Abstract
Soluble forms of the IL-6 receptor (sIL-6R) bind to the cytokine IL-6 with similar affinity as the membrane-bound IL-6R. IL-6·sIL-6R complexes initiate IL-6 trans-signaling via activation of the ubiquitously expressed membrane-bound β-receptor glycoprotein 130 (gp130). Inhibition of IL-6 trans-signaling has been shown to be favorable in numerous inflammatory diseases. Furthermore, different soluble forms of gp130 (sgp130) exist that, together with the sIL-6R, are thought to form a buffer for IL-6 in the blood. However, a functional role for the different sgp130 forms has not been described to date. Here we demonstrate that the metalloproteases ADAM10 and ADAM17 can produce sgp130 by ectodomain shedding of gp130, even though this mechanism only accounts for a minor proportion of sgp130 in the circulation. We further show that full-length sgp130 and the shorter forms sgp130-rheumatoid arthritis-associated peptide (RAPS) and sgp130-E10 are differentially expressed in a cell type- specific manner. Remarkably, full-length sgp130 is expressed by monocytes, but this expression is completely lost during differentiation into macrophages in vitro. Using genetically engineered murine pre-B cells that secrete different forms of sgp130, we found that these secreted sgp130 proteins are able to prevent trans-signaling-driven cell proliferation of the secreting cells, whereas conditioned supernatant from these cells failed to block IL-6 trans-signaling in other cells. Thus, our data suggest that the different sgp130 forms are released from cells into their immediate surroundings and appear to form cell-associated gradients to modulate their own susceptibility for IL-6 trans-signaling.
Keywords: interleukin 6 (IL-6), interleukin 6 receptor (IL6R), Janus kinase (JAK), proteolysis, STAT3, gp130, sgp130, trans-signaling
Introduction
IL-6 is a pleiotropic cytokine with pro- and anti-inflammatory functions (1–4). Classic signaling via the membrane-bound IL-6R3 accounts for the regenerative properties of IL-6; trans-signaling via sIL-6R is rather pro-inflammatory (2, 4). In both cases, IL-6 binds initially to the (s)IL-6R, and the IL-6·(s)IL-6R complex recruits two signal-transducing gp130 β-receptors. This final IL-6·(s)IL-6R/gp130 complex formation initiates the activation of intracellular signaling pathways, among them Jak/STAT, PI3K, MAPK, and Src/Yes-associated protein (YAP) (5).
Soluble forms of gp130 (sgp130) exist in human serum at concentrations of up to 400 ng/ml (6) and have been shown to act as natural inhibitors of IL-6 trans-signaling (7), although, at high concentrations, they can interfere with classic signaling as well (8). Interestingly, specific inhibition of trans-signaling with an Fc-dimerized version of sgp130 (sgp130Fc) has been shown to be beneficial in numerous inflammatory diseases and cancer (9, 10). sgp130Fc is currently in clinical development as a drug candidate to treat inflammatory bowel diseases (11).
gp130 is a transmembrane protein with three N-terminal Ig-like domains followed by three fibronectin-type III domains. sgp130 forms with molecular weights of 50, 90, and 110 kDa have been detected in human body fluids (6, 12, 13). Where and how these three proteins act mechanistically has not been explored in detail. The different forms of sgp130 most likely derive from differentially processing of the gp130 mRNA, although limited proteolysis of the membrane-bound gp130 is another possible mechanism for sgp130 generation (14). The smallest variant, comprising domains 1–3 of full-length gp130, is called sgp130-RAPS and corresponds to the 50-kDa variant (12). Because of a unique C terminus of sgp130-RAPS, the occurrence of the protein in vivo could be verified using monoclonal antibodies recognizing this C terminus (12). sgp130-E10, which is generated by alternative polyadenylation through the use of an intronic polyadenylation site, contains domains 1–4 and has also been shown to exist in human serum with the help of a selective monoclonal antibody, although it only accounts for 1–2% of total sgp130 (15). The identities of the 90- and 110-kDa variants are still unclear. Two different variants of full-length sgp130 have been described that contain all six extracellular domains of gp130 and would give rise to a soluble protein within this range (16, 17). However, sgp130 originating from limited proteolysis would most likely also contain all six extracellular domains and thus could be one of the described isoforms. Although recombinant proteins of the different sgp130 variants have been shown to inhibit IL-6 trans-signaling with varying efficacy, the expression and functions of these variants are not clear.
In this study, we found that limited proteolysis of gp130 is only a minor event and does not account for the majority of sgp130. We further show that primary human immune cells have different expression profiles of full-length sgp130, sgp130-E10, and sgp130-RAPS. Intriguingly, monocytes express high amounts of full-length sgp130, and this expression is completely lost when monocytes are differentiated into macrophages in vitro. Finally, we developed a novel in vitro assay to compare the ability of the different sgp130 variants to inhibit IL-6 trans-signaling in a cell-autonomous manner.
Results
gp130 Can Be Shed by ADAM10 and ADAM17, albeit with Low Efficiency
Limited proteolysis of the IL-6R by the metalloproteases ADAM10 and ADAM17 has been studied in much detail (18–21). Far less attention has been paid to a possible role for metalloproteases in the generation of sgp130, probably because this is generally believed to solely result from alternative mRNA splicing (2). However, recent studies showed that metalloproteases participate in the generation of soluble forms of the leukemia inhibitory factor receptor (LIFR) (22) and of the IL-27R WSX-1 (23), two other β receptors that are used by IL-6 family of cytokines (24).
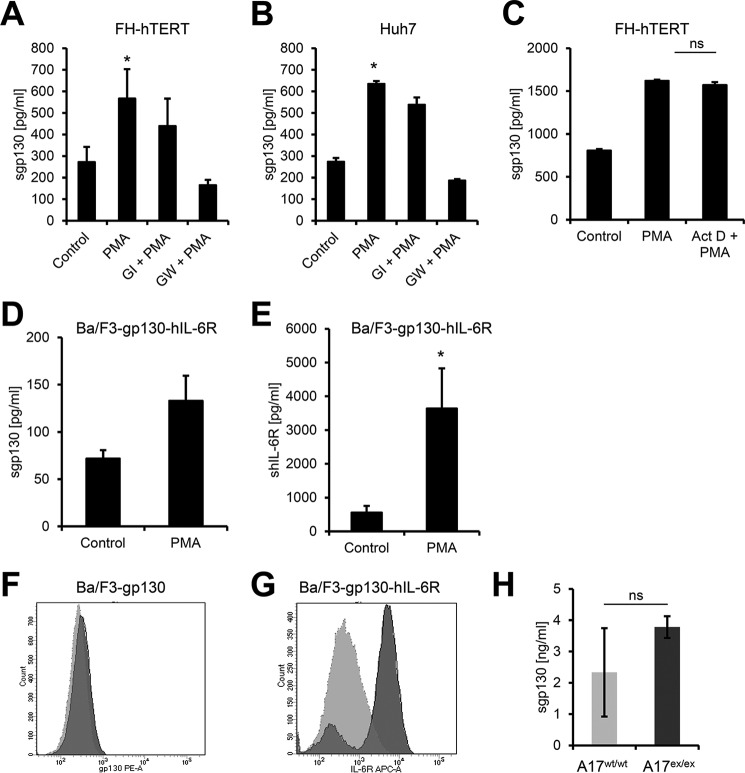
To address this question, we stimulated FH-hTERT cells with the phorbol ester PMA, which activates protein kinase C and is the strongest known activator of ADAM17, and measured sgp130 formation in the supernatant of the cells via ELISA. FH-hTERT cells are immortalized fetal hepatocytes that express gp130 endogenously. As shown in Fig. 1A, stimulation with 100 nm PMA led to a significant increase in sgp130. Although pretreatment of the cells with the ADAM10-specific hydroxamate inhibitor GI did not significantly alter sgp130 formation, chemical inhibition of both ADAM10 and ADAM17 with the double inhibitor GW reduced sgp130 levels back to baseline. We verified this observation with a second cell line that expresses gp130 endogenously and found a comparable PMA-induced increase in sgp130 released from Huh7 cells, a hepatocarcinoma cell line (Fig. 1B). To exclude a contribution of alternative splicing in these assays, we pretreated FH-hTERT cells with the transcription inhibitor actinomycin D and found no difference in gp130 shedding when the cells were stimulated with PMA (Fig. 1C).
