Abstract
Elmo2, a member of the Elmo protein family, has been implicated in the regulation of Rac1 and Akt activation. Recently, we found that Elmo2 specifically interacts with ClipR-59. Because Akt and Rac1 have been implicated in insulin dependent Glut4 membrane translocation, we hypothesize here that Elmo2 may play a role in insulin-dependent Glut4 membrane translocation. Accordingly, we found that overexpression of Elmo2 enhanced, whereas its knockdown suppressed, insulin-dependent Glut4 membrane translocation in both 3T3-L1 adipocytes and L6 skeletal muscle cells. We also examined whether Elmo2 contributes to the insulin-mediated activation of Rac1 and Akt. We found that Elmo2 is required for insulin-induced Rac1 GTP loading, but not AKT activation, in L6 cells induced by insulin. Instead, Elmo2 is required to promote the insulin-induced membrane association of Akt. Together, our studies demonstrate that Elmo2 is a new regulator of insulin-dependent Glut4 membrane translocation through modulating Rac1 activity and Akt membrane compartmentalization.
Keywords: adipocyte, Akt PKB, glucose transporter type 4 (GLUT4), Rac (Rac GTPase), trafficking
Introduction
Elmo (engulfment and cell motility) is a protein family that consists of three members, Elmo1, Elmo2, and Elmo3. Elmo proteins are characterized by the presence of a Ras GTPase-binding domain, a region that is present only in Elmo proteins and ElmoD protein (known as the ELMO domain), a pleckstrin homology (PH)3 domain, and a proline-rich region (1, 2). In addition, two regions in Elmo proteins were shown to mediate a closed and inactive conformation state, the Elmo-inhibitory domain and the Elmo autoregulation domain (3). The exact function of this autoregulation remains to be fully established.
Elmo proteins have no apparent enzymatic activity (4) and have been found to act as scaffolds for Dock (dedicator of cytokinesis) guanine nucleotide exchange factors (2). In addition, Elmo proteins have been found to interact with a panel of proteins, including BAI1–3 (5, 6), Gαi2 (7), RhoG (8) and Gβγ (9). A common feature of these proteins is that they all are associated with signaling at the plasma membrane. Through these protein interactions, Elmo proteins are ideal candidates to coordinate the activation of downstream effectors at the plasma membrane. For instance, the interaction of Elmo1/2 with BAI (brain-specific angiogenesis inhibitor) recruits them onto the proximal membrane to activate the small GTPase Rac1 through the Dock180 guanine nucleotide exchange factor to mediate engulfment of apoptotic cells or to promote muscle differentiation (5, 6). In addition, Elmo2 has been implicated in Akt activation, probably through the interaction with small GTPases, such as RhoG (9–11).
Glut4 (glucose transporter 4), which is predominantly expressed in adipose and muscle tissues, is a member of a facilitative glucose transporter family that consists of 12 members (12). Unlike other glucose transporters that constitutively reside on the plasma membrane, Glut4 is stored in intracellular compartments in the form of small membrane vesicles, termed Glut4 vesicles, at basal state and is only redistributed at the membrane upon insulin stimulation (13, 14). As an insulin-regulated glucose transporter, Glut4 plays a central role in the control of whole body glucose homeostasis and peripheral insulin sensitivity. In mice and humans, impaired Glut4 function results in hyperglycemia and insulin resistance, which are hallmarks of type II diabetes (15–17).
Insulin-dependent Glut4 membrane translocation depends on Akt membrane compartmentalization in adipocytes and on both Akt membrane compartmentalization and Rac1 activation in muscle cells (18). A protein that regulates insulin-induced Akt membrane compartmentalization is the membrane-associated ClipR-59 (Clip (cytoplasmic linker protein)-related 59 kDa) (19). Once at the membrane, Akt phosphorylates a key substrate, AS160 (Akt substrate 160 kDa), which promotes Glut4 membrane redistribution.
In the course of studying ClipR-59, we have identified Elmo2 as a novel interacting protein (20). By interacting with Elmo2, ClipR-59 modulates Rac1 activity in myoblast cells to promote muscle differentiation. Because ClipR-59 interacts with Akt and regulates its membrane compartmentalization, we have postulated that Elmo2 might also regulate Akt compartmentalization and thereby insulin-dependent Glut4 membrane translocation. In the present study, we found that Elmo2 modulates insulin-dependent Glut4 membrane translocation in both cultured adipocytes and skeletal muscle cells.
Results
The Interaction of Elmo2 with ClipR-59 Enhances the Association of Akt with ClipR-59
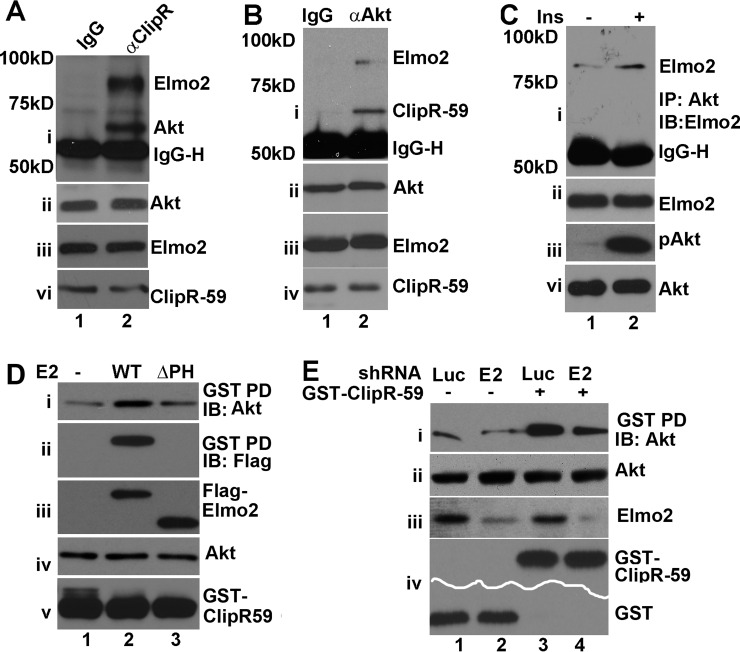
Recently, we reported that Elmo2 interacted with ClipR-59 (20). ClipR-59 interacts with Akt (19). We wondered whether Elmo2, ClipR-59, and Akt are in the same complex. To test this, a co-immunoprecipitation assay of total cell lysates from 3T3-L1 adipocytes was carried out with anti-ClipR-59 antibody, and the presence of Elmo2 and Akt in anti-ClipR-59 immunoprecipitates was assessed by immunoblotting with anti-Elmo2 and anti-Akt antibodies. As shown in Fig. 1A, a band around 60 kDa and the other one around 80 kDa, which correspond to the molecular mass of Akt and Elmo2 were detected in anti-ClipR-59 immunoprecipitates but not that of control IgG (Fig. 1A, i), although there were compatible levels of Akt (ii), Elmo2 (iii), and ClipR-59 (iv) in each sample.
FIGURE 1.
The interaction of Elmo2 with ClipR-59 enhances the association of Akt with ClipR-59 Akt. A, co-immunoprecipitation (IP) assay of 3T3-L1 adipocytes with anti-ClipR-59 antibody. The anti-ClipR-59 immunoprecipitates were analyzed by Western blotting (IB) with anti-Akt and Elmo2 antibodies (i). ii, iii, and vi, total cellular levels of Akt, Elmo2 and ClipR-59. Pre-S, pre-sera. IgG-H, IgG heavy chain. B, similar to A, except the co-immunoprecipitation was carried out with anti-Akt antibody. IgG, rabbit IgG. C, similar to B, except the cells were treated with 10 nm insulin for 10 min. D, GST pull-down assay to examine the impact of Elmo2 on the association of ClipR-59 with Akt. HEK293 cells were transiently co-transfected with GST-ClipR-59 expression vector with the vector coding for FLAG-Elmo2 or ΔPH-Elmo2. The proteins associated with GST beads were analyzed by Western blotting with anti-Akt (i), anti-FLAG (ii), and anti-GST (v) antibodies, respectively. iii and iv show the levels of FLAG-Elmo2 and Akt in total cell lysates (Tcl). E, GST pull-down assay to examine the impact of Elmo2 shRNA expression on the association of ClipR-59 with Akt. HEK293 cells were transiently co-transfected with expression vectors coding for GST or GST-ClipR-59 plus the expression vectors expressing either luciferase or Elmo2 shRNA. The proteins associated with GST beads were analyzed by Western blotting with anti-Akt (i) and anti-GST (vi), respectively. ii and iii show the levels of Akt and Elmo2 in total cell lysates. These experiments were repeated 3–4 times with similar results.
To examine this further, we next carried out the other co-immunoprecipitation assay with anti-Akt antibody and examined the presence of ClipR-59 and Elmo2 in anti-Akt immunoprecipitates. As shown in Fig. 1B, both ClipR-59 and Elmo2 were detected in anti-Akt immunoprecipitates but not that of control IgG (Fig. 1B, i). The presence of Elmo2 and ClipR-59 anti-Akt immunoprecipitates was not because of sample variations because compatible levels of Akt, Elmo2, and ClipR-59 were seen in each sample (ii, iii, and vi).
The interaction between Akt and ClipR-59 is regulated by insulin (19). We therefore examined whether the interaction between Akt and Elmo2 is regulated by insulin. As shown in Fig. 2C, insulin stimulation increased the amount (∼2.5-fold) of Elmo2 in anti-Akt immunoprecipitates (i) without altering the levels of total cellular Akt and Elmo2 (iii and v). In these studies, insulin is effective because it induced Akt phosphorylation (ii). Taken together, our data demonstrated that ClipR-59, Akt, and Elmo2 are in an insulin-regulated complex.
