBs, suggesting that NSs may be also involved in viral replication (23). However, the mechanism by which NSs targets and is translocated into lipid droplets remains largely unknown.
Lipid droplets are intracellular lipid storing organelles involved in lipid and carbohydrate metabolism (24, 25). They consist of a neutral lipid core surrounded by a phospholipid monolayer containing lipid droplet-associated proteins, which include proteins of the PAT family consisting of perilipin (26–28), adipose differentiation-related protein (ADRP) (28), and tail-interacting protein of 47 kDa (TIP47) (29, 30). Upon infection with SFTSV, the morphology of lipid droplets appears to be reconstructed in parallel to the expression and translocation of NSs onto the lipid vesicles (23). Similar morphological changes of IBs occur in rotavirus infection to form viroplasm for viral replication after viral NSP2 and NSP5 are translocated into lipid droplets (31). To date we know little about the mechanism by which viral proteins, such as NSs of SFTSV or NSP2 and NSP5 of rotavirus, are translocated onto lipid droplets and how these lipid vesicles are restructured to become IBs or viroplasm, the potential virus factories for viral replication and/or virion assembly (32).
In this study, we sought to characterize the mechanism by which NSs targets and transforms lipid droplets for building IBs, the unique structure for both viral replication and immune evasion. We identified synaptogyrin-2 (1), a non-neural member of the synaptogyrin family, as being highly up-regulated in response to SFTSV infection. Members of the synaptogyrin family are important in biogenesis and trafficking of synaptic vesicles, but the function of synaptogyrin-2 is largely unknown (3). Our data demonstrate that synaptogyrin-2 is translocated into lipid droplets through its interaction with NSs of SFTSV during infection. Altered expression of synaptogyrin-2 affects the number and size of the lipid droplet-based IBs and viral replication. We hypothesize that synaptogyrin-2 plays a critical role in reconstruction of lipid droplets in SFTSV infection, transforming them into IBs or virus factories as a potential enhancer in viral replication.
Results
Up-regulation of Synaptogyrin-2 in SFTSV-infected HepG2 Cells
During SFTSV infection the liver may be one of the severely affected organs, as indicated by increased levels of alanine and aspartate aminotransferases detected in the sera of fatal patients. Our previous studies showed that SFTSV-infected human liver epithelial cells HepG2 and unique cytoplasmic IBs were formed by NSs of SFTSV (21, 23, 33, 37). We have further discovered that the IBs were constructed based on lipid droplets and that NSs was the only viral protein that targets lipid droplets. Apparently lipid droplets underwent vigorous reconstruction because of changes in both size and number in infected cells during the infection and translocation of NSs. To further decipher the mechanism by which NSs may transform lipid droplets and whether there are cellular proteins that are involved in the process, we sought to profile global gene expression and screen potential host genes regulated in SFTSV-infected HepG2 cells with an Agilent expression microarray analysis. Among a number of significantly regulated host genes, we found that the mRNA transcript of synaptogyrin-2, a member of the synaptogyrin family involved in synaptic vesicle biogenesis and trafficking and neurotransmission, was up-regulated more than 100-fold starting from the early stage of infection (Fig. 1A). We confirmed the induction of the synaptogyrin-2 transcript by real time RT-PCR with synaptogyrin-2-specific primers and total RNA prepared from infected or non-infected control cells at various time points after infection. The levels of synaptogyrin-2 mRNA sharply increased up to 10.4-, 36.3-, and 275.0-fold at 12, 24, and 36 h.p.i., respectively, in HepG2 similar to the results from HeLa cells and HEK293 cells infected with SFTSV (Fig. 1B). Viral replication was confirmed by measuring fold change of the M gene copy numbers in HepG2, HeLa, and HEK293 cells with real time RT-PCR, using primers specific for the M gene as shown in Fig. 1C.
FIGURE 1.
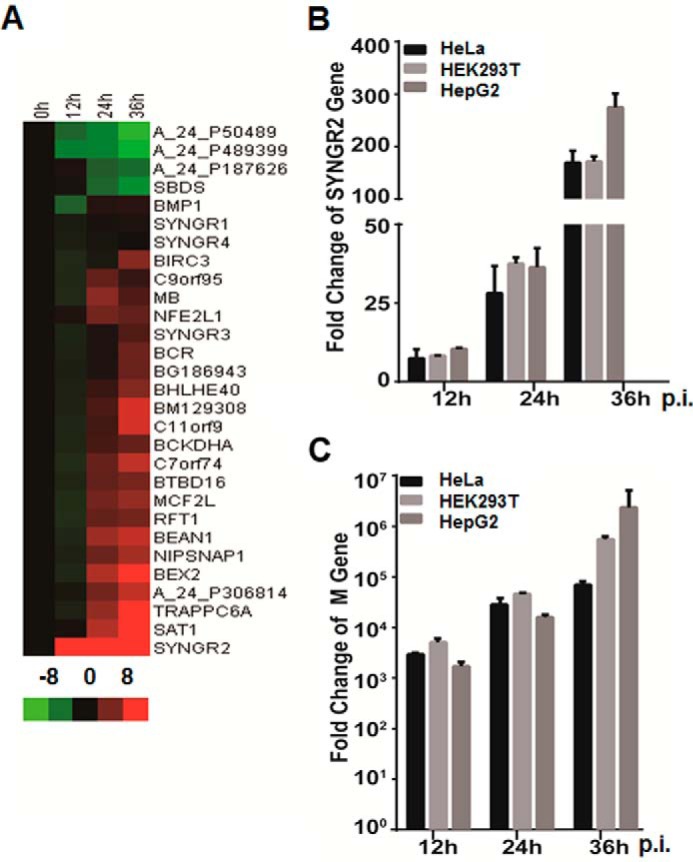
Up-regulation of synaptogyrin-2 in SFTSV-infected HepG2 cells. A, microarray profiling of transcriptionally regulated host genes. HepG2 cells were infected with SFTSV at an MOI of 1. Total RNA from non-infected and infected cells at 12, 24, and 36 h.p.i. was prepared, and cRNA was labeled with Cy3-CTP before being subjected to hybridization on A4 × 44 microarray slides (Agilent) and scanning. The GeneData Expressionist platform was used to calculate relative fold changes of genes. Log ratios are depicted in red (up-regulated) or green (down-regulated). B, fold change of synaptogyrin-2 mRNA transcripts. Total RNA from uninfected and infected cells was prepared for reverse transcription, and cDNA was amplified with SYNGR2 specific primers by real time PCR. C, replication of SFTSV in infected cells. Real time RT-PCR was performed to measure fold increase of the viral M segment RNA in infected cells.
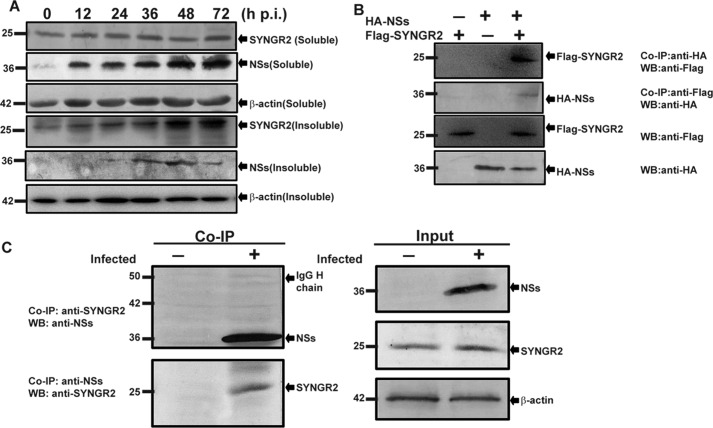
We examined the protein expression of synaptogyrin-2 in HepG2 cells infected with SFTSV with the cells lysed at various time points. Synaptogyrin-2 was constitutively expressed in non-infected cells. Interestingly, we were initially unable to detect an obvious increase of synaptogyrin-2 in the soluble fraction of the infected cells in response to SFTSV infection. Significant increase of synaptogyrin-2, however, was detected in the insoluble fraction (Fig. 2A), indicating that synaptogyrin-2 was up-regulated but that a large portion of the protein was translocated into an apparently insoluble structure. Viral nonstructural protein NSs was also detected in the insoluble fraction in infected HepG2 cells. It was not detected in large quantities, likely because of poor solubility of the insoluble structure.
FIGURE 2.