FIGURE 1.
The protease ADAM17 sheds membrane-bound gp130 with low efficiency. A and B, FH-hTERT (A) and Huh7 (B) cells were stimulated with 100 nm PMA for 2 h or treated with DMSO as a control. Cells were pretreated with 3 μm GI (a specific ADAM10 inhibitor) or 3 μm GW (a specific combined ADAM10/ADAM17 inhibitor) 30 min prior to PMA stimulation as indicated. The amount of sgp130 in the cell culture supernatant was analyzed by ELISA. C, FH-hTERT cells were either stimulated with 100 nm PMA for 2 h or treated with DMSO as a control. Where indicated, cells were pretreated with 1 μg/ml actinomycin D (Act D) 30 min prior to stimulation with PMA. The amount of sgp130 in the cell culture supernatant was analyzed by ELISA. D and E, Ba/F3-gp130-hIL-6R cells were stimulated with 100 nm PMA for 2 h or treated with DMSO as a control. The amount of sgp130 (D) or shIL-6R (E) in the cell culture supernatant was analyzed via specific ELISAs. F, analysis of proteolytic cleavage of gp130 in DMSO- (dark gray) or PMA-treated (light gray) Ba/F3-gp130 cells by flow cytometry. G, analysis of proteolytic cleavage of IL-6R in DMSO- (dark gray) or PMA-treated (light gray) Ba/F3-gp130-IL6R cells by flow cytometry. H, analysis of endogenous sgp130 in wild-type or ADAM17ex/ex mice (n = 3 each) by murine sgp130-ELISA. All ELISA data shown are the mean ± S.E. derived from three independent experiments. Flow cytometry data show one representative experiment of three independent experiments with similar outcomes. *, p < 0.05; ns, not significant.
However, these experiments only show that ADAM17 is, in principle, able to shed gp130 but not to which extent cellular gp130 is proteolytically released. To investigate this, we made use of Ba/F3-gp130-hIL-6R cells. Ba/F3-gp130-hIL-6R cells are stably transduced with gp130 and the human (h) IL-6R, a well established substrate of ADAM10 and ADAM17 (20, 21), but express neither protein endogenously. Stimulation with PMA resulted again in sgp130 formation (Fig. 1D), but the amounts of sIL-6R were much higher than those of sgp130 (Fig. 1E). Accordingly, we detected no reduction of the cell surface levels of gp130 by flow cytometry (Fig. 1F), whereas IL-6R surface levels were strongly reduced after PMA treatment (Fig. 1G). Furthermore, we found that endogenous sgp130 serum levels in hypomorphic ADAM17ex/ex mice, which have no detectable ADAM17 activity (25), were similar to those of wild-type mice (Fig. 1H).
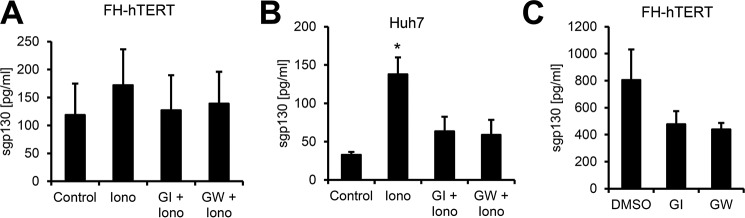
Besides ADAM17, the IL-6R can also be shed by ADAM10, and the ionophore ionomycin is a strong activator of this process (21, 26). To investigate a possible role for ADAM10 in sgp130 generation, we stimulated FH-hTERT and Huh7 cells with 1 μm ionomycin. Although we could not detect a significant increase in sgp130 released from FH-hTERT cells (Fig. 2A), stimulation of Huh7 cells with ionomycin led to a significant increase in sgp130 levels in the supernatant (Fig. 2B). Pretreatment of the cells with GI or GW reduced sgp130 to the level of the vehicle-treated controls, demonstrating that ADAM10 was also able to release sgp130 from cells. To elucidate whether ADAM10 or ADAM17 are also able to shed gp130 without any stimulus, we treated FH-hTERT cells with GI, GW, or the vehicle DMSO for 150 min and analyzed supernatants via ELISA. As shown in Fig. 2C, both metalloprotease inhibitors reduced sgp130 levels, which suggests that at least ADAM10 can cleave gp130 constitutively in this cell line.
FIGURE 2.
The protease ADAM10 is an inducible and constitutive sheddase of gp130. A and B, FH-hTERT (A) and Huh7 (B) cells were stimulated with 1 μm ionomycin (Iono) for 1 h or treated with DMSO as a control. Cells were pretreated with 3 μm GI (a specific ADAM10 inhibitor) or 3 μm GW (a specific combined ADAM10/ADAM17 inhibitor) 30 min prior to ionomycin stimulation as indicated. The amount of sgp130 in the cell culture supernatant was analyzed by ELISA. C, FH-hTERT cells were incubated with 3 μm GI, 3 μm GW, or DMSO as a control for 1 h. The amount of sgp130 in the cell culture supernatant was analyzed by ELISA. All ELISA data shown are the mean ± S.E. derived from three independent experiments. *, p < 0.05.
In conclusion, we show that gp130 can be shed by ADAM10 and ADAM17. However, gp130 appears to be a weak protease substrate compared with the IL-6R. At least cleavage by ADAM17 does not appear to play a significant role in sgp130 generation in vivo, as we found unaltered serum levels in ADAM17ex/ex mice.
The Three Different Forms of sgp130 Are Expressed in a Cell Type-specific Manner
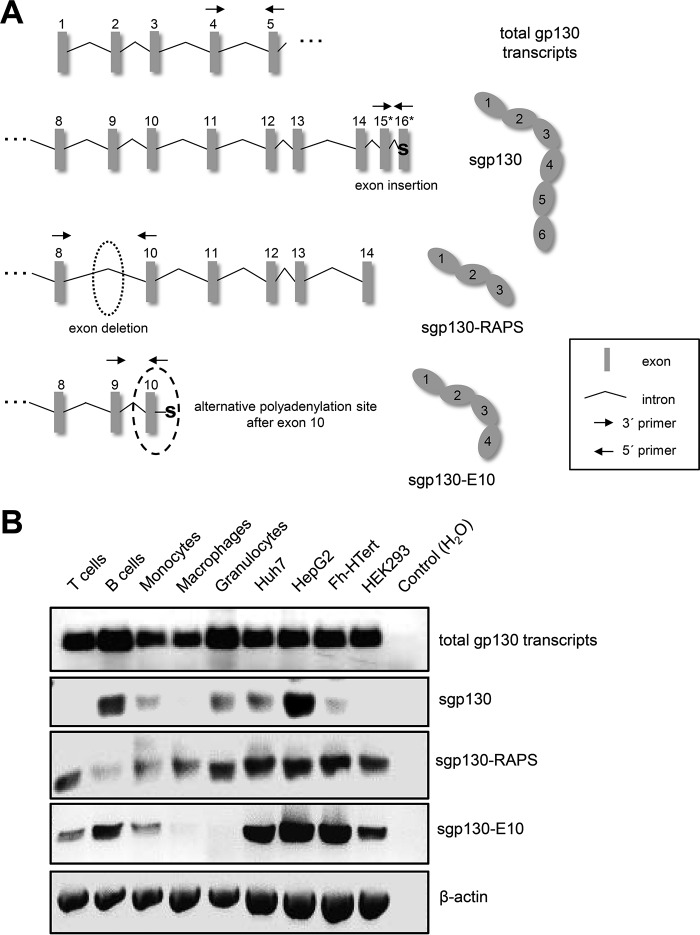
Having excluded limited proteolysis as the major mechanism of sgp130 generation, we sought to analyze alternative splicing of the gp130 mRNA in greater detail. To date, three different forms of sgp130 have been described (reviewed in Ref. 2). We developed an RT-PCR based assay that is able to distinguish between the mRNAs of the three soluble and the membrane-bound form of sgp130 (Fig. 3A). As an internal control, we designed a primer pair that binds within exons 4 and 5 and amplifies all gp130 transcript variants irrespective of their C terminus (Fig. 3A). The so-called full-length form of sgp130 contains all six extracellular domains of gp130 and is generated through incorporation of a novel exon 16* after the regular exon 15 (16). We designed an oligonucleotide pair that binds in exons 15 and 16* so that only the full-length form of sgp130 would be amplified but not any other variant (Fig. 3A). To detect sgp130-E10, which is generated through the usage of an alternative polyadenylation site within intron 10, we used oligonucleotides that bind in exon 9 and intron 10 (Fig. 3A, a strategy adopted from Ref. 15). sgp130-RAPS, which contains domains D1-D3 of gp130, is generated via deletion of exon 9 and a premature stop codon because of the resulting frameshift (12). Thus, we designed oligonucleotides spanning exons 8–10 to specifically detect mRNA encoding sgp130-RAPS (Fig. 3A). Importantly, this strategy excludes detection of genomic DNA or the unprocessed primary RNA transcript of gp130.