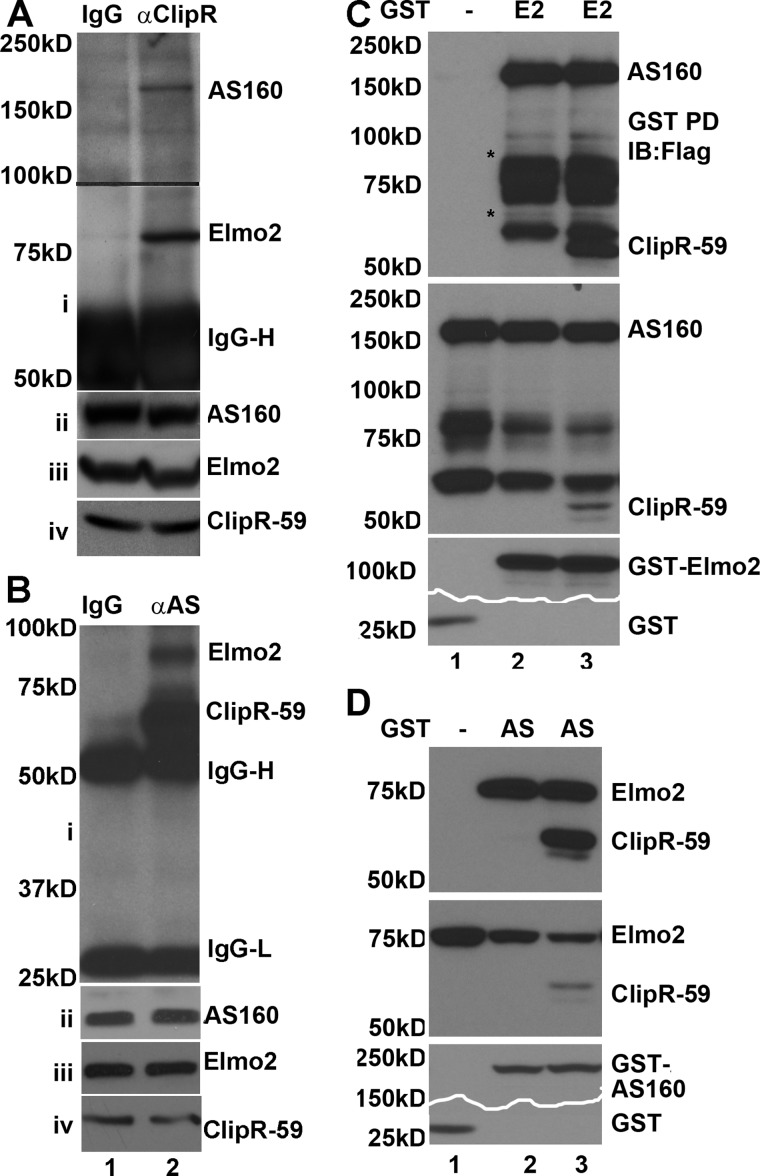
FIGURE 2.
Elmo2 is in the complex consisting of AS160 and ClipR-59. A, co-immunoprecipitation assay to show the interaction of with Elmo2 and AS160. Total cell lysates from differentiated 3T3-L1 adipocytes were immunoprecipitated with anti-ClipR-59 antibody. The anti-ClipR-59 immunoprecipitates were separated on SDS-PAGE and transferred onto membrane. The membrane was divided into two parts on the 100 kDa marker (indicated by a line) and analyzed by Western blotting with anti-AS1 antibody and anti-Elmo2 antibodies, respectively. The IgG heavy chain (IgG-H) is shown. The remaining panels show the levels of Elmo2, ClipR-59, and AS160 in total cell lysates. B, similar to A except that the co-immunoprecipitation assay was carried out with anti-AS160 (AS) antibody. C, GST pull-down assays with GST-Elmo2 to show that both AS160 and ClipR-59 are associated with Elmo2. GST-Elmo2 (E2) or GST prepared from 239T cells was incubated with cell lysates from COS-7 cells that were transiently co-transfected with expression vector coding for FLAG-AS160 or coding for FLAG-AS160 plus FLAG-ClipR-59. The proteins associated with GST beads were analyzed by Western blotting (IB) with anti-FLAG antibody (top). The middle panel shows the levels of FLAG-AS160 (AS) and FLAG-ClipR-59 in total cell lysates (Tcl). The bottom panels show the levels of GST fusion proteins associated with GST beads. Note that part of the bottom panel was removed to save. The bands marked with an asterisk likely represent degraded FLAG-AS160 as they reacted with anti-FLAG-antibody. D, GST pull-down assays with GST-AS160 to show that both ClipR-59 and Elmo2 are associated with AS160. The experiments were carried out similarly to those in C except that GST-AS160, FLAG-Elmo2, and FLAG-ClipR-59 were used. In all cases, the molecular mass markers are indicated on the right. These experiments were repeated three times with similar results.
After demonstrating that ClipR-59, Akt, and Elmo2 are present in the same complex, we next asked whether the interaction of Elmo2 with ClipR-59 affects the association of ClipR-59 with Akt. To address this, a GST pull-down assay was carried out with the lysates from HEK293 cells that were co-transfected with expression vectors coding for GST-ClipR-59 fusion protein and FLAG-Elmo2. The proteins that were associated with GST beads were analyzed by Western blotting with anti-Akt. As shown in Fig. 1D, the Akt associated with GST-ClipR-59 increased with ectopically co-expressed Elmo2 (i). We also examined FLAG-Elmo2 in GST-ClipR-59 beads. In agreement with our previous findings that Elmo2 interacts with ClipR-59 (20), FLAG-Elmo2 was readily detected in GST-ClipR-59 (Fig. 1D, iii). The enhancement of the association of Akt with GST-ClipR-59 was specific at a comparable level as comparable input levels of Akt and GST-ClipR-59 were seen in each sample (iv and v).
The PH domain of Elmo2 mediates the interaction of Elmo2 to ClipR-59 (20). To determine whether the enhancement of ClipR-59 association with Akt by Elmo2 depends on the PH domain, we examined the impact of ΔPH Elmo2, which does not interact with ClipR-59, on the association of ClipR-59 with Akt. As shown in Fig. 1D, co-expression of ΔPH Elmo2 with GST-ClipR-59 did not enhance the amount of Akt retained on GST-ClipR-59, suggesting that the interaction between ClipR-59 and Elmo2 is required for Elmo2 to enhance the association of ClipR-59 with Akt. Furthermore, in agreement with the notion that the PH of Elmo2 mediates the interaction between ClipR-59 and Elmo2 (20), no ΔPH Elmo2 was detected on GST-ClipR-59 beads.
To further evaluate the enhancement of ClipR-59 association with Akt by Elmo2, we examined the impact of Elmo2 knockdown on this interaction. HEK293 cells were co-transfected with pEBG-ClipR-59 plus either Elmo2 or luciferase (as a control) shRNA expression vector. As an additional control, the cells were also co-transfected with pEBG empty vector plus either Elmo2 or luciferase shRNA expression vector. Then GST pull-down assays were carried out, and the amounts of Akt associated with GST beads were analyzed with anti-Akt antibody. As shown in Fig. 1E, Akt was detected on GST-ClipR-59 beads, but not GST beads alone (i), demonstrating the specific association of Akt with ClipR-59. Co-expression of Elmo2 shRNA reduced the amounts of Akt associated with GST-ClipR-59 beads ∼50%. The Elmo2 shRNA was effective to suppress Elmo2 expressions because a reduction of Elmo2 in Elmo2 shRNA-expressing cells was observed (Fig. 1E, iii). Taken together, these data demonstrate that Elmo2 enhances the association of ClipR-59 with Akt.
Formation of a Multiple-protein Complex Composed of Elmo2, AS160, and ClipR-59
We previously showed that ClipR-59 interacts with AS160, a major Akt substrate, in an insulin-dependent manner to promote Glut4 membrane translocation (21). The interaction of Elmo2 with ClipR-59 promoted us to test whether ClipR-59, Elmo2, and AS160 are in the same protein complex. To investigate this, we carried out co-immunoprecipitation assays of 3T3-L1 with anti-ClipR-59 antibody. The presence of Elmo2 and AS160 in anti-ClipR-59 immunoprecipitates was examined. As shown in Fig. 2A, an 80 kDa anti-Elmo2 immunoreactive band and a 160 kDa anti-AS160 band were specifically detected in the anti-ClipR-59 immunoprecipitates but not that of control IgG (Fig. 2A, i). As a control, we verified that comparable levels of ClipR-59 (ii), Elmo2 (iii), and AS160 (iv) were present in each sample. Taken together, these results demonstrate that Elmo2, ClipR-59, and AS160 form a trimeric protein complex.
Next, we carried out a similar co-immunoprecipitation assay with anti-AS160 antibody. We observed that both ClipR-59 and Elmo2 were presented in anti-AS160 immunoprecipitates (Fig. 2B, i) but not IgG control. Again, as a control, we verified that comparable levels of ClipR-59 (ii), Elmo2 (iii), and AS160 (iv) were present in each sample.