Synaptogyrin-2 expression and interaction with NSs of SFTSV. A, induced expression of synaptogyrin-2 in infected HepG2 cells. Soluble and insoluble fractions of cell lysates were prepared from infected and uninfected HepG2 cells at indicated time points and resolved with 10–15% SDS-PAGE before proteins were transferred to PVDF membranes for Western blot analyses staining with either anti-NSs or anti-SYNGR2 antibodies. Analyses were performed at least three times, and representative results were presented. B, co-immunoprecipitation of synaptogyrin-2 and NSs in transfected cells. HEK293 cells were transfected with plasmids expressing HA-tagged NSs and FLAG-tagged synaptogyrin-2. After NSs or synaptogyrin-2 was immunoprecipitated with anti-HA or anti-FLAG antibodies in the cell lysates, the immunoprecipitates were analyzed by Western blot analysis with anti-FLAG or anti-HA antibodies, respectively. C, co-immunoprecipitation of synaptogyrin-2 and NSs in infected HepG2 cells. The cells were either mock infected or infected with SFTSV, and cell lysates were immunoprecipitated with anti-NSs or anti-synaptogyrin-2 antibodies. The immunoprecipitates were subjected to Western blot analyses with anti-synaptogyrin-2 or anti-NSs antibodies, respectively (top two panels). Input NSs and synaptogyrin-2 in the lysates were shown by Western blot analyses with anti-NSs or anti-synaptogyrin-2 antibodies, respectively (bottom two panels). We repeated the experiments at least twice, and representative data are presented for each panel. Co-IP, co-immunoprecipitation; WB, Western blotting.
Synaptogyrin-2 Interacted with NSs in Infected and Transfected Cells
To understand the role of synaptogyrin-2 in SFTSV infection, we sought to explore whether NSs interacted with synaptogyrin-2. First, we transfected HEK293 cells with plasmids pRK5-F-SYNGR2 and/or pRK5-HA-NSs expressing FLAG-tagged synaptogyrin-2 and HA-tagged NSs, respectively. Cell lysates were immunoprecipitated with either anti-HA or anti-FLAG antibodies, and the immunoprecipitates were analyzed with SDS-PAGE and Western blot analysis. The results indicated that synaptogyrin-2 was associated with NSs in the transfected cells as shown in Fig. 2B.
We confirmed the interaction of NSs and synaptogyrin-2 in HepG2 cells infected with SFTSV. Cells lysates were prepared from both infected and non-infected cells 24 h.p.i. and immunoprecipitated with either anti-SYNGR2 or anti-NSs antibodies, followed by SDS-PAGE and Western blot analysis. As shown in Fig. 2C, NSs interacted with synaptogyrin-2 in infected HepG2.
Synaptogyrin-2 occurred in the soluble fraction in non-infected cells or the cells without NSs overexpression. However, in infected or in NSs-transfected cells, synaptogyrin-2 appeared in the insoluble fraction (Fig. 2A), possibly because of its interaction with NSs and being transported into insoluble structures, such as IBs, in the cytoplasm.
NSs Co-localized with Synaptogyrin-2 in the Cytoplasmic IBs
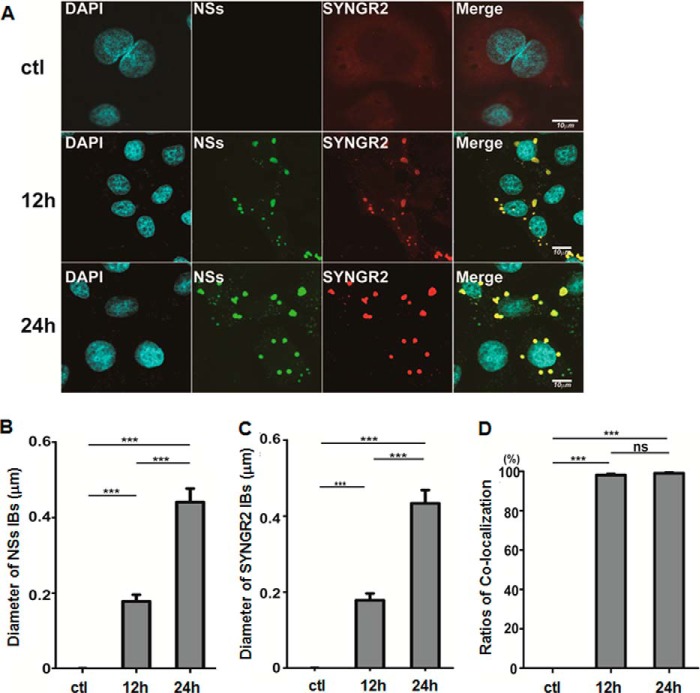
To ascertain the location of synaptogyrin-2 in the presence of NSs, we infected HeLa cells with SFTSV and fixed the cells at 12 and 24 h.p.i., followed by staining with either anti-NSs and/or anti-SYNGR2 antibodies in an immunofluorescence assay. We observed under a confocal fluorescence microscope that synaptogyrin-2 was dispersedly distributed in the cytoplasm, stained gloomily because of its low expression level, in non-infected cells (Fig. 3A). However, in SFTSV-infected cells NSs was shown in the unique IB structures, which grew larger in size throughout the infection (Fig. 3, A and B), whereas synaptogyrin-2 was translocated into the IBs (Fig. 3, A and C) and co-localized with NSs in the cytoplasm (Fig. 3, A and D). The IBs were present only in infected cells and increased in size while being stained either with anti-NSs or anti-SYNGR2 antibodies at 12 and 24 h.p.i. (Fig. 3, A–C). NSs and SYNGR2 were highly co-localized in the IBs (Fig. 3, A and D).
FIGURE 3.
NSs co-localized with synaptogyrin-2 in the cytoplasmic IBs. HepG2 cells were infected with SFTSV, and the cells were fixed and permeabilized at 12 and 24 h.p.i. The cells were co-stained with anti-NSs and anti-synaptogyrin-2 antibodies, which were further stained with Alexa Fluor 594- or 488-conjugated secondary antibodies. A, after washes, the cells were subjected to confocal microscopy. We repeated the experiments at least twice, and representative data are presented. B and C, the sizes of the IBs, stained with either anti-NSs (B) or anti-SYNGR2 (C) antibody, were measured in diameter with the ImageJ software in 50 randomly selected cells each from the control or 12 or 24 h.p.i., respectively. The data of the IB sizes in diameter were presented as the means ± S.D. by GraphPad Prism 5 with statistical analysis (***, p < 0.001). D, co-localization of NSs and SYNGR2 was counted in the cells from the control or 12 or 24 h.p.i. as well and presented as a percentage of the IBs stained by only anti-NSs or SYNGR2 antibody (Student's t test; ***, p < 0.001; ns, p > 0.05). ctl, control.
We confirmed the co-localization of NSs and synaptogyrin-2 in HeLa cells transfected with plasmids pRK5-F-SYNGR2 and pRK5-HA-NSs. In the absence of NSs, synaptogyrin-2 was distributed dispersedly in the cytoplasm, whereas NSs showed its typical location in IBs (Fig. 4A, top two rows). However, in co-transfected cells, synaptogyrin-2 changed its location by co-localizing with NSs in the IB structure at 12 and 24 h post co-transfection under a confocal microscope (Fig. 4A, bottom two rows). The number of the large IBs, or the IBs with a size larger than 0.2 μm in diameter, increased significantly at 12 and 24 h post-transfection (Fig. 4B).
FIGURE 4.
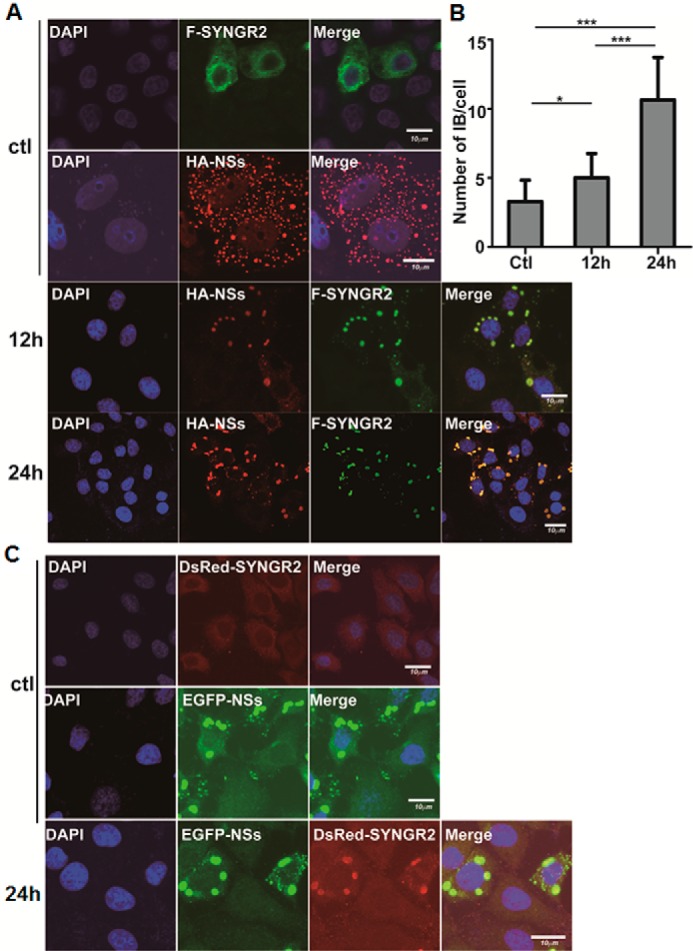
Co-localization of NSs and synaptogyrin-2 in transfected cells. A, co-localization of synaptogyrin-2 or NSs in HeLa cells. The cells were transfected alone with plasmids expressing FLAG-tagged synaptogyrin-2 or HA-tagged NSs (top two rows, 24 h post-transfection) or co-transfected with both. The cells were fixed and stained with either anti-HA, anti-FLAG, or both for staining NSs, synaptogyrin-2, or both. After further staining with conjugated secondary antibodies, the cells were subjected to observation under a confocal fluorescence microscope. B, quantitation of large IBs in transfected cells. The IBs with a diameter larger than 0.2 μm were counted with the ImageJ software in 50 randomly selected cells from either the control or 12 or 24 h post-transfection and presented as means ± S.D. in a single cell by GraphPad Prism 5 with a statistical analysis (Student's t test; ***, p < 0.001; *, p < 0.05). C, co-localization of synaptogyrin-2 or NSs observed in live HeLa cells. HeLa cells were co-transfected with plasmids expressing DsRed-SYNGR2 (top), or EGFP-NSs (middle), or both (bottom) and subjected to confocal fluorescence microscopy at 24 h post-transfection. We repeated the experiments at least twice, and representative data are presented for each panel. ctl, control.