FIGURE 3.
RNA expression of sgp130 variants. A, schematic of the exon-intron structure of the 5′ beginning of all gp130 transcripts and the individual 3′ ends of the three soluble forms of gp130, adapted from Ref. 40. Full-length sgp130 results from an exon insertion, whereas sgp130-RAPS is produced after an exon deletion. sgp130-E10 is produced when an alternative polyadenylation site after exon 10 is used. Binding sites of oligonucleotides used to specifically amplify the different transcripts are indicated by arrows. B, RNA expression of the total gp130 transcripts and the three soluble forms of gp130 was analyzed in different human primary cell types (T cells, B cells, monocytes, macrophages, and granulocytes) as well as in human cell lines (Huh7, HepG2, Fh-hTERT, and HEK293) as indicated. Amplification of β-actin was used as a control. One representative experiment of three independent experiments with similar outcomes is shown.
In the next step, we analyzed the expression of the three different sgp130 forms in primary human leukocytes and human liver cell lines. To this end, we obtained human peripheral blood mononuclear cells, followed by an isolation of different subtypes of leukocytes via magnetic cell separation. In addition, granulocytes were isolated via two different gradients. Analysis of total gp130 transcripts showed overall comparable levels, although the amount appeared to be slightly increased in B cells and granulocytes and slightly decreased in monocytes and macrophages (Fig. 3B). We observed expression of full-length sgp130 only in B cells, monocytes, and granulocytes but not in T cells or macrophages (Fig. 3B). Expression of sgp130-E10 was detected in T cells, B cells, and monocytes but not in macrophages or granulocytes, whereas sgp130-RAPS was expressed in all five leukocyte populations examined (Fig. 3B). All three forms of sgp130 were expressed in the three different hepatic cell lines Huh7, HepG2, and FH-hTERT (Fig. 3B). As a control, we analyzed sgp130 expression in HEK293 cells, a commonly used cell line originating from human embryonic kidney. HEK293 cells expressed sgp130-E10 and sgp130-RAPS but not the full-length form of sgp130 (Fig. 3B).
In conclusion, we observed a distinct expression pattern of the three different forms of sgp130 in primary human leukocytes but not in human hepatocyte-derived cell lines. The full-length form of sgp130 (and, to a lesser extent, also sgp130-E10) appears to be expressed in a cell type-specific manner, whereas sgp130-RAPS was expressed in all cells examined.
Down-regulation of Full-length sgp130 during Monocyte-to-Macrophage Transition Occurs Only in Primary Cells
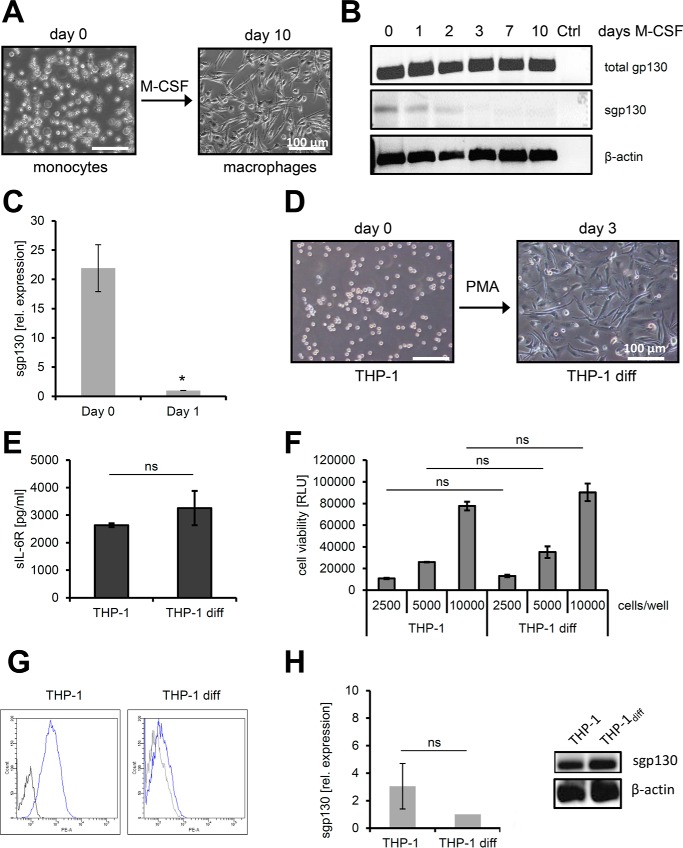
Circulating monocytes migrate to sites of inflammation to differentiate into macrophages, which is an important step in the resolution of inflammation (27). Therefore, we decided to investigate the loss of full-length sgp130 mRNA expression during the monocyte-to-macrophage transition (Fig. 3B) in more detail. To this end, we isolated monocytes from human peripheral blood and cultured them for up to 10 days in the presence of 100 ng/ml M-CSF, thereby inducing differentiation into macrophages (Fig. 4A). It has to be noted, however, that macrophages differentiated via M-CSF treatment may not fully resemble macrophages generated during inflammation in vivo. Semiquantitative analysis of mRNA by RT-PCR revealed sgp130 expression on day 0 of the differentiation process, which decreased during the first 2 days and was completely absent after 3, 7, and 10 days of M-CSF treatment (Fig. 4B). Quantitative (qPCR) analysis demonstrated a 21.9 ± 4.0-fold significantly higher expression of sgp130 mRNA on day 0 compared with day 1 of the differentiation process, underlining the rapid change of sgp130 mRNA expression during the monocyte-to-macrophage transition (Fig. 4C).
FIGURE 4.
Down-regulation of full-length sgp130 during monocyte-to-macrophage transition. A, differentiation of human monocytes to macrophages was induced by addition of 100 ng/ml M-CSF for 10 days and verified by bright-field microscopy. B, semiquantitative analysis of sgp130 RNA expression in human macrophages after different time points of differentiation by RT-PCR. Total gp130 transcripts and the specific full-length variant of sgp130 were analyzed. Amplification of β-actin was used as a control (Ctrl). C, quantitative analysis of sgp130 RNA expression in human monocytes before and after 1 day of differentiation to macrophages by qPCR. Expression was normalized to GAPDH expression (mean ± S.D., n = 3). D, differentiation of the monocytic cell line THP-1 to macrophages (THP-1 diff) was induced by addition of 100 nm PMA for 3 days and verified by bright-field microscopy. E, release of sIL-6R over 72 h by monocytic THP-1 and differentiated THP-1 cells was analyzed by ELISA (mean ± S.D., n = 3). F, cell viability of 2500, 5000, or 10,000 THP-1 cells compared with the same number of differentiated THP-1 cells was analyzed as described under “Experimental Procedures” (mean ± S.D., n = 3). RLU, relative light unit. G, cell surface expression of gp130 was analyzed on THP-1 cells and on differentiated macrophages on day 3. gp130 staining is shown in blue, and control staining to exclude unspecific antibody binding is shown in black. H, analysis of sgp130 RNA expression in THP-1 cells before and differentiation (diff) by qPCR, where expression was normalized to GAPDH (mean ± S.D., n = 3), and by RT-PCR, where β-actin served as a control. If not indicated otherwise, one representative experiment of three independent experiments with similar outcomes is shown. *, p < 0.05; ns, not significant.