To further validate that ClipR-59, AS160, and Elmo2 were in the same complex, a GST pull-down assay was carried out with GST-Elmo2 and FLAG-AS160. Specifically, purified GST-Elmo2 isolated from HEK293T cell lysate was incubated with total cell lysates of COS-7 cells that were transiently transfected with expression vector coding for FLAG-tagged AS160. As shown in Fig. 2C, AS160 was retained on GST-Elmo2 beads (top). The AS160 protein retained on the beads was specific to GST-Elmo2 because no detectable AS160 protein was found on GST beads alone. The variation in AS160 retained on GST-Elmo2 beads could not be ascribed to sample variations because comparable levels of AS160 proteins and GST proteins were observed. In addition, we also carried out a similar assay with total cell lysates of COS-7 cells that were transiently co-transfected with expression vectors coding for FLAG-tagged AS160 and FLAG-ClipR-59. As expected, both AS160 and ClipR-59 proteins were retained on GST-Elmo2 beads (Fig. 2C).
Next, we carried out the other GST pull-down assay with purified GST-AS160 from HEK293T cells and the lysates of COS-7 cells that were transiently transfected with expression vectors coding for FLAG-Elmo2 and FLAG-ClipR-59. As shown in Fig. 2D, Elmo2 and ClipR-59 proteins were retained on GST-AS160 beads (top). The Elmo2 protein retained on the beads was specific to GST-AS160 because there was no detectable Elmo2 and ClipR-59 in GST beads alone.
The Association of Elmo2 with ClipR-59 and AS160 Is Regulated by Insulin
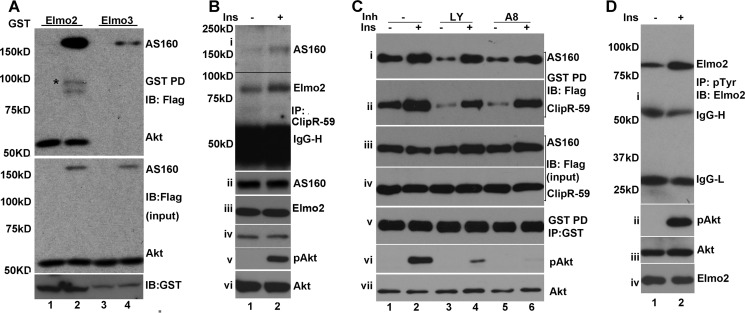
In the early studies, Akt was found to be present in the complex including AS160 and ClipR-59. In the preceding experiments (Fig. 1), we showed that Elmo2 is associated with Akt endogenously. Because Elmo2 interacts with AS160, we examined the possibility that Akt is complexed with Elmo2 and AS160 in a GST pull-down assay with GST-Elmo2 purified from 293 cells and the total cell lysates from COS-7 cells co-transfected with the vectors coding for FLAG-Akt and FLAG-AS160. FLAG-Akt either individually or combined with FLAG-AS160 was found on GST-Elmo2 beads (Fig. 3A, top). Furthermore, in agreement with the view that AS160 is associated with Elmo2, FLAG-AS160 was detected on GST-Elmo2 beads. As a control, we also carried out a similar GST pull-down assay using GST-Elmo3. FLAG-AS160, but not FLAG-Akt, was retained on GST-Elmo3 beads (Fig. 3A, top), indicating that whereas both Elmo2 and Elmo3 form a complex with AS160, it is Elmo2 that forms a complex with Akt. In these studies, we verified the expression of FLAG-Akt, FLAG-AS160 (Fig. 3A, middle), and GST-Elmo (Fig. 3A, bottom).
FIGURE 3.
Insulin regulates the association of Elmo2 with ClipR-59 and AS160. A, GST pull-down assay with GST-Elmo2 to show that Akt is in the complex with Elmo2. The experiments were carried out similarly to those in Fig. 2C. except that FLAG-Akt and GST-Elmo3 were used. The proteins associated with GST beads were analyzed by immunoblotting (IB) with anti-FLAG antibody (top). Middle and top panels show the total cellular levels of Akt, AS160, and GST fusion protein. The bands marked with an asterisk represent the degraded FLAG-AS160. B, co-immunoprecipitation (IP) assay of the total cell lysates of L6 cells treated with or without 100 nm insulin for 10 min with anti-ClipR-59 antibody. The anti-ClipR-59 immunoprecipitates were analyzed by immunoblotting with anti-Elmo2 and anti-AS160 antibodies. ii, iii, iv, v, and vi show the total cellular levels of AS160, Elmo2, ClipR-59, phospho-Akt, and Akt, respectively. C, GST pull-down assay similar to Fig. 2B except that the cells were pretreated with either DMSO (−) or PI3K inhibitor (Inh), LY294002 (LY), or Akt inhibitor VIII (A8) for 45 min, followed by 100 nm insulin for 10 min. The GST beads were analyzed by immunoblotting with anti-FLAG (i and ii) and anti-GST (v) antibodies. The total cellular levels of FLAG-AS160, FLAG-ClipR-59, phospho-Akt, and Akt are shown in iii, iv, vi, and vii, respectively. D, insulin induces Elmo2 phosphorylation at tyrosine residue(s). The total cell lysates in radioimmune precipitation assay buffer from 3T3-L1 adipocytes that were treated with or without 10 nm insulin were subjected to immunoprecipitation with anti-Tyr(P) antibody. The anti-Tyr(P) immunoprecipitates were analyzed by immunoblotting with anti-Elmo2 antibody (i). ii, iii, and iv show the total cellular levels of phospho-Akt, Akt, and Elmo2. IgG-H, IgG heavy chain. IgG-, IgG light chain.
In the preceding studies, we have demonstrated that Elmo2 is associated with ClipR-59 and AS160 endogenously. Previously, we showed that the interaction between ClipR-59 and Elmo2 was regulated by insulin (20). We wondered whether the complex consisting of these proteins is regulated by insulin. To investigate this possibility, we carried out an assay with L6 cell lysates treated with or without insulin and anti-ClipR-59 antibody and assessed the impact of insulin on the presence of ClipR-59 and Elmo2 in anti-ClipR-59 immunoprecipitates. As shown in Fig. 3B, insulin treatment increased the amounts of both Elmo2 and AS160 in the anti-ClipR-59 immunoprecipitates without altering the total cellular levels of Elmo2, AS160, and ClipR-59 (ii, iii, and iv). We also examined Akt phosphorylation. As expected, Akt phosphorylation was increased following insulin stimulation without altering total cellular levels of Akt (v and vi).
To verify that the association of Elmo2 with AS160 and ClipR-59 is regulated by insulin, we next carried out a GST pull-down assay with the total cell lysates from HEK293 cells that transiently co-transfected with expression vectors coding for GST-ClipR-59, FLAG-AS160, and FLAG-ClipR-59, respectively, treated with or without 100 nm insulin for 10 min. As shown in Fig. 3C, in agreement with the notion that insulin promotes the association of Elmo2 with AS160 and ClipR-59, insulin stimulation increased the amount of AS160 (i) and ClipR-59 (ii) associated with GST-Elmo2 beads. In the experiments, we also pretreated the cells with PI3K inhibitor LY294002 and Akt inhibitor VIII, respectively, and examined the role of PI3K and Akt in insulin-regulated association of ClipR-59 and AS160. No appreciable impact of both inhibitors on the association of Elmo2 with ClipR-59 and AS160 were observed (i and ii). As a control, we verified the expression of FLAG-AS160 (iii), FLAG-ClipR-59 (iv), and GST-Elmo2 (v) in total lysates. In addition, we also examined Akt phosphorylation. As expected, both inhibitors suppressed Akt phosphorylation by Akt (vi and vii). Taken together, these data demonstrated that insulin promotes the association of Elmo2 with ClipR-59 and AS160 independent of PI3K and Akt activity.
Recent studies revealed that Elmo2 is phosphorylated at tyrosine residue(s) (10). Proteomic studies of adipocyte tyrosine phosphoproteins implied that insulin could induce Elmo2 phosphorylation (22). Because insulin regulates the association of Elmo2 and ClipR-59 is independent of PI3K and Akt kinase, we wondered whether insulin induces Elmo2 tyrosine phosphorylation. To investigate this, 3T3-L1 adipocytes were treated with or without 10 nm insulin for 10 min. Total cell lysates in radioimmune precipitation assay buffer were immunoprecipitated by Tyr(P) antibodies, and the presence of Elmo2 in anti-Tyr(P) immunoprecipitates was examined by immunoblotting with anti-Elmo2 antibody. As shown in Fig. 3D, Elmo2 was easily detected in anti-Tyr(P) immunoprecipitates, and its levels were increased following insulin stimulation (i). The increased levels of Elmo2 in anti-Tyr(P) immunoprecipitates were not the result of sample variation because compatible levels of Elmo2 were seen in each sample (ii). We also examined Akt phosphorylation in total cell lysates. As expected, Akt phosphorylation increased following insulin stimulation (iii and iv). Taken together, out data demonstrated that insulin promotes Elmo2 phosphorylation.