When HeLa cells were co-transfected with plasmids encoding EGFP-NSs and DsRed-SYNGR2 for expressing fluorescence fusion proteins, nearly all synaptogyrin-2 was localized in NSs-formed IBs as well (Fig. 4C). In contrast, DsRed-SYNGR2 remained dispersedly distributed in the cytoplasm when the cells were transfected with the plasmid only expressing DsRed-SYNGR2 (Fig. 4C). All the confocal microscopy data as described above have been generated from observations of multiple cells in dozens of fields.
NSs Co-localized with Perilipin A and ADRP in IBs and Lipid Droplets
We have previously described that NSs-formed IB structure was based on lipid droplets, was stained positively with Nile Red in infected cells, and may play a role as virus factories in viral RNA replication (21). We sought to further characterize the nature of the IBs in SFTSV-infected cells, trying to identify lipid droplets-associated proteins together with synaptogyrin-2 in the IBs. HepG2 cells were infected with SFTSV, and the cells were fixed for staining with antibodies against perilipin A or ADRP, which were present specifically on lipid droplets (38), as well as the antibodies for NSs. In non-infected cells, ADRP and perilipin A were shown in lipid droplets, which were distributed dispersedly in the cytoplasm (Fig. 5A). In infected cells, however, NSs co-localized with ADRP (Fig. 5B) and perilipin A (Fig. 5C), confirming that NSs-formed IBs were based on lipid droplets. In addition to smaller and dispersedly distributed lipid droplets, there appeared to be large IB structures, which presumably were reconstructed and transformed from regular lipid droplets in the presence of NSs.
FIGURE 5.
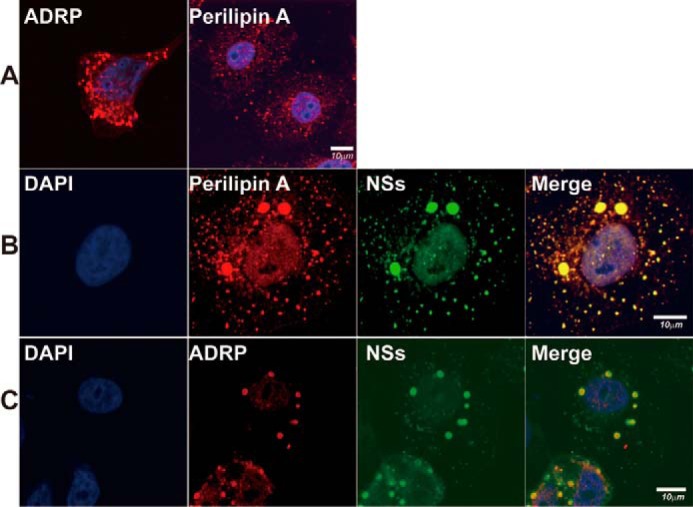
NSs co-localized with perilipin A and ADRP in IBs and lipid droplets. A, distribution of perilipin A and ADRP in lipid droplets. HepG2 cells were fixed and stained with anti-perilipin A and anti-ADRP antibodies to show their distribution in lipid droplets under a confocal microscope. B and C, HepG2 cells were infected with SFTSV, fixed at 24 h.p.i., and co-stained with anti-NSs and anti-ADRP (B) or anti-perilipin A (C) antibodies. After further staining with conjugated secondary antibodies, the cells were subjected to confocal fluorescence microscopy. We repeated the experiments at least twice, and representative data are presented for each panel.
Translocation of Synaptogyrin-2 by NSs into Lipid Droplets in Infected Cells
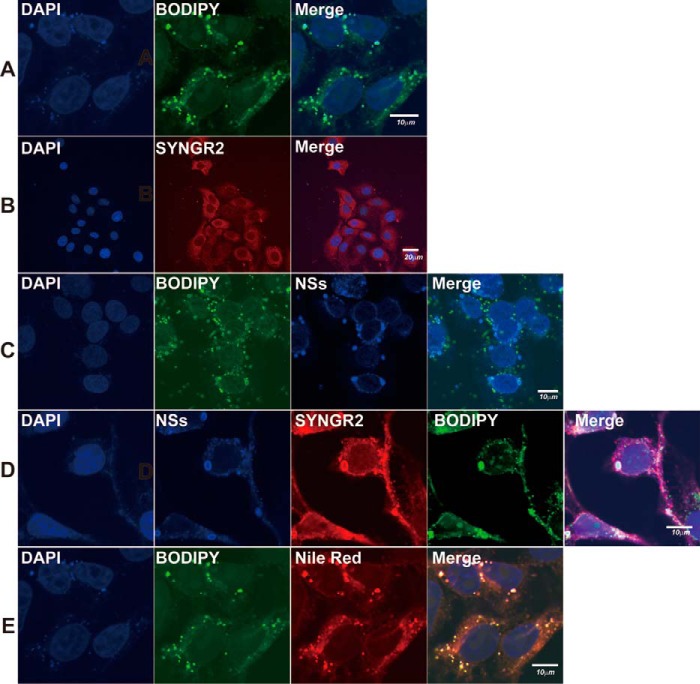
Presumably, synaptogyrin-2 was translocated into and present on lipid droplets, because it interacted with NSs and co-localized with NSs on lipid droplet-based IBs. HepG2 cells were stained with BODIPY 493/503, a lipophilic stain, and lipid droplets were shown scattered and evenly distributed in the cytoplasm (Fig. 6A), whereas synaptogyrin-2 was distributed dispersedly in the cytoplasm with anti-SYNGR2 antibody staining (Fig. 6B). In SFTSV-infected cells, NSs was shown in the IBs, which were positively stained with BODIPY 493/503 as lipid droplets (Fig. 6C). Co-staining of the infected cells with anti-NSs and anti-SYNGR2 antibodies, along with BODIPY 493/503, demonstrated that synaptogyrin-2 was translocated into lipid droplets, along with emergence of the large IB structures (Fig. 6D). Co-localization of synaptogyrin-2 and NSs in lipid droplets was also confirmed by staining the cells with Nile Red as shown in Fig. 6E.
FIGURE 6.
Translocation of synaptogyrin-2 by NSs into lipid droplets in infected cells. HepG2 cells were either mock infected (A, B, and E) or infected (C and D) with SFTSV at an MOI of 1 and incubated for 24 h before being fixed and permeabilized. A and B, the cells were stained with BODIPY 493/503 to show lipid droplets (A) or with anti-SYNGR2 antibodies to show distribution of synaptogyrin-2 (B) in non-infected cells. C, the infected cells were stained with both BODIPY 493/503 and anti-NSs antibodies, and co-localization of NSs with lipid droplets is shown. D, the infected cells were stained with anti-NSs and anti-SYNGR2 antibodies, and NSs and synaptogyrin-2 were co-localized. E, in non-infected HepG2 cells, BODIPY 493/503 and Nile Red co-stained lipid droplets. All the non-infected or infected cells, after staining with dyes and/or antibodies, were finally stained with DAPI for the nucleus, right before being subjected to confocal fluorescence microscopy. We repeated the experiments at least twice, and representative data are presented.