We sought to further analyze this process using a human monocytic cell line and stimulated THP-1 cells with 100 nm PMA, which is a well known stimulus to differentiate THP-1 cells into macrophages (Fig. 4D) (28). Both THP-1 cells and THP-1-derived differentiated macrophages released comparable amounts of sIL-6R, indicating that IL-6 signaling should not influence gp130 analysis (Fig. 4E). Furthermore, cell viability did not differ between the two cell types (Fig. 4F). Interestingly, the amount of gp130 at the cell surface of the differentiated THP-1-derived macrophages was reduced compared with the undifferentiated THP-1 cells, as judged by flow cytometry (Fig. 4G), but no significant difference in sgp130 mRNA expression was detected by qPCR or RT-PCR, suggesting different regulation of these two gp130 mRNAs (Fig. 4H). In conclusion, we show that the reduction of sgp130 mRNA during the monocyte-to-macrophage-differentiation induced by M-CSF is a rapid event in primary human cells but is not reproduced in the human monocytic cancer cell line THP-1.
Cell-autonomous Secretion of sgp130 Is Sufficient to Block Proliferation of Ba/F3-gp130 Cells
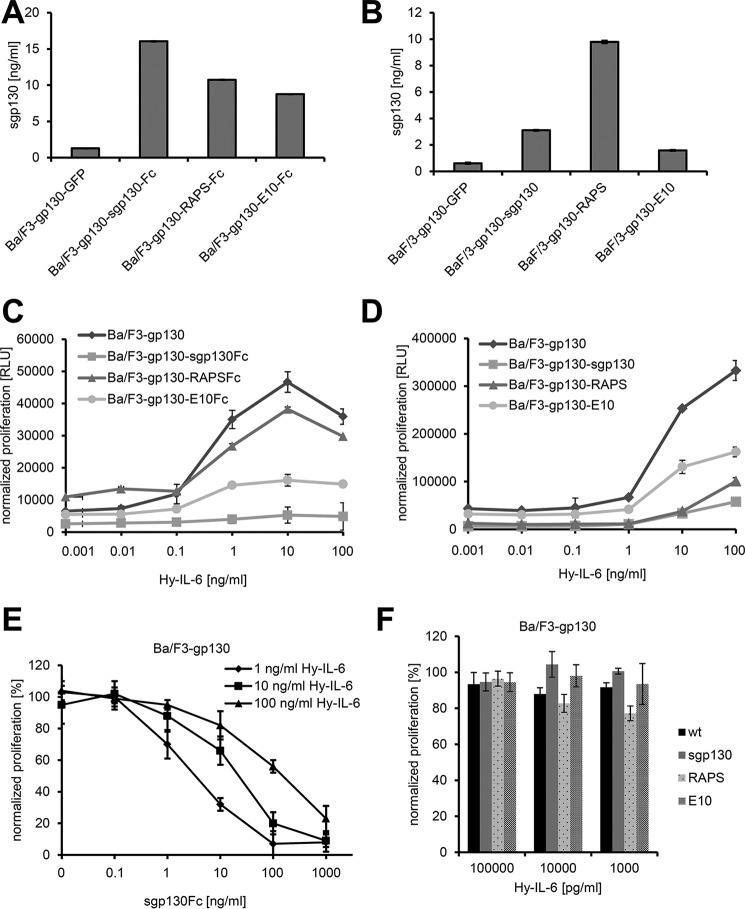
It is well established that exogenous addition of recombinant sgp130 or sgp130Fc is able to block proliferation of Ba/F3-gp130 that is induced by IL-6 trans-signaling (7, 8). However, our data with primary monocytes show that cells can secrete sgp130, possibly to protect themselves against trans-signaling. “Protection” as used here does not imply that IL-6 trans-signaling harms cells but that signaling events triggered by trans-signaling may be undesirable in a given context. To study this phenomenon in a controlled in vitro setting, we stably transduced Ba/F3-gp130 pre-B-cells with cDNAs encoding sgp130(Fc) (termed Ba/F3-gp130-sgp130(Fc) hereafter), sgp130-RAPS(Fc) (termed Ba/F3-gp130-RAPS(Fc) hereafter), or sgp130-E10(Fc) (termed Ba/F3-gp130-E10(Fc) hereafter). These cells express membrane-bound gp130 and therefore respond to stimulation by trans-signaling with hyper-IL-6 and furthermore secrete the different isoforms of sgp130 or sgp130Fc, which enabled comprehensive analysis of the inhibitory capacity of the individual sgp130(Fc) isoforms when secreted from cells. We first analyzed sgp130 secretion by ELISA (Fig. 5, A and B). All generated cell lines indeed secreted higher sgp130 amounts compared with the GFP-transfected controls (Fig. 5, A and B). However, the amounts of the three sgp130 variants differed between the individual cell lines. This might be due to the fact that all transduced cell lines are derived from individual transduced clones but also reflects that the sgp130 isoforms have different stabilities. Among the Fc-tagged variants, the smallest amount of sgp130 was detected in the supernatant of Ba/F3-gp130-E10Fc cells (Fig. 5A), which confirms previous results showing that the sgp130-E10 variant is rather unstable (15). The same was observed for the untagged protein variants, where similarly Ba/F3-gp130-E10 cells secreted the lowest amounts of sgp130 (Fig. 5B).
FIGURE 5.
Implications of the secretion of various sgp130 forms for cell proliferation. A and B, equal amounts of Ba/F3-gp130-GFP, Ba/F3-gp130-sgp130(Fc), Ba/F3-gp130-sgp130-RAPS(Fc), and Ba/F3-gp130-sgp130-E10(Fc) cells were incubated for 24 h. Supernatants were harvested, and the amount of sgp130 in the cell culture supernatant was quantified via ELISA (mean ± S.D., n = 3). C and D, Ba/F3-gp130 cells stably transduced with various forms of sgp130 were stimulated with increasing amounts of hyper-IL-6 (0.001–100 ng/ml), and cell proliferation was measured after 48 h. Data shown are the mean ± S.D. (n = 3 biological replicates) from one representative experiment of three performed with similar outcomes. RLU, relative light unit. E, Ba/F3-gp130 cells were incubated with either 1, 10, or 100 ng/ml hyper-IL-6 and increasing amounts (0–1000 ng/ml) of recombinant sgp130Fc, and cell proliferation was determined after 48 h. F, Ba/F3-gp130 cells stably transduced with sgp130, sgp130-RAPS, or sgp130-E10 as well as Ba/F3-gp130 cells (wild-type) were grown for 48 h, and their supernatants were used for 48-h cultivation of Ba/F3-gp130 cells in the presence of various amounts of hyper-IL-6. Cell proliferation was measured after this 48-h incubation with the supernatants. Data are the mean ± S.D. of three independent experiments (n = 3 biological replicates each).
Ba/F3-gp130 cells, which served as negative control, proliferated in a dose-dependent manner when stimulated with hyper-IL-6 (Fig. 5C). Ba/F3-gp130-RAPSFc cells showed a slightly reduced proliferation compared with Ba/F3-gp130 cells (Fig. 5C). In contrast, Ba/F3-gp130-E10Fc cells proliferated less when stimulated with the same amounts of hyper-IL-6, and Ba/F3-gp130-sgp130Fc cells did not proliferate at all at hyper-IL-6 concentrations up to 100 ng/ml (Fig. 5C).