Elmo2 Regulates Glut4 Membrane Distribution in 3T3-L1 Adipocytes
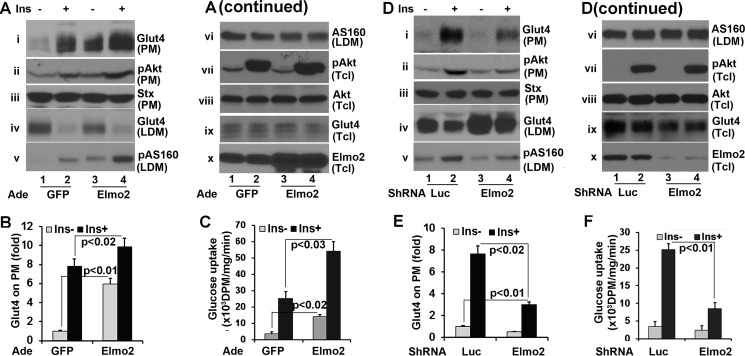
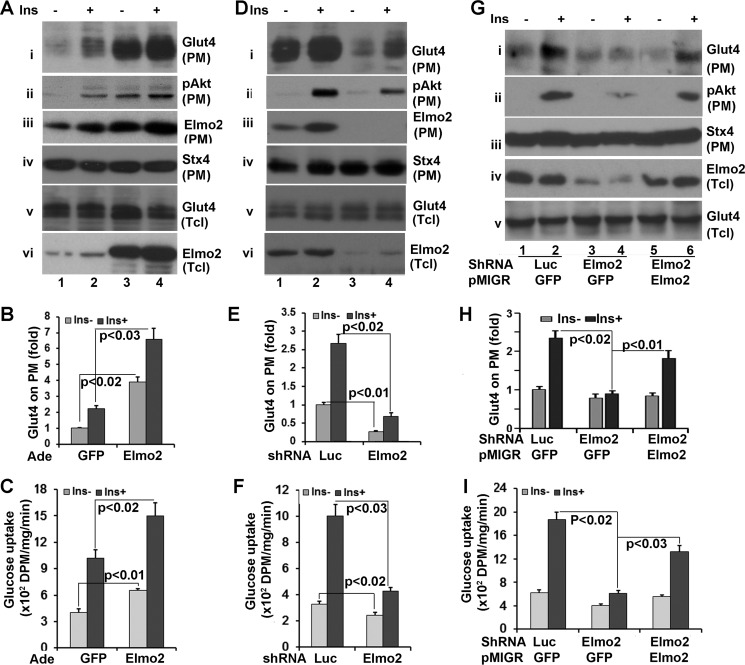
Both ClipR-59 (19) and AS160 are involved in insulin-dependent Glut4 membrane translocation (23). The view that Elmo2 is in the same complex that includes AS160 and ClipR-59 promoted us to examine whether Elmo2 regulates insulin-dependent Glut4 membrane translocation. To address this, differentiated 3T3-L1 adipocytes were transduced with adenoviral vectors coding either GFP or Elmo2. Then plasma membrane (PM) and low density microsomes (LDM) were isolated through a subcellular fractionation assay after the cells were treated with or with 25 nm insulin for 30 min. The levels of Glut4 on each fraction were then determined by Western blotting with an anti-Glut4 antibody. As shown in Fig. 4A, ectopic expression of Elmo2 increased the amount of Glut4 on PM by >5-fold (Fig. 4, A (i) and B) under basal conditions compared with that in GFP-expressing cells. Insulin induced a 7-fold increase of Glut4 on PM in GFP-expressing cells, whereas it induced a ∼10-fold increase in cells ectopically expressing Elmo2 (i). The recruitment of Glut4 at the PM correlated with its decrease in LDM (i and iv). In these experiments, equal levels of syntaxin 4 on PM (iii) and Glut4 in total cell lysates (ix) were detected in each sample as loading controls.
FIGURE 4.
Elmo2 regulates insulin-dependent Glut4 membrane translocation in adipocytes. A, ectopic expression of Elmo2 in 3T3-L1 adipocytes enhances insulin-dependent Glut4 membrane translocation in 3T3-L1 adipocytes. 3T3-L1 adipocytes were transduced with adenoviral (Ade) vectors coding for either GFP or FLAG-Elmo2. Transduced 3T3-L1 cells were serum-starved overnight followed by insulin treatment (25 nm) for 30 min. PM and LDM were prepared through a subcellular fractionation assay as described under “Experimental Procedures.” The PM was analyzed by Western blotting with anti-Glut4 (i), anti-phospho-Akt (ii), and anti-syntaxin 4 (iii) antibodies, respectively. The LDM was analyzed by Western blotting with anti-Glut4 (iv), anti-phospho-AS160 at Thr-652 (v), and anti-AS160 (vi) antibodies, respectively. vii, viii, ix, and x show the levels of phospho-Akt, Akt, Glut4, and Elmo2 in total cell lysates (Tcl). B, densitometric analysis of Glut4 on PM. The amount of Glut4 on PM from GFP-expressing cells without insulin treatment was set as 1 after normalization to PM syntaxin 4 and total cellular levels of Glut4. The bar graphs show means ± S.D. (error bars) (n = 3). In all cases, p < 0.03. C, ectopic expression of Elmo2 increases 3T3-L1 adipocyte glucose transport. 3T3-L1 adipocytes were transduced with adenoviral vectors coding for either GFP or FLAG-Elmo2. Transduced 3T3-L1 adipocytes were serum-deprived for 6 h and treated with 10 nm insulin for 30 min. The glucose uptake assay was carried out as described under “Experimental Procedures.” The bar graphs show means ± S.D. (n = 3). In all cases, p < 0.045. D, Elmo2 knockdown suppresses insulin-dependent Glut4 membrane translocation. The experiments were carried out essentially as in a, except 3T3-L1 adipocytes were transduced with adenoviral vectors that express either luciferase or Elmo2 shRNA. E, densitometric analysis of Glut4 on PM. The amount of Glut4 on PM from luciferase shRNA-expressing cells without insulin treatment was set as 1 after normalization to PM syntaxin 4 and total cellular levels of Glut4. The bar graphs show means ± S.D. (n = 3). In all cases, p < 0.022. F, Elmo2 knockdown decreases 3T3-L1 adipocyte glucose transport. 3T3-L1 adipocytes were transduced with adenoviral vectors coding for either luciferase (Luc) or Elmo2 shRNA. Transduced 3T3-L1 adipocytes were serum-deprived for 6 h and treated with 10 nm insulin for 30 min. The glucose uptake assay was carried out as described under “Experimental Procedures.” The bar graphs show means ± S.D. (n = 3). In all cases, p < 0.045.
We also found that the level of Akt on PM was also increased following ectopic expression of Elmo2 (Fig. 4A, ii). Ectopic expression of Elmo2 did not impact on the total cellular Akt activation induced by insulin (vii and viii). As a control, we verified that Elmo2 protein was ectopically expressed upon transduction (∼3-fold; Fig. 4A, x).
The key consequence of ClipR-59-regulated Akt compartmentalization is to increase AS160 phosphorylation in LDM (19). Hence, we examined AS160 phosphorylation at Thr-642 in LDM. As shown in Fig. 4A, an increase in AS160 at Thr-642 was seen in LDM from Elmo2 ectopically expressing cells (v and vi).
A primary function of insulin-dependent Glut4 membrane translocation is to transport glucose. To determine whether increased Glut4 on PM by Elmo2 expression is associated with adipocyte glucose transport, we carried out an adipocyte glucose uptake assay. As shown in Fig. 4C, compatible with the view that Elmo2 expression increased Glut4 on PM, it also enhanced adipocyte glucose uptake.
To further examine the impact of Elmo2 on insulin-dependent Glut4 membrane translocation, we next transduced 3T3-L1 adipocytes with adenoviral vectors coding for either Elmo2 shRNA or luciferase shRNA (control). Then subcellular fractionation assays were carried out to examine the Glut4 on each fraction. As shown in Fig. 4D, Elmo2 knockdown by shRNA reduced the amount of Glut4 at the PM by >50% under basal conditions and >60% following insulin stimulation (Fig. 4, D (i) and E), with corresponding changes in LDM (iii), in comparison with cells expressing the luciferase shRNA. As a control, we verified that equal levels of syntaxin 4 on PM (iii) and Glut4 in total cell lysates (ix) were detectable in each sample. In agreement with our hypothesis that Elmo2 modulates Akt membrane association, the levels of Akt on PM reduced by more than half (ii) without affecting the total cellular levels of phospho-Akt (vii and viii). We also examined AS160 phosphorylation at Thr-642 in LDM and found that it decreased in Elmo2 shRNA-expressing adipocytes. Elmo2 shRNA was effective in silencing Elmo2 expression as the levels of Elmo2 protein were reduced by more than 80% under Elmo2 shRNA expression (Fig. 4E, x).
We also examined the impact of Elmo2 shRNA expression on adipocyte glucose uptake. In agreement with the finding that Elmo2 shRNA expression suppressed Glut4 membrane translocation, it also reduced glucose uptake following insulin stimulation (Fig. 4F). Taken together, our data demonstrate that Elmo2 regulates insulin-dependent Glut4 membrane translocation in adipocytes.