Decreased Viral Replication in Synaptogyrin-2 Silenced Cells Infected with SFTSV
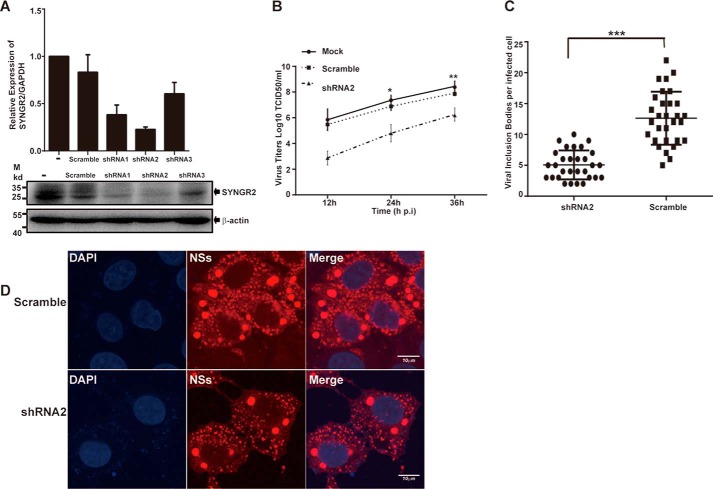
We hypothesized that synaptogyrin-2 was involved in the reconstruction of lipid droplets into IBs by interacting with NSs, suggesting that synaptogyrin-2 was also involved in SFTSV replication. We therefore explored the function of synaptogyrin-2 in viral replication by generating a synaptogyrin-2 knockdown cell line from HeLa cells through infection of a lentivirus expressing shRNA specifically targeting mRNA of synaptogyrin-2. We tested a panel of three lentiviruses (shRNA1, shRNA2, and shRNA3) for their silencing efficiency and found that shRNA2 was the most efficient to knockdown the synaptogyrin-2 mRNA (Fig. 7A) compared with the cells generated from a scramble shRNA control. Accordingly, the cell line stably expressing shRNA 2 was shown with reduced protein expression of synaptogyrin-2 (Fig. 7A).
FIGURE 7.
Decreased viral replication in synaptogyrin-2 knockdown cells infected with SFTSV. A, silence of synaptogyrin-2 gene expression in HeLa cells. Lentiviral vectors expressing synaptogyrin-2 specific shRNAs were used to transfect TLK293 cells to produce infectious recombinant lentiviruses. HeLa cells were infected with the lentiviruses, and shRNA-expressing cells were selected under puromycin. A real time RT-PCR for synaptogyrin-2 mRNA transcript levels was performed to show the efficacy of synaptogyrin-2 transcripts or proteins silenced by shRNA2. B, decreased viral replication in synaptogyrin-2 silenced cell culture. HeLa cells were infected with SFTSV virus at MOI of 1, the cultural media from the infected cells were sampled at 12, 24, 36 h.p.i., and infectious viral titers were titrated in Vero cells for TCID50. The titration was performed with duplicates, and the mean titers were exhibited. C and D, reduced number of NSs-formed IBs in synaptogyrin-2 silenced cells. HeLa cells, silenced by either the synaptogyrin-2 specific shRNA2 or a scramble shRNA, were infected with SFTSV. The cells were fixed and stained with anti-NSs at 24 h.p.i. The stained cells were subjected to confocal fluorescence microscopy (D), by which NSs-formed large IBs were counted (C). Circles (scramble) or squares (shRNA2) represent the number of viral IBs from randomly counted 30 cells infected with SFTSV (C). All values represent means ± S.D. from two independent experiments (Student's t test; *, p < 0.05; **, p < 0.01; ***, p < 0.001).
We infected the cells with silenced expression of synaptogyrin-2, caused by shRNA2 knockdown, with SFTSV at a multiplicity of infection (MOI) of 1. Cultural media were harvested at various time points of 12, 24, and 36 h.p.i. and subjected to infectious viral titration (50% tissue culture infective dose (TCID50)) in Vero cells. As shown in Fig. 7B, significantly lower titers were shown in the cells with silenced synaptogyrin-2, compared with those in normal or scramble shRNA-treated cells.
Synaptogyrin-2 Was Involved in Reconstruction of Lipid Droplets after Translocation with NSs
We further examined how synaptogyrin-2 plays a role as a promoter in viral replication and whether it was directly involved in the generation of the uniquely large IBs by NSs. We infected HeLa cell lines, stably expressing either shRNA2 or scramble shRNA, with SFTSV, and stained the cells with anti-NSs antibodies to show the IB structure. We counted and compared the numbers of the unique large IBs, only present in infection, in synaptogyrin-2 silenced cells or the scramble shRNA-treated controls. Our data showed that silencing of synaptogyrin-2 was associated with a significant decrease in number of the large IB (Fig. 7, C and D), which correlated with the reduced titers of the infectious virus in infection (Fig. 7B). These results demonstrate that the ability of NSs to form the uniquely large IBs is weakened in synaptogyrin-2-silenced cells, which could be one of the reasons for the subsequent decrease in viral replication.
Synaptogyrin-2 Enhances Viral Replication in Transfected Cells
To further confirm that synaptogyrin-2 is a promoter of viral replication, we transfected HeLa cells with plasmids overexpressing synaptogyrin-2, followed by infection with SFTSV at an MOI of 1. Culture media were collected at 12, 24, and 36 h.p.i. for infectious virus titration. The results showed a significant increase of viral titers in the cells with overexpressed synaptogyrin-2 compared with control cells (Fig. 8A). Ectopic expression of synaptogyrin-2 in transfected cells was shown in Fig. 8B.
FIGURE 8.
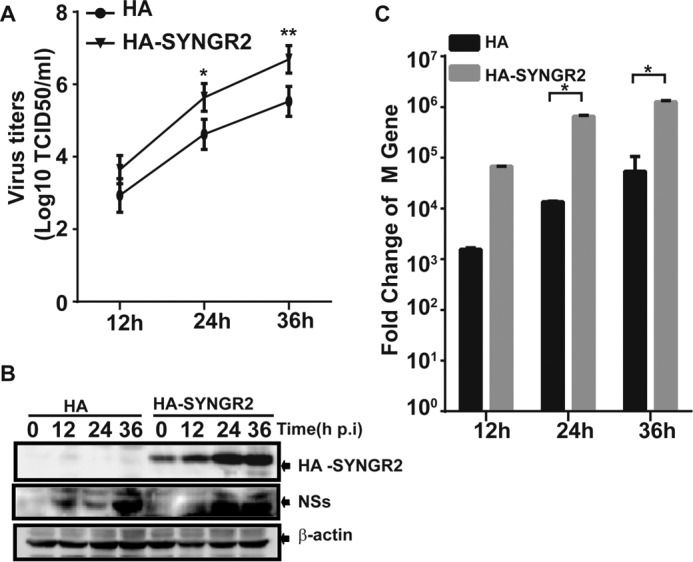
Synaptogyrin-2 enhanced viral replication in transfected cells. A, increased viral replication in synaptogyrin-2 overexpressing cells. HeLa cells were transfected with plasmids expressing synaptogyrin-2. After 24 h, the cells were infected with SFTSV. Cultural media were taken at various time points for infectious virus titration on Vero cells. HeLa cells were transfected with a blank plasmid as a control. Viral titers were presented as means ± S.D. of two independent assays (Student's t test; *, p < 0.05; **, p < 0.01.). B, ectopic expression of synaptogyrin-2 in transfected cells. Total lysates were prepared from the cells as described above for Western blot analyses with anti-HA or anti-NSs antibodies. C, increased viral gene copy numbers in synaptogyrin-2-overexpressing cells. Real time RT-PCR with specific primers to the viral M gene was performed to measure the M gene copy numbers in infected cells. Mean fold changes plus standard deviation of the viral M copy numbers were presented from three independent assays (Student's t test; *, p < 0.05).
We also examined the expression level of the viral M gene using real time RT-PCR. The transcripts of the M gene increased significantly following SFTSV infection in synaptogyrin-2-overexpressing cells compared with the controls (Fig. 8C). Collectively, these data demonstrate that viral replication in SFTSV-infected cells is enhanced with synaptogyrin-2 overexpression and that synaptogyrin-2 is a promoter of viral replication in SFTSV infection.
Discussion
Viral infections commonly lead to formation of IBs, which appear to be nuclear or cytoplasmic aggregates of viral proteins associated with cellular components. Studies have shown that some viruses make IBs functional as virus factories or viroplasm for viral replication and/or assembly (39–41). Our previous studies showed that SFTSV NSs may act as a scaffold protein to form IBs (21), a platform morphologically similar to viroplasm found in rotavirus-infected cells for supporting viral replication (31). The similar structures were also reported to be endosome-like (22), containing components including RIG-I (retinoic acid-inducible gene 1), the E3 ubiquitin ligase TRIM25, and Rab5 (RAS-related GTP-binding protein 5). Even though microtubule-associated protein LC3 (light chain 3) was also evidently co-localized with NSs in this structure, it was not a conventional autophogosome because Atg7 (autophagy-related gene 7) was irrelevant to the formation of the structure (22).