To not solely rely on the artificially dimerized Fc-tagged sgp130 proteins, we performed the same experiment with the Ba/F3-gp130 cells expressing the monomeric sgp130 forms, which had been shown to inhibit IL-6 trans-signaling, albeit with reduced efficacy (7). Here Ba/F3-gp130-RAPS cells proliferated less than Ba/F3-gp130-E10 cells (Fig. 5D), and Ba/F3-gp130-sgp130 cells, which express the longest isoform of sgp130, showed the strongest reduction of proliferation in comparison with the other three cell lines (Fig. 5D). Of note, all cell lines proliferated at high concentrations of 100 ng/ml hyper-IL-6, indicating that monomeric sgp130 is less efficient to block IL-6 trans-signaling compared with dimerized sgp130Fc, as has been shown before with recombinant proteins (7). Although we were able to block hyper-IL-6-induced proliferation of Ba/F3-gp130 cells with recombinant sgp130Fc in a dose-dependent manner (Fig. 5E), interestingly, we observed no inhibition of IL-6 trans-signaling when we transferred supernatant of Ba/F3-gp130-sgp130, Ba/F3-gp130-RAPS, and Ba/F3-gp130-E10 cells to Ba/F3-gp130 cells and analyzed proliferation in response to different concentrations of Hyper-IL-6, suggesting that secreted sgp130 forms some kind of buffer in close proximity to the cell membrane, which protects against IL-6 trans-signaling (Fig. 5F) but does not reach sufficient concentration in the whole supernatant.
In conclusion, we show that secreted sgp130 from Ba/F3-gp130 cells is sufficient to block hyper-IL-6-mediated proliferation of these cells. The three different isoforms vary in their efficacy to block IL-6 trans-signaling, with full-length sgp130 being the most potent inhibitor.
Discussion
Overshooting activities of IL-6 are known to occur in all inflammatory human diseases. Interestingly, IL-6 signals either via the membrane-bound or the soluble IL-6R, and these two modes of signaling have diametrical opposite effects. Inhibition of trans-signaling has been shown to be sufficient to block disease progression, leading to the conclusion that the trans-signaling branch of IL-6 signaling is mainly responsible for the detrimental chronic pro-inflammatory activities of IL-6 (5, 9).
These conclusions stem from the observation that sgp130Fc, which is an Fc-dimerized version of sgp130, specifically blocks IL-6 trans-signaling and is highly efficacious in diverse models of chronic inflammation (3, 5, 7). Interestingly, dimerization increased the efficacy to block signaling via IL-6/sIL-6R by a factor of 10–100 compared with monomeric sgp130 (7), which might be explained by the homodimeric nature of gp130 in the IL-6 signaling complex. Furthermore, gp130 exists as a dimeric preformed cytokine receptor complex even when no IL-6 is present (29). Classic signaling can only be influenced by sgp130 under certain conditions when sIL-6R is present in molar excess over IL-6 (8). However, correct dosing of sgp130Fc ensures exclusive blockade of IL-6 trans-signaling (8).
At least three different natural soluble isoforms of sgp130 have been described in human body fluids, which account for serum levels of up to 400 ng/ml (6). These isoforms have molecular masses of 110, 90, and 50 kDa (12, 16, 17). The latter one corresponds to sgp130-RAPS, which contains the extracellular domains D1-D3 and a unique C terminus that arises from alternative splicing of the gp130 mRNA (12). The 90-kDa isoform of sgp130 could represent sgp130-E10, which is generated through the usage of an alternative intronic polyadenylation sequence (15). sgp130-E10 contains domains D1-D4, which is again followed by an unique C-terminal peptide (15). The nature of the 110-kDa isoform has not been addressed biochemically, but it could represent the full-length isoform of sgp130, which comprises all extracellular domains. Like sgp130-RAPS, the full-length form of sgp130 arises from alternative mRNA splicing (16, 17). As shown in this study, ectodomain cleavage of membrane-bound gp130, which would result in a full-length sgp130 protein, is not a likely mechanism, as gp130 is only a weak substrate for the metalloproteases ADAM10 and ADAM17. It is possible, however, that gp130 can be cleaved more efficiently by other proteases.
The biological function of the endogenous sgp130 proteins is not known. When seen in combination with sIL-6R, they could likely represent a buffer system to prevent that local peaks of IL-6 have systemic effects. In this context, IL-6 binds to sIL-6R with 1 mm affinity, and this IL-6·sIL-6R complex binds to sgp130 with 10 pm affinity and is immediately neutralized rather than being able to activate a target cell because the IL-6·sIL-6R·sgp130 complexes form quickly and are biologically inactive (3, 5, 30). The capacity of this buffer is controlled by the amount of sIL-6R, as sgp130 is present in molar excess over sIL-6R (5). Indeed, individuals who are homozygous for the SNP rs2228145 have elevated sIL-6R levels and reduced C-reactive protein levels, which is accompanied with a slightly reduced risk of suffering from coronary heart diseases (31–33).
However, it has not been addressed why three different isoforms are present and what their biological impact in terms of inhibiting IL-6 signaling might be. In this study, we show for the first time a cell type-specific expression pattern of the three sgp130 isoforms in primary human immune cells and different established human cell lines. Most interestingly, although monocytes express full-length sgp130, this expression is completely lost during their M-CSF-induced differentiation into macrophages. It is tempting to speculate that the secreted sgp130 isoforms form an endogenous, cell-specific, and cell-autonomous protective system that can block activation of immune cells via IL-6 trans-signaling. Furthermore, through the expression of different sgp130 isoforms, cells can fine-tune this protection in terms of efficacy and stability.
Indeed, our novel Ba/F3-gp130 cell-based assay for sgp130 variant efficacy revealed different capacities of the three sgp130 isoforms to block IL-6 trans-signaling. Cells that were genetically engineered to secrete full-length sgp130Fc did not proliferate in response to hyper-IL-6, and even secretion of the monomeric sgp130 largely prevented hyper-IL-6-mediated proliferation. Secretion of sgp130-E10 and sgp130-RAPS also showed an inhibitory function, albeit less efficient compared with the full-length sgp130. Importantly, this protective effect was absent when conditioned supernatant was transferred to normal Ba/F3-gp130 cells, indicating that the amount of sgp130 was insufficient to systematically block IL-6 trans-signaling and that the cell-mediated secretion was important for the protective function. It can be envisaged that the secretion of specific sgp130 isoforms leads to a protective gradient in the glycocalyx of individual cells. In summary, we characterize the expression and function of three different endogenous sgp130 isoforms that fine-tune the cellular capacity to prevent activation by IL-6 trans-signaling.
Experimental Procedures
Cells and Reagents
HEK293, HepG2, THP-1, and Phoenix cells were obtained from DSMZ GmbH (Braunschweig, Germany), Ba/F3-gp130 cells from Immunex (Seattle, WA) (Gearing et al. (42)), and U-937 cells from the ATCC (Wesel, Germany). FH-hTERT cells were kindly provided by Henning Wege (University Hospital Eppendorf, Hamburg, Germany) (34), and Huh 7 cells were kindly provided by Gerald Rimbach (Institute of Human Nutrition and Food Science, Kiel University, Kiel, Germany).
HEK293, HepG2, THP-1, and Phoenix cells were cultured in high-glucose DMEM (PAA Laboratories, Cölbe, Germany) supplemented with 10% FBS (PAN Biotech) and penicillin (60 mg/liter)/streptomycin (100 mg/liter) (Sigma-Aldrich, Taufkirchen, Germany) under standard conditions (37 °C, 5% CO2, and water-saturated atmosphere). For Ba/F3-gp130 cells, 10 ng/ml recombinant hyper-IL-6 was added for cultivation. Expression and purification of hyper-IL-6 were performed as described previously (35, 36). For Ba/F3-gp130-IL-6R cells (8), 10 ng/ml recombinant IL-6 was added for cultivation, which was produced as described previously (37).
Huh7 cells were cultivated in RPMI 1640 medium (PAA Laboratories) supplemented with 10% FBS and penicillin (60 mg/liter)/streptomycin (100 mg/liter) under standard conditions. FH-hTERT cells were cultivated in high-glucose DMEM containing 10% FBS, penicillin (60 mg/liter)/streptomycin (100 mg/liter), 5 μg/ml insulin (Sigma-Aldrich), and 2.4 μg/ml hydrocortisone (Sigma-Aldrich).