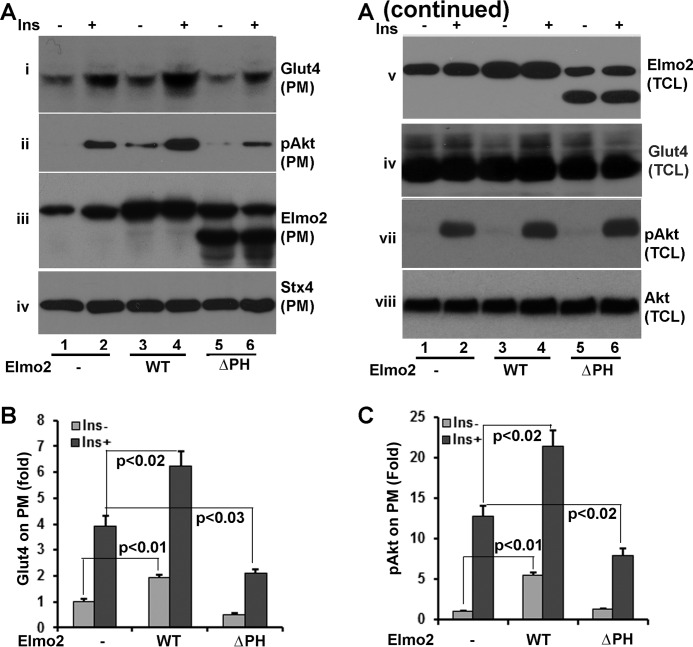
The interaction of Elmo2 with ClipR-59 is mediated by the PH domain of Elmo2 (20). To determine whether the interaction of Elmo2 with ClipR-59 is required for Elmo2 to promote insulin-dependent Glut4 membrane translocation, 3T3-L1 adipocytes were transduced with adenoviral vectors coding for GFP (control), Elmo2, and ClipR-59 interaction-defective ΔPH Elmo2, in which the PH domain of Elmo2 was deleted. Then subcellular fractionation assays were performed to examine the impact of ΔPH Elmo2 on Glut4 membrane translocation. As shown in Fig. 5A, ectopic expression of Elmo2 increased Glut4 membrane distribution (i). On the other hand, ectopic expression of ΔPH Elmo2 reduced the amount of Glut4 on PM ∼30% (Fig. 5, A (i) and B) without affecting the levels of syntaxin 4 on PM (iv) and total cellular levels of Glut4 (vi), an indication that the interaction between Elmo2 and ClipR-59 is required for Elmo2 to promote Glut4 membrane translocation induced by insulin. Moreover, in agreement with the finding that the interaction of Elmo2 with ClipR-59 enhances the association of ClipR-59 with Akt and Akt membrane association, ectopic expression of ΔPH Elmo2 reduced the amount of Akt on PM. As expected, the levels of Akt on PM were also reduced. Finally, we examined Elmo2 on PM (iii) and total cellular levels of Elmo2 (v). Both wild type and ΔPH Elmo2 proteins were ectopically expressed with the view that the levels of endogenous Elmo2 increased following insulin stimulation, raising the possibility that insulin may regulate endogenous Elmo2 subcellular localization.
FIGURE 5.
The atypical PH domain of Elmo2 is required for Elmo2 to promote insulin-dependent Glut4 membrane translocation. A, 3T3-L1 adipocytes were transduced with adenoviral expression vectors coding for either GFP, FLAG-Elmo2, or ΔPH Elmo2. Then the cells were serum-deprived overnight, followed by a 30-min insulin treatment, and PMs were prepared. PM was analyzed by Western blotting with anti-Glut4 (i), anti-phospho-Akt at Ser-473 (ii), anti-Elmo2 (iii), and anti-syntaxin 4 (iv), respectively. v, vi, vii, and viii show the levels of Elmo2, Glut4, phospho-Akt, and Akt in total cell lysates (Tcl), respectively. B, densitometric analysis of Glut4 on PM. The amount of Glut4 on PM from GFP-expressing cells without insulin treatment was set as 1 after normalization to PM syntaxin 4 and total cellular levels of Glut4. The bar graphs show means ± S.D. (error bars) (n = 3). In all cases, p < 0.03. C, densitometric analysis of Akt on PM. The amount of Akt on PM from GFP-expressing cells without insulin treatment was set as 1 after normalization to PM syntaxin 4 and total cellular levels of Akt. The bar graphs show means ± S.D. (n = 3). In all cases, p < 0.03.
Elmo2 Regulates Glut4 Membrane Distribution in L6 Skeletal Muscle Cells
As well as in adipocytes, insulin also induces Glut4 membrane translocation in skeletal muscle cells (24). We therefore next examined whether Elmo2 could modulate insulin-dependent Glut4 membrane translocation in skeletal muscle cells. To investigate this, L6 cells, a rat skeletal muscle cell line, were transduced with adenoviral vectors coding for either GFP (control) or Elmo2. After transduced cells were serum-deprived for overnight and treated with 100 nm insulin for 15 min, plasma membranes were prepared, and the levels of Glut4 on PM were assessed by Western blotting with an anti-Glut4 antibody. As shown in Fig. 6, A and B), treatment of L6 cells with insulin increased the amount of Glut4 on PM ∼2.5-fold (i). Forcing expression of Elmo2 increased the levels of Glut4 on PM by >4-fold under basal conditions and 7-fold under insulin stimulation (i). The differences in Glut4 on PM could not be ascribed to sample variation because comparable levels of syntaxin 4 on PM (iv) and total cellular levels of Glut4 were seen in each sample. In these experiments, we also examined phospho-Akt (ii) and Elmo2 (iii) on PM. Forcing expression of Elmo2 increased the levels of Akt on PM. We found that insulin stimulation increased the amount of Elmo2 on PM. As a control, we verified the efficiency of our transductions and found marked increases in Elmo2 expression (vi). In addition, we also carried out a glucose uptake assay to determine whether Elmo2 expression affects L6 uptake. As expected, forcing expression of Elmo2 in L6 adipocytes increased L6 cell glucose uptake in response to insulin (Fig. 6C).
FIGURE 6.
Elmo2 regulates insulin-dependent Glut4 membrane translocation in L6 skeletal muscle cells. A, ectopic expression of Elmo2 in L6 cells enhances insulin-dependent Glut4 membrane translocation. L6 cells were transduced with adenoviral vectors coding for either GFP or FLAG-Elmo2. Transduced L6 cells were serum-deprived for 6 h and treated with 100 nm insulin for 15 min. Then PM was prepared through a subcellular fractionation assay and analyzed by Western blotting with anti-Glut4 (i), anti-phospho-Akt (ii), and anti-syntaxin 4 (Stx4), respectively. v and vi show the levels of Glut4 and Elmo2 in total cell lysates, respectively. B, densitometric analysis of Glut4 on PM. The amount of Akt on PM from GFP-expressing cells without insulin treatment was set as 1 after normalization to PM syntaxin 4 and total cellular levels of Akt. The bar graphs show means ± S.D. (error bars) (n = 3). In all cases, p < 0.032. C, ectopic expression of Elmo2 enhances L6 glucose uptake. L6 cells were transduced with adenoviral vectors coding for either GFP or FLAG-Elmo2. Transduced L6 cells were serum-deprived for 6 h and treated with 100 nm insulin for 15 min. The glucose uptake assay was carried out as described under “Experimental Procedures.” The bar graphs show means ± S.D. (n = 3). In all cases, p < 0.033. D, Elmo2 knockdown in L6 cells suppresses insulin-dependent Glut4 membrane translocation. The experiments were carried out in essentially the same manner as in A, except adenoviral vectors that express either luciferase shRNA or Elmo2 shRNA were used. E, densitometric analysis of Glut4 on PM. The amount of Akt on PM from luciferase shRNA-expressing cells without insulin treatment was set as 1 after normalization to PM syntaxin 4 and total cellular levels of Akt. The bar graphs show means ± S.D. (n = 3). In all cases, p < 0.023. F, Elmo2 knockdown in L6 cells suppresses glucose uptake. The experiments were carried out in a manner similar to that described in C, except the cells were transduced with adenoviral vectors expressing either luciferase shRNA or Elmo2 shRNA. The bar graphs show means ± S.D. (n = 3). In all cases, p < 0.029. G, forcing expression of Elmo2 in Elmo2 shRNA-expressing cells rescues insulin-dependent Glut4 membrane translocation. L6 cells transduced with Elmo2 shRNA expressing adenoviral vectors were infected with pMigR1 retrovirus coding for either GFP or an Elmo2 shRNA-resistant Elmo2 (see “Experimental Procedures”). 48 h post-infection, the cells were serum-deprived for 6 h, followed by a 15-min insulin treatment. Then PM was prepared through a subcellular fractionation assay and analyzed by Western blotting with anti-Glut4 (i), anti-Akt (ii), and anti-syntaxin 4 antibodies, respectively. iv and v show the levels of Elmo2 and Glut4 in total cell lysates, respectively. H, densitometric analysis of Glut4 on PM. The amount of Glut4 on PM from GFP-expressing cells that were infected with GFP-expressing pMigR1 without insulin treatment was set as 1 after normalization to PM syntaxin 4 and total cellular levels of Glut4. The bar graphs show means ± S.D. (n = 3). In all cases, p < 0.022. I, forcing expression of Elmo2 in Elmo2 shRNA-expressing cells rescues the defect in glucose uptake. L6 cells were transduced with viral vectors as in G, and a glucose uptake assay was carried out. The bar graphs show means ± S.D. (n = 3). In all cases, p < 0.037.
To further examine the regulation of insulin-dependent Glut4 membrane translocation by Elmo2, we next examined how knockdown of Elmo2 in L6 cells affects Glu4 membrane translocation induced by insulin. To address this, L6 cells were transduced with adenoviral vectors coding for either a luciferase or Elmo2-specific shRNAs. PM were then prepared after the transduced cells were serum-deprived for overnight followed by insulin treatment for 15 min. As shown in Fig. 6D, expression of Elmo2 shRNA in L6 cells decreased Glut4 on PM by >50% under basal conditions and ∼80% following insulin stimulation (Fig. 6, D (i) and E), without affecting the levels of syntaxin 4 on PM (iii). In agreement with the notion that Elmo2 shRNA expression decreased insulin-dependent membrane translocation, the glucose uptake under Elmo2 shRNA expression decreased by >2-fold (Fig. 6F). In addition, insulin treatment increased Elmo2 on PM (Fig. 5C, iii). Again, Elmo2 shRNA was effective to silence Elmo2 expression because the amount of Elmo2 in Elmo2 shRNA-expressing cells was reduced by >80% (vi). Taken together, our data demonstrate that Elmo2 regulates Glut4 membrane translocation in muscle cells.