Viral IBs were reported for viral replication in other infections with paramyxoviruses such as human respiratory syncytial virus (hRSV) (42, 43), measles virus (44), and human metapneumovirus (45). In hRSV infection, viral NP, phosphoprotein, M2–1, polymerase L protein, and viral genomic RNAs were localized in IBs (42, 43, 46). We are currently examining whether the IBs formed by NSs in SFTSV infection are used for viral RNA replication because dsRNA and viral NP could be found in the cytoplasmic IBs in infected cells (23). Interestingly, IBs seem to be used by SFTSV for sequestering tank-binding kinase 1/IκB kinase ϵ/interferon regulatory factor 3 (TBK1/IKKϵ/IRF3) complex for evasion of host innate antiviral immunity (21), which has also been reported by other studies (22, 47). The IFN response could also be affected because of the interaction of NSs and STAT1/2, which sequesters STAT1/2 into the IBs (48). IBs could be used by SFTSV for both viral replication and immune evasion, whereas IBs formed in hRSV infection could have the same dual functions, in which MDA5 (melanoma differentiation-associated protein 5) and MAVS (mitochondrial antiviral-signaling protein) were sequestered into IBs (43).
Characterization of viral IBs has been performed with some viruses. IBs formed in cells infected with rotaviruses are based on lipid droplets (32, 50), subcellular organelles important for lipid storage and metabolism. Our previous study indicated that the IBs formed in SFTSV infection are built on the foundation of lipid droplets as well (23). In this study we confirm that the IBs were lipid droplet-based, which were positively stained not only by Nile Red but also by antibodies specific for ADRP or perilipin A (38), the cellular components present on lipid droplets (51–55). We also used BODIPY 493/503 (55), another lipophilic fluorescence dye, to positively stain the lipid droplet-based structures formed by NSs. Apparently lipid droplets were reconstructed with altered morphology, with their size increased and number decreased, after NSs was translocated onto them in infected cells. It has been unclear, however, how NSs reconstructs or transforms lipid droplets into the IBs, or viroplasm-like structures, for viral replication in infection. Indeed, viral proteins are vital in the initiation of the reconstruction in IBs. NP and P proteins were essential to forming IBs in hRSV infection, and the number of IBs was reduced, and size was increased through infection with an unknown mechanism (42, 46). Nonstructural NSP2 and NSP5 were sufficient enough to transform lipid droplets in rotavirus infection. However, little has been understood regarding how lipid droplets were reconstructed in the presence of NSP2 and NSP5 in rotavirus infection (32) or of NSs in SFTSV infection (23). No cellular proteins have been reported to be implicated in the process.
In this study, we demonstrate that synaptogyrin-2 is significantly up-regulated in response to SFTSV infection and interacts with NSs. The function of human synaptogyrin-2 is unknown, but studies of a similar rat protein suggest it may play a role in membrane trafficking as well. A recent study identified synaptogyrin-2 to be a potential lysosomal transporter because it was found in lysosomes and has multiple transmembrane domains (56).
What role might synaptogyrin-2 play in SFTSV infection? In this study, synaptogyrin-2 was identified through a high throughput screening of host proteins that are transcriptionally regulated in response to SFTSV infection. We further confirmed that synaptogyrin-2 was translocated onto viral IBs or restructured lipid droplets, apparently through its interaction with NSs. Our evidence, that support synaptogyrin-2 was present in the lipid droplet-based structures in infected cells, includes that the NSs-formed IBs or restructured lipid droplets were stained positively and unambiguously with an anti-SYNGR2 antibody (Figs. 3, 4 and 6). In addition, these structures were positively recognized by anti-NSs, anti-ADPR, and perilipin A staining or by Nile Red or BODIPY 493/503 staining.
Our data also indicated that synaptogyrin-2 appeared to be a promoter and played an important role in viral replication. We postulated that synaptogyrin-2 might function in targeting and reconstructing lipid droplets by NSs in several mechanisms. First, NSs might target lipid droplets through its interaction with synaptogyrin-2, which seems unlikely because synaptogyrin-2 is dispersedly localized in the cytoplasm by itself. Second, we highly suspect that synaptogyrin-2, containing four transmembrane domains, could be present in smaller vesicles in the cytosol. It is possible that NSs interacts with synaptogyrin-2 on vesicles, and the vesicles could subsequently fuse to lipid droplets. NSs can interact with each other, as shown in our previous study (23), which could allow synaptogyrin-2 to aggregate and its associated vesicles to fuse with each other or with lipid droplets to form large IBs. Third, NSs could carry synaptogyrin-2 as single molecule to lipid droplets through their interaction, where synaptogyrin-2 may initiate reconstruction of lipid droplets because of its nature as a vesicle membrane protein.
Although studies on synaptogyrin-2 have been limited and we know little about its function, a few lines of evidence may support the above-mentioned postulations. Members of the synaptogyrin family, also called vesicle-associated membrane proteins, such as synaptogyrin-1, are abundantly present in synaptic vesicles and are essential to presynaptic vesicle biogenesis, vesicle trafficking, and neuronal transmission (2, 57). A homolog of synaptogyrin-2 has also shown a similar roles in Caenorhabditis elegans (58). A study in Drosophila revealed that a synaptogyrin-1 homolog is involved in size control and biogenesis of synaptic vesicles as shown in knock-out mutant mice (59). Unlike other members of the family, human synaptogyrin-2 is not distributed in neurons but is rich in all other tissues (5) and has been identified as a lysosomal transporter protein in lysosomes (56), suggesting its existence in cytoplasmic vesicles. Its importance as a lysosomal transporter protein indicates its ability to fuse with endosomes or other vesicles to form phagosomes. In addition, a recent study shows that overexpression of synaptogyrin-2 could induce the formation of synaptic vesicles-like microvesicles in neuroendocrine cells (49). Aggregation of synaptogyrin-2 through interaction with NSs could trigger vesicle formation or reconstruction of existing vesicles, including membrane rearrangement or fusion in lipid droplets upon NSs. If it were eventually proved that synaptogyrin-2 is involved in driving reconstruction of lipid droplets, it would be essential to make IBs functional for both viral replication and immune evasion. Significant induction of synaptogyrin-2 in SFTSV infection apparently benefits the virus for making lipid droplets be reshaped as unique IBs or viroplasm-like structures.
In summary, we report a function of synaptogyrin-2, a member of the synaptogyrin family essential to presynaptic vesicle biogenesis and vesicle trafficking, in promoting a newly identified bunyavirus replication. Synaptogyrin-2 promotes viral replication through its interaction with viral NSs, which results in reconstructing the lipid droplet-based IB for becoming a functional viroplasm-like structure or virus factory. The role of synaptogyrin-2 in viral replication is unique, with a mechanism that may be important in many viral infections in which lipid droplets are involved.
Experimental Procedures
Cells, Viruses, and Reagents
HeLa cells, African green monkey kidney Vero cells, human embryonic kidney HEK293 cells, and human liver hepatocellular carcinoma cells HepG2, all from ATCC, were grown in Dulbecco's modified Eagle's medium (Gibco, Invitrogen) supplemented with 10% fetal bovine serum (Gibco), 1 mm sodium pyruvate (HyClone), and 1% antibiotic-antimycotic solution (Gibco). The cells were cultured at 37 °C with 5% CO2. A previously described SFTSV strain, JS-2010-014, was used in this study and propagated in cell culture as described (33). All viral aliquots were stored at −80 °C. The cDNA sequence of synaptogyrin-2 (SYNGR2) has been deposited at the GenBankTM at the National Center for Biotechnology Information; the accession number is AJ002308.1.
Antibodies for anti-SYNGR2 antibody, anti-ADRP antibody, anti-perilipin A, and secondary antibodies with fluorescence-labeled Alex Fluor 647 donkey anti-mouse and Alex Fluor 594 goat anti-rabbit IgG were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Other secondary antibodies used for immunoblots, confocal microscopy, and immunoprecipitation were obtained from Cell Signaling Technology (Danvers, MA) or Abcam (Cambridge, MA). Anti-FLAG M2 antibody, Nile Red, and DAPI were obtained from Sigma-Aldrich. Anti-β-actin antibody and anti-HA antibody were purchased from Santa Cruz Biotechnology. Rabbit anti-NSs antibody was obtained by immunizing rabbits with purified recombinant NSs protein of SFTSV. Protein G Plus/protein A-agarose beads were purchased from Millipore. PrimeScript (R036A) was obtained from Takara (Shiga, Japan) and used as reverse transcriptase for reverse transcription. TRIzol reagent and Lipofectamine 2000 reagents were purchased from Invitrogen.