Recombinant sgp130Fc was expressed and purified as described previously (7). For flow cytometry, gp130-phycoerythrin-labeled antibodies (Abcam, Cambridge, United Kingdom) or IL-6R-allophycocyanin-labeled antibody (Biolegend, Fell, Germany) were used. The ADAM10-specific inhibitor GI254023X and the combined ADAM10/ADAM17-specific inhibitor GW280264X were synthesized by Iris Biotech (Marktredwitz, Germany) (38, 39).
Construction of Plasmids Coding for sgp130, sgp130-RAPS, or Agp130-E10 and Fc-tagged Variants Thereof
For cloning sgp130-RAPS and sgp130-E10 into the human expression vector pcDNA3.1, optimized DNA fragments were amplified by using GATCAAGCTTAGAACCATGCTGACACTGCAGACATG as forward and GATCGCGGCCGCTCAAAAGGAGGCAATGTTGTC (RAPS) and GATCGCGGCCGCTCACAGATACAAACCCTGAAAAT (E10) as reverse primers. Cloning of the optimized fragment of sgp130 (16) is described in Ref. 40. Cloning of the Fc-tagged variant of sgp130 was achieved using the specific primers GATCAAGCTTAGAACCATGCTGACACTGCAGACATG (forward) and GATCGCGGCCGCCTCGCCCTGGGCGAACTT (reverse). Cloning of RAPS-Fc is described in Ref. 40) and cloning of sgp130-E10-Fc in Ref. 15. Subcloning into the vector pMOWS-puromycin was achieved by restriction of pcDNA3.1 plasmids using HindIII and NotI, followed by generation of blunt ends by Klenow fragment (Thermo Scientific) and restriction of pMOWS using PmeI (41).
Retroviral Transduction of Ba/F3-gp130 Cells
Retroviral transduction of the murine pre-B cell line Ba/F3-gp130 to obtain stably transfected cells producing the soluble proteins sgp130 (16), sgp130-RAPS, sgp130-E10, sgp130-Fc, sgp130-RAPS-Fc, or sgp130-E10-Fc, was described previously (41). After transduction, cells were resuspended in standard medium including 10 ng/ml hyper-IL-6. After 48 h, 1.5 μg/ml puromycin was added to select transduced clones.
ELISA
The ELISAs for human sgp130 and sIL-6R as well as the murine sgp130 were purchased from R&D Systems and performed according to the instructions of the manufacturer in ELISA plates (F96 Cert. NUNC MaxiSorp, Thermo Fisher Scientific). For all ELISAs, signal detection was performed after addition of the peroxidase substrate BM Blue POD (Sigma-Aldrich) by measurement of absorbance at 450 nm on a plate reader (Tecan Rainbow, Tecan, Crailsheim, Germany).
Cell Viability Assay
To analyze cell viability and proliferation, Ba/F3-gp130 cells stably transduced with sgp130 variants were washed three times with sterile PBS. Cells were resuspended in standard DMEM without hyper-IL-6 and seeded into microtiter plates (5000 cells/well). Hyper-IL-6 was added in various concentrations as indicated. Cells were cultured for 72 h in a final volume of 100 μl. Cell viability was measured using Cell Titer Blue Cell viability assay reagent (Promega, Mannheim, Germany) according to the instructions of the manufacturer. The resulting signal was detected on a Lambda Fluoro320 multiplate reader (excitation, 560 nm; emission, 590 nm; BioTek, Bad Friedrichshall, Germany). Alternatively, supernatants of Ba/F3-gp130 cells stably transduced with various forms of sgp130 were harvested and used as a culture medium for Ba/F3-gp130 cells after supplementation with different amounts of hyper-IL-6. Cell viability was measured as mentioned above. Furthermore, 2500, 5000 or 10,000 THP-1 cells/well (either undifferentiated or differentiated into macrophages) were seeded into 96-well plates, and cell viability was determined 48 h later as described above.
ADAM Protease Shedding Assays
Cells were seeded in 6-well plates and stimulated with either 100 nm phorbol 12-myristate 13-acetate (PMA, Calbiochem/Merck Millipore, Schwalbach, Germany) or 1 μm ionomycin (Sigma-Aldrich) for 2 h in serum-free medium. For protease inhibition studies, GI or GW (3 μm each (38)) were added 30 min prior to stimulation. For transcription inhibition experiments, actinomycin D (1 μg/ml, Sigma-Aldrich) and for translation-inhibition experiments, cycloheximide (100 μg/ml, Sigma-Aldrich) were added 2 h before stimulation. Supernatants were used for ELISA experiments. Cells were analyzed by flow cytometry.
Preparation and Differentiation of Primary Human Cells and Human Cell Lines
For T cell, B cell, and monocyte enrichment from human blood, peripheral blood mononuclear cells were isolated by gradient density centrifugation using Histopaque® 1077 (Sigma-Aldrich). Cells were washed several times using sterile-filtered Dulbecco's PBS (pH 7.2) (Biochrom, Berlin, Germany) including 2 mm EDTA. For preparation of different cell subtypes, cells were magnetically labeled and isolated using the MACS® technology (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the instructions of the manufacturer. Here T cells were labeled with CD3 beads, B cells with CD19 beads, and monocytes with CD14 beads. The purity of the resulting cell suspensions was confirmed by flow cytometry. For differentiation of human monocytes into macrophages, up to 107 cells were seeded in 10-cm plates in 10 ml of RPMI medium containing 10% FBS, 1 mm sodium pyruvate (Life Technologies/Thermo Fisher Scientific), non-essential amino acids (Sigma-Aldrich), 50 μm β-mercaptoethanol (Life Technologies/Thermo Fisher Scientific), penicillin (60 mg/liter)/streptomycin (100 mg/liter), and 10 ng/ml human M-CSF (eBioscience, Frankfurt am Main, Germany) and cultivated under standard conditions for 10 days. Every 3–4 days, 30–50% of the medium was replaced, including supplementation of fresh M-CSF.
Granulocytes were isolated by gradient centrifugation of human blood using 60/70% Percoll (GE Healthcare), followed by a second gradient centrifugation using 65/70/75/80/85% Percoll. Granulocytes were found between the 75% and 80% layer. Ethics approval for this study was obtained from the institutional review board of the Medical Faculty of Kiel University (study D 556/15). For in vitro differentiation of THP-1 and U-937 monocytic cell lines to macrophages, cells were incubated in standard medium including 100 nm PMA for 3 days.
Isolation of RNA and cDNA Synthesis
From all cells, RNA was isolated according to the instructions of the manufacturer using the NucleoSpin® RNA kit (Macherey-Nagel, Düren, Germany). 0.2–2 μg of RNA was used for cDNA synthesis by reverse transcription using random hexamer primers according to the instructions of the manufacturer (Thermo Fisher Scientific).
Analysis of sgp130 Variants by PCR
For analysis of various splice variants of soluble gp130, primers were designed according to Fig. 3A. The sequences were as follows: sgp130 (16), GCAGCATACACAGATGAAGGTG forward primer (FP) and TAAGCTGTAAGGTCCTCGTTGG reverse primer (RP); sgp130-RAPS, TCCACCCGATCTTCATTCACTG (FP) and AAGGAGGCAATGTTATCTTCATAGGT (RP); and sgp130-E10, TCCATCCCATACTCAAGGCTAC (FP) and TCACAGATACAAACCTTGAAAGTCAC (RP). All gp130 transcript variants were detected with the following primers: GAACAGCATCCAGTGTCACC (FP) and CATTTTCTTCCCCTCGTTCAC (RP). The primers for amplification of full-length gp130 were AACACATCTGGCCTAATGTTCC (FP) and TACCACTGCTGTGTCCTTCAGT (RP), and the primers for the IL-6R were GGGACCATGGAGTGGTAGC (FP) and ACTGGTCAGCACGCCTCT (RP). The correct sequences of all PCR products were verified by Sanger sequencing (GATC Biotech AG, Konstanz, Germany). Semiquantitative analysis was performed using 1–2 μl of cDNA and Dream Taq polymerase (Thermo Fisher Scientific) with a starting denaturation of 3 min at 95 °C. Other parameters, annealing temperatures, and elongation times depended on the amplified fragments and were chosen according to the recommendations of the manufacturer. Quantitative analysis by qPCR was performed according to the instructions of the manufacturer using SYBR® Green qPCR mix (Thermo Fisher Scientific) on a LightCycler® 480 SW 1.5 (Roche Diagnostics).