So far, we have shown that forcing expression of Elmo2 increased, whereas its knockdown decreased, insulin-dependent Glut4 membrane translocation. To further evaluate the regulation of Glut4 membrane translocation, we next examined whether reintroducing Elmo2 into Elmo2 shRNA-expressing L6 cells will rescue insulin-mediated Glut4 membrane translocation. To investigate this, we first generated an Elmo2 cDNA that is resistant to the shRNA against Elmo2 through site-directed mutagenesis. Briefly, the shRNA-targeted sequence of Elmo2 was mutated without introducing changes to Elmo2 amino acids (see “Experimental Procedures”). L6 cells transduced with adenoviral vector coding for Elmo2 shRNA were also infected with pMGR1 retroviruses coding for either Elmo2 (shRNA resistant) or GFP (as a control). 24 h later, the cells were serum-starved overnight and treated with insulin for 15 min. PM were then isolated for Western blotting with anti-Glut4 antibody. As shown in Fig. 6 (G and H), reintroducing Elmo2 into Elmo2 shRNA-expressing cells rescued Glut4 membrane distribution induced by insulin (i) with corresponding changes of glucose uptake (Fig. 6I). Re-expression of Elmo2 also rescued membrane-associated Akt (Fig. 6G, iii) without altering the levels of syntaxin 4 on PM or the total cellular levels of Glut4. Taken altogether, our data demonstrate that Elmo2 regulates Glut4 membrane translocation in muscle cells.
Elmo2 Modulates Insulin-induced Rac1 Activation
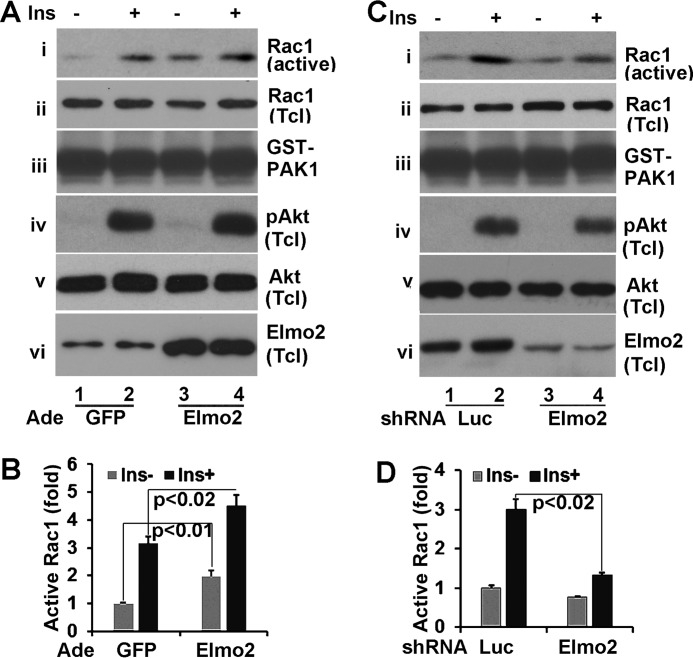
Herein, we have showed that Elmo2 regulates the insulin-induced Akt membrane association and Glut4 membrane translocation in L6 skeletal muscle cells. It is well established that insulin-induced Glut4 membrane translocation in muscle depends on Akt activation but also that of Rac1 induced by insulin (24, 25). Elmo2 has been shown to regulate Rac1 activity (26). This prompted us to examine whether Elmo2 is involved in Rac1 activation induced by insulin. To investigate this, we carried out a GST pull-down assay with GST-CRIB. GST-CRIB is a GST fusion protein that consists of the Cdc42/Rac1 interactive-binding domain (CRIB) of PAK1 (p21-activated kinase) and only interacts with active Rac1 or Cdc42 (27). Recombinant GST-CRIB was incubated with total cell lysates from L6 cells that were transduced with adenoviral vectors coding for either GFP or Elmo2 and treated with or without insulin as described in the legend to Fig. 7A. Then Rac1 proteins retained on GST-CRIB beads were assessed by Western blotting with anti-Rac1 antibody. As shown in Fig. 6A, insulin treatment increased the levels of active Rac1 ∼3-fold (Fig. 7, A (i) and B). When Elmo2 was ectopically expressed, the levels of active Rac1 increased ∼2-fold under basal conditions and ∼5-fold following insulin stimulation (Fig. 7, A (i) and B). The differences in the Rac1 protein retained on GST-CRIB beads were not due to sample variation because comparable levels of Rac1 (ii) and GST-CRIB (iii) were seen in each sample. In these experiments, we also examined Akt activation. No appreciable changes in Akt phosphorylation were seen (iv and v). The levels of Elmo2 in L6 cells that were transduced with adenoviral vector coding for Elmo2 were verified (vi).
FIGURE 7.
Elmo2 modulates Rac1 activation by insulin in L6 cells. A, ectopic expression of Elmo2 in L6 cells enhances Rac1 activity induced by insulin. L6 cells were transduced with adenoviral vectors coding for either GFP or FLAG-Elmo2. Then total cell lysates were prepared after these cells were serum-deprived for 6 h, followed by a 10-min insulin treatment, and subjected to GST pull-down assays with recombinant GST-CRIB. The levels of Rac1 associated with GST-CRIB beads were analyzed by Western blotting with anti-Rac1 antibody (i). iii shows the levels of GST-CRIB in each sample. ii, iv, v, and vi show the level of phospho-Akt, Akt, and Elmo2 in total cell lysates. B, densitometric analyses of Rac1 associated with GST-CRIB beads in A. The amount of Rac1 on GST-CRIB beads from GFP-expressing cells without insulin treatment was set as 1 after normalization to total cellular levels of Rac1. The bar graphs show means ± S.D. (error bars) (n = 3). In all cases, p < 0.034. C, knockdown of Elmo2 in L6 cells reduces Rac1 activity induced by insulin. The experiments were carried out in a manner similar to that described for A except that L6 cells were transduced with adenoviral vectors expressing either luciferase or Elmo2 shRNA. D, densitometric analysis of Rac1 associated with GST-CRIB beads in C. The amount of Rac1 on GST-CRIB beads from luciferase shRNA-expressing cells without insulin treatment was set as 1 after normalization to total cellular levels of Rac1. The bar graphs show means ± S.D. (n = 3). In all cases, p < 0.042.
To examine this further, we evaluated the impact of Elmo2 knockdown on Rac1 activation in L6 cells that were transduced with adenoviral vectors expressing either luciferase or Elmo2 shRNA. In GST-CRIB pull-down assays, we found that active Rac1 was markedly reduced upon Elmo2 shRNA expression (Fig. 7, C (i and vi) and D). Interestingly, Elmo2 knockdown appeared to decrease Akt activation induced by insulin (Fig. 7C, iv and v). Globally, our data demonstrate that Elmo2 is involved in Rac1 activation induced by insulin.
Discussion
Elmo proteins were identified based on their homology to Caenorhabditis elegans Ced-12, a protein that is required for apoptotic cell engulfment and cell migration (28). Based on this notion, the majority of Elmo studies have been focused on the role of Elmo proteins in cell migration and phagocytosis. However, mammalian Elmo proteins consist of additional structural features that are not present in Ced-12 (i.e. a tyrosine phosphorylation site (e.g. Tyr-713 in Elmo2)) (10). In addition, in addition to Rac1, several studies indicate that Elmo2 is also involved in Akt activation (9–11). This suggests that mammalian Elmo proteins probably have additional biological functions other than regulating cell migration. In the present studies, we have explored the possibility that Elmo2 is involved in insulin-dependent Glut4 membrane translocation in adipocyte and muscle cells, two major types of insulin-sensitive cells, and found that Elmo2 is involved in insulin-dependent Glut4 membrane translocation.
ClipR-59 interacts with Akt and AS160 (21). Because Elmo2 interacts with ClipR-59, we have examined whether the interaction of Elmo2 with ClipR-59 affects the association of Akt with ClipR-59. Our data from the co-immunoprecipitation assay and GST pull-down assay showed that the interaction of Elmo2 with ClipR-58 enhanced the association of Akt with ClipR-59 (Fig. 1). In addition, we also examined whether ClipR-59, Elmo2, and AS160 were in the same complex. In GST pull-down and co-immunoprecipitation assays, we found that ClipR-59, Elmo2, and AS160 were indeed presented in the same complex, making it possible for Elmo2 to modulate insulin-dependent Glut4 membrane translocation.
To obtain more information on the regulation of the association of Elmo2 with ClipR-59 and AS160, we have examined the impact of insulin on the association of Elmo2 with AS160 and ClipR-59. Our data indicated that insulin promotes the association of Elmo2 with AS160 and ClipR-59. Interestingly, this regulation is independent of PI3K and Akt, two major downstream kinases of insulin signaling (Fig. 3). Since insulin induced Elmo2 tyrosine phosphorylation (Fig. 3D), it is possible that insulin regulates the association of Elmo2 with AS160 and ClipR-59 through Elmo2 tyrosine phosphorylation. Elmo proteins are known to exist in an inhibitory state (3). Thus, it is possible that tyrosine phosphorylation of Elmo2 may relieve Elmo2 from the inhibitory state to interact with ClipR-59 and AS160, an issue that is under active investigation in our laboratory right now.
Because Akt, ClipR-59, and AS160 are involved in insulin-dependent Glut4 membrane translocation, we have examined how Elmo2 ectopic expression and Elmo2 knockdown impact insulin-dependent Glut4 membrane translocation in 3T3-L1 adipocytes and L6 skeletal muscle cells, two cell lines that are widely used to study the regulation of insulin-dependent Glut4 membrane translocation. Our data showed that in both cell lines, Elmo2 ectopic expression enhanced, whereas its knockdown suppressed, insulin-dependent Glut4 membrane translocation (Figs. 4 and 6), therefore demonstrating that Elmo2 is a novel regulator of Glut4 membrane translocation.