Construction of Plasmids
Synaptogyrin-2/SYNGR2 cDNA was amplified by PCR procedures from the reverse transcription product with indicated primers and then subcloned into a plasmid pRK5 with either a FLAG or HA tag for mammalian expression under a cytomegalovirus promoter. 5′ (EcoRI) and 3′ (BamHI) primers were used for HA-tagged pRK5 plasmid construction, whereas 5′ (BamHI) and 3′ (HindIII) primers were used for FLAG-tagged pRK5 plasmid construction. The primer sequences are as follows: 5′ primer EcoRI, 5′-CCGGAATTCATGGAGAGCGGGGCCT; 5′ primer BamHI, 5′-CGCGGATCCGCGATGGAGAGCGGGGCCT; 3′ primer BamHI, 5′-CGCGGATCCTCAGTACACAGGGGGC; and 3′ primer HindIII; 5′-CCCAAGCTTGGGTCAGTACACAGGGGGC. SYNGR2 cDNA was also subcloned into pDsRed-N1 for expression of DsRed-SYNGR2 fusion proteins. Plasmids pRK5-NSs and pEGFP-N3-NSs were described previously (21). HeLa or HEK293T cells were transfected with plasmids containing cDNA encoding and expressing synaptogyrin-2 or NSs with Lipofectamine 2000 reagents as instructed by the manufacturer.
shRNA Knockdown of SYNGR2 in HeLa Cells
Lentiviral vectors expressing shRNAs, specifically targeting synaptogyrin-2 gene, were used to obtain SYNGR2 knockdown cell lines. We chose three shRNAs targeting distinct sites in mRNA of human synaptogyrin-2 gene; the constructs were obtained from Open Systems via Thermo Fisher Scientific. To obtain infectious lentiviruses, we transfected the TLK293 packaging cells (Thermo) with pGIPZ-shRNA containing shRNA sequences, either targeting synaptogyrin-2 (knockdown) or scramble (control) RNA. Arrest-In transfection reagent (Thermo) was used following the manufacturer's procedure. After incubation for 48 h at 37 °C, the cultural medium was harvested and used to infect HeLa cells in 6-well plates. The medium was replaced 48 h later with fresh one containing 1.5 μg/ml of puromycin for selection of shRNA-expressing cells. The selected cells, with expression of shRNAs either specific for synaptogyrin-2 or against control scramble RNA, were tested for viability, which showed no difference between them before SFTSV infection. The HeLa cells were monitored under a Nikon fluorescence microscope daily until almost all the cells turned positive (green), which resulted from lentivirus expression of TurboGFP in the construct. The selected cells were lysed, and the lysates were subjected to SDS-PAGE and Western blot analyses with an anti-SYNGR2 antibody to measure knockdown efficiency. Meantime, total RNA extracted from selected cells with an RNeasy kit (Qiagen) was used for reverse transcription using a PrimeScript reagent kit (TAKARA, Shiga, Japan) to test knockdown efficiency. The cells with the highest knockdown efficiency were propagated and maintained in 1 μg/ml of puromycin for subsequent studies.
Immunoprecipitation and Western Blot Analysis
Cell lysates of transfected or co-transfected cells were incubated with specific or control antibodies at 4 °C for 2 h and then precipitated with protein A/G beads. After 2 h of incubation, the beads were washed four times with lysis buffer containing 25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 5% glycerol, 1 mm DTT, 1 mm PMSF, 2 mm NaF, 1 mm Na3VO4, and 1 μg of aprotinin/ml. The immunoprecipitates were subjected to SDS-PAGE and then transferred onto an Immunoblot PVDF (Millipore, Billerica, MA) membrane for primary antibody incubation overnight. Alkaline phosphatase- or HRP-conjugated secondary antibodies were used for further incubation with the membranes for 90 min, and signals on blots were developed by 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium or ECL reagents (Millipore, Billerica, MA).
To prepare soluble and insoluble fractions of cell lysates, HEK293T cells were cultured in 6-well plates for 14 h followed by transfection with 2 μg of pRK5-F-SYNGR2 and/or pRK5-F-NSs using Lipofectamine 2000 reagents for the expression of SYNGR2 and NSs proteins. 24–32 h post-transfection, the cells were rinsed with PBS and harvested by gentle scraping into 500 μl of ice-cold 1× Nonidet P-40 lysis buffer containing 1 mm DTT, 1 mm PMSF, 2 mm NaF, 1 mm Na3VO4, and 1 μg of aprotinin/ml. Cell lysates were incubated at −80 °C for 15 min, followed by centrifugation (19,000 × g for 10 min) at 4 °C for separation into two fractions: Nonidet P-40-soluble (supernatant) and Nonidet P-40-insoluble (pellet). The pellet was resuspended in a buffer containing 1% SDS and 10 mm Tris-EDTA (pH 7.5). Proteins from both fractions were quantified using the BCA protein assay kit (BCA kit; Pierce) before loading for SDS-PAGE analysis.
Quantitative Real Time PCR
200 ng of total RNA prepared from infected or non-infected HepG2 cells was used for reverse transcription with a Primescript reagent kit (Takara). Quantitative real time PCR was performed with 1 μl of cDNA in a total volume of 10 μl with SYBR Premix Ex TaqII (Takara) following the manufacturer's instructions. Relative expression values were standardized by an internal GAPDH control. Fold change of the M segment was calculated following the formula:2(ΔCt of gene-ΔCt of GAPDH) as described previously (34). The reactions were implemented in duplicate and repeated three times for each sample, and the mean values and standard deviations were calculated.
Immunofluorescence Assay and Virus Titration
HeLa cells, transfected with plasmids encoding synaptogyrin-2, NSs mutants, and NSs or infected with SFTSV were washed twice with PBS, fixed with 4% paraformaldehyde for 30 min, and permeabilized with 0.1% Triton X-100 for 10 min, followed by three washes with PBS. The coverslips were blocked with 5% bovine serum albumin (Sigma) in PBS for 30 min at 37 °C and then incubated with anti-NSs antibody at 1:50 to 1:100 dilution at 4 °C overnight. After three washes with PBS-Tween 20, the cells were incubated with a 1:200 dilution of Alex Fluor 488 donkey anti-rabbit and Alex Fluor 594 goat anti-mouse IgG (H+L) at 37 °C for 1 h, washed, and stained for 10 min with DAPI (1 μg/ml). To make sure there was no nonspecific fluorescence staining, we used purified IgG from normal rabbits or mouse ascites for staining controls. We also used mouse IgG2a-FITC or IgG2a-Tricolor for staining as antibody isotype controls.
For lipid droplets staining, unfixed HeLa cells infected with SFTSV were stained at 4–8 h.p.i. with Nile Red (35) (100 ng/ml) for 4 h at 4 °C and then fixed with 2% paraformaldehyde in PBS containing 10 mm CaCl2 for 1 h at 4 °C. Alternatively, HeLa cells were fixed with 3% paraformaldehyde for 30 min at room temperature and then incubated for 30 min with BODIPY 493/503 (1 μg/ml, diluted from 1 mg/ml stock in dimethyl sulfoxide; Molecular Probes) (27). Coverslips were then washed five times with PBS (10 min/wash) and counterstained with anti-NSs or anti-SYNGR2 antibodies for 1 h, followed by incubation with secondary antibody conjugated with Alexa Fluor 488 or 647. After washing, the coverslips were subjected to microscopy under an Olympus confocal microscope.
Cell culture medium was collected at various time points from SFTSV-infected Vero cells and titrated for infectious virus titration (TCID50) with indirect immunofluorescence as previously described (23, 33). Infectious virus titers (TCID50/ml) were calculated based on the Reed and Muench method (36).
Microarray Analysis
Total RNAs prepared at each time point from infected HepG2 cells was pooled in equivalent amounts and subjected to microarray analyses. For microarray experiments, cyanine 3-CTP-labeled cRNA probes were generated from 1 μg of total RNA with Quick Amp Labeling (Agilent) following Agilent's protocol for one-color microarray-based gene expression analysis. Human 4 × 44K slides (Agilent) were used for hybridization and scanning with an Agilent scanner (G2565BA). Each microarray experiment was performed with technical duplicates for infected or uninfected control samples. Changes in the levels of mRNA of any gene were marked significant only when the following two criteria were met: (i) the alteration in expression was statistically significant (p value for paired Student's t test of 0.05), and (ii) the change was at least 50% (equivalent to a 1.5-fold change where the value for no change is 0) above or below the baseline expression level. The baseline was calculated as the expression level of the 0 h (uninfected control) for a particular gene. Gene transcription data, which have been sent to the GenBankTM for deposition, were further analyzed with GeneData Expressionist for differential expression and heat map construction.
Size Measurement and Counting of the IBs and Statistical Analysis
The sizes of IBs were measured in diameter, and the IBs were counted with the ImageJ software (National Institutes of Health). Graphic presentation was prepared with GraphPad Prism 5 software (GraphPad Software). For statistical analysis, a two-tailed Student's t test was used to evaluate the data by either the SPSS software (IBM SPSS) or GraphPad Prism 5. An χ2 analysis was used to determine significant differences of the data in two or more groups, and a p value <0.05 or less was considered statistically significant.