Analysis of Stimulated Primary Human Cells
Monocytes or macrophages were stimulated using various amounts of IL-6 or hyper-IL-6 in serum-free RPMI 1640 medium for 15 min at 37 °C, followed by harvesting of the cells. After two washing steps with ice-cold PBS, cells were either lysed (buffer contained 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1% Triton X-100, and Complete protease inhibitor mixture from Roche) and boiled in Laemmli buffer for Western blotting analysis, or the cells were used for RNA preparation and cDNA synthesis (see above). Western blotting was performed as described above.
Flow Cytometry
After stimulation, 0.5–1 × 106 cells were washed twice with flow cytometry buffer (0.5% BSA/PBS), followed by staining against various antigens for 1 h on ice. Monocytes and macrophages were blocked using human TruStain FcX Fc-receptor blocking solution (Biolegend) prior to staining according to the instructions of the manufacturer. Antibodies were diluted in flow cytometry buffer according to the recommendations of the manufacturer. After three more washing steps and resuspension of cells in flow cytometry buffer, the presence of cell surface proteins was analyzed using a BD FACS-Canto II cytometer (BD Biosciences).
Statistical Analysis
Statistical analyses were performed using GraphPad Prism (GraphPad Software, La Jolla, CA). Data were analyzed by Student's t test. Significant differences are indicated (p < 0.05). Where appropriate, p values for multiple comparisons were corrected via the Bonferroni correction.
Author Contributions
C. G. conceived and coordinated the study and wrote the paper. J. W. performed and analyzed the experiments. G. H. W., T. M. R., and S. R. J. designed the experiments and analyzed data. A. C. and H. W. contributed reagents and analyzed data. All authors analyzed the results and approved the final version of the manuscript.
This work was funded by grants from the Deutsche Forschungsgemeinschaft (DFG) (SFB877 projects A1 (to A. C. and S. R. J.) and A10 (to C. G.)), from the Federal Ministry of Education and Research (BMBF, E:Bio-Modul II InTraSig project B) and by the Cluster of Excellence “Inflammation at Interfaces.” G. H. W. is employed by CONARIS Research Institute AG (a company commercially developing sgp130Fc proteins), S. R. J. is a shareholder of CONARIS, and both are inventors of patents owned by CONARIS. T. M. R. is employed at Ferring Pharmaceuticals A/S.
- IL-6R
- IL-6 receptor
- sIL-6R
- soluble IL-6 receptor
- gp
- glycoprotein
- sgp
- soluble glycoprotein
- RAPS
- rheumatoid arthritis-associated peptide
- PMA
- phorbol 12-myristate 13-acetate
- GI
- GI254023X
- GW
- GW280264X
- hIL-6R
- human IL-6 receptor
- qPCR
- quantitative PCR
- FP
- forward primer
- RP
- reverse primer
- ADAM
- a disintegrin and metalloprotease.
References
- 1. Garbers C., and Scheller J. (2013) Interleukin-6 and interleukin-11: same but different. Biol. Chem. 394, 1145–1161 [DOI] [PubMed] [Google Scholar]
- 2. Wolf J., Rose-John S., and Garbers C. (2014) Interleukin-6 and its receptors: a highly regulated and dynamic system. Cytokine 70, 11–20 [DOI] [PubMed] [Google Scholar]
- 3. Scheller J., Garbers C., and Rose-John S. (2014) Interleukin-6: from basic biology to selective blockade of pro-inflammatory activities. Semin. Immunol. 26, 2–12 [DOI] [PubMed] [Google Scholar]
- 4. Scheller J., Chalaris A., Schmidt-Arras D., and Rose-John S. (2011) The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim. Biophys. Acta 1813, 878–888 [DOI] [PubMed] [Google Scholar]
- 5. Garbers C., Aparicio-Siegmund S., and Rose-John S. (2015) The IL-6/gp130/STAT3 signaling axis: recent advances towards specific inhibition. Curr. Opin. Immunol. 34, 75–82 [DOI] [PubMed] [Google Scholar]
- 6. Narazaki M., Yasukawa K., Saito T., Ohsugi Y., Fukui H., Koishihara Y., Yancopoulos G. D., Taga T., and Kishimoto T. (1993) Soluble forms of the interleukin-6 signal-transducing receptor component gp130 in human serum possessing a potential to inhibit signals through membrane-anchored gp130. Blood 82, 1120–1126 [PubMed] [Google Scholar]
- 7. Jostock T., Müllberg J., Ozbek S., Atreya R., Blinn G., Voltz N., Fischer M., Neurath M. F., and Rose-John S. (2001) Soluble gp130 is the natural inhibitor of soluble IL-6R transsignaling responses. Eur. J. Biochem. 268, 160–167 [DOI] [PubMed] [Google Scholar]
- 8. Garbers C., Thaiss W., Jones G. W., Waetzig G. H., Lorenzen I., Guilhot F., Lissilaa R., Ferlin W. G., Grötzinger J., Jones S. A., Rose-John S., and Scheller J. (2011) Inhibition of classic signaling is a novel function of soluble glycoprotein 130 (sgp130), which is controlled by the ratio of interleukin 6 and soluble interleukin 6 receptor. J. Biol. Chem. 286, 42959–42970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jones S. A., Scheller J., and Rose-John S. (2011) Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J. Clin. Invest. 121, 3375–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chalaris A., Garbers C., Rabe B., Rose-John S., and Scheller J. (2011) The soluble Interleukin 6 receptor: generation and role in inflammation and cancer. Eur. J. Cell Biol. 90, 484–494 [DOI] [PubMed] [Google Scholar]
- 11. Waetzig G. H., and Rose-John S. (2012) Hitting a complex target: an update on interleukin-6 trans-signalling. Expert. Opin. Ther. Targets 16, 225–236 [DOI] [PubMed] [Google Scholar]
- 12. Tanaka M., Kishimura M., Ozaki S., Osakada F., Hashimoto H., Okubo M., Murakami M., and Nakao K. (2000) Cloning of novel soluble gp130 and detection of its neutralizing autoantibodies in rheumatoid arthritis. J. Clin. Invest. 106, 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J.-G., Zhang Y., Owczarek C. M., Ward L. D., Moritz R. L., Simpson R. J., Yasukawa K., and Nicola N. A. (1998) Identification and characterization of two distinct truncated forms of gp130 and a soluble form of leukemia inhibitory factor receptor chain in normal human urine and plasma. J. Biol. Chem. 273, 10798–10805 [DOI] [PubMed] [Google Scholar]
- 14. Müllberg J., Dittrich E., Graeve L., Gerhartz C., Yasukawa K., Taga T., Kishimoto T., Heinrich P. C., and Rose-John S. (1993) Differential shedding of the two subunits of the interleukin-6 receptor. FEBS Lett. 332, 174–178 [DOI] [PubMed] [Google Scholar]
- 15. Sommer J., Garbers C., Wolf J., Trad A., Moll J. M., Sack M., Fischer R., Grötzinger J., Waetzig G. H., Floss D. M., and Scheller J. (2014) Alternative intronic polyadenylation generates the interleukin-6 trans-signaling inhibitor sgp130-E10. J. Biol. Chem. 289, 22140–22150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Diamant M., Rieneck K., Mechti N., Zhang X. G., Svenson M., Bendtzen K., and Klein B. (1997) Cloning and expression of an alternatively spliced mRNA encoding a soluble form of the human interleukin-6 signal transducer gp130. FEBS Lett. 412, 379–384 [DOI] [PubMed] [Google Scholar]
- 17. Sharkey A. M., Dellow K., and Blayney M. (1995) Stage-specific expression of cytokine and receptor messenger ribonucleic acids in human preimplantation embryos. Biol. Reprod. 53, 955–962 [DOI] [PubMed] [Google Scholar]