It is noteworthy that Elmo2 shRNA expression in adipocytes decreased Glut4 expression (Fig. 4D). At present, the reason for this is not clear. A potential reason could be that inactivation of Elmo2 might cause Glut4 mistranslocation (e.g. to the lysosome), which leads to Glut4 degradation because the Elmo protein has been implicated in endosome trafficking (29). Alternatively, Elmo2 might affect the proteolytic pathway that controls Glut4 membrane trafficking to affect Glut4 expression because the proteolytic pathway plays a critical role in the Glut4 membrane trafficking in both adipocytes and muscle cells (30), an issue that requires further investigation.
Mechanistically, we suggest that Elmo2 regulates Akt membrane compartmentalization in adipocytes and Akt membrane compartmentalization and Rac1 activation in muscles. First, in both 3T3-L1 adipocytes and L6 cells, ectopic expression of Elmo2 increased, whereas its knockdown decreased, Akt associated with plasma membranes and altered AKT phosphorylation (Figs. 4 and 6). Second, ectopic expression of Elmo2 increased, whereas its knockdown decreased, Rac1 activity in L6 cells (Fig. 7).
We propose that the regulation of Akt membrane association by Elmo2 probably depends on the interaction between Elmo2 and ClipR-59 because ClipR-59 interaction-defective ΔPH Elmo2 lacked the ability to promote the association of Akt with ClipR-59 and Akt membrane association (Fig. 4). However, at present, we are unable to conclude that the regulation of Rac1 activity induced by insulin may also depend on the interaction between Dock180 and Elmo2, which is also dependent on Elmo2 PH domain (31–33). Further studies are required to clarify this issue.
The family of Elmo proteins includes three members, Elmo1, -2, and -3. In our study of the interaction between ClipR-59 and Elmo proteins, we have shown that ClipR-59 preferentially interacts with Elmo2 and Elmo3 (20). Our current studies indicated that Elmo3 is associated with AS160 but not Akt (Fig. 3D). This would suggest that Elmo3 may have a different functional role in insulin-dependent Glut4 membrane translocation, a question worthy of further study.
The other interesting point raised in our study is that insulin appears to regulate Elmo2 subcellular localization and membrane translocation (Figs. 4 and 5). At present, we do not understand the mechanism under which insulin promotes Elmo2 membrane trafficking. In our current studies, we have demonstrated that insulin induces Elmo2 tyrosine phosphorylation (Fig. 3D). Moreover, the interaction between Elmo2 and ClipR-59 is regulated by insulin (20) (Fig. 1 and 3). Thus, it will be of great interest to know whether Elmo2 tyrosine phosphorylation and the interaction between ClipR-59 are involved in Elmo2 subcellular localization. Nevertheless, the view that insulin regulates Elmo2 membrane translocation emphasizes the notion that Elmo2 is involved in insulin action in metabolic regulation.
In summary, we have presented evidence that Elmo2, by modulating Akt membrane compartmentalization and Rac1 activity, plays an important role in insulin-dependent Glut4 membrane translocation. Insulin-dependent Glut4 membrane translocation plays a central role in the maintenance of body glucose homeostasis. In addition, Elmo2 has been found to play a critical role in the regulation of muscle differentiation. All of these findings point out a possible scenario in which Elmo2 is a potential regulator of body glucose homeostasis, an important question to be addressed in the future.
Experimental Procedures
Reagents
Insulin, dexamethasone, 3-isobutyl-1-methylxanthine, monoclonal anti-FLAG antibody, and HRP-conjugated anti-FLAG and anti-HA antibodies were from Sigma. Mouse monoclonal anti-HA antibody was from Covance. Mouse monoclonal anti-Elmo2, anti-Glu4, anti-Rac1, and anti-GST were from Santa Cruz Biotechnology, Inc. Rabbit monoclonal anti-phospho-AS160 at Thr-642, normal rabbit IgG, anti-AS160, anti-Akt, and phospho-Akt antibodies were from Cell Signaling. Rabbit anti-ClipR-59 antibody and pre-sera have been described previously (19). 2-[3H]deoxyglucose was from (PerkinElmer Life Sciences).
Plasmids and Virus Production
ClipR-59 and its mutants as well as the FLAG-AS160 plasmids have been described (21). Elmo2 shRNA has been described (5). FLAG-tagged Elmo2, ΔPH Elmo2, and GST-Elmo2 expression vectors have been described (20). GST-CRIB was a gift from Dr. Rachel Nussbaum (Tufts Medical Center) (27). The adenoviral vectors for expressing Elmo2 shRNA and Elmo2 were generated according to the procedure described previously (21).
To generate AS160 expression vectors, full-length AS160 cDNA were fused to ClaI and KpnI sites of pEBG vectors. To generate retroviral expression vector for Elmo2 that was resistant Elmo2 shRNA, Elmo2 was subjected to site-directed mutagenesis with primers 5′-GGAATTGGCCTTCTCGATTCTCTATGATCCTGA-3′ (forward) and 5′-CATCAGGATCATACAGGATGGAGAAGGCCAATTCC-3′ (reverse) (the boldface nucleotides correspond to Elmo2 shRNA; the underlined nucleotides are mutated) and cloned into pMigR1 retroviral vector between the HpaI and XhoI sites. Elmo2 mutations were verified by sequencing. This site-directed mutagenesis altered Elmo2 cDNA sequence without changing Elmo2 peptide sequence. The viral particles were produced as described (34).
Cell Culture and Transfection
HEK293T and COS-7 cells were grown in high glucose DMEM supplemented with 10% (v/v) FBS, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). L6 cells were grown in the same DMEM but supplemented with 15% bovine serum instead of FBS. The differentiation of 3T3-L1 preadipocytes was as follows. 3T3-L1 preadipocytes were cultured for an additional 2 days after reaching 100% confluence and treated with differentiation medium (DMEM-high glucose containing 10% FBS, 2.5 μg/ml insulin, 0.5 mm 3-isobutyl-1-methylxanthine, 2.5 μm dexamethasone, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin) for 4 days. Then the medium was changed to regular medium. After 7 days of differentiation, the adipocytes were used for experiments. When the cells were infected with viruses, at least 75% of cells were transduced. Transient transfection was carried out with Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions.
Subcellular Fractionation Assay
L6 cells (in 15-cm dishes) and 3T3-L1 adipocytes (in 10-cm dishes), with or without insulin treatments, were suspended into HES I buffer (0.25 m sucrose, 20 mm Tris, pH 7.6, 1 mm EDTA plus a protease inhibitor mixture). The cells were homogenized by passing a 23-gauge needle 10 times, and then the homogenates were centrifuged at 19,000 × g for 20 min. To isolate the membrane fraction, the resultant pellets from the 19,000 × g centrifugation were layered on HES II buffer (1.12 m sucrose, 20 mm Tris, pH 7.6, 1 mm EDTA) and centrifuged at 100,000 × g for 60 min. The resulting pellets were designated as nuclear and mitochondrial fractions. The plasma membrane layers were removed from the sucrose cushion and suspended into HES I buffer and centrifuged at 41,000 × g for 20 min. The resultant pellets were the PM. To isolate microsomes, the resultant supernatant from the 19,000 × g centrifugation was centrifuged at 175,000 × g for 75 min, and the pellets were collected as LDM. The supernatants from the 175,000 × g centrifugations were saved and designated as cytosols.
Glucose Uptake Assay
The glucose uptake assay has been described (19). Briefly, adipocyte or L6 cells in a 12-well dish were serum-deprived overnight and then were treated with or without insulin for 30 min. Then the cells were washed twice with Krebs buffer (50 mm HEPES, pH 7.4, 136 mm NaCl, 4.7 mm KCl, 1.25 mm MgSO4, 1.25 mm CaCl2) and further incubated with Krebs buffer plus 100 mm 2-deoxyglucose, 1 μm 2-[3H]deoxyglucose (PerkinElmer Life Sciences). After 5 min of incubation, the cells were washed three times with PBS and lysed with 0.05 m NaOH. The amount of 2-[3H]deoxyglucose in the total cell lysates was counted. The final results were subtracted from the background glucose uptake, which was determined by the amount of 2-[3H]deoxyglucose in the lysates when the cells were pretreated with 10 μm Cytochalasin B (Sigma) and normalized to protein concentration in the lysates.
Immunoprecipitation Assays
Transfected COS-7 cells or 3T3-L1 adipocytes were extracted with immunoprecipitation buffer (150 mm NaCl, 25 mm Tris, pH 7.6, 0.5 mm EDTA, 10% glycerol, 0.5% Nonidet P-40 plus a protease inhibitor mixture). 500 μg of total proteins from the cell lysates were subjected to immunoprecipitation with the corresponding antibodies.
GST Pull-down Assays
GST-AS160 or Elmo2 fusion peptides were expressed in HEK293T cells from pEBG expression vector and purified with glutathione-Sepharose 4B according to the manufacturer's instructions. For examining Elmo2 and AS160 interaction, the purified GST-AS160 or Elmo2 was mixed with the total cell lysate of COS-7 cells that had been transiently transfected with proper expression vectors in immunoprecipitation buffer and incubated for 4 h. Then the beads were washed three times, and the proteins associated with GST beads were analyzed by Western blotting with the corresponding antibodies. Also, before washing, a 2% volume of the mixtures was taken out for Western blotting to determine the input level of each component. In some cases, cells were co-transfected with pEBG expression vectors with the indicated Elmo2 or ClipR-59 expression constructs.