Author Contributions
Q. S. and Z. X. conceived and coordinated the study. Q. S., X. Q., and X. W. designed, performed, and analyzed the experiments shown in Figs. 1–8. C. L., M. L., C. J. C., and D. L. provided reagents and technical assistance and contributed to completion of the studies. Q. S., C. J. C., and Z. X. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgment
We thank Sandy Shanks for dedicated and excellent work in editing the manuscript.
This work was supported by National Natural Science Foundation of China Grant 81571993, Grant 2014ZX10004001-002 from the Mega Infectious Diseases Program from the Ministry of Science and Technology of China (to ZX), 973 Grant (2011CB504705 from the Ministry of Science and Technology of China (to M. L. and D. L.), and Grant BK20131450 from the Natural Science Foundation of Jiangsu Province (to X. Q.). The authors declare that they have no conflicts of interest with the contents of this article.
- SYNGR2
- synaptogyrin-2
- SFTS
- severe fever with thrombocytopenia syndrome
- SFTSV
- SFTS bunyavirus
- IB
- inclusion body
- ADRP
- adipose differentiation-related protein
- h.p.i.
- hours postinfection
- MOI
- multiplicity of infection
- hRSV
- human respiratory syncytial virus
- BODIPY 493/503
- 4,4-difluoro-1,3,5,7,8 pentamethyl-4-bora 3a, 4a-diaza-s-indacene.
References
- 1. Janz R., and Südhof T. C. (1998) Cellugyrin, a novel ubiquitous form of synaptogyrin that is phosphorylated by pp60c-src. J. Biol. Chem. 273, 2851–2857 [DOI] [PubMed] [Google Scholar]
- 2. Belfort G. M., and Kandror K. V. (2003) Cellugyrin and synaptogyrin facilitate targeting of synaptophysin to a ubiquitous synaptic vesicle-sized compartment in PC12 cells. J. Biol. Chem. 278, 47971–47978 [DOI] [PubMed] [Google Scholar]
- 3. Hübner K., Windoffer R., Hutter H., and Leube R. E. (2002) Tetraspan vesicle membrane proteins: synthesis, subcellular localization, and functional properties. Int. Rev. Cytol. 214, 103–159 [DOI] [PubMed] [Google Scholar]
- 4. Valtorta F., Tarelli F. T., Campanati L., Villa A., and Greengard P. (1989) Synaptophysin and synapsin I as tools for the study of the exo-endocytotic cycle. Cell Biol. Int. Rep. 13, 1023–1038 [DOI] [PubMed] [Google Scholar]
- 5. Kedra D., Pan H. Q., Seroussi E., Fransson I., Guilbaud C., Collins J. E., Dunham I., Blennow E., Roe B. A., Piehl F., and Dumanski J. P. (1998) Characterization of the human synaptogyrin gene family. Hum. Genet. 103, 131–141 [DOI] [PubMed] [Google Scholar]
- 6. Elliott R. M., and Brennan B. (2014) Emerging phleboviruses. Curr. Opin. Virol. 5, 50–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yu X. J., Liang M. F., Zhang S. Y., Liu Y., Li J. D., Sun Y. L., Zhang L., Zhang Q. F., Popov V. L., Li C., Qu J., Li Q., Zhang Y. P., Hai R., Wu W., Wang Q., et al. (2011) Fever with thrombocytopenia associated with a novel bunyavirus in China. N. Engl. J. Med. 364, 1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu Y., Li Q., Hu W., Wu J., Wang Y., Mei L., Walker D. H., Ren J., Wang Y., and Yu X. J. (2012) Person-to-person transmission of severe fever with thrombocytopenia syndrome virus. Vector Borne Zoonotic Dis. 12, 156–160 [DOI] [PubMed] [Google Scholar]
- 9. Cui F., Cao H. X., Wang L., Zhang S. F., Ding S. J., Yu X. J., and Yu H. (2013) Clinical and epidemiological study on severe fever with thrombocytopenia syndrome in Yiyuan County, Shandong Province, China. Am. J. Trop. Med. Hyg. 88, 510–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gai Z. T., Zhang Y., Liang M. F., Jin C., Zhang S., Zhu C. B., Li C., Li X. Y., Zhang Q. F., Bian P. F., Zhang L. H., Wang B., Zhou N., Liu J. X., Song X. G., et al. (2012) Clinical progress and risk factors for death in severe fever with thrombocytopenia syndrome patients. J. Infect. Dis. 206, 1095–1102 [DOI] [PubMed] [Google Scholar]
- 11. Sun Y., Jin C., Zhan F., Wang X., Liang M., Zhang Q., Ding S., Guan X., Huo X., Li C., Qu J., Wang Q., Zhang S., Zhang Y., Wang S., et al. (2012) Host cytokine storm is associated with disease severity of severe fever with thrombocytopenia syndrome. J. Infect. Dis. 206, 1085–1094 [DOI] [PubMed] [Google Scholar]
- 12. Takahashi T., Maeda K., Suzuki T., Ishido A., Shigeoka T., Tominaga T., Kamei T., Honda M., Ninomiya D., Sakai T., Senba T., Kaneyuki S., Sakaguchi S., Satoh A., Hosokawa T., et al. (2014) The first identification and retrospective study of severe fever with thrombocytopenia syndrome in Japan. J. Infect. Dis. 209, 816–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim W. Y., Choi W., Park S. W., Wang E. B., Lee W. J., Jee Y., Lim K. S., Lee H. J., Kim S. M., Lee S. O., Choi S. H., Kim Y. S., Woo J. H., and Kim S. H. (2015) Nosocomial transmission of severe fever with thrombocytopenia syndrome in Korea. Clin. Infect. Dis. 60, 1681–1683 [DOI] [PubMed] [Google Scholar]
- 14. Park S. W., Song B. G., Shin E. H., Yun S. M., Han M. G., Park M. Y., Park C., and Ryou J. (2014) Prevalence of severe fever with thrombocytopenia syndrome virus in Haemaphysalis longicornis ticks in South Korea. Ticks Tick Borne Dis. 5, 975–977 [DOI] [PubMed] [Google Scholar]
- 15. Shimojima M., Fukushi S., Tani H., Yoshikawa T., Morikawa S., and Saijo M. (2013) [Severe fever with thrombocytopenia syndrome in Japan]. Uirusu 63, 7–12 [DOI] [PubMed] [Google Scholar]
- 16. Savage H. M., Godsey M. S. Jr., Lambert A., Panella N. A., Burkhalter K. L., Harmon J. R., Lash R. R., Ashley D. C., and Nicholson W. L. (2013) First detection of heartland virus (Bunyaviridae: Phlebovirus) from field collected arthropods. Am. J. Trop. Med. Hyg. 89, 445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McMullan L. K., Folk S. M., Kelly A. J., MacNeil A., Goldsmith C. S., Metcalfe M. G., Batten B. C., Albariño C. G., Zaki S. R., Rollin P. E., Nicholson W. L., and Nichol S. T. (2012) A new phlebovirus associated with severe febrile illness in Missouri. N. Engl. J. Med. 367, 834–841 [DOI] [PubMed] [Google Scholar]
- 18. Stubbs A. M., and Steele M. T. (2014) Heartland virus disease: United States, 2012–2013. Ann. Emerg. Med. 64, 314. [PubMed] [Google Scholar]
- 19. Niu G., Li J., Liang M., Jiang X., Jiang M., Yin H., Wang Z., Li C., Zhang Q., Jin C., Wang X., Ding S., Xing Z., Wang S., Bi Z., and Li D. (2013) Severe fever with thrombocytopenia syndrome virus among domesticated animals, China. Emerg. Infect. Dis. 19, 756–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li D. (2013) A highly pathogenic new bunyavirus emerged in China. Emerg. Microbes Infect. 2, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu X., Qi X., Qu B., Zhang Z., Liang M., Li C., Cardona C. J., Li D., and Xing Z. (2014) Evasion of antiviral immunity through sequestering of TBK1/IKKϵ/IRF3 into viral inclusion bodies. J. Virol. 88, 3067–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Santiago F. W., Covaleda L. M., Sanchez-Aparicio M. T., Silvas J. A., Diaz-Vizarreta A. C., Patel J. R., Popov V., Yu X. J., García-Sastre A., and Aguilar P. V. (2014) Hijacking of RIG-I signaling proteins into virus-induced cytoplasmic structures correlates with the inhibition of type I interferon responses. J. Virol. 88, 4572–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu X., Qi X., Liang M., Li C., Cardona C. J., Li D., and Xing Z. (2014) Roles of viroplasm-like structures formed by nonstructural protein NSs in infection with severe fever with thrombocytopenia syndrome virus. Faseb J. 28, 2504–2516 [DOI] [PubMed] [Google Scholar]
- 24. Brown D. A. (2001) Lipid droplets: proteins floating on a pool of fat. Curr. Biol. 11, R446–R449 [DOI] [PubMed] [Google Scholar]
- 25. Kondrup J. (1984) In vitro effect of ethanol on the synthesis of triacylglycerol in hepatic cytoplasmic lipid droplets. Dan. Med. Bull. 31, 109–120 [PubMed] [Google Scholar]
- 26. Londos C., Brasaemle D. L., Gruia-Gray J., Servetnick D. A., Schultz C. J., Levin D. M., and Kimmel A. R. (1995) Perilipin: unique proteins associated with intracellular neutral lipid droplets in adipocytes and steroidogenic cells. Biochem. Soc. Trans. 23, 611–615 [DOI] [PubMed] [Google Scholar]