- 18. Müllberg J., Oberthür W., Lottspeich F., Mehl E., Dittrich E., Graeve L., Heinrich P. C., and Rose-John S. (1994) The soluble human IL-6 receptor: mutational characterization of the proteolytic cleavage site. J. Immunol. 152, 4958–4968 [PubMed] [Google Scholar]
- 19. Müllberg J., Schooltink H., Stoyan T., Günther M., Graeve L., Buse G., Mackiewicz A., Heinrich P. C, and Rose-John S. (1993) The soluble interleukin-6 receptor is generated by shedding. Eur. J. Immunol. 23, 473–480 [DOI] [PubMed] [Google Scholar]
- 20. Baran P., Nitz R., Grötzinger J., Scheller J., and Garbers C. (2013) Minimal interleukin (IL-)6 receptor stalk composition for IL-6R shedding and IL-6 classic signaling. J. Biol. Chem. 288, 14756–14768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garbers C., Jänner N., Chalaris A., Moss M. L., Floss D. M., Meyer D., Koch-Nolte F., Rose-John S., and Scheller J. (2011) Species specificity of ADAM10 and ADAM17 proteins in interleukin-6 (IL-6) trans-signaling and novel role of ADAM10 in inducible IL-6 receptor shedding. J. Biol. Chem. 286, 14804–14811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wauman J., De Ceuninck L., Vanderroost N., Lievens S., and Tavernier J. (2011) RNF41 (Nrdp1) controls type 1 cytokine receptor degradation and ectodomain shedding. J. Cell Sci. 124, 921–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dietrich C., Candon S., Ruemmele F. M., and Devergne O. (2014) A soluble form of IL-27Rα is a natural IL-27 antagonist. J. Immunol. 192, 5382–5389 [DOI] [PubMed] [Google Scholar]
- 24. Garbers C., Hermanns H. M., Schaper F., Müller-Newen G., Grötzinger J., Rose-John S., and Scheller J. (2012) Plasticity and cross-talk of Interleukin 6-type cytokines. Cytokine Growth Factor Rev. 23, 85–97 [DOI] [PubMed] [Google Scholar]
- 25. Chalaris A., Adam N., Sina C., Rosenstiel P., Lehmann-Koch J., Schirmacher P., Hartmann D., Cichy J., Gavrilova O., Schreiber S., Jostock T., Matthews V., Häsler R., Becker C., Neurath M. F., et al. (2010) Critical role of the disintegrin metalloprotease ADAM17 for intestinal inflammation and regeneration in mice. J. Exp. Med. 207, 1617–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jones S. A., Horiuchi S., Novick D., Yamamoto N., and Fuller G. M. (1998) Shedding of the soluble IL-6 receptor is triggered by Ca2+ mobilization, while basal release is predominantly the product of differential mRNA splicing in THP-1 cells. Eur. J. Immunol. 28, 3514–3522 [DOI] [PubMed] [Google Scholar]
- 27. Shi C., and Pamer E. G. (2011) Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 11, 762–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Park E. K., Jung H. S., Yang H. I., Yoo M. C., Kim C., and Kim K. S. (2007) Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm. Res. 56, 45–50 [DOI] [PubMed] [Google Scholar]
- 29. Tenhumberg S., Schuster B., Zhu L., Kovaleva M., Scheller J., Kallen K.-J., and Rose-John S. (2006) gp130 dimerization in the absence of ligand: preformed cytokine receptor complexes. Biochem. Biophys. Res. Commun. 346, 649–657 [DOI] [PubMed] [Google Scholar]
- 30. Rose-John S. (2012) IL-6 trans-signaling via the soluble IL-6 receptor: importance for the pro-inflammatory activities of IL-6. Int. J. Biol. Sci. 8, 1237–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garbers C., Monhasery N., Aparicio-Siegmund S., Lokau J., Baran P., Nowell M. A., Jones S. A., Rose-John S., and Scheller J. (2014) The interleukin-6 receptor Asp358Ala single nucleotide polymorphism rs2228145 confers increased proteolytic conversion rates by ADAM proteases. Biochim. Biophys. Acta 1842, 1485–1494 [DOI] [PubMed] [Google Scholar]
- 32. Sarwar N., Butterworth A. S., Freitag D. F., Gregson J., Willeit P., Gorman D. N., Gao P., Saleheen D., Rendon A., Nelson C. P., Braund P. S., Hall A. S., Chasman D. I., Tybjærg-Hansen A., Chambers J. C., et al. (2012) Interleukin-6 receptor pathways in coronary heart disease: a collaborative meta-analysis of 82 studies. Lancet 379, 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Swerdlow D., Holmes M., Kuchenbaecker K., Engmann J., Shah T., Sofat R., Guo Y., Chung C., Peasey A., Pfister R., Mooijaart S., Ireland H., Leusink M., and Langenberg C. (2012) The interleukin-6 receptor as a target for prevention of coronary heart disease: a Mendelian randomisation analysis. Lancet 379, 1214–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wege H., Le H. T., Chui M. S., Liu L., Wu J., Giri R., Malhi H., Sappal B. S., Kumaran V., Gupta S., and Zern M. A. (2003) Telomerase reconstitution immortalizes human fetal hepatocytes without disrupting their differentiation potential. Gastroenterology 124, 432–444 [DOI] [PubMed] [Google Scholar]
- 35. Fischer M., Goldschmitt J., Peschel C., Brakenhoff J. P., Kallen K. J., Wollmer A., Grotzinger J., and Rose-John S. (1997) A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat. Biotechnol. 15, 145–145 [DOI] [PubMed] [Google Scholar]
- 36. Schroers A., Hecht O., Kallen K. J., Pachta M., Rose-John S., and Grötzinger J. (2005) Dynamics of the gp130 cytokine complex: a model for assembly on the cellular membrane. Protein Sci. 14, 783–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mackiewicz A., Schooltink H., Heinrich P. C., and Rose-John S. (1992) Complex of soluble human IL-6-receptor/IL-6 up-regulates expression of acute-phase proteins. J. Immunol. 149, 2021–2027 [PubMed] [Google Scholar]
- 38. Hundhausen C., Misztela D., Berkhout T. A., Broadway N., Saftig P., Reiss K., Hartmann D., Fahrenholz F., Postina R., Matthews V., Kallen K.-J., Rose-John S., and Ludwig A. (2003) The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell-cell adhesion. Blood 102, 1186–1195 [DOI] [PubMed] [Google Scholar]
- 39. Ludwig A., Hundhausen C., Lambert M. H., Broadway N., Andrews R. C., Bickett D. M., Leesnitzer M. A., and Becherer J. D. (2005) Metalloproteinase inhibitors for the disintegrin-like metalloproteinases ADAM10 and ADAM17 that differentially block constitutive and phorbol ester-inducible shedding of cell surface molecules. Comb. Chem. High Throughput Screen. 8, 161–171 [DOI] [PubMed] [Google Scholar]
- 40. Wolf J., Waetzig G. H., Reinheimer T. M., Scheller J., Rose-John S., and Garbers C. (2016) A soluble form of the interleukin-6 family signal transducer gp130 is dimerized via a C-terminal disulfide-bridge resulting from alternative mRNA splicing. Biochem. Biophys. Res. Commun. 470, 870–876 [DOI] [PubMed] [Google Scholar]
- 41. Ketteler R., Glaser S., Sandra O., Martens U. M., and Klingmüller U. (2002) Enhanced transgene expression in primitive hematopoietic progenitor cells and embryonic stem cells efficiently transduced by optimized retroviral hybrid vectors. Gene Ther. 9, 477–487 [DOI] [PubMed] [Google Scholar]
- 42. Gearing D. P., Ziegler S. F., Comeau M. R., Friend D., Thoma B., Cosman D., Park L., and Mosley B. (1994) Proliferative responses and binding properties of hematopoietic cells transfected with low-affinity receptors for leukemia inhibitory factor, oncostatin M, and ciliary neurotrophic factor. Proc. Natl. Acad. Sci. U.S.A. 91, 1119–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]