Rac1 Activation Assay
The assay to measure GTP-loaded Rac1 was carried out as described previously (27). Briefly, GST-CRIB was produced in Escherichia coli Bl21 and purified on glutathione beads. Total lysates from L6 cells that were transduced with the adenoviral vectors indicated in the figure legends were incubated with GST-CRIB beads. After 4 h of incubation, GST beads were washed extensively and analyzed by Western blotting with the proper antibodies.
Western Blotting
After treatments, cells were washed twice with PBS and extracted with cell lysis buffer (20 mm Tris, pH 7.6, 150 mm NaCl, 0.5 mm EDTA, 0.5 mm DTT, 10% glycerol, protease and phosphatase inhibitors). For cellular fractionation experiments, the cellular fractions were directly dissolved into lysis buffer. Equal amounts of protein were subjected to SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad). After blocking in 5% dry milk, the membranes were incubated with the indicated primary antibody, followed by incubation with a horseradish peroxidase-conjugated secondary antibody. The protein bands were visualized using the ECL detection system (Pierce). The quantification of the Western blotting was determined with ImageJ software.
Statistical Analysis
Means ± S.D. were calculated, and statistically significant differences among groups were determined by one-way analysis of variance followed by post hoc comparisons or by a two-tailed unpaired Student's t test between two groups as appropriate, with significance at p < 0.05.
Author Contributions
S. Y. participated in study design, performed experiments, and analyzed the data. J. F. C. participated in data analyses and wrote the manuscript. K. D. participated in study design, performed experiments, analyzed data, supervised the study, and wrote the manuscript.
Acknowledgment
We thank Dr. Rachel Buchsbaum for the GST-CRIB expression vector.
This work was supported by National Institutes of Health Grant RO1 DK084319 (to K. D.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PH
- pleckstrin homology
- CRIB
- Cdc24/Rac1 interactive binding domain of Pak1
- PM
- plasma membrane
- LDM
- low density microsome(s).
References
- 1. Ravichandran K. S., and Lorenz U. (2007) Engulfment of apoptotic cells: signals for a good meal. Nat. Rev. Immunol. 7, 964–974 [DOI] [PubMed] [Google Scholar]
- 2. Laurin M., and Côté J. F. (2014) Insights into the biological functions of Dock family guanine nucleotide exchange factors. Genes Dev. 28, 533–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Patel M., Margaron Y., Fradet N., Yang Q., Wilkes B., Bouvier M., Hofmann K., and Côté J. F. (2010) An evolutionarily conserved autoinhibitory molecular switch in ELMO proteins regulates Rac signaling. Curr. Biol. 20, 2021–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Patel M., Pelletier A., and Côté J. F. (2011) Opening up on ELMO regulation: new insights into the control of Rac signaling by the DOCK180/ELMO complex. Small GTPases 2, 268–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hamoud N., Tran V., Croteau L. P., Kania A., and Côté J. F. (2014) G-protein coupled receptor BAI3 promotes myoblast fusion in vertebrates. Proc. Natl. Acad. Sci. U.S.A. 111, 3745–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Park D., Tosello-Trampont A. C., Elliott M. R., Lu M., Haney L. B., Ma Z., Klibanov A. L., Mandell J. W., and Ravichandran K. S. (2007) BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450, 430–434 [DOI] [PubMed] [Google Scholar]
- 7. Li H., Yang L., Fu H., Yan J., Wang Y., Guo H., Hao X., Xu X., Jin T., and Zhang N. (2013) Association between Gαi2 and ELMO1/Dock180 connects chemokine signalling with Rac activation and metastasis. Nat. Commun. 4, 1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Katoh H., and Negishi M. (2003) RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature 424, 461–464 [DOI] [PubMed] [Google Scholar]
- 9. Fritsch R., de Krijger I., Fritsch K., George R., Reason B., Kumar M. S., Diefenbacher M., Stamp G., and Downward J. (2013) RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell 153, 1050–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abu-Thuraia A., Gauthier R., Chidiac R., Fukui Y., Screaton R. A., Gratton J. P., and Côté J. F. (2015) Axl phosphorylates elmo scaffold proteins to promote rac activation and cell invasion. Mol. Cell. Biol. 35, 76–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schäker K., Bartsch S., Patry C., Stoll S. J., Hillebrands J. L., Wieland T., and Kroll J. (2015) The bipartite rac1 guanine nucleotide exchange factor engulfment and cell motility 1/dedicator of cytokinesis 180 (elmo1/dock180) protects endothelial cells from apoptosis in blood vessel development. J. Biol. Chem. 290, 6408–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thorens B., and Mueckler M. (2010) Glucose transporters in the 21st Century. Am. J. Physiol. Endocrinol. Metab. 298, E141–E145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adekola K., Rosen S. T., and Shanmugam M. (2012) Glucose transporters in cancer metabolism. Curr. Opin. Oncol. 24, 650–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Leto D., and Saltiel A. R. (2012) Regulation of glucose transport by insulin: traffic control of GLUT4. Nat. Rev. Mol. Cell Biol. 13, 383–396 [DOI] [PubMed] [Google Scholar]
- 15. Graham T. E., and Kahn B. B. (2007) Tissue-specific alterations of glucose transport and molecular mechanisms of intertissue communication in obesity and type 2 diabetes. Horm. Metab. Res. 39, 717–721 [DOI] [PubMed] [Google Scholar]
- 16. Shulman G. I. (2000) Cellular mechanisms of insulin resistance. J. Clin. Invest. 106, 171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nandi A., Kitamura Y., Kahn C. R., and Accili D. (2004) Mouse models of insulin resistance. Physiol. Rev. 84, 623–647 [DOI] [PubMed] [Google Scholar]
- 18. Klip A., Sun Y., Chiu T. T., and Foley K. P. (2014) Signal transduction meets vesicle traffic: the software and hardware of GLUT4 translocation. Am. J. Physiol. Cell Physiol. 306, C879–C886 [DOI] [PubMed] [Google Scholar]
- 19. Ding J., and Du K. (2009) ClipR-59 interacts with Akt and regulates Akt cellular compartmentalization. Mol. Cell. Biol. 29, 1459–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun Y., Ren W., Côté J. F., Hinds P. W., Hu X., and Du K. (2015) ClipR-59 interacts with Elmo2 and modulates myoblast fusion. J. Biol. Chem. 290, 6130–6140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ren W., Cheema S., and Du K. (2012) The association of ClipR-59 protein with AS160 modulates AS160 protein phosphorylation and adipocyte Glut4 protein membrane translocation. J. Biol. Chem. 287, 26890–26900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schmelzle K., Kane S., Gridley S., Lienhard G. E., and White F. M. (2006) Temporal dynamics of tyrosine phosphorylation in insulin signaling. Diabetes 55, 2171–2179 [DOI] [PubMed] [Google Scholar]
- 23. Sakamoto K., and Holman G. D. (2008) Emerging role for AS160/TBC1D4 and TBC1D1 in the regulation of GLUT4 traffic. Am. J. Physiol. Endocrinol. Metab. 295, E29–E37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chiu T. T., Jensen T. E., Sylow L., Richter E. A., and Klip A. (2011) Rac1 signalling towards GLUT4/glucose uptake in skeletal muscle. Cell. Signal. 23, 1546–1554 [DOI] [PubMed] [Google Scholar]
- 25. Ishikura S., Koshkina A., and Klip A. (2008) Small G proteins in insulin action: Rab and Rho families at the crossroads of signal transduction and GLUT4 vesicle traffic. Acta Physiol. 192, 61–74 [DOI] [PubMed] [Google Scholar]
- 26. Côté J. F., and Vuori K. (2007) GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 17, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Buchsbaum R. J., Connolly B. A., and Feig L. A. (2002) Interaction of Rac exchange factors Tiam1 and Ras-GRF1 with a scaffold for the p38 mitogen-activated protein kinase cascade. Mol. Cell Biol. 22, 4073–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gumienny T. L., Brugnera E., Tosello-Trampont A. C., Kinchen J. M., Haney L. B., Nishiwaki K., Walk S. F., Nemergut M. E., Macara I. G., Francis R., Schedl T., Qin Y., Van Aelst L., Hengartner M. O., and Ravichandran K. S. (2001) CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107, 27–41 [DOI] [PubMed] [Google Scholar]
- 29. Sun L., Liu O., Desai J., Karbassi F., Sylvain M. A., Shi A., Zhou Z., Rocheleau C. E., and Grant B. D. (2012) CED-10/Rac1 regulates endocytic recycling through the RAB-5 GAP TBC-2. PLoS Genet. 8, e1002785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Belman J. P., Habtemichael E. N., and Bogan J. S. (2014) A proteolytic pathway that controls glucose uptake in fat and muscle. Rev. Endocr. Metab. Disord. 15, 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brugnera E., Haney L., Grimsley C., Lu M., Walk S. F., Tosello-Trampont A. C., Macara I. G., Madhani H., Fink G. R., and Ravichandran K. S. (2002) Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 4, 574–582 [DOI] [PubMed] [Google Scholar]
- 32. Laurin M., Fradet N., Blangy A., Hall A., Vuori K., and Côté J. F. (2008) The atypical Rac activator Dock180 (Dock1) regulates myoblast fusion in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 15446–15451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Komander D., Patel M., Laurin M., Fradet N., Pelletier A., Barford D., and Côté J. F. (2008) An α-helical extension of the ELMO1 pleckstrin homology domain mediates direct interaction to DOCK180 and is critical in Rac signaling. Mol. Biol. Cell 19, 4837–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kato S., and Du K. (2007) TRB3 modulates C2C12 differentiation by interfering with Akt activation. Biochem. Biophys. Res. Commun. 353, 933–938 [DOI] [PubMed] [Google Scholar]