- 27. Marcinkiewicz A., Gauthier D., Garcia A., and Brasaemle D. L. (2006) The phosphorylation of serine 492 of perilipin a directs lipid droplet fragmentation and dispersion. J. Biol. Chem. 281, 11901–11909 [DOI] [PubMed] [Google Scholar]
- 28. Londos C., Brasaemle D. L., Schultz C. J., Segrest J. P., and Kimmel A. R. (1999) Perilipins, ADRP, and other proteins that associate with intracellular neutral lipid droplets in animal cells. Semin. Cell Dev. Biol. 10, 51–58 [DOI] [PubMed] [Google Scholar]
- 29. Wolins N. E., Rubin B., and Brasaemle D. L. (2001) TIP47 associates with lipid droplets. J. Biol. Chem. 276, 5101–5108 [DOI] [PubMed] [Google Scholar]
- 30. Ploen D., Hafirassou M. L., Himmelsbach K., Schille S. A., Biniossek M. L., Baumert T. F., Schuster C., and Hildt E. (2013) TIP47 is associated with the Hepatitis C virus and its interaction with. Rab9 is required for release of viral particles. Eur. J. Cell Biol. 92, 374–382 [DOI] [PubMed] [Google Scholar]
- 31. Fabbretti E., Afrikanova I., Vascotto F., and Burrone O. R. (1999) Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J. Gen. Virol. 80, 333–339 [DOI] [PubMed] [Google Scholar]
- 32. Cheung W., Gill M., Esposito A., Kaminski C. F., Courousse N., Chwetzoff S., Trugnan G., Keshavan N., Lever A., and Desselberger U. (2010) Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J. Virol. 84, 6782–6798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qu B., Qi X., Wu X., Liang M., Li C., Cardona C. J., Xu W., Tang F., Li Z., Wu B., Powell K., Wegner M., Li D., and Xing Z. (2012) Suppression of the interferon and NF-κB responses by severe fever with thrombocytopenia syndrome virus. J. Virol. 86, 8388–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gao W., Sun W., Qu B., Cardona C. J., Powell K., Wegner M., Shi Y., and Xing Z. (2012) Distinct regulation of host responses by ERK and JNK MAP kinases in swine macrophages infected with pandemic (H1N1) 2009 influenza virus. PLoS One 7, e30328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Greenspan P., Mayer E. P., and Fowler S. D. (1985) Nile red: a selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 100, 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matumoto M. (1949) A note on some points of calculation method of LD50 by Reed and Muench. Jpn J. Exp. Med. 20, 175–179 [PubMed] [Google Scholar]
- 37. Sun Q., Jin C., Zhu L., Liang M., Li C., Cardona C. J., Li D., and Xing Z. (2015) Host responses and regulation by NFκB signaling in the liver and liver epithelial cells infected with a novel tick-borne bunyavirus. Sci. Rep. 5, 11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Walther T. C., and Farese R. V. Jr. (2012) Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81, 687–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Netherton C., Moffat K., Brooks E., and Wileman T. (2007) A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv. Virus Res. 70, 101–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Netherton C. L., and Wileman T. (2011) Virus factories, double membrane vesicles and viroplasm generated in animal cells. Curr. Opin. Virol. 1, 381–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Novoa R. R., Calderita G., Arranz R., Fontana J., Granzow H., and Risco C. (2005) Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell 97, 147–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. García J., García-Barreno B., Vivo A., and Melero J. A. (1993) Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology 195, 243–247 [DOI] [PubMed] [Google Scholar]
- 43. Lifland A. W., Jung J., Alonas E., Zurla C., Crowe J. E. Jr., and Santangelo P. J. (2012) Human respiratory syncytial virus nucleoprotein and inclusion bodies antagonize the innate immune response mediated by MDA5 and MAVS. J. Virol. 86, 8245–8258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Spehner D., Kirn A., and Drillien R. (1991) Assembly of nucleocapsidlike structures in animal cells infected with a vaccinia virus recombinant encoding the measles virus nucleoprotein. J. Virol. 65, 6296–6300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Derdowski A., Peters T. R., Glover N., Qian R., Utley T. J., Burnett A., Williams J. V., Spearman P., and Crowe J. E. Jr. (2008) Human metapneumovirus nucleoprotein and phosphoprotein interact and provide the minimal requirements for inclusion body formation. J. Gen. Virol. 89, 2698–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Carromeu C., Simabuco F. M., Tamura R. E., Farinha Arcieri L. E., and Ventura A. M. (2007) Intracellular localization of human respiratory syncytial virus L protein. Arch. Virol. 152, 2259–2263 [DOI] [PubMed] [Google Scholar]
- 47. Ning Y. J., Wang M., Deng M., Shen S., Liu W., Cao W. C., Deng F., Wang Y. Y., Hu Z., and Wang H. (2014) Viral suppression of innate immunity via spatial isolation of TBK1/IKKϵ from mitochondrial antiviral platform. J. Mol. Cell. Biol. 6, 324–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ning Y. J., Feng K., Min Y. Q., Cao W. C., Wang M., Deng F., Hu Z., and Wang H. (2015) Disruption of type I interferon signaling by the nonstructural protein of severe fever with thrombocytopenia syndrome virus via the hijacking of STAT2 and STAT1 into inclusion bodies. J. Virol. 89, 4227–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Belfort G. M., Bakirtzi K., and Kandror K. V. (2005) Cellugyrin induces biogenesis of synaptic-like microvesicles in PC12 cells. J. Biol. Chem. 280, 7262–7272 [DOI] [PubMed] [Google Scholar]
- 50. Khor V. K., Shen W. J., and Kraemer F. B. (2013) Lipid droplet metabolism. Curr. Opin. Clin. Nutr. Metab. Care 16, 632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sztalryd C., and Kimmel A. R. (2014) Perilipins: lipid droplet coat proteins adapted for tissue-specific energy storage and utilization, and lipid cytoprotection. Biochimie 96, 96–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Storey S. M., McIntosh A. L., Senthivinayagam S., Moon K. C., and Atshaves B. P. (2011) The phospholipid monolayer associated with perilipin-enriched lipid droplets is a highly organized rigid membrane structure. Am. J. Physiol. Endocrinol. Metab. 301, E991–E1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McIntosh A. L., Storey S. M., and Atshaves B. P. (2010) Intracellular lipid droplets contain dynamic pools of sphingomyelin: ADRP binds phospholipids with high affinity. Lipids 45, 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Listenberger L. L., Ostermeyer-Fay A. G., Goldberg E. B., Brown W. J., and Brown D. A. (2007) Adipocyte differentiation-related protein reduces the lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover. J. Lipid Res. 48, 2751–2761 [DOI] [PubMed] [Google Scholar]
- 55. Listenberger L. L., and Brown D. A. (2007) Fluorescent detection of lipid droplets and associated proteins. Curr. Protoc. Cell Biol. Chapter 24, Unit 24.2 [DOI] [PubMed] [Google Scholar]
- 56. Chapel A., Kieffer-Jaquinod S., Sagné C., Verdon Q., Ivaldi C., Mellal M., Thirion J., Jadot M., Bruley C., Garin J., Gasnier B., and Journet A. (2013) An extended proteome map of the lysosomal membrane reveals novel potential transporters. Mol. Cell. Proteomics 12, 1572–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bragina L., Giovedì S., Barbaresi P., Benfenati F., and Conti F. (2010) Heterogeneity of glutamatergic and GABAergic release machinery in cerebral cortex: analysis of synaptogyrin, vesicle-associated membrane protein, and syntaxin. Neuroscience 165, 934–943 [DOI] [PubMed] [Google Scholar]
- 58. Abraham C., Bai L., and Leube R. E. (2011) Synaptogyrin-dependent modulation of synaptic neurotransmission in Caenorhabditis elegans. Neuroscience 190, 75–88 [DOI] [PubMed] [Google Scholar]
- 59. Stevens R. J., Akbergenova Y., Jorquera R. A., and Littleton J. T. (2012) Abnormal synaptic vesicle biogenesis in Drosophila synaptogyrin mutants. J Neurosci 32, 18054–18067, 18067a [DOI] [PMC free article] [PubMed] [Google Scholar]